Abstract
Purpose
To map the phosphoproteome and identify changes in the phosphorylation patterns in the HIV-infected and uninfected brain using high-resolution mass spectrometry.
Experimental Design
Parietal cortex from brain of individuals with and without HIV infection were lysed and trypsinized. The peptides were labeled with iTRAQ reagents, combined, phospho-enriched by titanium dioxide chromatography, and analyzed by LC-MS/MS with high-resolution.
Results
Our phosphoproteomic workflow resulted in the identification of 112 phosphorylated proteins and 17 novel phosphorylation sites in all the samples that were analyzed. The phosphopeptide sequences were searched for kinase substrate motifs which revealed potential kinases involved in important signaling pathways. The site-specific phosphopeptide quantification showed that peptides from neurofilament medium polypeptide, myelin basic protein, and 2′–3′-cyclic nucleotide-3′ phosphodiesterase have relatively higher phosphorylation levels during HIV infection.
Clinical Relevance
This study has enriched the global phosphoproteome knowledge of the human brain by detecting novel phosphorylation sites on neuronal proteins and identifying differentially phosphorylated brain proteins during HIV infection. Kinases that lead to unusual phosphorylations could be therapeutic targets for the treatment of HIV-associated neurocognitive disorders (HAND).
Keywords: phosphorylation, proteomics, drug target, HIV-associated neurocognitive disease, myelin basic protein, protein kinase A
INTRODUCTION
Infection with the human immunodeficiency virus (HIV) may lead to a neurodegenerative condition called HAND. Since the introduction of combined antiretroviral therapy, the life expectancy of HIV-infected individuals has increased and milder forms of HAND have become prevalent. [1] The pathophysiology of HAND is only partially understood and no effective neuroprotective treatment has been established to date. The majority of the mass spectrometry-based proteomic analyses of samples from patients with HIV infection focus on changes in protein expression and abundance. [2–4] However, post-translational modifications of proteins have the ability to dramatically alter cellular function but besides nitration, other modifications have not been investigated in neural tissue from this patient population. [5–7]
Protein phosphorylation is a key modification in signal transduction that regulates diverse cellular processes. A finely tuned balance between kinase and phosphatase activity is essential for the physiological functioning of the cell. Dysregulation of this process can lead to a diseased state. Upon infection, HIV hijacks the host’s phosphorylation pathways to promote its own replication. [8] In response, a cascade of phosphorylation events activates the immune system. [9] HIV protein Tat can decrease the activity of cyclin E-cyclin-dependent kinase 2 activity in glial cells, [10] but increase the activity of protein kinase C in astrocytes, [11] focal adhesion kinase in brain endothelial cells [12] and lead to higher levels of phosphorylation on N-methyl-D-aspartate receptor in neurons. [13,14] The envelope protein of HIV, gp120, can also trigger the phosphorylation of double-stranded RNA-activated protein kinase, a stress kinase that localizes to the nucleus of neurons and leads to neuronal injury. [15] Moreover, redox-sensitive p38 mitogen-activated phosphokinase pathway has been implicated in HIV neuropathogenesis. [16] Specific phosphorylation sites have been identified in the HIV-infected brain. For example, S795 of the retinoblastoma protein is uniquely phosphorylated in vivo and in neurons following exposure to HIV-infected macrophages in vitro. [17]
Recent advances in mass spectrometry have permitted a comprehensive analysis of the brain phosphoproteome with precise phosphorylation site information. Orbitrap mass analyzers provide high resolution and high mass accuracy necessary for the confident identification of peptides and the specific modification sites. Additionally, titanium dioxide (TiO2) chromatography is a well-established technique for the isolation of serine/threonine-phosphorylated peptides, further enhancing phosphopeptide detection. We analyzed the phosphoproteome of the human brain parietal cortex, an area specifically involved with neuronal injury in HIV infected individuals. [18,19] We compared tissues to identify differential protein phosphorylation events that may be related with the pathophysiology of HAND. The samples were labeled with isobaric tags for relative and absolute quantitation (iTRAQ), then phospho-enriched by TiO2 chromatography and analyzed by high-resolution tandem mass spectrometry. Furthermore, we validated our results by Western blot analysis.
EXPERIMENTAL PROCEDURES
Sample preparation
Frozen postmortem human brains (parietal cortex) from individuals with HIV infection without encephalitis (HIV) (n=2), with HIV-encephalitis (n=2) (HIV-E) and matching sections of the brain (n=4) (Control) were analyzed (Supporting Table 1). The tissues were washed with ice-cold buffer containing 20 mM Tris/HCl, pH 6.8, 100 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, complete protease inhibitor cocktail and PhosSTOP phosphatase inhibitor cocktail (Roche) to remove any blood clots. Tissues were homogenized in the same buffer (1:5 w/v) with the addition of 1% (v/v) Triton-X 100, and 0.1% (w/v) SDS. The lysate was centrifuged at 18,000 × g for 20 min to remove insoluble cellular components. The protein concentrations were determined using the BCA assay (Pierce) according to the manufacturer’s directions.
Protein digestion and iTRAQ labeling
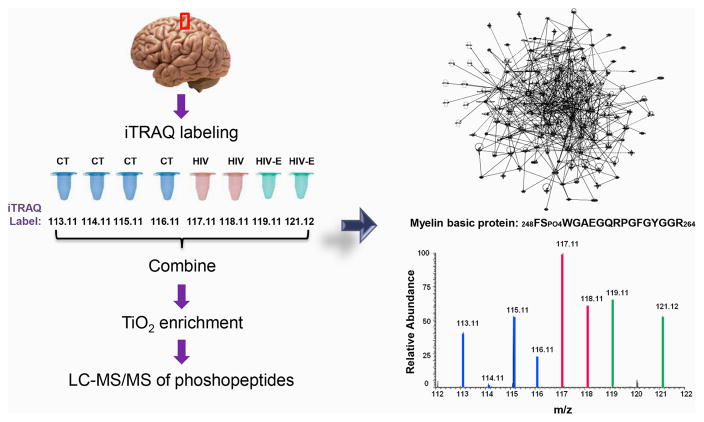
100 μg of protein from each brain sample was dried down and resuspended in 100 μL 8M urea/0.4 M ammonium bicarbonate for reduction and alkylation with a standard protocol [20] followed by overnight trypsin digestion at 37°C. The samples were cleaned using Oasis HLB 200 mg extraction cartridges (Waters) as reported previously. [21] The samples were dried by vacuum centrifugation and resuspended with the dissolution buffer from the 8-plex iTRAQ kit (AB Sciex). The manufacturer’s suggestions were followed for labeling each sample with a different iTRAQ reagent (Figure 1). The labeled peptides were pooled and diluted to 20 mL with 0.1% (v/v) trifluoroacetic acid (TFA). Oasis HLB 500 mg extraction cartridges (Waters) were utilized with the same protocol for clean-up.
Figure 1.
Experimental design. Control, HIV and HIV-encephalitis human brain lysate samples were reduced, alkylated, and trypsinized separately. Each sample was labeled with a different 8plex iTRAQ reagent and combined. The labeled peptides were enriched for phosphopeptides by TiO2 chromatography. The phosphopeptides were separated on a nanoLC system followed by online high-resolution tandem mass spectrometry.
Phosphopeptide enrichment
To enrich phosphopeptides, TiO2 chromatography with 1–200 μL TiO2 TopTips (Glygen) was utilized following a previously reported protocol [22]. The eluted sample was dried in a SpeedVac (Eppendorf) and reconstituted in 0.1% formic acid for analysis by LC-MS/MS.
LC-MS/MS analysis
The iTRAQ labeled and phosphopeptide enriched peptide fraction was separated on an Eksigent 2D nanoflow LC system coupled to an LTQ-Orbitrap Velos (Thermo Scientific) mass spectrometer. The reversed phase- LC system consisted of a peptide trap column (75 μm × 2 cm) and an analytical column (75 μm × 10 cm) packed with YMC gels ODS-A (10 μm, 120 Å) and Magic AQ C18 material (5 μm, 120 Å), respectively. Directly after separation on a 90 min linear gradient, the peptides were introduced to the LTQ-Orbitrap Velos at a flow rate of 300 nL/min and a capillary temperature of 200°C. Data dependent tandem MS analysis was employed in the Orbitrap with 30,000 resolution for MS and 15,000 resolution for MS/MS. Full scans were acquired from m/z 350–1700 with up to 10 masses isolated using a 1.2 Da window and fragmented using a collision energy of 45% in the HCD cell with a dynamic exclusion of 30 s. The minimum signal requirement for triggering an MS/MS scan was set to 10,000 and the first mass value was fixed at m/z 110. An ambient air lock mass was set at m/z 371.10123 for real time calibration. [23] For MS/MS analysis, the rejection of unassigned and singly charged ion criteria was enabled. The list of precursor ion values was exported and utilized as the exclusion list for a second LC-MS/MS run to detect the lower abundant ions.
Bioinformatics
MS and MS/MS data were searched using Proteome Discoverer v. 1.3.0.339 (Thermo Fisher Scientific) with both Mascot (v. 1.27) and SEQUEST (v. 1.2) algorithms. Database searching of MS/MS spectra was performed using a human protein reference sequence database released by the National Center for Biotechnology Information in 2011. Homo sapiens was selected for the taxonomy. The scan event filter had the following criteria: FTMS as the mass analyzer, MS2 for MS order, HCD for activation type, full for scan type and positive for polarity mode. Trypsin was selected as the enzyme with the allowance of 2 maximum missed cleavages. The static modifications were 8-plex iTRAQ (N-terminal, +304.205) and carbamidomethylation (C, +57.021). The variable modifications included oxidation (M, +15.995), deamidation (N, Q, +0.984), 8-plex iTRAQ (K, Y, +304.205), and phosphorylation (S, T, Y, +79.966). The precursor tolerance was set to 15 ppm and the fragment tolerance to 0.03 Da for the LTQ-Orbitrap Velos. Phospho RS was utilized for the localization of the phospho modification, and peptide validator was applied to the Mascot and SEQUEST results to search against a decoy database and obtain false discovery rates (strict: 0.01; relaxed: 0.05). The human brain parietal cortex phosphoproteome network was generated using the Ingenuity Knowledge Base with genes and endogenous chemical reference setting.
Statistical analysis
The absolute peak intensities of the iTRAQ reporter ions (Supporting Table 2) were directly used for the statistical tests. Quantitative difference were assessed by using the exact Wilcoxon Rank sum test for control vs. HIV comparison and the exact Kruskal-Wallis test for control vs. HIV vs. HIV-encephalitis for each phosphopeptide sequence because the data did not follow a Gaussian distribution. P-values less than 0.05 were considered statistically significant.
Immunoprecipitation and Western Blot Analysis
Immunoprecipitation was performed with mouse anti-myelin basic protein, clone S.961.3 (ThermoFisher) according to manufacturer’s protocols. Briefly, the antibody was incubated with 100 μg of brain lysate for 18 hours at 4°C. The antibody-antigen complex was captured by Dynabeads Protein G (Life Technologies). The complex was washed five times with 1x Tris-buffered saline (TBS)/0.01% Tween 20 (v/v) and the protein of interest was eluted with SDS-PAGE application buffer. The eluents were loaded on 12% Bis-Tris SDS-PAGE mini gel. The proteins were transferred to PVDF membrane using the iBlot system (Life Technologies). 5% non-fat dry milk (w/v) (American Bioanalytical) in 1x TBS/0.01% Tween 20 (v/v) was used as the blocking buffer. The primary and secondary antibodies used for western blot analysis are: rabbit anti-phospho (S/T) PKA substrate (Cell signaling) (1:1000) and peroxidase conjugated IgG fraction monoclonal mouse anti-rabbit IgG, light chain specific (Jackson ImmunoResearch) (1:5000), respectively. The binding was followed by three washes with 1x TBS/0.01% Tween 20 (v/v). The membrane was developed using SuperSignal West Dura Extended Duration Substrate kit (Thermo Scientific) and visualized by FluorChem M (Protein Simple).
RESULTS
Human brain parietal cortex phosphoproteome
Global phosphoproteomic analysis resulted in the identification of 112 phosphorylated proteins by Mascot and SEQUEST algorithms (Supporting Table 3) and 208 phosphorylated peptides (Supporting Table 4). The proteins were identified with at least one unique peptide. Peptides were filtered with peptide confidence false discovery rate <0.05 prior to protein identification. No proteins were present exclusively in the control or HIV-infected tissues.
The Ingenuity Pathway Analysis of the identified phosphoproteome (Supporting Figure 1) revealed that 45 of the phosphorylated proteins were involved in nervous system development and function and 40 of the proteins were implicated in various neurological diseases. Also represented were important phosphorylation-based canonical pathways such as protein kinase A, ERK/MAPK and 14-3-3 mediated signaling pathways that contained 15, 9 and 7 of the identified proteins, respectively. ERK1/2, Akt, actin and tubulin were the nodes with the most connectivity, suggesting a potential role of phosphorylation in influencing cytoskeleton dynamics.
Identification of novel phosphorylation sites on human proteins
Phosphoproteomics analysis of the brain samples identified 17 phosphorylation sites (highlighted in Supporting Table 4, Supporting Figures 2–18) that had not been previously reported in humans according to the Phosphosite database (www.phosphosite.org). No significant differences were found between the control and the HIV sample sets for these peptides. All the novel human phosphopeptides listed have Δmass (actual-observed) ≤12 ppm and are manually validated. The kinase binding motifs, potential kinases, protein binding motifs and potential phosphorylated motif binding domains of these recently discovered phosphosites were obtained from the Human Reference Database (www.hprd.org) utilizing the PhosphoMotif Finder function and are summarized in Table 1 and their functions are listed in Supporting Information.
Table 1.
Novel phosphorylation sites discovered in the human brain.a
| Sequence | Protein | UniProt Accession | Phospho site | Corresponding motif | Kinase Substrate motif | Binding motif | Binding domain |
|---|---|---|---|---|---|---|---|
| 1473KTEVQAHSPSR1483 | Microtubule-associated protein 2 | P11137 | S1480 | XXpSP, X[pS/pT]P, pSX[E/pS*/pT*] [pS/pT]PX[R/K], pSP, pSPX[R/K]X |
GSK3, ERK1, ERK2, CK II CDK1, 2, 4, 5, 6, Cdc2 Growth associated histone HI kinase |
[pS/pT]P | WW |
| 384GKVPVTYLELLS395 | SH3-domain GRB2-like endophilin B2 | Q9NR46 | S395 | [E/D]XX[pS/pT] | CK I | na | na |
| 255NVPATPPR262 | SH3-domain GRB2-like (endophilin) interacting protein 1 |
Q9BQI5 | T259 | PX[pS/pT]P, PX[pS/pT]PP X[pS/pT]P, X[pS/pT]XXX[A/P/S/T] [pS/pT]PX[R/K], PXpTP |
GSK3, ERK1, ERK2, CDK5 G protein-coupled receptor kinase 1 CDK1, 2, 4, 6, Cdc2 kinase, Growth associated histone HI kinase |
[pS/pT]P | WW |
| 210EPSLHEIGEK219 | fibroblast growth factor 12 | P61328 | S212 | [M/I/L/V/F/Y]XRXX[pS/pT] [M/I/L/V/F/Y]X[R/K]XX[pS/pT] [M/I/L/V/F/Y]XRXX[pS/pT][M/I/L/V/F/Y] [M/V/L/I/F]X[R/K]XX[pS/pT]XX [M/V/L/I/F]XRXX[pS/pT]XXX[M/V/L/I/F] RXXpS, RXX[pS/pT] [R/K]XX[pS/pT], R[K/E/R]XpS [E/D]X[pS/pT][I/L/V/M]X[E/D] P[pS/pT]X, pSXX[E/D] pSXX[E/pS*/pT*], [pS/pT]XX[E/D] [pS/pT]XX[E/D/pS*/pT*] |
Chk1 kinase Calmodulin-dependent protein kinase I, II, IV PKA, PKC, Plk1 kinase, CK II DNA dependent Protein kinase |
RXXpS | 14-3-3 |
| 921RGSPEEAAEREPR933 | calcium channel, voltage-dependent, N type, alpha 1B subunit | Q00975 | S923 | RXXpS, RXX[pS/pT], [R/K]X[pS/pT], RXpS, XXpSP, X[pS/pT]P, X[pS/pT]XXX[A/P/S/T], pSP, pSX[E/pS*/pT*], pSXX[E/D], pSXX[E/pS*/pT*], [pS/pT]XX[E/D], [pS/pT]XX[E/D/pS*/pY*] | PKC, PKA, GSK-3, ERK1, ERK2, CDK5, G-protein coupled receptor kinase 1, CK II, Calmodulin dependent protein kinase | RXXpS, [pS/pT]P | 14-3-3, WW |
| 214(S)(S)PAFGDRR222 | aminolevulinate dehydratase | P13716 | S214 or S215 | [R/K]X[pS/pT], X[pS/pT]P, XXpSP, pSP | PKA, PKC, GSK-3, ERK1/2, CDK5 | S[pS/pT]X, [pS/pT]P | MDC1 BRCT, Plk1 PBD, WW |
| 97ANCSPEDPCQETVSKPEVSK116 | proline rich transmembrane protein | Q7Z6L0 | S100 | KXXX[pS/pT], XXpSP, X[pS/pT]P X[pS/pT]XXX[A/P/S/T] XpSXXDXX, pSP pSX[E/pS*/pT*], pSXX[E/D] [pS/pT]XX[E/D] [pS/pT]XX[E/D/pS*/pT*] |
PKA, GSK-3, ERK1, ERK2, CDK5, G protein-coupled receptor kinase1, Pyruvate dehydrogenase kinase, ERK1/2, CKII | [pS/pT]P | WW |
| 793TEEERTPNHDGGK805 | neuronal cell adhesion molecule | P13591 | T798 | [E/D]XX[pS/pT], X[pS/pT]P, [R/K][pS/pT]P | Casein kinase I, GSK-3, ERK1, ERK2, CDK5, Growth associated histone HI kinase | [pS/pT]P | WW |
| 361NSAAATSPK369 | Ca2+/calmodulin-dependent protein kinase | Q13554 | S367 | XXpSP, X[pS/pT]P pSP, [pS/pT]P[R/K], [pS/pT]X[R/K], pSPXX[pS*/pT*] [pS/pT]XXX[S/T][M/L/V/I/F] |
GSK-3, ERK1/2, CDK5, PKA, PKC, CK I, Growth associated histone HI kinase | [pS/pT]P | WW |
| 185QTGNTCK191 | synaptophysin | P08247 | T186 | [R/K]X[pS/pT], [pS/pT]XX[S/T] | PKA, PKC, CK I | na | na |
| 297SGSPMAR303 | myelin basic protein | P02686 | S299 | pSXXX[pS/pT], pSXXXpS* RXXpS, RXX[pS/pT], [R/K]XX[pS/pT], XXpSP, X[pS/pT]P, pSP |
MAPKAPK2, GSK3, ERK1, ERK2, CDK5, Calmodulin-dependent protein kinase II, PKA, PKC | RXXpS, [pS/pT]P | 14-3-3, WW |
| 372G(S)(S)GVGL(T)AAVTTDQETGERR392 | DNA replication licensing factor MCM3 | P25205 | S373, S374 or T379 | X[pS/pT]XXX[A/P/S/T] | G-protein coupled receptor kinase1 | na | na |
| 137SRTAAYNTLK146 | V-type proton ATPase subunit C 2 | Q8NEY4 | Y142, T144 | [pS/pT]X[R/K], pYXX[L/I/V] | PKA, PKC, JAK2 | na | na |
| 16GWSTDEANTYFK27 | intelectin 1 | Q8WWA0 | Y25 | Na | na | na | na |
| 304(T)I(T)QKVLQK312 | olfactory receptor 10A7 | Q8NGE5 | T304 or T306 | [M/I/L/V]X[R/K]XX[pS/pT], [M/I/L/V/F/Y]XRXX[pS/pT], [M/V/L/I/F]X[R/K]XX[pS/pT]XX, [M/V/L/I/F][R/K/H]XXX[pS/pT]XXX[M/V/L/I/F], [pS/pT]X[R/K], [M/V/L/I/F]XRXX[pS/pT]XXX[M/V/L/I/F], [R/K]X[pS/pT], [R/K]XX[pS/pT] KXXX[pS/pT], RXX[pS/pT] [R/K]XX[pS/pT]X[R/K] |
Chk1 kinase, PKA, PKC, AMP-activated protein kinase Calmodulin-dependent protein kinase I, II, IV |
na | na |
| 1778EQASSLSDDRK1784 | Dynein heavy chain 3, axonemal | Q8TD57 | S1784 | pSXX[E/pS*/pT*] pSX[E/pS*/pT*] |
CK II | na | na |
| 628MKEPDGKLSPPK639 | teashirt zinc finger homeobox 3 | Q63HK5 | S636 | [R/K]X[pS/pT], XXpSP X[pS/pT]P, pSP [pS/pT]PX[R/K], pSPX[R/K]X |
PKA, PKC, GSK-3, ERK1/2, CDK1, 2, 4, 5, 6, Growth associated histone HI kinase, Cdc2 kinase | [pS/pT]P | WW |
Phosphorylated residues are underlined. Phosphosites with ambiguous localization appear in parenthesis. The phosphosites observed on these peptides are represented in bold on the motif, while the sites that are required to be phosphorylated first for the recognition of the motif are indicated with an asterisk. na: information not available.
Differential phosphorylation patterns on proteins in the brains from individuals with HIV and HIV-encephalitis
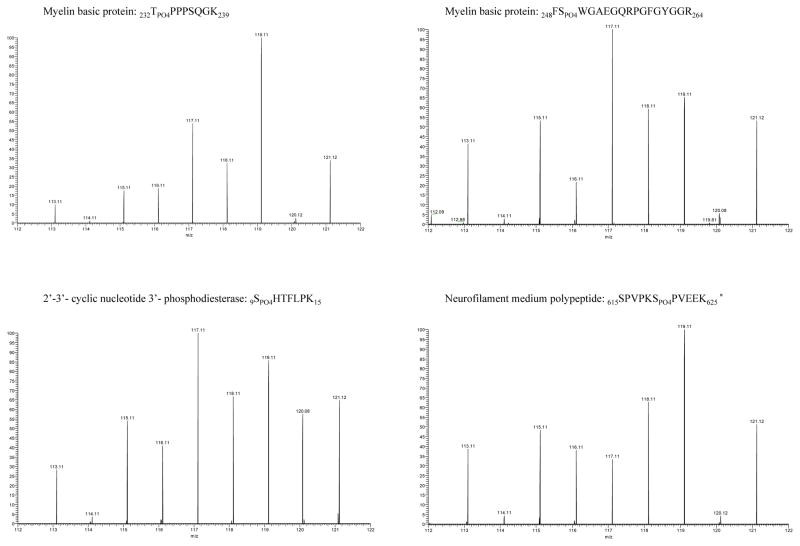
Site-specific quantitative phosphosite analysis was performed using the iTRAQ reporter ion intensities. Although fold change-based quantification is commonly used for relative protein quantification that involves multiple peptides per protein, we chose to rely on the statistical significance between the relative abundances of peptides in the control vs. HIV group and control vs. HIV vs. HIV-E group for determining the peptides that exhibited differential phosphorylation levels in different sample groups. The Wilcoxon Rank Sum and the Kruskal-Wallis tests identified four peptides that had statistically significant differences (p ≤ 0.05). Peptides 232TPO4PPPSQGK239 and 248FSPO4WGAEGQRPGFGYGGR264 were from myelin basic protein (MBP), 9SPO4HTFLPK15 from 2′–3′-cyclic nucleotide-3′ phosphodiesterase (CNP) and 615SPVPKSPO4PVEEK625 from neurofilament medium polypeptide (NFM). The SPVPKSPVEEK is a sequence within NMF that is repeated four times on the protein between residues 615-625, 628-638, 641-651, and 654-664, thus it is not possible to localize the exact phosphorylation site to S620, S633, S646 or S659 based on the identified tryptic peptides. The reporter ion regions of the MS/MS spectra are shown in Figure 2 for each of the four phosphopeptides with statistically significant differences among the sample groups (Supporting Figures 19–22). Although the phosphorylated peptides were detected in all of the samples, their relative abundance was higher in the HIV and the HIV-E samples as opposed to the control samples.
Figure 2.
iTRAQ reporter ion regions of phosphopeptide MS/MS spectra (p-value < 0.05). The labeling of post-mortem brain samples was as follows: control 113.11, 114.11, 115.11, 116.11; HIV 117.11, 118.11; and HIV-encephalitis 119.11, 121.12. * This sequence is termed as a KSP repeat and is found four times on the protein between residues 615-625, 628-638, 641-651, and 654-664.
The similarity of the amino acid sequences of the phosphorylated peptides indicated that they may share kinase substrate motifs and be phosphorylated by similar kinases. The MotifFinder search of the Human Protein Reference Database resulted in the potential kinases listed in Table 2. Both TPPPSQGK and SPVPKSPVEEK can potentially be phosphorylated by glycogen synthase kinase 3 (GSK3), ERK1/2, Cyclin-dependent kinase 5 (CDK5) kinases and growth-associated histone HI kinase. FSWGAEGQRPGFGYGGR and SHTFLPK can potentially be phosphorylated by casein kinase II, PKA and PKC. Casein kinase may also phosphorylate SPVPKSPVEEK. The TPPPSQGK and SPVPKSPVEEK peptides also contain WW binding domains indicating a potential role in protein-protein interaction. No binding domains were found on the FSWGAEGQRPGFGYGGR and SHTFLPK peptides.
Table 2.
The kinase substrate motifs and binding motifs for differentially phosphorylated peptides between control and HIV-infected brain samples. a
| Sequence | Protein | Corresponding Motif | Kinase Substrate Motif | Binding Motif | Binding Domain |
|---|---|---|---|---|---|
| 232TPO4PPPSQGK239 | Myelin basic protein | PXpTP, PX[pS/pT]P, PX[pS/pT]PP, X[pS/pT]P, [R/K][pS/pT]P | GSK3, ERK1, ERK2, CDK5 kinase, Growth associated histone HI kinase | [pS/pT]P | WW |
| 248FSPO4WGAEGQRPGFGYGGR264 | Myelin basic protein | pSXX[E/pS*/pT*], [R/K]X[pS/pT] [pS/pT]XX[E/D/pS/pY*], RXpS |
CK II, PKA, PKC | na | na |
| 9SPO4HTFLPK15 | 2′–3′-cyclic nucleotide 3′-phosphodiesterase | pSXX[E/pS/pT*], [pS/pT]XX[E/D/pS/pY*], RXpS, [R/K]X[pS/pT], KXX[pS/pT], [R/K]XX[pS/pT], pSX[E/pS*/pT*] | CK II, PKA, PKC | na | na |
|
615SPVPKSPO4PVEEK625 628SPVPKSPO4PVEEK638 641SPVPKSPO4PVEEK651 654SPVPKSPO4PVEEK664 |
Neurofilament medium polypeptide | X[pS/pT]P, [R/K][pS/pT]P, KpSPXXXK, pSP, pSXX[E/D], pSXX[E/pS/pT*], [pS/pT]XX[E/D], [pS/pT]XX[E/D/pS*/pY*] | Growth associated histone HI kinase, GSK3, ERK1, ERK2, CDK5 kinase, CKII | [pS/pT]P | WW |
The motifs were obtained from the Human Protein Reference Database. The phosphosites observed on these peptides are represented in bold on the motif while the sites that are required to be phosphorylated first for the recognition of the motif by the kinase are indicated with an asterisk, na: information not available.
Validation of differential phosphorylations of MBP on PKA substrate motifs
Unlike the site-specific data obtained from the high-resolution and high-mass-accuracy MS, the Western blot analysis of site-specific phosphorylations is limited by the availability of antibodies. MBP is a PKA substrate and can be phosphorylated on multiple PKA substrate motifs. We immunoprecipitated the MBP protein from the 8 brain lysates and performed Western blot analysis using the phospho-serine/threonine PKA substrate antibody. The highest signal was in the HIV and HIV-E samples (Figure 3). Due to the presence of multiple PKA substrate motifs on MBP, it is not possible to correlate the signal intensity in the Western blot to the iTRAQ intensity in the LC-MS/MS analysis. However, increased phosphorylation of MBP on PKA substrate motifs indicates an increase in PKA activity during HIV infection and is in agreement with the differential phosphorylation we observed on the PKA substrate motif 9SPO4HTFLPK15 on MBP.
Figure 3.
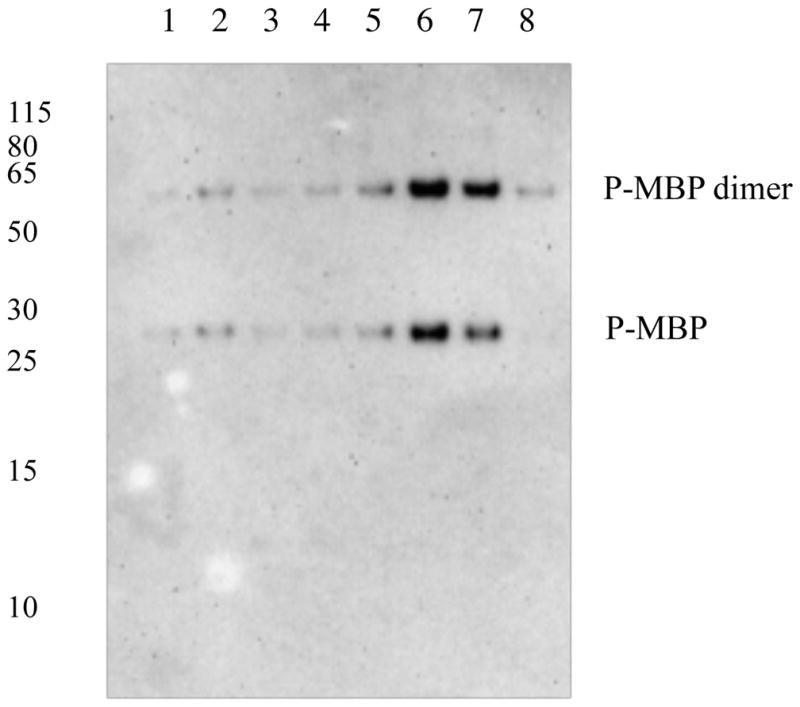
Phosphorylation of myelin basic protein (P-MBP) on PKA substrate motifs is increased with HIV infection of the brain. Myelin basic protein was immunoprecipitated and analyzed using the anti-phospho serine/threonine PKA substrate primary antibody. Lanes 1–4, brain lysates from healthy individuals; lanes 5–6, brain lysates from individuals with HIV infection without encephalitis; lanes 7–8, brain lysates from individuals with HIV infection and encephalitis.
DISCUSSION
Increase in site-specific phosphorylation of brain proteins may be the result of altered phosphorylation pathway proteins in HIV infection
Our study revealed that phosphopeptides 232TPO4PPPSQGK239 and 248FSPO4WGAEGQRPGFGYG264 from MBP, 9SPO4HTFLPK15 from CNP and SPVPKSPO4PVEEK (repeat motif) from NFM are found at significantly higher levels in the brains from individuals with HIV infection when compared to the controls. All the potential kinases other than growth-associated histone H1 kinase involved in the phosphorylation of these sites on the proteins have been shown to be activated during HIV infection and HIV neuropathogenesis, further supporting our findings.
The 248FSPO4WGAEGQRPGFGYG264 peptide from MBP and 9SPO4HTFLPK15 peptide from CNP share PKC, CKII and PKA kinase substrate motifs. During infection, HIV hijacks macrophage cellular machinery and activates PKC for the rearrangement of the actin cytoskeleton necessary for viral entry. [24] PKC is also involved in the expression of NF-κB that is required for viral gene transcription. [25] Its activity increases upon exposure to HIV-Tat protein. [11] Similarly, CKII is activated by HIV infection for the phosphorylation of viral proteins. Several HIV inhibitors target this protein, as reviewed previously. [26] Conversely, PKA is involved in triggering the immune response and hyperactivation of the cAMP/PKA system has been associated with HIV infection. [27]
Phosphopeptides 232TPO4PPPSQGK239 (MBP) and 9SPVPKSPO4PVEEK (NFM) contain kinase substrate motifs for ERK1/2, CDK5 and GSK3. The ERK1/2 pathway is involved in HIV replication and immune response. It has roles in cytokine production, HIV-1 reverse transcription and viral assembly. [9] Additionally, the HIV coat protein gp120 can activate ERK1/2 in human neurons. [28] A mechanism of fine-tuning the ERK1/2 system may be through reactive oxygen species that regulate the upstream signaling pathways such as PKC [29]. Additionally, cyclin-dependent kinase 5 (CDK5), a kinase highly expressed in the nervous system, is hyperactivated during HIV encephalopathy[30] and exhibits neurotoxic effects potentially through tau and β-catenin phosphorylation. [31–33] Finally, the GSK3 inhibitors prevent Tat-mediated HIV-1 replication and neurotoxicity [34] and GSK3 is a therapeutic target in HAND. [35]
As explained above, the relative increase in MBP, CNP, and NFM phosphorylations is in agreement with previous publications on the activation of specific kinases during HIV infection. A recent study with post-mortem human brain and HIV/gp120 transgenic mouse brain has also linked cysteine S-nitrosylation induced changes on kinase activity to alteration in neuronal function in HIV infection. [36] A similar finding is also present in Alzheimer’s disease through the activation of CDK5 upon this modification. [37] Our findings on the effect of altered phosphorylation pathways at the neuronal level has critical implications and opens the door for future studies to understand how changes in phosphorylation may impair neuronal activities. Furthermore, a better understanding of unusual phosphorylation events during HIV neuropathogenesis and identification of the upregulated kinases may present a new approach in treatment. GSK3 has already been proposed as a therapeutic target in HAND; thus inhibition of upregulated kinases may have important clinical implications.
Phosphorylation and protein-protein interactions in neuronal proteins
Phosphorylation has a role in protein-protein interaction, protein localization and signal transduction. MBP and CNP are the major proteins in the myelin sheath and are critical for facilitating nerve impulses and supporting the axon. NFM is a major protein in the axon of the neuron and is important for its structure. Since the myelin wraps around the axon, these proteins are intimately related. Upon the identification of relatively higher levels of phosphorylated peptides from MBP, CNP and NFM, we investigated the proteins that they interact with using the Ingenuity Pathway Analysis network (Supporting figure 1). Among the numerous direct and indirect interactions involved with these proteins, the interaction of MBP with actin and the interaction of CNP with tubulin were of particular interest due to the effect of phosphorylation resulting from these interactions. MBP interacts with actin through electrostatic interactions. Upon phosphorylation, the net positive charge is decreased and this interferes with its binding to actin and actin polymerization. [38] Similarly, CNP is involved in microtubule assembly through its binding to tubulin. Phosphorylation of CNP has been shown to interfere with its assembly promoting activity. [39] Although these studies were not performed using site-specific antibodies for the detection of phosphorylation on the proteins, the altered phosphorylation on myelin sheath proteins may have significant implications for neuronal function through altered cytoskeleton dynamics. Lastly, NFM is an important protein in axonal cytoskeleton and transport. Aberrant neurofilament phosphorylation and neurofibrillary accumulation in the neuronal cell body are found in various neurodegenerative diseases. Any alteration on the phosphorylation pattern may influence the binding of neurofilament cross-bridge formation and their association with motor proteins for axonal transport.
Limitations
We acknowledge that our study has some limitations, namely the small sample size. Although iTRAQ-based relative quantification permitted the direct comparison of eight samples in our study, larger sample sizes and different regions of the brain need to be studied to confirm our observations. Phosphorylation is a dynamic process that can occur very rapidly. Hence, the use of post-mortem tissue has the inherent issue of being a cross-sectional analysis and changes in the phosphorylation profile may occur during the post-mortem interval. However, post-mortem tissue is critical for confirming findings in model systems [40] and identifying stably phosphorylated proteins that can accumulate in pathological tissues, such as in Alzheimer’s disease. [41,42]
Conclusions
Taken together, our data demonstrate the presence of phosphorylations on important proteins in control and HIV-infected human brain. The novel phosphorylation sites identified on critical human brain proteins may be involved in signaling pathways and have critical functions. Increased phosphorylation of MBP, CNP and NFM during HIV infection may have important implications for the associated neurocognitive dysfunction.
Supplementary Material
CLINICAL RELEVANCE.
Human immunodeficiency virus (HIV) can penetrate the blood brain barrier and eventually lead to neurological impairment, collectively termed, HIV-associated neurocognitive disorders (HAND). Although combined antiretroviral therapy has extended the lifetime of HIV-infected individuals, mild forms of HAND persist. To understand the molecular basis of HAND we mapped the phosphoproteome of the HIV-infected and uninfected human brain using high resolution and high mass accuracy mass spectrometry. We discovered 112 phosphorylated proteins and 17 novel phosphorylation sites. These observations provide the basis for further studies to understand the role of these phosphorylation events in neuro-glial function under normal and pathological conditions. Specific phosphorylation patterns in HIV-infected brain could also reveal upregulated kinases that can be potential therapeutic targets.
Acknowledgments
RJC and LU were supported by grant R01NS039253 from the NINDS. AN and SA are supported by intramural NIH funds. We wish to thank the Johns Hopkins University School of Medicine Mass Spectrometry and Proteomics Facility for technical assistance, Dr. Wenxue Li for discussion and Drs. Ronald Schnaar and Stefani Thomas for the critical reading of the manuscript.
ABBREVIATIONS
- Akt
protein kinase B
- BRCT
breast cancer susceptibility protein C terminal domain
- Cdc2
cyclin dependent kinase 2
- Chk1
checkpoint kinase
- cGMP
cyclic guanosine monophosphate
- CDK5
cyclin-dependent kinase 5
- CNP
2′–3′-cyclic nucleotide-3′ phosphodiesterase
- CK II
casein kinase II
- ERK 1/2
extracellular-stimulated phosphoprotein-like protein
- GSK3
glycogen synthase kinase 3
- HAND
HIV-associated neurocognitive disorders
- HCD
higher energy collision dissociation
- HIV
human immunodeficiency virus
- IPA
Ingenuity Pathway Analysis
- ITMS
ion trap mass spectrometry
- JAK2
Janus kinase 2
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- MAPK
mitogen-activated protein kinase
- MAPKMAPK2
MAPK-activated protein kinase 2
- MBP
myelin basic protein
- MDC1
mediator of DNA-damage checkpoint 1
- NFM
neurofilament medium polypeptide
- PBD
protein binding domain
- PKA
protein kinase A
- PKC
protein kinase C
- Plk1
Polo-like kinase (serine/threonine-protein kinase PLK1)
- PTEN
phosphatase and tensin homolog
- SDS
sodium dodecyl sulfate
Footnotes
Declaration
The authors declare no competing financial interest.
Dedications
The authors dedicate this work to the memory of our beloved colleague Dr. Robert J. Cotter, who died on November 12, 2012.
References
- 1.Heaton RK, Franklin DR, Ellis RJ, McCutchan JA, et al. HIV-associated neurocognitive disorders before and during the era of combination antiretroviral therapy: differences in rates, nature, and predictors. J Neurovirol. 2011;17:3–16. doi: 10.1007/s13365-010-0006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bora A, Mohien CU, Chaerkady R, Chang L, et al. Identification of putative biomarkers for HIV-associated neurocognitive impairment in the CSF of HIV-infected patients under cART therapy determined by mass spectrometry. J Neurovirol. 2014 doi: 10.1007/s13365-014-0263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laspiur JP, Anderson ER, Ciborowski P, Wojna V, et al. CSF proteomic fingerprints for HIV-associated cognitive impairment. J Neuroimmunol. 2007;192:157–170. doi: 10.1016/j.jneuroim.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rozek W, Ricardo-Dukelow M, Holloway S, Gendelman HE, et al. Cerebrospinal fluid proteomic profiling of HIV-1-infected patients with cognitive impairment. J Proteome Res. 2007;6:4189–4199. doi: 10.1021/pr070220c. [DOI] [PubMed] [Google Scholar]
- 5.Uzasci L, Bianchet MA, Cotter RJ, Nath A. Identification of nitrated immunoglobulin variable regions in the HIV-infected human brain: implications in HIV infection and immune response. J Proteome Res. 2014;13:1614–1623. doi: 10.1021/pr401117m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uzasci L, Nath A, Cotter R. Oxidative Stress and the HIV-Infected Brain Proteome. J Neuroimmune Pharmacol. 2013;8:1167–1180. doi: 10.1007/s11481-013-9444-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beasley A, Anderson C, McArthur J, Sacktor N, et al. Characterization of Nitrotyrosine-Modified Proteins in Cerebrospinal Fluid. Clinical Proteomics. 2010:29–41. [Google Scholar]
- 8.Francis AC, Di Primio C, Allouch A, Cereseto A. Role of phosphorylation in the nuclear biology of HIV-1. Curr Med Chem. 2011;18:2904–2912. doi: 10.2174/092986711796150478. [DOI] [PubMed] [Google Scholar]
- 9.Furler RL, Uittenbogaart CH. Signaling through the P38 and ERK pathways: a common link between HIV replication and the immune response. Immunol Res. 2010;48:99–109. doi: 10.1007/s12026-010-8170-1. [DOI] [PubMed] [Google Scholar]
- 10.Kundu M, Sharma S, De Luca A, Giordano A, et al. HIV-1 Tat elongates the G1 phase and indirectly promotes HIV-1 gene expression in cells of glial origin. J Biol Chem. 1998;273:8130–8136. doi: 10.1074/jbc.273.14.8130. [DOI] [PubMed] [Google Scholar]
- 11.Conant K, Ma M, Nath A, Major EO. Extracellular human immunodeficiency virus type 1 Tat protein is associated with an increase in both NF-kappa B binding and protein kinase C activity in primary human astrocytes. J Virol. 1996;70:1384–1389. doi: 10.1128/jvi.70.3.1384-1389.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avraham HK, Jiang S, Lee TH, Prakash O, Avraham S. HIV-1 Tat-mediated effects on focal adhesion assembly and permeability in brain microvascular endothelial cells. J Immunol. 2004;173:6228–6233. doi: 10.4049/jimmunol.173.10.6228. [DOI] [PubMed] [Google Scholar]
- 13.King JE, Eugenin EA, Hazleton JE, Morgello S, et al. Mechanisms of HIV-tat-induced phosphorylation of N-methyl-D-aspartate receptor subunit 2A in human primary neurons: implications for neuroAIDS pathogenesis. Am J Pathol. 2010;176:2819–2830. doi: 10.2353/ajpath.2010.090642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haughey NJ, Nath A, Mattson MP, Slevin JT, et al. HIV-1 Tat through phosphorylation of NMDA receptors potentiates glutamate excitotoxicity. J Neurochem. 2001;78:457–467. doi: 10.1046/j.1471-4159.2001.00396.x. [DOI] [PubMed] [Google Scholar]
- 15.Alirezaei M, Watry DD, Flynn CF, Kiosses WB, et al. Human immunodeficiency virus-1/surface glycoprotein 120 induces apoptosis through RNA-activated protein kinase signaling in neurons. J Neurosci. 2007;27:11047–11055. doi: 10.1523/JNEUROSCI.2733-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medders KE, Kaul M. Mitogen-activated protein kinase p38 in HIV infection and associated brain injury. J Neuroimmune Pharmacol. 2011;6:202–215. doi: 10.1007/s11481-011-9260-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akay C, Lindl KA, Wang Y, White MG, et al. Site-specific hyperphosphorylation of pRb in HIV-induced neurotoxicity. Mol Cell Neurosci. 2011;47:154–165. doi: 10.1016/j.mcn.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valero IP, Baeza AG, Hernandez-Tamames JA, Monge S, et al. Cerebral volumes, neuronal integrity and brain inflammation measured by MRI in patients receiving PI monotherapy or triple therapy. J Int AIDS Soc. 2014;17:19578. doi: 10.7448/IAS.17.4.19578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfefferbaum A, Rogosa DA, Rosenbloom MJ, Chu W, et al. Accelerated aging of selective brain structures in human immunodeficiency virus infection: a controlled, longitudinal magnetic resonance imaging study. Neurobiol Aging. 2014;35:1755–1768. doi: 10.1016/j.neurobiolaging.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsudaira PT. A Practical guide to protein and peptide purification for microsequencing. 2. Academic Press; San Diego: 1993. [Google Scholar]
- 21.Dosemeci A, Jaffe H. Regulation of phosphorylation at the postsynaptic density during different activity states of Ca2+/calmodulin-dependent protein kinase II. Biochem Biophys Res Commun. 2010;391:78–84. doi: 10.1016/j.bbrc.2009.10.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J, Shakey Q, Liu W, Schuller A, et al. Global profiling of phosphopeptides by titania affinity enrichment. J Proteome Res. 2007;6:4684–4689. doi: 10.1021/pr070481m. [DOI] [PubMed] [Google Scholar]
- 23.Olsen JV, de Godoy LM, Li G, Macek B, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Harmon B, Ratner L. Induction of the Galpha(q) signaling cascade by the human immunodeficiency virus envelope is required for virus entry. J Virol. 2008;82:9191–9205. doi: 10.1128/JVI.00424-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cosentino-Gomes D, Rocco-Machado N, Meyer-Fernandes JR. Cell Signaling through Protein Kinase C Oxidation and Activation. Int J Mol Sci. 2012;13:10697–10721. doi: 10.3390/ijms130910697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.St-Denis NA, Litchfield DW. Protein kinase CK2 in health and disease: From birth to death: the role of protein kinase CK2 in the regulation of cell proliferation and survival. Cell Mol Life Sci. 2009;66:1817–1829. doi: 10.1007/s00018-009-9150-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torgersen KM, Vang T, Abrahamsen H, Yaqub S, et al. Molecular mechanisms for protein kinase A-mediated modulation of immune function. Cell Signal. 2002;14:1–9. doi: 10.1016/s0898-6568(01)00214-5. [DOI] [PubMed] [Google Scholar]
- 28.Lannuzel A, Barnier JV, Hery C, Huynh VT, et al. Human immunodeficiency virus type 1 and its coat protein gp120 induce apoptosis and activate JNK and ERK mitogen-activated protein kinases in human neurons. Ann Neurol. 1997;42:847–856. doi: 10.1002/ana.410420605. [DOI] [PubMed] [Google Scholar]
- 29.Runchel C, Matsuzawa A, Ichijo H. Mitogen-activated protein kinases in mammalian oxidative stress responses. Antioxid Redox Signal. 2011;15:205–218. doi: 10.1089/ars.2010.3733. [DOI] [PubMed] [Google Scholar]
- 30.Lee MH, Amin ND, Venkatesan A, Wang T, et al. Impaired neurogenesis and neurite outgrowth in an HIV-gp120 transgenic model is reversed by exercise via BDNF production and Cdk5 regulation. J Neurovirol. 2013;19:418–431. doi: 10.1007/s13365-013-0194-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu J, Li W, Mao Z. Cdk5: mediator of neuronal development, death and the response to DNA damage. Mech Ageing Dev. 2011;132:389–394. doi: 10.1016/j.mad.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patrick C, Crews L, Desplats P, Dumaop W, et al. Increased CDK5 expression in HIV encephalitis contributes to neurodegeneration via tau phosphorylation and is reversed with Roscovitine. Am J Pathol. 2011;178:1646–1661. doi: 10.1016/j.ajpath.2010.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dou H, Ellison B, Bradley J, Kasiyanov A, et al. Neuroprotective mechanisms of lithium in murine human immunodeficiency virus-1 encephalitis. J Neurosci. 2005;25:8375–8385. doi: 10.1523/JNEUROSCI.2164-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kehn-Hall K, Guendel I, Carpio L, Skaltsounis L, et al. Inhibition of Tat-mediated HIV-1 replication and neurotoxicity by novel GSK3-beta inhibitors. Virology. 2011;415:56–68. doi: 10.1016/j.virol.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dewhurst S, Maggirwar SB, Schifitto G, Gendelman HE, et al. Glycogen synthase kinase 3 beta (GSK-3 beta) as a therapeutic target in neuroAIDS. J Neuroimmune Pharmacol. 2007;2:93–96. doi: 10.1007/s11481-006-9051-1. [DOI] [PubMed] [Google Scholar]
- 36.Banerjee S, Liao L, Russo R, Nakamura T, et al. Isobaric tagging-based quantification by mass spectrometry of differentially regulated proteins in synaptosomes of HIV/gp120 transgenic mice: implications for HIV-associated neurodegeneration. Exp Neurol. 2012;236:298–306. doi: 10.1016/j.expneurol.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qu J, Nakamura T, Cao G, Holland EA, et al. S-Nitrosylation activates Cdk5 and contributes to synaptic spine loss induced by beta-amyloid peptide. Proc Natl Acad Sci US A. 2011;108:14330–14335. doi: 10.1073/pnas.1105172108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boggs JM, Rangaraj G, Gao W, Heng YM. Effect of phosphorylation of myelin basic protein by MAPK on its interactions with actin and actin binding to a lipid membrane in vitro. Biochemistry. 2006;45:391–401. doi: 10.1021/bi0519194. [DOI] [PubMed] [Google Scholar]
- 39.Bifulco M, Laezza C, Stingo S, Wolff J. 2′,3′-Cyclic nucleotide 3′-phosphodiesterase: a membrane-bound, microtubule-associated protein and membrane anchor for tubulin. Proc Natl Acad Sci US A. 2002;99:1807–1812. doi: 10.1073/pnas.042678799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rakhade SN, Fitzgerald EF, Klein PM, Zhou C, et al. Glutamate receptor 1 phosphorylation at serine 831 and 845 modulates seizure susceptibility and hippocampal hyperexcitability after early life seizures. J Neurosci. 2012;32:17800–17812. doi: 10.1523/JNEUROSCI.6121-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudrabhatla P, Grant P, Jaffe H, Strong MJ, et al. Quantitative phosphoproteomic analysis of neuronal intermediate filament proteins (NF-M/H) in Alzheimer’s disease by iTRAQ. FASEB J. 2010;24:4396–4407. doi: 10.1096/fj.10-157859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, Wang M, Turko IV. Mass spectrometry quantification of clusterin in the human brain. Mol Neurodegener. 2012;7:41-1326-7-41. doi: 10.1186/1750-1326-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.