Abstract
Feed efficiency (FE) is essential for pig production. In this study, 300 significantly differentially expressed (DE) transcripts, including 232 annotated genes, 28 cis-natural antisense transcripts (cis-NATs), and 40 long noncoding RNAs (lncRNAs), were identified between the liver of Yorkshire pigs with extremely high and low FE. Among these transcripts, 25 DE lncRNAs were significantly correlated with 125 DE annotated genes at a transcriptional level. These DE genes were enriched primarily in vitamin A (VA), fatty acid, and steroid hormone metabolism. VA metabolism is regulated by energy status, and active derivatives of VA metabolism can regulate fatty acid and steroid hormones metabolism. The key genes of VA metabolism (CYP1A1, ALDH1A2, and RDH16), fatty acid biosynthesis (FASN, SCD, CYP2J2, and ANKRD23), and steroid hormone metabolism (CYP1A1, HSD17B2, and UGT2B4) were significantly upregulated in the liver of high-FE pigs. Previous study with the same samples indicated that the mitochondrial function and energy expenditure were reduced in the muscle tissue of high-FE pigs. In conclusion, VA metabolism in liver tissues plays important roles in the regulation of FE in pigs by affecting energy metabolism, which may mediate fatty acid biosynthesis and steroid hormone metabolism. Furthermore, our results identified novel transcripts, such as cis-NATs and lncRNAs, which are also involved in the regulation of FE in pigs.
Keywords: Feed efficiency, liver, vitamin A metabolism, pig
Feed efficiency (FE) is an important economic trait in the pig industry. Feed cost accounts for > 60% of the total production costs. As such, FE should be improved to enhance the benefits of pig production. Thus far, two highly correlated indicators, i.e., feed conversion ratio (FCR) and residual feed intake (RFI), are used to measure FE, and a low FCR/RFI level denotes an improvement in FE (Do et al. 2013). Microarray analysis has revealed that lipogenesis and steroidogenesis in liver tissues are related to the FE of pigs (Lkhagvadorj et al. 2010). Selection on RFI in growing pigs indicates that catabolic pathway activities in liver and muscle increase in low-FE pigs (Le Naou et al. 2012). Meanwhile, the activities of lactate dehydrogenase and β-hydroxylacylCoA dehydrogenase in the liver increase in high-FE pigs (Le Naou et al. 2012). In our previous study, the expression of mitochondrial genes was downregulated in the muscle tissue of high-FE pigs (Jing et al. 2015). These studies have suggested that the energy metabolism of liver and muscle tissues are essential for the regulation of FE of pigs.
The liver maintains the homeostasis of metabolic processes, including energy and vitamin metabolism. Approximately 70–95% of vitamin A (VA) is stored in the liver of some mammals. VA and its metabolite retinoic acid (RA), which includes all-trans-RA (atRA), 9-cis-RA, and 13-cis-RA etc. (Heyman et al. 1992; Lampen et al. 2000), modulate energy balance by regulating the function of adipose cells and carbohydrate metabolism (Bonet et al. 2012; Obrochta et al. 2015). In mice, RA treatment can reduce body fat and improve insulin sensitivity (Berry and Noy 2009; Manolescu et al. 2010). In young rats, the synergistic effect of VA and high-fat diet increases adiposity (Redonnet et al. 2008). In young ferrets, chronic supplementation with β-carotene, which is a natural material for VA synthesis, increase their body weight and subcutaneous fat mass (Murano et al. 2005). The short-term atRA treatment in adulthood is associated with a decrease in adiposity in ferrets (Sánchez et al. 2009). The effect of VA and its metabolites on energy metabolism may be dependent on developmental stage.
Molecular mechanism studies have indicated that VA, especially its metabolites, can regulate several genes related to energy metabolism. atRA and 9-cis RA can bind to retinoic acid receptors (RARs) with high affinity in vitro. The 9-cis RA specifically binds to retinoid X receptors (RXRs) in vitro (Blomhoff and Blomhoff 2006). RAR:RXR heterodimers regulate typical RA target genes by binding to RA response elements, which include phosphoenolpyruvate carboxykinase (PEPCK) (Cadoudal et al. 2008) and stearoyl-CoA desaturase 1 (SCD1) (Samuel et al. 2001; Zolfaghari and Ross 2003). Moreover, RXR can form heterodimers with liver X receptor (LXR), which can positively regulate key lipogenic genes, including SREBP-1C, SCD, and FASN, in mouse liver cells and human hepatoma cells (Mukherjee et al. 1997; Roder et al. 2007). Therefore, VA and its metabolites play important roles in energy metabolism.
Long noncoding RNAs (lncRNAs) and natural antisense transcripts (NATs) have emerged as essential components of regulatory factors in several physiological processes. lncRNAs are involved in adipogenesis, hepatic lipid metabolism, energy balance, and so on (Zhao and Lin 2015). The knockdown of the liver-enriched lncRNA, lncLSTR, can reduce plasma triglyceride levels in a hyperlipidemic mouse model (Li et al. 2015). sno-lncRNAs from the Prader-Willi syndrome locus modulate energy balance in mice (Yin et al. 2012; Powell et al. 2013). In humans, the lncRNA HULC modulates lipid metabolism in hepatocellular carcinoma by activating the acyl-CoA synthetase subunit ACSL1 (Cui et al. 2015). In previous studies, NATs, which include trans- and cis- NATs, have been identified in humans, mice, and pigs, etc. (Katayama et al. 2005; Zhang et al. 2006; Zhao et al. 2016). In eukaryotic, NATs-mediated gene expression regulation mechanisms include chromatin remodeling, transcriptional interference, RNA masking, and double-stranded RNA-dependent mechanisms (Lapidot and Pilpel 2006; Faghihi and Wahlestedt 2009). The antisense transcript of apolipoprotein A1 (APOA1), referred to as ApoA1-AS, is an lncRNA that negatively regulates APOA1 expression (Halley et al. 2014). However, the roles of lncRNAs and NATs in the FE of pigs remain largely unknown.
On the basis of high-throughput RNA sequencing, we systematically analyzed differentially expressed (DE) annotated genes, lncRNAs, and cis-NATs in liver tissues between low- and high-FCR pigs. Gene ontology (GO) and pathway analysis revealed that the VA metabolism pathway in liver tissues was related to the variation of FE in pigs.
Materials and Methods
Animals, tissues, and RNA extraction
In this study, the feed intake of 236 purebred castrated boars from a Yorkshire pig population was detected using an ACEMA 64 automated individual feeding system at the Agricultural Ministry Breeding Swine Quality Supervision Inspecting and Testing Center (Wuhan, China) (Jing et al. 2015). The FCR of each individual was analyzed in the R environment using the following formula:
Furthermore, the FCR value between the three lowest pigs and the three highest pigs was significantly different (P < 0.05) (Jing et al. 2015). Those six castrated individuals were selected for RNA sequencing; the selected animals were not sibships. For tissue samples, pigs were slaughtered at 90 kg according to a standard procedure approved by guidelines from Regulation of the Standing Committee of Hubei People’s Congress (Hubei Province, P. R. China). Liver tissues were sampled and snap-frozen in liquid nitrogen within half an hour after slaughter before storage at –80°. Briefly, total RNA was isolated from frozen liver tissues with the TRIzol reagent (Invitrogen). All experimental protocols were approved by the Ethics Committee of Huazhong Agricultural University (HZAUMU2013-0005).
Library construction and sequencing
For each liver sample, equal quantities of RNA were sent to Genergy Biotechnology (Shanghai, China) for library construction. The RNA-seq library of each sample was prepared by with the TruSeq Stranded Total RNA Sample Preparation kit (Illumina). The Second Strand Marking Mix was also used during second strand cDNA synthesis to replace dTTP with dUTP to ensure strand specificity. After quality control, sequencing was performed with Illumina HiSeq2000.
RNA sequencing analysis
TopHat (version 2.0.9) (Trapnell et al. 2009) software was used to align reads to the pig reference genome (NCBI Sscrofa10.2). Up to two mismatches were allowed in reads mapping. Multiple-mapped reads were discarded; uniquely mapped reads were compared with the gff3 file of the Sscrofa10.2 genome with in-house Perl scripts. The reference genome sequence and its gff3 file were downloaded from the NCBI genome database (ftp://ftp.ncbi.nlm.nih.gov/genomes/Sus_scrofa/). Furthermore, mapped reads in the intergenic region were also compared to annotated lncRNAs (Zhou et al. 2014).
Genome-wide identification of cis-NATs
Mapped reads were pooled into one SAM file, which was used to identify novel transcripts by Cufflinks (Trapnell et al. 2012) with the option -library-type fr-firststrand. The transcripts identified by Cufflinks were compared with the gff3 file of the Sscrofa10.2 genome. Cis-NATs were identified on the basis of the following criteria: (i) located on the antisense strand of annotated genes, and (ii) novel transcripts but not belonging to the annotated genes. Cis-NATs were extracted from novel transcripts by in-house Perl scripts. The identified cis-NATs were used for further analysis.
Differential expression analysis
The count of reads located in the exon regions of each annotated gene, cis-NAT, and lncRNA was calculated using in-house Perl scripts. Genes with CPM (at least one count per million) > 1 in at least four samples were kept for further analysis. Both normalization of expression profiling for all expressed transcripts, and identification of DE transcripts were performed by edgeR (Robinson et al. 2010) in the R environment. Transcripts were determined as significantly DE with FDR < 0.05, and with an upregulated or downregulated fold-change (FC) of ≥ 2.5 between low-FCR and high-FCR pigs.
Correlation analysis
Pearson correlation analysis was performed to identify the correlatively expressed SA (sense–antisense) pairs and DE annotated gene–lncRNA pairs in the R environment. The cis-NATs, annotated genes, and lncRNAs that were detected at least in four samples were chosen for correlation analysis. The criteria for significantly correlated SA pairs was P < 0.05 and |R| > 0.8, whereas those for lncRNA-annotated gene pairs was P < 0.01 and |R| > 0.95.
q-PCR validation of DE genes
Total RNA was reverse-transcribed into cDNA using a PrimeScript RT reagent kit (Takara Bio Inc.). Oligonucleotide primers for six DE genes were designed with oligo7 software; the primer sequences are available in Supplemental Material, Table S1. Relative expression levels of these genes in liver were quantified by qPCR. The reactions were conducted on a BIO-RAD CFX384 Real-Time System with SYBR Green PCR Master Mix (Bio-Rad) as described in the manufacturer’s instruction manual. The 10 μl reaction mixture consisted of 1 μl cDNA, with 5 μl 2× SYBR Green PCR Master Mixture, 0.2 μl each of the forward and reverse primers, and 3.6 μl of RNase-free water. Samples were preincubated at 95° for 3 min, followed by 40 PCR amplification cycles (denaturation: 95° for 20 sec, annealing: 60° for 20 sec, elongation: 72° for 15 sec). A dissociation curve was generated at the end of the last cycle by collecting the fluorescence data from 60° to 95°. Relative gene expression levels were normalized to the RPL32 gene, which has stable expression in liver tissues (Ostrowska et al. 2014), by the 2−ΔΔCt method. Student’s t-test was used to analyze the expression difference between the low-FCR and high-FCR groups.
Gene ontology enrichment and pathway analysis
GO enrichment analysis was performed with DAVID Bioinformatics Resources 6.7 (Da Wei Huang and Lempicki 2008). DE genes were sorted by their involvement in significantly enriched biological process GO terms.
The pathway that involved the significant GO-terms-enriched genes was structured by referring to the KEGG pathway database and published articles. cis-NATs and DE lncRNAs that were correlated with the expression of genes involved in structured pathways, were also investigated and displayed. Visualization of pathway analysis was implemented in Cytoscape (Saito et al. 2012). The log2FC of annotated genes, cis-NATs, and lncRNAs was calculated as
where Low and High represented expression profile of the low-FCR and high-FCR groups, respectively.
Data availability
The raw data of RNA-seq were submitted to NCBI Sequence Read Archive database (SRA) under series SRP076030 which will be release in June 2017. DE transcripts list can be found in File S2.
Results
RNA sequencing data mapping and annotation
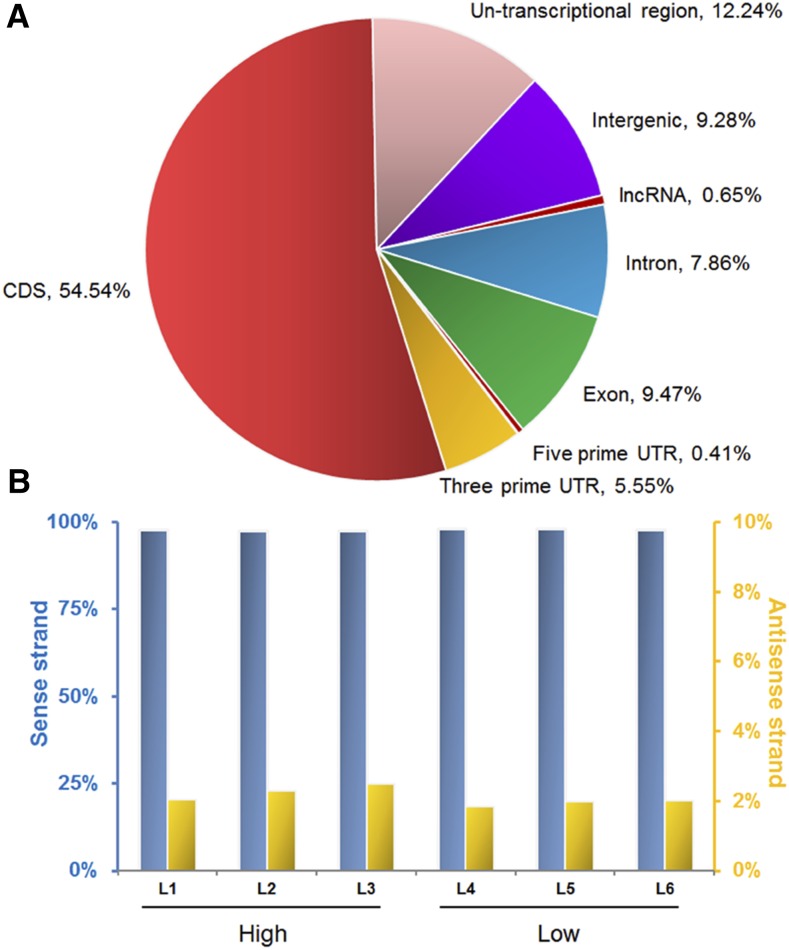
RNA of liver tissues of three high-FCR (H) and three low-FCR (L) pigs was isolated for RNA-seq. The RNA-seq raw data have been submitted to the NCBI Sequence Read Archive (SRA) under series SRP076030. After removing adaptors and filtering low quality reads, 7.1–10.0 million clean reads were yielded (Table 1). In these clean reads, ∼87% (6.1–9.6 million) were mapped on the Sscrofa10.2 genome, and over 90% of them were unique mappings (Table 1). The majority of uniquely mapped reads (90.72%) were located in the annotated gene region, whereas > 50% were located in the CDS region (Figure 1A). However, nearly 20% uniquely mapped reads were located in the intron region (9.28%), or other nontranscriptional regions (12.24%; Figure 1A). We also identified 1.8–2.5% uniquely mapped reads that were distributed on the antisense strand of annotated genes (Figure 1B).
Table 1. Summary of RNA-seq data from six liver samples.
| Group | Sample | Input | Mapped | Uniquely mapped | Multiple mapped |
|---|---|---|---|---|---|
| High | L1 | 11,028,888 | 9,662,607 (87.61%) | 9,023,627 (93.39%) | 638,980 (6.61%) |
| L2 | 8,901,564 | 7,827,119 (87.93%) | 7,076,183 (90.41%) | 750,936 (9.59%) | |
| L3 | 8,551,718 | 7,670,784 (89.70%) | 7,038,474 (91.76%) | 632,310 (8.24%) | |
| Low | L4 | 7,148,669 | 6,283,279 (87.89%) | 5,677,878 (90.36%) | 605,401 (9.64%) |
| L5 | 8,324,699 | 7,348,705 (88.28%) | 6,797,361 (92.50%) | 551,344 (7.50%) | |
| L6 | 7,155,132 | 6,191,448 (86.53%) | 5,741,224 (92.73%) | 450,224 (7.27%) |
Figure 1.
Annotation of the uniquely mapped reads of RNA-seq in liver tissue of pigs. (A) Distribution of uniquely mapped reads in the pig genome. The percentages in this pie chart represent the mean of all six RNA-seq data. On average, over 50% uniquely mapped reads were located in the CDS region. The untranscriptional regions were defined as genome region of annotated genes but without coverage of annotated transcripts. (B) Reads distributed on the sense and antisense strands of annotated genes. Approximately 2% reads were mapped on antisense strands of annotated genes.
Identification of cis-NATs
Based on the strand-specific RNA-seq reads, cis-NATs could be identified directly. Reads that were mapped on the antisense strand of the annotated gene were identified and defined as antisense reads. A total of 0.78 million antisense reads were identified from the uniquely mapped reads. Finally, 1769 cis-NATs were identified (File S1).
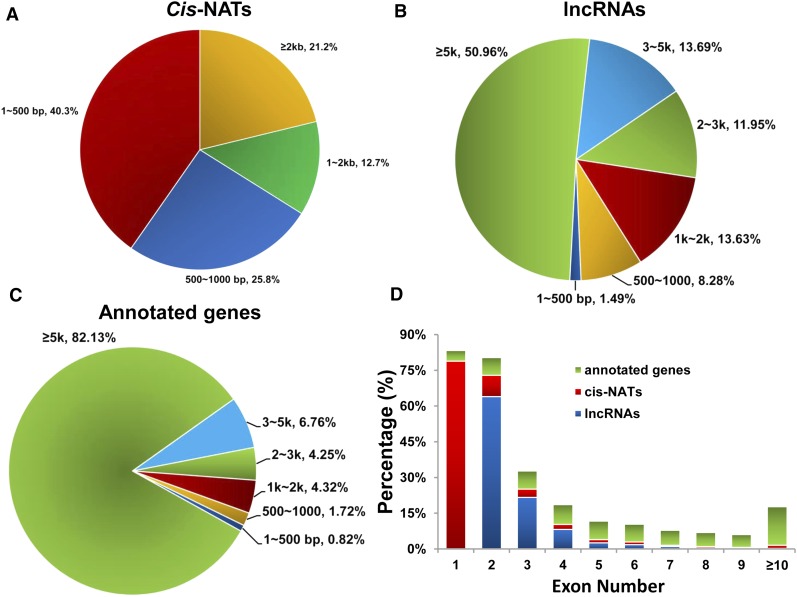
To understand the structural characteristics of the cis-NATs, we further investigated their length distribution and exon number. Statistical results showed that the majority (78.80%) of cis-NATs were shorter than 2 kb (Figure 2A). The statistical results of reported lncRNAs showed that ∼50% of the lncRNAs were shorter than 5 kb (Figure 2B). Both cis-NATs and lncRNAs differed with the detected annotated genes. For the annotated genes, > 80% were > 5 kb (Figure 2C). Moreover, the overwhelming majority of cis-NATs (91.22%) and lncRNAs (93.84%) contained fewer than three exons (Figure 2D). Besides, 78.83% of cis-NATs contained only one exon (Figure 2D). Most of the detected annotated genes contained more than three exons (Figure 2D). Therefore, the structural characteristics of cis-NATs were similar to those of lncRNAs, and both of them were different from the detected annotated genes.
Figure 2.
Characteristics of cis-NATs, lncRNAs, and annotated genes expressed in collected liver tissues of six pigs. (A) Length distribution of identified cis-NATs. The most enriched length of cis-NATs was < 500 bp. (B) Length distribution of expressed lncRNAs. Almost 50% expressed lncRNAs were < 5 kb. (C) Length distribution of expressed annotated genes. (D) Statistical results of exon number for expressed cis-NATs (red), lncRNAs (blue), and annotated genes (green).
DE liver transcripts between high and low FCR pigs
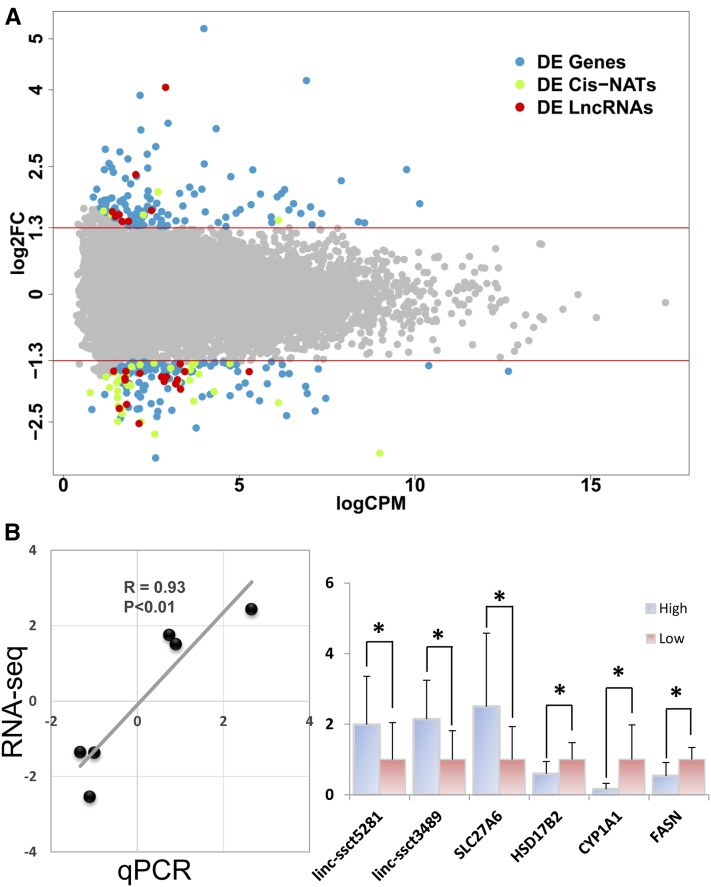
To compare the transcriptome alteration between high- and low-FCR pigs, the DE annotated genes, cis-NATs, and lncRNAs were determined using the edgeR package. A total of 300 significantly DE transcripts were identified, which included 232 annotated genes, 28 cis-NATs, and 40 lncRNAs, respectively (up or downregulated FC ≥ 2.5 and FDR < 0.05; File S2). Among these 232 annotated genes, 126 genes were upregulated, and 106 genes were downregulated in the low-FCR pigs (Figure 3 and File S2). Moreover, the majority of the DE cis-NATs (35 of 40) and lncRNAs (20 of 28) were downregulated in the low-FCR pigs (Figure 3 and File S2). The top 10 DE annotated genes cis-NATs and lncRNAs are listed in Table 2.
Figure 3.
DE transcripts between low- and high-FCR pigs. (A) Plot of the DE transcripts with FC ≥ 2.5 and FDR ≤ 0.05. Light blue, light green, and red dots represented the DE annotated genes, cis-NATs and lncRNAs, respectively. The x-axis and y-axis represent the logCPM and log2FC, respectively. (B) qPCR validation of DE transcripts in liver tissue of high-FCR (n = 10) and low-FCR (n = 10) pigs. The left plot is correlation analysis of RNA-seq and qPCR. For right bar plot, qPCR data were represented as mean ± SD; * P < 0.05.
Table 2. DE transcripts between low- and high-FCR groups (top 10).
| Source | ID | Gene Name | Log2FC (Low/High) | P-Value | FDR |
|---|---|---|---|---|---|
| Annotated genes | NM_001167795.1 | ACTA1 | 5.20 | 7.39E-31 | 4.98E-27 |
| NM_001243319.1 | IGLC | 4.18 | 1.40E-33 | 1.89E-29 | |
| NM_213855.1 | MYH7 | 3.89 | 5.04E-14 | 8.48E-11 | |
| XM_001924431.4 | ASAH2 | 3.35 | 4.71E-15 | 9.05E-12 | |
| XM_003126676.2 | MYBPC1 | 3.22 | 2.02E-11 | 1.43E-08 | |
| XM_001924841.4 | ELOVL2 | −2.38 | 8.84E-14 | 1.32E-10 | |
| XM_003359949.1 | TP53INP2 | −2.39 | 6.44E-09 | 2.22E-06 | |
| XM_003359189.2 | ERO1LB | −2.43 | 1.46E-06 | 2.46E-04 | |
| XM_003133832.3 | KIF1A | −2.62 | 7.90E-13 | 7.59E-10 | |
| XM_001929103.3 | PARP6 | −3.20 | 6.13E-13 | 6.87E-10 | |
| Cis-NATs | XM_001925338.4-AS1.1 | A2ML1 | 2.00 | 4.58E-07 | 8.98E-05 |
| XM_003127358.3-AS1.1 | LOC100524748 | 1.63 | 2.12E-03 | 4.75E-02 | |
| XM_003126577.2-AS1.1 | CCDC77 | 1.57 | 1.64E-03 | 4.03E-02 | |
| XM_003359961.2-AS1.1 | LOC100622161 | 1.55 | 1.48E-04 | 8.60E-03 | |
| NM_214056.1-AS3.1 | MT-III | 1.45 | 2.65E-06 | 3.81E-04 | |
| XM_003358629.1-AS2.3 | CCNL1 | −2.34 | 2.27E-06 | 3.40E-04 | |
| XM_001926397.3-AS7.1 | FAM135B | −2.49 | 1.40E-06 | 2.41E-04 | |
| XM_003480614.1-AS1.1 | LOC100736683 | −2.50 | 2.51E-08 | 6.75E-06 | |
| XM_003122843.3-AS1.1 | DGKZ | −2.74 | 1.54E-10 | 8.64E-08 | |
| XM_003481282.1-AS1.1 | LOC100739207 | −3.11 | 3.34E-22 | 1.50E-18 | |
| lncRNA | linc-ssct5500 | linc-ssct5500 | 4.05 | 1.75E-18 | 4.72E-15 |
| linc-ssct3976 | linc-ssct3976 | 2.34 | 2.31E-07 | 5.02E-05 | |
| linc-ssct3660 | linc-ssct3660 | 1.64 | 3.67E-05 | 3.12E-03 | |
| linc-ssct5436 | linc-ssct5436 | 1.62 | 1.10E-03 | 3.18E-02 | |
| linc-ssct5501 | linc-ssct5501 | 1.56 | 9.53E-04 | 2.99E-02 | |
| linc-ssct6319 | linc-ssct6319 | −1.76 | 1.56E-06 | 2.54E-04 | |
| linc-ssct6159 | linc-ssct6159 | −1.86 | 3.16E-07 | 6.54E-05 | |
| linc-ssct1651 | linc-ssct1651 | −2.16 | 5.59E-06 | 6.97E-04 | |
| linc-ssct6167 | linc-ssct6167 | −2.24 | 7.63E-06 | 8.91E-04 | |
| linc-ssct3489 | linc-ssct3489 | −2.53 | 2.40E-08 | 6.67E-06 |
qPCR was performed to validate the DE transcripts that were identified by RNA-seq. A total of 10 low-FCR individuals and 10 high-FCR individuals, which contained the individuals for RNA-seq, were chosen for qPCR analysis. The difference between these low- and high-FCR pigs was significant. Here, six DE transcripts were chosen for qPCR analysis. Three DE genes (HSD17B2, CYP1A1, and FASN) were selected from the GO enrichment analysis, and another three DE transcripts (SLC27A6, linc-ssct5281, and linc-ssct3489) were selected randomly from DE analysis results (File S2). The qPCR results shows that all six selected genes were validated as significantly DE genes in low-FCR vs. high-FCR. Moreover, the correlation coefficient of the log2FC values between RNA-seq and qPCR was 0.93 (P < 0.05).
Identification of correlated expression pairs
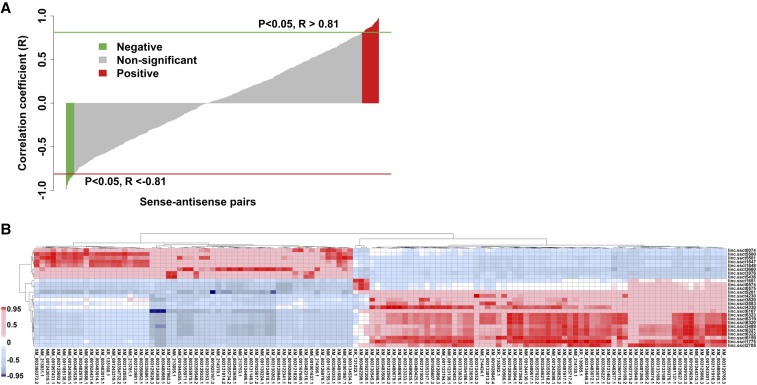
To understand function of cis-NATs, correlation analysis was performed between the expression of cis-NATs and their corresponding sense annotated genes. A total of 91 cis-NATs were significantly (P < 0.05 and |R| > 0.8) correlated with the expression of their sense annotated genes (Figure 4A and File S3). Among these pairs, 61 (67.4%) were positively correlated (Figure 4A and File S3).
Figure 4.
Representation of correlation analysis. (A) Correlation analysis between cis-NATs and their sense genes. The red and green bar plots represent the positively and negatively correlated cis-NATs and sense genes pairs, respectively. (B) Heat map of correlation analysis between DE lncRNAs and annotated genes. The red and blue fill colors of the heat map cells represent the positive and negative correlations, respectively. The color bar is shown on the left.
We also analyzed the correlation between the expression of DE lncRNAs and their annotated genes. In total, 337 significant correlation pairs were identified (P < 0.01 and |R| > 0.95), which included 26 DE lncRNAs and 126 DE annotated genes (Figure 4B and File S3). Furthermore, the majority (19 in 25) of the lncRNAs were correlated with at least two annotated genes (Figure 4B and File S3). Moreover, 333 of 337 correlated pairs (98.81%) were positively correlated.
Function annotation
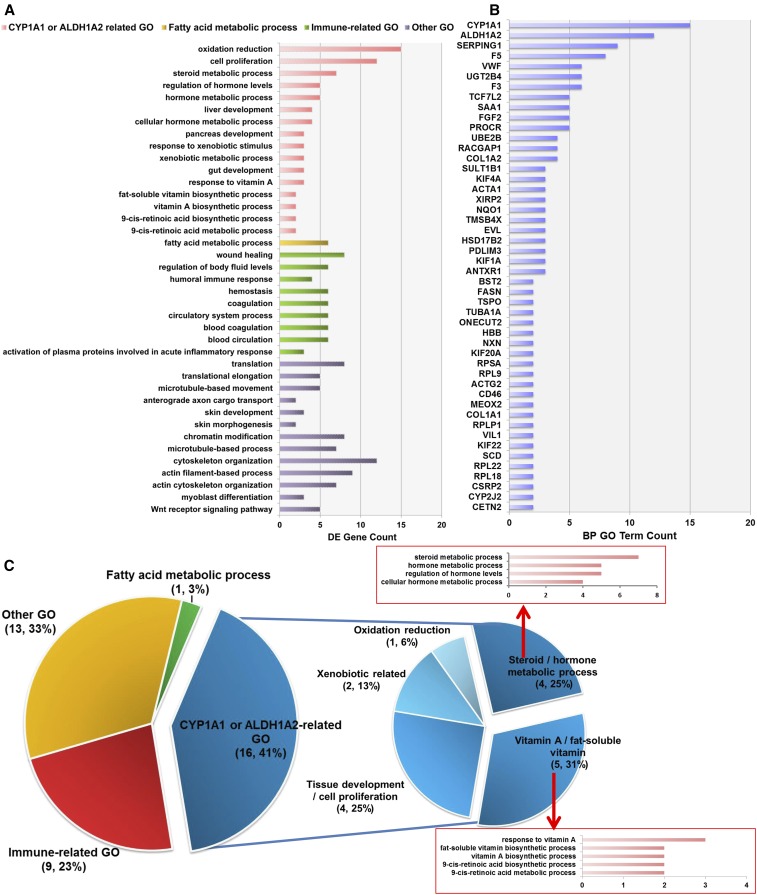
Function enrichment analysis was performed to detect significant biological processes GO terms of the DE annotated genes and cis-NATs according to the DAVID Bioinformatics Resources 6.7. A total of 39 significantly (EASE Score < 0.1) enriched GO terms were identified (Figure 5A and Table S2). The GO enrichment results showed the GO terms could be clustered based on the same DE annotated genes or similar biological functions (Figure 5, A and B). Based on that, the resultant 39 GO terms were clustered into four categories: CYP1A1- or ALDH1A2-related, fatty acid metabolic processes, immune-related, and other GO terms (Figure 5, A and C, left pie plot). Furthermore, 11 of 16 CYP1A1- or ALDH1A2-related GO terms contained both genes. Distribution investigation of annotated genes, among significantly GO terms, showed that CYP1A1 and ALDH1A2 were the top two most enriched genes (Figure 5B). Each was involved at least 12 GO terms (Figure 5B). Moreover, five of the 16 CYP1A1- or ALDH1A2-related GO terms were related to vitamin metabolism (Figure 5C).
Figure 5.
Significantly enriched GO terms for biological processes. (A) Significantly enriched biological process GO terms. The significant GO terms were determined by DAVID Bioinformatics Resources 6.7 (EASE Score < 0.1). Pink, orange, green, and purple colors represented the CYP1A1- or ALDH1A2-related, fatty acid metabolic process, immune-related, and other GO terms, respectively. (B) Distribution of DE genes in significantly enriched GO terms. Genes involved in at least two GO terms are listed. (C) Statistical analysis of the clustering results of GO terms. The most enriched category was the CYP1A1- or ALDH1A2-related GO terms (41%; left pie plot), which included five vitamin processes and four steroid-/hormone-related processes (right pie plot).
Pathway analysis of DE genes and lncRNAs
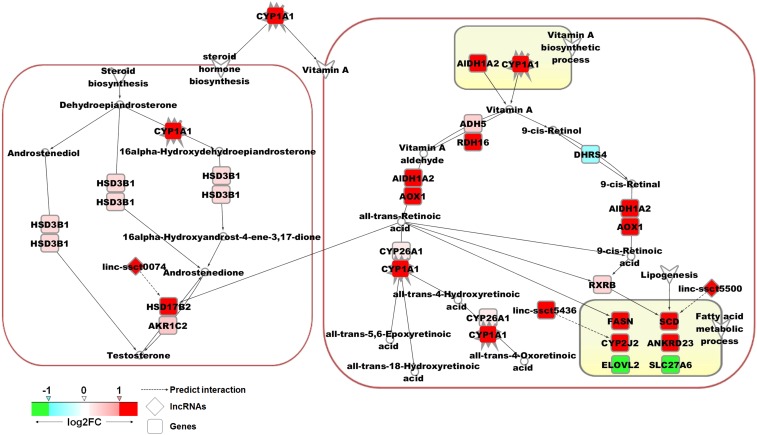
Based on functional enrichment analysis, CYP1A1 and ALDH1A2 were the two most important genes, mainly participating in VA- and hormone-related processes. Furthermore, we drew the VA and steroid hormone metabolism pathways according to the KEGG pathway database and previous studies. The results showed that CYP1A1, ALDH1A2, and RDH16 were both involved in VA anabolism and catabolism (Figure 6, right). Moreover, fatty acid and steroid hormone metabolic processes were located downstream of the VA metabolism pathway. These processes were regulated by RA, which is a metabolite of VA. The varied expression of the key genes and lncRNAs involved in these pathways were labeled by different colors. In Figure 6, genes in red and pink were upregulated in low-FCR pigs, whereas genes in green and blue were downregulated. The CYP1A1, ALDH1A2, RDH16, HSD17B2, UGT2B4, FASN, SCD, CYP2J2, and ANKRD23 genes were upregulated, in low-FCR pigs; the DE lncRNAs linc-ssct5500, linc-ssct5436, and linc-ssct0074 were also positively correlated with SCD, CYP2J2, and HSD17B2, respectively (Figure 6). Other VA metabolism-involved genes (ADH5, CYP26A1, AOX1, and RXRB) were also slightly upregulated in low-FCR pigs.
Figure 6.
Potential pathway of the annotated DE genes and lncRNAs in the liver tissues of high and low FCR pigs. Red and pink indicates upregulation in low FCR pigs (red, log2FC > 0.5; pink, 0 < log2FC ≤ 0.5); green and blue indicates downregulation in low FCR pigs (green, log2FC < −0.5; blue, −0.5 ≤ log2FC < 0).
Discussion
The improvement of FE is one of the most efficient ways to improve the benefits of pig production. Thus, investigation of the mechanisms of FE is very important for pig breeding. In this study, we systematically analyzed the transcription profiles of liver tissue in high- and low-FCR pigs. We found that the VA metabolism pathway in liver tissues was important for FE in pigs. The upregulation of genes related to VA metabolism in the liver tissue was positively correlated with FE in pigs.
The liver is one of the main tissues for VA storage in mammals. In this study, the key genes of VA metabolism, namely, CYP1A1, ALDH1A2, and RDH16, were all significantly upregulated in the liver of high-FE (low-FCR) pigs. This result indicated that the pathways of VA metabolism differed between high- and low-FE pigs. A recent study indicated that RA biosynthesis is regulated by energy status at the rate-limiting step catalyzed by retinol dehydrogenases (RDH) (Obrochta et al. 2015). The retinol dehydrogenase family members RDH1, RDH10, and RDH16 had significantly decreased transcriptional levels after refeeding, oral gavage with glucose, or injection with insulin (Obrochta et al. 2015). These results implied a negative relationship between RA biosynthesis and energy levels. Moreover, previous studies indicated that mitochondrial reactive oxygen species, and mitochondrial uncoupling reactions involving genes of high-FE pigs, were lower than those of low-FE pigs (Grubbs et al. 2013a; Jing et al. 2015). However, the antioxidant defenses of high-FE pigs were higher than those of low-FE pigs (Grubbs et al. 2013b). The oxidation and energy loss of high-FE pigs was lower than that of low-FE pigs. Thus, the upregulation of genes involving in VA metabolism in high-FE pigs was possibly caused by the relatively lower oxidation and energy loss in this study. Besides, the key genes of fatty acid biosynthesis SCD (Samuel et al. 2001; Zolfaghari and Ross 2003; Zhang et al. 2012; Weiss et al. 2014) and FASN (Zhou et al. 2010; Zhang et al. 2012) were induced by RA at the transcriptional level and significantly upregulated in high-FE pigs. SCD encodes stearoyl-CoA desaturase, which converts saturated fatty acids into monounsaturated fatty acids, and influences fatty acid partitioning in the liver by promoting fatty acid synthesis but decreasing oxidation (Flowers and Ntambi 2009). Furthermore, other genes related to fatty acid biosynthesis, namely, CYP2J2 and ANKRD23 were also upregulated. These results indicated that fatty acid biosynthesis in the liver was increased in high-FE pigs. Therefore, the VA pathway could affect FE by involving the energy metabolism of pigs and promoting fatty acid biosynthesis. Moreover, the moderate activity of the VA metabolic pathway in liver tissue may benefit FE in pigs.
Previous studies also indicated that RA could induce and maintain testosterone production (Tucci et al. 2008). Testosterone is a steroid hormone in vertebrates and belongs to the androgen group. This hormone plays a key role in male reproductive tissue development, as well as in increasing muscle mass, muscle strength, bone mass, and bone mineral density (van den Beld et al. 2000; Sinha-Hikim et al. 2002). In this study, genes involved in testosterone metabolism, namely, CYP1A1, HSD17B2, and UGT2B4, were significantly upregulated in low-FCR pigs. Moreover, HSD17B2 is positively regulated by RA (Ito et al. 2001). HSD3B1 and AKR1C2 are involved in testosterone metabolism. and were also slightly upregulated in high-FE pigs. These results suggested that the metabolism of testosterone was increased in high-FE pigs; VA may also affect FE by affecting steroid hormone metabolism.
In addition, 40 lncRNAs were differentially expressed between high- and low-FCR pigs. lncRNAs are involved in several bioprocesses, including energy metabolism (Yin et al. 2012; Powell et al. 2013; Cui et al. 2015; Li et al. 2015) and growth (Dey et al. 2014; Mueller et al. 2015). The DE lncRNAs linc-ssct5500, linc-ssct5436, and linc-ssct0074 were upregulated in the livers of low-FCR pigs, and positively correlated with SCD, CYP2J2, and HSD17B2, respectively. Previous studies indicated that SCD and CYP2J2 (Wang et al. 2004) are involved in fatty acid metabolism, whereas HSD17B2 is related to testosterone metabolism (Labrie et al. 1997). Therefore, these lncRNAs may also affect the FE of pigs by altering the metabolism of fatty acids and steroid hormones.
We also identified 1769 cis-NATs in pig liver tissue. Among them, 28 were significantly different between low and high FE pigs. Correlation analysis showed 61 (67.4%) SA pairs from 91 significantly correlated SA pairs were positively correlated. The coexpression (positively) between SA pairs was reported in humans (Chen et al. 2005), Drosophila melanogaster (Okamura et al. 2008), and pigs (Zhao et al. 2016). These results indicate that cis-NATs are involved in the regulation of FE of pigs. However, the mechanisms of the regulatory roles of cis-NATs in FE of pigs remained largely unknown. Furthermore, some immune-related signaling pathways were also identified to relate to FE. Therefore, the FE of pigs may also be regulated by ncRNAs or other pathways.
In summary, annotated DE genes, cis-NATs, and lncRNAs in the liver tissues of high- and low-FE pigs were analyzed comparatively. Our results revealed that different expression was enriched mainly in VA, fatty acid, and steroid hormone metabolism. VA metabolism in liver tissues involved regulation of FE in pigs by mediating fatty acid biosynthesis and steroid hormone metabolism. CYP1A1, ALDH1A2, and RDH16 are the key upstream genes of VA metabolism, and possible candidate genes for FE in pigs. Some lncRNAs and cis-NATs were also related to the FE of pigs.
Supplementary Material
Acknowledgments
This research was supported by the National Natural Science Foundation of China (31372291), the National High Technology Research and Development Program of China (863 Program, 2013AA102502 to SZ), and the National Special Foundation for Transgenic Species of China (2014ZX0800950B).
Author contributions: X.L. and S.Z. conceived and supervised the project. Y.Z., F.L., Y.L., A.L., and L.J. undertook the analysis. Y.Z. and YH wrote the paper. Y.H., C.Z., and Y.M. performed the tissue collection and PCR experiments. All authors read and approved the final manuscript. The authors declare that they have no competing interests.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.032839/-/DC1.
Communicating editor: D. J. de Koning
Literature Cited
- Berry D. C., Noy N., 2009. All-trans-retinoic acid represses obesity and insulin resistance by activating both peroxisome proliferation-activated receptor β/δ and retinoic acid receptor. Mol. Cell. Biol. 29: 3286–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomhoff R., Blomhoff H. K., 2006. Overview of retinoid metabolism and function. J. Neurobiol. 66: 606–630. [DOI] [PubMed] [Google Scholar]
- Bonet M. L., Ribot J., Palou A., 2012. Lipid metabolism in mammalian tissues and its control by retinoic acid. Biochim. Biophys. Acta 1821: 177–189. [DOI] [PubMed] [Google Scholar]
- Cadoudal T., Glorian M., Massias A., Fouque F., Forest C., et al. , 2008. Retinoids upregulate phosphoenolpyruvate carboxykinase and glyceroneogenesis in human and rodent adipocytes. J. Nutr. 138: 1004–1009. [DOI] [PubMed] [Google Scholar]
- Chen J., Sun M., Hurst L. D., Carmichael G. G., Rowley J. D., 2005. Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense-antisense transcripts. Trends Genet. 21: 326–329. [DOI] [PubMed] [Google Scholar]
- Cui M., Xiao Z., Wang Y., Zheng M., Song T., et al. , 2015. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 75: 846–857. [DOI] [PubMed] [Google Scholar]
- Da Wei Huang B. T. S., Lempicki R. A., 2008. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Dey B. K., Pfeifer K., Dutta A., 2014. The H19 long noncoding RNA gives rise to microRNAs miR-675–3p and miR-675–5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 28: 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do D. N., Strathe A. B., Jensen J., Mark T., Kadarmideen H. N., 2013. Genetic parameters for different measures of feed efficiency and related traits in boars of three pig breeds. J. Anim. Sci. 91: 4069–4079. [DOI] [PubMed] [Google Scholar]
- Faghihi M. A., Wahlestedt C., 2009. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 10: 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers M. T., Ntambi J. M., 2009. Stearoyl-CoA desaturase and its relation to high-carbohydrate diets and obesity. Biochimi. Biophys. Acta 1791: 85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubbs J. K., Fritchen A., Huff-Lonergan E., Dekkers J. C., Gabler N. K., et al. , 2013a Divergent genetic selection for residual feed intake impacts mitochondria reactive oxygen species production in pigs. J. Anim. Sci. 91: 2133–2140. [DOI] [PubMed] [Google Scholar]
- Grubbs J. K., Fritchen A. N., Huff-Lonergan E., Gabler N. K., Lonergan S. M., 2013b Selection for residual feed intake alters the mitochondria protein profile in pigs. J. Proteomics 80: 334–345. [DOI] [PubMed] [Google Scholar]
- Halley P., Kadakkuzha B. M., Faghihi M. A., Magistri M., Zeier Z., et al. , 2014. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Reports 6: 222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman R. A., Mangelsdorf D. J., Dyck J. A., Stein R. B., Eichele G., et al. , 1992. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell 68: 397–406. [DOI] [PubMed] [Google Scholar]
- Ito K., Suzuki T., Moriya T., Utsunomiya H., Sugawara A., et al. , 2001. Retinoid receptors in the human endometrium and its disorders: a possible modulator of 17 beta-hydroxysteroid dehydrogenase. J. Clin. Endocrinol. Metab. 86: 2721–2727. [DOI] [PubMed] [Google Scholar]
- Jing L., Hou Y., Wu H., Miao Y., Li X., et al. , 2015. Transcriptome analysis of mRNA and miRNA in skeletal muscle indicates an important network for differential Residual Feed Intake in pigs. Sci. Rep. 5: 11953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., et al. , 2005. Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566. [DOI] [PubMed] [Google Scholar]
- Labrie F., Luu-The V., Lin S.-X., Claude L., Simard J., et al. , 1997. The key role of 17β-hydroxysteroid dehydrogenases in sex steroid biology. Steroids 62: 148–158. [DOI] [PubMed] [Google Scholar]
- Lampen A., Meyer S., Arnhold T., Nau H., 2000. Metabolism of vitamin A and its active metabolite all-trans-retinoic acid in small intestinal enterocytes. J. Pharmacol. Exp. Ther. 295: 979–985. [PubMed] [Google Scholar]
- Lapidot M., Pilpel Y., 2006. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep. 7: 1216–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Naou T., Floc’h L., Louveau I., Gilbert H., Gondret F., 2012. Metabolic changes and tissue responses to selection on residual feed intake in growing pigs. J. Anim. Sci. 90: 4771–4780. [DOI] [PubMed] [Google Scholar]
- Li P., Ruan X., Yang L., Kiesewetter K., Zhao Y., et al. , 2015. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 21: 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lkhagvadorj S., Qu L., Cai W., Couture O. P., Barb C. R., et al. , 2010. Gene expression profiling of the short-term adaptive response to acute caloric restriction in liver and adipose tissues of pigs differing in feed efficiency. Am. J. Physiol. Regul. Integr. Comp. Physiol. 298: R494–R507. [DOI] [PubMed] [Google Scholar]
- Manolescu D.-C., Sima A., Bhat P. V., 2010. All-trans retinoic acid lowers serum retinol-binding protein 4 concentrations and increases insulin sensitivity in diabetic mice. J. Nutr. 140: 311–316. [DOI] [PubMed] [Google Scholar]
- Mueller A. C., Cichewicz M. A., Dey B. K., Layer R., Reon B. J., et al. , 2015. MUNC, a ong noncoding RNA that facilitates the function of MyoD in skeletal myogenesis. Mol. Cell. Biol. 35: 498–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee R., Davies P. J., Crombie D. L., Bischoff E. D., Cesario R. M., et al. , 1997. Sensitization of diabetic and obese mice to insulin by retinoid X receptor agonists. Nature 386: 407–410. [DOI] [PubMed] [Google Scholar]
- Murano I., Morroni M., Zingaretti M. C., Oliver P., Sánchez J., et al. , 2005. Morphology of ferret subcutaneous adipose tissue after 6-month daily supplementation with oral beta-carotene. Biochim. Biophy. Acta 1740: 305–312. [DOI] [PubMed] [Google Scholar]
- Obrochta K. M., Krois C. R., Campos B., Napoli J. L., 2015. Insulin regulates retinol dehydrogenase expression and all-trans-retinoic acid biosynthesis through FoxO1. J. Biol. Chem. 290: 7259–7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Balla S., Martin R., Liu N., Lai E. C., 2008. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat. Struct. Mol. Biol. 15: 998. [DOI] [PubMed] [Google Scholar]
- Ostrowska M., Zelazowska B., Sloniewski K., Kowalski Z. M., Zwierzchowski L., 2014. Technical note: selecting the best references in gene expression experiments in liver of cows receiving glucogenic supplements during the transition period. J. Dairy Sci. 97: 911–916. [DOI] [PubMed] [Google Scholar]
- Powell W. T., Coulson R. L., Crary F. K., Wong S. S., Ach R. A., et al. , 2013. A Prader–Willi locus lncRNA cloud modulates diurnal genes and energy expenditure. Hum. Mol. Genet. 22: 4318–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redonnet A., Ferrand C., Bairras C., Higueret P., Noël-Suberville C., et al. , 2008. Synergic effect of vitamin A and high-fat diet in adipose tissue development and nuclear receptor expression in young rats. Br. J. Nutr. 100: 722–730. [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roder K., Zhang L., Schweizer M., 2007. SREBP‐1c mediates the retinoid‐dependent increase in fatty acid synthase promoter activity in HepG2. FEBS Lett. 581: 2715–2720. [DOI] [PubMed] [Google Scholar]
- Saito R., Smoot M. E., Ono K., Ruscheinski J., Wang P. L., et al. , 2012. A travel guide to Cytoscape plugins. Nat. Methods 9: 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel W., Kutty R. K., Nagineni S., Gordon J. S., Prouty S. M., et al. , 2001. Regulation of stearoyl coenzyme A desaturase expression in human retinal pigment epithelial cells by retinoic acid. J. Biol. Chem. 276: 28744–28750. [DOI] [PubMed] [Google Scholar]
- Sánchez J., Fuster A., Oliver P., Palou A., Picó C., 2009. Effects of β-carotene supplementation on adipose tissue thermogenic capacity in ferrets (Mustela putorius furo). Br. J. Nutr. 102: 1686–1694. [DOI] [PubMed] [Google Scholar]
- Sinha-Hikim I., Artaza J., Woodhouse L., Gonzalez-Cadavid N., Singh A. B., et al. , 2002. Testosterone-induced increase in muscle size in healthy young men is associated with muscle fiber hypertrophy. Am. J. Physiol. Endocrinol. Metab. 283: E154–E164. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucci P., Cione E., Genchi G., 2008. Retinoic acid-induced testosterone production and retinoylation reaction are concomitant and exhibit a positive correlation in Leydig (TM-3) cells. J. Bioenerg. Biomembr. 40: 111–115. [DOI] [PubMed] [Google Scholar]
- van den Beld A. W., de Jong F. H., Grobbee D. E., Pols H. A., Lamberts S. W., 2000. Measures of bioavailable serum testosterone and estradiol and their relationships with muscle strength, bone density, and body composition in elderly men 1. J. Clin. Endocrinol. Metab. 85: 3276–3282. [DOI] [PubMed] [Google Scholar]
- Wang H., Zhao Y., Bradbury J. A., Graves J. P., Foley J., et al. , 2004. Cloning, expression, and characterization of three new mouse cytochrome p450 enzymes and partial characterization of their fatty acid oxidation activities. Mol. Pharmacol. 65: 1148–1158. [DOI] [PubMed] [Google Scholar]
- Weiss K., Mihaly J., Liebisch G., Marosvolgyi T., Garcia A. L., et al. , 2014. Effect of high vs. low doses of fat and vitamin A dietary supplementation on fatty acid composition of phospholipids in mice. Genes Nutr. 9: 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q.-F., Yang L., Zhang Y., Xiang J.-F., Wu Y.-W., et al. , 2012. Long noncoding RNAs with snoRNA ends. Mol. Cell 48: 219–230. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu X. S., Liu Q. R., Wei L., 2006. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res. 34: 3465–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li R., Li Y., Chen W., Zhao S., et al. , 2012. Vitamin A status affects obesity development and hepatic expression of key genes for fuel metabolism in Zucker fatty rats. Biochem. Cell Biol. 90: 548–557. [DOI] [PubMed] [Google Scholar]
- Zhao X.-Y., Lin J. D., 2015. Long noncoding RNAs: a new regulatory code in metabolic control. Trends Biochem. Sci. 40: 586–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Hou Y., Zhao C., Liu F., Luan Y., et al. , 2016. Cis-natural antisense transcripts are mainly co-expressed with their sense transcripts and primarily related to energy metabolic pathways during muscle development. Int. J. Biol. Sci. 12: 1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Yang J., Zhao H., Li H. J., 2010 [Effect of all-trans retinoic acid on the proliferation of and Fas protein expression in human malignant melanoma A375 cells]. Sichuan Da Xue Bao Yi Xue Ban 41: 464–466. [PubMed] [Google Scholar]
- Zhou Z.-Y., Li A.-M., Adeola A. C., Liu Y.-H., Irwin D. M., et al. , 2014. Genome-wide identification of long intergenic noncoding RNA genes and their potential association with domestication in pigs. Genome Biol. Evol. 6: 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolfaghari R., Ross A. C., 2003. Recent advances in molecular cloning of fatty acid desaturase genes and the regulation of their expression by dietary vitamin A and retinoic acid. Prostaglandins Leukot. Essent. Fatty Acids 68: 171–179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of RNA-seq were submitted to NCBI Sequence Read Archive database (SRA) under series SRP076030 which will be release in June 2017. DE transcripts list can be found in File S2.