Abstract
Clostridium difficile is the most commonly reported nosocomial pathogen in the United States and is an urgent public health concern worldwide1. Over the past decade, incidence, severity, and costs associated with C. difficile infection (CDI) have increased dramatically2. CDI is most commonly initiated by antibiotic-mediated disruption of the gut microbiota; however, non-antibiotic associated CDI cases are well documented and on the rise3,4. This suggests that unexplored environmental, nutrient, and host factors likely influence CDI. Here we show that excess dietary zinc (Zn) significantly alters the gut microbiota and in turn reduces the threshold of antibiotics needed to confer susceptibility to C. difficile infection. In mice colonized with C. difficile, excess dietary Zn severely exacerbates C. difficile-associated disease by increasing toxin activity and altering the host immune response. In addition, we show that the Zn binding S100 protein calprotectin is antimicrobial against C. difficile and an essential component of the innate immune response to CDI. Together, these data suggest that nutrient Zn levels play a key role in determining susceptibility to CDI and severity of disease, and that calprotectin-mediated metal limitation is an important factor in the host immune response to C. difficile.
C. difficile is a spore-forming gram-positive bacterium that causes a range of gastrointestinal disorders varying in severity from diarrhea to colitis5. The transmissible spore of C. difficile is abundant in health care facilities and resistant to disinfectants, making it a major nosocomial pathogen5–7. The primary risk factor for CDI is antibiotic use, which reduces colonization resistance to C. difficile by altering gut microbiota3,8,9. Non-antibiotic associated and community-acquired CDI cases are increasing, suggesting that other host and environmental factors impact susceptibility to C. difficile4. One of the most important environmental factors influencing the gut microbiota, and potentially susceptibility to CDI, is diet10. Specifically, dietary metals are associated with susceptibility to numerous infections11. Metal availability is a critical factor affecting the outcome of host-pathogen interactions and metal levels vary widely depending on host diet and environmental exposures12. Here, we sought to examine the impact of dietary Zn and Zn availability on susceptibility to and severity of CDI.
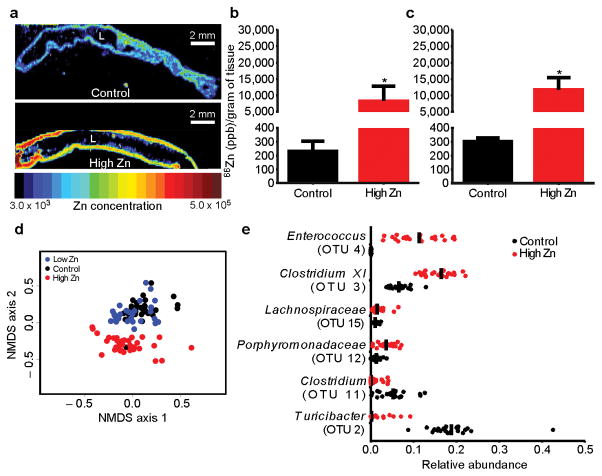
To determine the impact of dietary Zn on CDI, diets containing no Zn (low Zn diet; 0 mg/kg Zn), normal Zn (control diet; 29 mg/kg Zn), or excess Zn (high Zn diet; 1,000 mg/kg Zn) were synthesized. The high Zn diet was designed to model excess Zn supplementation in humans, with approximately 12-fold the Zn level of standard mouse chow (LabDiet 5K52/5K67; LabDiet 5100). Mice were fed altered Zn diets for five weeks, during which no signs of toxicity were observed. Mice fed the high Zn diet had increased levels of Zn in cecal and colonic tissue, and feces (Figure 1a–c; Supplementary Fig 1a). In contrast, mice fed the low Zn diet had significantly decreased levels of Zn in these compartments (Supplementary Fig 1a). These findings demonstrate that dietary Zn alterations impact cecal and colonic Zn levels.
Figure 1. Increased dietary Zn alters tissue level Zn and dramatically alters the gut microbiota.
Relative Zn levels following diet manipulation imaged using laser ablation inductively coupled plasma mass spectrometry (LA-ICP MS) in the colon (a). Colons were cleared of all contents prior to imaging. The lumen of each colon is denoted with “L”. Scale bars, 2 mm. Zn concentrations measured by ICP-MS following diet manipulation in the cecum (b) or colon (c) normalized to organ weight (n=5/group). Values are represented in parts per billion (ppb). ICP-MS data are represented as mean ± standard deviation. * P < 0.01; by Mann-Whitney test. Non-metric multidimensional scaling (NMDS) ordination plot showing fecal gut microbiota β-diversity during the five-week time course of dietary Zn manipulation measured by θYC (d). Significance between high Zn and control groups was determined by Analysis of Molecular Variance (AMOVA) (P < 0.001). Control and Low Zn groups were not significantly different. Strip charts represent the relative abundance of significantly altered members of the microbiota following dietary Zn alteration (e). Significance was determined using ANOVA. Median relative abundance is represented by a vertical black line.
Diet is an important mediator of microbial community structure13; however, the impact of dietary Zn on the gut microbiota has yet to be elucidated. To examine the impact of Zn on the gut microbiota, feces were collected throughout a five-week time course of dietary Zn manipulation. Mice fed the high Zn diet showed decreased microbial diversity and a marked shift in community structure compared to mice fed the control and low Zn diets, which did not significantly differ (Fig. 1d and Supplementary Fig. 1b). Zn-mediated community shifts were observed as early as one week following diet alteration (Supplementary Fig. 1d). Characterization of taxa from mice fed the high Zn diet revealed a significant decrease in operational taxonomic units (OTUs) affiliated with the Turicibacter (OTU 2) and Clostridium (OTU 11) genera, and increases in OTUs affiliated with the Enterococcus (OTU 4) and Clostridium XI (OTU 3) genera (Fig. 1e). Excess Zn did not have an impact on the bacterial burden (Supplementary Fig. 1e). These data demonstrate that excess dietary Zn alters the diversity and structure of the gut microbiota in mice.
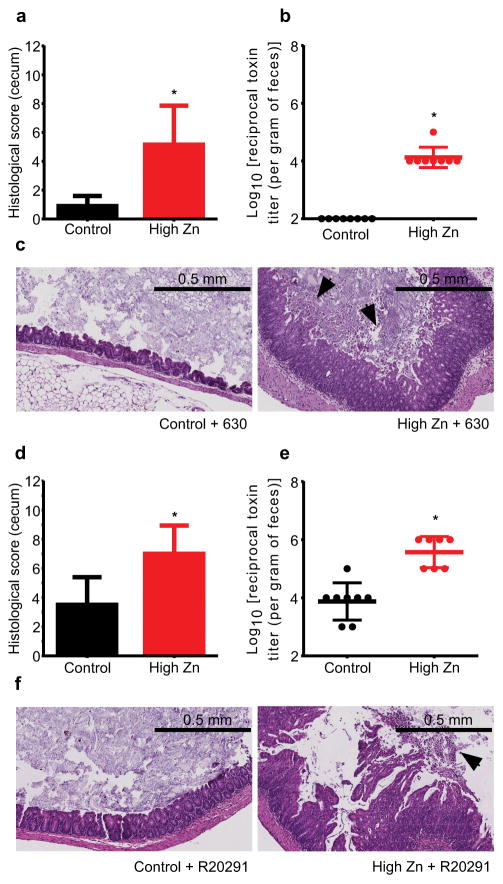
A number of C. difficile virulence factors have a Zn requirement; however, the impact of Zn on CDI has yet to be explored14–17. To test this, we used a mouse model of CDI that induces susceptibility to C. difficile through administration of cefoperazone (0.5 mg/ml)(Supplementary Fig. 2)18. Mice fed the high Zn diet showed exacerbation of disease, highlighted by increased inflammation, epithelial damage, and pseudomembrane formation (Fig. 2a,c,d,f; Supplementary Fig. 3b,d). Zn-mediated disease exacerbation was observed following infection with both C. difficile strains 63019 (ribotype 012) and R2029120 (ribotype 027) (Fig. 2a–f). Mice fed the low Zn diet did not exhibit increased disease and mice fed altered Zn diets showed no pathology prior to infection (Supplementary Fig. 4a–f). We did not observe significant differences in C. difficile burden between mice given control and high Zn diets (Supplementary Fig. 3a,c). To examine the impact of excess Zn on C. difficile virulence, C. difficile toxin A (TcdA) and B (TcdB) dependent toxicity was quantified using a cell-rounding assay18. Mice fed excess Zn showed increased toxin titers in feces following infection (Fig. 2b,e). Cytokine analysis revealed a significant increase in IL-1β levels in mice fed excess Zn and infected with R20291 (Supplementary Fig. 5). Moreover, the pro-inflammatory cytokines IL-6 and IL-12(p70) and the anti-inflammatory cytokine IL-10, were decreased in mice fed excess Zn (Supplementary Fig. 5). There were no significant increases in inflammation in mice fed excess Zn prior to infection (Supplementary Fig. 6). Loss of barrier function and microbiota translocation is associated with severe CDI and increased IL-1β production21,22. Therefore, the degree of microbiota translocation following infection was examined. Bacterial burdens from livers revealed a significant increase in microbiota translocation associated with CDI in mice fed excess Zn (Supplementary Fig. 7a,b). Livers from mice fed the control diet and infected with C. difficile were exclusively colonized with members of the Enterobacteriaceae. Livers from mice fed excess Zn were colonized with Enterobacteriaceae, Enterococcus, and non-C. difficile Clostridium spp. Together, these data demonstrate that excess Zn has a profound impact on C. difficile-associated disease.
Figure 2. Excess dietary Zn exacerbates C. difficile-associated disease.
Susceptibility to CDI was induced with 0.5 mg/ml cefoperazone treatment and mice were infected with either C. difficile strain 630 or R20291. Blinded histological scores from ceca of mice fed high Zn or control diets were determined four days post-infection with C. difficile strains 630 (a) or R20291 (d) (n=10/group). C. difficile toxin titer per gram of feces was determined in mice infected with C. difficile strains 630 (b) and R20291 (e). Representative H&E stained cecal sections from four days post-infection are shown for mice infected with C. difficile strain 630 (c) and R20291 (f). Images are representative of 10 ceca per group. Arrowheads denote pseudomembrane formation. All data are represented as mean ± standard deviation. * P < 0.01; by Mann-Whitney test.
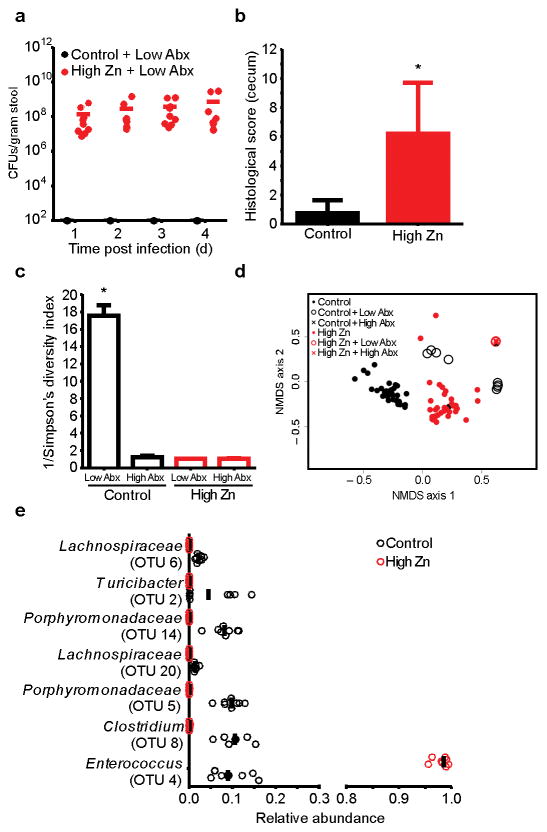
When we treated mice with antibiotics (0.5 mg/ml cefoperazone), C. difficile burden was not impacted by Zn; however, we reasoned that community shifts induced by Zn might lower the threshold of antibiotics needed to confer susceptibility to CDI (Supplementary Fig. 3). To test this, mice fed altered diets were treated with fifty-fold less cefoperazone (0.01 mg/ml) prior to infection. This low-level antibiotic treatment did not render mice fed the low Zn or control diets susceptible to CDI (Fig. 3a; Supplementary Fig. 8). In contrast, mice fed the high Zn diet were susceptible to CDI and exhibited severe disease (Fig. 3a–b). Mice that were not treated with antibiotics showed no C. difficile colonization for any of the diets. Examination of gut microbiota from mice fed the high Zn diet followed by low-level antibiotic treatment revealed a complete loss in diversity and a significant shift in the community that mirrored mice given high-level antibiotics (0.5 mg/ml) (Fig. 3c–e). In contrast, mice fed the control diet maintained high microbial diversity after low-level antibiotics, which may account for the robust resistance to CDI (Fig. 3c). These data suggest that excess Zn renders the gut microbiota vulnerable to low-level perturbations and decreases the threshold of antibiotics needed to diminish colonization resistance to C. difficile.
Figure 3. Excess dietary Zn decreases the threshold of antibiotics needed to confer susceptibility to CDI.
CFU analysis for C. difficile strain R20291 following low-level cefoperazone treatment (0.01 mg/ml cefoperazone) and oral gavage with 105 spores (a) (n=9/group). Solid bars represent mean colonization. Blinded histological scores quantified four days following low-level cefoperazone treatment and infection with R20291 (b) (n=9/group). Inverse Simpson’s diversity for mice fed control or high Zn diets following high-level (0.5 mg/ml) (high Abx; n=5/group) or low-level (0.01 mg/ml) (low Abx; n=4/control diet; n=5/high Zn diet) cefoperazone treatment (c). Histology and diversity data are represented as mean ± standard deviation. * P < 0.01; by Mann-Whitney test. NMDS plot depicting β-diversity of fecal microbiota measured by θYC for mice fed control (n=4) and high Zn (n=5) diets followed by high and low-level antibiotic treatment (d). Strip chart showing significantly altered members of the microbiota following dietary Zn alteration and subsequent treatment with low-levels of cefoperazone (0.01 mg/ml cefoperazone) (e). Significance was determined using ANOVA.
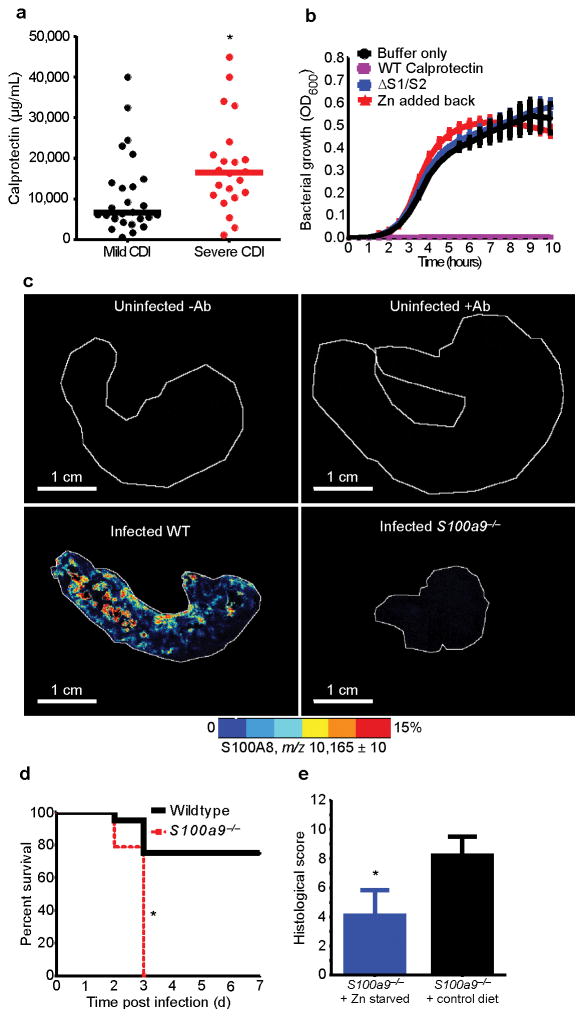
Calprotectin is a Zn binding protein that has antimicrobial activity against various pathogens23–26. Moreover, fecal calprotectin levels are predictive of disease severity in adults with CDI27,28. To further elucidate the association of calprotectin with CDI, fecal calprotectin was measured in twenty-five pediatric subjects with CDI. Fecal calprotectin levels were significantly higher in pediatric subjects that showed signs of severe CDI (Supplementary Table 1). Furthermore, increased serum calprotectin was associated with increased disease severity in forty-eight adults with CDI (Fig. 4a). However, the role of calprotectin during CDI has yet to be explored.
Figure 4. Calprotectin is essential for combating CDI.
Serum calprotectin levels were determined for forty-eight adult ICU patients infected with C. difficile (a). Cases of mild (n=26) and severe CDI (n=22) are represented. Solid bars represent median serum calprotectin levels. * P < 0.01; by Mann-Whitney test. C. difficile strain R20291 was grown in the presence of recombinant WT calprotectin (1 mg/ml) or a Zn binding deficient mutant (ΔS1/S2) (b). Ten μM ZnCl2 was supplemented in the medium prior to growth (red line). Representative MALDI-MS images from ceca of an uninfected C57BL/6, uninfected antibiotic treated (0.5 mg/ml cefoperazone) C57BL/6, infected C57BL/6, or infected calprotectin-deficient mouse (c). Each image is representative of five independent ceca. Scale bars, 1 cm. Percent survival for standard chow fed wildtype C57BL/6 and calprotectin-deficient C57BL/6 mice infected with C. difficile strain R20291 following cefoperazone treatment (0.5 mg/ml) (n=20/group) (d). * P < 0.01; by log-rank test. Blinded histological scores were determined following Zn starvation of calprotectin-deficient mice and subsequent infection with R20291 (n=10/group) (e). * P < 0.01; by Mann-Whitney test.
In culture, C. difficile growth was inhibited by the addition of recombinant calprotectin (1 mg/ml) (Fig. 4b). This inhibition was Zn-dependent, as treatment with a calprotectin mutant defective in Zn binding (ΔS1/S2) or supplementation with Zn (10 μM ZnCl2) reversed this effect (Fig. 4b). Supplementation with manganese (10 μM MnCl2) partially restored growth, but supplementation with iron (10 μM Fe(II)SO4) did not (Supplementary Fig. 9a,b). In mice infected with C. difficile, calprotectin was abundant at the site of infection (Fig. 4c). To examine the importance of calprotectin during CDI, calprotectin-deficient mice (S100a9−/−)29 were infected with C. difficile strain R20291. Calprotectin-deficient mice showed decreased survival and increased disease severity (Fig. 4d,e). In contrast to what was observed in culture, C. difficile burden was not altered in calprotectin-deficient mice (Supplementary Fig. 10). This suggests that the kinetics of calprotectin accumulation in the cecum are not sufficient to fully inhibit C. difficile growth in vivo. We hypothesized that exacerbation in disease associated with calprotectin-deficiency is mediated by a decreased capacity of these animals to limit Zn during CDI. Consistent with this, calprotectin-deficient mice harbor significantly higher levels of Zn in their feces compared to wildtype mice (Supplementary Fig. 10). To test the contribution of calprotectin-dependent Zn limitation during infection, calprotectin-deficient mice were fed the low Zn diet for 5-weeks prior to infection with C. difficile strain R20291. Under conditions of Zn starvation, calprotectin-deficient animals showed significantly less pathology following infection (Fig. 4e). These data demonstrate that calprotectin is an essential host factor for combating CDI, and the protective mechanisms of calprotectin are in part due to the ability to limit Zn during infection.
Collectively, these data reveal that dietary Zn plays an important role in modulating the gut microbiota and Zn-mediated shifts in microbial community structure lowers the threshold of antibiotics necessary to confer susceptibility to CDI. In addition, increased Zn levels intensify the severity of C. difficile-associated disease, and calprotectin is important for combating C. difficile by limiting Zn availability. This study advances our understanding of the environmental and host factors associated with CDI. Moreover, these findings provide the basis for shaping future prevention and treatment strategies for CDI. Alteration in diet and limitation of excess Zn intake may prove efficacious for the prevention of CDI in high-risk patients and help limit morbidity during CDI.
Online Materials & Methods
Mouse husbandry and model of C. difficile infection
Studies were conducted on adult (8 to 12 week old) age-matched male C57BL/6 (Jackson Laboratories) or C57BL/6 S100a9−/− mice29 (breeding colony) that were housed in groups of five. All animals were maintained at Vanderbilt University Medical Center Animal Facilities. Individual mice were not randomized upon arrival from Jackson Laboratories; however, multiple litters were included in each experimental group and all animal experiments were independently performed at least twice. Mice were maintained on a standard chow diet (LabDiets; Rodent Chow Diet 5001) unless specified otherwise. Mice were subjected to a previously described model of CDI18. Briefly, mice were given cefoperazone in their drinking water (0.5 mg/ml or 0.01 mg/ml) for five days. Mice were then given two days of recovery prior to administration of 105 spores of C. difficile in PBS via oral gavage. C. difficile strains R20291 and 630 were utilized, as noted. Prior to infection, each mouse was confirmed C. difficile culture negative. Following infection mice were monitored for signs of severe disease, including inappetence, diarrhea, hunching, and weight loss. Mice that exhibited severe disease or weight loss in excess of 20% were humanely euthanized. All animal experiments were approved and performed in compliance with the Institutional Animal Care and Use Committee (IACUC).
Dietary modulation
Diets containing altered metal levels were synthesized by Dyets Inc. using the AIN-93M standardized diet30. To minimize metal contamination, this L-amino acid defined AIN-93M base diet was formulated as a Zn free diet and the following concentration of Zn was supplemented: control diet: 29 mg/kg; low Zn diet: 0 mg/kg; high Zn diet: 1,000 mg/kg. Zn was added as carbonate salts, as specified by the AIN-93M diet30. Mice were fed an altered Zn diet starting at four weeks of age and diets were maintained for five weeks prior to infection and throughout the time-course of infection. For each separate diet treatment, two cages of mice from separate litters were run in parallel. The amount of diet that was consumed was monitored and each group was weighed during the time-course of diet manipulation to monitor for disease. Each group consumed equal amounts of diet and showed no signs of disease associated with dietary modulation.
Quantification of bacterial burden
C. difficile colony forming units (CFUs) were determined during the time course of infection using fecal samples and at the end point of infection using cecal contents. Samples were diluted in PBS and plated on Taurocholate Cycloserine Cefoxitin Fructose Agar (TCCFA) for quantification of C. difficile burden. Stool samples from antibiotic treated mice were plated on TCCFA prior to infection to ensure that mice were C. difficile culture negative prior to infection with spores. Bacterial burdens in livers were quantified at the end-point of infection. Livers were harvested from mice during necropsy and homogenized in 1 ml of sterile PBS. Liver homogenates were serially diluted in PBS and plated in parallel on 5% sheep blood and Luria-Bertani agar plates under aerobic and anaerobic conditions. Following overnight growth, CFUs were quantified and individual colonies were isolated. Genomic DNA was extracted from each cultivar and the 16S rRNA gene was sequenced for identification using the 27F and 1492R universal 16S rRNA gene primers.
Histological analysis
At necropsy, ceca and colons were harvested, fixed in 10% formalin solution, and embedded in paraffin. Sections were stained with hemotoxylin and eosin. Each section was given a disease score by a pathologist in a blinded fashion based on previously described criteria18. Histological scores were reported as a cumulative score of three independent scoring criteria: inflammation, edema, and epithelial cell damage.
C. difficile toxin cytotoxicity assay
C. difficile toxin specific cytotoxicity was determined using a Green African monkey kidney epithelial (Vero) cell-rounding cytotoxicity assay18,31. Briefly, fresh fecal samples were normalized to weight, homogenized in sterile PBS, and pelleted at 13,000 x g. Tenfold serial dilutions of supernatants were added to monolayers of Vero cells and cell-rounding was assessed for each dilution. The presence of C. difficile toxin A and toxin B were confirmed by neutralizing cell-rounding activity with a combined antitoxin (toxin A and toxin B) antisera (Techlab, cat# T5000). Cytotoxicity data are expressed as the log-ten reciprocal value of the highest dilution that rounded 100% of the Vero cells.
Cytokine quantification
Cytokine analysis was performed on mice prior to infection (n=5/group) and 2 days following infection (n=5/group) with C. difficile strain R20291. At necropsy, whole ceca from mice fed altered Zn diets were flash frozen in liquid nitrogen and frozen at −80°C. Ceca were thawed, thoroughly homogenized in 1 mL of Pierce IP Lysis Buffer (Thermo Scientific), and pelleted at 4°C. Supernatants were collected and normalized to total protein content using a Pierce BCA Protein Assay Kit (Thermo Scientific). Cytokine levels were measured using the Milliplex MAP Mouse Cytokine/Chemokine Magnetic Bead Panel (Millipore) and the Luminex Flexmap 3D platform (Luminex).
Preparation of tissue sections for mass spectrometry
For preparation of ICP-MS, each cecum or colon was aseptically removed, flushed of contents, and immediately flash-frozen in metal-free tubes. Samples were digested in Optima-grade nitric acid and heated until complete digestion was achieved. Samples were diluted in Milli-Q ultrapure water prior to ICP-MS analysis. For LA-ICP-MS, colon samples were flushed of all contents and filled with 25% Optimal Cutting Temperature compound (OCT)(Tissue-Tek) and then frozen in 100% OCT. Ceca were sectioned at 30 μm thickness using a Leica CM 3050S Cryostat (Leica Microsystems, Bannockburn, IL) and mounted to chilled nitric acid-washed polylysine-coated vinyl slides. For MALDI IMS, tissues were collected fresh and frozen in 25% OCT with all contents intact. Ceca were sectioned at 10 μm thickness and thaw-mounted onto chilled indium-tin oxide coated slides (Delta Technologies, Loveland, CO).
Mass spectrometry
LA-ICP-MS and ICP-MS were performed as previously described32. Briefly, quantitative elemental analysis was performed on a Thermo Element 2 high-resolution sector field inductively coupled plasma mass spectrometer (ICP-MS; Thermo Fisher Scientific, Bremen, Germany). Elemental imaging was performed using a laser ablation system (LA; LSX-213; CETAC. Omaha, NE) coupled with a high-resolution sector field ICP-MS. Slides containing ceca were ablated with a focused Nd:YAG laser beam and transported by helium gas to the ablation cell, mixed with argon gas and then introduced to the ICP-MS. LA-ICP-MS images are represented as relative Zn intensity. For ICP-MS, Zn concentrations were normalized to sample weight and are represented in parts per billion (ppb). For MALDI IMS, sections were sequentially washed to remove interfering lipids, salts, and OCT using 70% ethanol for thirty seconds, 100% ethanol for thirty seconds, 6:2:1 ethanol:chloroform:acetic acid for 2 minutes, 70% ethanol for thirty seconds, and 100% ethanol for thirty seconds. Slides were allowed to dry in a desiccator before MALDI matrix was applied. Fifteen mg/mL 2,5-dihydroxyacetophenone was prepared in 90% acetonitrile with 0.2% trifluoracetic acid and sonicated for ten minutes to dissolve all crystals. Matrix was applied using a TM-Sprayer (HTX Imaging, Carrboro, NC) operated at 1100 mm/min. Six passes of matrix were applied to the surface at a flow rate of 0.2 mL/min using 90% acetonitrile as a pushing solvent. The track spacing was set to 2 mm and the spray nozzle was heated to 80°C. The coating was rehydrated using 1 mL of 50 mM acetic acid in a sealed hydration chamber at 85°C for 3 minutes. IMS was performed using a rapifleX MALDI Tissuetyper (Bruker Daltonics, Billerica, MA) operated in linear positive ion mode. The laser was operated in single mode (beam width of ≈5μm) and pixels were set to be 50 by 50 μm. A total of five-hundred laser shots were captured per pixel with fifty laser shots at each position. The laser was operated at 10,000 hertz. Data were processed using fleXimaging version 4.1. Data were further analyzed using SCiLS Lab 2015b version 3.02.7774 (Bruker Daltonics). Spectra were normalized to total ion count and baseline subtracted using a top hat algorithm. The images are displayed without denoising but with interpolation turned on.
DNA extraction and 16S rRNA gene sequencing
Fecal samples were collected fresh from individual mice during the time-course of diet manipulation and immediately frozen for storage at −20°C. For analysis of the microbiota following antibiotic treatment, fecal samples were collected fresh during the two-day recovery period following cefoperazone treatment. Microbiota analysis was performed for randomly selected mice (n=5/group) representing both sets of cages in each diet group. Microbial genomic DNA was extracted using the PowerSoil DNA isolation kit (MO BIO Laboratories). The V4 region of the 16S rRNA gene from each sample was amplified and sequenced using the Illumina MiSeq Personal Sequencing platform as described elsewhere33. Sequences were curated as described previously using the mothur software package34. Briefly, we reduced sequencing and PCR errors, aligned the resulting sequences to the SILVA 16S rRNA sequence database35, and removed any chimeric sequences flagged by UCHIME36. After curation, we obtained between 1,436 and 67,633 sequences per sample (median=39,487), with a median length of 253 bp. We did not observe any correlation between treatment group and number of sequence reads. To limit effects of uneven sampling, we rarefied the dataset to 14,659 sequences per sample. FASTQ sequence data obtained in this study has been deposited to the Sequence Read Archive (SRA) at NCBI under accession number SRP067079.
Analysis of the gut microbiota
Sequences were clustered into OTUs based on a 3% distance cutoff calculated using the average-neighbor algorithm. All sequences were classified using the RDP training set (version 9) and OTUs were assigned a classification based on the taxonomy that had the majority consensus of sequences within each OTU using a naïve Bayesian classifier37. Sequences that were binned to OTUs that classified as Clostridium XI were individually classified and confirmed to be non-C. difficile members of the Clostridium XI genus. Microbial diversity (alpha-diversity) was calculated using the inverse Simpson index38. Species richness was calculated using Sobs (observed OTUs)34. β-diversity was calculated using the θYC distance metric with OTU frequency data39.
16S rRNA gene qPCR analysis
Relative bacterial load in the gut microbiota of mice fed altered Zn diets was measured using universal 16S rRNA gene qPCR primers (F, ACTCCTACGGGAGGCAGCAGT; R, ATTACCGCGGCTG CTGGC)40. Samples were normalized to host DNA using TNFα gene primers (F; GGCTTTCCGAATTCACTGGAG; R, CCCCGGCCTTCCAAATAAA)3. qPCR was performed on genomic DNA isolated from stool using IQ SYBR Green Supermix (Bio-Rad). Note that qPCR measures relative fold change of the 16S rRNA gene copy number in a sample, not overall bacterial numbers.
Growth in the presence of recombinant calprotectin
C. difficile growth curves were performed following a 1:50 dilution of overnight R20291 or 630 cultures. Recombinant calprotectin was supplemented into media at specified concentrations. Media consisted of 50% calprotectin buffer and 50% BHIS, supplemented with 3 mM calcium chloride25. A Zn and Mn binding mutant with inactivating mutations in both calprotectin metal binding sites41 was used as specified. Growth curves were quantified using optical density (OD600) at 37°C under anoxic conditions. Growth medium was supplemented with Zn (10 ZnCl2), Mn (MnCl2), or Fe(II) (FeSO4) to evaluate growth rescue. Metal supplementation of medium was performed immediately prior to addition of C. difficile and incubation for growth. Each growth curve was performed at least three independent times and representative growth curves were selected.
Measurement of calprotectin in human subjects
Serum calprotectin
Serum calprotectin levels were measured on previously collected serum samples that were collected on day two in the intensive care unit (ICU) from a subset of forty-eight adult CDI positive patients enrolled in the Validating Acute Lung Injury Biomarkers for Diagnosis (VALID) study. The VALID study is a 3,200 patient prospective cohort study that has been enrolling critically ill patients in the Vanderbilt Medical, Surgical, Trauma and Cardiovascular ICUs since 2006 (IRB 051065). Detailed inclusion and exclusion criteria and informed consent procedures for the VALID study have been published42. CDI was defined by a positive toxin antigen or amplification test. Patients ranged in age from nineteen to ninety-two years old, with a median age of fifty-five. Frozen serum samples were thawed, diluted 1,000-fold, and calprotectin levels were measured using the Human S100A8/S100A9 Heterodimer Quantikine ELISA kit (R&D Systems, Minneapolis, Mn). CDI infection severity was determined using previously defined criteria where 1 severity point is given for each of the following criteria: albumin levels ≤ 2.5 mg/dL, peripheral white blood cell count ≥ 15,000, and temperature ≥ 38.3°C27. Severe CDI was defined by a score ≥ 2 severity points or if CDI-associated surgical intervention was reported.
Fecal calprotectin
Fecal calprotectin levels were measured on twenty-five pediatric patients diagnosed with CDI. Subjects were enrolled at two tertiary-care children’s hospitals (Monroe Carrell Jr. Children’s Hospital, Nashville, Tennessee and Texas Children’s Hospital, Houston, Texas). The Institutional Review Boards of the participating institutions reviewed and approved the study (IRB 130315). Consent was obtained from all subjects in the study. Hospital laboratory records were used to identify individuals who tested positive for C. difficile at the time of diagnosis. Both hospitals employ molecular-based assays to test stool for the presence of toxigenic C. difficile. Children aged twelve months through eighteen years with CDI were eligible for enrollment. CDI was defined as a positive C. difficile diagnostic test and diarrhea. Subjects were excluded from the study if an alternative enteropathogen was identified by routine clinical testing requested by the primary medical team. Fecal samples were thawed, normalized to weight, and fecal calprotectin was measured using the Calprotectin ELISA Assay Kit (Eagle Biosciences, Nashua, NH).
Statistics
Statistical analyses were performed using GraphPad Prism (Version 6.0) and R (Version 3.1.0). The Mann-Whitney test was used for comparisons of continuous variables between two treatment groups. Analysis of molecular variance (AMOVA) was performed to determine significance between the community structures of different groups of samples based on θYC distance matrices43. The machine learning algorithm random forest was implemented using the randomForest R package to identify features (OTUs) differentially abundant in each dietary group and to further assess gut microbiota variation between dietary groups (http://CRAN.R-project.org)44. Repeated-measure paired treatment analysis of variance for each OTU was utilized to identify OTUs important for driving differences between dietary groups. We corrected for multiple comparisons using an experiment-wise error rate of 0.01 and controlled for false discovery rate using the Benjamani-Hochberg method45. For survival analysis, the log-rank test was used to evaluate significance between groups. A sample size of at least five per group was used for animal experiments. Power analyses were not performed prior to animal studies. Sample size for our human serum calprotectin study was determined by retrospectively selecting all available C. difficile positive subjects from the VALID study42. Samples size for our pediatric fecal calprotectin study was based on the anticipated and achieved collection of stool samples obtained during a Thrasher-funded fourteen-month study at two tertiary care children’s hospitals. Whenever feasible, blinding was performed during experimentation and analyses (DNA extraction from stool, 16S rRNA sequence curation and analysis, ICP-MS analysis, calprotectin ELISA analyses, survival analysis in calprotectin-deficient animals, and pathology scoring). To avoid Zn contamination during diet manipulation, diet treatment groups were not blinded. No mice were excluded from analysis.
Supplementary Material
Acknowledgments
We thank P. Schloss and J. Sorg for critical feedback on this study, and D. Aronoff and S. Walk for providing C. difficile strains. This research was supported by Merit Review Award # 1I01BX002482 from the United States (U.S.) Department of Veterans Affairs, NIH R01 AI101171 and P41 GM103391-05, and the Vanderbilt Digestive Disease Research Center (VDDRC) grant # P30DK058404. J.P.Z. was supported by T32DK007673 and F32AI120553. J.L.M. was supported by T32GM065086. M.R.N. was supported by the Thrasher Research Fund Early Career Award.
Footnotes
Accession codes
NCBI SRA: SRP067079.
Author Contributions
J.P.Z. and E.P.S. designed experiments and wrote manuscript with input from co-authors. J.P.Z. performed animal experiments and corresponding assays and analyses with assistance from A.T.J. J.P.Z. performed microbiota community analyses. L.J.J. designed altered metal diets. J.L.M. and R.M.C. performed MALDI-MS imaging and corresponding analyses. Y.Z. and R.M.C. performed ICP-MS. M.K.W. performed histological analyses. W.J.C. assisted in calprotectin assays. M.R.N. and J.D.C. enlisted pediatric patients and collected fecal samples. M.W.S., M.J.N., and L.B.W. collected adult serum and aided in analyses of calprotectin experiments.
Competing financial interests
The authors declare no competing financial interest.
References
- 1.Lessa FC, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly CP, LaMont JT. Clostridium difficile--more difficult than ever. N Engl J Med. 2008;359:1932–1940. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 3.Theriot CM, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 5.Leffler DA, Lamont JT. Clostridium difficile Infection. N Engl J Med. 2015;373:287–288. doi: 10.1056/NEJMc1506004. [DOI] [PubMed] [Google Scholar]
- 6.Gerding DN, Muto CA, Owens RC., Jr Measures to control and prevent Clostridium difficile infection. Clin Infect Dis. 2008;46(Suppl 1):S43–49. doi: 10.1086/521861. [DOI] [PubMed] [Google Scholar]
- 7.Fekety R, et al. Epidemiology of antibiotic-associated colitis; isolation of Clostridium difficile from the hospital environment. Am J Med. 1981;70:906–908. doi: 10.1016/0002-9343(81)90553-2. [DOI] [PubMed] [Google Scholar]
- 8.Schubert AM, Sinani H, Schloss PD. Antibiotic-Induced Alterations of the Murine Gut Microbiota and Subsequent Effects on Colonization Resistance against Clostridium difficile. MBio. 2015;6:e00974. doi: 10.1128/mBio.00974-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buffie CG, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker CF, Black RE. Zinc and the risk for infectious disease. Annu Rev Nutr. 2004;24:255–275. doi: 10.1146/annurev.nutr.23.011702.073054. [DOI] [PubMed] [Google Scholar]
- 12.Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chumbler NM, et al. Crystal structure of Clostridium difficile toxin A. Nature Microbiology. 2016;1:15002. doi: 10.1038/nmicrobiol.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cafardi V, et al. Identification of a novel zinc metalloprotease through a global analysis of Clostridium difficile extracellular proteins. PLoS One. 2013;8:e81306. doi: 10.1371/journal.pone.0081306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubino JT, et al. Structural characterization of zinc-bound Zmp1, a zinc-dependent metalloprotease secreted by Clostridium difficile. J Biol Inorg Chem. 2016;21:185–196. doi: 10.1007/s00775-015-1319-6. [DOI] [PubMed] [Google Scholar]
- 17.Hensbergen PJ, et al. Clostridium difficile secreted Pro-Pro endopeptidase PPEP-1 (ZMP1/CD2830) modulates adhesion through cleavage of the collagen binding protein CD2831. FEBS Lett. 2015;589:3952–3958. doi: 10.1016/j.febslet.2015.10.027. [DOI] [PubMed] [Google Scholar]
- 18.Theriot CM, et al. Cefoperazone-treated mice as an experimental platform to assess differential virulence of Clostridium difficile strains. Gut Microbes. 2011;2:326–334. doi: 10.4161/gmic.19142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wust J, Sullivan NM, Hardegger U, Wilkins TD. Investigation of an Outbreak of Antibiotic-Associated Colitis by Various Typing Methods. J Clin Microbiol. 1982;16:1096–1101. doi: 10.1128/jcm.16.6.1096-1101.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stabler RA, et al. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 2009;10:R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasegawa M, et al. Protective role of commensals against Clostridium difficile infection via an IL-1β-mediated positive-feedback loop. J Immunol. 2012;189:3085–3091. doi: 10.4049/jimmunol.1200821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seo SU, et al. Distinct Commensals Induce Interleukin-1β via NLRP3 Inflammasome in Inflammatory Monocytes to Promote Intestinal Inflammation in Response to Injury. Immunity. 2015;42:744–755. doi: 10.1016/j.immuni.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zackular JP, Chazin WJ, Skaar EP. Nutritional Immunity: S100 Proteins at the Host-Pathogen Interface. J Biol Chem. 2015;290:18991–18998. doi: 10.1074/jbc.R115.645085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hood MI, et al. Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLoS Pathog. 2012;8:e1003068. doi: 10.1371/journal.ppat.1003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kehl-Fie TE, et al. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe. 2011;10:158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaddy JA, et al. The host protein calprotectin modulates the Helicobacter pylori cag type IV secretion system via zinc sequestration. PLoS Pathog. 2014;10:e1004450. doi: 10.1371/journal.ppat.1004450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanania A, Jiang Zhi-Dong, Smiley Casey, Lasco Todd, Garey Kevin W, DuPont Herbert L. Fecal Calprotectin in the Diagnosis of Clostridium difficile Infection. Infectious Diseases in Clinical Practice. 2016;24:31–34. [Google Scholar]
- 28.Rao K, Santhosh K, Mogle JA, Higgins PD, Young VB. Elevated fecal calprotectin associates with adverse outcomes from Clostridium difficile infection in older adults. Infect Dis (Lond) 2016:1–7. doi: 10.1080/23744235.2016.1186832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hobbs JA, et al. Myeloid cell function in MRP-14 (S100A9) null mice. Mol Cell Biol. 2003;23:2564–2576. doi: 10.1128/MCB.23.7.2564-2576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reeves PG, Nielsen FH, Fahey GC., Jr AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J Nutr. 1993;123:1939–1951. doi: 10.1093/jn/123.11.1939. [DOI] [PubMed] [Google Scholar]
- 31.Reeves AE, et al. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile Infection. Gut Microbes. 2011;2:145–158. doi: 10.4161/gmic.2.3.16333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kehl-Fie TE, et al. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect Immun. 2013;81:3395–3405. doi: 10.1128/IAI.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zackular JP, Rogers MA, Ruffin MTt, Schloss PD. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila) 2014;7:1112–1121. doi: 10.1158/1940-6207.CAPR-14-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruesse E, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magurran AE, Henderson PA. Temporal turnover and the maintenance of diversity in ecological assemblages. Philos Trans R Soc Lond B Biol Sci. 2010;365:3611–3620. doi: 10.1098/rstb.2010.0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yue JC, Clayton MK. A similarity measure based on species proportions. Commun Stat-Theor M. 2005;34:2123–2131. [Google Scholar]
- 40.Vaishnava S, et al. The antibacterial lectin RegIII γ promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Damo SM, et al. Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc Natl Acad Sci U S A. 2013;110:3841–3846. doi: 10.1073/pnas.1220341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siew ED, et al. Urine neutrophil gelatinase-associated lipocalin moderately predicts acute kidney injury in critically ill adults. J Am Soc Nephrol. 2009;20:1823–1832. doi: 10.1681/ASN.2008070673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin AP. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl Environ Microbiol. 2002;68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cutler DR, Edwards TC, Beard KH, Cutler A, Hess KT. Random forests for classification in ecology. Ecology. 2007;88:2783–2792. doi: 10.1890/07-0539.1. [DOI] [PubMed] [Google Scholar]
- 45.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.