Abstract
Background/Objectives
Maternal obesity increases risk for childhood obesity, but molecular mechanisms are not well understood. We hypothesized that primary umbilical vein endothelial cells (HUVEC) from infants of overweight and obese mothers would harbor transcriptional patterns reflecting offspring obesity risk.
Subjects/Methods
In this observational cohort study, we recruited 13 lean (pre-pregnancy BMI <25.0 kg/m2) and 24 overweight-obese (‘ov-ob’, BMI ≥25.0 kg/m2) women. We isolated primary HUVEC, and analyzed both gene expression (Primeview, Affymetrix) and cord blood levels of hormones and adipokines.
Results
142 transcripts were differentially expressed in HUVEC from infants of overweight-obese mothers (false discovery rate, FDR <0.05). Pathway analysis revealed that genes involved in mitochondrial and lipid metabolism were negatively correlated with maternal BMI (FDR <0.05). To test whether these transcriptomic patterns were associated with distinct nutrient exposures in the setting of maternal obesity, we analyzed the cord blood lipidome and noted significant increases in levels of total free fatty acids (lean: 95.5 ± 37.1 ug/ml, ov-ob: 124.1 ± 46.0 ug/ml, P=0.049), palmitate (lean: 34.5 ± 12.7 ug/ml, ov-ob: 46.3 ± 18.4 ug/ml, P=0.03) and stearate (lean: 20.8 ± 8.2 ug/ml, ov-ob: 29.7 ± 17.2 ug/ml, P=0.04), in infants of overweight-obese mothers.
Conclusion
Prenatal exposure to maternal obesity alters HUVEC expression of genes involved in mitochondrial and lipid metabolism, potentially reflecting developmentally-programmed differences in oxidative and lipid metabolism.
Introduction
Evidence from human populations and animal models indicates that environmental exposures during early development are critical determinants of disease susceptibility throughout the lifespan, a phenomenon termed ‘developmental programming’ (1). A wide range of prenatal perturbations, including maternal undernutrition, obesity, diabetes, high-fat diet, and endocrine-disrupting chemicals, are now recognized as risk factors for chronic diseases including diabetes, obesity, and cardiovascular disease (2–4). Maternal obesity is of particular concern, as it is a potent risk factor for childhood obesity: offspring of mothers entering pregnancy with BMI >30 kg/m2 have a 1.5 to 4-fold higher risk of childhood obesity (5). Studies of siblings born before vs. after a mother’s weight loss surgery - which minimize the contribution of shared genetics - suggest that in utero exposure to maternal obesity per se can increase risk of childhood obesity >2-fold (6, 7).
Unfortunately, the molecular mechanisms by which maternal obesity increases metabolic risk in offspring remain incompletely understood. Previous rodent and primate studies indicate that maternal insulin resistance, which is tightly correlated with maternal obesity (8), may be one contributor to obesity-associated developmental programming (9, 10). Other mediators may include shared environmental risk factors, epigenetics, and/or hormonal and metabolic adaptations to an ‘obese’ intrauterine environment. Fewer mechanistic studies have examined metabolic phenotypes in humans, largely due to the practical and ethical challenges of obtaining cells and tissues from infants. However, umbilical cords, which are usually discarded after delivery, provide an accessible source of infant cells for translational studies. Interestingly, analysis of umbilical cord segments from infants of women with type 1 diabetes identified differences in expression of genes related to vascular development and function (11).
Primary human umbilical vein endothelial cells (HUVEC) are readily isolated, remain viable and metabolically active in culture, and are insulin-responsive, features leading to their wide use in vascular biology for over 40 years (12), and more recently, in studies of fetal adaptations to maternal diabetes and placental insufficiency. For example, maternal gestational diabetes is associated with reduced vasodilation (13) and increased leukocyte adhesion in HUVEC (14), potentially mediated by specific miRNAs (15). Moreover, increased eNos promoter methylation has been reported in HUVEC from infants with intrauterine growth restriction (IUGR) (16).
We therefore hypothesized that maternal obesity would alter metabolism in HUVEC in a cell-autonomous fashion. We now demonstrate that maternal obesity is associated with a dramatic transcriptional response in infant HUVEC, particularly within pathways related to lipid metabolism and mitochondrial structure/function, and is accompanied by increases in cord blood insulin, palmitate, and stearate.
Methods
Human subjects - Recife Cohort
Pregnant women were recruited during prenatal visits at Instituto de Medicina Integral Prof. Fernando Figueira, Recife, Brazil. Eligibility criteria included: age ≥18 years and known gestational age (based on date of last menstrual period (LMP) or ultrasound before 16 weeks). Exclusion criteria included: diabetes diagnosed prior to or during the current pregnancy, multiple gestation, hypertension, artificial reproductive techniques, HIV, syphilis, maternal disease requiring medications known to affect fetal growth or glucose metabolism, fetal congenital malformations, premature delivery (<36 weeks), and deliveries occurring outside the study hospital. Participants were grouped by early pregnancy BMI, defined as BMI at the first prenatal visit (<12 weeks gestation): lean (<25.0 kg/m2) vs. overweight and obese (‘ov-ob’, ≥25.0 kg/m2). The study protocol was approved by the Hospital Institutional Review Board and the National Committee for Ethics in Research at National Health Council (CONEP-Statement # 387/2011), Brasilia, Brazil, and by the Committee for Human Subjects at Joslin Diabetes Center, Boston, Massachusetts. All participants provided written informed consent.
Assessments during pregnancy – Recife Cohort
At the first prenatal visit, maternal medical history, including systemic disease, smoking, obstetric history, and clinical data (height, weight, blood pressure, date of LMP, and fasting glucose values) were collected. Maternal glucose metabolism was assessed twice: a 1-hour 75 g oral glucose screen during the second trimester (24 weeks ± 2 weeks), and a 2-hour 75 g oral glucose tolerance test (OGTT) during the third trimester (glucose and insulin at time 0 (fasting), 1, and 2 hours (30 weeks ± 2 weeks). Homeostasis method assessment of insulin resistance (HOMA-IR) was calculated as [(Glucose × Insulin) / 405], and homeostatic method assessment of beta cell secretion (HOMA-β) as [(360 × Insulin)/(Glucose − 63)], as described (17). Fasting cholesterol, HDL, LDL, VLDL, and triglycerides were also assessed during the third trimester GTT visit. The final maternal BMI was calculated during the last prenatal visit (36 – 40 weeks). Cord blood was collected at delivery and plasma aliquots frozen. Newborn weight, length, head circumference, and abdominal girth were measured within the first 24 hours of life by trained nurses. Newborn weight for gestational age and sex percentiles were derived from the growth curves of Olsen et al.(18); small for gestational age was defined as birth weight < 10th percentile, while large for gestational age was defined as birth weight >90th percentile. Medical records were abstracted to obtain data regarding prenatal, labor and delivery, postpartum, and neonatal course.
Laboratory analysis
Plasma glucose, hemoglobin A1c, and lipids were analyzed using a colorimetric method (Architect c8000, Abbott); LDL cholesterol was calculated using the Friedwald formula. Insulin was measured using an electrochemiluminescence immunoassay (Siemens). Urinalysis, HIV, and syphilis tests were performed using Symex XT-1800 (Roche), and URiSCAN Pro II, YD, AxSYM System (Abbott) and ELISA immunoassay (Elisys Quattro) respectively. Cord blood IL-1β, IL-6, IL-8, leptin, adiponectin, resistin, total PAI-1, MCP-1, HGF and TNF-α levels were determined by multiplex ELISA (Milliplex) in the Joslin DRC Specialized Assay Core.
HUVEC isolation
Umbilical cord segments (10 cm) were transported to the lab within 1 hour of delivery, and kept on ice in buffer containing PBS and antibiotics until HUVEC isolation; all isolations were performed within 2h of delivery. Umbilical segments were rinsed with PBS, and the umbilical vein was cannulated using a 3-way stopcock (Poly Medicure Limited (India), MODEL #1021). The umbilical vein was rinsed twice with 50 ml PBS before infusion with 25 ml 0.1% collagenase (Gibco, Cat. No.# 17100-017) dissolved in 37°C PBS. The cord was clamped at both ends and incubated at room temperature for 30 minutes. The cord was then unclamped and the vein flushed with vascular cell complete growth media (ATCC, PCS-100-030, containing 100 mg/dl glucose), and the effluent collected. The cells were then centrifuged and resuspended in complete growth media. Vascular cell growth media contains VEGF (5 ng/mL), EGF (5 ng/mL), FGF basic (5 ng/mL), IGF-1 (15 ng/mL), L-glutamine (10 mM), heparin sulfate (0.75 Units/mL), hydrocortisone (1 µg/mL), ascorbic acid (50 µg/mL) and fetal bovine serum (2%) (ATCC Endothelial Cell Growth Kit, PCS-100-041).
RNA isolation and microarray analysis
For each cell line, an aliquot of freshly isolated HUVEC was stored in RNA Later reagent (Life Technologies) at −80°C until RNA extraction, using Trizol (Life Technologies) according to the manufacturer’s protocol, with glycogen (AM9510, Life Technologies) added as a carrier. cRNA was prepared and hybridized to Affymetrix PrimeView microarrays. Data were normalized using Robust Multichip Average (RMA) (19) and log-2 transformed. Normalized microarray data were analyzed (p, FDR) using GenePattern Comparative Marker Selection module (Broad Institute). Heatmaps were created using GENE-E (Broad Institute). Pathway analyses were performed using Gene Set Enrichment Analysis (GSEA, Broad Institute) (20, 21) and DAVID (Bioinformatics Resources v6.7) (22).
Quantitative RT-PCR
Complementary DNA (cDNA) was generated using High-Capacity cDNA Reverse Transcription Kit (Life Technologies) with 500 ng input RNA. Expression of selected genes was assessed by qRT-PCR, with expression normalized to cyclophilin E (PPIE), using an ABI 7900 HT thermocycler. Primer sequences are available upon request.
Lipidomic analysis
Lipidomic analyses in cord plasma samples were performed at the Vanderbilt Lipid Core. Lipids were extracted (23), filtered, and recovered in the chloroform phase prior to separation by thin layer chromatography. Phospholipids, diacylglycerides, triglycerides and cholesteryl esters were scraped from the plates and methylated (24); methylated fatty acids were extracted and analyzed by gas chromatography (Agilent 7890A) with helium, and compared to known standards (C15:0, C17:0, C20:1, and cholesteryl eicosenoate (C20:1)). For total cholesterol, an internal standard (5-alpha-cholestane) was added to a portion of the lipid extract and saponified at 80 C in 1 N KOH in 90% methanol for 1 hour. The non-saponifiable sterol was extracted into hexane, concentrated under nitrogen, and solubilized in carbon disulfide to inject onto the gas chromatograph. For unesterified cholesterol, an internal standard is added to the lipid extract, concentrated under nitrogen and solubilized in carbon disulfide to inject onto the gas chromatograph (25). We calculated the ratio of palmitoleate (16:1n7) to palmitate (16:0), and of oleic acid (18:1n9) to oleate (18:0) as a readout of delta-9 desaturase activity, as described (26).
Statistical analysis
Data are reported as mean ± standard deviation. Between-group comparisons were performed by unpaired, unequal variance, two-sided T-tests for continuous data, and Fisher’s exact test for categorical data (JMP 10, SAS Corporation, Cary, NC). To adjust for multiple comparisons, false discovery rates were calculated using R software.
Results
Clinical, metabolic, and hormonal characteristics of mothers and newborns
Maternal age tended to be higher in the overweight/obese (‘ov-ob’) group (lean: 24.6 ± 5.4, ov-ob: 27.9 ± 5.9 years, P=0.1, Table 1). As expected, women in the ov-ob group had significantly higher first trimester BMI than lean women (lean: 21.9 ± 1.7 vs. ov-ob: 32.2 ± 5.7 kg/m2, P<0.0001), and higher BMI at the last prenatal visit (lean: 27.6 ± 3.4 vs. ov-ob: 36.2 ± 6.6 kg/m2, P<0.0001). Gestational weight gain was lower in the ov-ob group (lean: 11.5 ± 4.5 vs. ov-ob: 7.8 ± 4.5 kg, P=0.03), but the proportion of women with excessive gestational weight gain (2009 IOM criteria) (27) was numerically higher in ov-ob (lean: 1/13 or 7.7%, ov-ob: 6/24 or 25%, P=0.38). Diastolic blood pressure was higher in the ov-ob group (lean: 62 ± 7 mmHg, ov-ob: 69 ± 10 mmHg, P=0.02), but systolic blood pressure was unaltered. While fasting glucose did not differ at 24 or 30 weeks, blood glucose 1 hour after glucose challenge at 24 weeks tended to be higher in ov-ob (lean: 105.0 ± 26.8 vs. ov-ob: 126.4 ± 32.6 mg/dl, P=0.09), as was 2 hour glucose at 30 weeks (lean: 117.4 ± 14.2, ov-ob: 134.9 ± 28.2 mg/dl, P=0.02). Insulin resistance (HOMA-IR), insulin secretion index (HOMA-β) and insulin levels during OGTT were similar, as were lipid levels.
Table 1.
Maternal pregnancy characteristics according to early pregnancy weight category.
| Maternal Data | BMI < 25.0 kg/m2 | BMI ≥ 25.0 kg/m2 | P-value |
|---|---|---|---|
| Age (years) | 24.6 ± 5.4 (13) | 27.9 ± 5.9 (24) | 0.1 |
| Early BMI | 21.9 ± 1.7 (13) | 32.2 ± 5.7 (24) | <0.0001 |
| Final BMI | 27.6 ± 3.4 (13) | 36.2 ± 6.6 (24) | <0.0001 |
| Weight Gain (kg) | 11.5 ± 4.5 (13) | 7.8 ± 4.5(24) | 0.03 |
| Height (meters) | 1.60 ± 0.06 (13) | 1.59 ± 0.07 (24) | 0.56 |
| Systolic BP (mm Hg) | 105 ± 15 (13) | 109 ± 13 (24) | 0.41 |
| Diastolic BP (mm Hg) | 62 ± 7 (13) | 69 ± 10 (24) | 0.02 |
| Hematocrit (%) | 36.3 ± 2.7 (13) | 36.4 ± 3.5 (24) | 0.98 |
| Hemoglobin (g/dl) | 11.8 ± 1.0 (13) | 12.1 ± 1.3 (24) | 0.49 |
| Fasting Glucose 1st trimester (mg/dl) | 78.6 ± 8.4 (11) | 79.9 ± 7.2 (24) | 0.64 |
| Glucose 1h OGTT 24 weeks (mg/dl) | 105.0 ± 26.8 (9) | 126.4 ± 32.6 (22) | 0.09 |
| Fasting Glucose OGTT 30weeks (mg/dl) | 75.7 ± 5.4 (12) | 78.0 ± 10.2 (22) | 0.40 |
| Glucose 1h OGTT 30 weeks (mg/dl) | 134.9 ± 18.9 (12) | 139.0 ± 24.0 (22) | 0.61 |
| Glucose 2h OGTT 30 weeks (mg/dl) | 117.4 ± 14.2 (12) | 134.9 ± 28.2 (22) | 0.02 |
| Fasting Insulin OGTT 30 weeks (uU/ml) | 9.9 ± 3.5 (11) | 12.4 ± 7.0 (22) | 0.27 |
| Insulin 1h OGTT 30 weeks (uU/ml) | 106.9 ± 52.3 (11) | 112.3 ± 70.3 (21) | 0.82 |
| Insulin 2h OGTT 30 weeks (uU/ml) | 106.4 ± 64.2 (11) | 126.8 ± 83.4 (22) | 0.48 |
| HOMA-IR | 1.9 ± 0.7 (11) | 2.5 ± 1.6 (22) | 0.24 |
| HOMA-B (%) | 478 ± 640 (11) | 432 ± 347 (22) | 0.79 |
| Total Cholesterol (mg/dl) | 252.7 ± 31.4 (12) | 233.6 ± 41.5 (22) | 0.18 |
| HDL-Cholesterol (mg/dl) | 66.0 ± 10.8 (12) | 66.7 ± 16.6 (22) | 0.90 |
| LDL-Cholesterol (mg/dl) | 146.2 ± 28.0(12) | 121.4 ± 39.5 (22) | 0.06 |
| VLDL-Cholesterol (mg/dl) | 40.6 ± 11.4 (12) | 44.4 ±19.6 (22) | 0.55 |
| Triglycerides (mg/dl) | 202.9 ± 56.0 (12) | 200.8 ± 49.6 (22) | 0.91 |
| C-section (%) | 37.1 (13) | 31.6 (12) | 0.49* |
Mean ± SD; sample size indicated in parentheses
P-value refers to T-test, except for categorical data, where Fisher’s exact test was used (denoted by asterisk).
Gestational age at delivery, birth weight, and newborn adiposity measures, including ponderal index and abdominal circumference, were similar between offspring of lean vs. ov-ob mothers (Table 2). Cord blood insulin was increased 2-fold in ov-ob (lean: 4.6 ± 3.9, ov-ob: 9.0 ± 9.1 µU/mL, P=0.048), as were leptin levels (lean: 23.9 ± 19.4, ov-ob: 48.9 ± 43.6 µg/mL, P=0.02). Other hormones and adipokines in cord blood approached statistical significance, notably hepatocyte growth factor (HGF), IL-6, and TNF-α, which tended to be higher, and PAI-1, which tended to be lower, in infants of ov-ob mothers.
Table 2.
Newborn characteristics and cord blood adipokines according to maternal weight category
| Newborn Data | BMI < 25.0 kg/m2 | BMI ≥ 25.0 kg/m2 | P-value |
|---|---|---|---|
| Gestational Age (weeks) | 39.1 ± 1.3 (13) | 39.1 ± 1.3 (24) | 0.85 |
| Birth weight (grams) | 3280 ± 662 (13) | 3229 ± 508 (24) | 0.81 |
| Infant sex (M/F) | 10M/3F | 13M/11F | 0.28 |
| Weight for gestational age and sex (percentile) |
41.5 ± 27.1 (13) | 45.2± 26.5 (24) | 0.69 |
| Head circumference (cm) | 33.9 ± 1.3 (13) | 34.3 ± 2.1 (24) | 0.56 |
| Abdominal girth (cm) | 31.6 ± 3.0 (13) | 31.6 ± 2.0 (24) | 0.96 |
| Length (cm) | 48.9 ± 2.8 (13) | 48.8 ± 2.7 (24) | 0.93 |
| Ponderal Index (kg/m3) | 27.7 ± 2.4 (13) | 27.6 ± 2.6 (24) | 0.88 |
| Cord blood hormones and adipokines |
|||
| Insulin (µU/ml) | 4.6 ± 3.9 (12) | 9.0 ± 9.1 (24) | 0.048 |
| HGF (ng/ml) | 2.5 ± 1.0 (12) | 3.1 ± 1.0 (24) | 0.08 |
| Leptin (ng/ml) | 23.9 ± 19.4 (13) | 49.0 ± 43.6 (24) | 0.02 |
| Adiponectin (ug/ml) | 407 ± 377 (13) | 337 ± 296 (24) | 0.57 |
| Resistin (pg/ml) | 174 ± 97 (12) | 196 ± 110 (24) | 0.55 |
| IL-1β (pg/ml) | 0.12 ± 0.06 (12) | 0.17 ± 0.29 (23) | 0.50 |
| IL-6 (pg/ml) | 11 ± 10 (12) | 41.9 ± 77.7 (23) | 0.07 |
| IL-8 (pg/ml) | 61 ± 137 (12) | 85 ± 210 (24) | 0.68 |
| MCP-1 (pg/ml) | 386 ± 245 (13) | 314 ± 205 (23) | 0.38 |
| TNF-α (pg/ml) | 9.1 ± 2.5 (12) | 10.8 ± 3.7 (24) | 0.11 |
| Total PAI-1 (pg/ml) | 186 ± 62 (13) | 152 ± 32 (23) | 0.08 |
Mean ± SD; sample size is indicated in parentheses.
P-value refers to Student’s T-test, except for Infant sex, which was analyzed by Fisher’s exact test.
Maternal obesity alters gene expression patterns in HUVEC
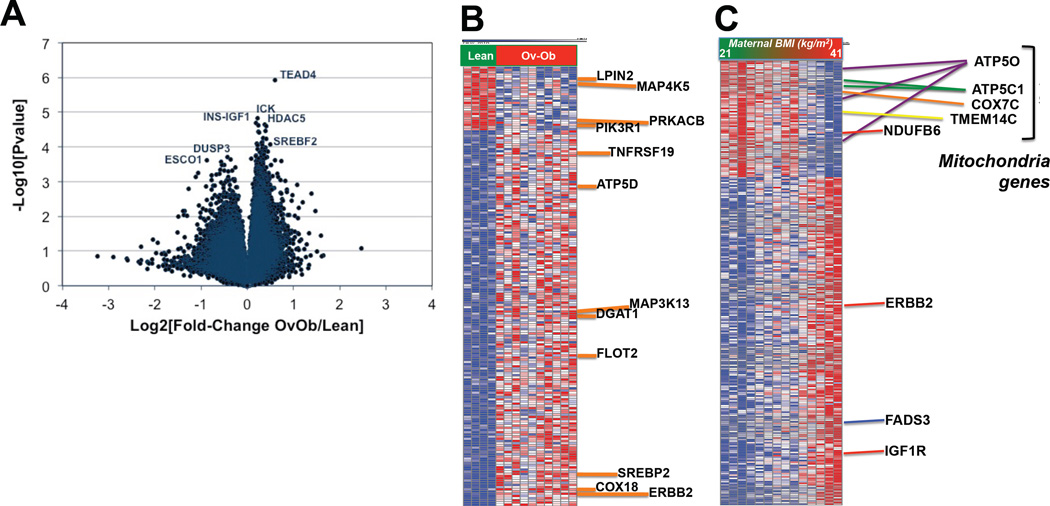
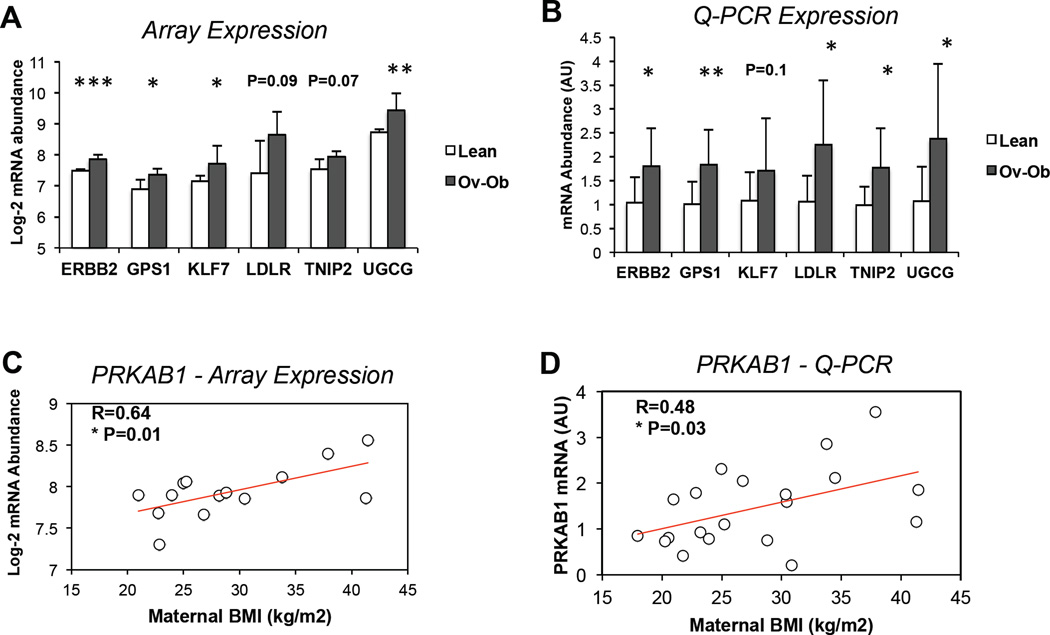
To test whether prenatal exposure to maternal obesity alters transcriptional patterns in infant cells, we analyzed global gene expression in a randomly selected subset of primary HUVEC (PrimeView microarray, Affymetrix). Characteristics of mothers and newborns included in the microarray analysis tended to be similar to the entire cohort, although cord blood levels of HGF, resistin, and TNF-α were more significantly increased in the array subset (Supplemental Table 1). Principal components analysis showed only modest separation of lean vs. ov-ob infants based on gene expression, with substantial overlap between groups (Supplemental Figure 1). 4916 transcripts were altered by maternal obesity (P<0.05), with 142 remaining different after adjustment for multiple comparisons (FDR<0.05), all of which were upregulated in ov-ob (complete list in Supplemental Table 2 A). As seen in the volcano plot in Figure 1A, more transcripts were upregulated by maternal obesity than downregulated. Strikingly, many of the top-ranking differentially expressed genes were related to lipid metabolism (e.g. LPIN2, MGAT2, DGAT1, SREBP2), insulin signaling (e.g. MAP4K5, PIK3R1, PRKACB, MAP3K13) and mitochondrial metabolism (e.g. COX18, ATP5D) (Figure 1B). Similarly, several genes related to lipid metabolism (e.g. LDLR, ANGPTL4, LMNA) (28, 29) were top-ranking in terms of fold-change (increased in Ov-Ob, Supplemental Table 2C–D). We next examined correlations between maternal BMI and gene expression, and found that several mitochondrial genes (e.g. ATP5O, ATP5C1, COX7C, NDUFB6, TMEM14C) were inversely correlated with maternal BMI, whereas the lipid metabolism genes FADS2 and FADS3, insulin-like growth factor 1 receptor (IGF1R) and receptor tyrosine-protein kinase erbB-2 (ERBB2) where strongly positively correlated with maternal BMI (Figure 1C). qRT-PCR confirmed significant upregulation of LDL receptor (LDLR), UDP-glucose ceramide glucosyltransferase (UGCG), erb-b2 receptor tyrosine kinase 2 (ERBB2), G protein pathway suppressor 1 (GPS1), Kruppel-like factor 7, and TNFAIP3 interacting protein 2 (TNIP2) in HUVEC from infants exposed to maternal obesity, consistent with microarray patterns. We also confirmed a significant, positive correlation between maternal BMI and PRKAB1 (Figure 2A/D).
Figure 1. Global gene expression patterns in HUVEC isolated from infants of overweight-obese mothers vs. controls.
A. Volcano plot.
B. Heatmap – top 200 differentially regulated probes, selected based on Comparative Marker Selection scores, GenePattern, Broad Institute.
C. Heatmap of top-ranking probesets correlating with maternal BMI, selected based on Pearson’s R≥0.75 and R≤0.75. The full gene list is available in Supplemental Table 2B.
Figure 2. Validation of HUVEC gene expression patterns identified by array data using Q-PCR.
A. Array expression of top-ranking differentially expressed genes in HUVEC from infants of Ov-Ob vs. lean mothers, including erb-b2 receptor tyrosine kinase 2 (ERBB2), G protein pathway suppressor 1 (GPS1), Kruppel-like factor 7 (KLF7), low density lipoprotein receptor (LDLR), TNFAIP3 interacting protein 2 (TNIP2), and UDP-glucose ceramide glucosyltransferase (UGCG). N=14.
B. qRT-PCR expression of ERBB2, GPS1, KLF7, LDLR, TNIP2, and UGCG, normalized to PPIE and to expression in lean controls. N=20.
C. Based on array analysis, HUVEC expression of protein kinase, AMP-activated, beta 1 non-catalytic subunit (PRKAB1) is positively correlated with maternal pre-pregnancy BMI. N=14.
D. qRT-PCR expression of PRKAB1, normalized to PPIE, is positively correlated with maternal pre-pregnancy BMI. N=20. * P<0.05, ** P<0.01, ***P<0.0001.
Mitochondrial and lipid metabolism transcriptional pathways are altered by maternal obesity
We next performed Gene Set Enrichment Analysis (GSEA) to analyze enrichment of Gene Ontology (GO) terms in genes correlating with maternal BMI. 98 GO terms were positively correlated with BMI (q<0.05), including pathways related to ion channels, transmembrane transporters, and neurotransmitter activity. 73 GO terms were inversely correlated (q<0.05). The top-ranking set inversely correlated with maternal BMI was “proton-transporting two sector ATPase complex”; many other inversely correlated pathways were related to mitochondria (e.g. mitochondrial small ribosomal subunit, matrix, aerobic respiration) and to lipid metabolism (e.g. fatty acid biosynthesis, beta-oxidation, lipid catabolism). Pathway analyses are shown in Supp Tables 3A/B.
Maternal obesity associated with reduced maternal LDL cholesterol and increased fatty acid and triglyceride species in cord blood
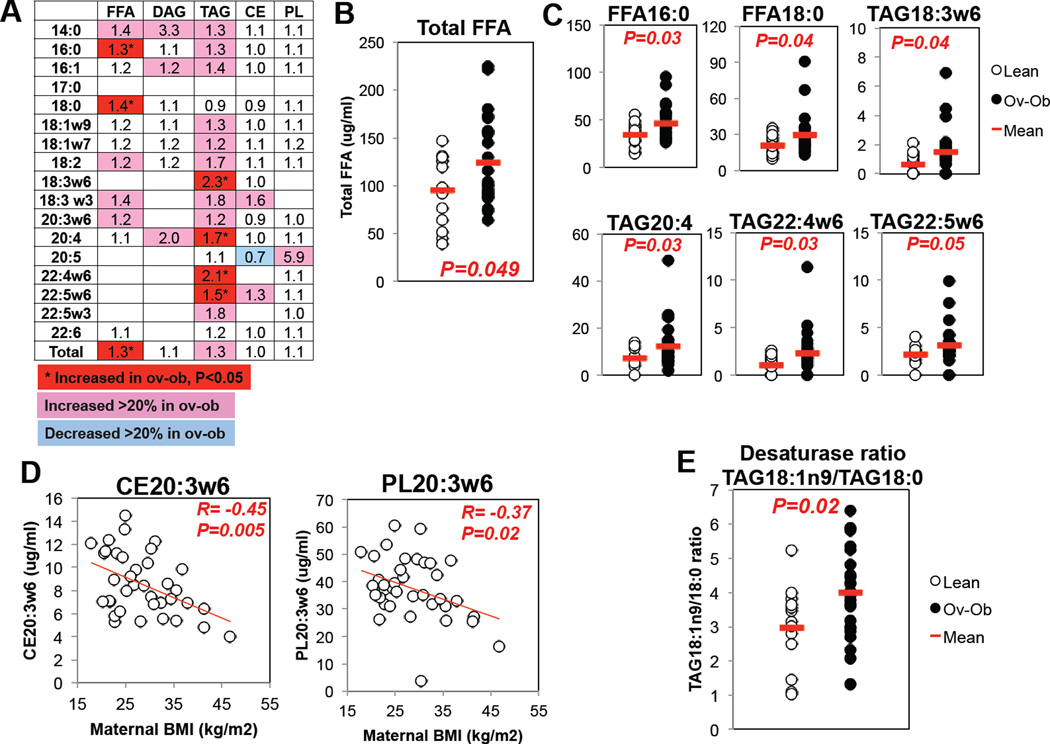
We next asked whether altered HUVEC gene expression, particularly within lipid and mitochondrial pathways, might reflect differences in nutrient exposure. Maternal LDL cholesterol tended to be lower in ov-ob mothers (lean: 146±28, ov-ob: 121±40 mg/dl, P=0.06), but levels of triglycerides and HDL were similar. In infant cord blood, there was a trend for increased levels of several triglyceride and fatty acid species from offspring of ov-ob mothers (Figure 4A). Notably, levels of total free fatty acids, and of two saturated fatty acids, palmitate (16:0) and stearate (18:0), were significantly increased (1.3, 1.3 and 1.4-fold, respectively, P<0.05, Figure 4B/C). Moreover, several triglyceride species containing long-chain and omega-6 fatty acids were increased by 1.5 to 2.3-fold in the ov-ob group (TAG18:3w6, TAG20:4, TAG22:4w6, and TAG22:5w6, P<0.05) (Figure 4C). The ratio of TAG18:1n9 to TAG18:0, a surrogate for desaturase activity, was significantly higher in ov-ob, suggesting increased SCD-mediated lipogenesis (Figure 4E). Interestingly, cholesterol esters and phospholipids containing the rare essential fatty acid 20:3w6 (dihomo-gamma-linolenic acid, DGLA) were negatively correlated with pre-pregnancy BMI (CE20:3w6: Pearson r= −0.45, P=0.005; PL20:3w6: r= −0.37, P=0.02, Figure 4D). Since levels of DGLA may be influenced by activity of delta-5 and delta-6 desaturase enzymes, it is interesting that expression of FADS2 (delta-6 desaturase) and FADS3 (delta-5 desaturase) were positively correlated with maternal BMI (Supplemental Figure 2). Strikingly, we noted strong correlations between levels of cord blood CE20:3w6 and expression of several transcripts related to lipid metabolism, including monoglycerate lipase (MGLL) and SREBP2, and the mitochondrial gene ATP5D (Supplemental Figure 3A). Cord blood palmitate (16:0) and stearate (18:0) also correlated with expression of PPARG2 (Supplemental Figure 3B) and several other transcripts dysregulated in ov-ob (e.g. correlation of 16:0 vs. NDUFB6 R=−0.62, P=0.02; KLF7, R=0.615, P=0.02; ARID4B, R=0.73, P=0.003). Together, these data raise the possibility that altered plasma lipid species might contribute to differential gene expression in HUVEC from infants of overweight-obese mothers.
Discussion
Given that HUVEC are exclusively fetal in origin, they provide a powerful model to test whether prenatal exposures such as maternal obesity result in cell autonomous transcriptional patterns in offspring. In this study, we perform the first analysis of transcriptional responses to maternal obesity in HUVEC. Strikingly, maternal obesity was associated with marked perturbations in gene expression in offspring HUVEC, with downregulation of numerous genes related to mitochondrial function and lipid metabolism. Transcriptional patterns in infant HUVEC were closely correlated with cord blood levels of the fatty acids palmitate and stearate, and with lipids containing 20:3w6, raising the possibility that plasma lipids may mediate altered gene expression patterns in HUVEC. Whether such transcriptional patterns might result from direct nutrient exposures, or indirect epigenetic mechanisms, will be a key question for future studies. Furthermore, it will be important to determine whether these expression patterns can serve as markers or mediators of metabolic disease risk in infants of overweight/obese mothers.
Many of the differentially expressed genes identified in our analysis have previously been linked to obesity, diabetes, or other metabolic traits. For example, decreased adipose expression of SREBP2 and other genes involved in lipolysis and triglyceride synthesis, has been reported in adipose tissue from women with GDM and/or obesity (30). Expression of the cholesterol regulatory gene LDLR (31), was increased in HUVEC of ov-ob mothers. For several other top-ranking genes identified in our analysis (e.g. LPIN2, KLF7, PPARG2), SNPs within these loci have been linked to obesity and type 2 diabetes risk (32–34). Intriguingly, ERBB2, which was upregulated in infants of ov-ob mothers, has previously been linked to maternal obesity; differences in methylation of ERBB2 in cord blood leukocytes have been linked to pre-pregnancy maternal BMI (35). It will thus be important for future studies to examine whether differences in infant expression of ERBB2 and other genes identified in our analysis, may be associated with differences in DNA methylation or other forms of epigenetic regulation.
Another striking pattern in HUVEC from infants exposed to maternal obesity is altered expression of individual genes and transcriptional pathways related to mitochondrial metabolism. A large body of evidence has linked diabetes and obesity to reduced adipose, muscle, and liver expression of genes involved in mitochondrial oxidation (21, 36). Moreover, functional defects in mitochondrial oxidation have been reported in humans with diabetes and obesity, and in rodent models of adverse prenatal exposures (37). In mice, maternal high fat diet reduces oocyte mitochondrial number and function (38). However, evidence from human infants is much more limited. Maternal obesity has been associated with reduced mitochondrial function in human placenta (39, 40), but given that the placenta contains maternal and fetal cells in close proximity, dissecting the cause of such transcriptional patterns is challenging. Our data in HUVEC, cells of purely fetal origin, suggest that altered mitochondrial gene expression may be an early marker of infant metabolic disease risk resulting from maternal obesity. Transcriptional patterns in HUVEC from infants of Ov-Ob mothers, moreover, appear distinct from previous analyses of HUVEC from infants of mothers with GDM (41), which highlighted differences in gene pathways related to extracellular matrix remodeling and insulin binding proteins. Gene expression patterns in the Ov-Ob group were somewhat more similar to a published analysis of transcriptional patterns in HUVEC exposed to acute insulin treatment, which identified “Electron Transport Chain” as a top-ranking significantly altered pathway (42).
In our analysis of individual genes altered by maternal obesity, we found that several genes critical for insulin signaling, including PIK3R1, MAP4K5, and MAP3K13, ranked among the most significantly differentially regulated transcripts. These data are intriguing in light of the recent study by Thakali et al. reporting increased phosphorylation of AKT, an insulin signaling node downstream of PI3-kinase, in umbilical cord segments from infants of overweight/obese mothers (43), and several reports of impaired PI3 kinase activation in rodent models of developmental programming (44–46). Together, these data raise the possibility that the PI3-kinase pathway may be susceptible to modulation by prenatal perturbations.
To test whether nutrient exposures might mediate transcriptional responses to maternal obesity in infant HUVEC, we analyzed the cord blood lipidome. Total free fatty acids, as well as individual lipids including palmitate (16:0), stearate (18:0), and triglycerides containing 18:3w6, 22:4w6, 22:5w6 and 20:4, were significantly increased in cord blood from infants of ov-ob women. Moreover, the ratio of 18:1n9 to 18:0, a surrogate for SCD1 delta-9 desaturase activity, was increased in the ov-ob group, similar to patterns in infants of women with gestational diabetes (47). By contrast, lipid species containing 20:3w6 fatty acids (e.g. CE20:3w6, PL20:3w6) were negatively correlated with maternal BMI. Previous reports have documented increases in plasma 20:3w6 (di-homo gamma linolenic acid, DGLA), and in phospholipids containing DGLA, in obese humans (reviewed in (48)). Moreover, maternal plasma levels of (20:3w6, or DGLA) during pregnancy have been associated with childhood obesity risk at age 7 years (49). By contrast, dietary DGLA supplementation in mice may protect against atherosclerosis (50). These data raise the intriguing possibility that reductions in plasma DGLA in infants of obese women might contribute to increased atherosclerosis risk. More broadly, the observation that transcriptional pathways involved in lipid metabolism, together with cord blood lipid levels, are altered by maternal obesity may have relevance to offspring vascular function given the recent discovery that fatty acid oxidation by endothelial cells is critical for proliferative capacity (51). Surprisingly, we did not observe any differences in plasma triglycerides in ov-ob vs. lean women, which stands in contrast to previous studies demonstrating associations between maternal BMI and serum triglycerides (52). It is difficult to assess whether the lack of association is due to the genetics, environment, or diet of this Brazilian group of women, as we unfortunately do not have information about these characteristics. This will be an important area for future studies, as maternal triglyceridemia may be an important contributor to fetal macrosomia, a key risk factor for future metabolic disease in offspring (53).
We acknowledge that our study has several limitations. First, our sample size is small, which limits our power to detect differences, and may limit the generalizability of our findings. Our limited sample size, moreover, prevented us from adjusting for offspring sex, mode of delivery, maternal age, and other covariates that might influence infant weight status or metabolism. Secondly, the infants were not observed beyond the newborn period, so we cannot assess whether the transcriptional patterns observed in HUVEC, or the hormonal/lipidomic differences in cord blood, were linked to the development of obesity or other metabolic diseases in offspring. Thirdly, we did not exclude infants with fetal growth restriction (SGA) or growth excess (LGA), so we cannot exclude the possibility that infant weight status, rather than maternal obesity, might drive the phenotypes we observed. Finally, we recognize that analyses of transcriptomic and metabolic variables in humans cannot prove causality. Nevertheless, as a readily isolated cell of purely fetal origin, HUVEC provide evidence for how prenatal perturbations such as maternal obesity may alter cellular responses in offspring. HUVEC provide a robust model for testing how maternal obesity may alter lipid metabolism, insulin signaling, nutrient sensing or other key cellular pathways in infants.
In summary, we tested whether prenatal exposure to maternal obesity influences transcriptional responses in infant cells. We found that maternal obesity alters expression of genes related to mitochondrial and lipid metabolism. Transcriptional patterns were associated with altered levels of cord blood lipids, notably palmitate, stearate, and lipids containing omega-6 fatty acids. These data suggest that altered lipid metabolism may be an early signature of mother-to-child transmission of obesity and metabolic disease risk.
Supplementary Material
Maternal and newborn characteristics, and cord blood adipokines for samples used in microarray analysis. Mean ± SD. N=14.
A. Top-ranking differentially expressed transcripts in HUVEC from infants of Ov-Ob vs. Lean mothers, based on FDR<0.05 and P<0.05. N=14.
B. Top-ranking transcripts whose expression in HUVEC correlates with maternal pre-pregnancy BMI at Pearson R> 0.75 and R<−0.75, P<0.01. These data are shown in heatmap format in Figure 1C. N=14.
C. List of 100 most upregulated probes in microarray analysis of HUVEC from infants of overweight-obese vs. lean mothers (N=14), ranked according to log-2 fold-change (ov-ob/lean).
D. List of 100 most downregulated probes in microarray analysis of HUVEC from infants of overweight-obese vs. lean mothers (N=14), ranked according to log-2 fold-change (ov-ob/lean).
A. Several pathways related to mitochondrial structure, mitochondrial metabolism, and fatty acid oxidation (shaded) are enriched among transcripts correlating negatively with maternal BMI. Table includes all gene sets with FDR q-value <0.05. N=14.
B. Several pathways related to ion channels and neurotransmitter activity are enriched among transcripts correlating positively with maternal BMI. Table includes all gene sets with FDR q-value <0.05. N=14.
Newborn anthropometrics and cord blood hormones and adipokines showed similar trends in male and female offspring. Data are shown as Mean ± SD (n). P-value refers to 2-sided Student’s T-test, except for cells in which there were 2 or fewer samples, for which no T-test was computed.
PCA does not clearly separate HUVEC gene expression patterns from infants of lean vs. ov-ob mothers.
A. HUVEC expression of several genes involved in mitochondrial oxidation negatively correlated with maternal pre-pregnancy BMI, including ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit (ATP5O; probe 1–3 refers to 3 distinct probe sets within this gene on Affymetrix PrimeView array), cytochrome c oxidase subunit VIIc (COX7C), ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 (ATP5C1), and NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa (NDUFB6). N=14.
B. HUVEC expression of several genes involved in lipid metabolism rank among the most strongly correlated with maternal BMI. Expression of lipin 2 (LPIN2) is negatively correlated with maternal BMI, whereas fatty acid desaturase 2 (FADS2) and fatty acid desaturase 3 (FADS3) are positively correlated with maternal BMI. Log-2 normalized Primeview array expression. N=14.
A. Cord blood CE20:3n6 levels were strongly correlated with HUVEC expression of several genes involved in lipid and mitochondrial metabolism, including monoglyceride lipase (MGLL; probe 1–3 refers to 3 distinct probe sets within this gene on Affymetrix PrimeView array), sterol regulatory element binding transcription factor 2 (SREBF2), acyl-CoA synthetase short-chain family member 2 (ACSS2), and ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit (ATP5D). N=14.
B. Associations of cord saturated fatty acids palmitate (16:0), left, and stearate (18:0), right, and HUVEC expression of peroxisome proliferator-activated receptor gamma (PPARG). N=14.
C. Expression of PPARG is higher in HUVEC from infants of ov-ob mothers. Log-2 normalized Primeview array expression. N=14.
Figure 3. Maternal obesity alters infant cord blood lipidome.
A. Overview of lipid composition of cord blood. Each column represents a class of lipids, and each row represents a fatty acid comprised within the lipid class. Each lipid subset was quantified by LC/MS; then normalized to the level in infants of lean mothers. Data are expressed as fold-change in ov-ob/lean. FFA: Free fatty acids; DAG: diacylglycerides; TAG: triacylglycerides; CE: cholesterol esters; PL: phospholipids.
B. Cord blood total fatty acids (ug/ml). N=37.
C. Cord blood FFA16:0, FFA18:0, omega-6 conjugated triglycerides and TAG20:4 are increased in infants of overweight/obese women. All lipid levels expressed as µg/ml. N=37.
D. 20:3w6-conjugated cholesterol esters and phospholipids are inversely correlated with maternal BMI. N=37.
E. The ratio of 18:1n9 to 18:0 triglycerides, a surrogate for stearoyl-coA desaturase 1 (SCD1) activity, is increased in cord blood from infants of Ov-Ob women. N=37.
Acknowledgments
We are thankful for our research support. EI was supported by NICHD K99/R00 Award HD064793. SMRC was supported by a Fullbright Fellowship. JMD and the Joslin Bioinformatics Core are supported by a Diabetes Research Center (DRC) grant (DK036836). We are grateful for the services of the Vanderbilt Lipid Core.
Footnotes
There are no conflicts of interest to disclose.
Author Contributions
SMRC designed the study, collected/analyzed data, and reviewed the manuscript; SMRC is the guarantor of the clinical data. EI analyzed data and wrote the manuscript; EI is the guarantor of transcriptomics and lipidomics analyses. TM, KH, GD, and JMD assisted with data analysis. GAPD supervised clinical study design. MEP supervised data analysis and reviewed/edited the manuscript.
References
- 1.Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med. 2008;359(1):61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woo M, Patti ME. Diabetes risk begins in utero. Cell Metab. 2008;8(1):5–7. doi: 10.1016/j.cmet.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Barker DJ, Gluckman PD, Godfrey KM, Harding JE, Owens JA, Robinson JS. Fetal nutrition and cardiovascular disease in adult life. Lancet. 1993;341(8850):938–941. doi: 10.1016/0140-6736(93)91224-a. [DOI] [PubMed] [Google Scholar]
- 4.Chamorro-Garcia R, Sahu M, Abbey RJ, Laude J, Pham N, Blumberg B. Transgenerational inheritance of increased fat depot size, stem cell reprogramming, and hepatic steatosis elicited by prenatal exposure to the obesogen tributyltin in mice. Environ Health Perspect. 2013;121(3):359–366. doi: 10.1289/ehp.1205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu Z, Han S, Zhu J, Sun X, Ji C, Guo X. Pre-pregnancy body mass index in relation to infant birth weight and offspring overweight/obesity: a systematic review and meta-analysis. PLoS One. 2013;8(4):e61627. doi: 10.1371/journal.pone.0061627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kral JG, Biron S, Simard S, Hould FS, Lebel S, Marceau S, et al. Large maternal weight loss from obesity surgery prevents transmission of obesity to children who were followed for 2 to 18 years. Pediatrics. 2006;118(6):e1644–e1649. doi: 10.1542/peds.2006-1379. [DOI] [PubMed] [Google Scholar]
- 7.Guenard F, Deshaies Y, Cianflone K, Kral JG, Marceau P, Vohl MC. Differential methylation in glucoregulatory genes of offspring born before vs. after maternal gastrointestinal bypass surgery. Proc Natl Acad Sci U S A. 2013;110(28):11439–11444. doi: 10.1073/pnas.1216959110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catalano PM, Ehrenberg HM. The short- and long-term implications of maternal obesity on the mother and her offspring. BJOG. 2006;113(10):1126–1133. doi: 10.1111/j.1471-0528.2006.00989.x. [DOI] [PubMed] [Google Scholar]
- 9.Isganaitis E, Woo M, Ma H, Chen M, Kong W, Lytras A, et al. Developmental programming by maternal insulin resistance: hyperinsulinemia, glucose intolerance, and dysregulated lipid metabolism in male offspring of insulin-resistant mice. Diabetes. 2014;63(2):688–700. doi: 10.2337/db13-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorn SR, Baquero KC, Newsom SA, El Kasmi KC, Bergman BC, Shulman GI, et al. Early life exposure to maternal insulin resistance has persistent effects on hepatic NAFLD in juvenile nonhuman primates. Diabetes. 2014;63(8):2702–2713. doi: 10.2337/db14-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koskinen A, Lehtoranta L, Laiho A, Laine J, Kaapa P, Soukka H. Maternal diabetes induces changes in the umbilical cord gene expression. Placenta. 2015;36(7):767–774. doi: 10.1016/j.placenta.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Lewis LJ, Hoak JC, Maca RD, Fry GL. Replication of human endothelial cells in culture. Science. 1973;181(4098):453–454. doi: 10.1126/science.181.4098.453. [DOI] [PubMed] [Google Scholar]
- 13.Westermeier F, Salomon C, Gonzalez M, Puebla C, Guzman-Gutierrez E, Cifuentes F, et al. Insulin restores gestational diabetes mellitus-reduced adenosine transport involving differential expression of insulin receptor isoforms in human umbilical vein endothelium. Diabetes. 2011;60(6):1677–1687. doi: 10.2337/db11-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giri H, Chandel S, Dwarakanath LS, Sreekumar S, Dixit M. Increased endothelial inflammation, sTie-2 and arginase activity in umbilical cords obtained from gestational diabetic mothers. PLoS One. 2013;8(12):e84546. doi: 10.1371/journal.pone.0084546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Floris I, Descamps B, Vardeu A, Mitic T, Posadino AM, Shantikumar S, et al. Gestational diabetes mellitus impairs fetal endothelial cell functions through a mechanism involving microRNA-101 and histone methyltransferase enhancer of zester homolog-2. Arterioscler Thromb Vasc Biol. 2015;35(3):664–674. doi: 10.1161/ATVBAHA.114.304730. [DOI] [PubMed] [Google Scholar]
- 16.Casanello P, Krause B, Torres E, Gallardo V, Gonzalez M, Prieto C, et al. Reduced l-arginine transport and nitric oxide synthesis in human umbilical vein endothelial cells from intrauterine growth restriction pregnancies is not further altered by hypoxia. Placenta. 2009;30(7):625–633. doi: 10.1016/j.placenta.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 17.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 18.Olsen IE, Groveman SA, Lawson ML, Clark RH, Zemel BS. New intrauterine growth curves based on United States data. Pediatrics. 2010;125(2):e214–e224. doi: 10.1542/peds.2009-0913. [DOI] [PubMed] [Google Scholar]
- 19.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 22.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226(1):497–509. [PubMed] [Google Scholar]
- 24.Morrison WR, Smith LM. Preparation of Fatty Acid Methyl Esters and Dimethylacetals from Lipids with Boron Fluoride--Methanol. J Lipid Res. 1964;5:600–608. [PubMed] [Google Scholar]
- 25.Rudel LL, Kelley K, Sawyer JK, Shah R, Wilson MD. Dietary monounsaturated fatty acids promote aortic atherosclerosis in LDL receptor-null, human ApoB100-overexpressing transgenic mice. Arterioscler Thromb Vasc Biol. 1998;18(11):1818–1827. doi: 10.1161/01.atv.18.11.1818. [DOI] [PubMed] [Google Scholar]
- 26.Attie AD, Krauss RM, Gray-Keller MP, Brownlie A, Miyazaki M, Kastelein JJ, et al. Relationship between stearoyl-CoA desaturase activity and plasma triglycerides in human and mouse hypertriglyceridemia. J Lipid Res. 2002;43(11):1899–1907. doi: 10.1194/jlr.m200189-jlr200. [DOI] [PubMed] [Google Scholar]
- 27.Rasmussen KM, Yaktine AL Institute of Medicine (U.S.) Weight gain during pregnancy : reexamining the guidelines. Washington, D.C.: National Academies Press; 2009. Committee to Reexamine IOM Pregnancy Weight Guidelines; p. xiv.p. 854. [PubMed] [Google Scholar]
- 28.Boschmann M, Engeli S, Moro C, Luedtke A, Adams F, Gorzelniak K, et al. LMNA mutations, skeletal muscle lipid metabolism, and insulin resistance. J Clin Endocrinol Metab. 2010;95(4):1634–1643. doi: 10.1210/jc.2009-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sukonina V, Lookene A, Olivecrona T, Olivecrona G. Angiopoietin-like protein 4 converts lipoprotein lipase to inactive monomers and modulates lipase activity in adipose tissue. Proc Natl Acad Sci U S A. 2006;103(46):17450–17455. doi: 10.1073/pnas.0604026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lappas M. Effect of pre-existing maternal obesity, gestational diabetes and adipokines on the expression of genes involved in lipid metabolism in adipose tissue. Metabolism. 2014;63(2):250–262. doi: 10.1016/j.metabol.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Austin MA, Hutter CM, Zimmern RL, Humphries SE. Familial hypercholesterolemia and coronary heart disease: a HuGE association review. Am J Epidemiol. 2004;160(5):421–429. doi: 10.1093/aje/kwh237. [DOI] [PubMed] [Google Scholar]
- 32.Zobel DP, Andreasen CH, Burgdorf KS, Andersson EA, Sandbaek A, Lauritzen T, et al. Variation in the gene encoding Kruppel-like factor 7 influences body fat: studies of 14 818 Danes. Eur J Endocrinol. 2009;160(4):603–609. doi: 10.1530/EJE-08-0688. [DOI] [PubMed] [Google Scholar]
- 33.Aulchenko YS, Pullen J, Kloosterman WP, Yazdanpanah M, Hofman A, Vaessen N, et al. LPIN2 is associated with type 2 diabetes, glucose metabolism, and body composition. Diabetes. 2007;56(12):3020–3026. doi: 10.2337/db07-0338. [DOI] [PubMed] [Google Scholar]
- 34.Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26(1):76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Chen Q, Tsai HJ, Wang G, Hong X, Zhou Y, et al. Maternal preconception body mass index and offspring cord blood DNA methylation: exploration of early life origins of disease. Environ Mol Mutagen. 2014;55(3):223–230. doi: 10.1002/em.21827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100(14):8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peterside IE, Selak MA, Simmons RA. Impaired oxidative phosphorylation in hepatic mitochondria in growth-retarded rats. Am J Physiol Endocrinol Metab. 2003;285(6):E1258–E1266. doi: 10.1152/ajpendo.00437.2002. [DOI] [PubMed] [Google Scholar]
- 38.Wu LL, Russell DL, Wong SL, Chen M, Tsai TS, St John JC, et al. Mitochondrial dysfunction in oocytes of obese mothers: transmission to offspring and reversal by pharmacological endoplasmic reticulum stress inhibitors. Development. 2015;142(4):681–691. doi: 10.1242/dev.114850. [DOI] [PubMed] [Google Scholar]
- 39.Lassance L, Haghiac M, Minium J, Catalano P, Hauguel-de Mouzon S. Obesity-induced down-regulation of the mitochondrial translocator protein (TSPO) impairs placental steroid production. J Clin Endocrinol Metab. 2015;100(1):E11–E18. doi: 10.1210/jc.2014-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mele J, Muralimanoharan S, Maloyan A, Myatt L. Impaired mitochondrial function in human placenta with increased maternal adiposity. Am J Physiol Endocrinol Metab. 2014;307(5):E419–E425. doi: 10.1152/ajpendo.00025.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ambra R, Manca S, Palumbo MC, Leoni G, Natarelli L, De Marco A, et al. Transcriptome analysis of human primary endothelial cells (HUVEC) from umbilical cords of gestational diabetic mothers reveals candidate sites for an epigenetic modulation of specific gene expression. Genomics. 2014;103(5–6):337–348. doi: 10.1016/j.ygeno.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 42.Di Camillo B, Sanavia T, Iori E, Bronte V, Roncaglia E, Maran A, et al. The transcriptional response in human umbilical vein endothelial cells exposed to insulin: a dynamic gene expression approach. PLoS One. 2010;5(12):e14390. doi: 10.1371/journal.pone.0014390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thakali KM, Saben J, Faske JB, Lindsey F, Gomez-Acevedo H, Lowery CL, Jr, et al. Maternal pregravid obesity changes gene expression profiles toward greater inflammation and reduced insulin sensitivity in umbilical cord. Pediatr Res. 2014;76(2):202–210. doi: 10.1038/pr.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ozanne SE, Dorling MW, Wang CL, Nave BT. Impaired PI 3-kinase activation in adipocytes from early growth-restricted male rats. Am J Physiol Endocrinol Metab. 2001;280(3):E534–E539. doi: 10.1152/ajpendo.2001.280.3.E534. [DOI] [PubMed] [Google Scholar]
- 45.Ozanne SE, Nave BT, Wang CL, Shepherd PR, Prins J, Smith GD. Poor fetal nutrition causes long-term changes in expression of insulin signaling components in adipocytes. Am J Physiol. 1997;273(1 Pt 1):E46–E51. doi: 10.1152/ajpendo.1997.273.1.E46. [DOI] [PubMed] [Google Scholar]
- 46.Shelley P, Martin-Gronert MS, Rowlerson A, Poston L, Heales SJ, Hargreaves IP, et al. Altered skeletal muscle insulin signaling and mitochondrial complex II-III linked activity in adult offspring of obese mice. Am J Physiol Regul Integr Comp Physiol. 2009;297(3):R675–R681. doi: 10.1152/ajpregu.00146.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yee JK, Mao CS, Ross MG, Lee WN, Desai M, Toda A, et al. High oleic/stearic fatty-acid desaturation index in cord plasma from infants of mothers with gestational diabetes. J Perinatol. 2014;34(5):357–363. doi: 10.1038/jp.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fekete K, Gyorei E, Lohner S, Verduci E, Agostoni C, Decsi T. Long-chain polyunsaturated fatty acid status in obesity: a systematic review and meta-analysis. Obes Rev. 2015;16(6):488–497. doi: 10.1111/obr.12280. [DOI] [PubMed] [Google Scholar]
- 49.de Vries PS, Gielen M, Rizopoulos D, Rump P, Godschalk R, Hornstra G, et al. Association between polyunsaturated fatty acid concentrations in maternal plasma phospholipids during pregnancy and offspring adiposity at age 7: the MEFAB cohort. Prostaglandins Leukot Essent Fatty Acids. 2014;91(3):81–85. doi: 10.1016/j.plefa.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 50.Takai S, Jin D, Kawashima H, Kimura M, Shiraishi-Tateishi A, Tanaka T, et al. Anti-atherosclerotic effects of dihomo-gamma-linolenic acid in ApoE-deficient mice. J Atheroscler Thromb. 2009;16(4):480–489. doi: 10.5551/jat.no430. [DOI] [PubMed] [Google Scholar]
- 51.Schoors S, Bruning U, Missiaen R, Queiroz KC, Borgers G, Elia I, et al. Fatty acid carbon is essential for dNTP synthesis in endothelial cells. Nature. 2015;520(7546):192–197. doi: 10.1038/nature14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scifres CM, Catov JM, Simhan HN. The impact of maternal obesity and gestational weight gain on early and mid-pregnancy lipid profiles. Obesity (Silver Spring) 2014;22(3):932–938. doi: 10.1002/oby.20576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olmos PR, Rigotti A, Busso D, Berkowitz L, Santos JL, Borzone GR, et al. Maternal hypertriglyceridemia: A link between maternal overweight-obesity and macrosomia in gestational diabetes. Obesity (Silver Spring) 2014;22(10):2156–2163. doi: 10.1002/oby.20816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maternal and newborn characteristics, and cord blood adipokines for samples used in microarray analysis. Mean ± SD. N=14.
A. Top-ranking differentially expressed transcripts in HUVEC from infants of Ov-Ob vs. Lean mothers, based on FDR<0.05 and P<0.05. N=14.
B. Top-ranking transcripts whose expression in HUVEC correlates with maternal pre-pregnancy BMI at Pearson R> 0.75 and R<−0.75, P<0.01. These data are shown in heatmap format in Figure 1C. N=14.
C. List of 100 most upregulated probes in microarray analysis of HUVEC from infants of overweight-obese vs. lean mothers (N=14), ranked according to log-2 fold-change (ov-ob/lean).
D. List of 100 most downregulated probes in microarray analysis of HUVEC from infants of overweight-obese vs. lean mothers (N=14), ranked according to log-2 fold-change (ov-ob/lean).
A. Several pathways related to mitochondrial structure, mitochondrial metabolism, and fatty acid oxidation (shaded) are enriched among transcripts correlating negatively with maternal BMI. Table includes all gene sets with FDR q-value <0.05. N=14.
B. Several pathways related to ion channels and neurotransmitter activity are enriched among transcripts correlating positively with maternal BMI. Table includes all gene sets with FDR q-value <0.05. N=14.
Newborn anthropometrics and cord blood hormones and adipokines showed similar trends in male and female offspring. Data are shown as Mean ± SD (n). P-value refers to 2-sided Student’s T-test, except for cells in which there were 2 or fewer samples, for which no T-test was computed.
PCA does not clearly separate HUVEC gene expression patterns from infants of lean vs. ov-ob mothers.
A. HUVEC expression of several genes involved in mitochondrial oxidation negatively correlated with maternal pre-pregnancy BMI, including ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit (ATP5O; probe 1–3 refers to 3 distinct probe sets within this gene on Affymetrix PrimeView array), cytochrome c oxidase subunit VIIc (COX7C), ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 (ATP5C1), and NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa (NDUFB6). N=14.
B. HUVEC expression of several genes involved in lipid metabolism rank among the most strongly correlated with maternal BMI. Expression of lipin 2 (LPIN2) is negatively correlated with maternal BMI, whereas fatty acid desaturase 2 (FADS2) and fatty acid desaturase 3 (FADS3) are positively correlated with maternal BMI. Log-2 normalized Primeview array expression. N=14.
A. Cord blood CE20:3n6 levels were strongly correlated with HUVEC expression of several genes involved in lipid and mitochondrial metabolism, including monoglyceride lipase (MGLL; probe 1–3 refers to 3 distinct probe sets within this gene on Affymetrix PrimeView array), sterol regulatory element binding transcription factor 2 (SREBF2), acyl-CoA synthetase short-chain family member 2 (ACSS2), and ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit (ATP5D). N=14.
B. Associations of cord saturated fatty acids palmitate (16:0), left, and stearate (18:0), right, and HUVEC expression of peroxisome proliferator-activated receptor gamma (PPARG). N=14.
C. Expression of PPARG is higher in HUVEC from infants of ov-ob mothers. Log-2 normalized Primeview array expression. N=14.