Abstract
Drosophila stocks bearing compound chromosomes, single molecules of DNA that carry the genomic complement of two chromosomes, are useful tools for studying meiosis and mitosis. However, these stocks cannot easily be crossed to stocks with regular chromosomes, due to the lethality of the resulting whole-chromosome aneuploidy. This prevents the examination of interesting genetic variants in a compound chromosome background. Methods to circumvent this difficulty have included the use of triploid females or nondisjunction (caused by either cold-induced microtubule depolymerization or meiotic mutants). Here, we present a new approach for crossing compound chromosomes that takes advantage of the nonhomologous segregations that result when multiple chromosomes in the same genome are prevented from meiotic crossing over by heterozygosity for balancer chromosomes. This approach gives higher yields of the desired progeny in fewer generations of crossing. Using this technique, we have created and validated stocks carrying both a compound-X and compound-2, as well as compound-2 stocks carrying the meiotic mutant nod.
Keywords: aneuploidy, C(2)EN, long chromosome, meiosis, segregation
Compound chromosomes, rearrangements in which two chromosomes share a common centromere, have been used in Drosophila to perturb both meiosis and mitosis in efforts to further understand both processes. While the most commonly used compound chromosomes, the compound Xs, have been studied for almost a century since their discovery by Lilian Morgan (Morgan 1922), compound chromosomes involving many different combinations of sex chromosomes and autosomes have been constructed and used in genetic studies (Novitski and Childress 1976; Holm 1976; Novitski et al. 1981). Compound chromosomes have been used in the identification and characterization of mutants defective in meiosis (Page et al. 2007), to examine the effect of extralong chromosome arms in the mitotic cleavage plane (Kotadia et al. 2012), and to examine the effect of ectopic heterochromatin blocks on cohesin distribution (Oliveira et al. 2014). Their ability to force a genome to break the normal rules for segregating homologs has also provided useful insights on the mechanisms underlying chromosome segregation (Gilliland et al. 2015). While it is relatively easy to introduce mutations and chromosome aberrations into strains with compound fourth chromosomes (due to the viability and fertility of flies trisomic for that small autosome), introducing genetic variants into strains with compound second or compound third chromosomes is more difficult. A recent paper (Martins et al. 2013) presented two approaches (cold-shock- and BubR1-induced nondisjunction) for introducing genetic variants into strains with C(2)EN, a compound chromosome with two entire second chromosomes sharing a single centromere (Novitski et al. 1981). While C(2)EN males or females give large numbers of viable progeny when mated with flies that also carry C(2)EN, they give very few viable progeny when crossed to wild-type flies. As shown in Figure 1, crosses of C(2)EN males to wild-type females produce mainly progeny that are either monosomic or trisomic for the second chromosome and that fail to survive to adulthood. The few surviving progeny from such crosses are the results of either chromosome loss or nondisjunction in the mother (diploid if only the second chromosomes nondisjoin, and triploid if all of the chromosomes fail to disjoin properly). It is through these rare survivors that genetic variants can be introduced into C(2)EN strains. A recent study used two different approaches to increase nondisjunction and/or chromosome loss in flies with wild-type second chromosomes (Martins et al. 2013). The first approach was to expose 300 virgin females to a prolonged cold-shock (10° for 7 d) before mating to C(2)EN males. While they were successful in introducing an X-linked mutation into the C(2)EN strain, they provided no data on the frequency of success. The second approach, which they describe as more successful, used males transheterozygous for two different alleles of the meiotic mutant bubR1 to generate high rates of nondisjunction during meiosis. The main drawback to this approach is the number of generations required to introduce the bubR1 alleles into strains with the genetic variants that one wants to introduce into the C(2)EN strain.
Figure 1.
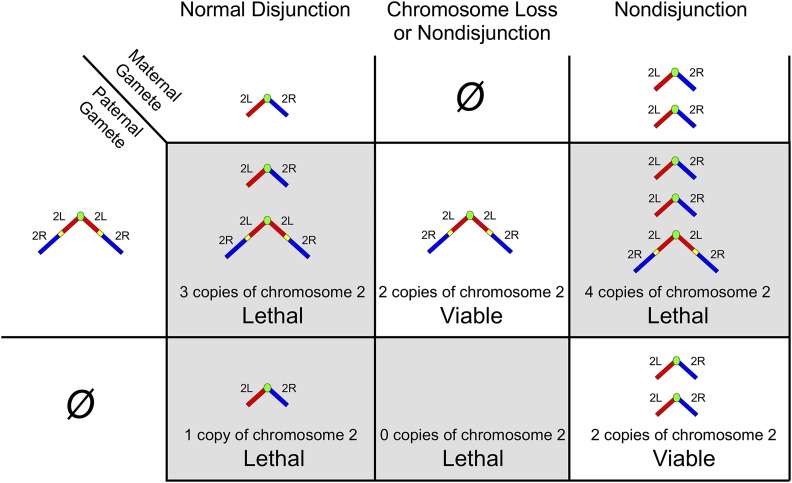
Progeny from crosses to C(2)EN. As C(2)EN males contribute gametes that carry the gene dosage of either two or zero copies of chromosome 2, only progeny that also inherit zero or two copies from the other parent will result in viable euploid progeny. Note that sex determination in Drosophila uses the ratio of X to autosomal chromosomes, and that the Y chromosome is not masculinizing as in mammals (Ashburner et al. 2005). This means that flies with one dose of X chromosome genes develop as males and flies with two doses of X chromosome genes (either free or attached) develop as females, while having three doses (including C(1)EN/X) is lethal.
We have developed a simpler approach to inducing nondisjunction that requires only one or two generations to introduce genetic variants into the C(2)EN strain. We generate females in which two different chromosome pairs fail to undergo meiotic exchange, allowing nonhomologous segregations to generate oocytes with either two second chromosomes, or no second chromosome. Since we wanted one of the chromosome pairs that fail to undergo meiotic exchange to be the second chromosome, we used females heterozygous for the multiply-inverted CyO balancer chromosome, which carries three inversions that are very effective in reducing meiotic exchange. The obvious choice for the other chromosome pair was the sex chromosomes, since half of the products of sex chromosome nondisjunction generate viable aneuploid progeny. All of our initial crosses were to males with wild-type sex chromosomes and C(2)EN, bw sp. Using this approach, we were able to generate a stock carrying two compound chromosomes (C(1)RM and C(2)EN), as well as both isosequential and balancer X chromosomes carrying the meiotic mutant nod. We were also able to use our method to introduce third and fourth chromosome markers into C(2)EN strains and to introduce X chromosome markers into C(3)EN strains. Our approach should simplify the use of compound chromosomes in Drosophila.
Materials and Methods
Starting stocks
Chromosomes used in this study were derived from the following stocks from the Bloomington Drosophila Stock Center: C(2)EN, bw sp/Ø (BDSC #1020), Ø/C(1)RM, y1 pn1/ C(1;Y)2, y1 P{lacW}elav5-45fD w* P{lacW}ogre5-45fP P{lacW}3-52d P{lacW}3-76a: y+ (BDSC #3710), C(1;Y)1, y1 v1 f1 B1: y+/C(1)RM, y2 su(wa)1 wa (BDSC #700), FM7a, nod2/ Dp(1;Y)y+/C(1)DX, y1 f1; svspa-pol (BDSC #2331), y1 w1 noda/C(1)DX, y1 f1/Dp(1;Y)y+; svspa-pol (BDSC #34510), Dp(1;Y)BSYy+/+; C(2)EN, bw sp/Ø (BDSC#1111), C(3)EN, th1 st1/Ø (BDSC #1114), C(3)EN, st1 cu1 es/Ø (BDSC #1117), and FM7a (BDSC #785). We additionally used the following stocks, which were created and maintained in the Kennison lab stock collection: FM7a; CyO/Sp, y1 w*; CyO/Sp, Ø/C(1;Y)1, y1, y1 w*; TM6C, cu1 Sb1 ca1/TM3, Ser1, and yd2 w1118 P{ry+t7.2 = ey-FLP.N}2; P{w+mC:PRE[Scr7-8]}Q1; Dp(1;4)193, y+ svspa-pol.
Oocyte chromosome preparations
Newly eclosed females were aged for either 2 d with males or 5 d as virgins, to enrich for oocytes in meiotic prometaphase I and metaphase I, respectively (Gilliland et al. 2009). Females were dissected in Robb’s buffer and fixed in a 1:1 mix of 16% formaldehyde and 2 × WHOoPaSS buffer (Gillies et al. 2013), then FISH was performed using 92° melting and 32° annealing temperatures as previously described. The chromosome-specific FISH probes used were as follows: X = TTTTCCAAATTTCGGTCATCAAATAATCAT (Ferree and Barbash 2009); 2L = (AATAG)6, and 2L-3L = (AATAACATAG)3 (Dernburg 2000). All probes are written 5′ to 3′ and were synthesized with fluorescent labels by IDTDNAcom and oocytes were mounted in Slowfade Gold (Molecular Probes).
Mitotic chromosome preparations
Mitotic chromosomes were prepared from third instar larvae using standard brain squash protocols (Sullivan et al. 2000), except that chromosomes were stained with SlowFade Gold antifade reagent plus DAPI (Molecular Probes).
Microscopy
All images were acquired on a Leica SPE II confocal microscope using LAS AF software (Leica) using the 63 × objective and zoomed to 1.7 × the Nyquist limit, followed by deconvolution in Huygens Essential (www.svi.nl) with an estimated PSF and default parameters (except for mounting media refractive index, which was 1.42 per manufacturer’s instructions).
Data availability
Strains are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Our first application of this method was to generate a stock carrying both compound-X and compound-2 chromosomes. This configuration is known to undergo heterologous C(1) ⇔ C(2) segregation of the two compounds, and had previously been examined during female meiotic prometaphase I (Dernburg et al. 1996). A recent study from our lab attempted to recreate this stock to study metaphase I arrest, but several attempts using both spontaneous nondisjunction and cold-shock-induced nondisjunction were unsuccessful (W. D. Gilliland and E. M. Colwell, unpublished data). We first crossed the CyO balancer into two different C(1)RM stocks, C(1)RM, y pn and C(1)RM, y2 su(wa) wa. These two compound X chromosomes differ not only in the visible X-linked mutations that they carry, but probably also in the amount and type of centric heterochromatin that each carries. C(1)RM, y2 su(wa) wa originally carried a bb allele (Parker and Hammond 1958), although the bb phenotype is no longer expressed. We then crossed approximately equal numbers of C(1)RM/Y; CyO/+ females and C(2)EN, bw sp/Ø males, transferring the parents to new vials every 2–3 d for several wk. The numbers of females tested and the number of days that they were allowed to lay eggs can be used as an estimate of the success rates.
As shown in Table 1, we recovered eight females with C(1)RM, y pn and C(2)EN, bw sp. To our surprise, none of these females laid eggs when mated to C(2)EN, bw sp males; we did not further examine oogenesis in these females. In contrast, from 50 parental C(1)RM, y2 su(wa) wa/Y; CyO/+ females, we recovered 56 daughters carrying both the C(1)RM, y2 su(wa) wa and C(2)EN, bw sp chromosomes, which were then crossed to their C(2)EN, bw sp brothers to generate a stable strain in which the females carry both compound chromosomes. We note it should also be relatively easy to introduce different marked Y chromosomes (Y*) into the C(2)EN strain using C(1)RM, y2 su(wa) wa/Y*; CyO/+ females.
Table 1. Summary of crosses to C(2)EN.
| Genotype | Parental Females | Days Laying | Desired Progeny | Other Progeny | Desired Progeny/Female | |
|---|---|---|---|---|---|---|
| ♀ | ♀ | ♂ | ||||
| C(1)RM, y pn/Y ; CyO/+ ♀ X | 33 | 13 | 8 C(1)RM; C(2)EN | 82 C(1)RM; CyO/+ | 7 C(2)EN | 0.24 |
| +/Y ; C(2)EN, bw sp ♂ | 251 CyO/+ | |||||
| C(1)RM, y2 su(wa)wa/Y ; CyO/+ ♀ X | 50 | 26 | 56 C(1)RM; C(2)EN | 129 C(1)RM; CyO/+ | 54 C(2)EN | 1.12 |
| +/Y ; C(2)EN, bw sp ♂ | 750 CyO/+ | |||||
| FM7a/C(1;Y)1, y ; CyO/+ ♀ X | 38 | 13 | 54 FM7a/C(1;Y); C(2)EN | 22 +/FM7a (or C(1;Y)); C(2)EN | 21 FM7a (or C(1;Y)); C(2)EN | 1.42 |
| +/Y ; C(2)EN, bw sp ♂ | 60 +/FM7a (or C(1;Y)); CyO/+ | 162 CyO/+ | ||||
| 40 FM7a (or C(1;Y)); CyO/+ | ||||||
| FM7a/y w ; CyO/+ ♀ X | 39 | 12 | 65 FM7a/y w; C(2)EN | 11 +/FM7a (or y w); C(2)EN | 6 FM7a (or y w); C(2)EN | 1.67 |
| +/Y ; C(2)EN, bw sp ♂ | 33 +/FM7a (or y w); CyO/+ | 167 CyO/+ | ||||
| 25 FM7a (or y w); CyO/+ | ||||||
| FM7a/y w noda; CyO/+ ♀ X | 30 | 7 | 21 FM7a/y w noda; C(2)EN | 9 +/FM7a (or y w noda); C(2)EN | 6 FM7a (or y w noda); C(2)EN | 0.70 |
| +/Y ; C(2)EN, bw sp ♂ | 12 +/FM7a (or y w noda); CyO/+ | 55 CyO/+ | ||||
| 17 FM7a (or y w noda); CyO/+ | ||||||
| FM7a, nod2/y w; CyO/+ ♀ X | 20 | 7 | 8 FM7a, nod2/y w; C(2)EN | 4 +/FM7a, nod2 (or y w); C(2)EN | 2 FM7a, nod2 (or y w); C(2)EN | 0.40 |
| +/Y ; C(2)EN, bw sp ♂ | 10 +/FM7a, nod2 (or y w); CyO/+ | 57 CyO/+ | ||||
| 11 FM7a, nod2 (or y w); CyO/+ | ||||||
| FM7a/yd2 w1118 P{ry+t7.2 = ey-FLP.N}2; CyO/+; P{w+mC:PRE[Scr7-8]}Q1/+; Dp(1;4)193, y+ svspa-pol/+ ♀ X | 25 | 5 | 12 FM7a/y w; C(2)EN; Q1/+ | 9 FM7a/y w; C(2)EN | 2 FM7a; C(2)EN | 0.76 |
| y w/Y ; C(2)EN, bw sp ♂ | 3 FM7a /y w; C(2)EN;Dp(1;4)/+ | 1 y w/y w; C(2)EN | 23 y w; CyO/+ | |||
| 3 FM7a /y w; C(2)EN;Q1/+; Dp(1;4)/+ | 1 FM7a/y w; CyO/+; Q1/+ | 19 y w; CyO/+; Q1/+ | ||||
| 1 y w/y w; C(2)EN;Dp(1;4)/+ | 3 y w/y w; CyO/+; Dp(1;4)/+ | 20 y w; CyO/+; Dp(1;4)/+ | ||||
| 2 y w/y w; CyO/+; Q1/+ ; Dp(1;4)/+ | 24 y w; CyO/+; Q1/+; Dp(1;4)/+ | |||||
| 2 y w/y w; CyO/+ | 3 FM7a; CyO/+ | |||||
| 1 y w/y w; CyO/+; Q1/+ | 3 FM7a; CyO/+; Dp(1;4)/+ | |||||
For each cross, flies were mated in fresh vials and transferred to new vials for the number of days indicated. While no females from the first cross were fertile, the other six crosses were sufficiently fertile to establish balanced stocks. Among the progeny from the last cross, we cannot distinguish a maternally-inherited X chromosome (yd2 w1118 P{ry+t7.2 = ey-FLP.N}2) from a paternally-inherited X chromosome (y w), so both are indicated only as y w. Q1 is the third chromosome transposon insertion, P{w+mC:PRE[Scr7-8]}Q1. Dp(1;4) is Dp(1;4)193, y+ svspa-pol.
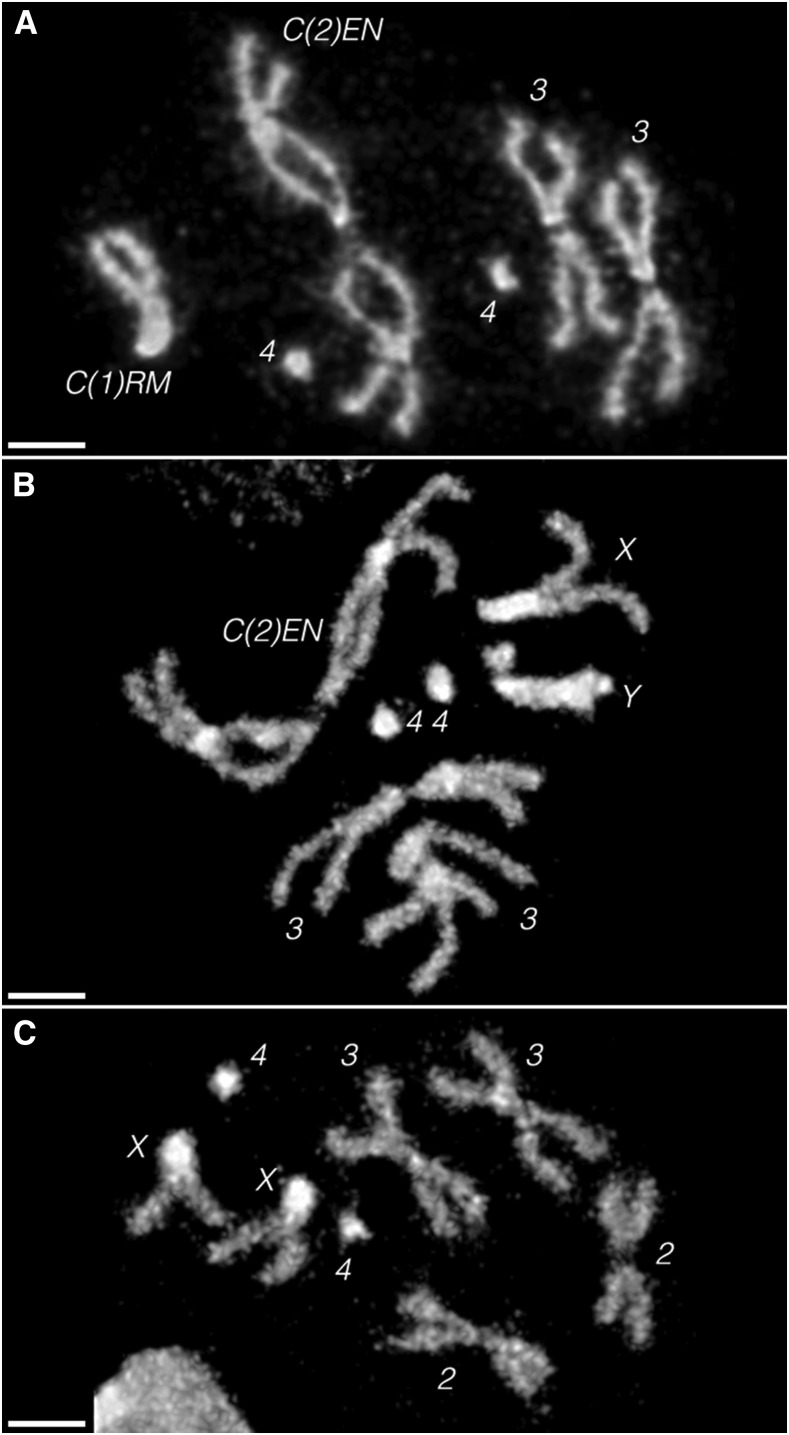
To validate this genotype, we performed brain squashes to visualize the mitotic chromosomes. As expected, female larvae clearly carried the C(1) and C(2) (Figure 2A), but no Y chromosomes were found in any of 10 female brains examined. This was a surprise, as the source stock for the C(2)EN is described as having normal sex chromosomes, and we expected males to segregate X ⇔ Y normally with the C(2) segregating at random, which should result in all surviving female progeny being C(1)RM/Y. We considered the possibility that the C(2)EN strain might actually contain an unmarked C(1;Y) chromosome, which could result in C(1;Y) ⇔ C(2)EN segregations in males that would explain the lack of a free Y in females. However, all brain squashes of male larvae from this stock revealed two normal-looking, independent sex chromosomes (Figure 2B). These results imply that males of this stock must be cosegregating the X and Y together, away from the C(2)EN, at high frequency. Therefore, males of this stock appear to get both X and Y chromosomes from the sperm and C(2)EN from the egg, while females of this stock appear to get C(2)EN from the sperm and C(1)EN from the egg. While we have not investigated the mechanism of this unusual male segregation pattern, similar patterns have been previously noted in this C(2)EN chromosome (see Discussion and Conclusion). To ensure that no C(1)RM, y2 su(wa) wa/Ø; C(2)EN, bw sp/Ø females were carrying unmarked Y chromosomes, we crossed several bottles of these females to males from a C(2)EN stock carrying a marked Y chromosome, Dp(1;Y)BSYy+/+; C(2)EN, bw sp/Ø. Over 90% of the male progeny (197/214) were Bar+, which were completely sterile when crossed to +/+; C(2)EN, bw sp females (0 progeny from 4 vials of > 20 males and females each), indicating that those B+ males were X/Ø and did not inherit an unmarked Y chromosome from the mother that could confer fertility. The remaining male progeny (17/214, about 8%) were BS, and must have received both the X and the Y chromosome from their fathers. Together, these data suggest that X ⇔ Y segregation occurs in over 90% of meioses in the Dp(1;Y)BSYy+/+; C(2)EN, bw sp/Ø males, whereas that segregation pattern must be quite rare in the +/Y; C(2)EN, bw sp/Ø males of our double-compound stock.
Figure 2.
Brain squashes of (A) a C(1)RM, y2 su(wa) wa/Ø; C(2)EN, bw sp/Ø female third instar larva, showing only six chromosomes, (B) a X/Y; C(2)EN, bw sp/Ø male larva, showing seven chromosomes and (C) an Oregon-R female larva, showing eight chromosomes. The constrictions in the middle of the arms of C(2)EN match previously published images of this chromosome in mitosis (Martins et al. 2013). These DAPI (4’,6-diamidino-2-phenylindole) images are representative of the karyosomes seen in at least 10 larvae of each sex. Note the lack of any free Y in the female, and that the sex chromosomes in the male are clearly not attached, indicating that males must undergo X/Y ⇔ C(2)EN segregation. All scale bars, 2 µm.
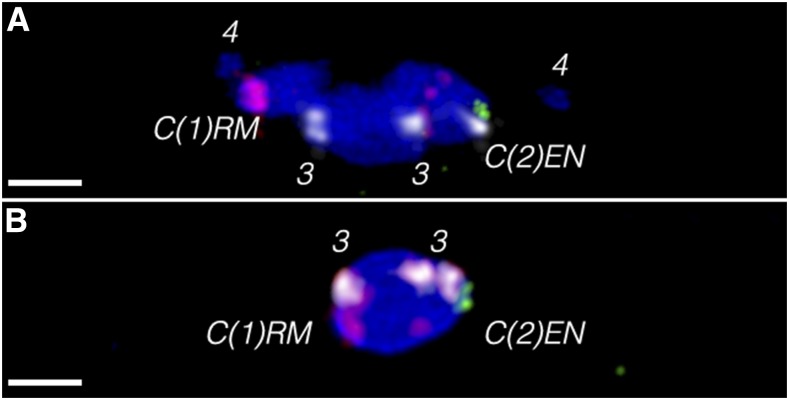
Having successfully generated C(1)/Ø; C(2)/Ø females, we then set out to assess chromosome coorientation at metaphase I arrest, which we recently examined for a number of other compound chromosome genotypes (Gilliland et al. 2015). Using FISH in aged virgin females, we found that 20/22 oocytes (91%) were in the heterologous C(1) ⇔ C(2) configuration (Figure 3). The two exceptions could not be unambiguously scored, but appeared to have maloriented both normal third chromosomes to the same pole, which would result in lethal aneuploidy in the progeny. These results are consistent with a cosegregation rate of > 99% based on analysis of surviving progeny (Dernburg et al. 1996), and confirms that (similar to other combinations of compound chromosomes) the heterologous segregation of these two compounds really occurs via coorientation and not by the death of nonheterologous progeny classes. We also note that Y chromosomes were not seen in these oocytes, consistent with the brain squash and genetic data above.
Figure 3.
FISH in C(1)RM, y2 su(wa) wa/Ø; C(2)EN, bw sp/Ø oocytes. (A) During prometaphase I, the two nonhomologous compounds can move to opposite sides of the prometaphase spindle, like the homologous nonexchange four chromosomes. This differs from FISH in meioses where paired free homologs are found out on either side of the spindle (Hughes et al. 2009). (B) At metaphase I arrest, the C(1) and C(2) chromosomes are cooriented with each other, indicating that they will undergo heterologous segregation to opposite poles once meiosis resumes. Probes are 2L3L (white), 2L (green), and X (red), with DAPI (blue). Note the C(2) is identified by a single spot of white probe adjacent to a single spot of green, and that the X probe also hybridizes to spots in the middle of the C(2) arm. Scale bars, 2 µm. DAPI, 4’,6-diamidino-2-phenylindole; FISH, fluorescent in situ hybridization.
Given our success with introducing the compound X chromosome into the C(2)EN strain, we next decided to introduce free X chromosomes carrying various mutations into the C(2)EN strain, using the multiply-inverted X chromosome balancer FM7a. As a general method, in the first generation FM7a; CyO/Sp females are crossed to males carrying genetic variants on the X, third, or fourth chromosomes. In the second generation, 20–30 daughters heterozygous for FM7a, CyO, and the genetic variants to be introduced into the C(2)EN strain are crossed to C(2)EN males. From this cross, sufficient females and males carrying C(2)EN and the genetic variants of interest are recovered to establish our desired strain. Since most of the surviving C(2)EN female progeny received both X chromosomes from their mothers and almost all of the surviving C(2)EN male progeny received one of these same X chromosomes, crossing them to each other immediately established a balanced strain. We elected to use the meiotic mutant nod to demonstrate this method; nod is a recessive mutant that primarily causes nonexchange chromosome loss in female meiosis (Zhang and Hawley 1990). As shown in Table 1, we were able to introduce five different X chromosomes into the C(2)EN, bw sp strain: (1) FM7a, (2) C(1;Y)1, y, (3) y w, (4) y w noda, and (5) FM7a, nod2. Previous cytological examination of metaphase I arrested oocytes from nod− females indicated that loss occurs by nonexchange chromosomes dissociating from the exchange chromosomes, resulting in high rates of oocytes with multiple chromosome masses at metaphase I arrest; the close agreement between the rate of cytological malorientation and genetically-measured aneuploidy indicates that the chromosomes in the separated masses are eventually lost (Gillies et al. 2013). As we lacked the markers to conduct a standard genetic nondisjunction assay, we validated the presence of nod through a similar cytological approach as used in Gillies et al. (2013). Our expectation was that oocytes from nod+ females would have all meiotic chromosomes in single mass, but nod− females would have multiple masses containing dissociated nonexchange chromosomes while exchange chromosomes would remain together. Consistent with this, nod+ oocytes from FM7a/y w noda; C(2)/Ø females had fully normal metaphase I arrested oocytes (20/20 oocytes with all chromosomes in a single mass, and all in the normal C(2)/3 ⇔ 3 coorientation). However, transheterozygous nod− oocytes from FM7 nod2/y w noda; C(2)EN, bw sp/Ø females had only 3/35 oocytes with all chromosomes in a single mass (8.6% single masses), comparable to the 5.5% observed in FM7 nod2/noda females without the C(2)EN (Gillies et al. 2013). These figures also showed that the exchange third chromosomes stayed together, while the X, C(2), and fourth chromosomes could all be widely separated (Figure 4). The defect was also less severe in females with exchange X chromosomes, as 10/20 oocytes scored from y w noda; C(2)EN/Ø females had all chromosomes in a single mass. These results demonstrate that the nod mutant alleles were successfully crossed into the C(2)EN background.
Figure 4.
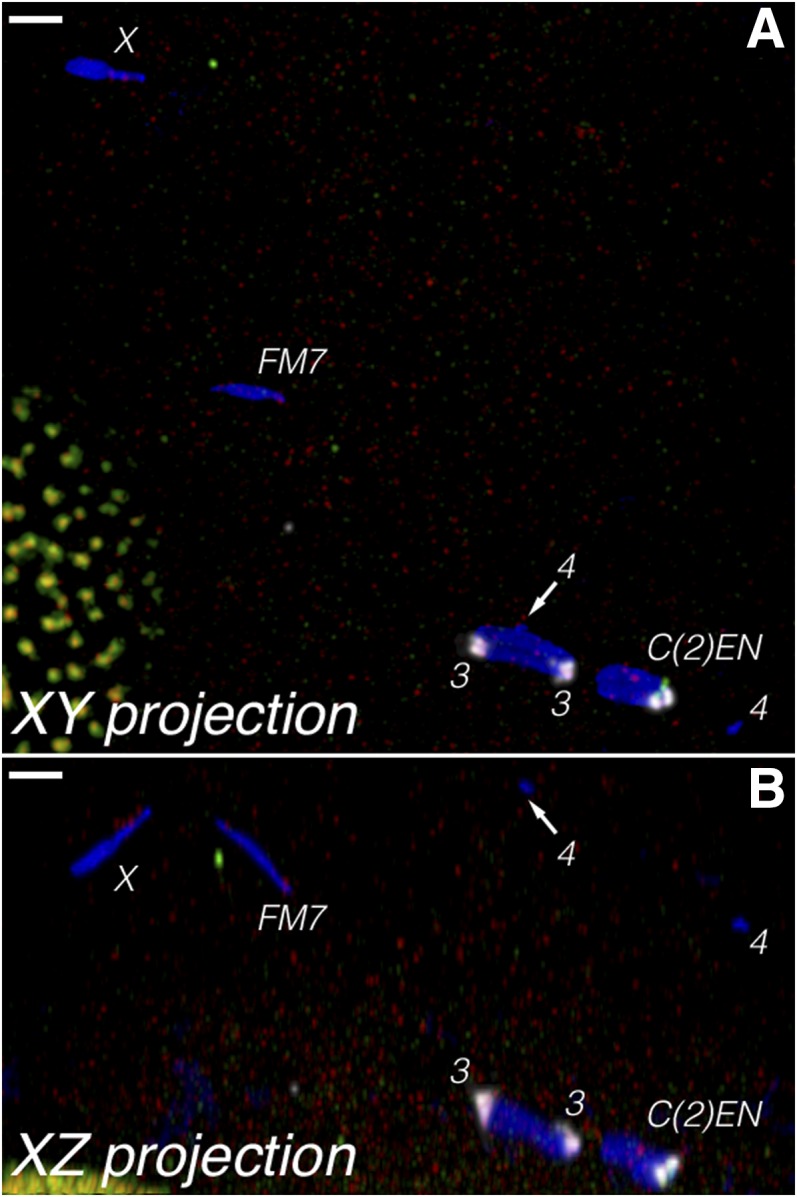
FISH in an oocyte from a FM7 nod2/y w noda; C(2)EN/Ø female in (A) normal projection and (B) orthogonal projection, showing the large separation of the nonexchange chromosomes in the oocyte, where only 3/35 oocytes (8.6%) reached a single mass. This is in contrast to the FM7 nod+/y w noda; C(2)EN/Ø control (not shown), where 20/20 oocytes had all chromosomes in single masses. This is similar to nod oocytes that have normal two chromosomes (Gillies et al. 2013). The differential staining allows the unambiguous identification of each chromosome. Note that even though the two arms of C(2)EN are isosequential and therefore can undergo recombination, this does not result in bipolar tension, and this chromosome dissociates like the nonexchange X and four chromosomes. Probes are 2L-3L (white), 2L (green), and X (red), with DAPI (blue). Scale bars, 2 µm. DAPI, 4’,6-diamidino-2-phenylindole; FISH, fluorescent in situ hybridization.
We wanted to test whether our method could be used to introduce genetic variants on the third and fourth chromosomes into C(2)EN. We crossed FM7a/yd2 w1118 P{ry+t7.2 = ey-FLP.N}2; CyO/+; P{w+mC: PRE[Scr7-8]}Q1/+; Dp(1;4)193, y+ svspa-pol/+ females to y w/Y ; C(2)EN, bw sp/Ø males. The results are shown in Table 1. We easily recovered both the third and the fourth chromosome variants with C(2)EN.
As a final test of our method, we wanted to introduce X chromosome genetic variants into C(3)EN. We tested both the TM3 and TM6C balancer chromosomes in combination with FM7a. We crossed females heterozygous for FM7a and one of the third chromosome balancers to males with two differently marked versions of C(3)EN [C(3)EN, th1 st1 and C(3)EN, st1 cu1 es]. We were successful with both third chromosome balancer chromosomes and with both versions of C(3)EN (Table 2). We would note that C(3)EN, st1 cu1 es has lower viability than C(3)EN, th1 st1, and that stocks carrying it are more difficult to maintain.
Table 2. Summary of crosses to C(3)EN.
| Genotype | Parental Females | Days Laying | Desired Progeny All with C(3)EN | Balancer/+ Progeny | Desired Progeny/Female | |
|---|---|---|---|---|---|---|
| ♀ | ♂ | |||||
| FM7a/y w; TM6C, cu Sb e ca/+♀ X | 57 | 6 | 17 FM7a/y w ♀ | 4 FM7a/y w | 220 + | 0.37 |
| +; C(3)EN, th st ♂ | 1 y w/+ ♀ | 19 y w/+ | 21 y w | |||
| 3 y w ♂ | 7 FM7a/+ | 3 FM7a | ||||
| FM7a/y w; TM3, Ser/+ ♀ X | 40 | 6 | 8 FM7a/y w ♀ | 12 y w/+ | 153 + | 0.35 |
| +; C(3)EN, th st ♂ | 3 y w/+ ♀ | 11 FM7a/+ | 6 y w | |||
| 2 FM7a/+ ♀ | 8 FM7a | |||||
| 1 y w ♂ | ||||||
| FM7a/y w; TM6C, cu Sb e ca/+♀ X | 17 | 8 | 11 FM7a/y w ♀ | 2 FM7a/y w | 72 + | 0.71 |
| +; C(3)EN, st cu es ♂ | 1 y w ♂ | 8 y w/+ | 8 y w | |||
| 4 FM7a/+ | 5 FM7a | |||||
| FM7a/y w; TM3, Ser/+ ♀ X | 15 | 8 | 12 FM7a/y w ♀ | 7 y w/+ | 88 + | 1.0 |
| +; C(3)EN, st cu es ♂ | 1 y w/+ ♀ | 3 FM7a/+ | 8 y w | |||
| 1 FM7a ♂ | 2 FM7a | |||||
| 1 + ♂ | ||||||
For each cross, flies were mated in fresh vials and transferred to new vials every 1–2 d for the total number of days indicated. The flies in the Desired Progeny column all carried the markers appropriate to the variant of C(3)EN in the cross, while the flies in the Balancer/+ columns all had the dominant markers appropriate to the balancers in the cross. We also note that C(3)EN, st1 cu1 es chromosome has lower viability than C(3)EN, th1 st1 and stocks carrying it are more difficult to maintain.
Discussion and Conclusion
The method we have presented here provides a straightforward method for performing crosses with compound autosomes that would otherwise be inviable. Crossing the variants of interest to a stock with multiple balancers, followed by mating the balanced progeny to the compound chromosome stock, allows the desired genotypes to be generated in only a few generations. With yields of around one desired progeny per parental female, this approach does not rely on rare segregation events that would require laborious amounts of fly pushing to guarantee success.
One limitation to the technique is that some chromosomes may be genetically incompatible. This was seen with the C(1)RM, y pn chromosome, where all 8 C(1) C(2) females that were produced were completely sterile, in contrast to the C(1)RM, y2 su(wa) wa chromosome, which had good fertility. The difference must be attributable to one or more uncharacterized differences between the two chromosomes. This idea has some precedent; a recent study examining chromosome 3 balancers by whole genome sequencing found that one of the inversions used to create TM3 bisected the conserved p53 tumor suppressor gene (Miller et al. 2016). As all compound chromosomes were generated by multiple rounds of irradiation, and have had decades of maintenance in stock to evolve, many changes of this nature could potentially have occurred. This highlights the need for better molecular characterization of these rearranged chromosomes, and suggests that the generation of new compound chromosomes via modern site-directed recombination techniques would potentially be very useful.
Our applications of this technique have already provided interesting data. We were able to confirm that heterologous segregation occurs in C(1)/Ø; C(2)/Ø females, a genotype we were previously unable to recreate by cold-induced nondisjunction (Gilliland et al. 2015). Curiously, we also found that males of this stock must be undergoing X/Y ⇔ C(2)EN heterologous segregation, a pattern not found in males of other stocks that just carry C(2)EN. While we have not attempted to unravel the cause of this segregation pattern, sex chromosome disjunction in males with C(2)EN is complex and varies with the sex chromosomes. Two studies of this chromosome from the early 1980s found that half to two-thirds of the progeny receiving C(2)EN from the father also received no paternal sex chromosome at all (Strommen 1982; Falk 1983). As this particular variant of C(2)EN is often used for its high rate of transmission through the male germline (Dernburg et al. 1996), it may be that this unusual disjunction contributes to this success. That C(2)EN was constructed with Y-chromosome heterochromatin on both arms suggests a potential pairing arrangement that could produce both the cosegregation and successful transmission of these chromosomes in males. While investigation of this segregation pattern is not the focus of this manuscript, we believe the tools we have generated here will facilitate more investigations into this problem.
In addition to confirming that nod alleles were introduced into the C(2)EN background, the cytological analysis of these genotypes has also showed that C(2) can become dissociated similar to the nonexchange chromosomes. This occurs even though the two arms of the compound autosome are isosequential, and can therefore undergo meiotic recombination. This confirms that the exchange chromosomes segregate normally in nod− because crossing over establishes bipolar tension, rather than an effect of the crossover event itself, similar to other cases where chromosomal rearrangements prevent crossing over from establishing tension (Jang et al. 1995).
In conclusion, the method presented here is a useful addition to the toolbox of methods for working with compound chromosomes. Our approach complements recent advancements in applying cold-shock- and meiotic mutant-induced nondisjunction to this problem (Martins et al. 2013), and should enable better characterization of the behavior of these unusual chromosomes in Drosophila meiosis and mitosis.
Acknowledgments
This work was supported by the National Institutes of Health (National Institute of General Medical Science award 1R15GM099054-01 to W.D.G.), the DePaul College of Science and Health, and the Intramural Research Program of the National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD). Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Footnotes
Communicating editor: A. Bashirullah
Literature Cited
- Ashburner M., Golic K. G., Hawley R. S., 2005. Drosophila: A Laboratory Handbook Second. CSH Press, New York. [Google Scholar]
- Dernburg A. F., 2000. In situ hybridization to somatic chromosomes in Drosophila Protocols, edited by Sullivan W., Ashburner M., Hawley R. S. CSH Press, New York. [Google Scholar]
- Dernburg A., Sedat J., Hawley R., 1996. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 86: 136–146. [DOI] [PubMed] [Google Scholar]
- Falk R., 1983. The effect of an unusual chromosome architecture on disjunction and non-disjunction in Drosophila. Genet. Res. 41(01): 17. [Google Scholar]
- Ferree P. M., Barbash D. A., 2009. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 7(10): e1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies S. C., Lane F. M., Paik W., Pyrtel K., Wallace N. T., et al. , 2013. Nondisjunctional segregations in Drosophila female meiosis I are preceded by homolog malorientation at metaphase arrest. Genetics 193(2): 443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland W. D., Hughes S. F., Vietti D. R., Hawley R. S., 2009. Congression of achiasmate chromosomes to the metaphase plate in Drosophila melanogaster oocytes. Dev. Biol. 325(1): 122–128. [DOI] [PubMed] [Google Scholar]
- Gilliland W. D., Colwell E. M., Lane F. M., Snouffer A. A., 2015. Behavior of aberrant chromosome configurations in Drosophila melanogaster female meiosis I. G3 (Bethesda) 5(2): 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm D. G., 1976. Compound autosomes, in The Genetics and Biology of Drosophila, edited by Ashburner M., Noviitski E. Academic Press, New York. [Google Scholar]
- Hughes S. E., Cotitta J. L., Takeo S., Collins K. A., Hawley R. S., et al. , 2009. Heterochromatic threads connect oscillating chromosomes during prometaphase I in Drosophila oocytes. PLoS Genet. 5(1): e1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J. K., Messina L., Erdman M. B., Arbel T., Hawley R. S., et al. , 1995. Induction of metaphase arrest in Drosophila oocytes by chiasma-based kinetochore tension. Science 268(5219): 1917–1919. [DOI] [PubMed] [Google Scholar]
- Kotadia S., Montembault E., Sullivan W., Royou A., 2012. Cell elongation is an adaptive response for clearing long chromatid arms from the cleavage plane. J. Cell Biol. 199(5): 745–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins T, Kotadia S., Malmanche N., Sunkel C. E., Sullivan W., 2013. Strategies for outcrossing and genetic manipulation of Drosophila compound autosome stocks. G3 (Bethesda) 3(1): 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. E., Cook K. R., Arvanitakis A. V., Hawley R. S., 2016. Third chromosome balancer inversions disrupt protein-coding genes and influence distal recombination events in Drosophila melanogaster. G3 (Bethesda, Md.) 6: 1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan L. V., 1922. Non-criss-cross inheritance in Drosophila melanogaster. Biol. Bull. 42(5): 267–274. [Google Scholar]
- Novitski E., Childress D., 1976. Compound Chromosomes Involving the X and the Y Chromosomes, edited by Novitski E., Ashburner M. Academic Press, New York. [Google Scholar]
- Novitski E., Grace D., Strommen C., 1981. The entire compound autosomes of Drosophila melanogaster. Genetics 98(2): 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira R. A., Kotadia S., Tavares A., Mirkovic M., Bowlin K., 2014. Centromere-independent accumulation of cohesin at Ectopic heterochromatin sites induces chromosome stretching during anaphase. PLoS Biol. 12(10): e1001962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S. L., Nielsen R. J., Teeter K., Lake C. M., Ong S., 2007. A germline clone screen for meiotic mutants in Drosophila melanogaster. Fly (Austin) 1(3): 172–181. [DOI] [PubMed] [Google Scholar]
- Parker D. R., Hammond A. E., 1958. The production of translocations in Drosophila oocytes. Genetics 43(1): 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strommen C. A., 1982. Paternal transmission of entire compounds of chromosome two in Drosophila melanogaster. Mol. Gen. Genet. 187(1): 126–131. [DOI] [PubMed] [Google Scholar]
- Sullivan W., Ashburner M., Hawley R. S., 2000. Drosophila Protocols. CSHL Press, New York. [Google Scholar]
- Zhang P., Hawley R. S., 1990. The genetic analysis of distributive segregation in Drosophila melanogaster. II. Further genetic analysis of the nod locus. Genetics 125(1): 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.