Abstract
Currently, there is no stable and flexible method to label and track cytotoxic T lymphocytes (CTLs) in vivo in CTL immunotherapy. We aimed to evaluate whether the sulfo-hydroxysuccinimide (NHS)-biotin–streptavidin (SA) platform could chemically modify the cell surface of CTLs for in vivo tracking. CD8+ T lymphocytes were labeled with sulfo-NHS-biotin under different conditions and then incubated with SA–Alexa647. Labeling efficiency was proportional to sulfo-NHS-biotin concentration. CD8+ T lymphocytes could be labeled with higher efficiency with sulfo-NHS-biotin in DPBS than in RPMI (P < 0.05). Incubation temperature was not a key factor. CTLs maintained sufficient labeling for at least 72 h (P < 0.05), without altering cell viability. After co-culturing labeled CTLs with mouse glioma stem cells (GSCs) engineered to present biotin on their surface, targeting CTLs could specifically target biotin-presenting GSCs and inhibited cell proliferation (P < 0.01) and tumor spheres formation. In a biotin-presenting GSC brain tumor model, targeting CTLs could be detected in biotin-presenting gliomas in mouse brains but not in the non-tumor-bearing contralateral hemispheres (P < 0.05). In vivo fluorescent molecular tomography imaging in a subcutaneous U87 mouse model confirmed that targeting CTLs homed in on the biotin-presenting U87 tumors but not the control U87 tumors. PET imaging with 89Zr-deferoxamine-biotin and SA showed a rapid clearance of the PET signal over 24 h in the control tumor, while only minimally decreased in the targeted tumor. Thus, sulfo-NHS-biotin–SA labeling is an efficient method to noninvasively track the migration of adoptive transferred CTLs and does not alter CTL viability or interfere with CTL-mediated cytotoxic activity.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-016-1911-9) contains supplementary material, which is available to authorized users.
Keywords: Biotin, Cytotoxic T lymphocytes, Tumor, Immunotherapy
Introduction
Antitumor treatment using cytotoxic (CD8+) T lymphocyte (CTL) adoptive immunotherapy is emerging to be an interesting area of research and potentially an important treatment option for cancer patients [1–3]. Tracking CTL migration would assist in analyzing the homing kinetics and treatment efficacy. Various in vivo imaging techniques are now available for monitoring adoptively transferred cells, including magnetic resonance imaging (MRI) [4–6], positron emission tomography (PET) [7–9] and optical imaging [10, 11]. Optical imaging such as fluorescent molecular tomography (FMT) has high sensitivity. It permits longitudinal assessment of fluorochrome concentrations in the whole body. Measurements are quantitative and three-dimensional [12] and can be utilized intraoperatively [13, 14].
In optical imaging, two principal strategies are used to label cells. One is to transfect cells to express heterologous fluorescent proteins or enzymes that digest and activate a fluorescent probe. Another way is to mark cells directly with external dyes. This is generally safer and more convenient than genetic modification. Red (~625–740 nm) to near-infrared (NIR, ~700–1000 nm) fluorescent molecules have been widely used for optical imaging because of their low tissue absorption and greater resolution [15–19], permitting real-time, dynamic imaging of biological processes [17, 20–22]. While cell trackers in this light spectral region partly resolve the problem of light absorption and scattering, cell trackers must also be considered for multi-photon excitation, biocompatibility and cellular retention.
In this study, we evaluated whether N-hydroxysuccinimide (NHS)-biotin–streptavidin (SA) can chemically modify the cell surface and be used as a cell tracker for in vivo imaging (Fig. 1). We tested the ability of this ester to label and remain on cells as well as its biocompatibility. We also illustrated its ability to report cell biodistribution in a mouse brain tumor model and performed proof-of-concept in vivo FMT imaging in a peripheral tumor mouse model and PET imaging in a brain tumor mouse model.
Fig. 1.
Overview of sulfo-NHS-biotin–streptavidin (SA)–Alexa647 labeling system. Upon vector-mediated delivery and expression, biotin acceptor peptide-transmembrane (BAP-TM) displays biotin on the cell surface. Cytotoxic T lymphocytes (CTLs) are chemically modified by sulfo-NHS-biotin by covalent binding to –NH2 on the cell surface. SA serves as a mediator and specifically binds to biotin on the surface of both tumor cells and CTLs. Alexa647 bound to SA serves as a reporter of this process, but other imaging probes may be used. Free Gluc is secreted by tumor cells and can be used to monitor the proliferation of tumor cells. FMT fluorescent molecular tomography, IVM intravital microscopy, BLI bioluminescence, PET positron emission tomography
Materials and methods
Generation of CD8+ T lymphocytes
Mouse spleen single cell suspensions were prepared by homogenizing the spleens and filtering through 70-µm cell strainers. CD8+ T lymphocytes were further isolated by magnetic negative isolation (anti-CD4/CD11b/B220/CD49b/Ter-119-PE, BioLegend, San Diego, CA, USA) and anti-PE-microbeads (Miltenyi Biotec, Auburn, CA, USA). Isolated CD8+ T lymphocytes were cultured in complete RPMI 1640 culture medium. CD8+ T lymphocytes were activated with Dynabeads (Mouse T-Activator CD3/CD28 for T Cell Expansion and Activation, Gibco, Grand Island, NY, USA), supplemented with rIL-2 (20 ng/mL, R&D Systems, Minneapolis, MN, USA) to obtain CD8+ CTLs.
Biotinylation of CD8+ T lymphocytes
Sulfo-NHS-biotin (sulfosuccinimidobiotin) ester powder (Pierce Biotechnology, Rockford, IL, USA) was firstly diluted in ddH2O to 1 mg/mL and then diluted further to different concentrations. A total of 5 × 106 freshly isolated CD8+ T lymphocytes were suspended in Dulbecco’s phosphate-buffered saline (DPBS) or RPMI 1640 culture medium and then incubated with sulfo-NHS-biotin (1:300 dilution) for 30 min, at room temperature or 37 °C separately. After labeling, cells were spun down (300G, 10 min, Thermo JOUAN CR3i, Waltham, MA, USA) to get rid of extra sulfo-NHS-biotin and then stained with SA–Alexa647 (Molecular Probes, Eugene, OR, USA) for 30 min. Labeling efficiency was analyzed on a flow cytometer (Becton-Dickinson, Franklin Lakes, NJ, USA), by comparing mean fluorescence intensity (MFI) with control CD8+ T lymphocytes under various conditions described below. Labeled CD8+ T lymphocytes were also examined under fluorescent microscopy (Nikon TE-2000, EX 628/40, DM 660, EM 692/40) for Alexa647.
Cell viability assays of biotinylation
Cell viability of biotinylation was assessed by quantitation of ATP present in metabolically active CTLs (CellTiter-Glo®Luminescent Cell Viability Assay, Promega, Madison, WI, USA). A total of 1 × 106 biotinylated and control CTLs/well in 96-well opaque-walled plates (with Dynabeads) were recorded for luminescence at 0, 24, 48 and 72 h after labeling, with a charge-coupled device camera. Wells containing culture medium and Dynabeads only were tested for background luminescence.
Efficiency and stability of biotinylation
To examine the efficiency of cell surface biotinylation, freshly isolated CD8+ T lymphocytes were labeled with increasing concentrations of sulfo-NHS-biotin (control, 1:600 dilution, 1:300 dilution, 1:30 dilution), then stained with SA–Alexa647. The MFIs were then compared. To determine the stability of biotinylation over time, sulfo-NHS-biotin-labeled CTLs (1:300 dilution, with Dynabeads) were collected at 0, 24, 48, 72 h after labeling with SA–Alexa647 and removal of Dynabeads. Cells were then analyzed on a FACS flow cytometer, and MFIs were compared to control CTLs at the same time points.
Generation of biotin-presenting GSCs/U87 cells
Mouse glioma stem cells (GSC-005) were cultured as previously described [23]. Briefly, GSCs were cultured in glioblastoma stem cell culture medium (EF20), which contains advanced Dulbecco’s modified Eagle’s medium (DMEM) F12 (Gibco, Carlsbad, CA, USA), 2 mM l-glutamine (Cellgro, Manassas, VA, USA), 1 % N2 (Gibco, Carlsbad, CA, USA), 2 μg/mL heparin (Sigma, St. Louis, MO, USA), 0.5× penicillin and streptomycin (Cellgro, Manassas, VA, USA). By co-transfecting GSCs with three lentivirus vectors [lenti-biotin acceptor peptide-transmembrane (BAP-TM)-inverted green fluorescent protein (IGFP), lenti-sshBirA-ImCherry and lenti-Gaussia luciferase (Gluc)] [24, 25], GSCs were genetically modified to present biotin on cell surface and secrete Gluc outside the cells [24]. Transfection of lenti-BAP-TM-IGFP and lenti-sshBirA-ImCherry was monitored by detecting tagged fluorescence under fluorescent microscopy. Expression of biotin on cell surface was determined by labeling transfected cells with SA–Alexa647 and analyzing cells with a flow cytometer. Expression of lenti-Gluc was determined by testing G-luciferase in conditioned medium from well-grown GSCs [26]. U87 cells (a primary human glioblastoma cell line, ATCC, Manassas, VA, USA) were also transfected to present biotin on cell surface in the same manner [25].
Biotinylated CTLs homing to GSCs in vitro
Transfected GSC spheres were formed by seeding 1 × 105 GSCs in serum-free EF20 on 24-well plates (Corning, Lowell, MA, USA) for 48 h. To test the specific targeting and cytotoxic activity of biotinylated CTLs to GSCs, GSC spheres were co-cultured with targeting CTLs (labeled with sulfo-NHS-biotin–SA–Alexa647, 1 × 107 per well, 1:300 dilution, with Dynabeads) for 24 h. GSC spheres co-cultured with non-targeting CTLs (labeled with sulfo-NHS-biotin only, without SA–Alexa647, 1 × 107 per well, 1:300 dilution, with Dynabeads) and GSC spheres cultured without CTLs were used as control. Gluc levels from medium of these groups were tested 24 h after co-culture to assess GSC proliferation.
Animal models
All animal studies were approved by the institutional animal care committee. GSC in situ mouse model: Mice (C57BL/6, 8 weeks, female, the Jackson laboratory; n = 5) were anesthetized with i.p. injection of ketamine (100 mg/mL, 80–100 mg/kg) and xylazine (100 mg/mL, 5–10 mg/kg), and fixed in a stereotactic head frame (David Kopf Instruments, Tujunga, CA, USA). A total of 6 × 104 of unmodified GSCs or biotin-presenting GSCs were injected into the deep frontal white matter (2.0 mm lateral and 1.2 mm anterior to the bregma) at a depth of 3.0 mm. U87 subcutaneous mouse model: Athymic nude mice (6–8 weeks, female, the Jackson laboratory; n = 3) were injected subcutaneously (s.c.) in the mammary fat pads with 1 × 107 biotin-presenting U87 cells on the left side and equal number of control U87 cells on the right side. Tumors were generally established after 14 days.
Tracking biodistribution of CTLs
Freshly isolated CD8+ T lymphocytes were activated with Dynabeads for 24 h and then labeled after removal of Dynabeads, as follows: targeting CTLs, with sulfo-NHS-biotin–SA–Alexa647, 1:300 dilution. These cells (2 × 107 per mouse) were i.v. injected into the biotin-presenting GSC brain tumor mice as described above. Single cell suspensions of each hemisphere of the brains, spleens and bone marrow were prepared and flow cytometry was used to test for Alexa647 positive cells to profile the CTL distribution in different organs.
Tracking CTLs in vivo (subcutaneous tumor)
To image CTLs targeting tumors in vivo, we implanted U87 cells in the mammary fat pads as described above. A total of 2 × 107 targeting CTLs (labeled with sulfo-NHS-biotin–SA–Alexa647, 1:300 dilution) were injected via tail vein; 24 h after injection, the mice were anesthetized under 2 % isoflurane gas anesthesia, placed in a cassette that contains fiducial markers, and the CTL accumulation at the tumor sites was imaged with an FMT 2500 scanner (Perkin Elmer, Billerica, MA, USA). In addition, computed tomography (CT) scans were also performed using the Siemen’s Inveon system with a 500 μA 80 kVp X-ray tube over 360 projections reconstructed via a cone beam modified Feldkamp reconstruction algorithm to give 3D images with an isotropic voxel size of 110 μm. The CT and FMT images were registered utilizing the fiducials on the cassette by a point-based rigid transformation in the Osirix software environment.
Tracking CTLs in vivo (brain glioma)
Synthesis of [89Zr]Zr-deferoxamine-biotin
To a solution of deferoxamine (56 mg, 1.0 equiv.) in DMSO (3 mL) was added triethylamine (28 μL, 2.0 equiv.) and NHS-biotin (44 mg, 1.3 equiv.). The reaction was stirred for 2 h. The reaction was filtered, and the solution underwent high-performance liquid chromatography (HPLC) (gradient H2O/CH3CN from 1/100 %) to give the desired compound as a white powder (28 mg, 35 %). Liquid chromatography mass spectroscopy (LCMS) found m/z 787.5: (M + 1). Deferoxamine-biotin (20 nmol, 2 μL of a 10 mM solution in DMSO) was mixed with 3.0 millicuries (111 megabecquerel) of [89Zr]Zirconium-oxalate (100 μL 1 M oxalic acid). The pH of the reaction was carefully adjusted to pH 7 by slow addition of a 1 M Na2CO3 solution. The reaction was shaken at 25 °C for 1 h, analyzed by radio-TLC (ITLC, 50 mM EDTA pH 7) and purified by HPLC. [89Zr]Zr-deferoxamine-biotin was isolated in 45.5 % radiochemical yield.
PET imaging
CTL isolation and transfection were performed in the same manner as the previous procedure except streptavidin (2 mg/mL) instead of streptavidin-AlexaFluor™ 647 conjugate was used to label the T cells after biotinylation of the cells. The cell suspension (200–300 μL) was injected in a mouse with biotin-expressing (BAP-TM) GSC brain tumor and a mouse with GSC brain tumor without biotin expression. After 24 h, the [89Zr]Zr-deferoxamine-biotin imaging agent was intravenously injected into the mice. The mice were scanned 3 and 20 h post-injection on a Siemens Inveon PET-CT scanner. PET imaging was acquired for 30 min and reconstructed with filtered back-projection using a ramp filter to maintain linearity. The CT was acquired similarly as the FMT-CT data set and registered to the PET data set with an affine transformation matrix. Note that we decided to inject the imaging agent after CTLs have been injected into the animals to avoid saturating the binding sites on the CTLs prior to targeting the tumor cells in vivo.
Histological analysis of CTL infiltration
Right after FMT imaging, mice were killed and tumors were removed, frozen and sectioned. To verify that the positive FMT fluorescent signal originated from targeting CTLs, 15-μm fresh frozen brain sections were examined directly under microscopy for SA–Alexa647-labeled cells.
Statistical analysis
Results were reported as mean ± standard error of measurement (SEM). The MFI for cell labeling efficiency and labeling kinetics between the labeled and control groups at different time points, the Gluc level of transfected GSCs and control GSCs, and CTL distributions in different organs were compared using the nonparametric Mann–Whitney U test. For CTL viability, the MFI of both control and labeled groups at 24, 48 and 72 h was firstly normalized to the average MFI (the average of control and labeled groups at 0 h) and then compared using the nonparametric Mann–Whitney U test. In the GSCs and CTLs co-culture study, the Gluc level of control, non-targeting and targeting groups at 24 h were firstly normalized to the average Gluc level of these three groups at 0 h and then compared with the nonparametric Kruskal–Wallis test. P < 0.05 was considered statistically significant. We used GraphPad Prism (v.5, GraphPad Software) for statistical analysis.
Results
CD8+ T lymphocytes can be biotinylated and cell viability was not affected
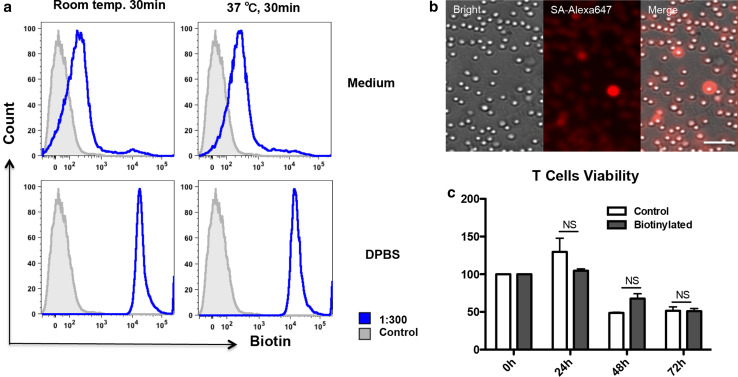
Labeling efficiency of CD8+ T lymphocytes in DPBS was significantly higher than that in RPMI 1640 culture medium (Fig. 2a; Table 1), suggesting that factors in the culture medium interfered with labeling. Labeling efficiency was not significantly different between cells labeled at room temperature or at 37 °C (Fig. 2a; Table 1). When detecting labeled CD8+ T lymphocytes under fluorescent microscopy, fluorescent signal could be detected around cells (Fig. 2b). Nearly all the T cells were labeled, though some cells exhibited stronger signal due to their closer position to the scope. This was because T cells in cell culture were not attached and were floating in the culture medium. We found no significant difference in cell viability at 0, 24, 48 and 72 h between labeled CTLs (100.0 ± 0.0, 129.6 ± 18.2, 48.7 ± 1.0, 51.5 ± 5.1, respectively; n = 3) and control CTLs (100.0 ± 0.0; 104.7 ± 2.4, P = 0.7000; 67.7 ± 6.4, P = 0.1000; 51.0 ± 3.6, P = 0.6579; respectively; n = 3, Fig. 2c).
Fig. 2.
Biotinylation of CD8+ cytotoxic T lymphocytes. a Labeling efficiency of CD8+ T lymphocytes in Dulbecco’s phosphate-buffered saline (DPBS) was higher than that in RPMI 1640 culture medium and was not different at room temperature or at 37 °C (MFI by flow cytometry, 1:300 dilution). b Strong streptavidin (SA)–Alexa647 fluorescent signal could be detected around CD8+ T lymphocytes under fluorescent microscopy. Bar 100 μm (1:300 dilution, original magnification ×200). c Cell viability by examining ATP in live CTLs at 0, 24, 48, 72 h separately. No significant difference between biotinylated CTLs (n = 3 for each time point in each group) and control CTLs (n = 3 for each time point in each group). NS not significant
Table 1.
Different labeling conditions of CD8+ T lymphocytes (n = 3)
| Room temp. 30 min | 37 °C 30 min | P value | |
|---|---|---|---|
| Medium | 6718 ± 721.7 | 5265 ± 943.5 | 0.1000 |
| DPBS | 21,517 ± 1564 | 17,939 ± 1400 | 0.2000 |
| P value | <0.05 | <0.05 |
CTLs biotinylation was highly efficient and stable
We found labeling efficiency was directly proportional to the concentration of sulfo-NHS-biotin (691.3 ± 27.28, control; 10,137 ± 1199, 1:600 dilution; 20,508 ± 1486, 1:300 dilution; 116,333 ± 6888, 1:30 dilution; n = 3, Fig. 3a). With any of these sulfo-NHS-biotin concentrations, the labeling efficiency was >90 % (Fig. 3b). In addition, when examining labeling stability at 0, 24, 48 and 72 h, the MFI was significantly different between the biotinylated CTLs (632.1 ± 46.0, 496.0 ± 27.2, 482.7 ± 9.8, 284.0 ± 11.4; n = 3) and unlabeled control CTLs (9.0 ± 0.5, P < 0.05; 9.8 ± 0.2, P < 0.05; 9.0 ± 0.9, P < 0.05; 10.9 ± 1.2, P < 0.05; n = 3, Fig. 3c). Thus, CTLs maintained sufficient labeling for at least 72 h, demonstrating that this would be suitable for in vivo tracking experiments. The decreasing signal over time in the biotinylated group is likely due to splitting of the CTLs.
Fig. 3.
Efficiency and stability of labeling. a MFI was proportional to the concentration of sulfo-NHS-biotin labeling CD8+ T lymphocytes (n = 3 in each group; sulfo-NHS-biotin dilution 1:30, 1:300, 1:600). b Labeling efficiency was >90 % with any of these concentrations (sulfo-NHS-biotin dilution 1:30, 1:300, 1:600). c CTLs maintained sufficient labeling for at least 72 h when detecting MFI at 0, 24, 48 and 72 h separately (n = 3 in each group at each time point). *P < 0.05
GSCs display biotin on the cell surface and express G-luciferase
Co-transfected GSCs expressed GFP and mCherry efficiently (Supplementary Figure 1a), demonstrating successful transfection of the lenti-BAP-TM-IGFP and lenti-sshBirA-ImCherry vectors. Compared to cells transfected with GFP only, there was positive SA–Alexa647 staining from cells transfected with lenti-BAP-TM-IGFP and lenti-sshBirA-ImCherry (Supplementary Figure 1b), demonstrating that biotin has been successfully presented on the surface of transfected GSCs. Significantly higher Gluc concentrations were detected from the media of GSCs transfected with lenti-Gluc (3141.0 ± 125.1, n = 6) than media from untransfected control GSCs (97.2 ± 10.1, n = 6, P < 0.01, Supplementary Figure 1c), confirming that transfected GSCs expressed and secreted Gluc outside the cells.
Biotinylated CTLs target and kill biotin-presenting GSCs in vitro
With SA–Alexa647 as a mediator and tracker, when examining co-culturing of labeled CTLs and biotin-presenting GSCs, we found that GSC spheres co-cultured with labeled CTLs were surrounded by CTLs bound to Dynabeads. On the other hand, scattered CTLs were found in the non-targeting group (without SA–Alexa647). Targeting CTLs also inhibited the formation of intact GSC spheres, as there were well-formed spheres in the control and non-targeting groups, while smaller, irregularly shaped clusters were observed in the targeting group (Fig. 4a); 24 h after co-culture, the Gluc concentration from biotin-presenting GSCs co-cultured with targeting CTLs (1.5 ± 0.3, n = 5) was lower than that from biotin-presenting GSCs with non-targeting CTLs (2.4 ± 0.3, n = 5) and that from control GSCs (4.2 ± 0.5, n = 5, P < 0.01, Fig. 4a). Thus, with SA–Alexa647 as a mediator, targeting CTLs prevented GSC sphere formation, destroyed intact GSC spheres, and inhibited GSC proliferation.
Fig. 4.
Biotinylated CTLs and biotin-presenting glioma stem cells (GSCs) in vitro. a Biotinylated CTLs (with Dynabeads, black and bright dots) gathered around the biotin-presenting GSC spheres and inhibited the formation of intact GSC spheres, but not in the control and non-targeting groups. Bar 100 μm (original magnification ×200); labeled CTLs exerted cytotoxic activity against GSCs by inhibiting proliferation of GSCs (low luciferase concentration, n = 5) more than GSCs co-cultured with non-targeting CTLs (n = 5) or control GSCs (n = 5). NS not significant, **P < 0.01. b Flow cytometry showed that targeting CTLs were found in the biotin-presenting glioma brain hemisphere (n = 3), while very few were found in the contralateral hemisphere (n = 3), the spleen (n = 3) or the bone marrow (n = 3). *P < 0.05
Tracking biotinylated CTLs by flow cytometry
In mice injected with targeting CTLs, more CTLs were found in the hemisphere bearing biotin-presenting GSC tumors (10.83 % ± 0.75, n = 3) compared to the contralateral hemisphere (1.82 % ± 0.17, n = 3, P < 0.05, Fig. 4b), the spleen (0.82 % ± 0.05, n = 3, P < 0.05, Fig. 4b) and bone marrow (0.17 % ± 0.03, n = 3, P < 0.05, Fig. 4b).
Tracking biotinylated CTLs in vivo
After the injection of targeting CTLs, FMT imaging of U87 tumor-bearing mice demonstrated that Alexa647 fluorescence signal increased in biotin-presenting U87 tumors, while not in control U87 tumors (Fig. 5a). Fluorescent staining confirmed that Alexa647 positive cells infiltrated only the biotin-presenting U87 tumor (Fig. 5b).
Fig. 5.
Tracking of biotinylated CTLs in vivo. a Alexa647 fluorescence signal increased in biotin-presenting U87 tumors (yellow dashed circle, L), while not in control U87 tumors (yellow dashed circle, R) on fluorescent molecular tomography (FMT) imaging. b Fluorescent staining confirmed streptavidin (SA)–Alexa647-positive cells (SA–Alexa647, arrow heads) infiltration only in biotin-presenting U87 tumor (GFP). Bar 100 μm (original magnification ×200). BAP-TM biotin acceptor peptide-transmembrane
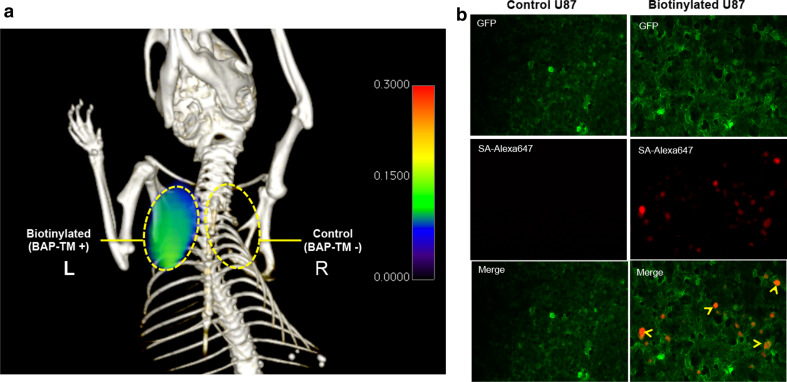
To test the ability of our platform for brain tumor imaging, we labeled biotinylated CTLs with streptavidin and injected these cells into mice bearing gliomas in the brain. In one mouse, the glioma expressed surface biotin via BAP-TM, while in the control mouse, the glioma did not express BAP-TM. We then injected a new PET agent, [89Zr]Zr-deferoxamine-biotin, into the mice. In the tumor lacking BAP-TM, there was a rapid clearance of the PET signal over 24 h, while in the targeted tumor, the PET signal only minimally decreased, consistent with binding of the PET agent to CTLs bound to the glioma (Fig. 6).
Fig. 6.
Tracking biotinylated cytotoxic T lymphocytes in vivo targeting brain gliomas. a Position emission tomography-computed tomography (PET/CT) imaging of CTLs targeting brain gliomas at 3 and 20 h after [89Zr]Zr-deferoxamine-biotin injection. Biotinylated CTLs with streptavidin were injected 24 h prior to the PET agent. The targeted [biotin acceptor peptide-transmembrane (BAP-TM)+] glioma expressed surface biotin, while the control tumor did not. The initial intensity of the control tumor appeared higher at the 3 h image due to the much larger size of the tumor compared to the targeted tumor, representing non-specific uptake of the PET agent due to leakage across a disrupted blood–brain barrier. However, over time, the control tumor showed a rapid loss of the PET signal while the targeted tumor showed only a minimal decrease in signal over time. b Quantification of the PET signal (tumor/normal brain, normalized to the 3-h time point)
Discussion
The highly specific and stable interaction of biotin with avidin or streptavidin has been used widely in many biological applications. Recently, streptavidin has been used as the core of different imaging probes, including magnetic nanoparticles, radiotracers and fluorescent dyes [25]. In our study, we labeled adoptively transferred CTLs with a sulfo-NHS-biotin–SA covalent binding platform and tracked the cells in vivo in biotin-presenting tumor mouse models with FMT imaging and PET imaging. Sulfo-NHS-biotin is one of the most popular types of biotinylation reagent. NHS-activated biotin can react efficiently with amino groups (–NH2) of proteins to form stable amide bonds. The primary amines on lysine residues and on the N terminus of cell surface proteins enable simple and efficient biotinylation of cells. Using this technology, we labeled CD8+ T lymphocytes and obtained a labeling efficiency >90 %. As shown in Fig. 3c, proliferating T cells could be well detected over a time course of 72 h, though the labeling efficiency decreased over time. Thus, within 72 h, cell proliferation should not be a problem for detection. Unlike fluorescent proteins that can be expressed in T cells through gene modification, in our method the signal decreased over time in dividing cells. Thus, our method is likely unsuitable for experiments addressing long-term persistence or rapid in vivo proliferation. Nonetheless, we found that this labeling method had no significant impact on CTL viability and cytotoxic functions. In vivo, imaging and flow cytometry analysis performed in immunocompetent mice demonstrated that labeled CTLs could target brain tumor. Furthermore, the relative small size of biotin (244 Da) allows conjugation to surface proteins without altering the biological activities or functions of CTLs. A previous study labeled whole blood T cells with biotinylated anti-CD5 antibody and streptavidin-coated micron-sized particles of iron oxide and tracked cells with MRI [4]. In that study, biotinylated anti-CD5 antibody was used to label cells instead of sulfo-NHS-biotin as the first step. A potential issue of that labeling method was activation of cells via anti-CD5 antibodies. CD5 is a T cell marker and activation of CD5 would trigger several signaling pathways that may alter production and release of a number of important immune regulatory cytokines.
There had been several prior studies on fluorescent imaging of T cells. Swirski et al. [17] demonstrated that murine splenocytes could be labeled with a biocompatible far-red ester fluorophore (VT680, Ex 670 nm, Em 688 nm) and tracked noninvasively in mice with high sensitivity by in vivo optical imaging. An ester dye with a longer wavelength (IRDye800CW, Ex 778 nm, Em 806 nm) was also examined by Foster et al. [27] for greater tissue penetration and sensitivity. Ester dyes also bear the NHS reactive group that allows the molecule to couple to free amino groups on proteins. However, some of these dyes not only labeled cell surface proteins but were also endocytosed. Consequently, there could be uneven distribution of dyes during cell division when dyes were endocytosed, which limited quantification of cell numbers. The sulfo-NHS-biotin used in our study dissolves readily in polar solutions and is charged by the sodium sulfoxide group on the succinimidyl ring, so it cannot penetrate the cell membrane. As long as the cell remains intact, only primary amines exposed on the surface will be biotinylated. This cell surface labeling assures even distribution of dyes, which makes quantification of cell accumulation more accurate. Youniss et al. [28] and Du et al. [11] also tried labeling cells directly on cell surface by interacting positively charged fluorescent dye with the negatively charged cell surface, but this method was limited to fluorescent imaging. A key advantage to our method is in its versatility. Our design provides a flexible platform that allows different imaging reporters and imaging modalities to be used, such as PET imaging, increasing the translational potential.
There were limitations to our study. We tested the in vivo tracking of biotinylated CTLs in a subcutaneous U87 mouse model, not the in situ GSC brain tumor model. We had attempted to perform FMT imaging in a brain glioma model, but the signal was weaker than we expected based on the results from the subcutaneous tumor model. This was likely due to the skull that scattered and diminished the fluorescent signal from the brain. However, since SA could be coupled to a wide range of imaging agents (including magnetic resonance nanoparticles, radiotracers and fluorochromes), we were able to perform PET imaging to overcome the scattering and sensitivity problems of optical imaging. Another limitation is that as a proof-of-principle study, we used genetically altered biotin-presenting tumor cells as targets, while naturally occurring tumors do not overexpress biotin on the surface. A universal, naturally occurring antigen for CTL targeting of tumors, though ideal, has not yet been identified. In this study, we described a new imaging strategy and that is not dependent on the availability of a specific cell surface antigen, which could restrict the applicability of the method to only a single tumor type. With our method, it may be possible to couple SA with a biotin-carrying moiety that is designed to bind to specific surface markers of different tumors as they are discovered. This could potentially enable targeting of different tumors in the translational setting. Future work using this proposed paradigm would shed light on the feasibility of this approach.
Conclusions
In summary, sulfo-NHS-biotin labeling is an efficient method to noninvasively image and track the migration of adoptively transferred cells and does not alter CTL viability or interfere with CTL-mediated cytotoxic activity.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by the US National Institute of Health (R01-NS070835 and R01-NS072167), National Natural Science Foundation of China (Grant 81271633). We thank the Memorial Sloan Kettering Cancer Center for providing [89Zr]Zr-oxalate.
Abbreviations
- BAP-TM
Biotin acceptor peptide-transmembrane
- BLI
Bioluminescence imaging
- CT
Computed tomography
- CTL
Cytotoxic T lymphocyte
- DPBS
Dulbecco’s phosphate-buffered saline
- FMT
Fluorescent molecular tomography
- Gluc
Gaussia luciferase
- GSC
Glioma stem cell
- HPLC
High-performance liquid chromatography
- IGFP
Inverted green fluorescent protein
- IVM
Intravital microscopy
- LCMS
Liquid chromatography mass spectroscopy
- NHS
N-hydroxysuccinimide
- NIR
Near infrared
- PET
Positron emission tomography
- SA
Streptavidin
- SEM
Standard error of measurement
- TLC
Thin liquid chromatography
- Zr
Zirconium
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Anning Li and Yue Wu have contributed equally to this work.
Contributor Information
Zhenwei Yao, Phone: 021-52888352, Email: aocnhnr@126.com.
John W. Chen, Phone: (617) 643-0500, Email: jwchen@mgh.harvard.edu
References
- 1.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348(6230):62–68. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Aquino MT, Malhotra A, Mishra MK, Shanker A. Challenges and future perspectives of T cell immunotherapy in cancer. Immunol Lett. 2015;166(2):117–133. doi: 10.1016/j.imlet.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vigneron N. Human tumor antigens and cancer immunotherapy. Biomed Res Int. 2015;2015:948501. doi: 10.1155/2015/948501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shapiro EM, Medford-Davis LN, Fahmy TM, Dunbar CE, Koretsky AP. Antibody-mediated cell labeling of peripheral T cells with micron-sized iron oxide particles (MPIOs) allows single cell detection by MRI. Contrast Media Mol Imaging. 2007;2(3):147–153. doi: 10.1002/cmmi.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lazovic J, Jensen MC, Ferkassian E, Aguilar B, Raubitschek A, Jacobs RE. Imaging immune response in vivo: cytolytic action of genetically altered T cells directed to glioblastoma multiforme. Clin Cancer Res. 2008;14(12):3832–3839. doi: 10.1158/1078-0432.CCR-07-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arbab AS, Janic B, Jafari-Khouzani K, Iskander AS, Kumar S, Varma NR, Knight RA, Soltanian-Zadeh H, Brown SL, Frank JA. Differentiation of glioma and radiation injury in rats using in vitro produce magnetically labeled cytotoxic T-cells and MRI. PLoS ONE. 2010;5(2):e9365. doi: 10.1371/journal.pone.0009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pittet MJ, Grimm J, Berger CR, Tamura T, Wojtkiewicz G, Nahrendorf M, Romero P, Swirski FK, Weissleder R. In vivo imaging of T cell delivery to tumors after adoptive transfer therapy. Proc Natl Acad Sci USA. 2007;104(30):12457–12461. doi: 10.1073/pnas.0704460104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doubrovin MM, Doubrovina ES, Zanzonico P, Sadelain M, Larson SM, O’Reilly RJ. In vivo imaging and quantitation of adoptively transferred human antigen-specific T cells transduced to express a human norepinephrine transporter gene. Cancer Res. 2007;67(24):11959–11969. doi: 10.1158/0008-5472.CAN-07-1250. [DOI] [PubMed] [Google Scholar]
- 9.Shu CJ, Radu CG, Shelly SM, Vo DD, Prins R, Ribas A, Phelps ME, Witte ON. Quantitative PET reporter gene imaging of CD8+ T cells specific for a melanoma-expressed self-antigen. Int Immunol. 2009;21(2):155–165. doi: 10.1093/intimm/dxn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charo J, Perez C, Buschow C, Jukica A, Czeh M, Blankenstein T. Visualizing the dynamic of adoptively transferred T cells during the rejection of large established tumors. Eur J Immunol. 2011;41(11):3187–3197. doi: 10.1002/eji.201141452. [DOI] [PubMed] [Google Scholar]
- 11.Du X, Wang X, Ning N, Xia S, Liu J, Liang W, Sun H, Xu Y. Dynamic tracing of immune cells in an orthotopic gastric carcinoma mouse model using near-infrared fluorescence live imaging. Exp Ther Med. 2012;4(2):221–225. doi: 10.3892/etm.2012.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ntziachristos V, Ripoll J, Wang LV, Weissleder R. Looking and listening to light: the evolution of whole-body photonic imaging. Nat Biotechnol. 2005;23(3):313–320. doi: 10.1038/nbt1074. [DOI] [PubMed] [Google Scholar]
- 13.Whitley MJ, Weissleder R, Kirsch DG. Tailoring adjuvant radiation therapy by intraoperative imaging to detect residual cancer. Semin Radiat Oncol. 2015;25(4):313–321. doi: 10.1016/j.semradonc.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schols RM, Connell NJ, Stassen LP. Near-infrared fluorescence imaging for real-time intraoperative anatomical guidance in minimally invasive surgery: a systematic review of the literature. World J Surg. 2015;39(5):1069–1079. doi: 10.1007/s00268-014-2911-6. [DOI] [PubMed] [Google Scholar]
- 15.Ballou B, Ernst LA, Waggoner AS. Fluorescence imaging of tumors in vivo. Curr Med Chem. 2005;12(7):795–805. doi: 10.2174/0929867053507324. [DOI] [PubMed] [Google Scholar]
- 16.Frangioni JV. In vivo near-infrared fluorescence imaging. Curr Opin Chem Biol. 2003;7(5):626–634. doi: 10.1016/j.cbpa.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Swirski FK, Berger CR, Figueiredo JL, Mempel TR, von Andrian UH, Pittet MJ, Weissleder R. A near-infrared cell tracker reagent for multiscopic in vivo imaging and quantification of leukocyte immune responses. PLoS ONE. 2007;2(10):e1075. doi: 10.1371/journal.pone.0001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Ke S, Wu Q, Charnsangavej C, Gurfinkel M, Gelovani JG, Abbruzzese JL, Sevick-Muraca EM, Li C. Near-infrared optical imaging of integrin alphavbeta3 in human tumor xenografts. Mol Imaging. 2004;3(4):343–351. doi: 10.1162/1535350042973481. [DOI] [PubMed] [Google Scholar]
- 19.Houston JP, Ke S, Wang W, Li C, Sevick-Muraca EM. Quality analysis of in vivo near-infrared fluorescence and conventional gamma images acquired using a dual-labeled tumor-targeting probe. J Biomed Opt. 2005;10(5):054010. doi: 10.1117/1.2114748. [DOI] [PubMed] [Google Scholar]
- 20.Ke S, Wen X, Gurfinkel M, Charnsangavej C, Wallace S, Sevick-Muraca EM, Li C. Near-infrared optical imaging of epidermal growth factor receptor in breast cancer xenografts. Cancer Res. 2003;63(22):7870–7875. [PubMed] [Google Scholar]
- 21.Gottschalk S, Edwards OL, Sili U, Huls MH, Goltsova T, Davis AR, Heslop HE, Rooney CM. Generating CTLs against the subdominant Epstein-Barr virus LMP1 antigen for the adoptive immunotherapy of EBV-associated malignancies. Blood. 2003;101(5):1905–1912. doi: 10.1182/blood-2002-05-1514. [DOI] [PubMed] [Google Scholar]
- 22.Kwon S, Ke S, Houston JP, Wang W, Wu Q, Li C, Sevick-Muraca EM. Imaging dose-dependent pharmacokinetics of an RGD-fluorescent dye conjugate targeted to alpha v beta 3 receptor expressed in Kaposi’s sarcoma. Mol Imaging. 2005;4(2):75–87. doi: 10.1162/15353500200505103. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W, Fulci G, Wakimoto H, Cheema TA, Buhrman JS, Jeyaretna DS, Stemmer Rachamimov AO, Rabkin SD, Martuza RL. Combination of oncolytic herpes simplex viruses armed with angiostatin and IL-12 enhances antitumor efficacy in human glioblastoma models. Neoplasia. 2013;15(6):591–599. doi: 10.1593/neo.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tannous BA, Grimm J, Perry KF, Chen JW, Weissleder R, Breakefield XO. Metabolic biotinylation of cell surface receptors for in vivo imaging. Nat Methods. 2006;3(5):391–396. doi: 10.1038/nmeth875. [DOI] [PubMed] [Google Scholar]
- 25.Niers JM, Chen JW, Weissleder R, Tannous BA. Enhanced in vivo imaging of metabolically biotinylated cell surface reporters. Anal Chem. 2011;83(3):994–999. doi: 10.1021/ac102758m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung E, Yamashita H, Au P, Tannous BA, Fukumura D, Jain RK. Secreted Gaussia luciferase as a biomarker for monitoring tumor progression and treatment response of systemic metastases. PLoS ONE. 2009;4(12):e8316. doi: 10.1371/journal.pone.0008316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foster AE, Kwon S, Ke S, Lu A, Eldin K, Sevick-Muraca E, Rooney CM. In vivo fluorescent optical imaging of cytotoxic T lymphocyte migration using IRDye800CW near-infrared dye. Appl Opt. 2008;47(31):5944–5952. doi: 10.1364/AO.47.005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Youniss FM, Sundaresan G, Graham LJ, Wang L, Berry CR, Dewkar GK, Jose P, Bear HD, Zweit J. Near-infrared imaging of adoptive immune cell therapy in breast cancer model using cell membrane labeling. PLoS ONE. 2014;9(10):e109162. doi: 10.1371/journal.pone.0109162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.