Abstract
Creation of wheat-alien disomic addition lines and localization of desirable genes on alien chromosomes are important for utilization of these genes in genetic improvement of common wheat. In this study, wheat-Agropyron cristatum derivative line 5113 was characterized by genomic in situ hybridization (GISH) and specific-locus amplified fragment sequencing (SLAF-seq), and was demonstrated to be a novel wheat-A. cristatum disomic 6P addition line. Compared with its parent Fukuhokomugi (Fukuho), 5113 displayed multiple elite agronomic traits, including higher uppermost internode/plant height ratio, larger flag leaf, longer spike length, elevated grain number per spike and spikelet number per spike, more kernel number in the middle spikelet, more fertile tiller number per plant, and enhanced resistance to powdery mildew and leaf rust. Genes conferring these elite traits were localized on the A. cristatum 6P chromosome by using SLAF-seq markers and biparental populations (F1, BC1F1 and BC1F2 populations) produced from the crosses between Fukuho and 5113. Taken together, chromosomal localization of these desirable genes will facilitate transferring of high-yield and high-resistance genes from A. cristatum into common wheat, and serve as the foundation for the utilization of 5113 in wheat breeding.
Introduction
Common wheat (Triticum aestivum, genome AABBDD) has a large number of wild relatives, which are considered as valuable gene resources for wheat genetic improvement [1]. Wheat-alien disomic addition lines are usually considered as key materials to transfer alien superior genes into common wheat, which play important roles in numerous applications, such as investigating genetic effects of alien chromosomes under wheat background, determining chromosomal localization of some desirable genes, purifying alien chromosomes or chromosome arms via flow sorting followed by construction of BAC libraries [2,3]. Therefore, creation of wheat-alien disomic addition lines and chromosomal localization of their elite genes are important for wheat genetic improvement. Thus far, plenty of wheat-alien addition lines derived from multiple species of Triticeae have been obtained. These Triticeae species include Hordeum vulgare L. [4–7], Secale cereale L. [8,9], Aegilops speltoides [10,11] and Haynaldia villosa [12,13] and so on. However, most wheat-alien disomic addition lines could not be directly used in wheat breeding due to the presence of linkage drag. Therefore, they are usually induced to produce various translocation or introgression lines, such as wheat-Psathyrostachys huashanica translocation line H9020-17-15 [14], wheat Haynaldia villosa translocation line 6VS/6AL [15], wheat-rye translocation line 1BL/1RS [16], and wheat-Thinopyrum pointicum introgression lines II-1-8 and II-2 [17]. Before addition lines are induced to produce translocation or introgression lines, localization of genes conferring elite agronomic traits were usually considered as a fundamental prerequisite.
Morphological, biochemical and molecular markers are indispensable in localizing alien genes on chromosomes. Compared with other markers, PCR-based molecular markers are much more popular due to their high efficiency and throughput. Specific-locus amplified fragment sequencing (SLAF-seq) marker is one of the PCR-based markers, which is developed from the SLAF-seq technology [18]. SLAF-seq markers have been widely used in chromosomal localization of alien genes [19,20], determination of homoeologous relationships for alien chromosomes [21,22], and genetic map constructions [23–25]. For example, 89 SLAF-seq markers specific to Th. elongatum 7E chromosome were successfully developed by the SLAF-seq technology [21]; the genetic maps of common carp (Cyprinus carpio L.), sesame (Sesamum indicum L.) and soybean (Glycine max (L.) Merr.) were constructed by the SLAF-seq technology [23–25]. In addition, a high-density genetic map of Agropyron Gaertn. was constructed by the SLAF-seq technology in our previous study [26].
Agropyron cristatum (L.) Gaertn. (2n = 4x = 28; genome PPPP) is a wild relative of wheat, and displays several superior traits, such as higher grain numbers, multiple fertile tiller numbers, and elevated resistance to biotic and abiotic stresses [27,28]. Pioneering work on the utilization of A. cristatum have been carried out nearly three decades ago, and a series of wheat-A. cristatum derivative lines have been obtained from the cross between common wheat cv. Fukuhokomugi (Fukuho, 2n = 6x = 42; AABBDD) and A. cristatum [29,30], including addition, translocation and introgression lines [31–35]. 5113 was one of the wheat-A. cristatum disomic addition lines, and the two added alien A. cristatum chromosomes were homoeologous to group 6 of common wheat. 5113 displays multiple elite agronomic traits such as higher yield and disease resistance. However, whether these elite traits were conferred by desirable genes located on 6P chromosomes has not been determined. Therefore, these desirable genes were localized using SLAF-seq markers and genetic populations in this study.
Materials and Methods
Materials
5113 was originally produced from the cross between A. cristatum accession Z559 and Fukuho, followed by self-pollination for six generations [31]. F1, BC1F1 and BC1F2 populations were then constructed from the crosses between 5113 and Fukuho, using Fukuho as the recurrent parent. Different types of wheat-A. cristatum disomic addition lines, each containing only one pair of P chromosomes, were used as controls to determine the homoelogy relationship of 5113 with wheat chromosomes. These wheat-A. criatatum disomic addition lines were as follows: II-3-1a (containing one pair of 1P chromosomes), II-9-3 (containing one pair of 2P chromosomes), II-11-1a (containing one pair of 3P chromosomes), II-21-2 (containing one pair of 4P chromosomes), II-11-1b (containing one pair of 5P chromosomes), II-5-1 (containing one pair of 7P chromosomes). Besides, Wheat-A. cristatum disomic 6P addition line 4844–12 was specially chosen as the control to compare the differences of 6P chromosomes. All the materials were provided by the Crop Germplasm Resources Research Center of the Institute of Crop Science, Chinese Academy of Agricultural Sciences (CAAS), Beijing, China.
Methods
GISH analysis and meiosis observation
Genomic in situ hybridization (GISH) was used to analyze the chromosomal composition of 5113 and the progenies of the genetic populations as previously described [33]. Genomic DNA was isolated using the modified CTAB method [36]. Agropyron cristatum DNA (labeled with Dig-Nick-Translation Mix) (Roche, Mannheim, Germany) and Fukuho genomic DNA was used as the probe and blocker, respectively. For meiosis studies, the procedures were described by Jauhar and Peterson [37]. Briefly, pollen mother cells (PMCs) at metaphase I (MI) stage were fixed in Carnoy’s solution (ethanol:chloroform:acetic acid, 6:3:1, by volume) for 24 h. Wheat and A. cristatum chromosomes were pseudo-colored as blue and red, respectively. All cytological images were taken under an OLYMPUS AX80 (Olympus Corporation, Tokyo, Japan) fluorescence microscope and captured with a CCD camera (Diagnostic Instruments, Sterling Heights, MI, USA).
SLAF library construction and data analysis
The SLAF library of 5113 was constructed as previously described by Sun et al. [23]. Briefly, 2 ug genomic DNA of 5113 was digested with NlaIII/MseI (New England Biolabs, Beverly, MA, USA). Fragments between 200 and 300 bp (not including adapter sequence indexes and adaptors) were isolated, and then subjected to PCR for paired-end sequencing (40bp each end) by Illumina HiSeq 2500 (Illumina, San Diego, CA, USA) at Biomarker Technologies Corporation in Beijing, China. Analysis of these SLAF-seq data were conducted as follows: Firstly, reads were filtered based on their quality, leaving high-quality clean reads; high-quality reads with high sequence depth (≧ 3) and similarity (≧ 95%) were clustered into single SLAF groups by using BLAT software (-tileSize = 10, -stepSize = 5); Secondly, six reads at most were selected as the representatives of each SLAF group (referred to as SLAF-tagI). Thirdly, all the SLAF-tagIs were blasted with wheat genomes, and those with higher identity (> 90%) were considered from wheat genome and then filtered out; only those with lower identity (< 90%) were referred to as SLAF-tagII. Fourthly, all the SLAF-tagIIs were blasted with the SLAF markers located on the genetic map of Agropyron Gaertn. published before [26], and only those with higher identity (> 90%) were considered as the SLAF-tags specific to A. cristatum 6P chromosome in 5113.
Development of SLAF-seq markers
In order to trace A. cristatum 6P chromosomal segments effectively, SLAF-seq markers were developed based on the sequences of SLAF-tags specific to A. cristatum 6P chromosome in 5113, PCR was conducted as previously described by Lu et al. [34], and the PCR products were separated on 0.8% agarose gel.
Evaluation of agronomic traits
All the materials were planted in 2.0 m rows, spaced 30 cm apart at Xinxiang experiment station in Henan province, China. A number of agronomic traits were evaluated, including plant height, uppermost internode, flag leaf size, grain number per spike, spikelets per spike, thousand-grain weight, and disease responses. A mixture of currently prevalent isolates of Blumeria graminis f. sp. tritici was used to evaluate the powdery mildew resistance of wheat plants at the adult stage, and disease responses were scored as described by Sheng et al. [38]. For leaf rust resistance, mixed P. triticina pathotypes isolates were used [39,40], and disease responses were recorded with 0–4 raking, where 0–2 were considered resistant and 3–4 susceptible [41]. Statistical analysis system software version 9.2 was used for statistical analyses (SAS Institute, Cary, NC, USA).
Results
Cytological characterization of 5113
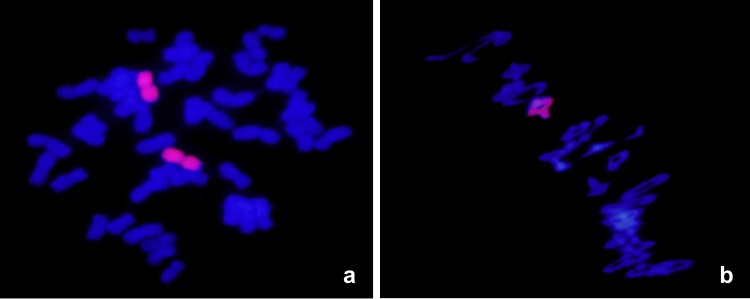
To cytologically characterize the genetic constitution of 5113, root cells and pollen mother cells (PMCs) were used (Fig 1 and Table 1). There were 44 chromosomes in the root cells of 5113, including 42 wheat chromosomes (shown in blue) and two A. cristatum chromosomes (shown in red) (Fig 1A). Chromosomal configuration at the metaphase I of 120 PMCs were investigated in 5113, showing 2n = 22 II, with averages of 0.6 univalents, 2.29 rod bivalents, and 19.41 ring bivalents (Table 1). Twenty-one wheat bivalents (shown in blue) and one A. cristatum bivalent (shown in red) were shown in Fig 1B, showing regular chromosome pairing behaviors. Taken together, all these results indicated that 5113 was a stable wheat-A. cristatum disomic addition line.
Fig 1. GISH analysis of the root cells and pollen mother cells of 5113.
(a), GISH analysis showing that 5113 contained 44 chromosomes including 42 wheat chromosomes (shown in blue) and two A. cristatum chromosomes (shown in red); (b), 5113 possessed 22 bivalents including 21 wheat bivalents (shown in blue) and one A. cristatum bivalent (shown in red) at metaphase I (MI) stage of the pollen mother cells (PMCs).
Table 1. Chromosome pairing during metaphase I in the pollen mother cells of 5113 and Fukuho.
| Materials | No. of chromosomes | No. of observed cells | Chromosome configuration | |||
|---|---|---|---|---|---|---|
| Univalent | Bivalent | |||||
| Rod | Ring | Total | ||||
| 5113 | 44 | 120 | 0.6 | 2.29 | 19.41 | 21.7 |
| (0–2) | (0–3) | (19–22) | (21–22) | |||
| Fukuho | 42 | 120 | 0.06 | 1.95 | 19.32 | 20.97 |
| (0–2) | (1–4) | (17–21) | (20–21) | |||
Development of SLAF-seq markers specific to 6P chromosome using the SLAF-seq technology
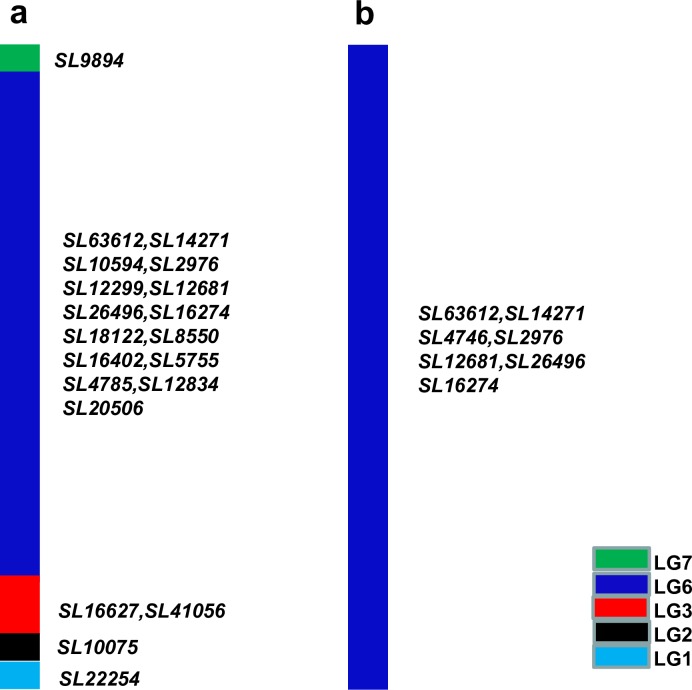
In order to acquire P chromosome-specific markers, the SLAF-seq technology was used on 5113. Meanwhile, 4844–12 was chosen as the control. 11,367,661 clean reads (clustered into 50,468 SLAF groups) and 747,660 clean reads (clustered into 27,379 SLAF groups) were acquired from 5113 and 4844–12, respectively. Six reads at most were selected from each SLAF group as representatives, and then 28,948 and 11,454 reads were acquired from 5113 and 4844–12, respectively (thereafter referred to as SLAF-tagI). All the SLAF-tagI sequences were used as queries to search against the wheat genome, and 4,295 and 1,091 tags with lower identity (< 90%) to their hits were referred to as SLAF-tagIIs. When SLAF-tagII sequences were used as queries to search SLAF markers located on the genetic map of Agropyron Gaertn., 20 and seven SLAF-tagIIs with high identity (≧ 90%) were obtained from 5113 and 4844–12, respectively (Fig 2). All the seven SLAF-tags from 4844–12 were located on the linkage group (LG) 6 of Agropyron Gaertn., indicating that 4844–12 was a 6P disomic addition line. The result was consistent with before [31]. 15 out of 20 SLAF-tags (accounting for 75%) from 5113 were localized on LG6, and others were localized on other different LGs (Fig 2A), such as one tag on LG1, one tag on LG2, two tags on LG3, and one tag on LG7. These results suggested that 5113 was also a 6P disomic addition line, but it contained 6P chromosomes different from those of 4844–12.
Fig 2.
SLAF-tags acquired from 5113 (a) and 4844–12 (b).
Evaluation of the agronomic traits of 5113, Fukuho, and 4844–12
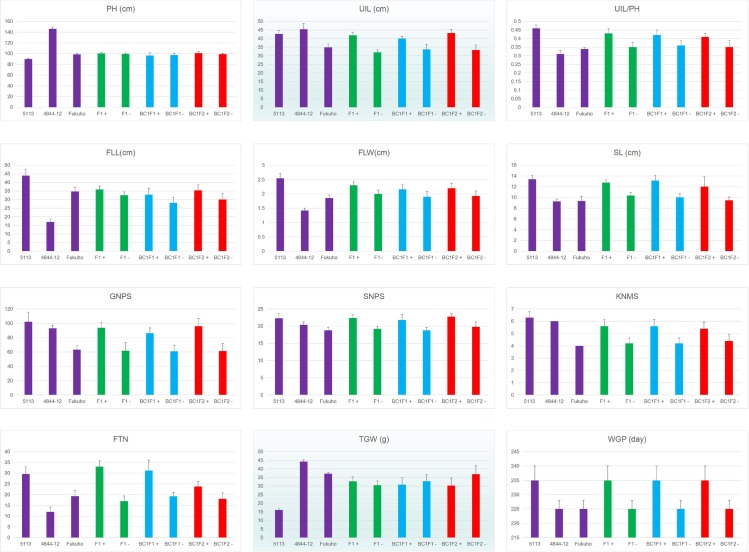
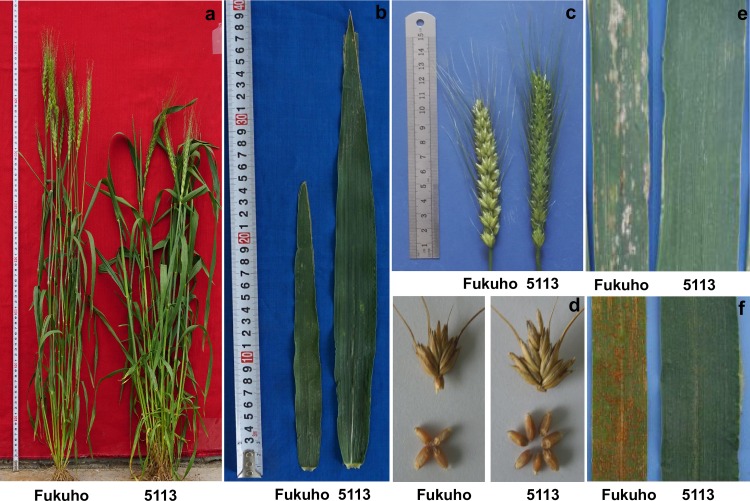
A number of important agronomic traits were investigated in 5113, common wheat cv. Fukuho and wheat-A. cristatum 6P addition line 4844–12. As shown in Fig 3 and S1 Table, 5113 showed obviously different agronomic traits from those of Fukuho and 4844–12. Compared with Fukuho, 5113 showed higher ratio between uppermost internode and plant height, larger flag leaf, longer spike length, more fertile tiller number per plant, more spikelet number per spike, more grains per spike and more kernels in the middle spikelet. Besides, 5113 displayed enhanced resistance to powdery mildew and leaf rust, lower thousand-grain weight, and longer whole growth period compared with Fukuho. Compared with the wheat-A. cristatum 6P addition line 4844–12, 5113 still exhibited higher ratio between uppermost internode and plant height, larger flag leaf, longer spike length, more spikelet number per spike, more fertile tiller number, lower thousand-grain weight and longer whole growth period. The morphologies of whole plants, flag leaves, spikes, spikelets with grains from 5113 and Fukuho were shown in Fig 4A–4D. The responses to powdery mildew and leaf rust were presented in Fig 4E and 4F. All these results suggested that 5113 was a novel wheat-A. cristatum disomic 6P addition line different from 4844–12, which possessed multiple desirable traits that could be used for wheat genetic improvement.
Fig 3. Histograms showing the agronomic traits of 5113, Fukuho, 4844–12 and genetic populations produced from the crosses between 5113 and Fukuho.
PH, Plant height; UIL, Uppermost internode length; FLL, Flag leaf length; FLW, Flag leaf width; SL, Spike length; GNPS, Grain number per spike; SNPS, Spikelet number per spike; KNMS, Kernel number in the middle spikelet; TGW, Thousand-grain weight; WGP, Whole growth period; FTN, Fertile tiller number per plant.
Fig 4. Morphological traits of 5113 and Fukuho.
whole plants (a), flag leaves (b), spikes (c), spikelets with grains (d), disease responses to powdery mildew (e) and leaf rust (f).
A. cristatum 6P chromosomal segments could be traced by SLAF-seq markers
Based on the sequences of 15 SLAF-tags on LG6, 15 SLAF-seq markers were developed. The primer sequences of these markers were listed in S2 Table. In order to test whether these markers were specific to A. cristatum 6P chromosome, they were used to amplify target fragments using the genomic DNA of A. cristatum, Fukuho, and different types of disomic addition lines as templates. All the 15 SLAF-seq markers were present in A. cristatum and 5113 but absent in Fukuho and other addition lines (Fig 5), suggesting that these markers were A. cristatum 6P chromosome-specific. In order to further test whether these SLAF-seq markers could trace the alien A. cristatum 6P chromosomal segments instead of GISH, 15 SLAF-seq markers were used to genotype multiple genetic populations derived from 5113 and Fukuho (F1, BC1F1 and BC1F2 populations, with Fukuho as the recurrent parent), followed by GISH analysis. All the progenies positive for the 15 SLAF-seq markers showed GISH signals, and all the progenies with GISH signals were positive for the 15 SLAF-seq markers (Fig 6). These results suggested that the 15 SLAF-seq markers could be applied to trace 6P chromosomal segments instead of GISH.
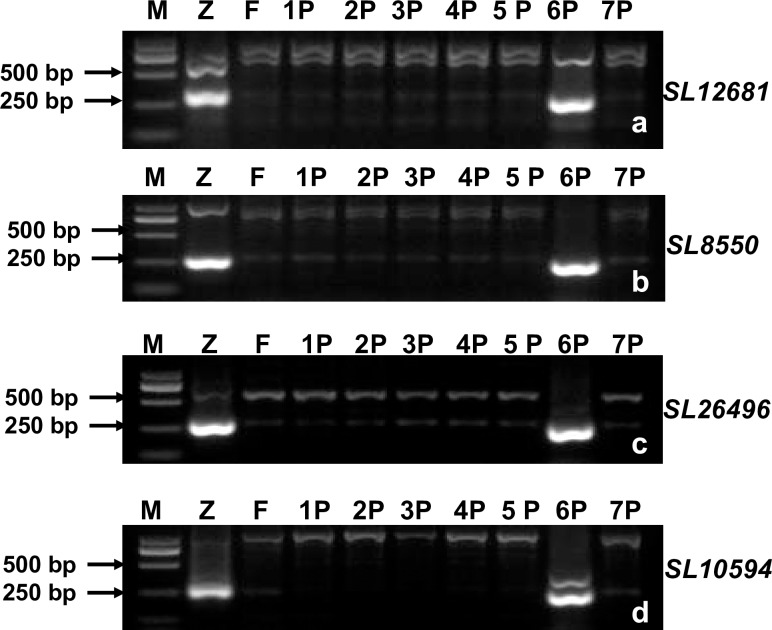
Fig 5. Development of SLAF-seq markers specific for A. cristatum 6P chromosome.
(a-d), PCR patterns of four SLAF-seq markers SL12681 (a), SL8550 (b), SL26496 (c) and SL10594 (d) M, Marker; Z, A. cristatum accession Z559; F, Fukuho; 1P-7P, wheat-A. cristatum disomic addition lines, each of which contained only one pair of P chromosomes (from 1P to 7P).
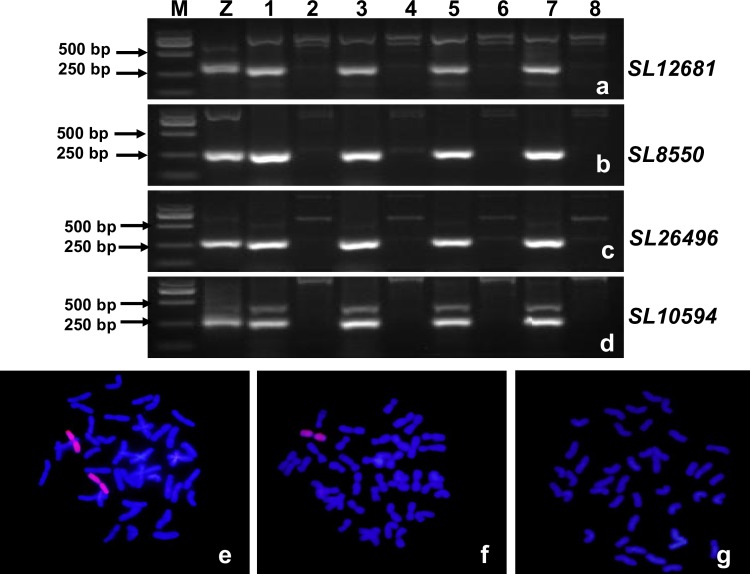
Fig 6. Genotyping of the progenies of the genetic populations using SLAF-seq markers and GISH.
(a-d), PCR patterns of four SLAF-seq markers SL12681 (a), SL8550 (b), SL26496 (c) and SL10594 (d) M, Marker; Z, A. cristutum accession Z559; 1, 5113; 2, Fukuho; 3, 5113 × Fukuho F1 plant with 6P chromosome; 4, 5113 × Fukuho F1 plant without 6P chromosome; 5, One progeny from the 5113 × Fukuho BC1F1 population with 6P chromosome; 6, One progeny from the 5113 × Fukuho BC1F1 population without 6P chromosome; 7, One progeny from the 5113 × Fukuho BC1F2 population with 6P chromosome; 8, One progeny from the 5113 × Fukuho BC1F2 population without 6P chromosome; e-g, GISH analysis of the progenies with three genotypes containing 44 (e), 43(f) and 42 (g) chromosomes, respectively.
Chromosomal localization of the desirable genes using genetic populations
In order to explore the source of the genes conferring the desirable traits in 5113, genotypic and phenotypic data were analyzed in F1, BC1F1 and BC1F2 populations. As shown in Fig 3 and S1 Table, progenies of three populations could be categorized into two types based on the genotypic data: progenies with SLAF-seq markers and progenies without SLAF-seq markers. The progenies positive for SLAF-seq markers displayed obvious differences from those negative for SLAF-seq markers in nine yield-related traits, including the ratio between uppermost internode length and plant height ratio, flag leaf length, flag leaf width, spike length, grain number per spike, spikelet number per spike, kernel number in the middle spikelet, fertile tiller number per plant and whole growth period. All the progenies with SLAF-seq markers and 5113 exhibited similar phenotypes in nine traits, while all the progenies negative for SLAF-seq markers and Fukuho exhibited similar phenotypes. All these results suggested that genes conferring these traits were located on the A. cristatum 6P chromosomes of 5113. Besides, genes conferring powdery mildew and leaf rust resistance were also proved to be located on the A. cristatum 6P chromosomes of 5113. However, there were no significant differences in two traits (plant height and thousand-grain weight) between plants positive and negative for SLAF-seq markers, suggesting that genes conferring these two traits were not located on A. cristatum 6P chromosomes of 5113.
Discussion
The alien chromosome-specific markers could be effectively developed by the SLAF-seq technology
After introduction of alien chromosomes into the wheat genome, cytological methods can be used to detect the alien chromosomal fragments. However, utilization of these methods has been limited by the fact that they are time-consuming and labor-intensive. Recently, SLAF-seq markers have been successfully applied in detecting alien chromosomal fragments, due to their high throughput, high accuracy and low-cost [23]. In this study, 15 SLAF-seq markers specific to A. cristatum 6P chromosome were developed by the SLAF-seq technology. In the 5113 × Fukuho genetic populations, the presence/absence of SLAF-seq markers were consistent with the presence/absence of GISH signals, indicating that SLAF-seq markers can be used to detect the existence of A. cristatum 6P chromosome instead of GISH. In our further studies, these SLAF-seq markers will be used to trace A. cristatum 6P chromosomal segments in the translocation and introgression lines induced from 5113.
5113 was a novel wheat-A. cristatum disomic 6P addition line different from 4844–12
Six wheat-A. cristatum disomic 6P addition lines have been acquired in our laboratory, including 5113, 4844–12, 5114, 5106, II-26, and II-29-2i [31]. Although all the added pairs of A. cristatum chromosomes in these addition lines were homoeologous with the homoeologous group 6 of wheat, their genetic constitutions exhibited varying degrees of differences. In this study, differences between 5113 and 4844–12 were revealed by both genotyping and phenotyping. Compared with Fukuho, both 5113 and 4844–12 showed higher grain number, and resistance to powdery mildew and leaf rust. However, 5113 showed higher ratio between uppermost internode and plant height, larger flag leaf, longer spike length, more spikelet number per spike, more fertile tiller number, lower thousand-grain weight and longer whole growth period, compared with 4844–12. These differences between 4844–12 and 5113 were mainly attributed to the genetic rearrangements of A. cristatum 6P chromosomes. Similar phenomena were also reported in other species, such as Leymus racemosus [42], Thinopyrum intermedium [43], and rye [44]. Therefore, 5113 was a novel wheat-A. cristatum disomic 6P addition line different from 4844–12, which could be used as a starting material to produce novel wheat germplasms (translocation and introgression lines).
5113 could be used to transfer high-yield and disease resistance genes from A. cristatum into common wheat
Wheat-alien disomic addition lines are usually used as the starting material to transfer desirable agronomic traits into common wheat through chromosome engineering [45–48]. In this study, genes conferring multiple elite agronomic traits were located on the A. cristatum 6P chromosome of 5113 by using three different genetic populations (F1, BC1F1 and BC1F2 populations). In order to transfer these desirable genes from A. cristatum into common wheat, 5113 will be induced by 60Co-γ rays or homoeologous pairing induction. Translocation lines with small 6P chromosomal fragments are preferred to transfer elite genes into wheat. There were several examples in which translocation lines were successfully induced from their corresponding addition lines. For example, wheat-Haynaldia villosa translocation line T4VS⋅4DL induced from the disomic chromosome addition line DA4V were reported to display high resistance to wheat spindle streak mosaic virus [49]; wheat-Haynaldia villosa translocation line T2VS⋅2DL induced from the T. durum-D. villosum amphiploid was found with longer spikes and more kernels [50]; wheat-Thinopyrum bessarabicum translocation line T2JS-2BS⋅2BL induced from the CS-Th. bessarabicum alien telosomic addition line TJ04 displayed more fertile spikes per plant, longer spikes, more grains per spike and higher yield per plant [51]. Successful applications of translocation lines mentioned above were based on the knowledge of their genetic constitutions and agronomic traits. Therefore, chromosomal localization of these desirable genes is a fundamental prerequisite for the utilization of 5113 to enrich the germplasm resources for wheat breeding.
Supporting Information
(XLS)
(XLS)
Acknowledgments
This work was funded by the National Key Technology Support Program of China (No. 2013BAD01B02), the National Natural Science Foundation of China (No. 31271714), and the CAAS Innovation Team Project.
Abbreviations
- Fukuho
Fukuhokomugi
- GISH
Genomic in situ hybridization
- LGs
Linkage groups
- MI
Metaphase
- PMCs
Pollen mother cells
- SLAF-seq
Specific-locus amplified fragment sequencing
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by the National Key Technology Support Program of China (No. 2013BAD01B02), the National Natural Science Foundation of China (No. 31271714), and the CAAS Innovation Team Project.
References
- 1.Riley R, Chapman V, Johnson R. Introduction of yellow rust resistance of Aegilops comosa into wheat by genetically induced homoeologous recombination. Nature. 1968; 217: 383–384. [Google Scholar]
- 2.Jaroslav Doležel, Marie Kubaláková, Jan Bartoš, Jiří Macas. Flow cytogenetics and plant genome mapping. Chromosom Res. 2004; 12 (1): 77–91. [DOI] [PubMed] [Google Scholar]
- 3.Suchánková P, Kubaláková M, Kovářová P, Bartoš J, Číhalíková J, Molnár-Láng M, et al. Dissection of the nuclear genome of barley by chromosome flow sorting. Theor Appl Genet. 2006; 113 (4): 651–659. 10.1007/s00122-006-0329-8 [DOI] [PubMed] [Google Scholar]
- 4.Murai K, Koba T, Shimada T. Effects of barley chromosome on heading characters in wheat-barley chromosome addition lines. Euphytica. 1997; 96 (2): 281–287. [Google Scholar]
- 5.Rubiales D, Moral A, Martin A. Chromosome location of resistance to septoria leaf blotch and common bunt in wheat-barley addition lines. Euphytica. 2001; 122 (2): 369–372. [Google Scholar]
- 6.Bilgic H, Cho S, Garvin DF, Muehlbauer GJ. Mapping barley genes to chromosome arms by transcript profiling of wheat-barley ditelosomic chromosome addition lines. Genome. 2007; 50: 898–906. 10.1139/g07-059 [DOI] [PubMed] [Google Scholar]
- 7.Darko E, Janda T, Majláth I, Szopkó D, Dulai S, Türkösi E, et al. Salt stress response of wheat-barley addition lines carrying chromosomes from the winter barley “Manas”. Euphytica. 2015; 203 (3): 491–504. [Google Scholar]
- 8.Khlestkina EK, Antonova EV, Pershina LA, Soloviev AA, Badaeva ED, Börner A, et al. Variability of Rc (red coleoptile) alleles in wheat and wheat-alien genetic stock collections. Cereal Res Commun. 2011; 39 (4): 465–474. [Google Scholar]
- 9.Nguyen V, Fleury D, Timmins A, Laga H, Hayden M, Mather D, et al. Addition of rye chromosome 4R to wheat increases anther length and pollen grain number. Theor Appl Genet. 2015; 128 (5): 953–964. 10.1007/s00122-015-2482-4 [DOI] [PubMed] [Google Scholar]
- 10.Schneider A, Molnár I, Molnár-Láng M. Utilisation of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica. 2008; 163 (1): 1–19. [Google Scholar]
- 11.Zhou JP, Yao CH, Yang EN, Yin MQ, liu C, Ren ZL. Characterization of a new wheat-Aegilops biuncialis addition line conferring quality-associated HMW glutenin subunits. Genet Mol Res. 2014; 13 (1): 660–669. 10.4238/2014.January.28.11 [DOI] [PubMed] [Google Scholar]
- 12.Chen Q, Conner RL, Laroche A. Molecular characterization of Haynaldia villosa chromatin in wheat lines carrying resistance to wheat curl mite colonization. Theor Appl Genet. 1996; 93 (5–6): 679–684. 10.1007/BF00224062 [DOI] [PubMed] [Google Scholar]
- 13.Li HJ, Conner RL, Chen Q, Jia X, Li H, Graf RJ, et al. Different reactions to the wheat curl mite and wheat streak mosaic virus in various wheat-Haynaldia villosa 6V and 6VS lines. Plant dis. 2002; 86 (4): 423–428. [DOI] [PubMed] [Google Scholar]
- 14.Cao ZJ, Deng ZY, Wang MN, Wang XP, Jing JX, Zhang XQ, et al. Inheritance and molecular mapping of an alien stripe-rust resistance gene from a wheat-Psathyrostachys huashanica translocation line. Plant Sci. 2008; 174: 544–549. [Google Scholar]
- 15.Qi LL, Cao MS, Chen PD, Li WL, Liu DJ. Identification, mapping, and application of polymorphic DNA associated with resistance gene Pm21 of wheat. Genome. 1996; 39 (1): 191–197. [DOI] [PubMed] [Google Scholar]
- 16.Cai XW, Liu DJ. Identification of a 1B/1R wheat-rye chromosome translocation. Theor Appl Genet. 1989; 77 (1): 81–83. 10.1007/BF00292320 [DOI] [PubMed] [Google Scholar]
- 17.Chen SY, Xia GM, Quan TY, Xiang FN, Jin Y, Chen HM. Introgression of salt-tolerance from somatic hybrids between common wheat and Thinopyrum ponticum. Plant Sci. 2004; 167: 773–779. [Google Scholar]
- 18.Hyten DL, Cannon SB, Song QJ, Weeks N, Fickus EW, Shoemaker RC, et al. High-throughput SNP discovery through deep resequencing of a reduced representation library to anchor and orient scaffolds in the soybean whole genome sequence. BMC Genom. 2010; 11 (1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang P, Zhu YQ, Wang LL, Chen LP, Zhou SJ. Mining candidate genes associated with powdery mildew resistance in cucumber via super-BSA by specific length amplified fragment (SLAF) sequencing. BMC Genom. 2015; 16 (1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao T, Jiang J, Liu G, He SS, Zhang H, Chen XL, et al. Mapping and candidate gene screening of tomato Cladosporium fulvum-resistant gene Cf-19, based on high-throughput sequencing technology. BMC Plant Biol. 2016; 16 (1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen SQ, Huang ZF, Dai Y, Qin SW, Gao YY, Zhang LL, et al. The development of 7E chromosome-specific molecular markers for Thinopyrum elongatum based on SLAF-seq technology. PLoS One. 2013; 8(6): e65122 10.1371/journal.pone.0065122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Tang ZX, Qiu L, Wang YY, Tang SY, Fu SL. Identification and physical mapping of new PCR-based markers specific for the long arm of rye (Secale cereale L.) chromosome 6. J Genet Genomics. 2016; 43: 209–216. 10.1016/j.jgg.2015.11.005 [DOI] [PubMed] [Google Scholar]
- 23.Sun XW, Liu DY, Zhang XF, Li WB, Liu H, Hong WG, et al. SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS One. 2013; 8 (3): e58700 10.1371/journal.pone.0058700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang YX, Wang LH, Xin HG, Li DH, Ma CX, Ding X, et al. Construction of a high-density genetic map for sesame based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Plant Biol. 2013; 13 (1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi ZM, Huang L, Zhu RS, Xin DW, Liu CY, Han X, et al. A high-density genetic map for soybean based on specific length amplified fragment sequencing. PLoS One. 2014; 9 (8): e104871 10.1371/journal.pone.0104871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Zhang JP, Huang L, Gao AN, Zhang J, Yang XM, et al. A high-density genetic map for P genome of Agropyron Gaertn. based on specific-locus amplified fragment sequencing (SLAF-seq). Planta. 2015; 242 (6): 1335–1347. 10.1007/s00425-015-2372-7 [DOI] [PubMed] [Google Scholar]
- 27.Dewey DR. The genomic system of classification as a guide to intergeneric hybridization with the perennial Triticeae Springer US; 1984. [Google Scholar]
- 28.Dong YC, Zhou RH, Xu SJ, Cauderon Y, Wang RC. Desirable characteristics in perennial Triticease collected in China for wheat improvement. Hereditas. 1992; 116 (s1): 175–178. [Google Scholar]
- 29.Li LH, Li XQ, Li P, Dong YC, Zhao GS. Establishment of wheat-Agropyron cristatum alien addition lines. I. Cytology of F3, F2 BC1, BC4, and BC3F1 progenies. Acta Genetica Sinica. 1997; 24: 154–159. [Google Scholar]
- 30.Li LH, Yang XM, Zhou RH, Li XQ, Dong YC. Establishment of wheat-Agropyron cristatum alien addition lines II. Identification of alien chromosomes and analysis of development approaches. Acta Genetica Sinica. 1998; 25: 538–544. [Google Scholar]
- 31.Han HM, Bai L, Su JJ, Zhang JP, Song LQ, Gao AN, et al. Genetic rearrangements of six wheat-Agropyron cristatum 6P addition lines revealed by molecular markers. PloS One. 2014; 9 (3): e91066 10.1371/journal.pone.0091066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li QF, Lu YQ, Pan CL, Zhang JP, Liu WH, Yang XM, et al. Characterization of a novel wheat-Agropyron cristatum 2P disomic addition line with powdery mildew resistance. Crop sci. 2016; 56 (5): 2390–2400. [Google Scholar]
- 33.Ye XL, Lu YQ, Liu WH, Chen GY, Han HM, Zhang JP, et al. The effects of chromosome 6P on fertile tiller number of wheat as revealed in wheat-Agropyron cristatum chromosome 5A/6P translocation lines. Theor Appl Genet. 2015; 128 (5): 797–811. 10.1007/s00122-015-2466-4 [DOI] [PubMed] [Google Scholar]
- 34.Lu YQ, Wu XY, Yao MM, Zhang JP, Liu WH, Yang XM, et al. Genetic mapping of a putative Agropyron cristatum-derived powdery mildew resistance gene by a combination of bulked segregant analysis and single nucleotide polymorphism array. Mol Breed. 2015; 35 (3): 1–13. [Google Scholar]
- 35.Lu YQ, Yao MM, Zhang JP, Song LQ, Liu WH, Yang XM, et al. Genetic analysis of a novel broad-spectrum powdery mildew resistance gene from the wheat-Agropyron cristatum introgression line Pubing 74. Planta. 2016; 1–11. [DOI] [PubMed] [Google Scholar]
- 36.Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 2006; 1: 2320–2325. 10.1038/nprot.2006.384 [DOI] [PubMed] [Google Scholar]
- 37.Jauhar PP, Peterson TS. Cytological analyses of hybrids and derivatives of hybrids between durum wheat and Thinopyrum bessarabicum, using multicolour fluorescent GISH. Plant Breed. 2006; 125:19–26. [Google Scholar]
- 38.Sheng BQ, Duan XY. Improvement of scale 0–9 method for scoring adult plant resistance to powdery mildew of wheat. Beijing Agricultural Sciences. 1991; 1: 38–39. [Google Scholar]
- 39.Zhao PP, Meng QF, Guo N, Zhang LY, Yan HF, Liu DQ. Analysis of virulence patterns of Puccinia triticina in Henan province in 2009–2011. Journal of Henan Agricultural Sciences. 2013; 42 (4): 91–94. [Google Scholar]
- 40.Song LQ, Lu YQ, Zhang JP, Pan CL, Yang XM, Li XQ, et al. Physical mapping of Agropyron cristatum chromosome 6P using deletion lines in common wheat background. Theor Appl Genet. 2016; 129 (5): 1023–1034. 10.1007/s00122-016-2680-8 [DOI] [PubMed] [Google Scholar]
- 41.Stakman EC, Stewart DM, Loegering WQ. Identification of physiologic races of Puccinia graminis var. tritici Washington, DC: USDA; 1962. [Google Scholar]
- 42.Kishii M, Yamada T, Sasakuma T. Production of wheat-Leymus racemosus chromosome addition lines. Theor Appl Genet. 2004; 109 (2): 255–260. 10.1007/s00122-004-1631-y [DOI] [PubMed] [Google Scholar]
- 43.Li G, Lang T, Dai G. Precise identification of two wheat-Thinopyrum intermedium substitutions reveals the compensation and rearrangement between wheat and Thinopyrum chromosomes. Mol Breed. 2015; 35 (1): 1–10. [Google Scholar]
- 44.Maestra B, De Jong J H, Shepherd K. Chromosome arrangement and behaviour of two rye homologous telosomes at the onset of meiosis in disomic wheat-5RL addition lines with and without the Ph1 locus. Chromosome Res. 2002; 10 (8): 655–667. [DOI] [PubMed] [Google Scholar]
- 45.Friebe B, Jiang J, Raupp WJ, Mclntosh RA, Gill BS. Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica. 1996; 91 (1): 59–87. [Google Scholar]
- 46.Xu SS, Faris JD, Cai X, Klindworth DL. Molecular cytogenetic characterization and seed storage protein analysis of 1A/1D translocation lines of durum wheat. Chromosom Res. 2005; 13 (6): 559–568. [DOI] [PubMed] [Google Scholar]
- 47.Qi LL, Friebe B, Zhang P, Gill BS. Homoeologous recombination, chromosome engineering and crop improvement. Chromosom Res. 2007; 15 (1): 3–19. [DOI] [PubMed] [Google Scholar]
- 48.Niu Z, Klindworth DL, Friesen TL, Chao SM, Jin Y, Cai XW, et al. Targeted introgression of a wheat stem rust resistance gene by DNA marker-assisted chromosome engineering. Genetics. 2011; 187 (4): 1011–1021. 10.1534/genetics.110.123588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang QP, Li Q, Wang XE, Wang HY, Lang SP, Wang YN, et al. Development and characterization of a Triticum aestivum-Haynaldia villosa translocation line T4VS·4DL conferring resistance to wheat spindle streak mosaic virus. Euphytica. 2005; 145 (3): 317–320. [Google Scholar]
- 50.Zhang RQ, Hou F, Feng YG, Zhang W, Zhang MY, Chen PD. Characterization of a Triticum aestivum-Dasypyrum villosum T2VS·2DL translocation line expressing a longer spike and more kernels traits. Theor Appl Genet. 2015; 128 (12): 2415–2425. 10.1007/s00122-015-2596-8 [DOI] [PubMed] [Google Scholar]
- 51.Qi ZJ, Du P, Qian BL, Zhuang LF, Chen HF, Chen TT, et al. Characterization of a wheat-Thinopyrum bessarabicum (T2JS-2BS·2BL) translocation line. Theor Appl Genet. 2010; 121 (3): 589–597. 10.1007/s00122-010-1332-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.