Abstract
Background
Elevated levels of C-reactive protein (CRP, determined by a high-sensitivity assay) indicate low-grade inflammation which is implicated in many age-related disorders. Epigenetic studies on CRP might discover molecular mechanisms underlying CRP regulation. We aimed to identify DNA methylation sites related to CRP concentrations in cells and tissues regulating low-grade inflammation.
Results
Genome-wide DNA methylation was measured in peripheral blood in 1,741 participants of the KORA F4 study using Illumina HumanMethylation450 BeadChip arrays. Four CpG sites (located at BCL3, AQP3, SOCS3, and cg19821297 intergenic at chromosome 19p13.2, P ≤ 1.01E-07) were significantly hypomethylated at high CRP concentrations independent of various confounders including age, sex, BMI, smoking, and white blood cell composition. Findings were not sex-specific. CRP-related top genes were enriched in JAK/STAT pathways (Benjamini-Hochberg corrected P < 0.05). Results were followed-up in three studies using DNA from peripheral blood (EPICOR, n = 503) and adipose tissue (TwinsUK, n = 368) measured as described above and from liver tissue (LMU liver cohort, n = 286) measured by MALDI-TOF mass spectrometry using EpiTYPER. CpG sites at the AQP3 locus (significant p-values in peripheral blood = 1.72E-03 and liver tissue = 1.51E-03) and the SOCS3 locus (p-values in liver < 2.82E-05) were associated with CRP in the validation panels.
Conclusions
Epigenetic modifications seem to engage in low-grade inflammation, possibly via JAK/STAT mediated pathways. Results suggest a shared relevance across different tissues at the AQP3 locus and highlight a role of DNA methylation for CRP regulation at the SOCS3 locus.
Introduction
Low-grade inflammation is thought to induce, promote or more generally influence human susceptibility to many age-related disorders such as coronary heart disease[1], type 2 diabetes[2], and several malignancies[3]. Modestly elevated concentrations of C-reactive protein (CRP), measured by a high-sensitivity assay, are a sensitive marker of low-grade inflammation. CRP is released into the systemic circulation in response to inflammatory stimuli as the final product of various inflammatory pathways. As an acute-phase reactant it is predominantly synthesized by hepatocytes and regulated via the transcription factors STAT3, C/EBP family members and NF-kappaB by the pro-inflammatory cytokines IL-6 and IL-1ß[4,5]. To a minor degree, extra-hepatic expression has been reported for adipose tissue and blood cells[5]. Systemic levels of CRP are known to be influenced by age, sex, environmental and life style conditions like smoking exposure and BMI as well as genetic determinants with substantial heritability estimates[6]. Recent research focusing on common sequence variants has only partially explained the molecular basis of systematically circulating CRP[7].
Epigenetic modifications such as changes in DNA methylation seem to have important regulatory functions in cellular processes[8], including inflammatory responses of the human body[9]. Previous studies suggested that non-genetic determinants of CRP like age[10–12], sex[13,14], diet[15] and exposure to cigarette smoking[16] as well as genetic factors[17,18] are associated with epigenetic modification. In addition, evidence indicated that epigenetic mechanisms are implicated in the development of several malignancies[19] as well as atherosclerosis[20] which are both characterized by aberrant inflammatory processes[21,22]. Finally, epigenetic modifications were observed in several inflammatory disease states[23–27]. Therefore, epigenetic modifications integrating both environmental as well as genetic factors might relevantly engage in low-grade inflammation as reflected by elevated levels of circulating CRP.
Epigenome-wide association studies (EWAS) on DNA methylation hold the potential to identify epigenetic modifications of CRP regulation across the genome. This could provide important clues to immune response pathways involved in the regulation of low-grade inflammation and might also be of relevance for related clinical entities. Therefore, we conducted an EWAS on CRP concentrations in a large population-based study using DNA methylation data in peripheral blood. To validate and to assess tissue specificity and relevance of the discovery findings, results were followed up in three independent studies using DNA methylation data derived from peripheral blood, adipose and liver tissue. In addition, gene expression panels for validated genes were generated, transcript levels of these genes were quantified in human liver samples, and enrichment analyses were conducted to ascertain functional properties of identified loci.
Results
Discovery of DNA methylation sites related to CRP in peripheral blood
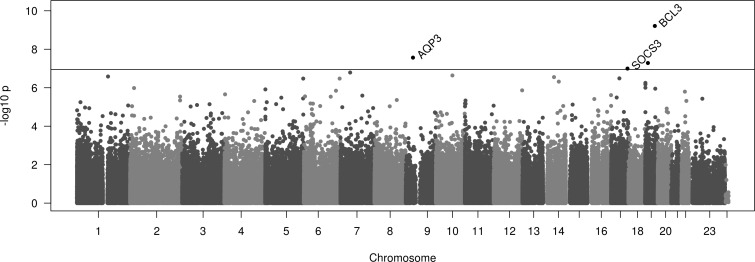
We observed epigenome-wide significant associations with systemic CRP concentrations for 4 CpG sites located intronic at B-cell lymphoma 3 (BCL3), and aquaporin 3 (AQP3), in exon 2 at Suppressor of cytokine signaling (SOCS3), and intergenic (12kb upstream of jun B proto-oncogene, JUNB) at chromosome 19p13.2 in the comprehensive model (p-values range 6.14E-10–1.01E-07, Table 1, Fig 1). There was an inverse association between DNA methylation and levels of CRP for all significant CpG sites with effects ranging from -0.023 to -0.031.
Table 1. Significant associations between CRP and DNA methylation sites in the discovery and validation panels.
| Locus | KORA F4 | EPICOR | TwinsUK | LMU liver cohort | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | chr | CpG | ß coef | se | p | ß coef | se | p | ß coef | se | p | ß coef | se | p |
| BCL3 | 19 | cg26470501 | -0.03 | 0.005 | 6.14E-10 | -0.02 | 0.013 | 1.21E-01 | -0.02 | 0.01 | 3.12E-01 | -0.01 | 0.01 | 3.24E-01 |
| AQP3 | 9 | cg02716826 | -0.03 | 0.005 | 2.72E-08 | -0.04 | 0.011 | 1.72E-03 | -0.02 | 0.01 | 2.44E-01 | 0.04 | 0.01 | 1.51E-03 |
| NA* | 19 | cg19821297 | -0.02 | 0.004 | 5.19E-08 | -0.02 | 0.01 | 6.56E-02 | -0.001 | 0.01 | 9.01E-01 | -0.02 | 0.01 | 8.62E-03 |
| SOCS3 | 17 | cg18181703 | -0.02 | 0.004 | 1.01E-07 | -0.01 | 0.011 | 1.89E-01 | -0.01 | 0.01 | 4.27E-01 | -0.01 | 0.01 | 1.12E-01 |
| SOCS3 | 17 | CpG_2.3 | NA | NA | NA | NA | NA | NA | NA | NA | NA | -0.04 | 0.01 | 4.36E-07 |
| SOCS3 | 17 | CpG_8.9 | NA | NA | NA | NA | NA | NA | NA | NA | NA | -0.03 | 0.01 | 2.82E-05 |
Significant associations between ln-transformed systemic CRP levels and beta values of DNA methylation sites were assessed using multivariate linear mixed effects models in KORA F4 (n = 1741), EPICOR (n = 503), and TwinsUK (n = 286) and multivariate linear models in the LMU liver cohort (n = 286) adjusting for various confounding variables and correcting for multiple testing according to Bonferroni. Significant p-values (1.13E-07, 1.25E-02, and 1.92E-03 in KORA F4, EPICOR and TwinsUK and the LMU liver cohort, respectively) are marked in bold font. chr: chromosome; Gene: UCSC reference gene according to USCS Genome Browser; ß coef: β coefficient; se: standard error
*no gene annotation for this CpG site according to the UCSC Genome Browser.
Fig 1. Manhattan Plot of the results of the genome-wide DNA methylation analysis on CRP conducted in the KORA F4 discovery sample.
The Manhattan plot displays all analyzed CpG sites with their calculated p-values. Threshold of epigenome-wide significance: P = 1.13E-07.
No statistically significant interaction term between methylation beta values of the 4 significant CpG sites and sex was observed (data not shown).
We identified correlations ≥ 0.8 with SNPs (listed in Table A in S1 File) for two CpG sites (cg26470501 and cg19821297). Associations remained similar when we repeated the analyses for these two loci with additional adjustment for correlated SNPs in participants of the KORA F4 study where genomic and DNA methylation data were available (Table A in S1 File). Likewise, associations remained within similar ranges when we repeated the analyses with additional adjustments for lipids, uric acid, leptin, fasting glucose, alcohol consumption, systolic blood pressure, or medication (Table A in S1 File).
Enrichment analysis
All unique genes identified in the EWAS at a 5% false discovery rate significance level (23 genes, File A and Table B in S1 File) were annotated in the Ingenuity Pathway Analysis (IPA) database. The enrichment analysis yielded 6 statistically significant canonical pathways with Benjamini-Hochberg corrected p-values < 0.05 (Table 2).
Table 2. Significant canonical pathways in the KOFA F4 discovery panel.
| Canonical pathways | Benjamini-Hochberg p-value | Genes |
|---|---|---|
| Role of JAK2 in Hormone-like Cytokine Signaling | 0.02 | SOCS3, SH2B2 |
| IL-9 Signaling | 0.02 | SOCS3, BCL3 |
| Role of JAK1 and JAK3 in Cytokine Signaling | 0.04 | SOCS3, SH2B2 |
| Growth Hormone Signaling | 0.04 | SOCS3, RPS6KA2 |
| Acute Myeloid Leukemia Signaling | 0.04 | RARA, PIM2 |
| Melanocyte Development and Pigmentation Signaling | 0.04 | RPS6KA2, SH2B2 |
Replication of significant loci in peripheral blood
One of the four significant CpG sites (cg02716826 at AQP3) was confirmed in the EPICOR peripheral blood cohort with the effect estimate being consistent with the one of the discovery analysis in direction and magnitude of effect (Table 1). The CpG site cg02716826 remained statistically significant after further adjustment for myocardial infarction case-control status (ß coefficient = -0.035, se = 0.011, p = 2.13E-3).
Validation of significant loci in adipose tissue and in liver samples
In the TwinsUK study, associations were consistent with those of the discovery analysis in direction and magnitude of effects but p-values were not statistically significant (Table 1).
In the LMU liver cohort, CpG sites representing 2 of the discovery loci (AQP3 and SOCS3) yielded statistically significant results in the validation analyses (Table 1). At the AQP3 locus the discovery CpG site cg02716826 was significantly associated with CRP. At the SOCS3 locus the discovery CpG site cg18181703 was not associated with CRP but adjacent CpG sites which were significantly correlated with the discovery CpG site (Spearman’s rank correlation coefficients = 0.48 and 0.52 for CpG_2.3 and CpG_8.9, respectively, each with P = 2.2E-16) yielded statistically significant associations. A full table of results of the LMU liver cohort is presented in Table C in S1 File.
Results of gene expression analyses
In the gene expression panels, both genes, AQP3 and SOCS3, were expressed not only in blood cells but also to different degrees in human tissues (Fig A in S1 File). While AQP3 was mainly expressed in kidney, lung, heart, and liver tissue, SOCS3 displayed highest gene activity in heart and adipose tissue.
In the LMU liver cohort, gene expression levels of SOCS3 were significantly correlated with CRP concentrations (Spearman’s rank correlation coefficient = 0.15, P = 0.01) but not with cg18181703 or any other CpG site covered by the corresponding amplicon (data not shown). Gene expression levels of AQP3 were not correlated with CRP concentrations but displayed a significant correlation with CpG_5 of the corresponding amplicon (Spearman’s rank correlation coefficient = -0.14, P = 0.01).
Discussion
In this study we report key findings from an epigenetic study on low-grade inflammation as reflected by concentrations of CRP in cells and tissues regulating low-grade inflammation.
DNA methylation sites were associated with CRP in peripheral blood independent of other risk factors
In a first step, we conducted an EWAS in a large population-based study and identified four loci at chromosomes 9, 17 and 19 inversely associated with concentrations of CRP. A lower degree of DNA methylation at some CpG sites was also observed in conditions related to elevated levels of CRP such as older age[12], adiposity[28], and smoking[16,29,30]. However, in the present study the association was independent of various CRP determinants including age, sex, BMI, and cigarette smoking. A general inverse association is consistent with findings of a previous EWAS conducted in hypertensive African Americans using DNA methylation data derived from leucocytes[31].
Functional properties of inflammation-related epigenetic signatures in peripheral blood
Six statistically significant pathways were identified in enrichment analyses including top CRP related genes identified in the EWAS. Janus kinase (JAK)/ Signal Transducer and Activator of Transcription (STAT) signaling plays a major role in almost all of these pathways. The highly conserved JAK/STAT signaling pathway is part of the orchestrated cascade during the acute-phase response transmitting extra-cellular signals through the cell membrane into gene promoters[32]. Thereby, it modulates transcription of genes like the acute-phase proteins fibrinogen, serum amyloid A, and CRP which are all responsive to STAT3. The importance of JAK/STAT signaling in the regulation of the immune system was further highlighted by studies reporting immune deficiency syndromes following disruption or dysregulation of JAK/STAT functionality[33]. Thus, from a biological perspective a conceivable mechanism via JAK/STAT signaling seems to be plausible.
DNA methylation signatures at AQP3 and SOCS3 were associated with CRP across different tissues
In order to validate the discovery findings as well as to assess whether the identified epigenetic effects are specific or shared across different tissues relevant for CRP expression results were followed up in three independent studies using DNA methylation data derived from peripheral blood, liver and adipose tissues. In these tissue-specific analyses we found significant associations at two loci, AQP3 and SOCS3. Both genes seem to be active in different human tissues according to our gene expression panels.
At the AQP3 locus, associations between DNA methylation and CRP were statistically significant in peripheral blood and liver tissue. In addition, we observed a significant correlation between DNA methylation at AQP3 and AQP3 transcript levels in human liver samples. Main functions of the aquaporin 3 protein are the transport of water, glycerol and small solutes such as urea and glycerol across the plasma membrane but it also seems to be involved in functions related to the immune system like wound healing[34] and the activation of the skin immune system at birth[35]. Furthermore, an up-regulation of AQP3 was observed under chronic inflammatory conditions such as present in periodontitis[36] and gastritis[37]. Interestingly, at cg02716826 which was measured in all four studies CRP effects were negative in the peripheral blood samples as well as in adipose tissue while they were positive in liver tissue. A complex and tissue specific relationship was also observed in different cell line studies investigating AQP3 expression in response to cytokine signaling. In one of these studies, using gingival epithelial cells gene expression was increased in response to TNF-alpha[36] while other studies report decreased expression levels in response to TNF-alpha in keratinocytes[38,39]. Our study seems to confirm tissue specific effects at this locus. However, as BMI or other measures of adiposity were not available in the liver study, we cannot exclude that effect differences might be caused by confounding. Hence, epigenetic mechanisms at the AQP3 locus seem to have a shared relevance in low-grade inflammation across different tissues, but further studies are warranted to evaluate tissue specificity of effects and the influence of adiposity in liver tissue.
The second region at SOCS3 was significantly associated with systemic levels of CRP in the peripheral blood discovery analysis as well as in the liver samples. Furthermore, SOCS3 transcript levels were significantly correlated with CRP in human liver tissue. SOCS3 is a negative feedback regulator of cytokine signaling along the JAK/STAT pathway. Growing evidence supports a role of epigenetic mechanisms at SOCS3 in several cancers including liver, lung, pancreatic and prostate cancer, as well as other malignancies[40–45]. Given well confirmed evidence of the presence of inflammatory cells in tumor microenvironment[3,21] our finding supports a role of epigenetic mechanisms at SOCS3 in conditions related to inflammation and suggests a novel link between DNA methylation at SOCS3 and systemic CRP in tissues in which immune mediators are expressed.
As the approach taken in this study is observational in nature it is not possible to draw causal inferences. Therefore, it might be possible that not the genes themselves, but small regulatory elements might engage in the identified associations between epigenetic modifications and CRP regulation. This is most likely the case as there has been evidence of regulatory elements like relevant transcription factor binding sites such as C/EBP and NF-kappaB as well as STAT3 in the AQP3 and the SOCS3 region, respectively (http://genome.ucsc.edu). The identification of causal inferences and molecular pathways underlying the relation between CpG sites and CRP levels represents promising targets for future functional studies.
Limitations and strengths
Two limitations of our study have to be mentioned. Firstly, epigenetic modifications are cell-type specific and peripheral blood constitutes a heterogeneous admixture of different cell populations. Observed changes in DNA methylation profiles might therefore reflect a differential representation of the cell types in the sample leading to false positive results[46]. However, for low-grade inflammation peripheral blood is a tissue of interest and a valuable source of information. Therefore, we used peripheral blood derived DNA methylation data in the discovery sample and in one of the validation samples. In addition, to diminish the risk of cell type confounding we adjusting the peripheral blood analyses for white blood cell composition estimated using algorithms developed on the basis of cell-type specific DNA methylation markers identified from cell-sorted reference profiles of specific cell populations[47]. Secondly, validation studies exhibited moderate sample sizes as well as differences in study design or availability of adjustment variables. In particular, further studies are warranted to investigate the effect of BMI on DNA methylation in liver tissue and epigenetic modifications in male adipose tissue. However, the present study had enough power to identify novel epigenetic patterns related to low-grade inflammation with a shared relevance across different tissue. Findings were plausible from a biologic perspective, and may promote future research on the regulation of low-grade inflammation and mechanisms contributing to related clinical disorders. The identified epigenetic patterns may be used not only in functional studies to provide further insights into molecular mechanisms of inflammatory processes but also in biomarker studies using whole blood to improve the prediction of inflammation related clinical disorders or events. In addition, gene expression panels suggest further tissues which seem to be relevant for the identified genes and promising for in-depth investigations with respect to clinical disorders affecting these tissues. Thereby, the identified epigenetic loci might present useful targets for the prevention and / or treatment of these diseases.
Conclusions
Using an epigenetic approach with DNA derived from different trait targeted tissues the present study identified epigenetic loci which seem to engage in low-grade inflammation independent of various confounders and conceivably via JAK/STAT signaling. In addition, validation results suggest novel evidence for an epigenetic mode at AQP3 with possible tissue specific effects but a shared relevance for CRP regulation and extend previous evidence of the importance of epigenetic modifications at SOCS3 with respect to inflammatory processes.
Materials and Methods
Study design
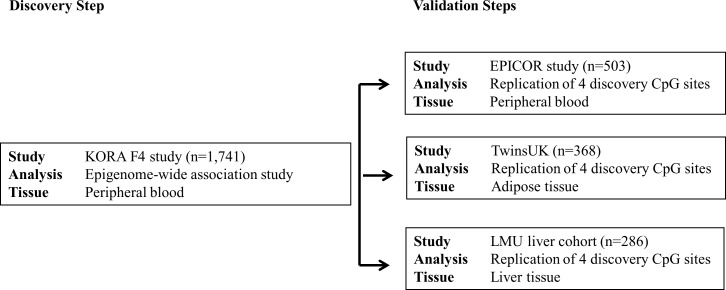
The present study incorporated data from four different studies and followed a two-stage design (Fig 2). In the discovery step, we assessed the association between CRP and the degree of DNA methylation using peripheral blood from 1,741 participants of the population-based Cooperative Health Research in the Region of Augsburg (KORA) F4 study. Subsequent follow-up of results was performed in three independent validation panels: the cardiovascular section of the Italian European Prospective Investigation into Cancer and Nutrition cohort (EPICOR, n = 503), the TwinsUK cohort (TwinsUK, n = 368 female participants), and a cohort from the Ludwig-Maximilians-University Munich (LMU liver cohort, n = 286) using DNA methylation data derived from peripheral blood, adipose, and liver tissue, respectively. For the present study, subjects with elevated levels of CRP indicating acute infection (CRP > 10 mg/L) and/ or missing data on CRP were excluded. All studies were approved by the local ethics committees. In detail, the KORA study and the LMU study were approved by the Ethics Committee of the Bavarian Medical Association (Bayerische Landesärztekammer); the EPICOR study was approved by the Ethical Committee of the Human Genetics Foundation (Turin, Italy); and the TwinsUK study has ethical approval from the Guy’s and St Thomas’ (GSTT) Ethics Committee. Written informed consent was obtained from all participants. Study populations are described in File B in S1 File. Baseline characteristics of participants of the four studies are provided in Table 3.
Fig 2. Outline of the present study.
Table 3. Baseline characteristics of study participants.
| KORA F4 | EPICOR | TwinsUK | LMU cohort | |
|---|---|---|---|---|
| N (%) | 1,741 | 503 | 368 | 286 |
| Age (years) / | 60.9 (8.89) | 52.5 (7.33) | 60.6 (8.04) | |
| Age (10 yrs) | 5.7 (1.36) | |||
| Male (%) | 48.8 | 62 | 0 | 52.4 |
| BMI (kg/m2) | 28.1 (4.68) | 26.6 (3.89) | 26.6 (4.67) | NA |
| current/former/never smoker (%) | 15 / 41 / 44 | 32 / 31 / 37 | 10 / 38 / 52 | 22 / 0 / 61 |
| fasting (%) | 99.5 | 29 | NA | 100 |
| CRP mg/L* | 1.25 (0.62–2.47) | 1.07 (0.49–2.19) | 1.57 (0.72–3.37) | 0.4 (0.2–0.98) |
Data with normal and skewed distribution (indicated by *) are given as mean (SD) and median (interquartile range) of the variables.
Measurement of CRP
CRP was measured in all four studies using high-sensitivity tests (File C in S1 File). The intra- and inter-assay coefficients of variation were below 6% in all four studies.
Assessment of DNA methylation data
Genome-wide DNA methylation in KORA F4, EPICOR, and TwinsUK was assessed using the Illumina HumanMethylation450 BeadChip. In brief, genomic data was bisulfite converted. Subsequently, the bisulfite converted samples were amplified and after enzymatic fragmentation and application of the samples the arrays were fluorescently stained and scanned. Beta values representing the percentage of DNA methylation of a cytosine were calculated as the ratio of the methylated signal over the sum of the methylated and unmethylated signals. Further details on DNA methylation measurement, data preprocessing, and quality assessments of the four studies are presented in File D in S1 File. Annotations are based on UCSC Genome Browser on Human Feb. 2009 (GRCh37/hg19) Assembly (https://genome.ucsc.edu/).
Replication in the LMU liver cohort samples was carried out by MALDI-TOF mass spectrometry using EpiTYPER by MassARRAY (Sequenom, San Diego, CA)[48]. Four amplicons covering 26 CpG sites were selected. Target regions were amplified at 58°C using the prime pairs described in Table D in S1 File. The chip was read by Sequenom MALDI-TOF MS Compact Unit and visualized using MassARRAY EpiTyper v1.2 software (Sequenom). Beta values were determined by comparing the signal intensities between the mass signals of methylated and non-methylated templates.
Assessment of gene expression data
In order to assess cell type and tissue specificity of the validated results we generated gene expression panels in which we quantified the expression of AQP3 and SOCS3 in different human tissues and blood cell types. Primers and probes for quantitative PCRs (qPCRs) of AQP3 and SOCS3 are given in Table E in S1 File. Total RNA (1 μg) from human liver, brain, heart, lung, kidney, small intestine, adipose tissue, skeletal muscle, peripheral blood mononuclear cells (PBMC), CD14-, CD19-, CD3-, CD4-, CD8-positive cells, and regulatory T-cells (pool of 2–3 donors; Clontech) was reverse transcribed and qPCRs were performed in quadruplicate as described[49]. Absolute copies were determined using plasmid standard curves and normalized to μg input RNA. RNA from human livers (n = 304; HTCR Stiftung[50]) was isolated with TRIzol according to the manufacturer’s instructions and 2 μg RNA was reverse transcribed according to published protocols[49]. Normalization was performed using house-keeping gene expression of ACTB and GAPDH[49]. Subjects with DNA methylation data and data on CRP concentrations ≤ 10 mg/L were included (n = 286).
Statistical analyses
Discovery analysis
In KORA F4 analysis natural log-transformed concentrations of CRP were modeled using linear mixed effects models with DNA methylation beta values, age, sex, BMI, fasting status (two categories: fasting for > 8 hours /non-fasting), and cigarette smoking (ever/former/never smoker) as fixed effects and technical variables (plate and position on plate) as random effects. In addition, because peripheral blood constitutes a heterogeneous admixture of different cell types which may be methylated in a cell-type specific way principal components of white blood cell components estimates[47] were added as fixed effects to adjust for cell type confounding.
Sensitivity analyses
Sensitivity analyses with various degrees of adjustment were performed and the following covariates were added to the statistical model: lipids (lipid ratio defined as total cholesterol levels divided by high-density lipoprotein cholesterol, triglycerides, and low-density lipoprotein cholesterol), uric acid, leptin, fasting glucose, alcohol consumption [g/day], systolic blood pressure, or systemic hormone therapy (yes/no/male) and other medication including, regular intake of corticoids or non-steroidal anti-inflammatory drugs, antidiabetic medication, intake of antihypertensive and lipid lowering drugs (yes/no).
In addition, effect modifications by sex were assessed by adding an interaction term in the multivariate models.
To control for genomic confounding, correlations between CRP related CpG sites identified in the KORA F4 discovery sample and SNPs with minor allele frequency > 0.05 were assessed using genomic data previously acquired in the KORA F4 study (File B in S1 File). Subsequently, for CpG sites which were correlated with one or more common sequence variants (correlation coefficient ≥ 0.8) the analysis was repeated, this time including principal components derived from the correlated SNPs in the multivariate linear mixed effects models.
Enrichment analyses
All CpG sites which were significant at a false discovery rate level in the KORA F4 study were included in pathway analyses. Pathway analyses were performed using the Ingenuity Pathway Analyses (IPA) software tool (IPA build version 338830M, content version: 23814503, release date 2015-03-23, analysis date 2015-04-20; http://www.ingenuity.com/). Gene enrichment in canonical pathways was assessed in the core analysis module using Fisher’s exact test right tailed with Benjamini-Hochberg corrected level of significance.
Validation analyses
To replicate the findings in another study using DNA methylation data from peripheral blood, we assessed the association between CRP concentrations and methylation beta values of significant CpG sites in the EPICOR study using the statistical model of the discovery analysis. Study center reflecting the fasting state of participants was used as proxy for fasting status.
To assess tissue specificity of the discovery findings results were also assessed in the TwinsUK study and the LMU liver cohort. In TwinsUK, all subjects were female and some subjects were twin pairs. Therefore, sex was not a covariate but family and zygosity were included as random effects in the model. Furthermore, CRP concentrations were not measured at the time of DNA extraction and the difference in years between measurements was used as fixed effect covariate. TwinsUK data analysis also considered other covariates such as age, BMI, and smoking status (ever/former/never smoker) as fixed effects and technical covariates (plate, bisulfite conversion levels and bisulfite conversion efficiency) as random effects. For the analysis in the LMU liver cohort, natural log-transformed levels of CRP were modeled in a linear model using DNA methylation beta values, 10 years age groups, sex, smoking (yes/no), experimental plate, information on chemotherapy (yes/no), and indication of surgery (metastasis of hepatocellular, cholangiocellular or colorectal carcinomas; other metastasis; benign liver tumor; other) as covariates in the model.
Gene expression analyses
Correlations between normalized transcript levels of AQP3 and SOCS3 and CRP as well as between normalized transcript levels of AQP3 and SOCS3 and CpG sites of corresponding amplicons were assessed using Spearman’s rank correlation coefficient.
Multiple testing
Results were corrected for multiple testing and thresholds of significance were adapted according to Bonferroni in all four studies. P-values below 1.13E-07 (KORA F4), 1.25E-02 (EPICOR and TwinsUK), and 1.92E-03 (LMU liver cohort) were considered significant.
All statistical analyses were carried out using the software R version 3.0.2 (http://www.r-project.org/).
Supporting Information
File A in S1 File. Significant associations (Benjamini Hochberg corrected) between CRP and DNA methylation sites in the KORA F4 discovery study. File B in S1 File. Study Populations. File C in S1 File. Measurement of CRP. File D in S1 File. Assessment of DNA methylation data in KORA F4, EPICOR, and TwinsUK using the Illumina HumanMethylation450 BeadChip. Table A in S1 File. Associations between CRP and DNA methylation sites in the KORA F4 discovery study (n = 1741) after additional adjustments. Table B in S1 File. Significant associations (Benjamini Hochberg corrected) between CRP and DNA methylation sites in the KORA F4 discovery study. Table C in S1 File. Associations between CRP and DNA methylation sites in the LMU liver cohort. Table D in S1 File. Sequences of PCR tagged primers used for EpiTYPER methylation analysis, product size of each amplicon, and informative CpG sites per amplicon. Table E in S1 File. Primers and probes for quantitative PCRs. Figure A in S1 File. Expression of AQP3 and SOCS3 (normalized to μg input RNA) in different human tissues (human brain, heart, lung, kidney, small intestine, adipose tissue, skeletal muscle) and blood cell types (peripheral blood mononuclear cells (PBMC), CD14-, CD19-, CD3-, CD4-, CD8-positive cells, and regulatory T-cells).
(DOCX)
Acknowledgments
The authors are grateful to all members of the Helmholtz Zentrum München, the field staff in Augsburg, and the Augsburg registry team who were involved in the planning, organization, and conduct of the KORA studies as well as to Mrs Gerlinde Trischler from the Biomarker Laboratory of the Department of Internal Medicine II at the University of Ulm Medical Center for expert technical assistance. In addition, the authors wish to thank all who collaborated with EPICOR and the Italian AVIS blood donor organization.
Finally, the authors express their appreciation to all study participants.
Abbreviations
- AQP3
Aaquaporin 3
- BCL3
B-cell lympohoma 3
- CRP
C-reactive protein
- EPICOR
the cardiovascular section of the Italian European Prospective Investigation into Cancer and Nutrition cohort
- EWAS
Epigenome-wide association studies
- hs
high-sensitivity
- IPA
Ingenuity Pathway Analyses
- JAK
Janus kinase
- JUNB
jun B proto-oncogene
- KORA
Cooperative Health Research in the Region of Augsburg
- LMU
Ludwig-Maximilians-University Munich
- SOCS3
Suppressor of cytokine signaling (SOCS3)
- STAT
Signal Transducer and Activator of Transcription
Data Availability
The informed consent given by KORA, EPIC-Italy, and LMU study participants does not cover data posting in public databases. However, data are available upon request. In detail, KORA data are available from KORA-gen (http://www.helmholtz-muenchen.de/kora-gen) by means of a project agreement. Requests should be sent to kora.passt@helmholtz-muenchen.de and are subject to approval by the KORA Board. Requests for LMU data should be sent to Lesca.Holdt@med.uni-muenchen.de. EPICOR data are available upon request from HuGeF (http://www.hugef.org) by means of a project agreement. Requests should be sent to info@hugef-torino.org and are subject to approval by the EPICOR-HuGeF Board. The ArrayExpress accession number for the TwinsUK adipose methylation data set is E-MTAB-1866.
Funding Statement
The KORA study was initiated and financed by the Helmholtz Zentrum München –German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria. Furthermore, KORA research was supported within the Munich Center of Health Sciences (MC-Health), Ludwig-Maximilians-Universität, as part of LMUinnovativ. The research leading to these results has received funding from the Helmholtz Association (Helmholtz-Russia Joint Research Group 310), the European Union’s Seventh Framework Programme (FP7/2007-2013) under grant agreements number 261433 (BioSHaRE-EU), number603288 (SysVasc) and HEALTH-F2-2013-602736 (PAIN-OMICS), as well as the European Union's Seventh Framework Programme (FP7-Health-F5-2012) under grant agreement number305280 (MIMOmics) and BMBF e:Med project: e:AtheroSysMed—Systems medicine of myocardial infarction and stroke, grant number01ZX1313A‐2014. In addition, part of this work was financed by the German Center for Diabetes Research (DZD) funded by the BMBF, by the German National Genome Research Network (NGFNplus, project number 01GS0834), and through additional funds from the University of Ulm. EPICOR: This work was supported by the Compagnia di San Paolo for the EPIC-Italy and EPICOR projects (to SP, VK, RT, LI, CS, GM), the Human Genetics Foundation-HuGeF (to GM), and the MIUR ex60% grant (to GM). EPIC-Italy is further supported by grant number IG2013 N.14410 from the Associazione Italiana per la Ricerca sul Cancro (AIRC, Milan) (to SP, CS). TwinsUK: Funding support for this project was obtained from the European Research Council (project number 250157), the Economic and Social Research council (grant number ES/N000404/1), and in part by TwinsUK, which is funded by the Wellcome Trust; European Community’s Seventh Framework Programme (FP7/2007-2013). The study also receives support from the National Institute for Health Research (NIHR)- funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust in partnership with King's College London. Laboratory work was performed by The Wellcome Trust Sanger Institute and National Eye Institute via NIH/CIDR. TDS is the holder of an ERC Advanced Principal Investigator award (ERC 250157). LMU liver cohort: We acknowledge the support of the non-profit foundation HTCR, which holds human tissue on trust, making it broadly available for research on an ethical and legal basis.The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Koenig W, Sund M, Frohlich M, Fischer HG, Lowel H, Doring A, et al. (1999) C-Reactive protein, a sensitive marker of inflammation, predicts future risk of coronary heart disease in initially healthy middle-aged men: results from the MONICA (Monitoring Trends and Determinants in Cardiovascular Disease) Augsburg Cohort Study, 1984 to 1992. Circulation 99: 237–242. [DOI] [PubMed] [Google Scholar]
- 2.Marzi C, Huth C, Herder C, Baumert J, Thorand B, Rathmann W, et al. (2013) Acute-phase serum amyloid A protein and its implication in the development of type 2 diabetes in the KORA S4/F4 study. Diabetes Care 36: 1321–1326. 10.2337/dc12-1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balkwill F, Mantovani A (2001) Inflammation and cancer: back to Virchow? Lancet 357: 539–545. 10.1016/S0140-6736(00)04046-0 [DOI] [PubMed] [Google Scholar]
- 4.Arnaud C, Burger F, Steffens S, Veillard NR, Nguyen TH, Trono D, et al. (2005) Statins reduce interleukin-6-induced C-reactive protein in human hepatocytes: new evidence for direct antiinflammatory effects of statins. Arterioscler Thromb Vasc Biol 25: 1231–1236. 10.1161/01.ATV.0000163840.63685.0c [DOI] [PubMed] [Google Scholar]
- 5.Black S, Kushner I, Samols D (2004) C-reactive Protein. J Biol Chem 279: 48487–48490. 10.1074/jbc.R400025200 [DOI] [PubMed] [Google Scholar]
- 6.MacGregor AJ, Gallimore JR, Spector TD, Pepys MB (2004) Genetic effects on baseline values of C-reactive protein and serum amyloid a protein: a comparison of monozygotic and dizygotic twins. Clin Chem 50: 130–134. 10.1373/clinchem.2003.028258 [DOI] [PubMed] [Google Scholar]
- 7.Dehghan A, Dupuis J, Barbalic M, Bis JC, Eiriksdottir G, Lu C, et al. (2011) Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation 123: 731–738. 10.1161/CIRCULATIONAHA.110.948570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waterland RA, Michels KB (2007) Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr 27: 363–388. 10.1146/annurev.nutr.27.061406.093705 [DOI] [PubMed] [Google Scholar]
- 9.Stenvinkel P, Karimi M, Johansson S, Axelsson J, Suliman M, Lindholm B, et al. (2007) Impact of inflammation on epigenetic DNA methylation—a novel risk factor for cardiovascular disease? J Intern Med 261: 488–499. 10.1111/j.1365-2796.2007.01777.x [DOI] [PubMed] [Google Scholar]
- 10.Bell JT, Tsai PC, Yang TP, Pidsley R, Nisbet J, Glass D, et al. (2012) Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLOS Genet 8: e1002629 10.1371/journal.pgen.1002629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentilini D, Mari D, Castaldi D, Remondini D, Ogliari G, Ostan R, et al. (2013) Role of epigenetics in human aging and longevity: genome-wide DNA methylation profile in centenarians and centenarians' offspring. Age (Dordr) 35: 1961–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuke C, Shimabukuro M, Petronis A, Sugimoto J, Oda T, Miura K, et al. (2004) Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann Hum Genet 68: 196–204. 10.1046/j.1529-8817.2004.00081.x [DOI] [PubMed] [Google Scholar]
- 13.El-Maarri O, Becker T, Junen J, Manzoor SS, Diaz-Lacava A, Schwaab R, et al. (2007) Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Human Genetics 122: 505–514. 10.1007/s00439-007-0430-3 [DOI] [PubMed] [Google Scholar]
- 14.Sarter B, Long TL, Tsong WH, Koh WP, Yu MC, Laird PW (2005) Sex differential in methylation patterns of selected genes in Singapore Chinese. Human Genetics 117: 402–403. 10.1007/s00439-005-1317-9 [DOI] [PubMed] [Google Scholar]
- 15.Anderson OS, Sant KE, Dolinoy DC (2012) Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem 23: 853–859. 10.1016/j.jnutbio.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeilinger S, Kuhnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, et al. (2013) Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLOS ONE 8: e63812 10.1371/journal.pone.0063812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, et al. (2008) Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet 40: 904–908. 10.1038/ng.174 [DOI] [PubMed] [Google Scholar]
- 18.Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, et al. (2011) DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol 12: R10 10.1186/gb-2011-12-1-r10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karsli-Ceppioglu S, Dagdemir A, Judes G, Ngollo M, Penault-Llorca F, Pajon A, et al. (2014) Epigenetic mechanisms of breast cancer: an update of the current knowledge. Epigenomics 6: 651–664. 10.2217/epi.14.59 [DOI] [PubMed] [Google Scholar]
- 20.Grimaldi V, Vietri MT, Schiano C, Picascia A, De Pascale MR, Fiorito C, et al. (2015) Epigenetic reprogramming in atherosclerosis. Curr Atheroscler Rep 17: 476 10.1007/s11883-014-0476-3 [DOI] [PubMed] [Google Scholar]
- 21.Coussens LM, Werb Z (2002) Inflammation and cancer. Nature 420: 860–867. 10.1038/nature01322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Libby P (2002) Inflammation in atherosclerosis. Nature 420: 868–874. 10.1038/nature01323 [DOI] [PubMed] [Google Scholar]
- 23.Ballestar E (2011) Epigenetic alterations in autoimmune rheumatic diseases. Nat Rev Rheumatol 7: 263–271. 10.1038/nrrheum.2011.16 [DOI] [PubMed] [Google Scholar]
- 24.Durham A, Chou PC, Kirkham P, Adcock IM (2010) Epigenetics in asthma and other inflammatory lung diseases. Epigenomics 2: 523–537. 10.2217/epi.10.27 [DOI] [PubMed] [Google Scholar]
- 25.Burrell AM, Handel AE, Ramagopalan SV, Ebers GC, Morahan JM (2011) Epigenetic mechanisms in multiple sclerosis and the major histocompatibility complex (MHC). Discov Med 11: 187–196. [PubMed] [Google Scholar]
- 26.Ho SM (2010) Environmental epigenetics of asthma: an update. J Allergy Clin Immunol 126: 453–465. 10.1016/j.jaci.2010.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kucukali CI, Kurtuncu M, Coban A, Cebi M, Tuzun E (2014) Epigenetics of Multiple Sclerosis: An Updated Review. Neuromolecular Med. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Zhu H, Snieder H, Su S, Munn D, Harshfield G, et al. (2010) Obesity related methylation changes in DNA of peripheral blood leukocytes. BMC Med 8: 87 10.1186/1741-7015-8-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H (2011) Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet 88: 450–457. 10.1016/j.ajhg.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joubert BR, Haberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, et al. (2012) 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect 120: 1425–1431. 10.1289/ehp.1205412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun YV, Lazarus A, Smith JA, Chuang YH, Zhao W, Turner ST, et al. (2013) Gene-specific DNA methylation association with serum levels of C-reactive protein in African Americans. PLOS ONE 8: e73480 10.1371/journal.pone.0073480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aaronson DS, Horvath CM (2002) A road map for those who don't know JAK-STAT. Science 296: 1653–1655. 10.1126/science.1071545 [DOI] [PubMed] [Google Scholar]
- 33.Kiu H, Nicholson SE (2012) Biology and significance of the JAK/STAT signalling pathways. Growth Factors 30: 88–106. 10.3109/08977194.2012.660936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hara-Chikuma M, Verkman AS (2008) Aquaporin-3 facilitates epidermal cell migration and proliferation during wound healing. J Mol Med (Berl) 86: 221–231. [DOI] [PubMed] [Google Scholar]
- 35.Marchini G, Stabi B, Kankes K, Lonne-Rahm S, Ostergaard M, Nielsen S (2003) AQP1 and AQP3, psoriasin, and nitric oxide synthases 1–3 are inflammatory mediators in erythema toxicum neonatorum. Pediatr Dermatol 20: 377–384. [DOI] [PubMed] [Google Scholar]
- 36.Tancharoen S, Matsuyama T, Abeyama K, Matsushita K, Kawahara K, Sangalungkarn V, et al. (2008) The role of water channel aquaporin 3 in the mechanism of TNF-alpha-mediated proinflammatory events: Implication in periodontal inflammation. J Cell Physiol 217: 338–349. 10.1002/jcp.21506 [DOI] [PubMed] [Google Scholar]
- 37.Mei WX, Lao SX, Yu N, Zhou Z, Huang LP, Hu B (2010) Relationship between gene expressions of aquaporin 3 and 4 and various degrees of spleen-stomach dampness-heat syndrome in chronic superficial gastritis. Zhong Xi Yi Jie He Xue Bao 8: 111–115. [DOI] [PubMed] [Google Scholar]
- 38.Nakahigashi K, Kabashima K, Ikoma A, Verkman AS, Miyachi Y, Hara-Chikuma M (2011) Upregulation of aquaporin-3 is involved in keratinocyte proliferation and epidermal hyperplasia. J Invest Dermatol 131: 865–873. 10.1038/jid.2010.395 [DOI] [PubMed] [Google Scholar]
- 39.Horie I, Maeda M, Yokoyama S, Hisatsune A, Katsuki H, Miyata T, et al. (2009) Tumor necrosis factor-alpha decreases aquaporin-3 expression in DJM-1 keratinocytes. Biochem Biophys Res Commun 387: 564–568. 10.1016/j.bbrc.2009.07.077 [DOI] [PubMed] [Google Scholar]
- 40.Molavi O, Wang P, Zak Z, Gelebart P, Belch A, Lai R (2013) Gene methylation and silencing of SOCS3 in mantle cell lymphoma. Br J Haematol 161: 348–356. 10.1111/bjh.12262 [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Zhou H, Han Y, Liu X, Wang M, Wang X, et al. (2014) SOCS3 methylation in synergy with Reg3A overexpression promotes cell growth in pancreatic cancer. J Mol Med (Berl) 92: 1257–1269. [DOI] [PubMed] [Google Scholar]
- 42.Isomoto H, Mott JL, Kobayashi S, Werneburg NW, Bronk SF, Haan S, et al. (2007) Sustained IL-6/STAT-3 signaling in cholangiocarcinoma cells due to SOCS-3 epigenetic silencing. Gastroenterology 132: 384–396. 10.1053/j.gastro.2006.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niwa Y, Kanda H, Shikauchi Y, Saiura A, Matsubara K, Kitagawa T, et al. (2005) Methylation silencing of SOCS-3 promotes cell growth and migration by enhancing JAK/STAT and FAK signalings in human hepatocellular carcinoma. Oncogene 24: 6406–6417. 10.1038/sj.onc.1208788 [DOI] [PubMed] [Google Scholar]
- 44.Tischoff I, Hengge UR, Vieth M, Ell C, Stolte M, Weber A, et al. (2007) Methylation of SOCS-3 and SOCS-1 in the carcinogenesis of Barrett's adenocarcinoma. Gut 56: 1047–1053. 10.1136/gut.2006.111633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pierconti F, Martini M, Pinto F, Cenci T, Capodimonti S, Calarco A, et al. (2011) Epigenetic silencing of SOCS3 identifies a subset of prostate cancer with an aggressive behavior. Prostate 71: 318–325. 10.1002/pros.21245 [DOI] [PubMed] [Google Scholar]
- 46.Jaffe AE, Irizarry RA (2014) Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol 15: R31 10.1186/gb-2014-15-2-r31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Houseman EA, Molitor J, Marsit CJ (2014) Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics 30: 1431–1439. 10.1093/bioinformatics/btu029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, et al. (2005) Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A 102: 15785–15790. 10.1073/pnas.0507816102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, et al. (2013) Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLOS Genet 9: e1003588 10.1371/journal.pgen.1003588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfeiffer L, Wahl S, Pilling LC, Reischl E, Sandling JK, Kunze S, et al. (2015) DNA methylation of lipid-related genes affects blood lipid levels. Circ Cardiovasc Genet 8: 334–342. 10.1161/CIRCGENETICS.114.000804 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File A in S1 File. Significant associations (Benjamini Hochberg corrected) between CRP and DNA methylation sites in the KORA F4 discovery study. File B in S1 File. Study Populations. File C in S1 File. Measurement of CRP. File D in S1 File. Assessment of DNA methylation data in KORA F4, EPICOR, and TwinsUK using the Illumina HumanMethylation450 BeadChip. Table A in S1 File. Associations between CRP and DNA methylation sites in the KORA F4 discovery study (n = 1741) after additional adjustments. Table B in S1 File. Significant associations (Benjamini Hochberg corrected) between CRP and DNA methylation sites in the KORA F4 discovery study. Table C in S1 File. Associations between CRP and DNA methylation sites in the LMU liver cohort. Table D in S1 File. Sequences of PCR tagged primers used for EpiTYPER methylation analysis, product size of each amplicon, and informative CpG sites per amplicon. Table E in S1 File. Primers and probes for quantitative PCRs. Figure A in S1 File. Expression of AQP3 and SOCS3 (normalized to μg input RNA) in different human tissues (human brain, heart, lung, kidney, small intestine, adipose tissue, skeletal muscle) and blood cell types (peripheral blood mononuclear cells (PBMC), CD14-, CD19-, CD3-, CD4-, CD8-positive cells, and regulatory T-cells).
(DOCX)
Data Availability Statement
The informed consent given by KORA, EPIC-Italy, and LMU study participants does not cover data posting in public databases. However, data are available upon request. In detail, KORA data are available from KORA-gen (http://www.helmholtz-muenchen.de/kora-gen) by means of a project agreement. Requests should be sent to kora.passt@helmholtz-muenchen.de and are subject to approval by the KORA Board. Requests for LMU data should be sent to Lesca.Holdt@med.uni-muenchen.de. EPICOR data are available upon request from HuGeF (http://www.hugef.org) by means of a project agreement. Requests should be sent to info@hugef-torino.org and are subject to approval by the EPICOR-HuGeF Board. The ArrayExpress accession number for the TwinsUK adipose methylation data set is E-MTAB-1866.