Abstract
Yeast cells enter and undergo gametogenesis relatively asynchronously, making it technically challenging to perform stage-specific genomic and biochemical analyses. Cell-to-cell variation in the expression of the master regulator of entry into sporulation, IME1, has been implicated to be the underlying cause of asynchronous sporulation. Here, we find that timing of IME1 expression is of critical importance for inducing cells to undergo sporulation synchronously. When we force expression of IME1 from an inducible promoter in cells incubated in sporulation medium for 2 hr, the vast majority of cells exhibit synchrony during premeiotic DNA replication and meiotic divisions. Inducing IME1 expression too early or too late affects the synchrony of sporulation. Surprisingly, our approach for synchronous sporulation does not require growth in acetate-containing medium, but can be achieved in cells grown in rich medium until saturation. Our system requires solely IME1, because the expression of the N6-methyladenosine methyltransferase IME4, another key regulator of early sporulation, is controlled by IME1 itself. The approach described here can be combined easily with other stage-specific synchronization methods, and thereby applied to study specific stages of sporulation, or the complete sporulation program.
Keywords: gametogenesis, sporulation, synchrony, budding yeast, DNA replication, meiotic divisions, IME1, IME4, temporal
Gametogenesis—the process of gamete formation—is an essential cell differentiation program for all sexually reproducing organisms. During gametogenesis, diploid cells undergo a single round of DNA replication, followed by double-strand break formation, homologous recombination, and two consecutive nuclear divisions called meiosis I and II, to generate progeny containing half the number of chromosomes of the diploid parent cell (Marston and Amon 2004). The products of meiotic divisions are subsequently packaged to form gametes or spores in yeasts.
In budding yeast, initiation of gametogenesis, or sporulation, is controlled by a single master regulatory transcription factor called inducer of meiosis 1 (IME1) (Kassir et al. 1988). Ime1 activates transcription of the early genes during sporulation (Honigberg and Purnapatre 2003). Extrinsic and intrinsic signals control IME1 expression via the IME1 promoter (van Werven and Amon 2011). For efficient IME1 induction, glucose and nitrogen needs to be absent from the growth medium, and cells need to express both mating types (MATa and MATα). Another gene important for entry into gametogenesis is IME4, which encodes an enzyme that methylates the adenosine base of mRNAs to form N-6-methyladenosine (m6a) (Shah and Clancy 1992; Clancy et al. 2002). Previous work indicated that Ime4 promotes the accumulation of IME1 transcripts, suggesting that there is positive regulation between the two genes during entry into sporulation (Shah and Clancy 1992).
Yeast cells undergo gametogenesis relatively asynchronously, making it challenging to perform stage-specific analyses using whole cell population based assays. Nachman et al. (2007) showed that cell-to-cell variability in IME1 levels causes variation in timing of meiotic events in a population of cells. In contrast, nutritional history, cell cycle stage, or cell size did not affect timing of meiotic divisions (Nachman et al. 2007). We reveal new insights into IME1 signaling, and describe a method to synchronize premeiotic DNA replication and meiotic divisions. First, we demonstrate that timed expression of IME1, but not IME1 expression by itself, is sufficient to drive synchronous sporulation. In addition, we find that IME1 expression regulates IME4 expression, explaining our observation that cells readily enter sporulation highly synchronously when IME1 is induced alone. Surprisingly, our system does not require growth in acetate-containing medium, but synchronous sporulation can be induced directly in cells grown in rich medium until saturation before shifting to sporulation medium. Finally, we show that the approach described here can be combined with other stage-specific synchronization methods to study specific stages of sporulation, or the complete sporulation program.
Materials and Methods
Yeast strains
All yeast strains used in this paper were derived from the sporulation proficient SK1 strain background, and genotypes are listed in Supplemental Material, Table S1. The CUP1 promoter fusion with IME1 (pCUP-IME1) strain was made as follows: a region of the pFA6a-KanMX6-pCUP1A-3HA plasmid was amplified using the primers 5′-GCATTGATATTTTCAAACTTATATAATTAATAATAATTAATAGCGCTTAGTTTAAAGAAgaattcgagctcgtttaaac-3′ and 5′-GAAACCATCTTCTAAGGCAGCGTGAAGTTTTCCATG CATATCCGCTTGCATgcactgagcagcgtaatctg-3′. Uppercase letters refer to IME1-specific sequences, while lowercase letters refer to the sequences for amplifying the CUP1 promoter with N-terminal 3HA tags (Longtine et al. 1998). For the untagged version of pCUP-IME1, the pFA6a-KanMX6-pCUP1A-3HA plasmid was amplified using the primers 5′-GCATTGATATTTTCAAACTTATATAATTAATAATAATTAATAGCGCTTAGTTTAAAGAAgaattcgagctcgtttaaac-3′ and 5′-CTAAGGCAGCGTGAAGTTTTCCATGCATATCCGCTTGCATtttatgtgatgattgattgattg-3′. The strains were generated by a one-step promoter replacement protocol as described by Longtine et al. (1998). Subsequently, the haploid transformants were backcrossed, from which homozygous diploid cells were generated.
Growth and conditions for synchronous sporulation
Cells were grown in YPD [1% yeast extract, 2% peptone, 2% glucose, and supplemented with tryptophan (9.6 mg/l), uracil (2.4 mg/l), and adenine (1.2 mg/l)] and grown to exponential phase (OD600 < 2.0) at 30° and 300 rpm. While developing the protocol, we found that supplemented tryptophan can be left out from the YPD. For optimal aeration, the ratio of the total volume of the flask to the volume of medium was at least 10:1. Approximately 0.05 OD of exponentially growing yeast were inoculated into a new flask containing reduced glucose YPD [1% yeast extract, 2% peptone, 1% glucose, and supplemented with uracil (2.4 mg/l), and adenine (1.2 mg/l)]. Cultures reached OD600 = 11.0–12.0 after 16–18 hr, and the majority of the cells (∼90%) were single, unbudded cells as observed under a light microscope. The cells were then pelleted by centrifugation (2000 × g, 3 min, room temperature). The pellets were washed with sterile Mili-Q water, centrifuged again (2000 × g, 3 min, room temperature), and suspended in sporulation medium [1.0% (w/v) potassium acetate, supplemented with adenine/uracil (40 mg/l each), histidine/leucine/tryptophan (20 mg/l each), and 0.02% (w/v) raffinose)] at OD600 of 2.5. After 2 hr, copper (II) sulfate (50 µM) was added to induce expression from the CUP1 promoter and initiate sporulation synchronously. For some experiments sporulation was induced by growing cells in acetate containing pre-sporulation medium (BYTA) followed by shifting cells to sporulation medium as described previously (Berchowitz et al. 2013).
Budding index determination
Cells were grown in regular YPD [1% yeast extract, 2% peptone, 2% glucose, and supplemented with tryptophan (9.6 mg/l), uracil (2.4 mg/l), and adenine (1.2 mg/l)] and grown to exponential phase (OD600 < 2) at 30° and 300 rpm. Cells were transferred to new flasks (OD600 = 0.05) containing reduced glucose YPD [1% yeast extract, 2% peptone, 1% glucose, and supplemented with uracil (2.4 mg/l) and adenine (1.2 mg/l)] or regular YPD with 2% glucose. After 16–18 hr, budded and unbudded cells were counted under a light microscope.
Flow cytometry analysis
Premeiotic DNA replication was monitored by flow cytometry analysis (BD LSRFortessa, BD Biosciences). Cells were pelleted by centrifugation (∼2400 × g, 1 min, room temperature), and fixed in 80% (v/v) ethanol for at least 60 min before further processing. Fixed cells were pelleted by centrifugation (∼2400 × g, 1 min), and resuspended in 50 mM Tris-HCl pH 7.5. Cells were sonicated for a few seconds before treatment with 0.2 mg/ml ribonuclease A in 50 mM Tris-HCl pH 7.5 at 37° overnight. After ribonuclease A digestion, cells were stained with 50 µg/ml propidium iodide in FACs buffer (200 mM Tris-HCl pH 7.5, 211 mM NaCl and 78 mM MgCl2) for 1 hr at room temperature before flow cytometry analysis. Propidium iodide stained cells were excited with a 561 nm yellow-green laser, and signals were detected using a 610/20 yellow filter. Pulse shape analysis (pulse height against pulse area) was used to exclude clumps and doublets. DNA content from single cells was estimated with a histogram of counts against pulse area. At least 50,000 cells were used for the analysis of each sample.
Nuclei/DAPI counting
To monitor meiotic divisions by DAPI staining, cells were pelleted by centrifugation (∼2400 × g, 1 min, room temperature), and fixed in 80% (v/v) ethanol for at least 60 min before further processing. Subsequently, samples were pelleted by centrifugation (∼2400 × g, 1 min) and resuspended in PBS with DAPI (1 µg/ml). Cells were sonicated for a few seconds, and left in the dark at room temperature for at least 5 min. After DAPI staining, the proportion of cells containing one, two, three, or four DAPI masses were counted using a fluorescent microscope.
Computing the synchrony of meiotic divisions
The synchrony of meiotic divisions for each time course experiment was approximated by fitting a linear trend line from the first time point when meiotic divisions were detected to the first time point when 75% or more of the cells completed meiotic divisions. From these analyses, we calculated the period, or time taken for 75% of the cells to complete meiotic divisions. A more synchronously dividing population would take a shorter time to complete meiotic divisions. The average values from three independent experiments, and the SEM are included in the figures. To illustrate the statistical significance we used one way ANOVA, post hoc multiple comparison testing, and two tailed t-tests where appropriate (Prism 6, Graphpad). A p-value ≤ 0.05 was considered significant.
Western blotting
Levels of hemagglutinin (HA) epitope-tagged Ime1 and Ime4 were determined by western blotting using the procedures as described previously (Berchowitz et al. 2013). In brief, cells were pelleted by centrifugation (∼2400 × g, 1 min, room temperature), and resuspended in cold 5% trichloroacetic acid (TCA) for at least 10 min. The pellets were then washed with acetone, mixed with lysis buffer [50 mM Tris pH 7.5, 1 mM EDTA, 2.75 mM dithiothreitol (DTT)], and cells were broken using a mini beadbeater (BioSpec). Lysates were mixed with SDS loading buffer (187.5 mM Tris pH 6.8, 6% v/v β-mercaptoethanol, 30% v/v glycerol, 9% w/v SDS, 0.05% w/v Bromophenol Blue), and boiled for 5 min for denaturation. Proteins were separated by PAGE, and transferred onto PVDF membranes using the Mini Trans-Blot Cell (Bio-Rad). The membranes were blocked for 60 min in blocking buffer (1% w/v BSA, 1% w/v milk), before incubation with mouse anti-HA (12CA5, Sigma-Aldrich) at a 1:1000 dilution overnight at 4°. Membranes were washed in phosphate buffered saline with 0.01% Tween-20 (PBST), and incubated with anti-mouse HRP secondary antibodies (GE Healthcare) at a 1:5000 dilution. Membranes were imaged using Imagequant 600 RGB (GE Healthcare). We also monitored Hxk1 levels using an anti-hexokinase antibody (H2035, Stratech) at a 1:8000 dilution overnight at 4°. The IRDye 680RD donkey anti-rabbit secondary antibody (LI-COR) was used at a 1:15,000 dilution. Hxk1 levels were detected using an Odyssey Imager (LI-COR).
RT-qPCR
Total RNA was treated with DNAse and purified (Macherey-Nagel); 750 ng of total RNA was used for the reverse transcription reaction using Superscript III (Life Technologies), and single-stranded cDNA were quantified by real-time PCR using SYBR green mix (Life Technologies). To measure IME1 mRNA levels, random primers were used for the reverse transcription reaction. Since IME4 has antisense transcription, we used an IME4 sense-strand specific primer (5′-ATTCTGCTTGGCCTCAGCAT-3′), and an ACT1 sense-strand specific primer (5′-TTAGAAACACTTGTGGTGAA-3′) during the reverse transcription reaction. The IME1 and IME4 signals were normalized to ACT1 transcript levels. The qPCR primer sequences used for IME1 were: 5′-CAACGCCTCCGATAATGTATATG-3′ and 5′-ACGTCGAAGGCAATTTCTAATG-3′. The qPCR primer sequences used for IME4 were: 5′-ACCCATGCCAGAAAACTAGAGA-3′ and 5′-CGTAAATGCAATTTCCTGTCAA-3′. The qPCR primer sequences used for ACT1 were: 5′-GTACCACCATGTTCCCAGGTATT-3′ and 5′-AGATGGACCACTTTCGTCGT-3′.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) experiments were performed as described previously (van Werven et al. 2012). In short, cells were fixed with formaldehyde (1% for 20 min), which was quenched with glycine (125 mM). After breaking cells using a mini beadbeater (BioSpec), crosslinked chromatin was sheared by sonication using Bioruptor (Diagenode, six cycles of 30 sec on/off). Extracts were incubated with anti-V5 agarose beads (Sigma) for 2 hr, and beads were washed. Ume6-V5 binding was measured by real-time PCR using SYBR green mix (Life Technologies), and primers corresponding to the IME4 promoter (5′-CGTCTTTAGGCGGCTTTTGG-3′ and 5′-ACCGATCTTCCAGAATGCCG-3′) on a 7500 Fast Real-Time PCR system (Life Technologies). The mating type locus HMR (5′-ACGATCCCCGTCCAAGTTATG-3′ and 5′-CTTCAAAGGAGTCTTAATTTCCCTG-3′) was used as a nonbinding control (van Werven et al. 2012).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Synchronous sporulation requires specific timing of IME1 and IME4 induction
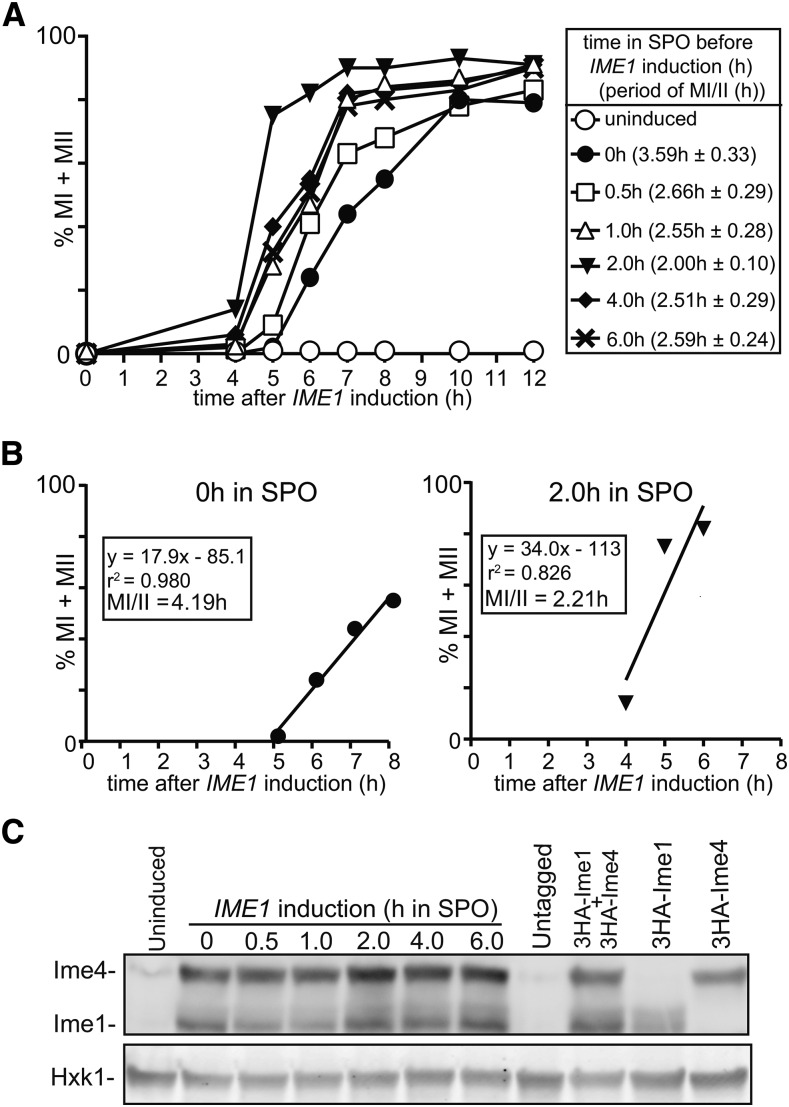
Whereas many of the laboratory yeast strains sporulate poorly, the sporulation-proficient strain background SK1 can undergo premeiotic DNA replication and meiosis division with a certain degree of synchrony (Cha et al. 2000; Stuart 2008). Nevertheless, to study specific stages of sporulation or meiosis, a highly synchronous cell population is desirable, which led to the development of different strategies to further improve the synchrony of sporulation (Wan et al. 2006; Carlile and Amon 2008; Berchowitz et al. 2013). Previous work showed that expressing IME1 together with IME4 from the inducible CUP1 promoter (CUP-IME1 and CUP-IME4) drives cells to undergo gametogenesis more synchronously compared to wild-type SK1 (Berchowitz et al. 2013). For this procedure, cells were grown in rich medium (YPD) until saturation, shifted to presporulation medium (BYTA), then transferred to sporulation medium (SPO). Subsequently, cells were incubated in SPO for 2 hr before IME1 and IME4 were induced with copper (II) sulfate. We speculated that the timing of IME1 and IME4 induction in SPO could be an important factor in regulating sporulation, since the expression of these genes are tightly regulated (Chu et al. 1998; Primig et al. 2000; van Werven and Amon 2011). To examine this, we expressed IME1 and IME4 at different times, and quantified the percentage of cells that completed meiotic divisions for a series of time points (Figure 1A). From these data, we estimated the synchrony of meiotic divisions by computing the time or period taken for 75% of the cells to complete meiotic divisions (see Materials and Methods for details) (Figure 1, A and B). The shorter the time or period, the more synchronous the meiotic divisions. We also conducted a one-way ANOVA and a post hoc Tukey’s test to compare the effect of expressing IME1 and IME4 at different times on the period taken to complete meiotic divisions. The ANOVA showed that the effect of expressing IME1 and IME4 at different times was statistically significant, F(5,12) = 3.82, P = 0.0265. We observed an improvement in the synchrony of meiotic divisions significantly when IME1 and IME4 were induced at 2 hr after shifting to SPO instead of at 0 hr (4.19 hr compared to 2.21 hr, P = 0.0112) (Figure 1B). Interestingly, inducing IME1 and IME4 either earlier or later resulted did not improve the synchrony significantly (P > 0.05), suggesting that there is an optimal period to induce the two master regulators. The differences in kinetics cannot be explained by Ime1 and Ime4 protein levels since they were comparable between the different samples (Figure 1C). In conclusion, our result shows that the timing of IME1 and IME4 induction in sporulation medium contributes to synchronous meiotic divisions.
Figure 1.
Synchronous sporulation requires specific timing of IME1 and IME4 induction. (A) Kinetics of meiotic divisions in diploid cells harboring CUP1 promoter fusions with IME1 and IME4 (pCUP-IME1/pCUP-IME4) (FW1810). Cells were grown overnight in rich medium (YPD), diluted to presporulation medium (BYTA), and grown for another 16 hr. Subsequently cells were transferred to sporulation medium (SPO), and IME1 and IME4 were induced at 0, 0.5, 1, 2, 4, and 6 hr in SPO. Samples were collected at 4 hr after induction up to 12 hr with a 1-hr interval, fixed in ethanol, nuclei were stained with DAPI, and DAPI masses were counted. Cells that harbored two, three, or four DAPI masses were classified as cells undergoing meiosis I or meiosis II (% MI + MII, y-axis). For each time point, at least 200 cells were counted. The time after IME1/IME4 induction is plotted on the x-axis. From each time course experiment, we also computed the time or period taken for 75% of the cells to complete meiotic divisions (see Materials and Methods for details). This number is displayed in brackets next to the legend, and represents the mean number of hours from three independent experiments followed by the SEM. (B) Graph to illustrate how we determined the time or period taken to complete meiotic divisions when IME1 and IME4 were induced at 0 or 2 hr after shifting cells to SPO medium as described in (A). A linear trend line was fitted from the first time point where meiotic divisions were detected, to the time point where 75% or more of the cells completed meiotic divisions. From the function, we calculated the period or time taken for 75% of the cells to complete meiotic divisions (MI/II). (C) Western blot showing Ime1 and Ime4 protein levels in cells described in (A). Samples were taken at 2 hr after inducing IME1 and IME4. Ime1 and Ime4 levels were detected by anti-hemagglutinin (HA) antibodies. We also measured Ime1 and Ime4 in an untagged control (FW1511), and in cells that contain HA-tagged IME1 (FW2444), or IME4 (FW2480) alone. To control for loading, Hxk1 levels were also determined.
Cells do not require growth in acetate-containing medium prior to induction of synchronous sporulation
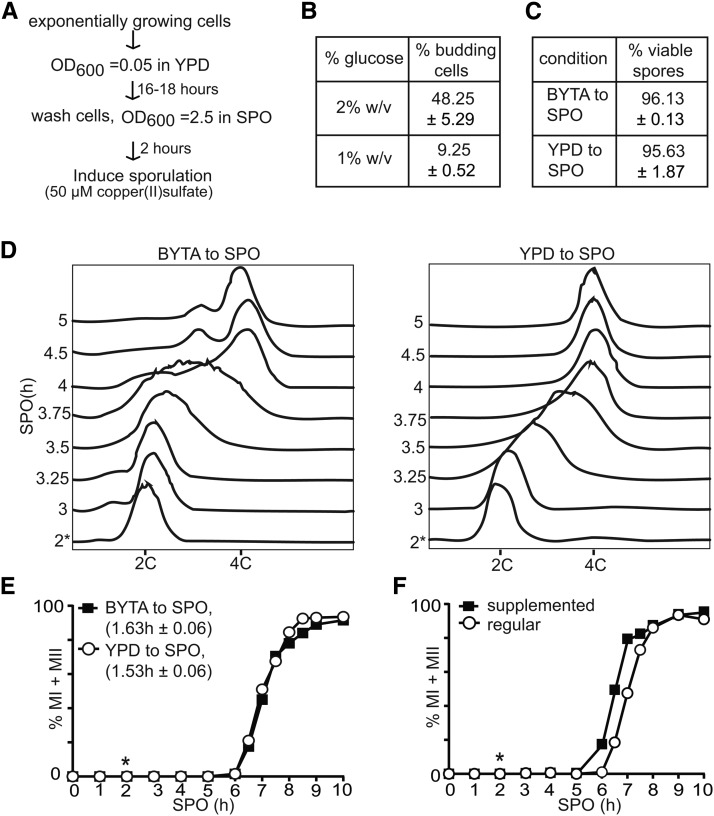
Efficient IME1 transcription requires glucose and nitrogen starvation, and the presence of a nonfermentable carbon source in the growth medium (Kassir et al. 1988). To obtain high levels of IME1 in SPO, cells are usually pregrown in acetate-containing medium. In contrast, induction of transcription from the CUP1 promoter requires solely the presence of copper ions in the medium. If variability in the onset of meiotic divisions is dependent largely on IME1 and IME4 levels, then pregrowth in acetate-containing medium should be dispensable when entry into sporulation is induced from the CUP1 promoter. Hence, we tested if the pCUP-IME1/pCUP-IME4 system can induce gametogenesis synchronously when cells were pregrown in glucose containing medium (YPD), and shifted to SPO directly (Figure 2A). To ensure that all cells were arrested as unbudded cells, we grew cells in YPD with reduced glucose (1% w/v instead of 2% w/v) for 16–18 hr. Whereas the majority of cells were budding when grown in medium with standard glucose levels, the reduced glucose condition enriched for unbudded cells (∼90%) after an overnight culture (Figure 2B). Cells pregrown in YPD or BYTA also gave rise to viable spores (Figure 2C). Next, we shifted cells to SPO medium, incubated cells for 2 hr, subsequently induced IME1 and IME4 with copper (II) sulfate, and measured the kinetics of meiotic S-phase and meiotic divisions. We observed that cells pregrown in BYTA or YPD both completed premeiotic DNA replication in ∼60 min (Figure 2D). Remarkably, in the YPD to SPO condition, the DNA profile showed intermediate peaks for several time points (between 2C and 4C) indicating that the population of cells underwent DNA replication with a high degree of synchrony (Figure 2D). When the cells were pregrown in BYTA, these intermediate peaks were less pronounced. In addition, we also measured the rate of meiotic divisions, and found the majority of cells completed meiotic divisions within comparable periods in both conditions (Figure 2E). We conducted a two-tailed t-test, and found that the time taken to complete meiotic divisions under both conditions was not statistically significantly different (P > 0.05). Overall, our results show that synchronous DNA replication and meiotic divisions can be induced from cells precultured until saturation in nutrient-rich medium containing glucose.
Figure 2.
Cells do not require growth in acetate-containing medium prior to induction of synchronous sporulation. (A) Flowchart for inducing synchronous sporulation. Diploid pCUP-IME1/pCUP-IME4 (FW1810) cells were grown to exponential phase for 6–7 hr in YPD. Cells were subsequently diluted to YPD medium with 1% glucose and grown for 16–18 hr to an OD600 of 11–12 to obtain mostly unbudded G1 cells. Cells were then pelleted by centrifugation, washed with sterile water and resuspended to a final OD600 of 2.5 in SPO; 50 µM copper (II) sulfate was added 2 hr after the cells were transferred to SPO to induce IME1 and IME4. (B) Budding index of cells cultured for 16 hr in YPD with different glucose concentrations. The mean and SEM from three independent experiments is shown, and n = 400 cells were counted for each repeat. (C) Spore viability of the pCUP-IME1/pCUP-IME4 strain. Cells were grown overnight in YPD, and induced to sporulate in SPO after transfer from YPD or presporulation media (BYTA). Sporulation was induced using standard protocols (BYTA to SPO), or by using the method described in (A) (YPD to SPO). Copper (II) sulfate was added 2 hr after the cells were transferred to SPO. Tetrads were collected 24 hr after IME1 induction, dissected, and assayed for viability (n = 160 spores). The mean value of three independent experiments plus the SEM is shown. (D) Flow cytometry analysis of DNA content of cells cultured in either reduced glucose YPD or presporulation medium (BYTA) before shifting to SPO. Samples were taken at indicated time points, fixed, and DNA content was measured by propidium iodide staining. At least 50,000 cells were analyzed at each time point. (E) Kinetics of meiotic divisions in cells as described in (C) and (D). For determining the kinetics of meiotic divisions, samples were taken at the indicated time point, fixed, and DAPI masses were counted. Cells that harbored two, three, or four DAPI masses were classified as cells undergoing meiosis I or meiosis II (% MI + MII). For each time point, at least 200 cells were counted. We also computed the time or period taken for 75% of the cells to complete meiotic divisions (see Materials and Methods for details). This number is displayed in brackets next to the legend, and represents the mean number of hours followed by the SEM of three independent experiments. (F) Kinetics of meiotic divisions of the pCUP-IME1/pCUP-IME4 strain as described (A) and (E), except that sporulation was induced in either regular SPO or supplemented SPO (see Materials and Methods). The graph displays a representative experiment from three repeated experiments. *, time of induction of IME1, 2 hr after the cells were transferred to SPO.
We also examined whether the composition of SPO medium influences the synchrony of meiotic divisions of cells pregrown in YPD. To do so, we increased the level of acetate in SPO from 0.3 to 1.0% w/v, and the medium was supplemented with amino acids and adenine (see Materials and Methods for details). Cells started meiotic divisions slightly earlier when using the supplemented SPO instead of the regular SPO (Figure 2F). Hence, we decided to use the supplemented SPO for the experiments described in the remainder of the manuscript.
Induction of IME1 is sufficient to induce gametogenesis synchronously
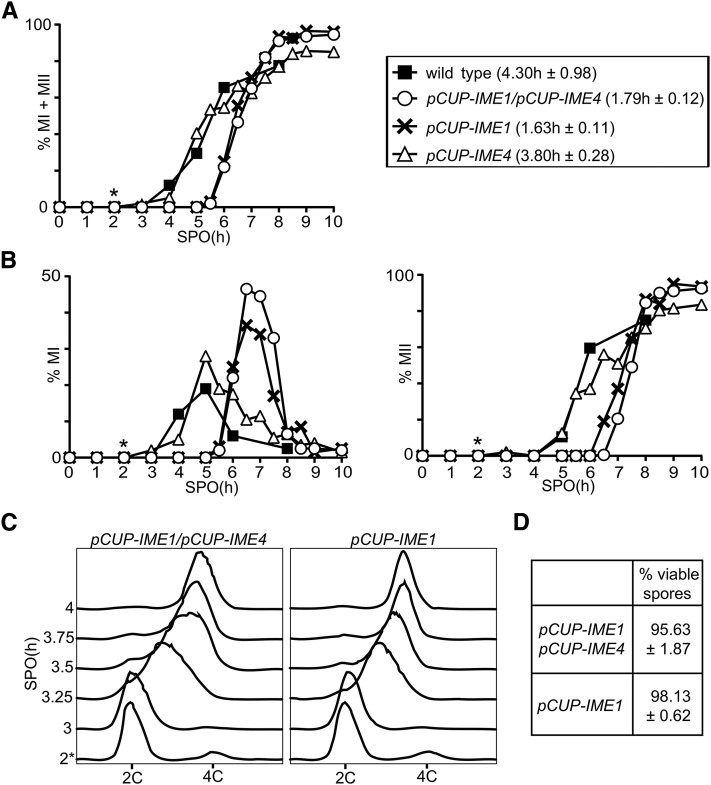
Both IME1 and IME4 have been shown to promote entry into sporulation (van Werven and Amon 2011). Whereas ime4 deletion mutants do not sporulate in certain strain backgrounds, but do in others, IME1 is essential for sporulation in Saccharomyces cerevisiae (Kassir et al. 1988; Shah and Clancy 1992; Hongay et al. 2006). IME4 has been implicated to positively regulate IME1 expression (Shah and Clancy 1992). We hypothesized that, if IME1 and IME4 regulate each other, then perhaps synchronous sporulation should require controlled expression of either IME1 or IME4 alone. To test this, we measured the period taken to complete meiotic divisions when IME1, IME4, or both, were induced from the CUP1 promoter. One-way ANOVA showed that there was a statistically significant difference between the group means, F(3,8) = 6.97, P = 0.0127. We found that the kinetics of meiotic divisions of cells that express pCUP-IME4 only was comparable to wild-type control cells (Figure 3A). In contrast, the kinetics of meiotic division in cells harboring pCUP-IME1 alone, or both pCUP-IME1 and pCUP-IME4, significantly improved when compared to the wild-type control, which was confirmed by a post hoc Dunnett’s test (cf. 1.63 hr and 1.79–4.30 hr, P = 0.0166 and P = 0.0223, respectively). The results were similar when we examined the kinetics of meiosis I and meiosis II separately (Figure 3B). To investigate more closely whether induction of pCUP-IME1 alone is sufficient for synchronous sporulation, we also monitored the kinetics of premeiotic DNA replication (Figure 3C). We found that cells harboring either pCUP-IME1 or pCUP-IME1/pCUP-IME4, underwent premeiotic DNA replication synchronously within ∼45 min, and gave rise to viable spores (Figure 3D). In conclusion, temporal expression of IME1 alone is sufficient to induce synchronous sporulation.
Figure 3.
Induction of IME1 is sufficient to induce gametogenesis synchronously. (A) Kinetics of meiotic divisions in wild-type cells (FW1511), cells harboring IME1 and IME4 fused to the CUP1 promoter (pCUP-IME1/pCUP-IME4) (FW1810), pCUP-IME1 (FW2444), or pCUP-IME4 (FW2480). Cells were grown overnight in YPD, and shifted to SPO (1.0% w/v potassium acetate supplemented with adenine, uracil, and amino acids); 50 µM copper (II) sulfate was added 2 hr after the cells were transferred to SPO. Samples were taken at the indicated time point, fixed in ethanol, nuclei were stained with DAPI, and DAPI masses were counted. Cells that harbored two, three, or four DAPI masses were classified as cells undergoing meiosis I or meiosis II (% MI + MII). For each time point, at least 200 cells were counted. We also computed the time or period taken for 75% of the cells to complete meiotic divisions (see Materials and Methods for details). This number is displayed in brackets next to the legend, and represents the mean number of hours followed by the SEM of three independent experiments. (B) Similar to A except that the percentages of bi- (left panel), tri-, and tetra-nucleate (right panel) cells are shown. (C) Flow cytometry analysis of DNA content of pCUP-IME1/pCUP-IME4 (FW1810) and the pCUP-IME1 (FW2444) cells that were induced to sporulate as described in (A). Samples were taken at indicated time points, fixed, and DNA content was measured by propidium iodide staining; 50 µM copper (II) sulfate was added 2 hr after the cells were transferred to SPO. At least 50,000 cells were analyzed at each time point. (D) Spore viability of the pCUP-IME1/pCUP-IME4 (FW1810) and the pCUP-IME1 (FW2444) cells that were induced to sporulate as described in (A). Tetrads were collected 24 hr after induction, dissected, and assayed for viability (n = 160 spores). The average result and the SEM of three independent experiments is shown. *, time of induction.
IME1 directly regulates the expression of IME4
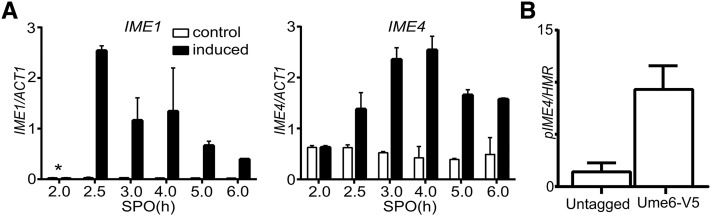
The observation that temporal expression of IME1, but not of IME4, generates a high degree of synchrony during DNA replication and meiotic divisions, prompted us to revisit how the two genes regulate each other. We hypothesized that IME1 directly or indirectly regulates IME4 expression. To examine this possibility, we measured IME4 transcript levels in cells harboring pCUP-IME1 in the presence or absence of copper (II) sulfate. As expected, IME1 transcript levels increased when copper ions were added to the SPO medium (Figure 4A). Since IME4 is also regulated by an antisense transcript, we specifically quantified IME4 sense mRNA using a transcript-specific primer in the reverse transcription reaction (Hongay et al. 2006; Gelfand et al. 2011). We found that IME4 transcript levels significantly increased when IME1 was induced, suggesting that Ime1 stimulates IME4 transcription (Figure 4B). Data from a genome-wide study indicated that IME4 is regulated directly by the repressor Ume6 (Williams et al. 2002). During early sporulation, Ime1 interacts with Ume6 to form a transcription-activating complex for the expression of early meiotic genes (Bowdish et al. 1995; Rubin-Bejerano et al. 1996). To test whether Ume6 indeed binds the IME4 promoter, we identified the canonical URS1 motif (TAGGCGGC) sequence at −234 bp upstream in the IME4 promoter. More importantly, we found that Ume6 was bound directly to the IME4 promoter, as shown by ChIP (Figure 4C). In conclusion, our results show that IME1 directly regulates the expression of IME4, explaining why IME1 can single-handedly induce synchronous sporulation. These results also suggest that IME1 and IME4 act in a positive feedback loop to stimulate the expression of each other.
Figure 4.
IME1 directly regulates the expression of IME4. (A) Induction of IME1 promotes IME4 expression. Diploid cells harboring pCUP-IME1 (FW2444) were transferred to SPO. Cells were either untreated (control) or treated with copper (II) sulfate (induced), and samples were collected at different time points. Total RNA was isolated, reverse transcribed, and IME1 (left panel) or IME4 (right panel) mRNA levels were measured by quantitative PCR. To quantify IME4 levels, primers specific for the IME4 and ACT1 sense strand were used in the reverse transcription reaction. Signals were normalized to ACT1 levels. The values and error bars represent two independent experiments. (B) Ume6 binds to the promoter of IME4. Diploid cells harboring Ume6 tagged with the V5 epitope (FW1208) and a wild-type control strain (FW1511) were grown in YPD to saturation. Cells were fixed with formaldehyde, and cells were processed for ChIP analyses (see Materials and Methods for details). DNA fragments specific to the IME4 promoter (pIME4) were amplified and quantified by qPCR. Signals were normalized to the HMR locus. The error bars represent the SE of at least three experiments. *, time of induction of IME1—2 hr after the cells were transferred to SPO.
Combining the pCUP-IME1 system with another synchronization method
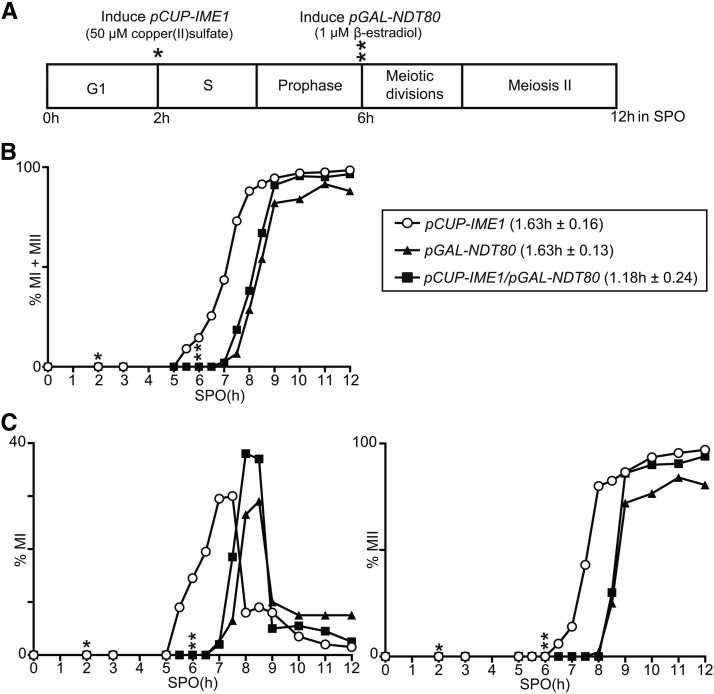
Different genetic approaches have been used to synchronize cells at different stages of sporulation (Benjamin et al. 2003; Wan et al. 2006; Carlile and Amon 2008). One approach makes use of controlled expression of the transcription factor NDT80, and, as a result, cells undergo meiotic divisions synchronously (Benjamin et al. 2003; Carlile and Amon 2008). The Ndt80 transcription factor promotes the expression of numerous genes that regulate meiotic divisions, also known as middle genes (Xu et al. 1995; Chu et al. 1998). Effective induction of NDT80 is achieved by controlling its expression from the GAL1 promoter (pGAL-NDT80) and the transcription factor GAL4-ER, consisting of the Gal4 DNA binding domain fused to the estrogen receptor binding domain. In the presence of β-estradiol, pGAL-NDT80 cells induce NDT80, and exit from pachytene arrest to undergo meiotic divisions (Benjamin et al. 2003; Carlile and Amon 2008). The pGAL-NDT80 system specifically synchronizes meiotic divisions during gametogenesis, but, unlike the pCUP-IME1 system, this method does not synchronize the events prior to meiotic chromosome segregation. To examine whether it is possible to combine the pCUP-IME1 system with the pGAL-NDT80 system, we generated a diploid strain with both synchronization systems. The early and middle stages of sporulation were initiated by IME1 at 2 hr, and NDT80 at 6 hr, after shifting cells to SPO, respectively (Figure 5A). We found that the pCUP-IME1 and pGAL-NDT80 cells had a similar degree of synchrony of meiotic divisions (Figure 5B). The pCUP-IME1/pGAL-NDT80 strain showed a minor improvement in synchrony, which was not statistically significant (P > 0.05), when compared to cells expressing pCUP-IME1 or pGAL-NDT80 alone (1.18 hr compared to 1.63 and 1.63 hr) (Figure 5B). We observed a similar trend when we examined meiosis I and meiosis II divisions separately, showing that the pCUP-IME1 or pGAL-NDT80 systems can be combined (Figure 5C). With the pCUP-IME1/pGAL-NDT80 system, we can synchronize cells at the level of premeiotic DNA replication until completion of meiotic divisions, with the added advantage of being able to control entry into the early and middle stages of gametogenesis.
Figure 5.
pCUP-IME1 system can be combined with the pGAL-NDT80 system to improve synchrony of sporulation. (A) Scheme of experimental setup. The diploid cells harboring IME1 fused to CUP promoter (FW2444), NDT80 expressed from the GAL promoter together with Gal4 fused to the estrogen receptor (GAL4.ER pGAL-NDT80) (FW1541) or a strain expressing both (pCUP-IME1 and GAL4.ER pGAL-NDT80) (FW2795) were grown in YPD overnight. Cells harboring GAL4.ER GAL-NDT80 (FW1541) were transferred to presporulation medium (BYTA). Subsequently, cells were pelleted by centrifugation, washed with sterile water and resuspended to a final OD600 of 2.5 in SPO; 50 µM copper (II) sulfate was added 2 hr after the cells were transferred to SPO, and 1 µM β-estradiol was added 6 hr after transfer to SPO. (B) Kinetics of meiotic divisions in strains, using procedures described in (A). Samples were taken at the indicated time points, fixed in ethanol, nuclei were stained with DAPI, and DAPI masses were counted. Cells that harbored two, three, or four DAPI masses were classified as cells undergoing meiosis I or meiosis II (% MI + MII). For each time point, at least 200 cells were counted. We also computed the time or period taken for 75% of the cells to complete meiotic divisions (see Materials and Methods for details). This number is displayed in brackets next to the legend, and represents the mean number of hours followed by the SEM of three independent experiments. (C) Similar to B except that the percentages of bi- (left panel), or tri- and tetra-nucleate (right panel) cells (n = 200 cells) of strains described in (A) were determined. *, time of IME1 induction; **, time of NDT80 induction.
Discussion
Here, we demonstrate that temporal expression of a single gene, IME1, induces sporulation synchronously in budding yeast. Our approach requires neither preculturing in acetate-containing medium nor expressing IME4 from a heterologous promoter. The system described here can be combined easily with other synchronization methods, and will be of use for studying specific stages of sporulation, or the complete sporulation program.
Our data indicate that induction of synchronous sporulation requires a specified timing of IME1 induction. Interestingly, in wild-type cells, the IME1 promoter, like in synchronous sporulation, is active at 2 hr after shifting to sporulation medium (Inai et al. 2007; Nachman et al. 2007). Given that nutrient availability is an important trigger for IME1 and sporulation, perhaps nutrients also control timing of sporulation after IME1 induction (Jambhekar and Amon 2008; van Werven and Amon 2011) . Inducing IME1 too late could affect sporulation, because cells have been starved for prolonged times, whereas inducing IME1 too early does not result in optimal sporulation because cells are not ready. In line with this hypothesis, in a recent report we showed that a certain level of nutrient-sensing target of rapamycin complex (TORC1) activity is needed for sporulation (Weidberg et al. 2016). Too much or too little TORC1 affects sporulation negatively. Perhaps, TORC1 activity is most optimal for synchronous sporulation at 2 hr in SPO. Another explanation is that downstream targets of IME1 are not properly activated because the meiosis promoting kinases Rim11 and Rim15 are not active (Bowdish et al. 1994; Rubin-Bejerano et al. 1996; Vidan and Mitchell 1997; Pedruzzi et al. 2003; Sarkar et al. 2014). This can explain why inducing IME1 too early did not result in synchronous sporulation, but cannot explain the reduced synchrony when IME1 is induced too late. More work is needed to pinpoint why timing of IME1 expression is critical for synchronous sporulation.
Our work sheds light on how IME1 and IME4 regulate each other in wild-type cells. It has been shown that ime4 mutant cells have diminished levels of IME1 (Shah and Clancy 1992). In addition, one report showed that IME1 transcripts contain the m6A modification, suggesting that Ime4 controls IME1 directly (Bodi et al. 2010). However, genome-wide sequencing of m6A did not identify the modification in IME1 (Schwartz et al. 2013). In this study, we demonstrate that IME1 can also directly regulate IME4 expression. Our data show that IME4 levels increased when IME1 was induced. We also find that the Ume6 repressor is bound to the promoter of IME4. Others have shown that IME4 transcripts accumulate later in sporulation than IME1, which also supports the idea that IME4 can be downstream of IME1 (Primig et al. 2000; Nachman et al. 2007). We propose that IME1 and IME4 can positively regulate each other. The advantage of such dynamic regulation is that it allows for rapid accumulation of both transcripts when cells are ready to undergo sporulation.
Several other approaches have been used to synchronize cells throughout, or at specific stages of, sporulation. First, the sporulation-proficient SK1 strain background can undergo sporulation efficiently, and with some degree of synchrony, when specific growth conditions are adopted (Kane and Roth 1974; Falk et al. 2010; Borner and Cha 2015). We show that the pCUP-IME1 strain reaches a much better synchrony in comparison to wild-type SK1. Second, mutations that cause cells to arrest at specific stages of gametogenesis are also used to synchronize cell populations. For example, ime2 mutants arrest prior to DNA replication, whereas ndt80 mutants arrest in meiotic prophase I (Xu et al. 1995; Dirick et al. 1998). Although these approaches are useful for studying specific stages, they have several limitations. For example, only one stage per mutant can be studied, and not all stages can be arrested. Third, there are other “block and release” genetic approaches that reversibly arrest, and then synchronize, cell populations in certain stages of sporulation. For example, the pGAL-NDT80 system synchronizes meiotic divisions (see previous section) (Carlile and Amon 2008). Another example of stage-specific synchronization is the analog-sensitive allele of CDC7 (cdc7-as3), which is used to arrest cells following premeiotic S-phase, and synchronizes cells through homologous recombination and meiosis I (Wan et al. 2006; Lo et al. 2008; Wan et al. 2008). However, these approaches synchronize cells only for a selective part of gametogenesis. Our data show that the pCUP-IME1 system can achieve a high degree of synchrony during premeiotic DNA replication and meiotic divisions. The pCUP-IME1 system also dispenses with the need to use presporulation medium, which shortens the procedure to 2 d. The pCUP1-IME1 system can be used alone, or combined with other synchronization systems, as a tool to profile gene expression or protein production patterns throughout gametogenesis. Finally, the high degree of synchrony achieved by our method will be useful in dissecting the different stages in finer detail, or to study temporal coordination and regulation of events during gametogenesis.
Supplementary Material
Acknowledgments
We are grateful to members of the Van Werven lab for their critical reading of the manuscript. This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001203), the UK Medical Research Council (FC001203), and the Wellcome Trust (FC001203).
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.034983/-/DC1.
Communicating editor: M. C. Zetka
Literature Cited
- Benjamin K. R., Zhang C., Shokat K. M., Herskowitz I., 2003. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 17: 1524–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz L. E., Gajadhar A. S., van Werven F. J., De Rosa A. A., Samoylova M. L., et al. , 2013. A developmentally regulated translational control pathway establishes the meiotic chromosome segregation pattern. Genes Dev. 27: 2147–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodi Z., Button J. D., Grierson D., Fray R. G., 2010. Yeast targets for mRNA methylation. Nucleic Acids Res. 38: 5327–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner, G. V., and R. S. Cha, 2015 Induction and analysis of synchronous meiotic yeast cultures. Cold Spring Harb. Protoc. 2015: 908–913.pdb prot085035. [DOI] [PubMed] [Google Scholar]
- Bowdish K. S., Yuan H. E., Mitchell A. P., 1994. Analysis of RIM11, a yeast protein kinase that phosphorylates the meiotic activator IME1. Mol. Cell. Biol. 14: 7909–7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowdish K. S., Yuan H. E., Mitchell A. P., 1995. Positive control of yeast meiotic genes by the negative regulator UME6. Mol. Cell. Biol. 15: 2955–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile T. M., Amon A., 2008. Meiosis I is established through division-specific translational control of a cyclin. Cell 133: 280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha R. S., Weiner B. M., Keeney S., Dekker J., Kleckner N., 2000. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 14: 493–503. [PMC free article] [PubMed] [Google Scholar]
- Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., et al. , 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705. [DOI] [PubMed] [Google Scholar]
- Clancy M. J., Shambaugh M. E., Timpte C. S., Bokar J. A., 2002. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 30: 4509–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick L., Goetsch L., Ammerer G., Byers B., 1998. Regulation of meiotic S phase by Ime2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science 281: 1854–1857. [DOI] [PubMed] [Google Scholar]
- Falk J. E., Chan A. C., Hoffmann E., Hochwagen A., 2010. A Mec1- and PP4-dependent checkpoint couples centromere pairing to meiotic recombination. Dev. Cell 19: 599–611. [DOI] [PubMed] [Google Scholar]
- Gelfand B., Mead J., Bruning A., Apostolopoulos N., Tadigotla V., et al. , 2011. Regulated antisense transcription controls expression of cell-type-specific genes in yeast. Mol. Cell. Biol. 31: 1701–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongay C. F., Grisafi P. L., Galitski T., Fink G. R., 2006. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127: 735–745. [DOI] [PubMed] [Google Scholar]
- Honigberg S. M., Purnapatre K., 2003. Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J. Cell Sci. 116: 2137–2147. [DOI] [PubMed] [Google Scholar]
- Inai T., Yukawa M., Tsuchiya E., 2007. Interplay between chromatin and trans-acting factors on the IME2 promoter upon induction of the gene at the onset of meiosis. Mol. Cell. Biol. 27: 1254–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A., Amon A., 2008. Control of meiosis by respiration. Curr. Biol. 18: 969–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane S. M., Roth R., 1974. Carbohydrate metabolism during ascospore development in yeast. J. Bacteriol. 118: 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassir Y., Granot D., Simchen G., 1988. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell 52: 853–862. [DOI] [PubMed] [Google Scholar]
- Lo H. C., Wan L., Rosebrock A., Futcher B., Hollingsworth N. M., 2008. Cdc7-Dbf4 regulates NDT80 transcription as well as reductional segregation during budding yeast meiosis. Mol. Biol. Cell 19: 4956–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Marston A. L., Amon A., 2004. Meiosis: cell-cycle controls shuffle and deal. Nat. Rev. Mol. Cell Biol. 5: 983–997. [DOI] [PubMed] [Google Scholar]
- Nachman I., Regev A., Ramanathan S., 2007. Dissecting timing variability in yeast meiosis. Cell 131: 544–556. [DOI] [PubMed] [Google Scholar]
- Pedruzzi I., Dubouloz F., Cameroni E., Wanke V., Roosen J., et al. , 2003. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol. Cell 12: 1607–1613. [DOI] [PubMed] [Google Scholar]
- Primig M., Williams R. M., Winzeler E. A., Tevzadze G. G., Conway A. R., et al. , 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26: 415–423. [DOI] [PubMed] [Google Scholar]
- Rubin-Bejerano I., Mandel S., Robzyk K., Kassir Y., 1996. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional represssor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol. Cell. Biol. 16: 2518–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Dalgaard J. Z., Millar J. B., Arumugam P., 2014. The Rim15-endosulfine-PP2ACdc55 signalling module regulates entry into gametogenesis and quiescence via distinct mechanisms in budding yeast. PLoS Genet. 10: e1004456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Agarwala S. D., Mumbach M. R., Jovanovic M., Mertins P., et al. , 2013. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155: 1409–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J. C., Clancy M. J., 1992. IME4, a gene that mediates MAT and nutritional control of meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 1078–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D., 2008. The meiotic differentiation program uncouples S-phase from cell size control in Saccharomyces cerevisiae. Cell Cycle 7: 777–786. [DOI] [PubMed] [Google Scholar]
- van Werven F. J., Amon A., 2011. Regulation of entry into gametogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366: 3521–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven F. J., Neuert G., Hendrick N., Lardenois A., Buratowski S., et al. , 2012. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell 150: 1170–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidan S., Mitchell A. P., 1997. Stimulation of yeast meiotic gene expression by the glucose-repressible protein kinase Rim15p. Mol. Cell. Biol. 17: 2688–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L., Zhang C., Shokat K. M., Hollingsworth N. M., 2006. Chemical inactivation of cdc7 kinase in budding yeast results in a reversible arrest that allows efficient cell synchronization prior to meiotic recombination. Genetics 174: 1767–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L., Niu H., Futcher B., Zhang C., Shokat K. M., et al. , 2008. Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev. 22: 386–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H., Moretto F., Spedale G., Amon A., van Werven F. J., 2016. Nutrient control of yeast gametogenesis is mediated by TORC1, PKA and energy availability. PLoS Genet. 12: e1006075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. M., Primig M., Washburn B. K., Winzeler E. A., Bellis M., et al. , 2002. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc. Natl. Acad. Sci. USA 99: 13431–13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Ajimura M., Padmore R., Klein C., Kleckner N., 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 6572–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.