Summary
Bacteria readily colonize kitchen surfaces, and the exchange of microbes between humans and the kitchen environment can impact human health. However, we have a limited understanding of the overall diversity of these communities, how they differ across surfaces, and sources of bacteria to kitchen surfaces. Here we used high-throughput sequencing of the 16S rRNA gene to explore biogeographical patterns of bacteria across >80 surfaces within the kitchens of each of four households. In total, 34 bacterial and two archaeal phyla were identified, with most sequences belonging to the Actinobacteria, Bacteriodetes, Firmicutes and Proteobacteria. Genera known to contain common food-borne pathogens were low in abundance but broadly distributed throughout the kitchens, with different taxa exhibiting distinct distribution patterns. The most diverse communities were associated with infrequently cleaned surfaces such as fans above stoves, refrigerator/freezer door seals, and floors. In contrast, the least diverse communities were observed in and around sinks, which were dominated by biofilm-forming gram-negative lineages. Community composition was influenced by conditions on individual surfaces, usage patterns, and dispersal from source environments. Human skin was the primary source of bacteria across all kitchen surfaces, with contributions from food and faucet water dominating in a few specific locations. This study demonstrates that diverse bacterial communities are widely distributed in residential kitchens and that the composition of these communities is often predictable. These results also illustrate the ease with which human- and food-associated bacteria can be transferred in residential settings to kitchen surfaces.
Introduction
Of all locations in our homes, kitchens are among the most heavily colonized by bacteria, and are likely where we are exposed to the broadest diversity of microbes (Scott et al., 1982; Ojima et al., 2002a; Ojima et al., 2002b; Sinclair and Gerba, 2011). Microbial exposures arise both directly, from handling, preparing, and eating food, and indirectly, from contact with surfaces that harbor microbes derived from a range of potential sources, including humans, food, and aerosolized water (Scott, 2000; Mattick et al., 2003; Medrano-Felix et al., 2011). Although it is well known that food items can harbor pathogenic bacteria (e.g. Campylobacter, Salmonella, Listeria) (Heaton and Jones, 2008; Luber, 2009; Berger et al., 2010), and that proper kitchen hygiene is critical for minimizing the spread of such disease-causing organisms (Rusin et al., 1998; Cogan et al., 1999; Scott, 2000; Cogan et al., 2002), the vast majority of bacteria on kitchen surfaces are likely harmless. However, the full extent of bacterial diversity in kitchens remains largely unknown as most previous studies of kitchen microbes focused on pathogen detection and relied upon cultivation-dependent techniques that preclude in-depth community characterization (Scott et al., 1982; Ojima et al., 2002a; Ojima et al., 2002b; Sinclair and Gerba, 2011). Nevertheless, from these studies it is apparent that both gram-negative and gram-positive bacteria can readily be cultivated from a variety of kitchen surfaces, with moist surfaces typically yielding the greatest number of colony-forming units.
Recently, high-throughput sequencing of the 16S rRNA gene has been used to study the bacterial communities of other indoor environments including public restrooms (Flores et al., 2011), hospitals (Kembel et al., 2012) and office buildings (Hewitt et al., 2012). In addition to revealing an enormous and unexpected amount of bacterial diversity in indoor settings, these studies have begun to identify important sources of bacteria in the built environment. For example, human skin was found to be the primary source of bacteria in each of these locations, whereas other environmental sources such as soil and outdoor air were much less important. In the kitchen environment, both humans and raw foods brought into the kitchen are likely major contributors of bacteria, although their relative importance as sources has not previously been studied.
To gain a deeper understanding of the biogeography of kitchen surface communities and to examine how bacterial communities vary across the wide range of surface types and environments present within kitchens, we characterized the bacterial communities of >80 surfaces in each of four residential kitchens using high-throughput sequencing of the 16S rRNA gene. Communities associated with surfaces from different households were expected to be dissimilar both because of behavioral (e.g. diet, disinfection frequency, cleaning products used, usage patterns) and intrinsic factors (e.g. surface material, differences in skin communities of residents, location of residence, kitchen design). Therefore, our intent here was not to control for these differences or investigate any individual factor in detail, but rather to determine bacterial community composition of “typical” kitchens, and whether these communities exhibited distribution patterns that were predictable across the wide array of surface types present within kitchens. We also sought to determine the relative importance of different sources of bacteria to the kitchen environment, because kitchen bacterial communities are likely composed of bacteria drawn from a range of potential sources including raw foods, faucet water and the humans that inhabit the home.
Results and Discussion
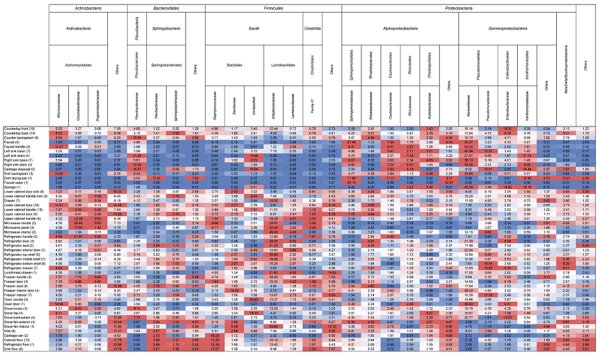
To explore the biogeographic patterns of bacteria in residential kitchens, we sampled over 80 surfaces per kitchen (Table S1) in the homes of four families living in Boulder, Colorado. We characterized the bacterial and archaeal communities using high-throughput sequencing of the 16S rRNA gene on the Illumina HiSeq platform, and, after removal of sequences of poor quality are rarefying each sample to 10,000 reads, analyzed the communities of 248 of the 332 original samples. From this unique data set, we identified 34 bacterial and two archaeal phyla, with the overwhelming majority of sequences (≈ 98% of all sequences) belonging to only four bacterial phyla: Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria (Fig. 1). Previous studies have also identified these as the dominant bacterial phyla in indoor environments (McManus and Kelley, 2005; Lee et al., 2007; Rintala et al., 2008; Aydogdu et al., 2010; Flores et al., 2011; Kembel et al., 2012). Within these dominant phyla, the most abundant families were the Micrococcaceae (≈ 6%), Flavobacteriaceae (≈ 4%), Streptococcaceae (≈ 10%) and Moraxellaceae (≈ 14%). Organisms from these families are found in a wide range of environments, including the human body (Wilson, 2008; Costello et al., 2009; Grice and Segre, 2011) and foods (Eribo and Jay, 1985; Jooste and Hugo, 1999; Rodriguez-Alonso et al., 2009). In addition, these families contain genera that are known to survive on surfaces for extended periods of time (Kramer et al., 2006; Santo et al., 2010). For example, several Acinetobacter spp. (Moraxellaceae) have been successfully cultivated from a variety of dry and wet surfaces (e.g. stainless steel, ceramic, rubber, copper coins) up to two weeks after inoculation (Getchell-White et al., 1989; Wendt et al., 1997; Santo et al., 2010). Our results thus show that the major bacterial taxa found on kitchen surfaces are qualitatively similar to those found in other indoor environments, and many of the dominant taxa appear to be able to persist on surfaces for extended periods of time.
Figure 1.
Heat map showing the average relative abundances of the dominant bacterial taxa across all kitchen surfaces. Each column is colored so that taxa with high relative abundances are red, intermediate abundances white, and low abundances blue.
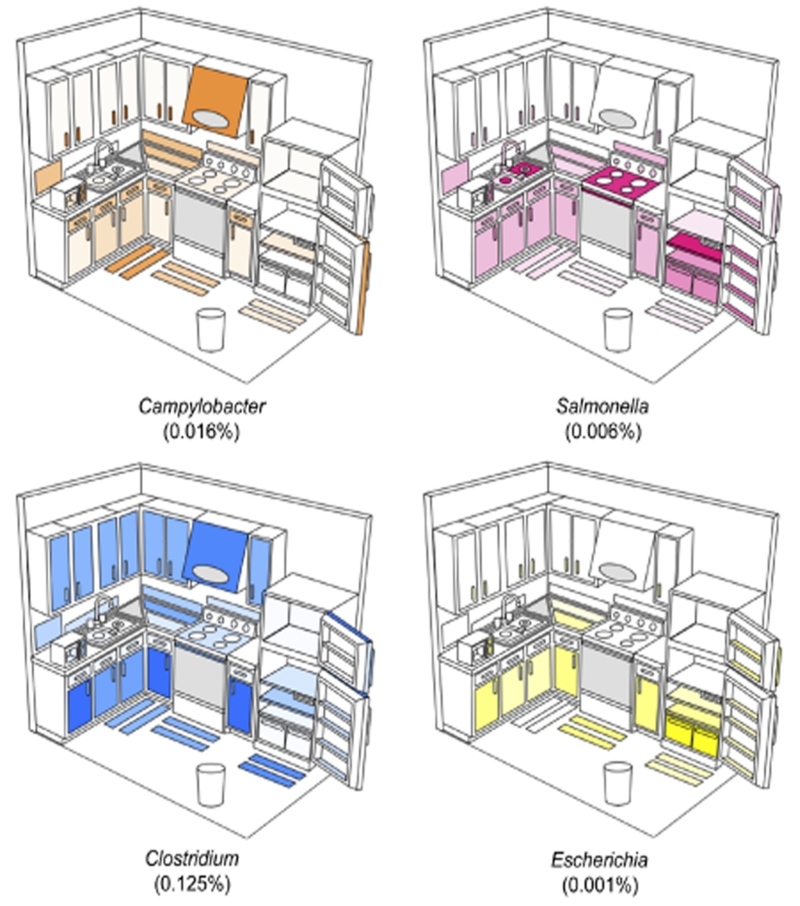
A major concern in kitchen microbiology is the potential transmission of pathogens from raw food items brought into the kitchen. The most common culprits of bacterial food-borne illnesses in industrialized nations are Campylobacter spp., Salmonella spp., Clostridium perfringens and various strains of E. coli (Mead et al., 1999). Although our techniques lack the phylogenetic resolution to detect pathogens, and not all members of these genera are pathogenic, these genera were not dominant members of the communities in any kitchen, suggesting that pathogens in the average kitchen are relatively rare (Fig. 2). These genera were, however, widely distributed in the four kitchens examined, even in areas where direct contact with raw food is unlikely. The relative abundance of Campylobacter, for example, was greatest on surfaces above counter tops, including upper cabinet handles and the microwave panel (Fig. 2). Because Campylobacter contamination in kitchens is typically attributed to raw poultry (Cogan et al., 1999; Luber, 2009), one possible explanation for these patterns is that contamination occurred from the hands of an individual who had handled raw poultry. Although the relative abundance of Campylobacter and other potentially pathogenic genera is low, these distribution patterns illustrate the ease with which food-associated bacteria are dispersed throughout residential kitchens. It is important to note, however, that the relative abundance data do not tell us if the distribution patterns are a product of differences in survivability, growth or frequency of transmission of certain bacteria on particular surface types.
Figure 2.
Distribution and average relative abundances of bacterial genera commonly associated with food-borne illnesses. Darker shades of color in each panel indicate a relatively higher abundance of the bacterial genus on that individual surface. Note that while each genus was observed in each kitchen, these patterns are a composite of the four kitchens studied and do not reflect the distribution patterns in any single kitchen. Grey indicates surfaces that were only successfully sampled in one kitchen while white surfaces were not sampled. Numbers in parentheses indicate the percentage of all sequences assigned to each genus. The triangle next to the sink represents the dish-drying rack while the oval on the stove exhaust fan represents the interior of the fan.
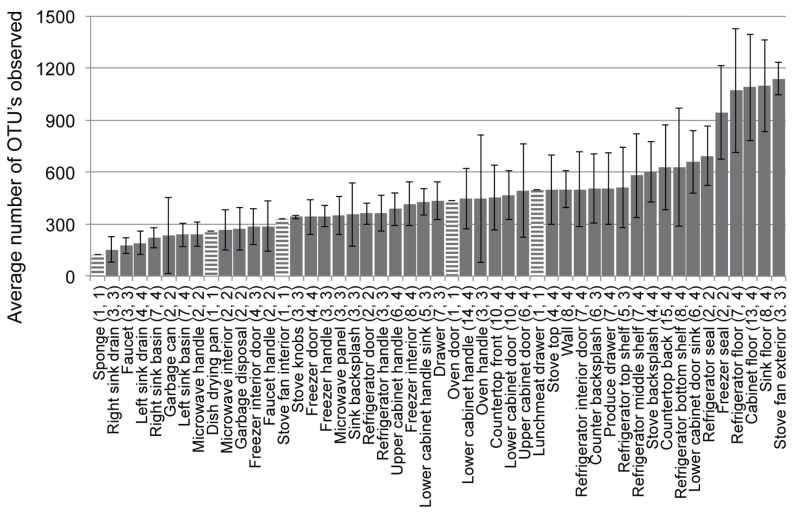
The kitchen surfaces that hosted the most diverse bacterial communities were the outside of the stove exhaust fans and the various floor surfaces (Fig. 3, Table S1). The relatively high bacterial diversity on the stove exhaust fan is likely a product of both passive (i.e. settling) and active (e.g. fan forcing) deposition of particulates, coupled with infrequent cleaning. Other surfaces that are likely cleaned infrequently, including floors and freezer door seals, also harbored diverse communities, pointing to surface hygiene as an important factor influencing bacterial alpha diversity in residential kitchens. The least diverse communities observed were associated with metallic surfaces around the kitchen sink and included the faucet, faucet handles, sink drains and sink basins (three of four sinks were stainless steel). These communities tended to be dominated by gram-negative bacteria, including several known biofilm-forming organisms such as the Sphingomonadaceae (Fig. 1) (Kelley et al., 2004). Interestingly, previous cultivation-based studies have found moist surfaces around sinks to harbor among the highest densities of cultivable bacteria (Scott et al., 1982; Rusin et al., 1998; Ojima et al., 2002a; Ojima et al., 2002b; Sinclair and Gerba, 2011). Thus, although the diversity of sink surfaces may be low, the proportion of cultivable organisms harbored by these environments is actually high. It is also likely that sinks are among the few surfaces in our kitchens where microorganisms are actively growing because moisture is more available than on other surfaces.
Figure 3.
Average number of bacterial OTUs observed on different residential kitchen surfaces determined from 10,000 randomly selected sequences from each sample. Bars are ordered from least to most diverse. Numbers in parentheses indicate the total number of samples characterized and the number of homes from which samples originated. Error bars are ± one standard deviation. Striped bars are surfaces for which only one sample yielded quality sequence data and thus, do not have error bars.
Kitchen surface communities were generally different based on the kitchen of origin (ANOSIM Global R = 0.175, p = 0.001), indicating that communities from the same kitchen were, on average, more similar than communities from different kitchens. This result may arise because the inhabitants of each home possess unique skin communities, eat different foods, have different surface disinfection routines, and have a number of other different behaviors likely to influence surface-associated bacterial communities. Furthermore, individual kitchens are not identical with respect to their design, usage patterns, surface materials, and environmental conditions (e.g. temperature, moisture, and ventilation rates) and any of these factors could influence the composition of bacterial communities found on kitchen surfaces. Surface communities also differed based on general sampling area (ANOSIM Global R = 0.282, p = 0.001) (Fig. S1), and were driven by the abundances of the two most abundant phyla, the Proteobacteria and Firmicutes, both of which were both strongly correlated with the first axis of the principal coordinates plot (r = −0.926 and r = 0.923, respectively) (Fig. S2). The gram-negative Proteobacteria were more abundant on moist surfaces, while the gram-positive, spore-forming Firmicutes were more abundant on dry (e.g. floors, cabinets, microwaves) and/or cold (e.g. refrigerator/freezer interior) surfaces. Again, these distribution patterns could be indicative of differences in the survivability, growth or frequency of transmission of particular organisms as moist surfaces could lead to the preferential growth of certain taxa and spore-formers may survive longer on dry or cold surfaces.
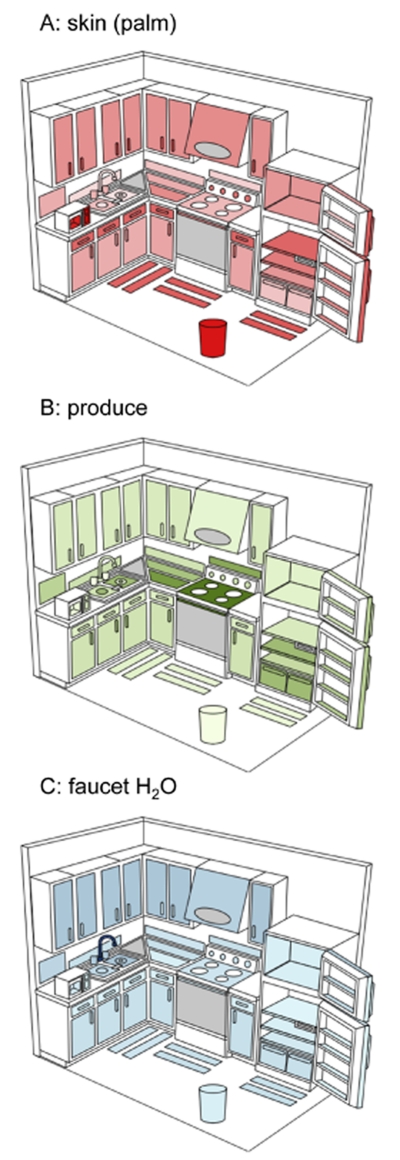
To determine the relative importance of different sources of bacteria on kitchen surfaces, we identified three indicator taxa from raw produce (Enterobacteriaceae, Microbacteriaceae, and Bacillales), four from human skin (Propionibacteriaceae, Corynebacteriaceae, Staphylococcaceae and Streptococcaceae) and three from faucet water samples (Sphingomonadaceae, Methylobacteriaceae and Gallionellaceae) using SIMPER (Clarke and Gorley, 2006) (Fig. S3). The most important source across all surfaces was clearly human skin with relatively larger contributions to those surfaces routinely touched with our hands (Tables S2-S4, Fig. 4). This is similar to what has previously been observed in public restrooms where skin was also the principal source of bacteria (Flores et al., 2011). Contributions from the other sources (faucet water and produce) tended to follow predictable patterns as illustrated by the relatively larger contribution of faucet water to sink surfaces and the importance of food associated bacteria on surfaces that regularly come into contact with food (e.g. counter/stove tops, refrigerator shelves, produce drawer) (Fig. 4). Indicator taxa from faucet water were also found to be relatively abundant on upper cabinets, possibly suggesting transport of bacteria in aerosolized water. We confirmed these overall patterns using the community-level SourceTracker approach, which, despite its different mathematical and conceptual basis, exhibited similar relative contributions of sources to the various kitchen surfaces (Fig. S4). Although these results mirror patterns we might expect based on usage patterns within the kitchen or proximity to bacterial sources, they do highlight the spread of bacteria from human skin to surfaces in our kitchens and for the potential transfer of bacteria from other sources to the human body after touching contaminated surfaces.
Figure 4.
Average relative abundances of indicator taxa from three potential sources of bacteria to residential kitchen surfaces. Figures are independent of each other and are colored so that the darker shades indicate a relatively higher abundance of bacteria derived from that particular source. For information about what taxa were used as indicators, see text or Supplementary Figure 1. Figures not drawn to scale.
Conclusions
This study shows that kitchen surfaces host remarkably diverse bacterial communities that differ both between kitchens and across surface types found within kitchens. Bacteria belonging to genera that include common pathogens comprise only a very small fraction of these communities (and even within these genera, most of the bacteria detected are probably not pathogenic, underscoring the rarity of pathogens as members of kitchen-associated microbial communities). Moreover, these results demonstrate that these bacterial communities are structured in ways that likely reflect expected dispersal from bacterial sources (i.e., human skin, food and faucet water) in ways related to how each surface is used or its proximity to each type of source. The baseline characterization of kitchen-associated microbial communities presented here, in combination with future experimental studies that show how specific behaviors or treatments affect the survivability, growth and transmission of bacteria on particular kitchen surfaces, will further improve our understanding of the microbial ecology of residential kitchens.
Experimental Procedures
Sampling, DNA extraction and sequencing
At least 82 kitchen surfaces (Table S1) in the homes of four families living in Boulder, Colorado were sampled in September 2011 using sterile cotton-swabs as described previously (Fierer et al., 2008; Fierer et al., 2010; Flores et al., 2011). Because each kitchen was unique in design and contained surfaces of different dimensions, we sampled surfaces based on general features (e.g. countertops, stoves, sinks, etc.) rather than by strict surface area measurements. The duration of swabbing individual surfaces, however, was uniform across kitchens at 10-15 seconds per location. One liter of cold and hot faucet water was also collected from each home, and filtered through a 0.2 μm bottle top filters (Nalgene, Rochester, NY, USA) to capture bacteria from this potential source. Kitchens A and B were in second story apartments, kitchen C was in a single-family home, and kitchen D was in a garden-level apartment of a different single-family home, providing a range of representative building types.
Genomic DNA was extracted from both cotton swabs and filters using the MoBio PowerSoil DNA isolation kit following the protocol of Fierer et al. (Fierer et al., 2008). A portion of the 16S rRNA gene spanning the variable region 4 (V4) was amplified using the barcoded, universal primer set (515F/806R), PCR mixture and thermal cycling conditions described in Caporaso et al. (Caporaso et al., 2012). This primer set was designed to cover a wide diversity of both Archaea and Bacteria with few biases (Bates et al., 2011; Bergmann et al., 2011). PCR reactions were performed in triplicate 25 μL reactions that were subsequently combined and quantified using the PicoGreen dsDNA assay kit (Invitrogen, Carlsbad, CA, USA). Quantified reactions were pooled in equimolar concentrations and cleaned using MoBio UltraClean PCR Clean-up Kit following the manufacturer’s instructions. Pools were submitted for sequencing on an Illumina HiSeq2000 instrument at The University of Colorado Biofrontiers Institute Advanced Genomics Facility. All sequence data and sample metadata are publicly available through the European Bioinformatics Institute (EBI) under ERP001751.
Sequence analysis
Because Illumina amplicon sequencing generates tens of millions of sequences, de novo picking of operational taxonomic units (OTUs, defined by pairwise sequence similarity) was not practical. Instead, we used a closed reference-based OTU picking approach as previously described (Hamady et al., 2010; Caporaso et al., 2011; Caporaso et al., 2012) using QIIME (Caporaso et al., 2010). Briefly, after sequences were demultiplexed and quality filtered, OTUs at the 97% sequence similarity level were picked using UCLUST (Edgar, 2010) against the Greengenes database pre-clustered at 97% identity (DeSantis et al., 2006). Sequences were assigned to OTUs with corresponding taxonomy based on their best match to a sequence in the Greengenes reference database. Sequences were discarded if they had less than a 97% match to any sequence in the database (34% of the sequences were discarded by this filter). For phylogenetic diversity metrics, the Greengenes tree was used.
After removing sequences of poor quality and those that did not match the Greengenes reference database, a total of 18,989,626 partial 16S rRNA gene sequences remained. An additional 3,537,685 sequences were determined to be from chloroplasts and were removed prior to further analysis, because we were only interested in the bacterial and archaeal communities. The remaining sequences were rarefied to 10,000 sequences per sample, resulting in a total of 248 kitchen samples used in all downstream analyses (Table S1). From these sequences, a total of 9,049 unique OTUs (range 62-1,557/sample) were observed. Of these, approximately 20% (1,838/9,049) were singletons (observed only once).
To determine which surfaces hosted the most diverse communities, both OTU-based (taxonomic richness) and phylogenetic-based (Phylogenetic Diversity – PD (Faith and Baker, 2006)) alpha diversity metrics were calculated for ten resampling events of 10,000 sequences per sample using QIIME (Caporaso et al., 2010). Values corresponding to surfaces present in multiple kitchens were then averaged across kitchens. Because both metrics showed similar patterns, only taxonomic richness results are presented here. The taxonomic compositions of the different surface communities were averaged across kitchens at multiple taxonomic levels and the relative abundances of individual taxa were projected onto a generic kitchen diagram using SitePainter (Gonzalez et al., 2012). To compare the surface-associated communities both across kitchens and by general kitchen area, we used the phylogenetically-based weighted UniFrac metric (Lozupone and Knight, 2005). The resulting distance matrix was visualized using principal coordinates analysis (PCoA), and sample categories were tested to determine whether they harbored distinct communities using analysis of similarity (ANOSIM) in PRIMER v6 (Clarke and Gorley, 2006). Pearson correlations between taxon abundances and principal coordinates were also conducted in PRIMER v6.
In order to determine the relative importance of potential sources of bacteria to the kitchen environment, we used two complementary approaches: a traditional indicator taxon approach, which examines individual taxa that are common in one environment while rare in others, and a Bayesian community-level source tracking approach, which uses features of the entire community with a Dirichlet model to take into account variability in individual source environments (Knights et al., 2011). For both approaches, OTUs were picked using the same referenced-based approach described above for a variety of produce items (n=192, Table S5), human palms (Costello et al., 2009) (n=64) and faucet water (this study; n=5). Indicator taxa were identified using the family level taxonomy determined from 200 randomly selected sequences from each source environment using SIMPER in Primer v6 (Clarke and Gorley, 2006). SIMPER identifies taxa that are far more abundant in one source environment than in other source environments under consideration (Fig. S3). Contributions of sources to kitchen surface communities were determined based on the average abundance of these indicator taxa across the four kitchens (Tables S2-S4). For the Bayesian approach, a composite OTU table of each environmental sample and source samples (produce, faucet water, skin) was used as input for the SourceTracker algorithm (Knights et al., 2011) in R (Ihaka and Gentleman, 1996). Output from SourceTracker was averaged for each surface type across all kitchens for each source environment. Relative contributions determined from both approaches were mapped onto a generic kitchen diagram using SitePainter (Gonzalez et al., 2012).
Supplementary Material
Acknowledgements
We thank Jessica Henley, Donna Lyons, Gaddy Bergmann and other members of the Fierer laboratory for their assistance with this project. Funding for this work was provided by grants to N.F. and R.K. from the Alfred P. Sloan Foundation’s Microbiology of the Built Environment program.
References
- Aydogdu H, Asan A, Tatman Otkun M. Indoor and outdoor airborne bacteria in child day-care centers in Edirne City (Turkey), seasonal distribution and influence of meteorological factors. Environ Monit Assess. 2010;164:53–66. doi: 10.1007/s10661-009-0874-0. [DOI] [PubMed] [Google Scholar]
- Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011;5:908–917. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger CN, Sodha SV, Shaw RK, Griffin PM, Pink D, Hand P, Frankel G. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ Microbiol. 2010;12:2385–2397. doi: 10.1111/j.1462-2920.2010.02297.x. [DOI] [PubMed] [Google Scholar]
- Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, et al. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem. 2011;43:1450–1455. doi: 10.1016/j.soilbio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1624–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K, Gorley R. PRIMER v6. User manual/tutorial. Plymouth Mariner Laboratory; Plymouth: 2006. [Google Scholar]
- Cogan TA, Bloomfield SF, Humphrey TJ. The effectiveness of hygiene procedures for prevention of cross-contamination from chicken carcases in the domestic kitchen. Lett Appl Microbiol. 1999;29:354–358. doi: 10.1046/j.1472-765x.1999.00656.x. [DOI] [PubMed] [Google Scholar]
- Cogan TA, Slader J, Bloomfield SF, Humphrey TJ. Achieving hygiene in the domestic kitchen: the effectiveness of commonly used cleaning procedures. J Appl Microbiol. 2002;92:885–892. doi: 10.1046/j.1365-2672.2002.01598.x. [DOI] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinform. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Eribo BE, Jay JM. Incidence of Acinetobacter spp. and other gram-negative, oxidase-negative bacteria in fresh and spoiled ground beef. Appl Environ Microbiol. 1985;49:256–257. doi: 10.1128/aem.49.1.256-257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith DP, Baker AM. Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evol Bioinform Online. 2006;2:121–128. [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Lauber CL, Zhou N, McDonald D, Costello EK, Knight R. Forensic identification using skin bacterial communities. Proc Natl Acad Sci U S A. 2010;107:6477–6481. doi: 10.1073/pnas.1000162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores GE, Bates ST, Knights D, Lauber CL, Stombaugh J, Knight R, Fierer N. Microbial biogeography of public restroom surfaces. PLoS One. 2011;6:e28132. doi: 10.1371/journal.pone.0028132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getchell-White SI, Donowitz LG, Groschel DH. The inanimate environment of an intensive care unit as a potential source of nosocomial bacteria: evidence for long survival of Acinetobacter calcoaceticus. Infect Control Hosp Epidemiol. 1989;10:402–407. doi: 10.1086/646061. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Stombaugh J, Lauber CL, Fierer N, Knight R. SitePainter: a tool for exploring biogeographical patterns. Bioinform. 2012;28:436–438. doi: 10.1093/bioinformatics/btr685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton JC, Jones K. Microbial contamination of fruit and vegetables and the behaviour of enteropathogens in the phyllosphere: a review. J Appl Microbiol. 2008;104:613–626. doi: 10.1111/j.1365-2672.2007.03587.x. [DOI] [PubMed] [Google Scholar]
- Hewitt KM, Gerba CP, Maxwell SL, Kelley ST. Office space bacterial abundance and diversity in three metropolitan areas. PLoS One. 2012;7:e37849. doi: 10.1371/journal.pone.0037849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- Jooste PJ, Hugo CJ. The taxonomy, ecology and cultivation of bacterial genera belonging to the family Flavobacteriaceae. Int J Food Microbiol. 1999;53:81–94. doi: 10.1016/s0168-1605(99)00162-2. [DOI] [PubMed] [Google Scholar]
- Kelley ST, Theisen U, Angenent LT, St Amand A, Pace NR. Molecular analysis of shower curtain biofilm microbes. Appl Environ Microbiol. 2004;70:4187–4192. doi: 10.1128/AEM.70.7.4187-4192.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kembel SW, Jones E, Kline J, Northcutt D, Stenson J, Womack AM, et al. Architectural design influences the diversity and structure of the built environment microbiome. ISME J. 2012;6:1469–1479. doi: 10.1038/ismej.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, et al. Bayesian community-wide culture-independent microbial source tracking. Nat Methods. 2011;8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Schwebke I, Kampf G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis. 2006;6:130. doi: 10.1186/1471-2334-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L, Tin S, Kelley S. Culture-independent analysis of bacterial diversity in a child-care facility. BMC Microbiol. 2007;7:27. doi: 10.1186/1471-2180-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luber P. Cross-contamination versus undercooking of poultry meat or eggs - which risks need to be managed first? Inter J Food Microbiol. 2009;134:21–28. doi: 10.1016/j.ijfoodmicro.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Mattick K, Durham K, Hendrix M, Slader J, Griffith C, Sen M, Humphrey T. The microbiological quality of washing-up water and the environment in domestic and commercial kitchens. J Appl Microbiol. 2003;94:842–848. doi: 10.1046/j.1365-2672.2003.01904.x. [DOI] [PubMed] [Google Scholar]
- McManus C, Kelley S. Molecular survey of aeroplane bacterial contamination. J Appl Microbiol. 2005;99:502–508. doi: 10.1111/j.1365-2672.2005.02651.x. [DOI] [PubMed] [Google Scholar]
- Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, et al. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano-Felix A, Martinez C, Castro-del Campo N, Leon-Felix J, Peraza-Garay F, Gerba CP, Chaidez C. Impact of prescribed cleaning and disinfectant use on microbial contamination in the home. J Appl Microbiol. 2011;110:463–471. doi: 10.1111/j.1365-2672.2010.04901.x. [DOI] [PubMed] [Google Scholar]
- Ojima M, Toshima Y, Koya E, Ara K, Kawai S, Ueda N. Bacterial contamination of Japanese households and related concern about sanitation. Inter J Environ Health Res. 2002a;12:41–52. doi: 10.1080/09603120120110040. [DOI] [PubMed] [Google Scholar]
- Ojima M, Toshima Y, Koya E, Ara K, Tokuda H, Kawai S, et al. Hygiene measures considering actual distributions of microorganisms in Japanese households. J Appl Microbiol. 2002b;93:800–809. doi: 10.1046/j.1365-2672.2002.01746.x. [DOI] [PubMed] [Google Scholar]
- Rintala H, Pitk‰ranta M, Toivola M, Paulin L, Nevalainen A. Diversity and seasonal dynamics of bacterial community in indoor environment. BMC Microbiol. 2008;8:56. doi: 10.1186/1471-2180-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Alonso P, Fernandez-Otero C, Centeno JA, Garabal JI. Antibiotic resistance in lactic acid bacteria and Micrococcaceae/Staphylococcaceae isolates from artisanal raw milk cheeses, and potential implications on cheese making. J Food Sci. 2009;74:M284–293. doi: 10.1111/j.1750-3841.2009.01217.x. [DOI] [PubMed] [Google Scholar]
- Rusin P, Orosz-Coughlin P, Gerba C. Reduction of faecal coliform, coliform and heterotrophic plate count bacteria in the household kitchen and bathroom by disinfection with hypochlorite cleaners. J Appl Microbiol. 1998;85:819–828. doi: 10.1046/j.1365-2672.1998.00598.x. [DOI] [PubMed] [Google Scholar]
- Santo CE, Morais PV, Grass G. Isolation and characterization of bacteria resistant to metallic copper surfaces. Appl Environ Microbiol. 2010;76:1341–1348. doi: 10.1128/AEM.01952-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott E. Relationship between cross-contamination and the transmission of foodborne pathogens in the home. Pediatr Infect Dis J. 2000;19:S111–113. doi: 10.1097/00006454-200010001-00005. [DOI] [PubMed] [Google Scholar]
- Scott E, Bloomfield SF, Barlow CG. An investigation of microbial contamination in the home. J Hyg. 1982;89:279–293. doi: 10.1017/s0022172400070819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair RG, Gerba CP. Microbial contamination in kitchens and bathrooms of rural Cambodian village households. Lett Appl Microbiol. 2011;52:144–149. doi: 10.1111/j.1472-765X.2010.02978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt C, Dietze B, Dietz E, Ruden H. Survival of Acinetobacter baumannii on dry surfaces. J Clin Microbiol. 1997;35:1394–1397. doi: 10.1128/jcm.35.6.1394-1397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. Bacteriology of humans: an ecological perspective. Blackwell Publishing Ltd; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.