Abstract
The availability of genomes across the tree of life is highly biased toward vertebrates, pathogens, human disease models, and organisms with relatively small and simple genomes. Recent progress in genomics has enabled the de novo decoding of the genome of virtually any organism, greatly expanding its potential for understanding the biology and evolution of the full spectrum of biodiversity. The increasing diversity of sequencing technologies, assays, and de novo assembly algorithms have augmented the complexity of de novo genome sequencing projects in nonmodel organisms. To reduce the costs and challenges in de novo genome sequencing projects and streamline their experimental design and analysis, we developed iWGS (in silico Whole Genome Sequencer and Analyzer), an automated pipeline for guiding the choice of appropriate sequencing strategy and assembly protocols. iWGS seamlessly integrates the four key steps of a de novo genome sequencing project: data generation (through simulation), data quality control, de novo assembly, and assembly evaluation and validation. The last three steps can also be applied to the analysis of real data. iWGS is designed to enable the user to have great flexibility in testing the range of experimental designs available for genome sequencing projects, and supports all major sequencing technologies and popular assembly tools. Three case studies illustrate how iWGS can guide the design of de novo genome sequencing projects, and evaluate the performance of a wide variety of user-specified sequencing strategies and assembly protocols on genomes of differing architectures. iWGS, along with a detailed documentation, is freely available at https://github.com/zhouxiaofan1983/iWGS.
Keywords: genome sequencing, high-throughput sequencing, de novo assembly, experimental design, simulation, nonmodel organism
Whole genome sequences are rich sources of information about organisms that are superbly useful for addressing a wide variety of evolutionary questions, such as measuring mutation rates (Kumar and Subramanian 2002), characterizing the genomic basis of adaptation (Roux et al. 2014), and building the tree of life (Rokas et al. 2003; Salichos and Rokas 2013). Until now, however, organismal diversity has been highly unevenly covered, and most sequenced genomes correspond to model organisms, organisms of medical or economic importance, or ones that have relatively small and simple genomes (Reddy et al. 2015).
The rapid advance of DNA sequencing technologies has dramatically reduced the labor and cost required for genome sequencing, which is evidenced by the burst of large-scale genome projects in recent years that includes, for example, the 1000 Fungal Genomes (1KFG) Project (Grigoriev et al. 2011), the Yeast 1000 Plus (Y1000+) Project (Hittinger et al. 2015), the Insect 5K Project (Robinson et al. 2011), and the Genome 10K Project (Genome 10K Community of Scientists 2009). Some of these projects have already begun to fuel important discoveries in evolution and other fields (Zhang et al. 2014). Equally importantly, high-throughput DNA sequencing has made it possible for single investigators to perform de novo genome sequencing in virtually any organism they are interested in (Rokas and Abbot 2009). Such sequencing efforts may target various organisms with a large diversity of genome architectures. Therefore, to achieve optimal results, the choice of sequencing strategy (i.e., the combination of sequencing technology [e.g., Illumina or Pacific Biosciences (PacBio)], sequencing assay (e.g., paired-end or mate-pair), and other variables, such as sequencing depth and assembly protocols (e.g., assemblers and the associated parameters) should ideally be tailored to the characteristics of a given genome, such as size and GC/repeat content (Nagarajan and Pop 2013).
The vast majority of de novo sequenced genomes have been generated using the Illumina technology, either solely or in combination with other technologies (Reddy et al. 2015). This is largely due to the Illumina technology’s ability to quickly generate tens to hundreds of millions of highly accurate short sequence reads of up to 300 bases per run at very low per base cost (Glenn 2011). Additionally, the Illumina technology offers two powerful sequencing assays, paired-end (PE) and mate-pair (MP), which generate sequence read pairs that span short (hundreds of base-pairs) and relatively long (thousands of base-pairs) genomic regions, respectively. Mixing multiple PE and MP libraries with different insert sizes allows for highly flexible sequencing strategies, and several state-of-the-art assembly algorithms have been developed that exploit all these advantages. For instance, the de novo genome assembler ALLPATHS-LG can generate high quality draft assemblies for mammalian-size genomes using only Illumina short-read data by including both MP and overlapping PE libraries (Gnerre et al. 2011). On its own, however, the Illumina technology performs less well for more complex genomes, mainly due to the short lengths of Illumina sequence reads and the technology’s bias against certain genomic regions (e.g., GC-rich regions) (Ross et al. 2013).
The PacBio technology generates sequence reads that are substantially longer and have much less sequencing bias, albeit at the cost of a substantially lower per-read accuracy; the average read length increases to above 10 kb with the latest chemistry but displays only ∼87% accuracy (Koren and Phillippy 2015). Thus, this technology is particularly useful for the sequencing of complex genomes (Koren and Phillippy 2015). Recent developments in both sequencing chemistry and assembly algorithms have enabled PacBio-only de novo assembly for microbial genomes (Koren et al. 2013), but the high sequence coverage required for this approach remains cost-prohibitive for large eukaryotic genomes. Nevertheless, in combination with more affordable Illumina short-read data, PacBio long reads—even at low coverage—can lead to significantly improved assemblies (Utturkar et al. 2014; McIlwain et al. 2016).
De novo genome sequencing projects are further complicated by the large array of assembly software tools, which differ in many aspects, such as algorithmic design, supported/required data types, and computational efficiency (Nagarajan and Pop 2013; Simpson and Pop 2015). Systematic evaluations of assembly programs show that no single assembler is the best across all circumstances; rather, an assembler’s performance critically depends on genome complexity and the sequencing strategy adopted (Earl et al. 2011; Bradnam et al. 2013). Moreover, many assemblers use adjustable parameters (e.g., the k-mer size for de Bruijn assemblers), the values of which can critically affect the assembly quality. In practice, such parameters are often selected intuitively or through the time-consuming process of testing multiple values.
The great number of possible ways to combine sequencing technologies, assays, and assembly algorithms poses a great challenge for the experimental design and data analysis in de novo genome sequencing projects, which in turn can sometimes lead to poor quality or downright incorrect assemblies (Denton et al. 2014). As a consequence, several pipelines have been developed to automate specific steps in the process; for example, the recently developed iMetAMOS (Koren et al. 2014) and RAMPART (Mapleson et al. 2015) have been specifically designed to automate genome assembly. However, as de novo genome sequencing is increasingly adopted by single investigator laboratories, there is an urgent need for streamlined approaches that enable investigators to not only efficiently generate high-quality draft genome assemblies but also to predict (via simulation) and identify the most suitable design(s) [i.e., the most suitable combination(s) of sequencing strategy and assembly protocol] currently available for a specific genome.
To address this need, we have developed an automated pipeline for the design and execution of de novo genome sequencing projects that we name iWGS (in silico Whole Genome Sequencer and Analyzer). To approximate the performance of different sequencing strategies and assembly protocols, iWGS simulates high-throughput genome sequencing on user-provided reference genomes (e.g., genomes that closely represent the characteristics of the real targets), facilitating the identification of optimal experimental designs. iWGS allows users to experiment with various combinations of sequencing technologies, assays, assembly tools, and relevant parameters in a single run. iWGS is also designed to work with real data and can be used as a convenient tool for automated selection of the best assembly or genome assembler. Finally, using three case studies, each one focused on specific challenges frequently encountered in de novo genome sequencing studies (e.g., high repeat content and biased nucleotide composition, etc.), we illustrate how iWGS can be applied to guiding the design and analysis of de novo genome sequencing studies.
Results
The design of iWGS
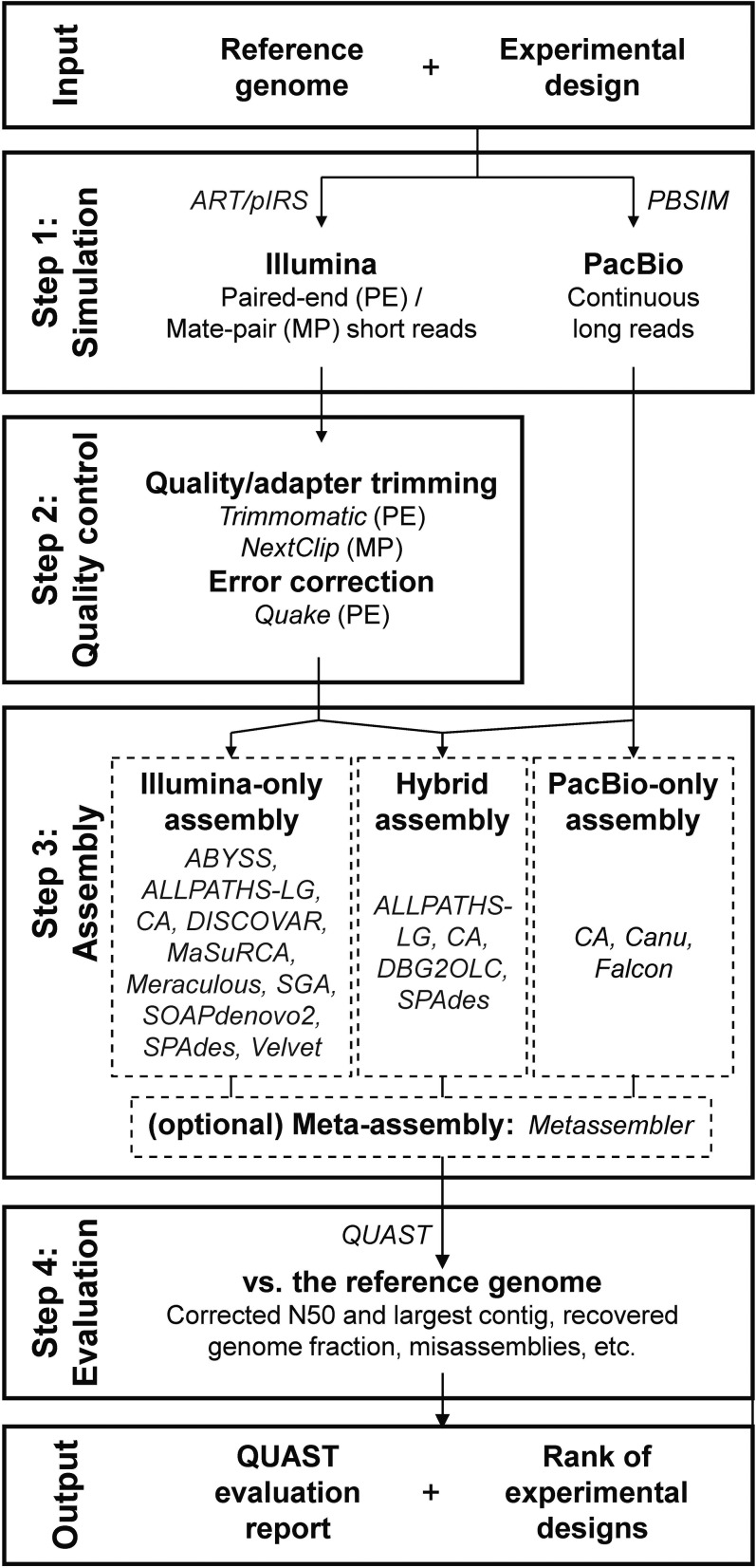
iWGS encompasses all major steps of a typical de novo genome sequencing study, including the generation of sequence reads, data quality control, de novo assembly, and evaluation of assemblies (Figure 1).
Figure 1.
iWGS workflow. A typical iWGS analysis consists of four steps: (1) data simulation (optional); (2) preprocessing (optional); (3) de novo assembly; and (4) assembly evaluation. iWGS supports both Illumina short reads and PacBio long reads, and a wide selection of assemblers to enable de novo assembly using either or both types of data. Users can start the analysis simulating data drawn from a reference genome assembly or, alternatively, use real sequencing data as input and skip the simulation step. iWGS, in silico Whole Genome Sequencer and Analyzer; MP, mate-pair; PacBio, Pacific Biosciences; PE, paired-end.
Simulation:
iWGS uses the realistic high-throughput sequencing (HTS) read simulators ART (Huang et al. 2012), pIRS (Hu et al. 2012), and PBSIM (Ono et al. 2013) to generate Illumina and PacBio sequence reads from a given user-specified genome. These programs can simulate all popular data types, including Illumina PE and MP sequence reads, as well as PacBio continuous long sequence reads. The distributions of read quality and read length are easily adjustable for both Illumina and PacBio data. Furthermore, these simulators mimic sequencing errors and nucleotide composition biases in real data by using empirical profiles of these artifacts, which can be easily customized to stay current with upgrades in sequencing technologies. For instance, we have created a quality-score frequency profile learned from sequence reads generated by the latest PacBio chemistry to better reflect the improved sequence read accuracy. This simulation step can be omitted when the goal is the analysis of real data. Alternatively, the users may choose to perform only the simulation and use the simulated data for other analyses.
Quality control:
HTS data generated by all technologies contain errors and artifacts, which may sometimes substantially compromise the quality of the assembly (Zhou and Rokas 2014). Therefore, iWGS includes an optional step to perform preprocessing of the data, including trimming of low-quality bases, removal of adapter contaminations, and correction of sequencing errors. Since some assemblers [e.g. ALLPATHS-LG (Ribeiro et al. 2012)] have their own preprocessing modules, iWGS automatically determines for each assembly protocol whether to use the original or the processed data.
Assembly:
To maximize users’ flexibility in experimental design, iWGS supports 15 de novo genome assembly tools [ABYSS (Simpson et al. 2009), ALLPATHS-LG (Ribeiro et al. 2012), Celera Assembler (Myers et al. 2000; Berlin et al. 2015), Canu (Koren et al. 2016), DBG2OLC (Ye et al. 2014), DISCOVAR (Weisenfeld et al. 2014), Falcon (Chin et al. 2016), MaSuRCA (Zimin et al. 2013), Meraculous (Chapman et al. 2011), Minia (Salikhov et al. 2013), Platanus (Kajitani et al. 2014), SGA (Simpson and Durbin 2012), SOAPdenovo2 (Luo et al. 2012), SPAdes (and a diploid-aware version called dipSPAdes) (Bankevich et al. 2012), and Velvet (Zerbino and Birney 2008)], most of which have participated in recent large-scale assembler comparisons (Bradnam et al. 2013; Magoc et al. 2013). These supported assemblers allow users to carry out de novo assembly using only Illumina short-read data (e.g., SOAPdenovo2) and only PacBio long-read data (e.g., Canu and Falcon), or to perform hybrid assembly that uses both (e.g., SPAdes and DBG2OLC). To achieve the best possible results while avoiding the computationally expensive process of testing multiple combinations of parameters, iWGS takes advantage of successful assembly recipes (i.e., recommended settings for each assembler) established in studies such as Assemblathon 2 (Bradnam et al. 2013) and GAGE-B (Magoc et al. 2013), and uses KmerGenie to determine the optimal k-mer size (Chikhi and Medvedev 2014). In addition, assemblies generated from different underlying data and/or assembly algorithms can be merged using Metassembler (Wences and Schatz 2015) to achieve a potentially better final assembly.
Evaluation:
iWGS uses QUAST (Gurevich et al. 2013) to evaluate all generated assemblies. In addition to providing basic statistics like N50 (the largest contig/scaffold size wherein half of the total assembly size is contained in contigs/scaffolds no shorter than this value), QUAST compares each assembly against the reference genome (in the case of simulations) and generates a number of highly informative quality matrices, such as misassemblies, assembled sequences not present in the reference (and vice versa), and genes recovered in the assembly if the reference genome is annotated. At the end, iWGS ranks all assemblies based on selected matrices in the QUAST report using a previously described weighting strategy (Abbas et al. 2014). This ranking, along with the detailed QUAST report, helps users to identify the best overall assembly, as well as the corresponding combination of sequencing strategy and assembly protocol. REAPR, which utilizes the sequence data itself for assembly evaluation, is also implemented to better suit real data analysis (Hunt et al. 2013).
iWGS is designed with flexibility and ease-of-use in mind to allow users to readily examine various experimental designs; each data set may be used multiple times in different assembly protocols, and each assembler may be run repeatedly with different input data sets. Multiple sequencing strategies and assembly protocols can be specified in a straightforward fashion in a single configuration file; only a few parameters are required for each strategy/protocol, while other settings (e.g., quality profiles for read simulation) are globally shared across strategies/protocols of the same type. Alternatively, advanced users can opt to customize the strategies/protocols so that, for example, each sequencing data set is simulated with different quality settings. Furthermore, iWGS rigorously checks the configurations for issues such as the compatibility between sequencing strategies and assembly protocols.
iWGS is a lightweight pipeline written in Perl. The source code, detailed documentation, and example test sets are freely available at https://github.com/zhouxiaofan1983/iWGS. Like many other bioinformatics pipelines, iWGS inevitably relies on a number of third-party software tools to carry out individual analyses such as data simulation and genome assembly. However, most of the tools, including at least one for each of the four major steps aforementioned, either have precomplied executables or can be compiled locally with ease. For the convenience of users, we also include in the package scripts to automate the acquisition and installation of most software dependencies. The users can also customize the selection of tools to install according to their own needs and computational environments.
Case studies
To demonstrate the use of iWGS and provide examples of its utility, we developed three case studies where iWGS was used to guide the selection of sequencing strategy for genomes representing a wide range of sizes and complexity levels (Supplemental Material, Table S1). The competing strategies were selected to enable both Illumina-only and PacBio-only assemblies, as well as hybrid assembly of the two data types (Table 1). To examine the effectiveness of the simulation step of our approach, we also analyzed real sequencing data that largely match our simulation settings.
Table 1. Sequencing strategies (top) and assembly protocols (bottom) evaluated in the three case studies.
| Name | Read Type | Parameters for Read Simulation |
|---|---|---|
| LIB1 | Illumina PE | Depth: 50 ×; read length: 100 bp; insert size: 180 ± 9 bp |
| LIB2 | Illumina MP | Depth: 50 ×; read length: 100 bp; insert size: 8000 ± 400 bp |
| LIB3 | Illumina PE | Depth: 50 ×; read length: 250 bp; insert size: 450 ± 23 bp |
| LIB4 | PacBio CLR | Depth: 60 ×; read accuracy: 0.87 ± 0.03; read length: 11,500 ± 8000 bp |
| LIB5 | PacBio CLR | Depth: 10 ×; read accuracy: 0.87 ± 0.03; read length: 11,500 ± 8000 bp |
| Name | Assembler | Sequencing strategies used for assembly |
| ILMN1 | ABYSS | LIB1, LIB2 (Illumina-only) |
| ILMN2 | ALLPATHS-LG | |
| ILMN3 | MaSuRCA | |
| ILMN4 | SGA | |
| ILMN5 | SOAPdenov2 | |
| ILMN6 | SPAdes | |
| ILMN7 | Velvet | |
| META | Metassembler | |
| ILMN8 | DISCOVAR | LIB3 (Illumina-only) |
| PACB1 | Celera Assembler | LIB4 (PacBio-only) |
| PACB2 | Canu | |
| PACB3 | FALCON | |
| HYBR1 | SPAdes | LIB1, LIB2, LIB5 (Hybrid) |
| HYBR2 | DBG2OLCa | LIB1, LIB5 (Hybrid) |
PE, paired-end; MP, mate pair; PacBio, Pacific Biosciences; CLR, continuous long-read.
SparseAssembler (Ye et al. 2012) was used to assemble LIB1 into contigs, which in turn were then used as input for DBG2OLC.
Case study I (repeat-content issue):
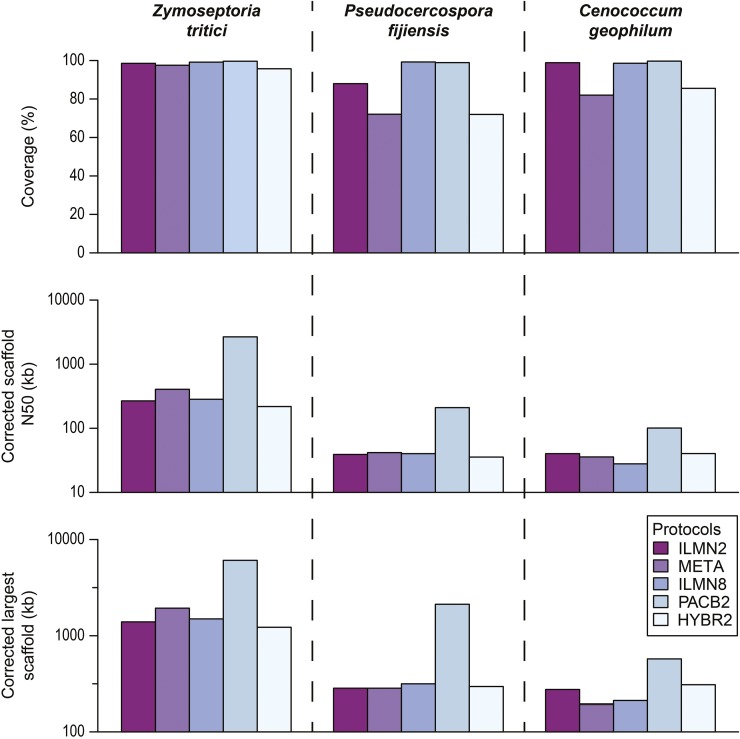
We first compared the sequencing of two fungi, Zymoseptoria tritici (synonym: Mycosphaerella graminicola) (Goodwin et al. 2011) and Pseudocercospora fijiensis (synonym: M. fijiensis) (Ohm et al. 2012), which both belong to the class Dothideomycetes yet have dramatically different repeat contents; the estimated repeat contents are ∼15 and ∼50% for the two genomes, respectively. Our simulations showed that, while good quality assemblies can be obtained for Z. tritici using either data type, the PacBio-only assembly for Ps. fijiensis vastly outperforms assemblies based on Illumina data alone or in combination with low-coverage PacBio data (Figure 2). The results are consistent with the notion that PacBio long reads are particularly powerful in resolving repeats (Koren et al. 2013). We then further tested if these results are informative for guiding the sequencing of another highly repetitive Dothideomycetes genome, Cenococcum geophilum, which has a repeat content of ∼76% (http://genome.jgi.doe.gov/Cenge3). For C. geophilum, the PacBio-only assembly was again found to be the best, while the hybrid assembly using DBG2OLC and the Illumina-only assembly using ALLPATHS-LG were next in rank (Figure 2 and Table S2), nicely recapitulating the results of Ps. fijiensis. We also performed meta-assembly of Illumina-only assemblies ILMN1 to ILMN7 (Table 1) on all three genomes using Metassembler. While the meta-assembly approach substantially improved the assembly continuity for Z. tritici, no improvement was observed for Ps. fijiensis and C. geophilum (Figure 2 and Table S2). These results suggest that the use of iWGS would provide critical information to help end users choose a successful sequencing of highly repetitive genomes that share similar characteristics. Importantly, since simulated assemblies are recoverable, the likely impact of the different assembly strategies on genes, gene families, or pathways of interest could also be examined in detail.
Figure 2.
Performance comparison of five representative experimental designs on three Dothideomycetes genomes. The five designs shown include three Illumina-only designs (ILMN2: ALLPATHS-LG, META: Metassembler, and ILMN8: DISCOVAR), the best performing PacBio-only design (PACB2: Canu), and the best performing hybrid design (HYBR2: DBG2OLC) for each genome. The statistics on the assembled fraction of the reference genome, scaffold N50, and largest scaffold size are all after correction for assembly errors using the reference genome as reported by QUAST in GAGE mode. By default, QUAST (in GAGE mode) corrects contigs/scaffolds by breaking them at assembly errors larger than 5 bp. Scaffold N50 and largest scaffold size are shown in log10 scale.
Case study II (GC-content and mtDNA assembly issue):
We next examined the de novo assembly of mitochondrial genomes from whole genome sequencing data of Saccharomyces cerevisiae (Mewes et al. 1997; Foury et al. 1998). Yeast mitochondrial genomes are valuable resources for evolutionary and functional studies (Freel et al. 2015), yet the acquisition of finished mitochondrial genome assemblies is not trivial because of their very low GC-content (∼17%). We simulated a genome sequencing experiment using the nuclear and mitochondrial genomes of S. cerevisiae. We tested two ratios of nuclear to mitochondrial genome copy numbers representing low (1:50) and high (1:200) mitochondrial contents, respectively (Solieri 2010). iWGS analysis showed that the S. cerevisiae mitochondrial genome was fully recovered at both low and high mitochondrial contents using Illumina data (Table 2). Consistent with recent observations made during the assembly of the S. eubayanus genome, only certain assemblers performed well; for example, ALLPATHS-LG performed surprisingly poorly, while SPAdes performed quite well (Baker et al. 2015). Importantly, the complete mitochondrial genome can be obtained as a single contig using only Illumina or only PacBio data, or using both data types (Table 2). Similarly, both Illumina and PacBio data resulted in good quality assemblies of the nuclear genome (Table S2). At the same time, different assemblers exhibited widely different performances even with the same input data (Table 2).
Table 2. Performance of all experimental designs evaluated in case study II.
| Nuclear:Mitochondrial Genome Ratio | Performance of Strategiesa | |||
|---|---|---|---|---|
| Complete, Single Contig Assembly of the Mitochondrial Genome | Assembled Fraction of Mitochondrial Genome ≥ 99% | 20% ≤ Assembled Fraction of Mitochondrial Genome < 99% | Assembled Fraction of Mitochondrial Genome < 20% | |
| 1:50 (low mitochondrial content) | ILMN1, ILMN6, ILMN8, PACB2, HYBR1, HYBR2 | ILMN7 | ILMN2, ILMN4, ILMN5, PACB1, PACB3 | ILMN3 |
| 1:200 (high mitochondrial content) | PACB2, HYBR1, HYBR2 | ILMN6, ILMN7 | ILMN1, ILMN8, PACB1, PACB3 | ILMN2, ILMN3, ILMN4, ILMN5 |
The de novo assembly generated by each strategy was compared against the reference mitochondrial genome of S. cerevisiae using both QUAST and BLASTN. Unless a single contig was found to represent the complete mitochondrial genome, the assembled fraction of mitochondrial genome was determined based on the number of “missing reference bases” reported by QUAST, and further confirmed by the BLASTN result.
Case study III (genomic architecture issue):
Lastly, we applied iWGS to three model eukaryotic genomes from different kingdoms and with different genomic architectures. Specifically, we analyzed Drosophila melanogaster (Adams et al. 2000) and Arabidopsis thaliana (Arabidopsis Genome Initiative 2000), which are medium-sized animal and plant genomes, respectively, as well as Plasmodium falciparum 3D7 (Gardner et al. 2002), a smaller protist genome with extremely low GC-content (∼19%). For all three genomes, the best assembly was generated by using only PacBio data (Table 3). In D. melanogaster and A. thaliana, several Illumina-only assemblies were of relatively high-quality (i.e., corrected scaffold N50 ≥ 100 kb; Table S2), among which the best two were generated by ALLPATHS-LG and DISCOVAR (Table 3). However, all Illumina-only assemblies of Pl. falciparum 3D7 had considerably lower corrected scaffold N50 values, except for DISCOVAR whose sequencing strategy is unique in requiring a PE library with a limited insert size.
Table 3. Summary of top-ranking assemblies generated in case study III.
| Organism (Genome Size) | Best Assembly from Each Sequencing Strategy | Assembly Statisticsa | ||
|---|---|---|---|---|
| Scaffold N50 (kb) | Largest Scaffold (kb) | Assembled Fraction of the Reference Genome | ||
| D. melanogaster (137.55 Mb) | ILMN2 | 169.7 | 1,007.9 | 89.1% |
| ILMN8 | 155.0 | 1,007.7 | 91.8% | |
| PACB2 | 5107.5 | 13,108.3 | 99.3% | |
| HYBR2 | 279.3 | 1,536.8 | 89.7% | |
| A. thaliana (119.15 Mb) | ILMN2 | 307.0 | 1,789.4 | 97.3% |
| ILMN8 | 266.6 | 2,533.4 | 98.5% | |
| PACB2 | 2065.7 | 8,552.9 | 99.7% | |
| HYBR2 | 289.3 | 1,412.2 | 97.4% | |
| Pl. falciparum 3D7 (23.29 Mb) | ILMN2 | 28.0 | 146.2 | 96.7% |
| ILMN8 | 222.0 | 729.9 | 98.4% | |
| PACB2 | 282.9 | 1,378.8 | 99.6% | |
| HYBR2 | 15.5 | 91.5 | 97.4% | |
| Pl. falciparum IT (real data) | ILMN2 | 167.1 | 641.7 | >100% |
| ILMN8 | 141.2 | 631.4 | 97.6% | |
| PACB3 | 1574.7 | 3,355.3 | 97.7% | |
| HYBR2 | 198.4 | 602.1 | 92.2% | |
The statistics for simulation-based analysis of D. melanogaster, A. thaliana, and Pl. falciparum 3D7 are after correction for assembly errors using the reference genome, as reported by QUAST in GAGE mode. By default, QUAST (in GAGE mode) corrects contigs/scaffolds by breaking them at assembly errors larger than 5 bp. The statistics for real data based analysis of Pl. falciparum IT are calculated from the original de novo assemblies.
To examine how well the simulation-based predictions made by iWGS are supported by empirical data, we collected four real genome sequencing data sets from a previous study of Pl. falciparum IT [one overlapping 100 bp PE library, one overlapping 250 bp PE library, one MP library, and one PacBio library from (Otto et al. 2014); Table S1] that were a good match to our simulated data sets, and ran the same set of assembly protocols. The best assembly was again generated by PacBio data alone, and the assemblies generated by ALLPATHS-LG, DISCOVAR (both are Illumina-only), and DBG2OLC (hybrid) were ranked next, while all other Illumina-only assembly protocols performed poorly (Table 3 and Table S2). The results are largely consistent with our simulation study, suggesting that our simulation-based approach is indeed informative.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Discussion
The design and analysis of de novo genome sequencing experiments is not trivial. On the design front, one has to balance between the complexity of the target genome, the strengths and weaknesses of each sequencing technology, and, importantly, the cost. Analysis is also challenging, as one is faced with multiple different algorithms and dozens of parameters. Although substantial efforts have been made to benchmark different approaches for genome assembly (Earl et al. 2011; Salzberg et al. 2012; Bradnam et al. 2013; Magoc et al. 2013), much less attention has been paid to investigating start-to-finish optimal sequencing strategies for a given genome [see (Chakraborty et al. 2016) for one example].
iWGS is an automated tool that allows users to explicitly compare alternative experimental designs by using simulated sequencing data, even allowing users to estimate costs when these are known for the generation of each data type. We have illustrated the utility of iWGS in several case studies on mitochondrial and nuclear genomes with varying levels of complexity. For instance, our simulations suggest that Illumina-only sequencing strategies may be economical choices for the sequencing of relatively simple genomes (e.g., Z. tritici; Table S2), whereas PacBio data would be highly desirable for genomes of greater complexity (e.g., Ps. fijiensis, C. geophilum, and Pl. falciparum). Although not done here, iWGS could also be used to evaluate different combinations of sequencing assays (e.g., PE and MP libraries), read quality, read lengths, and sequencing depths. Empirical studies of both short- and long-read data have shown that these parameters are critical determinants of the quality of de novo genome assemblies (Utturkar et al. 2014; Chakraborty et al. 2016).
One key function of iWGS is the use of simulation data generated from a related reference genome to inform the experimental design for organisms lacking genomic data. A similar concept was previously used to evaluate sequencing strategies for cacao by using the rice genome as the reference (Haiminen et al. 2011). In principle, one could apply iWGS on one or more related reference genomes that resemble the characteristics (e.g., genome size, repeat content, and sequence composition) of the sequencing target. However, if such reference genome is lacking, one solution is to start with a closely related reference genome and tune it toward the target (e.g., adjust GC- and repeat contents) by using third-party tools that simulate genome-wide evolution (Arenas and Posada 2014) before running iWGS. Alternatively, one may simply use iWGS with reference genomes that are of comparable complexity (e.g., similar in size and repeat content) regardless of the evolutionary relatedness. As suggested by previous studies, these factors not only influence the difficulty of genome assembly, but can also be excellent predictors of the assembly quality (Lee et al. 2014). Therefore, iWGS could also be informative in evaluating the performance of alternative experimental designs on genomes with similar characteristics to the sequencing target.
Other important features of iWGS include the support for both Illumina short, and PacBio long, sequence reads and, correspondingly, a wide selection of software tools compatible with these data types, as well as the ability to analyze real data. In comparison, the support for third generation sequencing data are relatively limited in iMetAMOS and currently lacking in RAMPART. Given the increasing importance of long sequence reads in de novo genome assembly, iWGS aims to allow users to fully exploit the strength of long-read data and explore alternative ways of data analysis. Along these lines, several further developments can be envisioned. First, support for additional sequencing technologies, such as Oxford Nanopore, can be added as technologies become commercially available. In fact, the Celera Assembler, Canu, and SPAdes assemblers, which are supported by iWGS, can already utilize nanopore reads (Bankevich et al. 2012; Berlin et al. 2015). Similarly, realistic simulation of nanopore data will be possible once the patterns of errors and biases are better characterized using real data. Second, iWGS will continue to expand its functionality to achieve better assemblies. For instance, a number of assembly polishing tools can be integrated in iWGS to improve the quality of the final output, including Pilon (Walker et al. 2014), Quiver (Chin et al. 2013), and Nanopolish (Loman et al. 2015), which use Illumina, PacBio, and nanopore data, respectively. In addition, iWGS currently uses Metassembler for meta-assembly; in the future, other meta-assembly tools that support assemblies based on PacBio data alone, such as quickmerge (Chakraborty et al. 2016), could be added. Lastly, it would be beneficial to enable users to add new software tools to iWGS in order to stay up-to-date with the rapid advances in genome assembly and other aspects of HTS data analysis. We intend to provide periodic updates, and the expert user can edit iWGS on their own. In summary, iWGS is a flexible, expandable, and easy to use pipeline that will aid in the design and execution of genome assembly experiments across the tree of life.
Supplementary Material
Acknowledgments
We thank Branden Timm for technical support on the WEI cluster. We thank Francis Martin for providing access to unpublished genome data for the C. geophilum genome produced by the U.S. Department of Energy Joint Genome Institute, a Department of Energy (DOE) Office of Science User Facility, supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University and the Wisconsin Energy Institute (WEI) cluster at the University of Wisconsin–Madison. This work was funded by the National Science Foundation (DEB-1442113 to A.R.; DEB-1253634 and DEB-1442148 to C.T.H.), in part by the DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494 to C.T.H.), the U.S. Department of Agriculture (USDA) National Institute of Food and Agriculture (Hatch Project 1003258 to C.T.H.), and the National Institutes of Health (NIAID AI105619 to A.R.). C.T.H. is an Alfred Toepfer Faculty Fellow, supported by the Alexander von Humboldt Foundation. C.T.H. is a Pew Scholar in the Biomedical Sciences, supported by the Pew Charitable Trusts. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. USDA is an equal opportunity provider and employer.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.034249/-/DC1.
Communicating editor: K. Thornton
Literature Cited
- Abbas M. M., Malluhi Q. M., Balakrishnan P., 2014. Assessment of de novo assemblers for draft genomes: a case study with fungal genomes. BMC Genomics 15(Suppl. 9): S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., et al. , 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative , 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Arenas M., Posada D., 2014. Simulation of genome-wide evolution under heterogeneous substitution models and complex multispecies coalescent histories. Mol. Biol. Evol. 31: 1295–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker E., Wang B., Bellora N., Peris D., Hulfachor A. B., et al. , 2015. The genome sequence of Saccharomyces eubayanus and the domestication of Lager-Brewing yeasts. Mol. Biol. Evol. 32: 2818–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., et al. , 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin K., Koren S., Chin C. S., Drake J. P., Landolin J. M., et al. , 2015. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 33: 623–630. [DOI] [PubMed] [Google Scholar]
- Bradnam K. R., Fass J. N., Alexandrov A., Baranay P., Bechner M., et al. , 2013. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience 2: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M., Baldwin-Brown J. G., Long A. D., Emerson J. J., 2016. Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nucl. Acids Res. DOI: 10.1093/nar/gkw654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J. A., Ho I., Sunkara S., Luo S., Schroth G. P., et al. , 2011. Meraculous: de novo genome assembly with short paired-end reads. PLoS One 6: e23501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhi R., Medvedev P., 2014. Informed and automated k-mer size selection for genome assembly. Bioinformatics 30: 31–37. [DOI] [PubMed] [Google Scholar]
- Chin C. S., Alexander D. H., Marks P., Klammer A. A., Drake J., et al. , 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10: 563–569. [DOI] [PubMed] [Google Scholar]
- Chin, C.-S., P. Peluso, F. J. Sedlazeck, M. Nattestad, G. T. Concepcion et al., 2016 Phased diploid genome assembly with single molecule real-time sequencing. bioRxiv DOI: http://dx.doi.org/10.1101/056887. [DOI] [PMC free article] [PubMed]
- Denton J. F., Lugo-Martinez J., Tucker A. E., Schrider D. R., Warren W. C., et al. , 2014. Extensive error in the number of genes inferred from draft genome assemblies. PLOS Comput. Biol. 10: e1003998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl D., Bradnam K., St John J., Darling A., Lin D., et al. , 2011. Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome Res. 21: 2224–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F., Roganti T., Lecrenier N., Purnelle B., 1998. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 440: 325–331. [DOI] [PubMed] [Google Scholar]
- Freel K. C., Friedrich A., Schacherer J., 2015. Mitochondrial genome evolution in yeasts: an all-encompassing view. FEMS Yeast Res. 15: fov023. [DOI] [PubMed] [Google Scholar]
- Gardner M. J., Hall N., Fung E., White O., Berriman M., et al. , 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419: 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genome 10K Community of Scientists 2009. Genome 10K: a proposal to obtain whole-genome sequence for 10,000 vertebrate species. J Hered. 100: 659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn T. C., 2011. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 11: 759–769. [DOI] [PubMed] [Google Scholar]
- Gnerre S., Maccallum I., Przybylski D., Ribeiro F. J., Burton J. N., et al. , 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. USA 108: 1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S. B., M’Barek S. B., Dhillon B., Wittenberg A. H., Crane C. F., et al. , 2011. Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 7: e1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I. V., Cullen D., Goodwin S. B., Hibbett D., Jeffries T. W., et al. , 2011. Fueling the future with fungal genomics. Mycology 2: 192–209. [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G., 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29: 1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiminen N., Kuhn D. N., Parida L., Rigoutsos I., 2011. Evaluation of methods for de novo genome assembly from high-throughput sequencing reads reveals dependencies that affect the quality of the results. PLoS One 6: e24182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger C. T., Rokas A., Bai F. Y., Boekhout T., Goncalves P., et al. , 2015. Genomics and the making of yeast biodiversity. Curr. Opin. Genet. Dev. 35: 100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Yuan J., Shi Y., Lu J., Liu B., et al. , 2012. pIRS: profile-based Illumina pair-end reads simulator. Bioinformatics 28: 1533–1535. [DOI] [PubMed] [Google Scholar]
- Huang W., Li L., Myers J. R., Marth G. T., 2012. ART: a next-generation sequencing read simulator. Bioinformatics 28: 593–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt M., Kikuchi T., Sanders M., Newbold C., Berriman M., et al. , 2013. REAPR: a universal tool for genome assembly evaluation. Genome Biol. 14: R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani R., Toshimoto K., Noguchi H., Toyoda A., Ogura Y., et al. , 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24: 1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Phillippy A. M., 2015. One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Curr. Opin. Microbiol. 23: 110–120. [DOI] [PubMed] [Google Scholar]
- Koren S., Harhay G. P., Smith T. P., Bono J. L., Harhay D. M., et al. , 2013. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 14: R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Treangen T. J., Hill C. M., Pop M., Phillippy A. M., 2014. Automated ensemble assembly and validation of microbial genomes. BMC Bioinformatics 15: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren, S., B. P. Walenz, K. Berlin, J. R. Miller, and A. M. Phillippy, 2016 Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. bioRxiv DOI: http://dx.doi.org/10.1101/071282. [DOI] [PMC free article] [PubMed]
- Kumar S., Subramanian S., 2002. Mutation rates in mammalian genomes. Proc. Natl. Acad. Sci. USA 99: 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H., J. Gurtowski, S. Yoo, S. Marcus, W. R. McCombie et al., 2014 Error correction and assembly complexity of single molecule sequencing reads. bioRxiv DOI: http://dx.doi.org/10.1101/006395.
- Loman N. J., Quick J., Simpson J. T., 2015. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods 12: 733–735. [DOI] [PubMed] [Google Scholar]
- Luo R., Liu B., Xie Y., Li Z., Huang W., et al. , 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T., Pabinger S., Canzar S., Liu X., Su Q., et al. , 2013. GAGE-B: an evaluation of genome assemblers for bacterial organisms. Bioinformatics 29: 1718–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapleson D., Drou N., Swarbreck D., 2015. RAMPART: a workflow management system for de novo genome assembly. Bioinformatics 31: 1824–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain S. J., Peris D., Sardi M., Moskvin O. V., Zhan F., et al. , 2016. Genome sequence and analysis of a stress-tolerant, wild-derived strain of Saccharomyces cerevisiae used in biofuels research. G3 (Bethesda) 6: 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewes H. W., Albermann K., Bahr M., Frishman D., Gleissner A., et al. , 1997. Overview of the yeast genome. Nature 387: 7–65. [DOI] [PubMed] [Google Scholar]
- Myers E. W., Sutton G. G., Delcher A. L., Dew I. M., Fasulo D. P., et al. , 2000. A whole-genome assembly of Drosophila. Science 287: 2196–2204. [DOI] [PubMed] [Google Scholar]
- Nagarajan N., Pop M., 2013. Sequence assembly demystified. Nat. Rev. Genet. 14: 157–167. [DOI] [PubMed] [Google Scholar]
- Ohm R. A., Feau N., Henrissat B., Schoch C. L., Horwitz B. A., et al. , 2012. Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog. 8: e1003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y., Asai K., Hamada M., 2013. PBSIM: PacBio reads simulator–toward accurate genome assembly. Bioinformatics 29: 119–121. [DOI] [PubMed] [Google Scholar]
- Otto T. D., Rayner J. C., Bohme U., Pain A., Spottiswoode N., et al. , 2014. Genome sequencing of chimpanzee malaria parasites reveals possible pathways of adaptation to human hosts. Nat. Commun. 5: 4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy T. B., Thomas A. D., Stamatis D., Bertsch J., Isbandi M., et al. , 2015. The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta)genome project classification. Nucleic Acids Res. 43: D1099–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro F. J., Przybylski D., Yin S., Sharpe T., Gnerre S., et al. , 2012. Finished bacterial genomes from shotgun sequence data. Genome Res. 22: 2270–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G. E., Hackett K. J., Purcell-Miramontes M., Brown S. J., Evans J. D., et al. , 2011. Creating a buzz about insect genomes. Science 331: 1386. [DOI] [PubMed] [Google Scholar]
- Rokas A., Abbot P., 2009. Harnessing genomics for evolutionary insights. Trends Ecol. Evol. 24: 192–200. [DOI] [PubMed] [Google Scholar]
- Rokas A., Williams B. L., King N., Carroll S. B., 2003. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425: 798–804. [DOI] [PubMed] [Google Scholar]
- Ross M. G., Russ C., Costello M., Hollinger A., Lennon N. J., et al. , 2013. Characterizing and measuring bias in sequence data. Genome Biol. 14: R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux J., Privman E., Moretti S., Daub J. T., Robinson-Rechavi M., et al. , 2014. Patterns of positive selection in seven ant genomes. Mol. Biol. Evol. 31: 1661–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salichos L., Rokas A., 2013. Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497: 327–331. [DOI] [PubMed] [Google Scholar]
- Salikhov K., Sacomoto G., Kucherov G., 2013. Using cascading Bloom filters to improve the memory usage for de Brujin graphs, pp. 364–376 in Algorithms in Bioinformatics, edited by Darling A., Stoye J. Springer, Berlin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg S. L., Phillippy A. M., Zimin A., Puiu D., Magoc T., et al. , 2012. GAGE: a critical evaluation of genome assemblies and assembly algorithms. Genome Res. 22: 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J. T., Durbin R., 2012. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 22: 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J. T., Pop M., 2015. The theory and practice of genome sequence assembly. Annu. Rev. Genomics Hum. Genet. 16: 153–172. [DOI] [PubMed] [Google Scholar]
- Simpson J. T., Wong K., Jackman S. D., Schein J. E., Jones S. J., et al. , 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19: 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solieri L., 2010. Mitochondrial inheritance in budding yeasts: towards an integrated understanding. Trends Microbiol. 18: 521–530. [DOI] [PubMed] [Google Scholar]
- Utturkar S. M., Klingeman D. M., Land M. L., Schadt C. W., Doktycz M. J., et al. , 2014. Evaluation and validation of de novo and hybrid assembly techniques to derive high-quality genome sequences. Bioinformatics 30: 2709–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., et al. , 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenfeld N. I., Yin S., Sharpe T., Lau B., Hegarty R., et al. , 2014. Comprehensive variation discovery in single human genomes. Nat. Genet. 46: 1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wences A. H., Schatz M. C., 2015. Metassembler: merging and optimizing de novo genome assemblies. Genome Biol. 16: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C., Ma Z. S., Cannon C. H., Pop M., Yu D. W., 2012. Exploiting sparseness in de novo genome assembly. BMC Bioinformatics 13(Suppl. 6): S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C., Hill C., Wu S., Ruan J., Z. S Ma. 2016. DBG2OLC: efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci. Rep. 6: 31900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D. R., Birney E., 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Li C., Li Q., Li B., Larkin D. M., et al. , 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346: 1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Rokas A., 2014. Prevention, diagnosis and treatment of high-throughput sequencing data pathologies. Mol. Ecol. 23: 1679–1700. [DOI] [PubMed] [Google Scholar]
- Zimin A. V., Marcais G., Puiu D., Roberts M., Salzberg S. L., et al. , 2013. The MaSuRCA genome assembler. Bioinformatics 29: 2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.