Abstract
The initiation of sexual development in the important edible and medicinal mushroom Flammulina velutipes is controlled by special genes at two different, independent, mating type (MAT) loci: HD and PR. We expanded our understanding of the F. velutipes mating type system by analyzing the MAT loci from a series of strains. The HD locus of F. velutipes houses homeodomain genes (Hd genes) on two separated locations: sublocus HD-a and HD-b. The HD-b subloci contained strain-specific Hd1/Hd2 gene pairs, and crosses between strains with different HD-b subloci indicated a role in mating. The function of the HD-a sublocus remained undecided. Many, but not all strains contained the same conserved Hd2 gene at the HD-a sublocus. The HD locus usually segregated as a whole, though we did detect one new HD locus with a HD-a sublocus from one parental strain, and a HD-b sublocus from the other. The PR locus of F. velutipes contained pheromone receptor (STE3) and pheromone precursor (Pp) genes at two locations, sublocus PR-a and PR-b. PR-a and PR-b both contained sets of strain-specific STE3 and Pp genes, indicating a role in mating. PR-a and PR-b cosegregated in our experiments. However, the identification of additional strains with identical PR-a, yet different PR-b subloci, demonstrated that PR subloci can recombine within the PR locus. In conclusion, at least three of the four MAT subloci seem to participate in mating, and new HD and PR loci can be generated through intralocus recombination in F. velutipes.
Keywords: mating type, homeodomain, pheromone receptor, Flammulina velutipes, Genetics of Sex
Sexual development in basidiomycetous fungi is controlled by specialized genes that are located at distinct loci called mating type (MAT) loci (Raper 1966; Brown and Casselton 2001; Raudaskoski and Kothe 2010; Raudaskoski 2015). In the majority of mushroom-forming fungi (homobasidiomycetes), two different and unlinked MAT loci can be found: MAT-A and MAT-B. Recently, MAT-A and MAT-B loci were named HD and PR loci, respectively (Kües 2015; Maia et al. 2015). Upon mating, the two parental sets of HD and PR loci can recombine into four sexually distinct mating types—a system referred to as the tetrapolar mating type system. The origin of the tetrapolar mating type system has been argued to be ancient for basidiomycetes (Burnett 1975; Fraser et al. 2007; Hsueh and Heitman 2008; Maia et al. 2015), and the genomes of tetrapolar, as well as bipolar and homothallic mushroom-forming species are known to contain both the HD and the PR locus (James et al. 2006; Aimi et al. 2005; Yi et al. 2009; Morin et al. 2012; Bao et al. 2013; Chen et al. 2016). Further developments in mushroom-forming fungi have resulted in often elaborate HD and PR systems that consist of multiple HD and PR subloci, each containing a series of possible alleles (Casselton and Olesnicky 1998; James et al. 2004b; Whitehouse 1949; Raper 1966; Casselton 1978; Badrane and May 1999).
The basic model of the HD locus encompasses a divergently transcribed pair of homeodomain1 (Hd1) and homeodomain2 (Hd2) genes that form a functional group, flanked by the mitochondrial intermediate peptidase (MIP) gene on one side, and the β-flanking gene on the other (James et al. 2004a; Kües et al. 2011). In the case of multiple HD subloci, several of these functional groups will be positioned between the MIP gene and the β-flanking gene (Brown and Casselton 2001; James 2007; Kües et al. 2011). The products encoded by the Hd1 and Hd2 gene pairs interact exclusively with HD proteins encoded by different alleles (nonself) of the same sublocus (Kües and Casselton 1992; Kües et al. 1994; Banham et al. 1995). HD subloci can be functionally redundant, and one compatible Hd1 and Hd2 gene are sufficient to mediate activation of the HD pathway. Upon interaction, an HD1 and HD2 protein will form a heterodimeric transcription factor that activates the sexual developmental pathway under the control of the HD locus (Brown and Casselton 2001). While the characteristic DNA binding motifs (the HD1 or HD2 domain) of the HD proteins are mostly conserved, allelic variation and mating compatibility is determined by the highly variable N-terminal regions (Kronstad and Leong 1990; Yee and Kronstad 1993; Banham et al. 1995; Wu et al. 1996; Badrane and May 1999; Coelho et al. 2010). In addition, divergence of the C-terminal parts of the HD-encoding genes has been argued to be useful in preventing the recombination of alleles on the same sublocus, which could otherwise result in self-activation (for a discussion see Kües 2015, and references herein). The HD locus and the surrounding genomic region are highly syntenic in most mushroom-forming fungi, and synteny is often maintained when gene clusters or HD subloci are transferred to other positions in the genome (James et al. 2006; Niculita-Hirzel et al. 2008; Bao et al. 2013; van Peer et al. 2011).
The basic PR locus consists of an archetypal subunit that contains a single pheromone receptor gene (STE3) in association with one or several pheromone precursor (Pp) genes (Vaillancourt et al. 1997; Casselton and Olesnicky 1998; O’Shea et al. 1998; Halsall et al. 2000; Casselton and Challen 2006; James 2007). Multiple of these subunits can form PR subloci. Pheromone receptors and pheromones within a subunit are incompatible, and only interact with products of different allelic subunits from corresponding subloci. PR subloci can be functionally redundant, yet not all nonself combinations are always active. In case of a compatible interaction, a pheromone peptide will bind the extracellular domain of a pheromone receptor, triggering activation of the PR pathway (Olesnicky et al. 2000; Brown and Casselton 2001). The fungal pheromone receptors are class D, G-protein coupled receptors that belong to the Ste3 subfamily (GPCRs, http://pfam.xfam.org/family/PF02076; Finn et al. 2016). They contain seven transmembrane (7-TM) domains, extracellular N-termini, and intracellular C-terminal regions (Dohlman et al. 1991; Caldwell et al. 1995). Pheromone precursors encode 40–100 amino acid long prepeptides. They mature through farnesylation of the C-terminal CAAX motif, and presumed splicing at a conserved position with two charged amino acids (e.g., “ER,” “ED,” or “EH”) located 10–15 amino acids upstream of the C-terminus (Bölker and Kahmann 1993; Caldwell et al. 1995; O’Shea et al. 1998; Fowler et al. 2001; Kües 2015). The PR loci and the surrounding genomic region are poorly conserved (James 2007), and PR subloci can be separated by distances of up to 170 kb (van Peer et al. 2011).
In addition to Hd, STE3, and Pp genes, many mushroom-forming fungi contain pheromone receptor like genes, which share considerable sequence similarity with the pheromone receptor genes on the PR loci (Niculita-Hirzel et al. 2008; van Peer et al. 2011, Kües 2015). Yet, pheromone receptor like genes are conserved (there are no different alleles in different mating types), and are not associated with pheromone precursor genes (Niculita-Hirzel et al. 2008; van Peer et al. 2011). To determine which genes belong to PR (sub)loci, and which PR and HD subloci partake in mating, it is necessary to identify and compare the PR and HD loci of different mating types. Flammulina velutipes (Curt. ex Fr.) Sing, also known as Winter Mushroom or Enokitake, is an important edible and medicinal mushroom that is cultivated on a large scale (Tonomura 1978; Leifa et al. 2001; Wang et al. 2012; Zhang et al. 2012; Sekiya et al. 2013). F. velutipes has a tetrapolar mating type system that generates basidiospores of four possible mating types. Although single mating types have been known to produce fruiting bodies under severe stress, they are typically self-infertile and controlled by the HD and PR pathways (Uno and Ishikawa 1971; Esser et al. 1979; Wessels 1993; Yi et al. 2007). The first HD and PR loci of F. velutipes were identified recently, indicating that the HD locus consisted of two subloci, HD-a and HD-b, and that the PR locus also consisted of two subloci, PR-a and PR-b (van Peer et al. 2011). As few mating compatible HD or PR loci have been identified, it remained unclear if (a) the reported organization of the HD and PR subloci was representative for F. velutipes, and (b) which of these subloci participated in mating. Only PR-a had been deduced to be involved in mating. In this study, we identified and compared the structure and content of a series of HD and PR subloci, using whole genome sequences as well as subcloned regions from a variety of F. velutipes strains.
Materials and Methods
Strains and media
Monokaryotic F. velutipes strains W23 (A2B5) and L11 (A1B1), dikaryotic strain 1123 (a cross of strain L11 and strain W23), and F. velutipes strains F0010, F0012, F0020, F0027, and F0025 (Jiang et al. 2009), were provided by the Fujian Edible Fungi Germplasm Resource Collection Center of China (Supplemental Material, Table S1). All strains were maintained on Potato Dextrose Agar medium (PDA; 200 g/l potato; 20 g/l glucose; 20 g/l agar) at 25°.
To produce fruiting bodies for spore isolation, strains were cultivated in sterile plastic bags containing growth substrate (cottonseed hulls 52.5%, wheat bran 25%, sawdust 15%, corn flour 5%, gypsum 2%, and ground limestone 0.5%, with a moisture content of 61%). Strains were grown at 23° for 30 d, followed by cold-stimulation at 16° and 90% humidity until primordia occurred (1 wk). Cultures were maintained at low temperature (16° and 75% humidity) to allow full fruiting body development.
For single-spore cultures, spores were collected by placing a paper under the pilei of harvested, mature fruiting bodies. Spores were washed with sterile ddH2O, inoculated on PDA, and incubated at 25° for 24–48 hr until spore germination occurred. Single spore isolates (SSIs) were selected under an optical microscope (Motic) and subcultured individually on PDA at 25°.
For DNA extraction, mycelium of F. velutipes strains was grown for 2 wk at 25° on PDA overlaid with a cellophane membrane. Harvested mycelium was stored at −80° until use.
Genome and transcriptome sequences of F. velutipes strains
The genome sequence of strain KACC42780 (Park et al. 2014) was provided by W.S. Kong of the Rural Development Administration (RDA) in Korea. Genome sequences of F. velutipes strain L11 and strain W23 were obtained from our lab (Zeng et al. 2015; Liu et al. 2015; Wang et al. 2015b). Genomic DNA was extracted from mycelium using a CTAB method (Stajich et al. 2010). A paired-end (2 × 90 bp) DNA library with 500 bp inserts was generated and sequenced (de novo sequencing) on a Illumina Cluster Station and Illumina GAII platform by Zhejiang California International NanoSystems Institute (Hangzhou, China). Over 6 GB clean reads were obtained for the genomes of strain L11 and W23, amounting to ∼170× coverage.
For transcriptomics, mycelium of monokaryotic strains L11 and W23, and dikaryotic strain 1123, was collected and flash frozen in liquid nitrogen. RNA was extracted and used for library construction (length of paired-end reads, 2 × 90 bp; insert size, 500 bp), and sequencing (Illumina Cluster Station, Illumina GAII platform, http://www.genomics.cn) at BGI (Shenzhen, China) (Liu et al. 2014; Wang et al. 2015a). Transcriptomics data were used to correct intron-exon boundaries of predicted gene models by using ZOOMlite software (Lin et al. 2008; Zhang et al. 2010).
Identification of mating type genes and analysis of predicted proteins
The genomes of F. velutipes strain L11 and W23 were screened for mating type genes using BLASTP 2.2.26 homology searches (Altschul et al. 1997) with known F. velutipes mating type proteins (van Peer et al. 2011). Gaps in the sequences of newly found mating type genes were filled by sequencing of PCR segments (Sangon, Shanghai, China). Pheromone precursor genes were identified as described before (van Peer et al. 2011).
Nuclear localization signals (NLS), dimerization motifs (Di), and homeodomains (HD) of homeodomain proteins were predicted by PSORT II Prediction (http://psort.hgc.jp/form2.html; Horton et al. 2007), COILS Server (http://embnet.vital-it.ch/software/COILS_form.html; Windows width = 14; Lupas et al. 1991), and MOTIF Search (http://www.genome.jp/tools/motif/). Transmembrane helices of pheromone receptors were predicted with the TMHMM Server (http://www.cbs.dtu.dk/services/TMHMM-2.0/) and the TMpred Server (http://www.ch.embnet.org/software/TMPRED_form.html). Multiple sequence alignments of the HD and PR mating type genes and subloci were performed with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Synteny maps were drawn using Chromomapper 1.0.10.26 (Niculita-Hirzel and Hirzel 2008).
Phylogenetic analysis of pheromone receptors and pheromone-receptor-like proteins
Pheromone receptors and pheromone-receptor-like proteins of Laccaria bicolor (Niculita-Hirzel et al. 2008), Schizophyllum commune, Pleurotus djamor, Coprinopsis cinerea, Cryptococcus neoformans (James et al. 2004b), together with pheromone receptors CcRcb2.43 (GenBank accession number: AAQ96345), CcRcb2.44 (GenBank accession number: AAQ96344), and ScBar8 (GenBank accession number: AAR99618) were selected for multiple alignment and tree building using the online tool “Phylogeny.fr” with “One click” stream (http://phylogeny.lirmm.fr/phylo_cgi/index.cgi, Dereeper et al. 2008). In the first step, protein sequences were aligned using MUSCLE 3.7 (“Find diagonals” option disabled; Maximum number of iterations: 16, Edgar 2004). Next, ambiguous regions (i.e., gapped and/or poorly aligned regions) were removed using Gblocks (v0.91b/minimum length of a block after gap cleaning was 10/no gap positions were allowed in the final alignment/all segments with contiguous nonconserved positions > 8 were rejected/minimum number of sequences for a flank position was 85%, Castresana 2000). Hereafter, the resulting 230 amino acids sequences (alignment of pheromone receptors and pheromone-receptor-like proteins of F. velutipes only), and 197 amino acids (alignment of pheromone receptors and pheromone-receptor-like proteins of multiple mushroom-forming fungi), were used to assemble phylogenetic trees. The supplemental materials file “Fv_STE3-Ori.txt” contains the original protein sequences, file “Fv_STE3-align_cured.txt” contains the processed sequences. Trees were reconstructed using maximum likelihood, as implemented in the PhyML program v3.0 (Guindon et al. 2010). The WAG substitution model was selected, assuming an estimated proportion of invariant sites and four gamma-distributed rate categories to account for rate heterogeneity across sites. The gamma shape parameter was estimated directly from the data. Reliability of internal branches was assessed using LRT testing (SH-Like) (Anisimova and Gascuel 2006). Graphical representation and editing of the phylogenetic tree were performed with TreeDyn v198.3 (Chevenet et al. 2006).
Crossing of strains and segregation analysis of mating-type genes
Genomic DNA from F. velutipes SSIs was extracted as described (van Peer et al. 2011). Mating type gene-specific primers (Table S2A) were used to determine the presence or absence of these genes. PCR was performed in volumes of 30 µl premix Taqkit (Takara, China), using 35 cycles and an initial melting step of the genomic DNA at 94° for 5 min. To establish the site of recombination between the HD-a and HD-b subloci, specific primers were designed that flanked single nucleotide polymorphisms (SNPs) located between the HD-a and HD-b subloci in strain L11 and W23 (Table S2B).
SSIs of F. velutipes strains F0010, F0012, F0020, F0025, and F0027 (Table S1) were crossed to distinguish the four different mating types of each strain, followed by PCR amplification (primers: Table S2C), and sequencing of the genes at the respective HD-a and HD-b subloci (Table S3). Compatibility of HD-b subloci was determined by crossing F. velutipes strain L11 with other strains containing different HD-b and similar HD-a subloci.
Data availability
The data from this study were deposited in NCBI’s GenBank under the accession numbers: HQ630588.1, HQ630589.1, HQ630597.1, HQ630590.1, HQ630591.1, HQ630592.1, HQ630593.1, HQ630594.1, HQ630595.1, HQ630596.1, BK009409.1, KC208604.1, KC208603.2, KC208605.1, KC208611.1, KC208606.2, KC208607.1, KC208608.2, KC208609.2, KC208610.1, KT808674.1, KC208594.1, KC208595.2, KC208612.1, KC208596.1, KC208597.1, KC208598.2, KC208599.1, KC208600.2, KC208601.2, KC208602.1, KT808675.1, KT808673.1, KT808672.1, KT808676.1, KT808677.1, KT808678.1 and KT808679.1. Genome sequences have been deposited at DDBJ/EMBL/GenBank (http://www.ncbi.nlm.nih.gov/) under accession nos. APHZ00000000 (W23; BioProject: 191864) and APIA00000000 (L11; BioProject: 191865). Transcriptomics data has been uploaded to the NCBI Sequence Read Archive (SRA), and is accessible through accession numbers: SRP072148–SRP072150 (http://www.ncbi.nlm.nih.gov/sra/).
Results
Identification of mating type genes in F. velutipes strain L11, W23, and KACC42780
New HD and PR loci of F. velutipes were identified using the draft genomes of two mating-compatible strains; F. velutipes strain L11 and F. velutipes strain W23 (Liu et al. 2015; Wang et al. 2015b; Zeng et al. 2015). The draft genome of strain L11 consisted of 1858 scaffolds covering 34.75 Mb and 11,526 predicted genes. The draft genome of strain W23 consisted of 1866 scaffolds, covering 35.13 Mb and 11,071 predicted genes (Table S4 and Table S5). A complete list of the identified mating type genes and their gene accession numbers is given (Table S3).
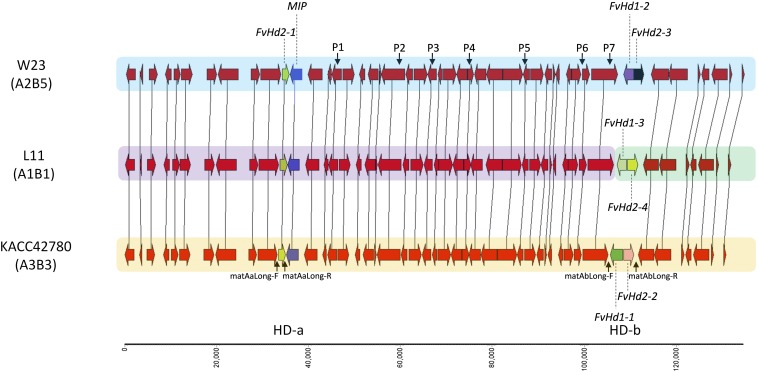
To identify HD loci, the genomes of strain L11 and strain W23 were screened for homeodomain genes based on homology searches with F. velutipes homeodomain proteins FvHD1-1, FvHD2-1, and FvHD2-2 (van Peer et al. 2011). Strain L11 contained three homeodomain genes, one FvHd2 gene on scaffold 1208, and an FvHd2/FvHd1 gene pair on scaffold 336 (Figure 1). The genome of strain W23 contained three homeodomain genes on scaffold 63, one single FvHd2 gene, and an FvHd2/FvHd1 gene pair (Figure 1). Reciprocal BLAST searches with the newly identified FvHd genes did not reveal any additional homeodomain genes in the genomes of strain L11, W23, or in the genome of strain KACC42780.
Figure 1.
Synteny map of the HD loci of F. velutipes strains W23, L11, and KACC42780. Names and mating types of strains are indicated at the left, the relative size of the regions is indicated in nucleotides at the bar at the bottom. The Mitochondrial Intermediate Peptidase (MIP) gene (blue arrow), and the different Hd genes (green, yellow, purple, and black arrows) are indicated separately. Locations of the primer sets that were used to determine the site of recombination between the HD-a and the HD-b sublocus in SSI No.24 are indicated with P1–P7. The binding sites for the conserved primers that were used to amplify the HD-a and HD-b subloci of F. velutipes strains are indicated under the HD region of strain KACC42780.
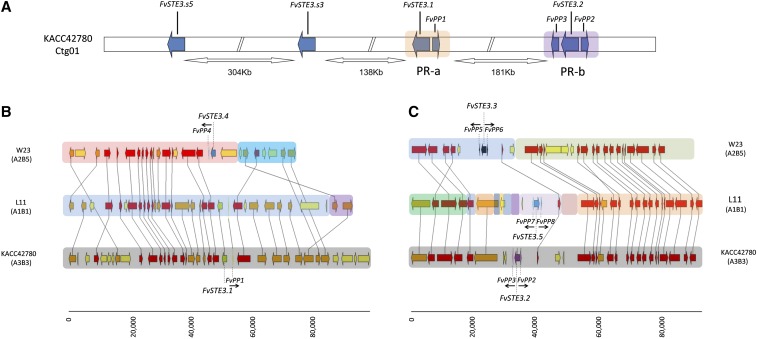
For identification of PR loci, pheromone receptor genes were identified in the genomes of strain L11 and strain W23 based on homology searches with mating type specific pheromone receptor proteins FvSTE3.1 and FvSTE3.2 of F. velutipes strain KACC42780 (van Peer et al. 2011). F. velutipes strain L11 contained two pheromone receptor genes located on scaffolds 263 and 789 (Figure 2, B and C). Two pheromone receptor genes were also identified in the genome of strain W23, on scaffolds 1147 and 1065 (Figure 2, B and C). Pheromone precursor genes were identified through analysis of the flanking regions of the new pheromone receptor genes (van Peer et al. 2011). The pheromone receptor gene on the PR-a sublocus of strain L11 was accompanied by a single pheromone precursor gene (Figure 2B). The pheromone receptor gene on the PR-b sublocus of strain L11 was associated with two pheromone precursor genes (Figure 2C). Similarly, the pheromone receptor gene at the PR-a sublocus of strain W23 was associated with one pheromone precursor gene (Figure 2B), while the second pheromone receptor gene at the PR-b sublocus was associated with two pheromone precursor genes (Figure 2C).
Figure 2.
(A) General organization of the PR-a and PR-b subloci of F. velutipes according to the updated genome sequence (Park et al. 2014). Pheromone receptor genes (FvSTE3.1 and FvSTE3.2), pheromone precursor genes (FvPp1, FvPp2, and FvPp3), and pheromone receptor like genes (FvSTE3.s3 and FvSTE3.s5) are indicated. (B, C) Synteny map of respectively the PR-a (B) and PR-b (C) sublocus regions of F. velutipes strains W23, L11, and KACC42780. Strain names and their mating types are indicated at the left (B) or right (C) of the depicted regions. The positions and directions of pheromone receptor genes (FvSTE3, blue, green, purple, and black arrows), and pheromone precursor genes (FvPP, black arrows below or above the gene name) are depicted. The lower bar indicates the relative distance between genes in nucleotides.
Next, the genomes of strain L11 and W23 were screened for nonmating type specific pheromone receptor genes (STE3.s), hereafter called “pheromone receptor like genes” based on homology searches with five STE3.s proteins from F. velutipes strain KACC42780 (van Peer et al. 2011). All five pheromone receptor like genes (Table S3) were represented in the genomes of strain L11 (scaffolds 13, 188, 383, 409, and 1473), and strain W23 (scaffolds 247, 275, 353, 576, and 1302). In addition, a new and sixth pheromone receptor like gene was detected in the genomes of strain L11 and W23 (Table S3). This sixth pheromone receptor like gene was subsequently identified in the genome of strain KACC42780 (ctg11_2-8_draft).
Analysis of homeodomain genes and proteins
The predicted gene models (intron and exon boundaries) of the homeodomain genes of strain L11 and W23 were verified using transcriptomics data of strain L11, W23, and dikaryotic strain 1123. Alignment of the homeodomain genes showed that strain L11, W23, and KACC42780 contained the same FvHd2-1 gene on the HD-a sublocus, with DNA sequence similarities of 99.77% over 1580 bp (Figure S1). The corresponding FvHD2-1 proteins showed 100% amino acid (AA) sequence identity. The FvHD2-1 proteins contained one predicted NLS. No Di motifs were detected in FvHD2-1 (Figure 3 and Figure S2). The FvHd2-1 genes and proteins on the HD-a subloci were clearly dissimilar from the FvHd2 genes and proteins on the HD-b subloci (Figure S2). DNA sequence alignment of the homeodomain genes on the HD-b subloci of strain L11, W23, and KACC42780 indicated that each strain contained a pair of unique FvHd1/FvHd2 genes (Figure S3, Figure S4, and Figure S5). DNA sequence polymorphism was restricted to the N-terminal portions of the FvHd1 and FvHd2 genes, while the remaining parts were highly conserved. The short intergenic region between the FvHd1 and FvHd2 genes on the HD-b subloci also revealed high DNA sequence polymorphism. In accordance with the DNA sequences, the N-termini of the FvHD1 and FvHD2 proteins on the HD-b sublocus were strongly dissimilar (Figure S6 and Figure S7). The remaining four-fifths of the total protein lengths of the HD proteins were conserved. These included most of the Di and NLS motifs (Figure 3). The FvHD1 and FvHD2 proteins on the HD-b sublocus of strain KACC42780 did not contain predicted Di motifs in the variable N-termini, in contrast to the FvHD1 and FvHD2 proteins on the HD-b subloci of strain L11 and strain W23. The FvHD2 proteins of the HD-b subloci of strain KACC42780 (688 AA), L11 (688 AA), and W23 (693 AA) were longer than the FvHD2-1 proteins (475 AA) of the HD-a sublocus. Conservation between the HD proteins from strain L11, W23, KACC42780, and the HD proteins of additionally identified HD-b subloci (see below) was high (Figure S8 and Figure S9).
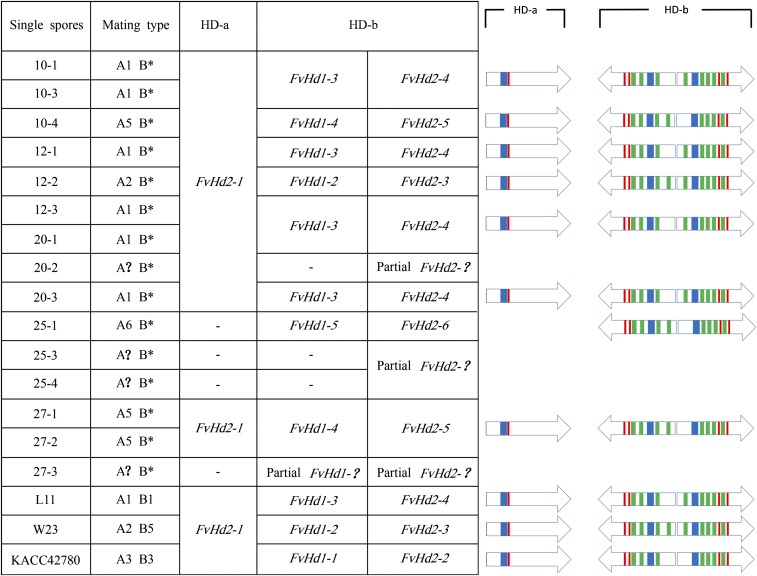
Figure 3.
Mating types of strains L11, W23, KACC42780, and the SSIs of which HD subloci were determined by PCR. PR mating types are named in the second column when known, or indicated with (*) when unknown. Schematic representations of the corresponding HD proteins (arrows) indicated the orientation of the gene on each sublocus, the position of the homeodomains (blue), NLSs (red), and the Di motifs (green). HD-a subloci for which no PCR products could be obtained, and HD-b subloci for which the obtained PCR products could not be fully sequenced, are indicated with “−” and are not represented by a schematic drawing. HD mating types were assigned to the F. velutipes strains in agreement with van Peer et al. (2011), applying HD3 (previously A3) for strain KACC42780, and preserving “HD4” (previously A4) for strain KACC43777.
The strong conservation of FvHD1 and FvHD2 proteins at the HD-b subloci in F. velutipes (86.06–89.81%) was unexpected. Interestingly, comparison of HD1 and HD2 sequences from other mushrooms, of which multiple alleles of HD subloci had been reported, revealed large variations in the conservation levels of these proteins. HD1 and HD2 proteins of Lentinula edodes (Au et al. 2014) showed high similarity (76.46–77.9%, Figure S10 and Figure S11). In contrast, HD1 and HD2 proteins in S. commune showed considerably lower similarity (38.85–58.89%, Figure S12, Figure S13, and Figure S14), while HD1 and HD2 proteins of different subloci in C. cinerea (Badrane and May 1999) showed strongly varying conservation levels, depending on the sublocus (39.26–70.3%, Figure S15 and Figure S16).
Comparison of the HD subloci and genomic regions
The genomic region containing the HD subloci is generally conserved in mushroom-forming fungi. In F. velutipes, this region had been indicated to contain structural deviations from the conserved gene arrangement (van Peer et al. 2011). To verify these differences, we compared the HD-a and HD-b sublocus-containing regions of strain L11 and W23, with that of the latest version of the F. velutipes KACC42780 genome that consisted of 11 chromosomes (35.6 Mb, 12,218 predicted genes; Park et al. 2014). The homeodomain genes of strain KACC42780 were located on chromosome 8 (2011_ctg03_draft). The HD-a and HD-b subloci on chromosome 8 were confirmed to be separated by ∼70 kb as previously reported, and no β-flanking gene was present near the HD-a or HD-b subloci. Alignment of a 134 kb section of chromosome 8 of strain KACC42780, with scaffold 63 of strain W23, and scaffolds 1208 and 336 of strain L11, revealed absolute synteny (Figure 1). The three HD-a subloci of strain L11, W23 and KACC42780, each contained the same single FvHd2-1 gene, without an associated FvHd1 gene. The HD-a sublocus was flanked by the MIP gene, which was located between the HD-a and the HD-b sublocus (Figure 1). All three HD-b subloci contained a pair of similarly arranged FvHd1 and FvHd2 genes.
Identification of additional HD subloci
Previously, the HD-a sublocus of strain KACC43777 had been reported not to contain the conserved FvHd2-1 gene. To further explore allelic variation of the HD subloci in F. velutipes, SSIs of five dikaryotic strains were selected for cloning and analysis of their respective HD-a and HD-b subloci (Figure 3 and Table S1). To ensure that all possible HD loci of the five strains were included, three SSIs with different mating types were analyzed from each dikaryon (15 in total). Specific primers were designed for the HD-a and the HD-b subloci, using flanking regions of the HD subloci that were conserved between the genomes of strain L11, W23, and KACC42780 (Figure 1 and Table S2C). Of the 15 SSIs that were analyzed, 11 contained a copy of the conserved FvHd2-1 gene found at the HD-a sublocus in strain L11, W23, and KACC42780 (Figure 3 and Figure S1). Complete sequences of HD-b subloci were obtained for 11 SSIs as well. Six SSIs contained the same HD-b sublocus as strain L11, and one contained the same HD-b sublocus as strain W23. Three SSIs contained a new HD-b sublocus with an FvHd1-4/FvHd2-5 gene pair, and one SSI contained a new HD-b sublocus with an FvHd1-5/FvHd2-6 gene pair (Figure 3, Figure S4, and Figure S5). The HD-b subloci of the four remaining SSIs were subcloned successfully, yet their sequences could be only partially determined. It remained unknown if the FvHd genes at these HD-b subloci were different from already identified genes, due to the high conservation of the parts of the FvHd genes that were represented by the partial sequences. Overall, allelic variation was strongest at the HD-b sublocus. However, the absence of FvHd2-1 in four SSIs might indicate variation (i.e., no gene or a different gene instead of the FvHd2-1 gene) at the HD-a sublocus.
Analysis of pheromone receptor, pheromone receptor like, and pheromone precursor genes and their proteins
The predicted gene models of the pheromone receptor genes and pheromone receptor like genes of strain L11 and W23 were verified based on transcriptomics data of strain L11, W23, and dikaryotic strain 1123. Comparisons with the genes of KACC42780 revealed a few differences in the intron and exon boundaries of gene FvSTE3.s2 of KACC42780 (van Peer et al. 2011). The gene model of FvSTE3.s2 from strain KACC42780 was updated following the transcriptomics data of strain L11, W23, and dikaryotic strain 1123. TMHMM and TMpred prediction software showed that proteins FvSTE3.1 to FvSTE3.5 contained 7-TM domains that are characteristic of pheromone receptors (Table S6). Alignment of the pheromone receptor genes and pheromone precursor genes revealed that strain L11 and strain KACC42780 contained the same genes on the PR-a sublocus. The pheromone that was encoded by the FvPp1 gene at the PR-a sublocus of KACC42780 had previously been reported to contain an additional tryptophan (W) behind the C-terminal CAAX box (CAAXW*). The FvPp1 gene of strain L11 also encoded this additional tryptophan (Figure S17). Subcloning and resequencing of the FvPp1 gene (primers Table S2D) confirmed that the sequence indeed encoded the extra tryptophan. The additional tryptophan was further supported by the transcriptomics data of strain L11, W23, and 1123 (Figure S18). Strain W23 contained different pheromone receptor (FvSTE3.4) and pheromone precursor (FvPp4) genes on the PR-a sublocus (Figure 2). Interestingly, the predicted mature FvPP4 pheromone on the PR-a sublocus of W23 (ERHGQGMAYTC), was identical to predicted mature pheromone FvPP1 except for the additional tryptophan at the C-terminus of FvPP1 (Figure S17). The pheromone receptor genes and pheromone precursor genes on the PR-b subloci of strain L11, W23, and KACC42780 were all strain specific (Figure 2). However, the mature FvPP3 pheromone from the PR-b sublocus of strain KACC42780, and the mature FvPP6 pheromone from the PR-b sublocus of strain W23, contained identical protein sequences (ERAGDPTFRGGAC). This indicated that pheromones FvPP3 and FvPP6 would be incompatible with pheromone receptors FvSTE3.2 and FvSTE3.3, respectively (Figure S17). Pheromone receptor like proteins FvSTE3.s1–FvSTE3.s6, which had been identified in the genomes of strain L11, W23 and KACC42780, each contained 7-TM domains (Table S6). None of the corresponding pheromone receptor like genes was accompanied by a pheromone precursor gene.
Phylogenetic analysis of pheromone receptors and pheromone receptor like proteins
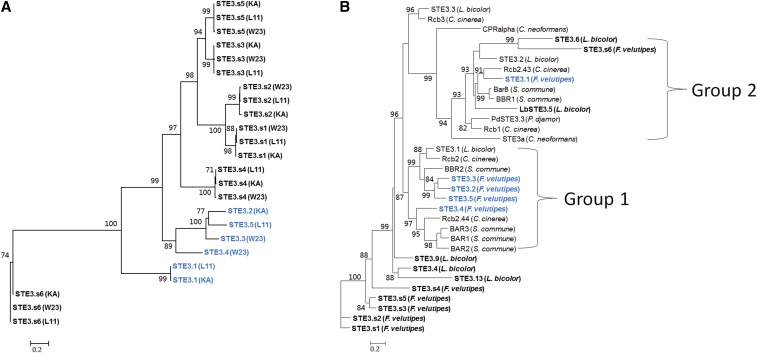
The phylogenetic origin of pheromone receptors and pheromone-receptor-like proteins has remained confused, and pheromone receptors from the same sublocus do not always cluster together (Riquelme et al. 2005; Kües 2015). To examine the relationship between the pheromone receptors and the pheromone-receptor-like proteins in F. velutipes, two analyses were performed. The first analysis comprised a comparison of all identified pheromone receptor and pheromone-receptor-like protein sequences from F. velutipes (Figure 4A). The second analysis compared the pheromone receptor and pheromone-receptor-like protein sequences from F. velutipes with that of other model mushroom-forming fungi (Figure 4B). Pheromone receptor and pheromone-receptor-like protein sequences from F. velutipes that were represented by multiple copies, like, for example, the FvSTE3.s proteins, were included only once in the second analysis. In brief, proteins were aligned using MUSCLE, and processed with Gblocks to remove poorly aligned regions. The resulting 230 amino acid sequences (first analysis), and 197 amino acid sequences (second analysis), were compiled into a maximum likelihood tree. For both analyses, the graphics of the resulting trees were adjusted with TreeDyn. The pheromone receptors of F. velutipes were divided into two main groups (Figure 4A). Comparison with other fungal pheromone receptors placed FvSTE3.1 in main group “Group2,” and FvSTE3.2 to FvSTE3.5 in main group “Group1” (Figure 4B). Pheromone receptors FvSTE3.2, FvSTE3.3, and FvSTE3.5 were closely related, in accordance with their localization on the same PR-b sublocus. FvSTE3.4 appeared less related to FvSTE3.2, FvSTE3.3, and FvSTE3.5 (Figure 4A). This separation was confirmed by comparison of the pheromone receptors from different fungi (Figure 4B). It is interesting to note that FvSTE3.4 is located on the PR-a sublocus, like FvSTE3.1, instead of on the PR-b sublocus with FvSTE3.2, FvSTE3.3, and FvSTE3.5. The presence of two pheromone receptors from different main phylogenetic groups (i.e., FvSTE3.1 and FvSTE3.4) on the same PR-a sublocus indicated that recombination of pheromone receptors from different subloci must have occurred. The pheromone-receptor-like proteins clustered in two groups that were clearly separated from the pheromone receptors of F. velutipes. One small group contained the FvSTE3.s6 proteins, and a second, larger group contained the FvSTE3.s1 to FvSTE3.s5 proteins (Figure 4A). Within the larger group of pheromone-receptor-like proteins, FvSTE3.s1 and FvSTE3.s2 were indicated to be related more closely, as were FvSTE3.s3 and FvSTE3.s5 (Figure 4A). When compared to other fungal pheromone receptors and pheromone-receptor-like proteins, FvSTE3.s1 to FvSTE3.s5 remained distant from the main pheromone receptor groups “Group1” and “Group2” (Figure 4B). No pheromone-receptor-like proteins clustered in pheromone receptor main group “Group1.” However, FvSTE3.s6 was included in pheromone receptor main group “Group2,” together with pheromone-receptor-like proteins from L. bicolor (STE3.5 and STE3.6).
Figure 4.
(A) Phylogenetic tree of the pheromone receptors (blue) and pheromone-receptor-like proteins (black) of F. velutipes strains L11, W23, and KACC42780. The corresponding strain is indicated behind each protein. (B) Phylogenetic tree of F. velutipes (blue) and S. commune, C. cinerea, L. bicolor, and C. neoformans (black) pheromone receptors. F. velutipes and L. bicolor pheromone-receptor-like proteins are given in black, bold font. Species names are indicated behind each protein. KA, strain KACC42780.
Comparison and analysis of PR loci
The PR loci of strain L11 and W23 were compared with the latest version of the F. velutipes KACC42780 genome (Park et al. 2014). Mapping of the KACC42780 pheromone receptor genes, pheromone precursor genes, and pheromone receptor like genes showed that the PR-a (FvSTE3.1 and FvPp1) and the PR-b (FvSTE3.2, FvPp2 and FvPp3) subloci were located on one chromosome. Pheromone receptor like genes FvSTE3.s3 and FvSTE3.s5 were located on the same chromosome as the PR subloci. Pheromone receptor like genes FvSTE3.s1, FvSTE3.s2, FvSTE3.s4, and FvSTE3.s6 were located on different chromosomes. The distance between the PR-a and PR-b subloci on the chromosome was similar to that of the previously reported draft genome, 181 vs. 177 kb. The distance between the PR-a sublocus and pheromone receptor like gene FvSTE3.s3 was found to be 43 kb less (138 vs. 181 kb). The distance between pheromone receptor like gene FvSTE3.s3 and FvSTE3.s5 was found to be much larger, at 304 kb compared to 184 kb (Figure 2, cf. Figure 1 in van Peer et al. 2011). The FvSTE3.1 and FvPp1 genes on the PR-a subloci of strain KACC4270 and L11 were arranged in the same way. Although some of the neighboring genes of the PR-a sublocus were arranged similarly, overall synteny of the PR-a sublocus region was not high.
The orientation of the pheromone receptor gene and the pheromone precursor genes on the PR-b sublocus of KACC42780 was found to be different from the reported draft genome. The pheromone precursor genes contained an outward instead of an inward orientation respective to the PR-b sublocus, and pheromone receptor gene FvSTE3.2 was directed toward FvPp3 instead of toward FvPp2 (Figure 2, compare to Figure 1 in van Peer et al. 2011). This new arrangement of pheromone receptor genes and pheromone precursor genes on the PR-b sublocus of KACC42780 was similar to the arrangement of the pheromone receptor and pheromone precursor genes on the PR-b subloci of strain L11 and W23 (Figure 2C). Interestingly, strain L11 and strain KACC42780 contained the same PR-a sublocus, but different PR-b subloci. This indicated that the PR-a and PR-b subloci could recombine independently.
Segregation analysis of HD and PR mating-type genes
Thirty one SSIs and F1 progeny of dikaryotic strain 1123 were examined to assess the occurrence of independent recombination between HD subloci and between PR subloci (Table S7). One of the 31 SSIs, no.24, showed a new combination of an HD-a and HD-b sublocus. SNPs in the FvHd2-1 gene at the HD-a sublocus were specific for strain L11, while the HD-b sublocus contained the W23-specific FvHd1-2 and FvHd2-3 genes. To determine the site of recombination, seven different regions with SNPs specific for strain L11 or W23, located in between the HD-a and the HD-b subloci (Figure 1), were PCR amplified and sequenced. Specific SNPs for strain L11 were found in the PCR products of regions 1–6. Region seven contained specific SNPs for strain W23. Recombination therefore occurred between SNP-specific region six and seven. Unfortunately, sequence homology between strains L11 and W23 prevented further specification of the recombination site. It remained undetermined if recombination had occurred between the first and the second gene that flanked the HD-b sublocus, or within one of those genes.
Independent recombination of PR-a and PR-b was apparent from the genome data of strains L11 and KACC42780. However, no such recombination was observed in the 31 SSIs of strain 1123.
Crossing of strains with different HD subloci
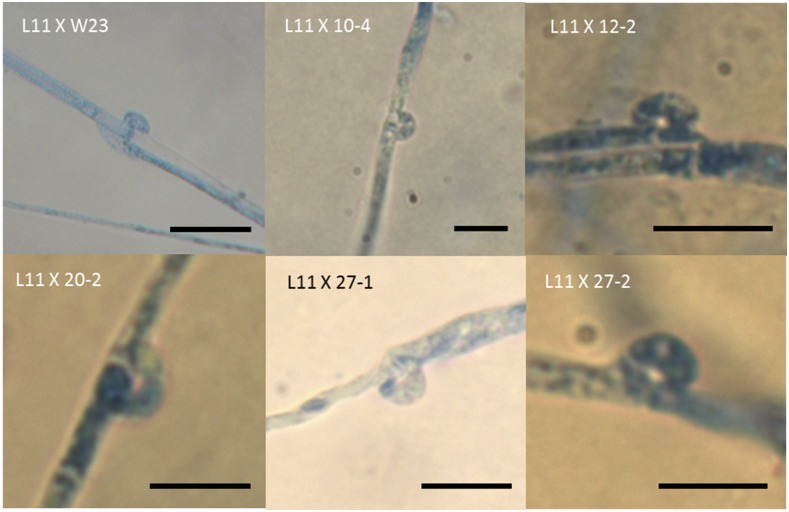
Single spore isolate (SSI) no.24, which contained the HD-a sublocus of strain L11 and the HD-b sublocus of strain W23, was crossed with parental strain L11. This combined two identical HD-a subloci (SSI no.24 and L11 contained the FvHd2-1 gene from L11) and two different HD-b subloci (SSI no.24 contained the FvHd1-2 and FvHd2-3 genes, and L11 contained the FvHd1-3 and FvHd2-4 genes, Figure 3). This new no.24/L11 dikaryon produced regular clamp connections, which demonstrated that the HD-b sublocus likely controled the HD pathway. Crossing of SSI no.24 and strain L22, which contained a different FvHd2-1 gene (SNPs) at the HD-a sublocus, and the same FvHd1-2 and FvHd2-3 genes at the HD-b sublocus, did not result in colonies that formed proper clamps. Combinations of HD-a subloci with different copies (SNPs) of the FvHd2-1 gene could not activate the HD pathway. Further crossing of strain L11 and a series of monokaryons with a FvHd2-1 gene at the HD-a sublocus and different HD-b subloci confirmed that the HD pathway could be activated by multiple combinations of FvHd1-3/FvHd2-4 (L11) with FvHd1-2/FvHd2-3 (strains W23 and 12-2), FvHd1-4/FvHd2-5 (strain 10-4, 27-1 and 27-2), and strain 20-2, of which the HD-b sublocus remained undetermined (Figure 5).
Figure 5.
Microscopic images of clamp connections in F. velutipes colonies from compatible crosses between strain L11 with strains W23, 10-4, 12-2, 20-2, 27-1, and 27-2. The different strains that were mated with L11 generated different combinations of HD-b subloci. Bars in the pictures represent 10 µm.
Poor conservation of the regions surrounding the PR-a and PR-b subloci had prevented the identification of PR subloci in addition to that of strain L11, W23, and KACC42780. Activity of the separate PR-a and PR-b subloci could therefore not be tested by specific crossings.
Discussion
The identification of the first HD and PR mating type loci of F. velutipes in the draft genome of strain KACC42780, had indicated that the HD and PR mating-type loci consisted of two subloci, HD-a and HD-b and PR-a and PR-b, respectively (van Peer et al. 2011). In contrast to most mushroom-forming fungi, the HD-a and HD-b subloci of F. velutipes were found to be separated by 73 kb. This separation was explained by inversions in the genomic region that contained the HD locus. In addition, the β-flanking gene that is commonly associated with the HD locus, was not found near the HD-a or the HD-b subloci in F. velutipes. This particular organization of the F. velutipes HD subloci was supported by this study. A newer version of the F. velutipes genome of strain KACC42780 (Park et al. 2014), and the genomes of F. velutipes strain L11 and strain W23, all showed the same organization of the HD locus. It had remained unclear if the HD-a, the HD-b, or both HD subloci of F. velutipes were involved in mating. The segregation analysis in this study had shown that independent recombination is possible between the HD-a and HD-b subloci. If both HD subloci were active, new HD mating types could be generated this way.
The FvHd2-1 gene of the HD-a sublocus had been reported to be specific for strain KACC42780 (i.e., was not detected in a mating compatible strain), but no compatible HD-a sublocus was identified (van Peer et al. 2011). The present study found that the FvHd2-1 gene of the HD-a sublocus was actually conserved in a large number of tested F. velutipes strains; however, not all F. velutipes strains contained the conserved FvHd2-1 gene (Figure 3). Together with the previous report, where no FvHd2-1 gene had been detected in strain KACC43777, the absence of the FvHd2-1 gene at the HD-a subloci of multiple F. velutipes strains indicated that there is variation at this sublocus. To understand the regulation of the HD pathway in F. velutipes, it will be important to determine of what this variation consists. If no FvHd genes are present at other HD-a subloci, the HD-a sublocus might be degenerate. However, if only a single FvHd1 gene (or a FvHd1/FvHd2 gene pair) would be present, the HD-a sublocus might still be active. Functional HD subloci that contain a single homeodomain gene are not uncommon (for a review, see Kües 2015).
The data from a previous study concerning the HD-b subloci of strain KACC42780 and mating compatible strain KACC43777 were unclear. While the FvHd1 gene on the HD-b sublocus of KACC42780 was also detected in strain KACC43777, the FvHd2 gene on the HD-b sublocus of KACC42780 (i.e., of the same functional pair), was not detected in strain KACC43777. This suggested a highly unlikely recombination within an Hd1/Hd2 gene couple, which would complicate the function of HD-b (van Peer et al. 2011). In this study, we revealed a strong conservation over the largest parts (four-fifths) of the FvHd1 and FvHd2 genes. It was found that the PCR primers that had been used to detect the FvHd1-1 gene in strain KACC42780 and KACC43777 bound within the conserved region of this gene. In contrast, the primers that were used to test for the presence of the FvHd2-2 gene in strain KACC42780 and KACC43777, did not bind within the conserved region of FvHd2-2. This could well explain why the FvHd1-1 primers generated highly similar sequences from strain KACC42780 and KACC4377, suggesting the presence of the same FvHd1 gene, while the sequence for the FvHd2-2 gene was specific for strain KACC42780. Most likely, therefore, is that the HD-b sublocus of strain KACC43777 contains a specific Hd1/Hd2 gene couple, different from the FvHd1 and FvHd2 genes on the HD-b sublocus of strain KACC42780. Unfortunately we did not have strain KACC43777 or KACC42780 to test this hypothesis. The high conservation of the FvHd genes might challenge the importance of divergence in the non N-terminal parts of HD-encoding genes in their role of preventing recombination between alleles (Hd1 genes or Hd2 genes) on the same sublocus (Kües 2015, and references herein). Selection in favor of Hd genes encoding highly divergent C-terminus sequences seems to be absent (or failing) in F. velutipes. Yet, none of the new HD-b subloci (Figure 3) indicated separate recombination between FvHd1 or FvHd2 genes. In addition, no recombination of L11 FvHd1 with W23 FvHd2 genes had been observed in the HD-b subloci of the 31 SSIs that were used for the segregation analysis.
A total of four new types of HD-b subloci was identified in addition to the HD-b sublocus of KACC42780. Each of the, in total five, HD-b subloci contained a specific Hd1/Hd2 gene couple (Figure 3). A function in activation of the HD pathway was derived for three of these subloci, using crosses between strain L11 and strains with similar HD-a, but different HD-b subloci (Figure 3 and Figure 5). Clearly, the HD-b sublocus is active.
The arrangement of the PR-a and PR-b subloci had been found to be unusual. The PR-a and PR-b subloci were separated by a very large, 177 kb genomic region. This large distance was supported (181 kb in this study) by the newest version of the genome sequence of strain KACC42780 (Park et al. 2014), as well as by the genomes of strain L11 and strain W23. Despite the large distance that separated the PR-a and the PR-b subloci, we did not observe independent recombination in a segregation population. However, the genomes of strain KACC42780 and strain L11 contained the same PR-a sublocus (Figure 2), while their PR-b subloci were different.
It had been deduced that the PR-a sublocus mediated activation of the PR pathway in a cross between mating compatible strains KACC42780 and KACC43777 (the same three FvSTE3.2, FvPp2, and FvPp3 genes had been detected on the PR-b subloci of both strains; van Peer et al. 2011). Our data indicated that the PR-a subloci of the mating compatible strains L11 and W23 might be inactive in a cross between strain L11 and W23. Although the pheromone receptors at the PR-a subloci of L11 (FvSTE3.1) and W23 (FvSTE3.4) were clearly different, the predicted mature pheromones FvPP1 (L11) and FvPP4 (W23) would be identical (Figure S17). We might speculate that the mutation in FvPP1 could have inactivated this pheromone, preventing self-activation of the L11 PR-a sublocus, and allowing activation of pheromone receptor FvSTE3.1 by pheromone FvPP4. Possibly, recombination within the PR-a sublocus of L11 created an initially self-active locus with FvSTE3.1 and FvPp1, followed by a mutation in FvPP1 that restored mating type specificity. Alternatively, FvSTE3.1 might have recombined with FvPp1 in the L11 PR-a sublocus after FvPP1 had lost its function. The possibility of within PR sublocus recombination is supported by studies on C. cinerea (Kües 2015, and references therein), and by the phylogenetic analysis that indicated that pheromone receptors FvSTE3.1 and FvSTE3.4 belong to different evolutionary clades (this study; James et al. 2004b, 2006; Riquelme et al. 2005; Kües et al. 2011).
The PR-b subloci of strains L11, W23, and KACC42780 each contained a unique pheromone receptor gene and two unique pheromone precursor genes (Figure 2). Crosses between strain L11 and W23 (which are known to generate proper dikaryons) would introduce two nonself pheromones to the pheromone receptor of strain L11, and two nonself pheromones to the pheromone receptor of W23. It is highly likely that at least one of these four combinations would be compatible, and thus that the PR-b sublocus is active.
Based on this study, we conclude that F. velutipes has two HD subloci. The HD-b sublocus is active, while the HD-a sublocus might be active and requires additional analysis. We further conclude that the PR locus consists of two subloci. The PR-a sublocus was previously confirmed to be active, and our data strongly indicate that the PR-b sublocus is active. Both the HD and the PR subloci have been found to be able to recombine, and new mating types could be generated. We are currently using transformation assays to confirm the function of individual mating-type genes, and continue to identify additional HD and PR subloci. This should help to establish if all four MAT subloci can be active, and which particular MAT genes can interact.
Supplementary Material
Acknowledgments
We are grateful to Dr. W.S. Kong of the Rural Development Administration in Korea for providing the F. velutipes KACC42780 genome sequence. The authors thank the Fujian Edible Fungi Engineering Technology Research Center and the National Fungi Breeding Center (Fujian Division) for providing the experimental facilities. This work was supported by grants from the Natural Science Foundation of Fujian Province (2015J01080), and the National Key Basic Research Program of China (2014CB138302).
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.034637/-/DC1.
Communicating editor: R. B. Brem
Literature Cited
- Aimi T., Yoshida R., Ishikawa M., Bao D., Kitamoto Y., 2005. Identification and linkage mapping of the genes for the putative homeodomain protein (hox1) and the putative pheromone receptor protein homologue (rcb1) in a bipolar basidiomycete, Pholiota nameko. Curr. Genet. 48: 184–194. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., et al. , 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M., Gascuel O., 2006. Approximate likelihood ratio test for branches: a fast, accurate and powerful alternative. Syst. Biol. 55: 539–552. [DOI] [PubMed] [Google Scholar]
- Au C. H., Wong M. C., Bao D., Zhang M., Song C., et al. , 2014. The genetic structure of the A mating-type locus of Lentinula edodes. Gene 535: 184–190. [DOI] [PubMed] [Google Scholar]
- Badrane H., May G., 1999. The divergence–homogenization duality in the evolution of the b1 mating-type gene of Coprinus cinereus. Mol. Biol. Evol. 16: 975–986. [DOI] [PubMed] [Google Scholar]
- Banham A. H., Asante-Owusu R. N., Gottgens B., Thompson S., Kingsnorth C. S., et al. , 1995. An N-terminal dimerization domain permits homeodomain proteins to choose compatible partners and initiate sexual development in the mushroom Coprinus cinereus. Plant Cell 7: 773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao D., Gong M., Zheng H., Chen M., Zhang L., et al. , 2013. Sequencing and comparative analysis of the straw mushroom (Volvariella volvacea) genome. PLoS One 8(3): e58294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölker M., Kahmann R., 1993. Sexual pheromones and mating responses in fungi. Plant Cell 5: 1461–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. J., Casselton L. A., 2001. Mating in mushrooms: increasing the chances but prolonging the affair. Trends Genet. 17: 393–400. [DOI] [PubMed] [Google Scholar]
- Burnett J. H., 1975. Mycogenetics. John Wiley & Sons, Hoboken. [Google Scholar]
- Caldwell G. A., Naider F., Becker J. F., 1995. Fungal lipopeptide mating pheromones: a model system for the study of protein prenylation. Microbiol. Rev. 59: 406–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casselton L. A., 1978. Dikaryon formation in higher basidiomycetes, pp. 275–297 in The Filamentous Fungi, edited by Smith J. E., Berry D. R. Edward Arnold, London. [Google Scholar]
- Casselton L. A., Challen M. P., 2006. The mating type genes of the basidiomycetes, pp. 357–374 in Growth, Differentiation and Sexuality, edited by Kües U., Fischer R. Springer, Berlin. [Google Scholar]
- Casselton L. A., Olesnicky N. S., 1998. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol. Mol. Biol. Rev. 62: 55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J., 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Chen B., van Peer A. F., Yan J., Li X., Xie B., et al. , 2016. Fruiting body formation in Volvariella volvacea can occur independent from its A controlled bipolar mating system, enabling homothallic and heterothallic life cycles. G3 6: 2135–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevenet F., Brun C., Banuls A. L., Jacq B., Chisten R., 2006. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7: 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho M. A., Sampaio J. P., Goncalves P., 2010. A deviation from the bipolar-tetrapolar mating paradigm in an early diverged basidiomycete. PLoS Genet. 6: 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., et al. , 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Web Server issue): W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman H. G., Thorner J., Caron M. G., Lefkowitz R. J., 1991. Model systems for the study of seven-transmembrane-segment receptors. Annu. Rev. Biochem. 60: 653–688. [DOI] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser K., Saleh F., Meinhardt F., 1979. Genetics of fruit body production in higher basidiomycetes II. Monokaryotic and dikaryotic fruiting in Schizophyllum commune. Curr. Genet. 1: 85–88. [DOI] [PubMed] [Google Scholar]
- Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., et al. , 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44: D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler T. J., Mitton M. F., Vaillancourt L. J., Raper C. A., 2001. Changes in mate recognition through alterations of pheromones and receptors in the multisexual mushroom fungus Schizophyllum commune. Genetics 158: 1491–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J., Hsueh Y., Findley K., Heitman J., 2007. Evolution of the mating-type locus: the basidiomycetes, pp. 19–34 in Sex in Fungi: Molecular Determination and Evolutionary Implications, edited by Heitman J., Kronstad J., Taylor J., Casselton L. ASM Press, Washington. [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., et al. , 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Halsall J. R., Milner M. J., Casselton L. A., 2000. Three subfamilies of pheromone and receptor genes generate multiple B mating specificities in the mushroom Coprinus cinereus. Genetics 154: 1115–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P., Park K. J., Obayashi T., Fujita N., Harada H., et al. , 2007. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35(Suppl. 2): W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh Y. P., Heitman J., 2008. Orchestration of sexual reproduction and virulence by the fungal mating-type locus. Curr. Opin. Microbiol. 11: 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James T. Y., 2007. Analysis of mating-type locus organization and synteny in mushroom fungi: beyond model species, pp. 317–332 in Sex in Fungi: Molecular Determination and Evolutionary Implications, edited by Heitman J., Kronstad J. W., Taylor J. W., Casselton L. A. ASM Press, Washington. [Google Scholar]
- James T. Y., Kües U., Rehner S. A., Vilgalys R., 2004a Evolution of the gene encoding mitochondrial intermediate peptidase and its cosegregation with the A mating-type locus of mushroom fungi. Fungal Genet. Biol. 41: 381–390. [DOI] [PubMed] [Google Scholar]
- James T. Y., Liou S. R., Vilgalys R., 2004b The genetic structure and diversity of the A and B mating-type genes from the tropical oyster mushroom, Pleurotus djamor. Fungal Genet. Biol. 41: 813–825. [DOI] [PubMed] [Google Scholar]
- James T. Y., Srivilai P., Kües U., Vilgalys R., 2006. Evolution of the bipolar mating system of the mushroom Coprinellus disseminatus from its tetrapolar ancestors involves loss of mating-type-specific pheromone receptor function. Genetics 172: 1877–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Wang X., Liu W., Xie B., Zhu J., 2009. Incompatibility factors of Flammulina velutipes. Acta Edulis Fungi. 16: 16–19. [Google Scholar]
- Kronstad J. W., Leong S. A., 1990. The b mating-type locus of Ustilago maydis contains variable and constant regions. Genes Dev. 4: 1384–1395. [DOI] [PubMed] [Google Scholar]
- Kües U., 2015. From two to many: multiple mating types in Basidiomycetes. Fungal Biol. Rev. 29: 126–166. [Google Scholar]
- Kües U., Casselton L. A., 1992. Molecular and functional analysis of the A mating-type genes of Coprinus cinereus, pp. 251–268 in Genetic Engineering: Principles and Methods, edited by Setlow J. K. Plenum Press, New York. [DOI] [PubMed] [Google Scholar]
- Kües U., Tymon A. M., Richardson W. V., May G., Gieser P. T., et al. , 1994. A mating-type factors of Coprinus cinereus have variable numbers of specificity genes encoding two classes of homeodomain proteins. Mol. Gen. Genet. 245: 45–52. [DOI] [PubMed] [Google Scholar]
- Kües U., James T. Y., Heitman J., 2011. Mating-type in basidiomycetes: unipolar, bipolar, and tetrapolar patterns of sexuality, pp. 97–160 in The Mycota XIV: Evolution of Fungi and Fungal-like Organisms, edited by Pöggeler S., Wöstemeyer J. Springer, Berlin. [Google Scholar]
- Leifa F., Pandey A., Soccol C. R., 2001. Production of Flammulina velutipes on coffee husk and coffee spent-ground. Braz. Arch. Biol. Technol. 44: 205–212. [Google Scholar]
- Lin H., Zhang Z. F., Zhang M. Q., Ma B., Li M., 2008. ZOOM! Zillions of oligos mapped. Bioinformatics 24: 2431–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Wang W., Xie B. G., 2014. Comparison of gene expression patterns in the mycelium and primordia of Flammulina velutipes strain 1123. Acta Edulis Fungi. 21: 1–7. [Google Scholar]
- Liu F., Wang W., Chen B. Z., Xie B. G., 2015. Homocitrate synthase expression and lysine content in fruiting body of different developmental stages in Flammulina velutipes. Curr. Microbiol. 70: 821–828. [DOI] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J., 1991. Predicting coiled coils from protein sequences. Science 252: 1162–1164. [DOI] [PubMed] [Google Scholar]
- Maia T. M., Lopes S. T., Almeida J. M., Rosa L. H., Sampaio J. P., et al. , 2015. Evolution of mating systems in basidiomycetes and the genetic architecture underlying mating-type determination in the yeast Leucosporidium scottii. Genetics 201: 75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin E., Kohler A., Baker A. R., Foulongne-Oriol M., Lombard V., et al. , 2012. Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc. Natl. Acad. Sci. USA 109: 17501–17506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculita-Hirzel H., Hirzel A. H. 2008. Visualizing the gene order conservation among genomes with ChromoMapper. Int. J. Comput. Intell. Bioinformatics Syst. Biol. 1:. [Google Scholar]
- Niculita-Hirzel H., Labbé J., Kohler A., le Tacon F., Martin F., et al. , 2008. Gene organization of the mating type regions in the ectomycorrhizal fungus Laccaria bicolor reveals distinct evolution between the two mating type loci. New Phytol. 180: 329–342. [DOI] [PubMed] [Google Scholar]
- Olesnicky N. S., Brown A. J., Honda Y., Dyos S. L., Dowell S. J., et al. , 2000. Self-compatible B mutants in Coprinus with altered pheromone-receptor specificities. Genetics 156: 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea S. F., Chaure P. T., Halsall J. R., Olesnicky N. S., Leibbrandt A., et al. , 1998. A large pheromone and receptor gene complex determines multiple B mating type specificities in Coprinus cinereus. Genetics 148: 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. J., Baek J. H., Lee S., Kim C., Rhee H., et al. , 2014. Whole genome and global gene expression analyses of the model mushroom Flammulina velutipes reveal a high capacity for lignocellulose degradation. PLoS One 9: e93560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper J., 1966. Sexuality of Higher Fungi. The Roland Press, New York. [Google Scholar]
- Raudaskoski M., 2015. Mating-type genes and hyphal fusions in filamentous basidiomycetes. Fungal Biol. Rev. 29: 179–193. [Google Scholar]
- Raudaskoski M., Kothe E., 2010. Basidiomycete mating type genes and pheromone signaling. Eukaryot. Cell 9: 847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme M., Challen M. P., Casselton L. A., Brown A. J., 2005. The origin of multiple B mating specificities in Coprinus cinereus. Genetics 170: 1105–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya S., Yamada M., Shibata K., Okuhara T., Yoshida M., et al. , 2013. Characterization of a gene coding for a putative adenosine deaminase-related growth factor by RNA interference in the Basidiomycete Flammulina velutipes. J. Biosci. Bioeng. 115: 360–365. [DOI] [PubMed] [Google Scholar]
- Stajich J. E., Wilke S. K., Ahrén D., Au C. H., Birren B. W., et al. , 2010. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc. Natl. Acad. Sci. USA 107: 11889–11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonomura H., 1978. Flammulina velutipes, pp. 409–421 in The Biology, Cultivation of Edible Mushrooms, edited by Chang S. T., Hayes W. A. Academic Press, London. [Google Scholar]
- Uno I., Ishikawa T., 1971. Chemical and genetical control of induction of monokaryotic fruiting bodies in Coprinus macrorhizus. Mol. Gen. Genet. MGG. 113: 228–239. [Google Scholar]
- Vaillancourt L. J., Raudaskoski M., Specht C. A., Raper C. A., 1997. Multiple genes encoding pheromones and a pheromone receptor define the Bβ1 mating-type specificity in Schizophyllum commune. Genetics 146: 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Peer A. F., Park S. Y., Shin P. G., Jang K. Y., Kong W. S., et al. , 2011. Comparative genomics of the mating-type loci of the mushroom Flammulina velutipes reveals widespread synteny and recent inversions. PLoS One 6(7): e22249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Chou T. S., Liu F., Yan J. J., Wu T. J., et al. , 2015a Comparison of gene expression patterns between the monokaryotic and dikaryotic mycelia of Flammulina velutipes. Mycosystema 34: 683–693. [Google Scholar]
- Wang W., Liu F., Jiang Y., Wu G., Guo L., et al. , 2015b The multigene family of fungal laccases and their expression in the white rot basidiomycete Flammulina velutipes. Gene 563: 142–149. [DOI] [PubMed] [Google Scholar]
- Wang Y., Bao L., Yang X., Li L., Li S., et al. , 2012. Bioactive sesquiterpenoids from the solid culture of the edible mushroom Flammulina velutipes growing on cooked rice. Food Chem. 132: 1346–1353. [DOI] [PubMed] [Google Scholar]
- Wessels J. G. H., 1993. Fruiting in the higher fungi. Adv. Microb. Physiol. 34: 147. [DOI] [PubMed] [Google Scholar]
- Whitehouse H. L. K., 1949. Heterothallism and sex in the fungi. Biol. Rev. Camb. Philos. Soc. 24: 411–447. [DOI] [PubMed] [Google Scholar]
- Wu J., Ullrich R. C., Novotny C. P., 1996. Regions in the Z5 mating gene of Schizophyllum commune involved in Y-Z binding and recognition. Mol. Gen. Genet. 252: 739–745. [DOI] [PubMed] [Google Scholar]
- Yee A. R., Kronstad J. W., 1993. Construction of chimeric alleles with altered specificity at the b incompatibility locus of Ustilago maydis. Proc. Natl. Acad. Sci. USA 90: 664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R. R., Cao H., Pan Y. J., 2007. Monokaryons fruiting and fruit body color of Flammulina velutipes. Acta Edulis Fungi. 14: 37–40. [Google Scholar]
- Yi R., Tachikawa T., Ishikawa M., Mukaiyama H., Bao D., et al. , 2009. Genomic structure of the A mating-type locus in a bipolar basidiomycete, Pholiota nameko. Mycol. Res. 113: 240–248. [DOI] [PubMed] [Google Scholar]
- Zeng X., Liu F., Chen J., Wang W., Xie B. G., et al. , 2015. Genomic sequencing and analysis of genes related to terpenoid compound biosynthesis of Flammulina velutipes. Mycosystema 34: 670–682. [Google Scholar]
- Zhang A., Xiao N., Deng Y., He P., Sun P., 2012. Purification and structural investigation of a water-soluble polysaccharide from Flammulina velutipes. Carbohydr. Polym. 87: 2279–2283. [Google Scholar]
- Zhang Z. F., Lin H., Ma B. 2010. Zoom lite: next-generation sequencing data mapping and visualization software. Nucleic Acids Res. 38(Web Server issue): W743–W748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data from this study were deposited in NCBI’s GenBank under the accession numbers: HQ630588.1, HQ630589.1, HQ630597.1, HQ630590.1, HQ630591.1, HQ630592.1, HQ630593.1, HQ630594.1, HQ630595.1, HQ630596.1, BK009409.1, KC208604.1, KC208603.2, KC208605.1, KC208611.1, KC208606.2, KC208607.1, KC208608.2, KC208609.2, KC208610.1, KT808674.1, KC208594.1, KC208595.2, KC208612.1, KC208596.1, KC208597.1, KC208598.2, KC208599.1, KC208600.2, KC208601.2, KC208602.1, KT808675.1, KT808673.1, KT808672.1, KT808676.1, KT808677.1, KT808678.1 and KT808679.1. Genome sequences have been deposited at DDBJ/EMBL/GenBank (http://www.ncbi.nlm.nih.gov/) under accession nos. APHZ00000000 (W23; BioProject: 191864) and APIA00000000 (L11; BioProject: 191865). Transcriptomics data has been uploaded to the NCBI Sequence Read Archive (SRA), and is accessible through accession numbers: SRP072148–SRP072150 (http://www.ncbi.nlm.nih.gov/sra/).