Abstract
There are now over 25 million volunteer donors registered worldwide for patients in need of a life-saving hematopoietic cell transplant to cure blood disorders. Although an HLA-matched donor remains the preferred stem cell source for transplantation, the use of a donor with limited HLA mismatching may be considered. Significant advances in clinical and basic research have been instrumental in furthering the understanding of donor-recipient HLA mismatches that are better tolerated than other mismatches. An increased appreciation of the role of regulatory region variation that affects the level of HLA expression provides new approaches for the selection of HLA-mismatched donors.
Introduction
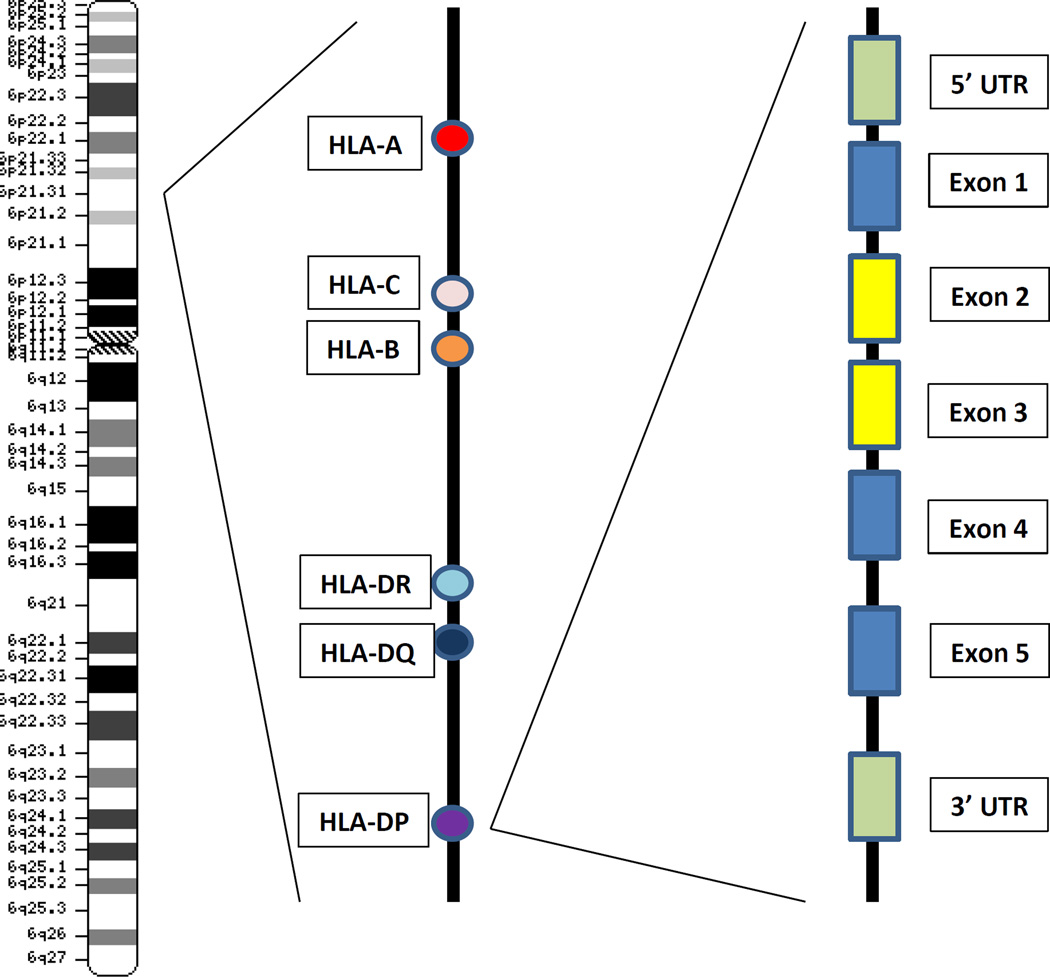
Advances in immunogenomics have propelled the field of allogeneic transplantation into a new era of clinical practice and have furthered understanding of the dynamic range of genes within the major histocompatibility complex (MHC). The best studied MHC-region genes remain the classical HLA genes, HLA-A, B, C, DRB1, DQB1 and DPB1 (Figure 1)1; however, new information on the extent of human MHC diversity and the implications of regulatory region variation has uncovered important biological implications relevant to the alloimmune response in transplantation.
Figure 1.
Map of the Human Major Histocompatibility Complex. The MHC is encoded on chromosome 6 at the 6p21.3 marker. Three classical class I genes HLA-A, C and B and three classical class II genes HLA-DR, DQ and DP are transplantation determinants. A schema for the classical class II gene organization is illustrated for HLA-DPB1: 5’ and 3’ untranslated regions (UTRs; denoted by green boxes); exon 1 (leader); exon 2 and exon 3 (extracellular domains denoted by yellow boxes); exon 4 (transmembrane domain); exon 5 (cytoplasmic tail). Map is not drawn to scale. Chromosome drawing retrieved from the National Library of Medicine.1
As a multi-gene complex, the HLA genes are amongst the most polymorphic genes in the human genome. As of January, 2016, the World Health Organization (WHO) Nomenclature Committee on Factors of the HLA System recognizes 3285 HLA-A, 4077 HLA-B, 2801 HLA-C, 1825 HLA-DRB1, 876 HLA-DQB1 and 587 HLA-DPB1 alleles.2 The preservation en bloc of genes with shared immunologic function ensures that the immune system is equipped with the armamentarium for maximal responses against foreign pathogens from infectious agents and transplanted tissues from allogeneic blood stem cells.
A hallmark of HLA genes is the extended conservation of haplotypes of physically-linked alleles. Known as positive linkage disequilibrium or LD, haplotypes both help and hinder the identification of suitable donors for transplantation. When the patient’s haplotypes carry strongly linked common alleles, the probability of identifying HLA-matched donors is much higher than when a patient’s haplotypes are less common or when linkage has been abrogated through a recombination event. Nevertheless, with over 25 million registered unrelated donors in worldwide registries,3 it is now feasible for approximately 70% of patients of Caucasian background to identify HLA-A, B, C, DRB1-matched (HLA “8/8”) donors. For patients of non-European backgrounds, the likelihood of identifying HLA 8/8-matched donors is significantly lower than for European white patients, and the prospect of finding donors with one mismatch (HLA 7/8) depends on their ethnic background.4 Recent advances in immunogenetics may provide much needed approaches for the selection of HLA-mismatched donors for these patients.
Basic Principles of HLA Mismatching
Three basic concepts of HLA mismatching have emerged from extensive clinical research in the field of allogeneic transplantation: 1) the vector of incompatibility between the donor and the recipient; 2) the number of HLA disparities, and 3) the specific locus/loci that are mismatched. HLA matching is the most consistent, predictive factor for outcome after hematopoietic cell transplantation from unrelated donors. Certain combinations of HLA mismatches that raise the risk of graft-versus-host disease (GVHD) may be compensated by a lower risk of post-transplant recurrence of the patient’s underlying disease (“graft-versus-leukemia”, or GVL). Finally, in the era of growth factor mobilized peripheral blood stem cell (PBSC) transplantation,5 comparison of HLA-associated risks appear to parallel historic data from bone marrow (BM) transplantation (Table 1).
Table 1.
The importance of allele and antigen mismatches on outcome after HLA-mismatched unrelated donor hematopoietic cell transplantation from marrow and peripheral blood stem cell graft sources.6 – 11 Low, intermediate and high risk refer to the relative risks associated with mismatching at the locus of interest.
| Locus | Marrow Transplant |
PBSC Transplant | Risks Associated with Allele versus Antigen Mismatch |
|---|---|---|---|
| HLA-A | High risk6 | Intermediate risk7 |
BM and PBSC: In general, allele mismatches are as risky as antigen mismatches6,7 |
| HLA-B | Intermediate risk6,7 |
Intermediate risk6,7 |
BM: In general, allele mismatches are as risky as antigen mismatches6 PBSC: limited numbers7 |
| HLA-C | Intermediate risk6,7 High risk8 |
High risk6,7 | In general, allele mismatches have lower risks than antigen mismatches6,7,8 |
| HLA- DRB1 |
High risk6 | The limited numbers prevent definitive analysis of allele versus antigen. |
|
| HLA- DQB1 |
Low risk6,7 | Low risk6,7 | Isolated mismatching at HLA-DQB1 is not associated with higher risks. |
| HLA- DPB1 |
High risk9–11 | High risk9–11 | The allele and antigen definitions are not applicable to HLA-DP typing and nomenclature.2 |
BM, bone marrow
PBSC, peripheral blood stem cells
The Vector of Incompatibility of an HLA Mismatch
Also known as the “direction” of the HLA mismatch, the vector of incompatibility is viewed from the perspective of the donor and from that of the patient. From the donor’s perspective, the presence of recipient HLA-A, B, C, DRB1 differences not shared by the donor stimulates donor-anti-host or GVH recognition that is associated with higher risks of GVHD and increased mortality compared with complete matching. Conversely, from the patient’s perspective, the presence of donor disparity not shared by the recipient provokes host-versus-graft (HVG) recognition that is associated with an increased risk of graft failure. Each HLA vector of incompatibility therefore represents what one individual recognizes in the other as different than self.
“Bidirectional” HLA mismatches involve both HVG and the GVH vectors of incompatibility; this is observed when the patient and the donor are each heterozygous at the locus in question (for example, HLA- A1,2 versus HLA- A1,3). HLA homozygosity occurs when an individual has inherited the same HLA allele(s) from each parent. Donor-recipient mismatching at a homozygous locus are “unidirectional” HVG or GVH. For example, an HLA-A1,1 patient and HLA-A1,2 donor have a unidirectional HVG mismatch; in contrast, an HLA-A1,2 patient and HLA-A1,1 donor have a unidirectional mismatch for GVH.
The clinical significance of the vector of incompatibility was originally described in high-risk transplant recipients at a single institution.12 In that historic series, patients who were homozygous at the mismatched HLA locus had a lower rate of grades III-IV acute GVHD (18% versus 52%) but a higher rate of graft failure (15% versus 6%) than patients who were heterozygous at the same locus. These sentinel observations have been validated in several large independent studies, the most recent conducted by the Center for International Blood and Marrow Transplant Research (CIBMTR).13 In a comparison of outcomes of HLA 8/8, HLA 7/8 (one HLA-A, B, C or DRB1 mismatch) bidirectional, HLA 7/8 unidirectional GVH and HLA 7/8 unidirectional HVG mismatched unrelated donor transplants, HLA 7/8 transplants with 1 HVG unidirectional mismatch had a lower risk of GVHD than HLA 7/8 transplants with a GVH vector mismatch. These results confirm that HVG vector mismatches are well-tolerated with respect to GVHD, and that the direction of incompatibility has biological consequences.
Transplant Outcome Depends on the Total Number of Mismatches and the Locus that is Mismatched
The total number of HLA mismatches as well as locus-specific effects are both important in shaping clinical outcome after HLA-mismatched transplantation (Table 2). As the number of HLA mismatches increases, the risks of GVHD and mortality also increase.10,12,14,15 The clinical significance of additive deleterious effects of multi- locus HLA mismatching was demonstrated by a recent retrospective analysis.14 In the class II region, a series of HLA-DRB genes named HLA-DRB3, DRB4 and DRB5 are encoded on specific HLA-DRB1 haplotypes: HLA-DRB3 is linked to HLA-DR3, DR5 and DR6 haplotypes; DRB4 to HLA-DR4, DR7 and DR9 haplotypes, and DRB5 to DR2 haplotypes. When a patient and donor are mismatched at HLA-DRB1, the probability of additional mismatching at DRB3/4/5 is increased on these specific HLA-DR haplotypes. The cumulative impact of multi-loci mismatching inclusive of DRB3/4/5 on mortality was significant, particularly when 3 or more mismatches were present. These data suggest that when an HLA-DRB1-mismatched donor is identified, additional characterization of HLA-DRB3/4/5 on relevant HLA-DR haplotypes, is warranted, and efforts to lower the total number of HLA-DRB mismatches will lower post-transplant risks to the patient.
Table 2.
| Mismatched Locus |
GVHD | GVL Effect |
Survival | Approach to Donor Selection |
|---|---|---|---|---|
| HLA-A | ↑ | ↓ | In most series, the presence of either an HLA-A allele or antigen mismatch increases post-transplant risks. In some studies, however, HLA-A antigen mismatches impart higher risks than allele mismatches. Bottom Line: If only HLA-A mismatched donors are available, avoid HLA-A antigen mismatches. |
|
| HLA-B | ↑ | ↓ | There currently are insufficient data on the relative importance of HLA-B allele and antigen mismatches. Bottom Line: If the donor choices include HLA-B- mismatched donors, assure that there is no linked HLA-C mismatch. |
|
| HLA-C | ↑ | Yes | ↓ | HLA-C mismatching has consistently been shown to increase mortality after transplantation. In most series, HLA-C antigen mismatches carry higher risks than allele mismatches. In a recent study from Japan, the higher risk of GVHD associated with HLA-C mismatching was accompanied by lower risk for relapse (GVL effect). Risks are also higher when the patient’s mismatched HLA-C allotype is expressed at high levels. Bottom Line: If the donor choices include HLA-C mismatched donors, assure there is no linked HLA-B mismatch. Select HLA-C allele mismatches over HLA-C antigen mismatches. Avoid mismatching for a high- expression HLA-C allotype in the patient. |
| HLA-DRB1 | ↑↓ | ↓ | There are insufficient data on the relative risks associated with HLA-DRB1 allele and antigen mismatches. HLA-DRB1 and DQB1 mismatching confer additive risks, as do DRB1/3/4/5 mismatches. Bottom Line: If the donor choices include HLA-DRB1- mismatched donors, assure there is no linked HLA-DRB3 (for DR3, 5, 6); DRB4 (for DR4, 7, 9); DRB5 (for DR2) or HLA- DQB1 mismatch. |
|
| HLA-DQB1 | ↓ | ↑ | Of all the classical HLA genes, only mismatching at HLA- DQB1 does not appear to increase risks, but only if truly a single locus mismatch. HLA-DRB1 and DQB1 mismatching confer additive risks. Bottom Line: If HLA-DQB1-mismatched donors are considered, assure there is no linked HLA-DRB1 mismatch. |
|
| HLA-DPB1 | ↑ | Yes | ↓ | Mismatching at HLA-DPB1 increases GVHD but lowers relapse. Among HLA-DPB1 mismatched transplants, T cell epitope and/or level of HLA-DP expression can define high- risk mismatches which should be avoided. Bottom Line: If matching for HLA 12/12 alleles is not feasible, consider the use of T cell epitope and expression to avoid high-risk HLA-DPB1 mismatches. |
increased risk
decreased risk
GVHD, graft-versus-host disease
GVL, graft-versus-leukemia
The powerful negative effects of HLA mismatching have recently been evaluated in a side-by-side comparison of unrelated donor and cord blood transplantation from the Japanese Society of Hematopoietic Cell Transplantation and the Transplant Registry Unified Management Program.16 This analysis included 2472 myeloablative transplants (1001 HLA 8/8; 656 HLA 7/8; 815 cord blood transplants). Overall, recipients of HLA 7/8 matched unrelated donor transplantation had lower overall survival, higher GVHD and higher non-relapse mortality compared to HLA 8/8 matched unrelated and cord blood transplant recipients. The risks of severe acute GVHD and non-relapse mortality after cord blood transplantation were comparable to those after HLA 8/8 matched unrelated donor transplantation, but the risks were higher for HLA 7/8 matched unrelated donor transplants. Chronic extensive GVHD was lower following cord blood compared to HLA 8/8 or 7/8 matched unrelated donor transplantation, with similar overall survival for cord blood and HLA 8/8 unrelated donor recipients; recipients of HLA 7/8 unrelated donor allografts had lower overall survival compared to cord blood and HLA 8/8 unrelated donor transplants.
In the largest analysis of unrelated donor transplants performed by the Japan Marrow Donor Program (JMDP) to date, the incidence of acute GVHD was evaluated according to the number of mismatches at six loci HLA-A, B, C, DRB1, DQB1, DPB1 typed with high resolution molecular methods.10 Overall, 1 – 5 HLA mismatches were associated with stepwise higher acute GVHD and lower survival compared to no HLA mismatching. The dose effect for class II genes was different than for class I genes: the presence of one HLA-DRB1 or one HLA-DQB1 was tolerable, but mismatching at both HLA-DRB1 and DQB1 was not. Compared to matching at both HLA-DRB1 and DQB1, the relative risk (RR) of grades III–IV acute GVHD risk associated with one HLA-DRB1 mismatch was 0.98; one HLA-DQB1 mismatch RR 0.92, but combined double locus HLA-DRB1 and DQB1 mismatch was RR 1.32 (P < .001). For survival, the risks were 1.04, 1.04 and 1.17 (P < .001), respectively.
The elucidation of HLA-DR/DQ mismatching lead to the grouping of HLA-DRB1/DQB1 mismatches together to re-define risks associated with mismatching at other HLA loci. The total number of HLA-A, B, C, DPB1 and DR/DQ mismatches was associated with increased risk of grades III-IV acute GVHD: RR 1.29 (P .001); 1.42 (P .001); 1.63 (P < .001) and 1.23 (P .001) for HLA-A, B, C, DPB1 mismatching, respectively. For survival, those risks were RR 1.29 (P < .001); 1.27 (P < .001), and 1.21 (P < .0010) for HLA-A, B, C, respectively.
A new analysis by the CIBMTR has confirmed the importance of the number of HLA mismatches on transplant outcome.15 The main objective of this analysis was to define variables that influence outcomes after unrelated donor transplantation for hematologic malignancy. This analysis used two cohorts of patients receiving transplants between 1988 and 2006 (n = 6349; “training cohort”) and between 2007 and 2011 (n = 4690; “validation cohort”). All donor-recipient transplant pairs were defined using molecular methods that discriminate HLA-A, B, C and DRB1 alleles at high resolution. Survival was better for recipients from young donors (aged 18 – 32 years) who were HLA matched (P < .001). This observation was validated in the 2007 – 2011 cohort. For every 10 year increment in donor age, there was a 5.5% increase in the hazard of mortality. Increasing numbers of HLA mismatches was also associated with worsening outcome. Importantly, other factors including sex, parity, and CMV serostatus were not associated with survival.
Compared to an HLA 8/8-matched transplant, the hazard ratio (HR)of mortality increased with increasing numbers of donor-recipient mismatches: HLA 7/8 HR 1.24; HLA 6/8 HR 1.62; HLA 5/8 or lower HR 1.89 (P < .001 for each). For non-relapse mortality, those hazards were 1.38, 1.85, and 2.16 for HLA 7/8, HLA 6/8, and HLA 5/8, respectively (P < .001). For grades II–IV acute GVHD, the relative risks were 1.23, 1.26, 1.46 (P < .001), respectively. No HLA associations were noted for relapse or chronic GVHD.
The JMDP and CIBMTR studies validate the importance of the number of mismatches, and the loci that contribute to risk. Taken together, these studies confirm that when an HLA-matched donor is not available for transplantation, then selection of a young donor with the fewest HLA mismatches will likely be associated with lower risks to the patient after transplantation. Avoidance of a double HLA-DRB1/ DQB1 mismatch will also help to lower the risks of GVHD.
GVL and HLA Mismatching
Early observations noted an overall lower risk of post-transplant disease recurrence among patients who had experienced clinical acute GVHD.17 Known as the graft-versus-leukemia (GVL) effect, several historical studies have confirmed that the deleterious effect of HLA mismatching on GVHD risk may be balanced by lower risk of relapse. Recently, GVL protective effects with HLA mismatching have been observed in elegant studies of the HLA-DP locus.9,18 In T-cell depleted transplantation, concurrent mismatching for HLA-DP and other loci was associated with better outcomes in patients with more advanced disease (OR 0.5 for mortality, P 0.001).18
The role for HLA-DP in GVL has been most recently elucidated in an updated analysis by the JMDP, where not only was a beneficial effect of GVL confirmed, but new information strongly suggests that each HLA mismatch does not necessarily confer the same beneficial GVL effect.10 When HLA-A, B, C, DRB1, DQB1, and DPB1 mismatches were analyzed side-by-side, only HLA-C and HLA-DPB1 mismatches were associated with a lower risk of relapse (Table 2). Specifically, HLA-C mismatches were associated with a protective effect against relapse (HR 0.70, P < 0.001) and similar protective risks were seen for HLA-DPB1 mismatches (HR 0.69, P < 0.001) compared to matching at those loci. These observations strongly suggest that future investigation into the underlying mechanisms responsible for GVL involving T and/or natural killer cell-mediated effects will facilitate understanding of GVL in transplantation.
HLA Mismatching in the Setting of Reduced Intensity Conditioning
The vast majority of the literature on the importance of HLA matching has been gleaned from the substantial worldwide experience in traditional ablative conditioning regimens. Strong evidence also points to the importance of donor-recipient HLA mismatching in the setting of reduced intensity conditioning. In one recent analysis, HLA 8/8-matched unrelated transplants had better adjusted 1 and 3 year survival rates of 54.7% versus 48.8% (P .01 at one year), and 37.4% versus 30.9% (P .005 at 3 years) compared to HLA 7/8-matched transplants.19 Furthermore, the HLA 7/8-matched transplants had a higher risk of grades II-IV acute GVHD (RR 1.29, P .0034); higher transplant-related mortality (RR 1.52, P< .0001); lower disease-free survival (RR 1.12, P .0015) and lower overall survival (RR 1.25, P .0001). No differences in chronic GVHD or relapse were appreciated. These results strongly suggest that the same principles of additive effects of HLA mismatching observed after ablative conditioning for unrelated donor transplantation, occur also with the use of reduced intensity conditioning regimens.
Choosing Among HLA-Mismatched Donors
Many patients will only have HLA-mismatched unrelated donors.4 Three major models have been investigated as a means to identify specific combinations of HLA mismatches that appear to be better tolerated than other combinations. They are based on the level of resolution of the HLA mismatch (allele or antigen), the presence of mismatching for specific amino acid residues, and the level of expression of the patient’s and the donor’s mismatched HLA allotype.
Alleles and Antigens
The classical definition of an HLA allele mismatch is a different between two HLA allotypes that can only be detected with high resolution molecular methods. HLA antigen mismatches occur between HLA allotypes that are detectable with alloantisera directed against epitopes of the expressed HLA protein. Although the serologic definition of HLA-C gene products has been hampered by the overall low level of expression of HLA-C allotypes, HLA-C has served as an important model locus in the investigation of risks associated alleles and antigens.
A high frequency HLA-C mismatch combination observed among Japanese patients and unrelated donors sheds light on the immunogenicity of specific HLA-C alleles.20 In this retrospective analysis of 6967 Japanese transplant recipients of T-replete unrelated donor bone marrow, patients and/or donors who possessed the HLA-B*51:01 allotype had significantly higher post-transplant risks (patient HR 1.37, P < .001; donor HR 1.35, P < .001). Furthermore, patients who were HLA-C*14:02-positive had increased risk of severe acute GVHD (HR 1.35, P < .001). Compared to HLA-C matched patients, HLA-C mismatched patients with an HLA-C*14:02 allele had the highest risk of severe acute GVHD (HR 3.61, P < .001) and transplant-related mortality (HR 2.53, P < .001). These results strongly point to a special feature(s) of the HLA-B*51:01 and HLA-C*14:02 allotypes that provoke strong alloimmune responses from the donor.
In Caucasian populations, differences in graft rejection, GVHD and mortality observed in early studies have provided the foundation for exploring differences between allele and antigen mismatches.6–8 In particular, the commonly observed HLA-C*03:03/03:04 mismatch has been of interest, because this combination does not elicit in vitro cytotoxic T cell responses. In a recent analysis, patients mismatched for the C*03:03/03:04 allele combination had similar outcomes as HLA 8/8-matched recipients.21 The basis for the lower risks associated with this particular mismatch compared to other HLA-C mismatches, has recently been elucidated and is described below in the section entitled “HLA Expression”.
Mismatching for Specific HLA Amino Acid Residues
This model tests the hypothesis that the presence of specific amino acid residues of the expressed HLA protein may be immunogenic and leads to donor-anti-host recognition that clinically is manifested by increased risks of acute GVHD and mortality (Table 3). Recent evidence for the role of mismatching at specific HLA residues has been provided by an analysis of HLA class I HLA-A, B, C22 and HLA-DP9 mismatches. The class I model identified 3 residues that form key anchors of the class I peptide binding groove: residues 116 and 99 of HLA-C, and residue 9 of HLA-B.22 Mismatching for amino acids a residue 116 of HLA-C was associated with a significantly higher risk of acute GVHD (HR 1.45) and mismatching at residue 99 with higher risk of transplant-related mortality (HR 1.37 for a cysteine versus tyrosine mismatch) compared to matching at these residues. At HLA-B, mismatching at residue 9 was associated with higher risk of chronic GVHD (HR 2.28) compared to matching at residue 9.
Table 3.
Donor-recipient mismatching at certain amino acid residues is permissive.
| Mismatched HLA Locus |
GVHD | Relapse | Survival |
|---|---|---|---|
| HLA-A | None identified22; Residue 9 [aGVHD23] |
None identified22 |
None identified22 |
| HLA-B | Residue 9 [cGVHD22] | None identified22 | None identified22 |
| HLA-C | Residue 116 [aGVHD22]; Residues 9, 77, 80, 99, 116, 156 [aGVHD23] |
Residues 9, 99, 156 24 |
Residue 99 [TRM 22] |
| HLA-DPB1 | TCE model [aGVHD9] |
*GVL24; *TCE model9 |
No [GVH and GVL balanced9] |
Evidence for a critical role of HLA-DP residues in GVHD and GVL responses has been provided by an elegant in vitro model based on the degree of cytotoxicity between T cell clones that recognize epitopes in the hypervariable regions of the HLA-DPβ protein encoded by the polymorphic exon 2.9 In an international study of 5428 HLA 10/10-matched unrelated donor transplants, a T cell epitope (TCE)-based model defined single HLA-DPB1 mismatches between the donor and recipient as “permissive” HLA 11/12 (ie., do not elicit an in vitro cytotoxic response), “non-permissive” HLA 11/12 (ie., do elicit strong in vitro cytotoxicity) and six-locus matched (HLA 12/12) transplants. A comparison between HLA 12/12 versus HLA 11/12 permissive transplants showed improved overall survival for HLA 12/12 but also increased risk of relapse, confirming a GVL effect associated with HLA-DPB1 mismatching. When HLA 11/12 permissive mismatches were compared to HLA 11/12 non-permissive mismatches, significantly increased mortality was observed for non-permissive mismatches with similar relapse rates. Among HLA 11/12 transplants with an HLA-DPB1 mismatch in the GVH vector, the non-permissive mismatches had a lower risk of relapse compared to permissive mismatches, also consistent with a GVL effect. When one HLA-A, B, C, DRB1 or DQB1 mismatch was also present, non-permissive HLA-DPB1 mismatches had increased risk of grades III-IV acute GVHD (OR 1.37, P 0.002]) compared to permissive mismatches, but no apparent beneficial decrease in relapse. These results are consistent with additive negative effects of mismatching at HLA-DP with other HLA loci.
The findings by Fleischhauer and colleagues were recently validated.25 Among 5449 HLA 8/8 transplants, there was lower GVHD (OR 0.7, P .007) but higher relapse (OR 1.4, P < 0.001) compared to mismatched transplants; non-permissive HLA-DPB1 mismatches had higher transplant-related mortality (HR 1.4, P < 0.001]) but similar relapse compared to the permissive mismatches and HLA-DPB1 matched transplants.
HLA Expression
HLA-C and HLA-DP expression affect the control of infection (HIV-AIDs and hepatitis B)26, 27 and the risk of autoimmune disease (Crohns).26 In the infectious disease model, high levels of HLA expression may facilitate the presentation of foreign antigens leading to improved immune surveillance and lower risk of disease progression (lower viral set point in the case of HIV-AIDs; lower progression of hepatitis B infection). In contrast, the same high HLA expression may provoke autoimmune attack of host tissues leading to autoimmunity in the case of Crohn’s disease.
It has long been recognized that the level of expression of HLA-DRB3, DRB4, DRB5, DQB1 gene products is generally lower than that of HLA-A, B, C and DRB1. In a recent analysis of HLA 8/8-matched transplants, a low-expression mismatch was not associated with higher risks; however, in the setting of an HLA 7/8-matched transplant, the presence of 3 or more low-expression mismatches at HLA-DRB1/ 3/4/5/DQ) was associated with higher overall mortality and transplant-related mortality.14
In transplantation, mismatching for low-expression HLA-C and HLA-DP allotypes in the transplant patient is better tolerated than mismatching for high-expression allotypes. The median fluorescent intensity value for each HLA-C allotype was instrumental in defining the role of HLA-C expression in risks after HLA-C-mismatched unrelated donor transplantation.28 Among unrelated donor transplants with only a single HLA-C mismatch, as the level of expression of the patient’s mismatched HLA-C allotype increased, the risks of acute GVHD and non-relapse mortality also increased. Mismatches involving HLA-C*03 and 07 (allotypes expressed at the lowest levels) and HLA-C*01 and 14 (allotypes expressed at the highest levels) showed increased risks for high-expression mismatches when the HLA-C mismatch was also mismatched at residue 116 and for natural killer immunoglobulin-like receptor (KIR) ligands. Residue 116 and KIR ligand mismatches that were low-expression had similar risks as HLA-C matches. These data strongly support a role for the level of HLA-C expression in immunogenicity related to graft-versus-host alloresponses.
Similar to HLA-C, HLA-DP has historically been considered a low-expression locus. Whereas each HLA-C allotype has a representative expression level,26 HLA-DP allotypes are expressed at high or low levels which is defined by the rs9277534 variant that resides within the 3’ untranslated region (UTR) of the HLA-DPB1 gene.11, 27 In a retrospective analysis of 3505 individuals, high HLA-DP allotype expression was confirmed for rs9277534G-linked HLA-DPB1 alleles and low expression for rs9277534A-linked alleles.11 In this retrospective analysis of2029 transplants, the risk associated with a single HLA-DPB1 mismatch (HLA 11/12) was evaluated according to the rs9277534 allele associated with the patient’s and the donor’s mismatched HLA-DPB1 allele. Patients with rs9277534G-linked HLA-DPB1 mismatches had higher risk of acute GVHD than patients with rs9277534A-linked mismatches. These results are consistent with the hypothesis that highly expressed mismatched patient HLA-DP allotypes may be potent targets for donor-anti-host recognition; low-expression patient HLA-DP allotypes may be sufficiently non-immunogenic and permit the use of such donors when fully-matched donors are not available. These data demonstrate that polymorphisms that reside within regulatory regions of HLA genes have clinical significance, and underscore a need for a more complete understanding of HLA expression in defining permissible HLA mismatches. Taken together, the results offer a new approach for understanding HLA-mediated immune responses in transplantation, infectious diseases, and autoimmune disorders.
Future Perspectives
There are substantial validated data that support the use of selected HLA mismatched unrelated donors for transplantation when fully compatible donors are not available (Table 4). The judicious selection may include consideration for: a single HLA mismatch; avoidance of two-locus HLA-DRB1/DQB1 mismatching; avoidance of simultaneous mismatching at HLA-DRB3/4/5 when there is an existing HLA-DRB1 mismatch, and avoidance of mismatching for a high-expression HLA-C and HLA-DP allotype in the patient. Future research endeavors focused on a comprehensive evaluation of both common and uncommon HLA mismatches, together with a more complete understanding of non-HLA variation within the MHC, may open new avenues for the use of HLA mismatched donors for transplantation. In this way, the availability of HLA mismatched unrelated donor transplantation as curative therapy may be increased, without concomitantly increasing risks to the patient.
Table 4.
Summary of HLA Mismatching in Unrelated Donor Hematopoietic Cell Transplantation
Validated data suggest that when HLA-matched donors are not available:
|
Emerging research suggests additional features may prove to be important in the future:
|
Acknowledgments
Dr. Petersdorf is supported by grants CA18029, CA100019, CA162194, and AI069197 from the National Institutes of Health, USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Dr. Petersdorf has no conflict of interest.
References
- 1. http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?TAXID=9606&CHR=6&MAPS=ideogr[0.00:10100.00]
- 2. http://www.ebi.ac.uk/imgt/hla. [Google Scholar]
- 3. www.worldmarrow.org. [Google Scholar]
- 4.Gragert L, Eapen M, Williams E, et al. HLA match likelihoods for hematopoietic stem-cell grafts in the U.S. registry. N Engl J Med. 2014;371:339–348. doi: 10.1056/NEJMsa1311707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anasetti C, Logan BR, Lee SJ, et al. Peripheral-blood stem cells versus bone marrow from unrelated donors. N Engl J Med. 2012;367:1487–1496. doi: 10.1056/NEJMoa1203517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SJ, Klein J, Haagenson M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110:4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 7.Woolfrey A, Klein JP, Haagenson M, et al. HLA-C antigen mismatch is associated with worse outcome in unrelated donor peripheral blood stem cell transplantation. Biol Blood Marrow Transplant. 2011;17:885–892. doi: 10.1016/j.bbmt.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersdorf EW, Hansen JA, Martin PJ, et al. Major-histocompatibility-complex class I alleles and antigens in hematopoietic-cell transplantation. N Engl J Med. 2001;345:1794–1800. doi: 10.1056/NEJMoa011826. [DOI] [PubMed] [Google Scholar]
- 9.Fleischhauer K, Shaw BE, Gooley T, et al. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol. 2012;13:366–374. doi: 10.1016/S1470-2045(12)70004-9. [Erratum appears in Lancet Oncol. 2012 Apr;13(4):e134–5] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morishima Y, Kashiwase K, Matsuo K, et al. Biological significance of HLA locus matching in unrelated donor bone marrow transplantation. Blood. 2015;125:1189–1197. doi: 10.1182/blood-2014-10-604785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersdorf EW, Malkki M, O’hUigin C, et al. High HLA-DP expression and graft-versus-host disease. N Engl J Med. 2015;373:599–609. doi: 10.1056/NEJMoa1500140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anasetti C, Hansen JA. Effect of HLA incompatibility in marrow transplantation from unrelated and HLA-mismatched related donors. Transfus Sci. 1994;15:221–230. doi: 10.1016/0955-3886(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 13.Hurley CK, Woolfrey A, Wang T, et al. The impact of HLA unidirectional mismatches on the outcome of myeloablative hematopoietic stem cell transplantation with unrelated donors. Blood. 2013;121:4800–4806. doi: 10.1182/blood-2013-01-480343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez-Vina MA, Klein JP, Haagenson M, et al. Multiple mismatches at the low expression HLA loci DP, DQ, and DRB3/4/5 associate with adverse outcomes in hematopoietic stem cell transplantation. Blood. 2013;121:4603–4610. doi: 10.1182/blood-2013-02-481945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kollman C, Spellman S, Zhang M, et al. The effect of donor characteristics on survival after unrelated donor transplantation for hematologic malignancy. Blood. 2016;127:260–267. doi: 10.1182/blood-2015-08-663823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terakura S, Atsuta Y, Tsukada N, et al. Comparison of outcomes of 8/8 and 7/8 allele-matched unrelated bone marrow transplantation and single-unit cord blood transplantation in adults with acute leukemia. Biol Blood Marrow Transplant. 2016;22:330–338. doi: 10.1016/j.bbmt.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Horowitz MM, Gale RP, Sondel PM, et al. Graft-versus-leukemia reactions after bone marrow transplantation. Blood. 1990;75:555–562. [PubMed] [Google Scholar]
- 18.Shaw BE, Mayor NP, Russell NH, et al. Diverging effects of hLA-DPB1 mismatching status on outcome following unrelated donor transplantation depending on disease stage and the degree of matching for other HLA alleles. Leukemia. 24:58–65. doi: 10.1038/leu.2009.239. 201. [DOI] [PubMed] [Google Scholar]
- 19.Verneris MR, Lee SJ, Ahn KW, et al. HLA mismatch is associated with worse outcomes after unrelated donor reduced-intensity conditioning hematopoietic cell transplantation an analysis from the Center for International Blood and Marrow Transplant Research. Biol Blood Marrow Transplant. 2015;21:1783–1789. doi: 10.1016/j.bbmt.2015.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morishima S, Kashiwase K, Matsuo K, et al. High risk HLA alleles for severe acute graft-versus-host disease and mortality in unrelated donor bone marrow transplantation. Haematologica. 2016;101:491–498. doi: 10.3324/haematol.2015.136903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez-Vina MA, Wang T, Lee SJ, et al. Identification of a permissible HLA mismatch in hematopoietic stem cell transplantation. Blood. 2014;123:1270–1278. doi: 10.1182/blood-2013-10-532671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pidala J, Wang T, Haagenson M, et al. Amino acid substitution at peptide-binding pockets of HLA class I molecules increases risk of severe acute GVHD and mortality. Blood. 2013;122:3651–3658. doi: 10.1182/blood-2013-05-501510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawase T, Morishima Y, Matsuo K, et al. High-risk HLA allele mismatch combinations responsible for severe acute-graft-versus-host disease and implication for its molecular mechanism. Blood. 2007;110:2235–2241. doi: 10.1182/blood-2007-02-072405. [DOI] [PubMed] [Google Scholar]
- 24.Kawase T, Matsuo K, Kashiwase K, et al. HLA mismatch combinations associated with decreased risk of relapse: implications for the molecular mechanism. Blood. 2009;113:2851–2858. doi: 10.1182/blood-2008-08-171934. [DOI] [PubMed] [Google Scholar]
- 25.Pidala J, Lee SJ, Ahn KW, et al. Nonpermissive HLA-DPB1 mismatch increases mortality after myeloablative unrelated allogeneic hematopoietic cell transplantation. Blood. 2014;124:2596–2606. doi: 10.1182/blood-2014-05-576041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apps R, Qi Y, Carlson JM, et al. Influence of HLA-C expression level on HIV control. Science. 2013;340:87–91. doi: 10.1126/science.1232685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas R, Thio CL, Apps R, et al. A novel variant marking HLA-DP expression levels predicts recovery from hepatitis B virus infection. J Virol. 2012;86:6979–6985. doi: 10.1128/JVI.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petersdorf EW, Gooley TA, Malkki M, et al. HLA-C expression levels define permissible mismatches in hematopoietic cell transplantation. Blood. 2014;124:3996–4003. doi: 10.1182/blood-2014-09-599969. [DOI] [PMC free article] [PubMed] [Google Scholar]