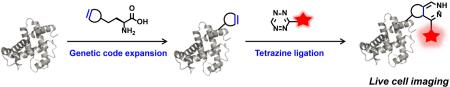
Abstract
Over the past years, fluorescent proteins (e.g., GFP) have been widely utilized to visualize recombinant protein expression and localization in live cells. Although powerful, fluorescent protein tags are limited by their relatively large size and potential perturbation to protein function. Alternatively, site-specific labeling of proteins with small-molecule organic fluorophores using bioorthogonal chemistry may provide a more precise and less perturbing method. This approach involves site-specific incorporation of unnatural amino acids (UAAs) into proteins via genetic code expansion, followed by bioorthogonal chemical labeling with small organic fluorophores in living cells. While this approach has been used to label extracellular proteins for live cell imaging studies, site-specific bioorthogonal labeling and fluorescence imaging of intracellular proteins in live cells is still challenging. Herein, we systematically evaluate site-specific incorporation of diastereomerically pure bioorthogonal UAAs bearing stained alkynes or alkenes into intracellular proteins for inverse–electron–demand Diels–Alder cycloaddition (IEDAC) reactions with tetrazine-functionalized fluorophores for live cell labeling and imaging in mammalian cells. Our studies show that site-specific incorporation of axial diastereomer of trans-cyclooct-2-ene-lysine (2’-aTCOK) robustly affords highly efficient and specific bioorthogonal labeling with monosubstituted tetrazine-fluorophores in live mammalian cells, which enabled us to image the intracellular localization and real-time dynamic trafficking of IFITM3, a small membrane-associated protein with only 137 amino acids, for the first time. Our optimized UAA incorporation and bioorthogonal labeling conditions also enabled efficient site-specific fluorescence labeling of other intracellular proteins for live cell imaging studies in mammalian cells.
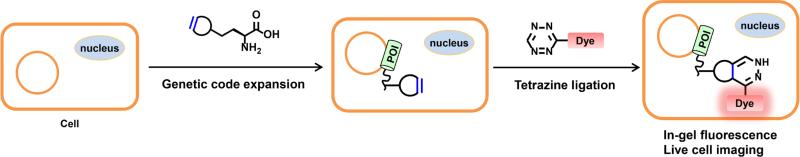
Graphical abstract
INTRODUCTION
The discovery and use of fluorescent proteins has illuminated and transformed biology.1-3 Fusion of fluorescent proteins to a protein of interest through classic genetic engineering allows for quantitative analysis and monitoring of protein expression, localization, and dynamic behaviors at molecular level in real time inside living cells and even whole organisms. This technique thus serves as the most popular and powerful strategy for protein tagging in biology.4 Nevertheless, fluorescent protein tags are not without limitations.5-7 The biggest concern about fluorescent proteins is their relatively large size, i.e., ~27 kDa, which may significantly perturb the expression, cellular localization, activity, or function of the protein they are attached to, especially when the protein of interest is small and membrane-anchored. Because of their large size, fluorescent proteins are normally only fused to the N- or C-terminus of an interested protein. By contrast, organic fluorophores are much smaller, i.e., ~0.5 kDa, and may therefore only minimally influence the cellular behavior and function of tagged protein. Therefore, organic fluorophores can even be attached on the internal sequence of a protein, without having to limit to N- or C-terminus. In addition, organic fluorophores may have better photophysical properties in photostability, photoswitchability, and color availability, which are often required for multiplexing, super-resolution, and single-molecule fluorescence imaging.7-9
To overcome the limitations of fluorescent protein tags and harness the advantages of organic fluorophores, several methods have been developed to directly attach a small-molecule organic fluorophore onto a protein of interest in live cells.6,7,10-13 Some of these methods utilize small peptide or protein tags, such as FlAsH-tag,14-17 SANP-tag,18,19 HaloTag,20 TMP-tag,21,22 LplA-tag,23 Sortasetag,24,25 PYP-tag,26 and dithiol-tag,27 that are firstly genetically fused to the proteins of interest and then specifically modified with functionalized organic fluorophores via self-labeling or enzyme-mediated labeling under physiological conditions.
Alternatively, advances in bioorthogonal chemistry28-30 have enabled a metabolic labeling and chemical tagging strategy for imaging proteins in live cells.5,31 This two-step approach involves incorporation of a unique bioorthogonal chemical functionality, such as azide, strained alkyne or alkene, into the target protein and a subsequent bioorthogonal reaction, such as strain–promoted alkyne–azide cycloaddition (SPAAC)32,33 or inverse–electron–demand Diels–Alder cycloaddition (IEDAC).34 This allows for specific attachment of various functionalized organic fluorophores onto the chemically tagged protein in live cells. In order to specifically install the bioorthogonal chemical functionality into a protein of interest, several approaches have emerged, including HaloTag technology,35 Lipoic acid ligase-mediated ligation,36-38 and genetic code expansion.39-41 Among them, the genetic incorporation of bioorthogonal unnatural amino acid (UAA) via genetic code expansion technology that utilizes orthogonal aminoacyl-tRNA synthetase/tRNA pairs and alterative codons (e.g., UAG amber stop codon) has several unique advantages. First, the structure of target protein is minimally altered with only one amino acid change and no additional tag required, which therefore ensures minimal perturbation to protein localization and function. Second, the UAA and therefore the organic fluorophore can in principle be incorporated into the target protein at any position site-specifically. Furthermore, this strategy is highly modular, providing a general system to introduce various fluorophores with desired photophysical properties into the target protein for different imaging applications. Amongst the different bioorthogonal reactions,42,43 the inverse–electron–demand Diels–Alder cycloaddition (IEDAC) between strained alkynes or alkenes with tetrazines, or “tetrazine ligation”,44,45 has emerged as the reaction of choice for labeling proteins in live cells, due to fast second-order rate constants (in the range of 102~106 M−1 s−1) and compatibility with live cells.34,46,47
Although the strategy of combining site-specific incorporation of UAAs with bioorthogonal reactions for protein labeling and imaging in live cells is advantageous (Figure 1), there are many challenges and requirements associated.6,48 For instance, the UAA should be stable for prolonged incubation time in cell culture medium under physiological temperature, and should be highly efficiently incorporated into proteins of interest in mammalian cells. In addition, the organic fluorophores should be carefully selected, as their cellular properties, such as membrane permeability, intracellular distribution, and retention, can affect protein labeling efficiency and specificity. Furthermore, the bioorthogonal UAA position should be readily accessible to fluorophores in order for efficient labeling. As such, although cellular labeling of UAA-containing GFP has been realized in bacteria using this strategy,49-52 its prior uses for protein labeling and imaging in live mammalian cells have largely been limited to cell surface proteins.6,48,53-57 Nonetheless, advances made by the Chin laboratory have enabled efficient site-specific live cell labeling and fluorescence imaging of a nuclear protein Jun54 and cytosolic proteins such as actin.58
Figure 1.
Scheme for site-specific fluorescence labeling and imaging of intracellular proteins of interest (POI) in live cells using unnatural amino acid incorporation via genetic code expansion and bioorthogonal tetrazine ligation reaction.
As part of ongoing studies in our laboratory to understand the mechanisms of action and regulation of interferon (IFN)-inducible transmembrane protein 3 (IFITM3),59-62 a relative small vesicle-associated membrane protein with only 137 amino acids (i.e., ~15 kDa) that is involved in host restriction of influenza virus and many other pathogenic viruses,59,60,63-65 our laboratory has been interested in imaging its localization and trafficking in live mammalian cells. However, it has not been possible to image IFITM3 in live cells, as N- or C-terminal fusions with fluorescent proteins (e.g., GFP or mCherry) show disrupted IFITM3 cellular localization and antiviral activity (unpublished data). This is perhaps not surprising, since the sizes of fluorescent proteins are nearly twice of that of IFITM3. Nevertheless, this experimental limitation has precluded us from live cell imaging experiments to directly visualize whether IFITM3 is recruited to virus-containing vesicles and shuttles pathogens into lysosomes for degradation. To overcome this technical limitation, herein we report the systematic comparison of IEDAC reactions between strained alkynes or alkenes and tetrazine-fluorophores for site-specific fluorescent labeling and imaging of IFITM3 in live mammalian cells (Figure 1). After systematic evaluation of different lysine derivatives bearing a strained alkyne or alkene and a series of tetrazine-fluorophores, we found that site-specific incorporation of the axial diastereomer of trans-cyclooct-2-ene-lysine (2’-aTCOK) into IFITM3 via the genetic code expansion technology robustly afforded efficient and specific bioorthogonal labeling with monosubstituted tetrazine-fluorophores in live mammalian cells (Figure 1). Our optimized conditions enabled site-specific fluorophore labeling and live cell imaging of IFITM3 for the first time and are providing new opportunities for its mechanistic studies in live cells. More broadly, we also demonstrate that the systematic investigation and optimized conditions are general and can be used for site-specific labeling and fluorescence imaging of other intracellular proteins in live cells.
RESULTS AND DISCUSSION
Selection and Synthesis of Bioorthogonal UAAs and Fluorophores
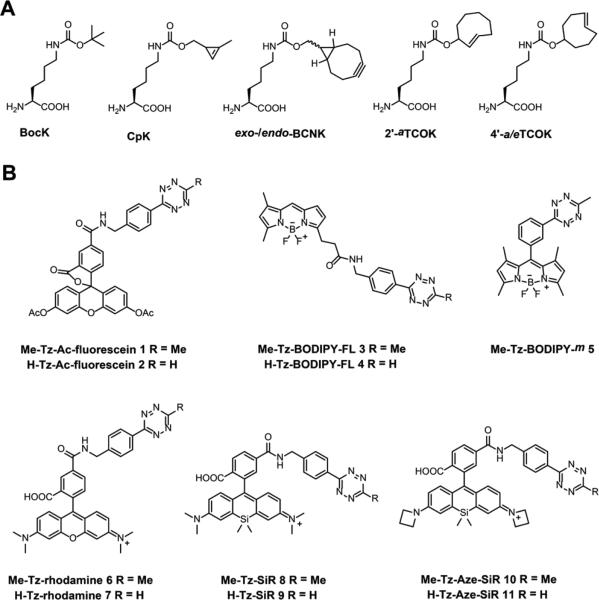
We first sought to establish a systematic and unbiased investigation of different bioorthogonal UAAs and fluorophores to examine bioorthogonal reactions in the context of live cells and optimize conditions for live cell imaging of individual proteins of interest. We focused on bioorthogonal IEDAC reactions because of their fast reaction rates (up to 106 M−1 s−1)66,67 and excellent biocompatibility. In this regard, we chose UAAs bearing strained alkyne or alkene functionalities (Figure 2A), including 1,3-disubstituted cyclopropene-lysine (CpK),68 bicyclo[6.1.0]nonyne-lysine (BCNK),54,69 trans-cyclooct-4-ene-lysine (4’-TCOK),51,54 and axial trans-cyclooct-2-ene-lysine (2’-aTCOK),55,70,71 all of which can be site-specifically incorporated into proteins expressed in mammalian cells using variants of the pyrrolysyl-tRNA synthetase (PylRS)/Pyl-tRNACUA pair and react with tetrazines chemoselectively. Diastereomeric mixtures of BCNK, 4’-TCOK, and 2’-TCOK have previously been used for live cell imaging of cell surface proteins.51,54,55,71 However, pure diastereomers of these UAAs have not been directly compared for labeling and imaging of intracellular proteins in live cells. Furthermore, CpK has not been reported for site-specific labeling of individual proteins in live cells. Recently it has been shown that the two diastereomers of 2’-TCOK, i.e., axial and equatorial diastereomers, can be separated by simple flash chromatography,70,71 and that the axial diastereomer, i.e., 2’-aTCOK (Figure 2A), is more stable than the classical trans-cyclooct-4-ene-lysine (4’-TCOK)51,54 and reacts faster with tetrazines than the equatorial isomer.70,71 Therefore, we prepared CpK, diastereomerically pure BCNK and TCOK (i.e., endo-BCNK, exo-BCNK, 2’-aTCOK, 4’-aTCOK, and 4’-eTCOK, Figure 2A) for a systematic comparison on site-specific protein labeling in live cells.
Figure 2.
Structures of (A) unnatural amino acids and (B) tetrazine-fluorophores 1-11 used in this study.
With regard to reactive fluorophores, we generated a series of cell membrane permeable tetrazine-conjugated fluorophores derived from fluorescein,72 BODIPY,73,74 rhodamine,37 and silicon-rhodamine (SiR)50,75 with varying fluorescence colors ranging from green to orange and red (Figure 2B). Generally, these tetrazine fluorophores were readily prepared by coupling the core fluorophores with methyl-disubstituted or monosubstituted tetrazine moieties76 (Me-Tz and H-Tz, respectively) through amide bonds. A unique BODIPY-based tetrazine-fluorophore Me-Tz-BODIPY-m 5 was also prepared (Figure 2B), as it exhibits dramatic fluorescence increase upon cycloaddition reaction with strained alkenes.74 In addition, we also prepared tetrazine derivatives of azetidine-silicon-rhodamines (Aze-SiR) that were recently reported to have improved brightness and photostability for fluorescence imaging (Figure 2B).77 Notably, all of these tetrazine fluorophores, including the newly synthesized tetrazine-Aze-SiR fluorophores (Figure S1 in Supporting Information (SI)), exhibit substantial fluorescence increases upon cycloaddition with strained alkynes or alkenes or after attachment onto proteins, facilitating protein imaging in live cells after brief washout of excess fluorophores.
Incorporation of UAAs into IFITM3 via Genetic Code Expansion
We first examined the incorporation efficiency of various UAAs (Figure 2A) into GFP model protein containing an amber codon at residue Y39 in the presence of a PylRS/Pyl-tRNACUA expression plasmid that contains a single copy of wild-type or mutant PylRS from Methanosarcina mazei (Mm) or Methanosarcina barkeri (Mb) under an EF1α constitutive promoter and four copies of Pyl-tRNACUA (Figure S2 in SI).78 Among the seven UAAs tested, BocK and CpK can be efficiently recognized by both wild-type Mm-PylRS and a previously reported Mm-PylRS double mutant (Y306A, Y384F) (Figure S3A and B in SI),55,79,80 while BCNK, 2’-aTCOK, and 4’-TCOK can only be incorporated into GFP-Y39TAG by the Mm-PylRS-AF mutant or other Mb-PylRS mutants (i.e., Mb-BCNKRS and Mb-TCOKRS)54 that have larger substrate binding pockets than wild-type (Figure S3 in SI).79 Interestingly, the Mm-PylRS-AF mutant can robustly incorporate endo-BCNK, exo-BCNK, and 2’-aTCOK (Figure S3C and D in SI), but is incapable of recognizing 4’-aTCOK and 4’-eTCOK (Figure S3E and F in SI), whose incorporation into GFP-Y39TAG specifically requires the presence of Mb-TCOKRS (Figure S3E and F in SI). Therefore, throughout our subsequent studies wild-type Mm-PylRS was used for incorporating BocK and CpK, while Mm-PylRS-AF mutant was chosen for endo-BCNK, exo-BCNK, and 2’-aTCOK. For the incorporation of 4’-aTCOK and 4’-eTCOK, Mb-TCOKRS was specifically utilized.
We then focused our investigation on IFITM3 for bioorthogonal labeling and imaging in live cells, as this protein has been refractory but intriguing for live cell imaging. We selected several amino acid positions on IFITM3 that are predicted to face the cytoplasm and mutated these amino acid codons into the amber codon TAG for UAA incorporation (data not shown). To evaluate IFITM3-TAG mutants in mammalian cells for intracellular labeling, we co-transfected HEK293T cells with HA-tagged IFITM3-TAG constructs and PylRS/Pyl-tRNACUA expression plasmids (Figure S2 in SI) in the presence of corresponding UAA. To our delight, the F8TAG mutant of HA-IFITM3 afforded optimal full length protein expression dependent on the presence of UAAs and corresponding optimal PylRS, as judged by western blot analysis (Figure S4 in SI). We also examined HA-IFITM3-F8TAG expression with different UAA concentrations in the culture medium (Figure S5 in SI). Appropriate amounts of UAAs are critical, as removal of excess UAAs may be necessary to minimize background fluorescence labeling in live cells. Notably, we found that 50 μM of UAA is sufficient for efficient incorporation of CpK, exo-BCNK, and 2’-aTCOK, whereas the incorporation of 4’-aTCOK and 4’-eTCOK is largely concentration-dependent, requiring much higher UAA concentrations (e.g., up to 1 mM, Figure S5 in SI). Similarly, the concentration-independence for 2’-aTCOK incorporation was also observed in the expression of GFP-Y39TAG (Figure S6A and B in SI). By contrast, the incorporation efficiency of 4’-aTCOK and 4’-eTCOK into GFP-Y39TAG again largely relies on the concentration of the UAAs (Figure S6C and D in SI), suggesting that this phenomenon is not protein-specific. With these results, we chose to use 50 μM for the incorporation of CpK, enco-BCNK, exo-BCNK, and 2’-aTCOK, whereas applied 1 mM of 4’-aTCOK and 4’-eTCOK for their incorporation throughout our subsequent studies.
Boorthogonal Labeling of IFITM3 in Live Cells
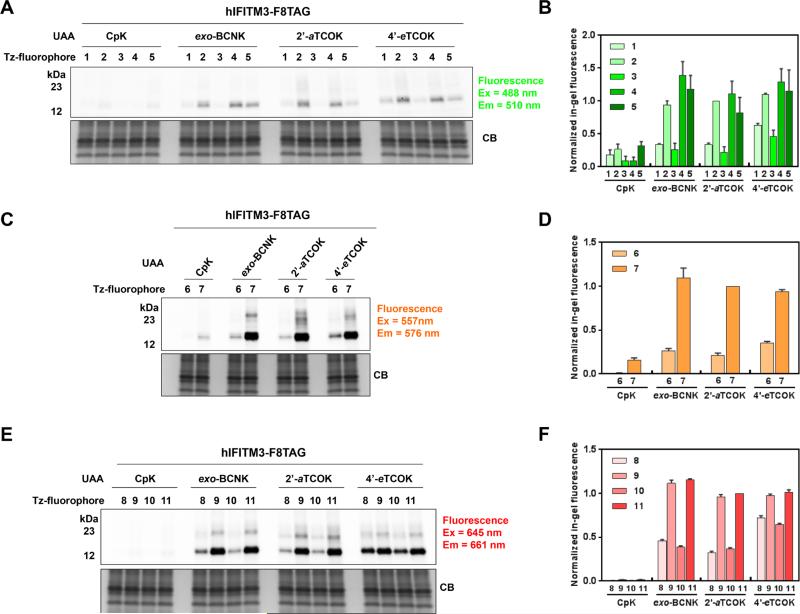
We then proceeded to test whether HA-IFITM3-F8UAA can be site-specifically labeled by tetrazine-fluorophores inside live cells. For this purpose, we expressed HA-IFITM3-F8TAG in the presence of different UAAs (i.e., CpK, exo-BCNK, endo-BCNK, 2’-aTCOK, 4’-aTCOK, and 4’-eTCOK) for 16 h, depleted the UAAs with complete growth media for 6 h, and then labeled the intact cells with 250-500 nM tetrazine-fluorophores 1-11 for 0.5 h. After brief washout of excess fluorophores, the cells were lysed with detergents containing excess amount of bicyclo[6.1.0]non-4-yn-9-ylmethanol (BCN) to minimize post-lysis labeling. Analysis of cell lysates by in-gel fluorescence scanning indicated that HA-IFITM3-F8UAA was specifically labeled by different tetrazine-fluorophores to varying levels, as judged by the fluorescent bands at around 15 kDa (Figure 3, Figure S7, and S8 in SI). A side-by-side comparison of BCNK and 4’-TCOK pure diastereomers suggested that exo-BCNK and 4’-eTCOK are generally superior to their corresponding diastereomeric counterparts for all tetrazine-fluorophores tested (Figure S7), probably due to distinct reaction rates and/or intracellular stability of the diastereomers. We therefore focused our comparative investigation on CpK, exo-BCNK, 2’-aTCOK, and 4’-eTCOK (Figure 3 and Figure S8 in SI). Notably, we only observed low non-specific fluorescence labeling on the whole proteome level (Figure S8), indicating minimal UAA incorporation into endogenous amber codons compared to HA-IFITM3-F8TAG and limited cross-reactivity of tetrazine-fluorophores with other abundant cellular proteins given the low fluorophore concentration and short labeling time used. We quantified the fluorescence intensity of every HA-IFITM3 band in the fluorescence gels and normalized the intensity of every band relative to that of the most intense band in each gel. We then plotted these relative fluorescence intensities from three independent replicates into a bar graph to quantitatively compare the labeling efficiency of various UAA and fluorophore combinations and explore the structure-reactivity relationship (Figure 3B, D, and F). We observed varying labeling efficiency of HA-IFITM3 with different UAAs tested. Generally, exo-BCNK, 2’-aTCOK, and 4’-eTCOK offers comparable fluorescence labeling, while all of these three are superior to CpK in absolute labeling efficiency for all tetrazine-fluorophores examined (Figure 3B, D, and F). This is in agreement with reaction rate constants previously reported (reactivities of exo-BCNK, 2’-aTCOK, and 4’-eTCOK with tetrazines are much higher than that of CpK).42 It is worth noting that monosubstituted tetrazines (H-Tz) are generally superior to methyl-disubstituted tetrazines (Me-Tz) in terms of HA-IFITM3 labeling efficiency, which is in accordance to the higher reactivity of H-Tz versus Me-Tz towards strained alkynes and alkenes (Figure 3B, D, and F).76 The only exception is Me-Tz-BODIPY-m 5 that provided much stronger fluorescence labeling of HA-IFITM3 than Me-Tz-BODIPY-FL 3, even to a comparable level with H-Tz-BODIPY-FL 4 (Figure 3B). We reasoned that the very hydrophobic Me-Tz-BODIPY-m 5 preferably distributes into intracellular membrane and therefore might be in close proximity to IFITM3, which could enhance the reaction rate between Me-Tz-BODIPY-m 5 and the UAA on HA-IFITM3. This phenomenon has also been noted in a recent report in which the authors demonstrated that azides situated on protein transmembrane surfaces react much faster with hydrophobic dibenzocyclooctyne (DIBO) labeling reagents than those on exposed protein surfaces.81 The unusual reactivity of Me-Tz-BODIPY-m 5 might also be attributed to the twisted orientation of the pendant phenyl ring as noted previously.74 Interestingly, tetrazine derivatives of Aze-SiR (10 and 11) showed similar or slightly higher fluorescence labeling of HA-IFITM3 compared to their counterparts of SiR (9 and 10, Figure 3C and Figure S5C in SI). We also quantified the fluorescence intensity of every HA-IFITM3 band in the fluorescence gels relative to the intensity of corresponding anti-HA band in the western blotting (Figure S8B, D, and F in SI), and found that exo-BCNK generally exhibits higher relative fluorescence labeling efficiency compared to 2’-aTCOK and 4’-eTCOK, which may be attributed to the higher reactivity of exo-BCNK55 and the low expression levels of HA-IFITM3-F8TAG in the presence of exo-BCNK.
Figure 3.
Comparative evaluation of UAAs and tetrazine-fluorophores for fluorescence labeling of IFITM3 in live cells using ingel fluorescence analysis. Hela cells expressing HA-IFITM3-F8UAA were labeled with (A, B) green tetrazine-fluorophores 1-5 (λex = 488 nm, λem = 510 nm), (C, D) orange tetrazine-fluorophores 6-7 (λex = 557 nm, λem = 576 nm), or (E, F) red tetrazine-fluorophores 8-11 (λex = 645 nm, λem = 661 nm) and lysed for in-gel fluorescence analysis (top panel) and Commassie Blue staining (CB, bottom pannel) after brief wash. 50 μM concentrations of CpK, exo-BCNK, and 2’-aTCOK were used, while 4’-eTCOK was supplemented into the media at 1 mM concentration. (B, D, and F) Bar graphs showing the fluorescence labeling efficiency of HA-IFITM3-F8UAA with tetrazine-fluorophores. Fluorescence intensity of every IFITM3 band was quantified and normalized to the most intense band of the corresponding gel, the intensity of which is set to 1. Data from three independent replicates were quantified and averaged for plotting the graphs. Data are mean ± S.E.M., n = 3. Representative gels are shown in (A), (C), and (E).
In addition to IFITM3, we also evaluated the generality of our site-specific bioorthogonal labeling conditions with other intracellular proteins. We chose two benchmark proteins, mCherry-Nucleus and mCherry-Rab5, that have defined intracellular localizations in nuclei and early endosome, respectively, and genetically introduced TAG codons into the mCherry tag. A similar comparative evaluation on fluorescence labeling of these two benchmark proteins by in-gel fluorescence profiling was conducted to characterize the optimal conditions for specific and efficient labeling (Figure S9-11 and S12-14 in SI). From the bar graphs plotted for labeling mCherry-Nucleus and mCherry-Rab5 (Figure S9-11 and S12-14 in SI), we noted that i) monosubstituted tetrazine-fluorophores always offered stronger fluorescence labeling than methyl-disubstituted tetrazine counterparts for both proteins, ii) 2’-aTCOK generally provided more intense absolute fluorescence labeling than CpK, exo-BCNK, and 4’-eTOCK, and that iii) exo-BCNK again displayed generally higher relative fluorescence labeling when taken anti-mCherry western blotting signals into consideration, which is in agreement with what we noted with IFITM3 labeling (Figure S8B, D, and F in SI).
Together, our comparative investigation of different UAA and fluorophore combinations for live cell labeling of HA-IFITM3, mCherry-Nucleus, and mCherry-Rab5 indicates our optimal two-step labeling conditions can yield efficient and specific protein labeling for potential live cell imaging studies. In addition, our results suggest that the reaction rate of UAA with tetrazine and the intracellular distribution of tetrazine-fluorophores (determined by their hydrophobicity) might be key determinants for efficient protein labeling in live cells.
Bioorthogonal Fluorescence Imaging of IFITM3 in Live Cells
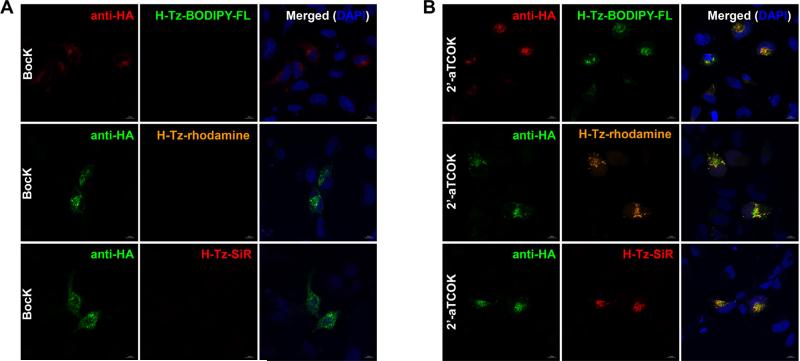
Our systematic evaluation of PylRSs, UAAs and fluorophores on HA-IFITM3 as well as mCherry-Nucleus and mCherry-Rab5 provided us the optimal conditions for protein labeling in live cells. Generally, 2’-aTCOK is most efficiently incorporated into all proteins we examined in terms of higher protein expression level compared to exo-BCNK and lower required concentration compared to 4’-eTCOK (i.e., 50 μM vs. 1 mM). Moreover, 2’-aTCOK robustly provided brighter, or at least comparable, site-specific fluorescence labeling of proteins versus other UAAs for almost all tetrazine-fluorophores tested. Therefore, we chose to use 2’-aTCOK in our subsequent studies for protein labeling and imaging in live cells. To validate the labeling conditions and further confirm the labeling efficiency and specificity, we performed immunofluorescence microscopy of HA-tagged IFITM3 in parallel with tetrazine-fluorophore labeling. Briefly, Hela cells expressing HA-IFITM3-F8UAA were labeled with tetrazine-fluorophores and briefly washed before fixation, permeabilization, and staining with fluorophore-conjugated anti-HA antibody for confocal fluorescence imaging (Figure 4). When HA-IFITM3-F8TAG was expressed in the presence of 2’-aTCOK, fluorescence puncta that are characteristic to IFITM3-containing vesicles were observed inside cells in the tetrazine-fluorophore channels (i.e., H-Tz-Ac-fluorescein, H-Tz-BODIPY-FL, Me-Tz-BODIPY-m, H-Tz-rhodamine, H-Tz-SiR, and H-Tz-Aze-SiR; Figure 4B and Figure S15 in SI). In addition, tetrazine-fluorophore signals were only observed in anti-HA fluorescence-positive cells and importantly overlapped with anti-HA fluorescence signals (Figure 4B and Figure S15 in SI). By contrast, when HA-IFITM3-F8TAG was expressed in the presence of BocK, a non-reactive lysine analog, no fluorescence signals were observed in the tetrazine-fluorophore channels for all tetrazine-fluorophores tested (Figure 4A and Figure S15 in SI). Together, these immunofluorescence results further confirm the bioorthogonal labeling efficiency and specificity, and thus validate our optimized conditions for IFITM3 labeling.
Figure 4.
Bioorthogonal fluorescence imaging of HA-IFITM3-F8UAA with tetrazine-fluorophores versus immunofluorescence staining. Hela cells expressing HA-IFITM3-F8UAA in the presence of (A) BocK (50 μM) or (B) 2’-aTCOK (50 μM) were labeled with H-Tz-BODIPY-FL, H-Tz-rhodamine, or H-Tz-SiR (500 nM, 0.5 h) under physiological conditions, briefly washed, and subjected to anti-HA immunofluorescence staining. DAPI (blue) was used to stain nuclei. Images were acquired with confocal microscopy. Scale bars = 10 μm.
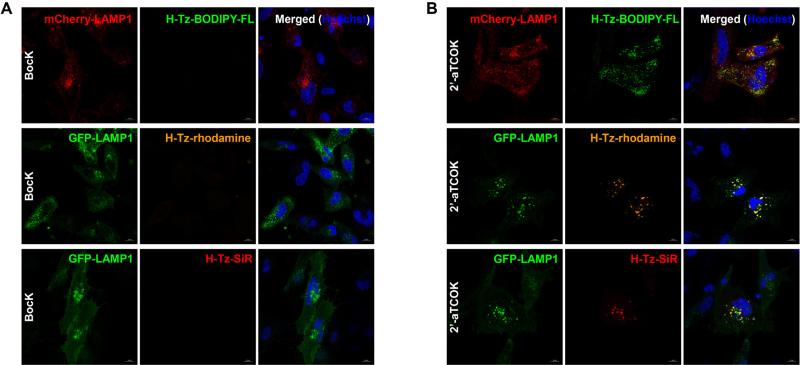
We then moved on to examine live cell imaging of IFITM3 using this method. Hela cells were transfected and labeled with tetrazine-fluorophores as described above. After washing out excess dyes, cells were directly analyzed with confocal fluorescence microscopy. As shown in Figure 5B, S16, and S17 in SI, clear intracellular fluorescent puncta were observed in tetrazine-fluorophore channels (e.g., H-Tz-BODIPY-FL, Me-Tz-BODIPY-m, H-Tz-rhodamine, H-Tz-SiR, and H-Tz-Aze-SiR), when IFITM3-F8TAG was expressed in the presence of 2’-aTCOK. These intracellular puncta, similar to those observed in immunofluorescence staining (Figure 4B and Figure S15 in SI), are characteristic to the morphology of IFITM3-positive vesicles and largely co-localized with LAMP1, a lysosome marker typically used for IFITM3 immunofluorescence co-localization studies,59 which is also consistent with the localization of endogenous IFITM3 probed by immunofluorescence imaging with an antibody against IFITM3 (Figure S18 in SI). When IFITM3-F8TAG was expressed in the presence of BocK, we did not observe any non-specific fluorescence signals inside cells in the tetrazine-fluorophore channels (Figure 5A), further validating the specificity of our bioorthogonal IFITM3 imaging in live cells.
Figure 5.
Live cell bioorthogonal fluorescence imaging of HA-IFITM3-F8UAA with tetrazine-fluorophores. Hela cells expressing HA-IFITM3-F8UAA and GFP- or mCherry-LAMP1 were labeled with H-Tz-BODIPY-FL, H-Tz-rhodamine, or H-Tz-SiR (500 nM, 0.5 h) under physiological conditions, briefly washed, and directly analyzed with confocal microscopy. (A) HA-IFITM3-F8UAA was expressed in the presence of BocK (50 μM). (B) HA-IFITM3-F8UAA was expressed in the presence of 2’-aTCOK (50 μM). Hoechst (blue) was used to stain nuclei. Scale bars = 10 μm.
It is worth noting that among all tetrazine-fluorophores we examined, including H-Tz-Ac-fluorescein, H-Tz-BODIPY-FL, Me-Tz-BODIPY-m, H-Tz-rhodamine, H-Tz-SiR, and H-Tz-Aze-SiR, H-Tz-Ac-fluorescein was not so efficient in live cell imaging and required stronger laser intensity than H-Tz-BODIPY-FL and Me-Tz-BODIPY-m (Figure S17B in SI). Meanwhile, a higher fluorescence background was observed inside the cells for H-Tz-Acfluorescein resulting in low signal-to-background contrast (Figure S17B in SI). Interestingly, H-Tz-Ac-fluorescein only provided slightly weaker fluorescence signals than H-Tz-BODIPY-FL and Me-Tz-BODIPY-m in our fixed cell imaging experiments (Figure S17A in SI). This discrepancy, in line with a previous report,35 may reflect the difference between live cell imaging and fixed cell imaging, as in fixed cell imaging experiments large excess tetrazine-fluorophores that contribute to significant intracellular fluorescence background can be easily washed out from fixed and permeabilized cells. Owing to the high labeling efficiency of 2’-aTCOK and fluorogenicity of SiR,50,82,83 no-wash imaging of FITM3 can also be achieved using this combination (Figure S19 in SI). Taken together, these results suggest that live cell imaging of IFITM3 can be optimally achieved through site-specific incorporation of 2’-aTCOK and its bioorthogonal reactions with tetrazine-fluorophores.
Fluorescence Imaging of IFITM3 Localization and Trafficking in Live Cells
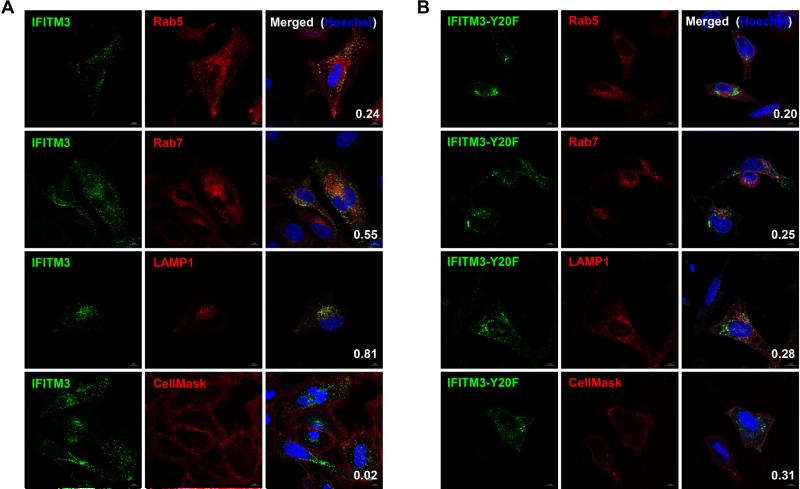
With the optimal conditions for live cell IFITM3 imaging, we sought to interrogate the localization of IFITM3 in live cells by fluorescence imaging. To this end, we focused on using H-Tz-BODIPY-FL, as this fluorophore is readily synthesized, observable with microscope eyepieces, and highly fluorogenic upon reaction with 4’-TCO.73 Endocytic vesicle markers including early endosome marker Rab5, late endosome marker Rab7, and lysosome marker LAMP1 were used for two-color imaging experiments to assess the steady-state distribution of IFITM3 in live cells. As shown in Figure 6A, IFTIM3 only partially co-localized with Rab5 and Rab7, with Pearson's correlation coefficients of 0.24 and 0.55, respectively. By contrast, IFITM3 largely co-localizes with lysosome marker LAMP1, in line with our immunofluorescence studies on the localization of overexpressed and endogenous IFITM3 (Figure S18 in SI). We also examined the localization of an inactive IFITM3-Y20F mutant, which was suggested to be enriched in the plasma membrane due to loss of endocytic sorting signal by immunofluorescence analysis of fixed cells.84,85 Indeed, our live cell imaging experiments demonstrate that IFITM3-Y20F is partially redistributed to the plasma membrane and no longer co-localizes with Rab5, Rab7, or LAMP1 (Figure 6B). Further imaging experiments with a selective plasma membrane stain CellMask also confirmed the redistribution of IFITM3-Y20F onto plasma membrane compared to wild-type IFITM3 (Figure 6 and S20 in SI).
Figure 6.
Live cell bioorthogonal fluorescence imaging of HA-IFITM3-F8UAA localization with H-Tz-BODIPY-FL. Hela cells were transfected with (A) HA-IFITM3-F8TAG or (B) HA-IFITM3-F8TAG-Y20F and mCherry-tagged endocytic markers in the presence of 2’-aTCOK (50 μM), labeled with H-Tz-BODIPY-FL (250 nM, 0.5 h) under physiological conditions, briefly washed, and directly analyzed with confocal microscopy. Rab5, Rab7, LAMP1, and CellMask were used as early endosome, later endo-some, lysosome, and plasma membrane markers, respectively. For plasma membrane staining, cells were transfected only with IFITM3 plasmids, labeled, and stained with CellMask before imaging. Hoechst (blue) was used to stain nuclei. Pearson's correlation coefficients were shown in the merged images to evaluate the co-localization of IFITM3 (green) with markers (red). 10-20 cells were used to calculate Pearson's correlation coefficients for each sample. Scale bars = 10 μm.
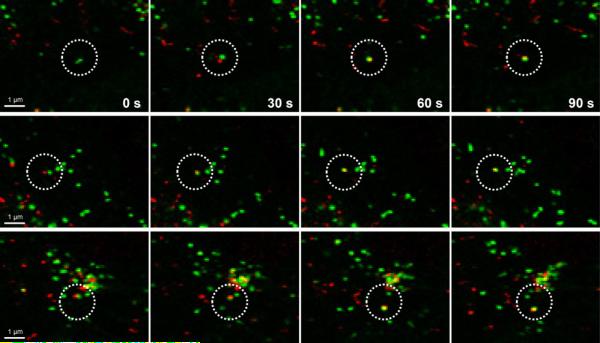
To explore the trafficking of IFITM3 in live cells, we proceeded to evaluate the dynamics of IFITM3 with exogenously added fluorescent cargoes that are internalized into endocytic vesicles. Our initial live cell imaging experiments indicated that IFITM3 partially co-localizes with exogenous endocytic cargoes such as dextran and EGF (Figure S21 in SI). For live cell trafficking experiments, IFITM3-F8aTCOK was labeled with green fluorophore H-Tz-BODIPY-FL, while dextran particles as exogenous endocytosis cargoes were labeled with pHrodo Red, a pH sensitive red fluorophore. The uptake and trafficking of dextran and IFITM3-containing vesicles were recorded by time-lapse confocal fluorescence microscopy. Our results show that red dextran particles were internalized into cells and then fused with BODIPY-labeled green IFITM3-residing puncta to yield yellow vesicular structures, implicating IFITM3 traffics to the same location as exogenously acquired cargoes after their internalization in the endocytic pathway (Figure 7 and Video S1 in SI). Moreover, our time-lapse imaging suggests that this fusion process may be completed within several minutes (Figure 7). Together, these results demonstrate that our imaging method can be used to directly monitor the localization and real-time dynamic trafficking of IFITM3 in live cells and affords new opportunities to monitor dynamic interactions of this important pathogen restriction factor during virus or bacterial infections.
Figure 7.
Time-lapse imaging of the fusion process of IFITM-containing vesicle with dextran particles. IFITM3-F8aTCOK was labeled with H-Tz-BODIPY-FL and dextran particles are labeled with pHrodo Red. Images were acquired every 30 s. The time points shown on the figure are relative to the first image in the series. Scale bars = 1 μm.
Bioorthogonal Fluorescence Imaging of Other Intracellular Proteins in Live Cells
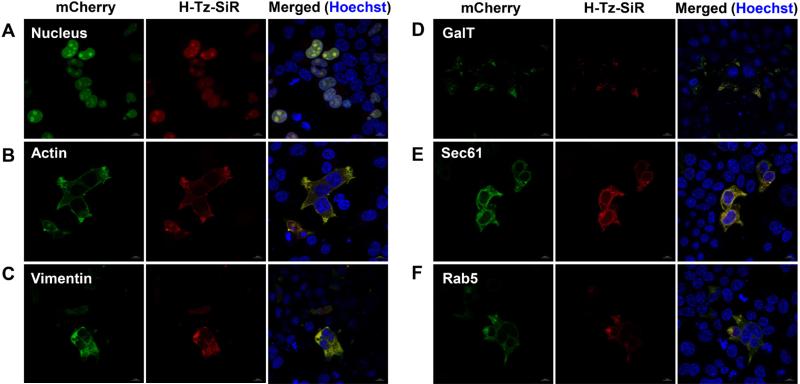
In addition to IFITM3, we also evaluated the generality of our site-specific bioorthogonal live cell imaging conditions with other intracellular proteins. We chose several proteins that have defined intracellular localizations, and genetically fused each protein with a mCherry tag bearing a TAG codon to confirm protein expression and co-localization with the mCherry signal by fluorescence microscopy. 2’-aTCOK in combination with H-Tz-SiR was used for live cell imaging studies, as this combination is very robust for labeling all benchmark proteins we examined and H-Tz-SiR is compatible with mCherry for multiplexing fluorescence imaging. As shown in Figure 8, a variety of benchmark proteins can be clearly labeled with H-Tz-SiR in live cells when expressed in the presence of 2’-aTCOK, including mCherry-Nucleus, cytoskeleton proteins mCherry-Actin, mCherry-Vimentin, and intracellular membrane proteins mCherry-GalT (Golgi marker), mCherry-Sec61 (ER marker), and mCherry-Rab5 (early endosome marker). Moreover, for all proteins H-Tz-SiR signals are perfectly overlapped with mCherry signals by confocal fluorescence microscopy. Furthermore, no background H-Tz-SiR fluorescence is observed in mCherry-negative cells or when the mCherry-tagged protein is expressed in the presence of BocK (Figure 8, S22, and S23 in SI). We also investigated exo-BCNK and H-Tz-Aze-SiR in live cell imaging of mCherry-Nucleus and mCherry-Actin (Figure S24 and S25 in SI), and found that exo-BCNK is also capable of imaging these two proteins in live cells, albeit with generally weaker intracellular fluorescence signals and lower signal-to-noise ratios compared to 2’-aTCOK (Figure S24 and S25 in SI). In addition, H-Tz-Aze-SiR can also be used for the live cell imaging of these two proteins (Figure S24 and S25 in SI). Finally, we found another PylRS/Pyl-tRNACUA expression plasmid86 that contains one copy of Pyl-tRNACUA and a CMV promoter for PylRS expression (Figure S2D in SI) can also be used for imaging IFITM3 and mCherry-Nucleus in live cells (Figure S26 in SI). Taken together, these results confirm the excellent specificity of this bioorthogonal imaging method in live cells and suggest our optimized labeling conditions can be generalized to other intracellular proteins, including soluble and membrane proteins.
Figure 8.
Live cell bioorthogonal fluorescence imaging of benchmark intracellular proteins. HEK293T cells were transfected with mCherry-tagged plasmids as indicated in the presence of 2’-aTCOK (50 μM), labeled with H-Tz-SiR (500 nM, 0.5 h) under physiological conditions, briefly washed, and directly analyzed with confocal microscopy. Hoechst (blue) was used to stain nuclei. Scale bars = 10 μm.
CONCLUSIONS
In summary, we present a systematic evaluation of bioorthogonal reactions between site-specifically incorporated UAAs and exogenously added fluorescent probes for fluorescence labeling and imaging of intracellular proteins in live cells. The diastereomerically pure bioorthogonal UAAs bearing a strained alkyne or alkene including CpK, exo-BCNK, endo-BCNK, 2’-aTCOK, 4’-aTCOK, and 4’-eTCOK are introduced into target proteins via the genetic code expansion technology, and the exogenous fluorescent probes are tailor-designed to contain tetrazine moieties with distinct chemical and photophysical properties. The Diels–Alder cycloaddition reaction of alkyne or alkene with tetrazine results in fluorescence labeling of the target protein in live cells, thus allowing for live cell imaging of the protein. We applied the systematic investigation to IFITM3, an important IFN-effector in mammals, and other intracellular proteins like chimeric mCherry-Nucleus and mCherry-Rab5 for optimizing their imaging conditions. Using this bioorthogonal imaging method and our optimal imaging conditions, we were able to directly visualize the localization and dynamic trafficking of IFITM3 in live mammalian cells for the first time and moreover achieve live cell imaging of other intracellular proteins.
The systematic evaluation of various diastereomerically pure UAA and tetrazine-fluorophore combinations for fluorescence labeling of IFITM3, mCherry-Nucleus, and mCherry-Rab5 by in-gel fluorescence analysis has provided important insights into successful labeling and imaging of intracellular proteins in live cells. The reaction rate between bioorthogonal UAA and fluorophore is the most critical factor determining the labeling efficiency and specificity. In this regard, we found monosubstituted tetrazine should be generally better than methyldisubstituted tetrazine, because of its faster reaction rate than the latter. Other unsymmetrical diaryl-tetrazines reported previously53 may have higher reactivities toward dienophiles than monosubstituted tetrazines and may warrant investigation as well. However, additional aryl ring on tetrazine may affect the chemical and physical properties of the core fluorophore. The intracellular stability of UAA also determines the labeling efficiency and specificity. It has been recently reported that 2’-aTCOK is more stable than classical 4’-TCOK,71 which is subjected to isomerization to the unreactive cis-isomer.87,88 Indeed, our imaging studies suggest that 2’-aTCOK is relatively stable even after incorporated into proteins for over 20 h and its intracellular stability seems to be organelle independent, as we have successfully imaged intracellular proteins in different organelles including the nucleus, ER, Golgi, and endosome using 2’-aTCOK. By contrast, BCNK is well-known to be covalently modified by cysteine and other intracellular thiols,89 likely leading to decreased BCNK-dependent protein expression as we observed. A potential concern about using 2’-aTCOK for imaging purpose is that the bicyclic reaction adducts of 2’-aTCOK with tetrazines may undergo elimination under certain conditions to release the free lysine, a lysine decaging reaction recently used to release prodrug or control protein activity.70,90 Nevertheless, our labeling and imaging results as well as studies from others71 suggest that the ligation product is still relatively stable and dominant, perhaps due to conjugation of the bulky fluorophores. Interestingly, due to the higher reactivity of exo-BCNK with tetrazines than 2’-aTCOK,48,55 the relative fluorescence labeling efficiency of exo-BCNK after normalization with protein expression level is generally higher than 2’-aTCOK. Therefore, when low protein expression level is desired, exo-BCNK may be used, although the overall fluorescence signal might be dimmer under fluorescence microscopy (Figure S24 and S25). Nevertheless, 2’-aTCOK seems to be the first choice to test, due to the robustness, strong overall fluorescence signal, and high signal-to-noise ratio it provides (Figure S24 and S25).
The chemical properties of fluorophores, such as membrane permeability, retention, and distribution, also significantly affect the labeling results. An “ideal” fluorophore for intracellular imaging should readily cross cell membrane of live cells, distribute evenly inside the cells, react specifically with the chemically tagged protein but not with others, and be easily washed away or highly fluorogenic. Although such an “ideal” fluorophore does not exist yet, H-Tz-SiR seems to be very robust in labeling many intracellular proteins as demonstrated in our studies. The most suitable fluorophore for an individual protein varies case by case and should be carefully optimized. For example, hydrophobic BODIPY fluorophores, e.g., H-Tz-BODIPY-FL and Me-Tz-BODIPY-m, are not suitable for labeling soluble proteins, but may be harnessed for labeling membrane proteins. Another crucial aspect affecting labeling efficiency is the UAA position that should be readily accessible on the protein surface and not affect the protein expression. For optimal labeling efficiency, multiple UAA sites should be tested (Figure S27 in SI).
Our systematic evaluation serves as an unbiased platform to optimize imaging conditions for individual proteins. With the optimal imaging conditions in hand, we have demonstrated the live cell imaging of IFITM3, which has not been possible due to its small size. Our imaging studies on IFITM3 have started to provide valuable information about IFITM3 localization and trafficking in live cells. Further efforts will be focused on time-lapse imaging of IFITM3 dynamic trafficking in cells infected with fluorescently labeled virus particles to determine whether IFITM3 contributes to alter virus trafficking pathway and/or accelerate the degradation of virus-containing vesicles by lysosomes. Additionally, bioorthogonal labeling of IFITM3 with H-Tz-SiR is providing a unique opportunity to examine whether this protein directly interacts with virus particles by single-molecule and super-resolution imaging.50,58,75 These imaging studies should enable more precise studies of IFITM3 antiviral mechanism in the future.
Generalization of our bioorthogonal imaging conditions to several benchmark intracellular proteins is also notable, as previous studies have been largely limited to cell surface proteins. Our studies thus significantly expand the scope of site-specific bioorthogonal imaging of intracellular proteins in live cells.
Supplementary Material
ACKNOWLEDGMENT
We thank Prof. Jason Chin at MRC Laboratory of Molecular Biology and Prof. Peng R. Chen at Peking University for PylRS/Pyl-tRNA plasmids. We also thank Hang lab members for helpful discussions and Bio-imaging Resource Center at The Rockefeller University for assistance in confocal fluorescence microscopy. HCH acknowledges support from NIHNIGMS R01 GM087544 grant and Starr Cancer Consortium I7-A717. TP acknowledges financial support from Shenzhen Peacock Plan (KQTD2015032709315529).
Footnotes
ASSOCIATED CONTENT
Supporting Information. Synthesis of UAAs and fluorophores, detailed experimental procedures, and supplementary figures are available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- 1.Tsien RY. Annu. Rev. Biochem. 1998;67:509. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 2.Shaner NC, Patterson GH, Davidson MW. J. Cell Sci. 2007;120:4247. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 3.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Physiol. Rev. 2010;90:1103. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 4.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 5.Prescher JA, Bertozzi CR. Nat. Chem. Biol. 2005;1:13. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 6.Lang K, Chin JW. Chem. Rev. 2014;114:4764. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
- 7.Zhang G, Zheng S, Liu H, Chen PR. Chem. Soc. Rev. 2015;44:3405. doi: 10.1039/c4cs00393d. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez-Suarez M, Ting AY. Nat. Rev. Mol. Cell Biol. 2008;9:929. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- 9.Linde S. v. d., Heilemann M, Sauer M. Annu. Rev. Phys. Chem. 2012;63:519. doi: 10.1146/annurev-physchem-032811-112012. [DOI] [PubMed] [Google Scholar]
- 10.Marks KM, Nolan GP. Nat. Meth. 2006;3:591. doi: 10.1038/nmeth906. [DOI] [PubMed] [Google Scholar]
- 11.Lin MZ, Wang L. Physiology. 2008;23:131. doi: 10.1152/physiol.00007.2008. [DOI] [PubMed] [Google Scholar]
- 12.Jing C, Cornish VW. Acc. Chem. Res. 2011;44:784. doi: 10.1021/ar200099f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue L, Karpenko IA, Hiblot J, Johnsson K. Nat. Chem. Biol. 2015;11:917. doi: 10.1038/nchembio.1959. [DOI] [PubMed] [Google Scholar]
- 14.Griffin BA, Adams SR, Tsien RY. Science. 1998;281:269. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 15.Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. J. Am. Chem. Soc. 2002;124:6063. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 16.Halo TL, Appelbaum J, Hobert EM, Balkin DM, Schepartz A. J. Am. Chem. Soc. 2009;131:438. doi: 10.1021/ja807872s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheck RA, Schepartz A. Acc. Chem. Res. 2011;44:654. doi: 10.1021/ar2001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. Nat. Biotech. 2003;21:86. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 19.Gautier A, Juillerat A, Heinis C, Corrêa IR, Jr, Kindermann M, Beaufils F, Johnsson K. Chem. Biol. 2008;15:128. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. ACS Chem. Biol. 2008;3:373. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 21.Miller LW, Cai Y, Sheetz MP, Cornish VW. Nat. Meth. 2005;2:255. doi: 10.1038/nmeth749. [DOI] [PubMed] [Google Scholar]
- 22.Chen Z, Jing C, Gallagher SS, Sheetz MP, Cornish VW. J. Am. Chem. Soc. 2012;134:13692. doi: 10.1021/ja303374p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uttamapinant C, White KA, Baruah H, Thompson S, Fernández-Suárez M, Puthenveetil S, Ting AY. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10914. doi: 10.1073/pnas.0914067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Nat. Chem. Biol. 2007;3:707. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- 25.Antos JM, Chew G-L, Guimaraes CP, Yoder NC, Grotenbreg GM, Popp MW-L, Ploegh HL. J. Am. Chem. Soc. 2009;131:10800. doi: 10.1021/ja902681k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hori Y, Ueno H, Mizukami S, Kikuchi K. J. Am. Chem. Soc. 2009;131:16610. doi: 10.1021/ja904800k. [DOI] [PubMed] [Google Scholar]
- 27.Girouard S, Houle M-H, Grandbois A, Keillor JW, Michnick SW. J. Am. Chem. Soc. 2005;127:559. doi: 10.1021/ja045742x. [DOI] [PubMed] [Google Scholar]
- 28.Sletten EM, Bertozzi CR. Angew. Chem., Int. Ed. 2009;48:6974. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramil CP, Lin Q. Chem. Commun. (Camb.) 2013;49:11007. doi: 10.1039/c3cc44272a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shih H-W, Kamber DN, Prescher JA. Curr. Opin. Chem. Biol. 2014;21:103. doi: 10.1016/j.cbpa.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Grammel M, Hang HC. Nat. Chem. Biol. 2013;9:475. doi: 10.1038/nchembio.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Debets MF, van Berkel SS, Dommerholt J, Dirks AJ, Rutjes FPJT, van Delft FL. Acc. Chem. Res. 2011;44:805. doi: 10.1021/ar200059z. [DOI] [PubMed] [Google Scholar]
- 33.Sletten EM, Bertozzi CR. Acc. Chem. Res. 2011;44:666. doi: 10.1021/ar200148z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selvaraj R, Fox JM. Curr. Opin. Chem. Biol. 2013;17:753. doi: 10.1016/j.cbpa.2013.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murrey HE, Judkins JC, am Ende CW, Ballard TE, Fang Y, Riccardi K, Di L, Guilmette ER, Schwartz JW, Fox JM, Johnson DS. J. Am. Chem. Soc. 2015;137:11461. doi: 10.1021/jacs.5b06847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernandez-Suarez M, Baruah H, Martinez-Hernandez L, Xie KT, Baskin JM, Bertozzi CR, Ting AY. Nat. Biotech. 2007;25:1483. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu DS, Tangpeerachaikul A, Selvaraj R, Taylor MT, Fox JM, Ting AY. J. Am. Chem. Soc. 2012;134:792. doi: 10.1021/ja209325n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao JZ, Uttamapinant C, Poloukhtine A, Baskin JM, Codelli JA, Sletten EM, Bertozzi CR, Popik VV, Ting AY. J. Am. Chem. Soc. 2012;134:3720. doi: 10.1021/ja208090p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Xie J, Schultz PG. Annu. Rev. Biophys. Biomol. Struct. 2006;35:225. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 40.Liu CC, Schultz PG. Annu. Rev. Biochem. 2010;79:413. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 41.Chin JW. Annu. Rev. Biochem. 2014;83:379. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 42.Lang K, Chin JW. ACS Chem. Biol. 2014;9:16. doi: 10.1021/cb4009292. [DOI] [PubMed] [Google Scholar]
- 43.Patterson DM, Nazarova LA, Prescher JA. ACS Chem. Biol. 2014;9:592. doi: 10.1021/cb400828a. [DOI] [PubMed] [Google Scholar]
- 44.Blackman ML, Royzen M, Fox JM. J. Am. Chem. Soc. 2008;130:13518. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Devaraj NK, Weissleder R, Hilderbrand SA. Bioconjugate Chem. 2008;19:2297. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devaraj NK, Weissleder R. Acc. Chem. Res. 2011;44:816. doi: 10.1021/ar200037t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Šečkutė J, Devaraj NK. Curr. Opin. Chem. Biol. 2013;17:761. doi: 10.1016/j.cbpa.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikić I, Kang JH, Girona GE, Aramburu IV, Lemke EA. Nat. Protocols. 2015;10:780. doi: 10.1038/nprot.2015.045. [DOI] [PubMed] [Google Scholar]
- 49.Seitchik JL, Peeler JC, Taylor MT, Blackman ML, Rhoads TW, Cooley RB, Refakis C, Fox JM, Mehl RA. J. Am. Chem. Soc. 2012;134:2898. doi: 10.1021/ja2109745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lukinavičius G, Umezawa K, Olivier N, Honigmann A, Yang G, Plass T, Mueller V, Reymond L, Corrêa IR, Jr, Luo Z-G, Schultz C, Lemke EA, Heppenstall P, Eggeling C, Manley S, Johnsson K. Nat. Chem. 2013;5:132. doi: 10.1038/nchem.1546. [DOI] [PubMed] [Google Scholar]
- 51.Plass T, Milles S, Koehler C, Szymański J, Mueller R, Wießler M, Schultz C, Lemke EA. Angew. Chem., Int. Ed. 2012;51:4166. doi: 10.1002/anie.201108231. [DOI] [PubMed] [Google Scholar]
- 52.Blizzard RJ, Backus DR, Brown W, Bazewicz CG, Li Y, Mehl RA. J. Am. Chem. Soc. 2015;137:10044. doi: 10.1021/jacs.5b03275. [DOI] [PubMed] [Google Scholar]
- 53.Lang K, Davis L, Torres-Kolbus J, Chou C, Deiters A, Chin JW. Nat. Chem. 2012;4:298. doi: 10.1038/nchem.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lang K, Davis L, Wallace S, Mahesh M, Cox DJ, Blackman ML, Fox JM, Chin JW. J. Am. Chem. Soc. 2012;134:10317. doi: 10.1021/ja302832g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nikić I, Plass T, Schraidt O, Szymański J, Briggs JAG, Schultz C, Lemke EA. Angew. Chem., Int. Ed. 2014;53:2245. doi: 10.1002/anie.201309847. [DOI] [PubMed] [Google Scholar]
- 56.Yang Y, Lin S, Lin W, Chen PR. ChemBioChem. 2014;15:1738. doi: 10.1002/cbic.201400057. [DOI] [PubMed] [Google Scholar]
- 57.Nikić I, Lemke EA. Curr. Opin. Chem. Biol. 2015;28:164. doi: 10.1016/j.cbpa.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 58.Uttamapinant C, Howe JD, Lang K, Beránek V, Davis L, Mahesh M, Barry NP, Chin JW. J. Am. Chem. Soc. 2015;137:4602. doi: 10.1021/ja512838z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yount JS, Moltedo B, Yang Y-Y, Charron G, Moran TM, López CB, Hang HC. Nat. Chem. Biol. 2010;6:610. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yount JS, Karssemeijer RA, Hang HC. J. Biol. Chem. 2012;287:19631. doi: 10.1074/jbc.M112.362095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng T, Hang HC. J. Am. Chem. Soc. 2015;137:556. doi: 10.1021/ja502109n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Percher A, Ramakrishnan S, Thinon E, Yuan X, Yount JS, Hang HC. Proc. Natl. Acad. Sci. U. S. A. 2016;113:4302. doi: 10.1073/pnas.1602244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. Cell. 2009;139:1243. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, Wise HM, Kane L, Goulding D, Digard P, Anttila V, Baillie JK, Walsh TS, Hume DA, Palotie A, Xue Y, Colonna V, Tyler-Smith C, Dunning J, Gordon SB, Smyth RL, Openshaw PJ, Dougan G, Brass AL, Kellam P. Nature. 2012;484:519. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Diamond MS, Farzan M. Nat. Rev. Immunol. 2013;13:46. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor MT, Blackman ML, Dmitrenko O, Fox JM. J. Am. Chem. Soc. 2011;133:9646. doi: 10.1021/ja201844c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Darko A, Wallace S, Dmitrenko O, Machovina MM, Mehl RA, Chin JW, Fox JM. Chemical Science. 2014;5:3770. doi: 10.1039/C4SC01348D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Elliott TS, Townsley FM, Bianco A, Ernst RJ, Sachdeva A, Elsasser SJ, Davis L, Lang K, Pisa R, Greiss S, Lilley KS, Chin JW. Nat. Biotech. 2014;32:465. doi: 10.1038/nbt.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Borrmann A, Milles S, Plass T, Dommerholt J, Verkade JMM, Wießler M, Schultz C, van Hest JCM, van Delft FL, Lemke EA. ChemBioChem. 2012;13:2094. doi: 10.1002/cbic.201200407. [DOI] [PubMed] [Google Scholar]
- 70.Li J, Jia S, Chen PR. Nat. Chem. Biol. 2014;10:1003. doi: 10.1038/nchembio.1656. [DOI] [PubMed] [Google Scholar]
- 71.Hoffmann J-E, Plass T, Nikić I, Aramburu IV, Koehler C, Gillandt H, Lemke EA, Schultz C. Chem. Eur. J. 2015;21:12266. doi: 10.1002/chem.201501647. [DOI] [PubMed] [Google Scholar]
- 72.Yang KS, Budin G, Reiner T, Vinegoni C, Weissleder R. Angew. Chem., Int. Ed. 2012;51:6598. doi: 10.1002/anie.201200994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Devaraj NK, Hilderbrand S, Upadhyay R, Mazitschek R, Weissleder R. Angew. Chem., Int. Ed. 2010;49:2869. doi: 10.1002/anie.200906120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carlson JCT, Meimetis LG, Hilderbrand SA, Weissleder R. Angew. Chem., Int. Ed. 2013;52:6917. doi: 10.1002/anie.201301100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Erdmann RS, Takakura H, Thompson AD, Rivera-Molina F, Allgeyer ES, Bewersdorf J, Toomre D, Schepartz A. Angew. Chem., Int. Ed. 2014;53:10242. doi: 10.1002/anie.201403349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karver MR, Weissleder R, Hilderbrand SA. Bioconjugate Chem. 2011;22:2263. doi: 10.1021/bc200295y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grimm JB, English BP, Chen J, Slaughter JP, Zhang Z, Revyakin A, Patel R, Macklin JJ, Normanno D, Singer RH, Lionnet T, Lavis LD. Nat. Meth. 2015;12:244. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmied WH, Elsässer SJ, Uttamapinant C, Chin JW. J. Am. Chem. Soc. 2014;136:15577. doi: 10.1021/ja5069728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S. Chem. Biol. 2008;15:1187. doi: 10.1016/j.chembiol.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 80.Yanagisawa T, Hino N, Iraha F, Mukai T, Sakamoto K, Yokoyama S. Mol. BioSyst. 2012;8:1131. doi: 10.1039/c2mb05321g. [DOI] [PubMed] [Google Scholar]
- 81.Tian H, Sakmar TP, Huber T. ChemBioChem. 2015;16:1314. doi: 10.1002/cbic.201500030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lukinavicius G, Reymond L, D'Este E, Masharina A, Gottfert F, Ta H, Guther A, Fournier M, Rizzo S, Waldmann H, Blaukopf C, Sommer C, Gerlich DW, Arndt H-D, Hell SW, Johnsson K. Nat. Meth. 2014;11:731. doi: 10.1038/nmeth.2972. [DOI] [PubMed] [Google Scholar]
- 83.Lukinavicius G, Blaukopf C, Pershagen E, Schena A, Reymond L, Derivery E, Gonzalez-Gaitan M, D'Este E, Hell SW, Wolfram Gerlich D, Johnsson K. Nat. Commun. 2015:6. doi: 10.1038/ncomms9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jia R, Xu F, Qian J, Yao Y, Miao C, Zheng Y-M, Liu S-L, Guo F, Geng Y, Qiao W, Liang C. Cell. Microbiol. 2014;16:1080. doi: 10.1111/cmi.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chesarino NM, McMichael TM, Hach JC, Yount JS. J. Biol. Chem. 2014;289:11986. doi: 10.1074/jbc.M114.557694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li J, Yu J, Zhao J, Wang J, Zheng S, Lin S, Chen L, Yang M, Jia S, Zhang X, Chen PR. Nat. Chem. 2014;6:352. doi: 10.1038/nchem.1887. [DOI] [PubMed] [Google Scholar]
- 87.Rossin R, van den Bosch SM, ten Hoeve W, Carvelli M, Versteegen RM, Lub J, Robillard MS. Bioconjugate Chem. 2013;24:1210. doi: 10.1021/bc400153y. [DOI] [PubMed] [Google Scholar]
- 88.Yang J, Šečkutė J, Cole CM, Devaraj NK. Angew. Chem., Int. Ed. 2012;124:7594. [Google Scholar]
- 89.van Geel R, Pruijn GJM, van Delft FL, Boelens WC. Bioconjugate Chem. 2012;23:392. doi: 10.1021/bc200365k. [DOI] [PubMed] [Google Scholar]
- 90.Versteegen RM, Rossin R, ten Hoeve W, Janssen HM, Robillard MS. Angew. Chem., Int. Ed. 2013;52:14112. doi: 10.1002/anie.201305969. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.