Abstract
Microglia, the only lifelong resident immune cells of the central nervous system (CNS), are highly specialized macrophages which have been recognized to play a crucial role in neurodegenerative diseases such as Alzheimer’s, Parkinson’s and Adrenoleukodystrophy (ALD). However, in contrast to other cell types of the human CNS, bona fide microglia have not yet been derived from cultured human pluripotent stem cells. Here we establish a robust and efficient protocol for the rapid production of microglia-like cells from human embryonic stem (ES) and induced pluripotent stem (iPS) cells that uses defined serum-free culture conditions. These in vitro pluripotent stem cell-derived microglia-like cells (termed pMGLs) faithfully recapitulate the expected ontogeny and characteristics of their in vivo counterparts and resemble primary fetal human and mouse microglia. We generated these cells from multiple disease-specific cell lines, and find that pMGLs derived from MeCP2 mutant hES cells are smaller than their isogenic controls. We further describe a culture platform to study integration and live behavior of pMGLs in organotypic 3D-cultures. This modular differentiation system allows the study of microglia in highly defined conditions, as they mature in response to developmentally relevant cues, and provides a framework to study the long-term interactions of microglia residing in a tissue-like environment.
INTRODUCTION
Microglia are the resident macrophages of the central nervous system (CNS) 1. Expanding on previous developmental studies 2,3, recent genetic lineage tracing in the mouse determined that microglia take up residence in the developing neural tube shortly before closure of the blood-brain barrier 4. They emigrate as erythromyeloid precursors or primitive macrophages from the yolk sac, before further hematopoiesis takes place in the periphery. In contrast to other adult tissue monocyte-derived macrophages, microglial identity is a combination of unique mesodermal ontogeny (primitive myelopoiesis) and lifelong residence in an immune-privileged neuro-glial environment. Their phenotype is shaped by the developmental stages of the nervous system as it transitions from neurogenesis and gliogenesis, to synaptogenesis and network maturation 5, to a largely post-mitotic aging tissue in absence of injury.
Microglia have emerged in recent years as primary modulators of both development and pathogenesis of the nervous system, urgently requiring study. However, unlike bone marrow and monocyte-derived macrophages, primary human microglia remain poorly characterized as they differ greatly from mature mouse microglia, are not easily accessible and current methods employed for isolation likely alter their defining characteristics6. In vitro approaches to model and study CNS disorders using human iPS and ES derived cultures are currently lacking this important component. Robust protocols have been developed to induce human ES or iPS cells to differentiate into the different neuroectodermal cell types of the CNS including neurons, astrocytes and oligodendrocytes 7,8. Protocols recapitulating classical hematopoiesis, followed by expansion of monocytes have shown that myeloid cells can be generated from mouse and human ES cells 9. However, generation of microglia-like cells matching the recently identified signatures of acutely isolated microglia 10,11, and following their non-monocytic yolk sac ontogeny 4, has not been reported from pluripotent human cells.
Here, we describe an efficient method to generate microglia-like cells from human ES and iPS cells representing a collection of control and disease subjects. Using recent insights into microglial differentiation 12, we show that microglia-like cells can be efficiently generated and enriched from multiple ES and iPS cell lines. Expression signatures of these pluripotent stem cell-derived microglia resemble those of purified human fetal microglia maintained in the same culture conditions and recapitulate the recently defined consensus signature of microglia compared to other macrophages 13,10,11. They perform the functions of professional phagocytes, are positive for microglial markers such as TMEM119, and react to canonical stimuli. Our results provide a system to study the maturation and steady-state behavior of human microglia in a highly defined cellular environment, and their involvement in disease onset, propagation and resolution.
RESULTS
A basal medium for the maintenance of human and murine fetal microglia
Serum-based media have been the mainstay of microglial isolation techniques over the years but these undefined culture conditions may unpredictably alter their characteristics. Building on recent refinements of the work of Brewer and colleagues 14 we have developed a fully-defined, serum-free Neuro-Glial Differentiation media (referred to as NGD), the component concentrations of which were adjusted to match those of human cerebrospinal fluid (Supplementary Table 1). Compared to the complexity of other published culture conditions, NGD constitutes a single defined serum-free medium allowing culture of multiple neural and glial cell types, as well as other cells of interest. Neural differentiation can be initiated in NGD by addition of a small molecule inhibitor of TGFβ signaling (dorsomorphin) 15. Neural progenitors can be expanded by addition of FGF2 to the NGD base and differentiation into neurons and glia can be subsequently triggered by FGF2 removal.
To selectively culture microglia, we used commercially available modified polystyrene (Primaria) plates that allow adherent maintenance of mouse neonatal microglia and human fetal microglia (mNMG and hFMG, respectively) while maintaining the possibility of enzymatic passaging (Supplementary Fig. 1a). In contrast, neuroglial derivatives from neural progenitors are poorly adherent, and thus selected against on this substrate (not shown). IL-34 and CSF1g, important for microglia differentiation and maintenance in vivo 16,17, were added to the medium. Both hFMGs and mNMGs died in the absence of any CSF1R agonist, with hFMGs adopting a more ramified morphology in the presence of high concentrations of IL-34 (Supplementary Fig 1a, b). Primary cells could be maintained in this medium for more than a month without passaging, suggesting this formulation may be generally useful for primary myeloid cultures.
Induction of primitive microglia from human pluripotent cells
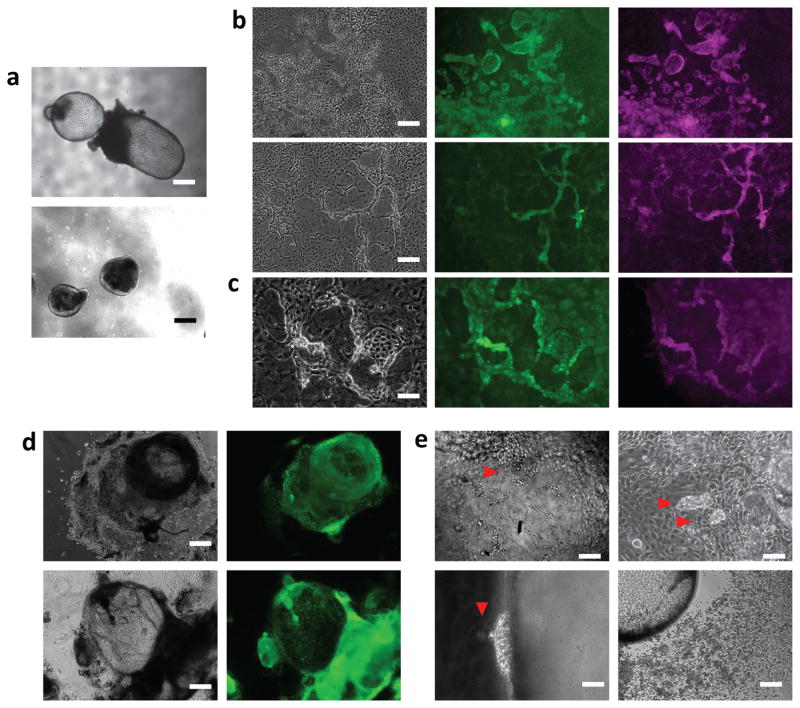
Human ES or iPS cells were grown in hES medium on feeder layers of murine embryonic fibroblasts. Following enzymatic passaging, uniform cell clusters, free of single cells, were used for initial embryoid body seeding. These clusters were re-suspended directly in NGD (without stepwise adaptation) containing 10ng/mL CSF1 and IL-34 (microglial differentiation medium, MGdM) in Corning ultra-low adherence plates. After one week, we observed the formation of two types of structures: dense neuralized spheroids appearing alongside embryoid bodies forming cystic structures, bound by a single cell layer (Fig. 1a). When plated at these early stages on poly-D-lysine, the neuralized EBs flattened into typical rosette-forming neuroepithelium, while the small cystic EBs flattened into cell lawns reminiscent of endothelial cells 18 (Supplementary Fig. 1b). Island whorls could be seen giving rise to three-dimensional clusters of round cells at their borders, organizing themselves in ropes (Fig. 1b). Proliferation and growth of these structures after attachment is CSF1R-ligand dependent, as they arrested and devolved in absence of CSF1 and IL-34 (not shown).
Figure 1. induction of primitive myelogenesis from human pluripotent stem cells.
(a) top panel: example image depicting the typical formation of cystic EBs bound by a single cell layer. lower panel: example of neuralized spheroid EB structures. Scale bars: 200μm. (b) typical appearance of endothelial lawns emerging from plated cystic EBs (on PDL coated plastic). Phase panels display the island formations with raised edges (left), and the progressive merger of such edges into raised ropes. Subsequent panels depict staining for VE-Cadherin (green), and c-kit (magenta). Scale Bars: 80μm. (c) higher magnification of raised structures surrounding islands (phase), staining for CD41 (green), and CD235a (magenta). Scale bar: 25μm. (d) after 2 weeks in suspension culture, cystic EBs can be plated to PDL coated plastic (left, phase contrast. Scale bar: 200μm), and large domains stain positive for the nucleus-localized transcription factor PU.1 (right, green). (e) Delamination of grape-like structures towards the luminal side from YS-EBs (red arrowheads, top left and right, scale bars:40 μm and 25μm, respectively). Putative myeloid cells are seen delaminating outward into the suspension medium (red arrowhead, bottom left, scale bar: 25μm). Homogeneous population of round motile cells seen delaminating and spreading away from the source YS-EB (Bottom right, scale bar: 80μm).
The cells at the island borders and in the three-dimensional ropes were positive for VE-Cadherin, c-kit CD41 and CD235a (Fig. 1b,c), which have been identified in mouse as markers of early yolk sac myelogenesis before definitive hematopoiesis or the establishment of embryonic circulation (<E8.5) 19. This immuno-histochemical panel is consistent with these cells representing the in vitro counterparts of early haemogenic endothelia 20. Based on this pattern of marker expression in the cystic EBs, we refer to them hereafter as Yolk Sac-EBs (YS-EBs). Large domains of the YS-EBs were positive for the master myeloid transcription factor PU.1 (Fig. 1d), which is necessary for microglial differentiation and maintenance 21,22 and can induce macrophages from fibroblasts 23. Under phase-contrast microscopy, clusters of cells were seen to delaminate towards the lumen of the cysts (Fig. 1e) as well as directly into the outer medium or on the plastic culture surface.
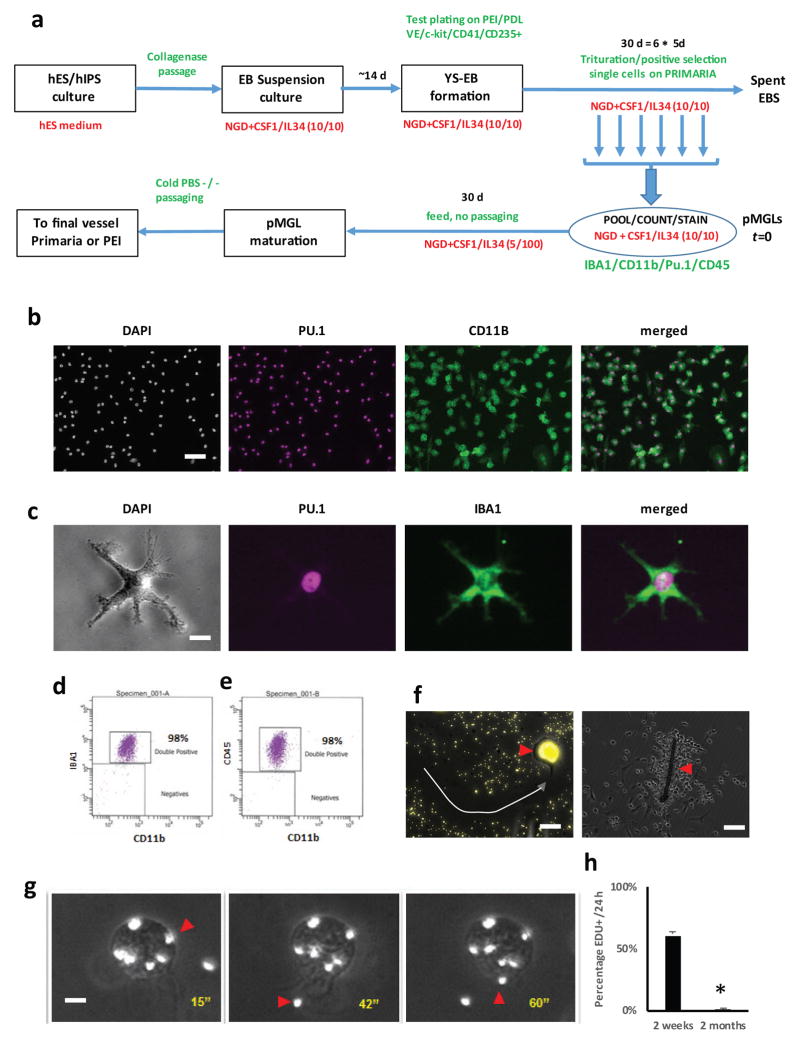
Fig. 2a summarizes the protocol and the timeline used to generate microglia-like cells from human ES and iPS cells. When harvested and plated separately on Primaria, semi-adherent cells appear vacuolated, round and motile, and stain positive for PU.1, as well as CD11b and IBA1 (AIF1) ((Fig. 2b,c, Supplementary Movie 1). These markers have been extensively used to label or define microglia in various systems 24. These cells can be further sorted by FACS and are homogeneously positive for IBA1, CD45 and CD11b (Fig 2d, e), with the majority of cells physically selected by the growth conditions positive for all markers. We went on to characterize these immature cells and their progeny, which we refer to as pluripotent stem cell-derived microglia-like cells (pMGLs). The immature pMGLs initially delaminating from YS-EBs are round yet already highly phagocytic, as observed by the uptake of fluorescent latex beads and the appearance of filopodia and membrane ruffles, allowing the capture of nearby corpuscles such as inert plastic beads (Fig. 2f, g and Supplementary Movies 1, 2). The cells are highly motile and will readily tax to and encapsulate foreign bodies such as fibers present in the well (Fig. 2f).
Figure 2. characterization of phagocytes delaminating from cystic YS-EBs.
(a) differentiation protocol schematic showing the suspension culture (top row), and the selective adherent conditions (bottom row). (b) low magnification view of delaminated lawn after plating stained for nuclear DAPI (grey scale), nuclear PU.1 (magenta) and membrane CD11b/ITGAM (green pseudocolor). Merged PU.1 and CD11b channels are depicted in the right panel. Scale bar: 25μm. (c) high magnification view of ramified cell in resting culture viewed under phase contrast (grey scale) and stained for nuclear PU.1 (magenta) and cytoplasmic IBA1/AIF1. Merged PU.1 and IBA1 channels are depicted in the right panel. Scale bar: 5μm (d, e) FACS scatter plots of harvested pMGLs for CD11b and IBA1 (left) or CD45 (right). (f) left panel, example image depicting the migratory path (white dashed arrow) of a single pMGL on a fluorescent bead lawn (yellow), as well as intracellular accumulation of phagocytosed fluorescent beads (arrow head) Scale bar: 10μm. Right panel, example image depicting a cotton fiber opsonized by pMGLs (scale bar: 25μm). (g) extracted frames from supplementary movie 2 depicting fluorescent beads (red arrowheads) taken up by a single pMGL Scale bar: 3μm. h: Quantification of pMGL EdU incorporation, measured at 2 weeks and 2 months (mean ± s.e.m. of 2 biological replicates, t-test, P<0.05).
Initially, EdU incorporation was seen in 60% of nuclei from early pMGLs (2 weeks from onset of production). After 2 months in culture, EdU incorporation showed that less than 5% of pMGLs kept in MGdM were mitotic within a 24-hour period (Fig. 2h). Importantly, both primary fetal microglia and pMGLs could be maintained for several months in culture despite ultimately limited renewal potential, demonstrating their longevity.
To assess the reproducibility of the differentiation protocol, we induced microglia from 20 different ES and iPS cell lines representing a panel of healthy and disease genotypes (Supplementary Table 2), and successfully generated pMGLs from all lines. Supplementary table 2 reports quantitative output at an early cutoff of 4 weeks, matching the cutoff for NPCs. The cumulative yields are significantly higher. For example, after 8 weeks of differentiation, hES-wibr1, iPS-wt1, iPS-wt4, iPS-ALD4 and iPS-ALD1 respectively generated 1×106, 1×106, 3×106, 2×106, and 8×106 microglia-like cells, from 2×106 pluripotent stem cells at the onset. We conclude that the differentiation protocol is efficient and robust, though the genetic background of the pluripotent donor affects the kinetics and yield of the microglia-like cells.
Phenotypic and functional characterization of pMGLs
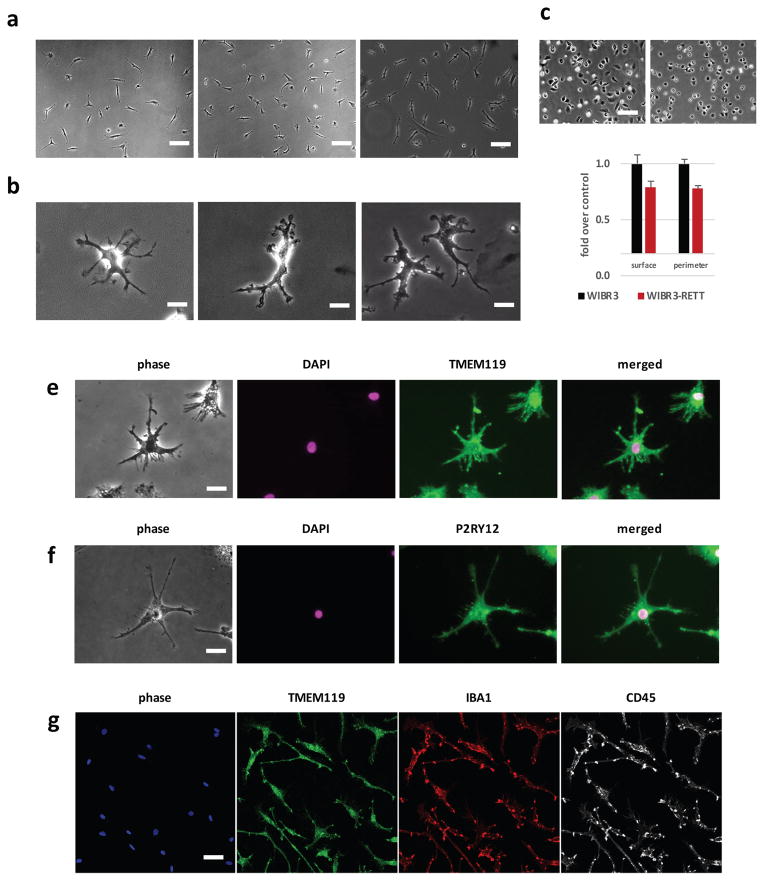
In culture, monocyte-derived macrophages adopt either a “fried-egg” morphology, or an axial bipolar spread reflecting the macrophage polarization and activation status 25. In contrast, a typical characteristic of microglia is to become highly ramified in vivo. To assess the phenotypic characteristics of the cells, we cultured pMGLs in a maintenance medium (MGM) containing high concentrations of IL-34 (100ng/mL) and low concentration of CSF1 (5ng/mL), in NGD base. As shown in Fig. 3a, b and Supplementary Movie 3 and 4, primary human FMGs and pMGLs derived from hES or hiPS cells adopted an identical morphology, characterized by multiple thin first-order branching terminated by actively motile membrane ruffles, around an immobile soma.
Figure 3. pMGLs adopt ramified morphologies over time and express specific markers of microglia.
(a) example phase contrast images of depicting morphologies of human fetal microglia (hFMG; left) and pMGLs derived from human ES cells (hES pMGLs, middle) or induced pluripotent stem cells (iPS pMGLs, right). Scale bars: 25μm. (b) example high magnification images depicting morphologies of mouse primary neonatal microglia (mNMG, left), hFMG (middle) and hiPS pMGLs (right). scale bars: 10μm. (c) Example of phenotypic output in freshly plated MECP2 mutant (right panel) and isogenic wild-type cells (left panel, scale bar: 25μm). Average cell spread (surface) and perimeter are significantly lower in MECP2 mutant microglia (mean ± s.e.m. for 2 biological replicates, TTEST, p<0.05). (e–f) example images of ramified pMGLs (phase) stained for DAPI (magenta) and either TMEM119 (e, green) or P2RY12 (f, green). Scale bars: 10μm (g) Example confocal image representing an optical slice of wider field-of-view, stained for IBA1 (red), TMEM119 (green) and CD45 (white). Scale bar: 20μm.
Microglia were missing from our study of isogenic MECP2 wild type and mutant human ES-derived neural cells 26, yet their involvement in Rett syndrome has become in recent year the focus of great interest 27, following controversial findings that transplanted wild-type phagocytes derived from bone marrow could rescue features of Rett syndrome 28. pMGLs have the potential to clarify some of the cell autonomous and non-cell autonomous aspects of Rett microglial biology. We derived pMGLs from isogenic male wild-type and MECP2 mutant cells, and observed that mutant cells were significantly smaller than the wild-type cells (Fig. 3c).
Mature pMGLs stained positive for markers of murine and human microglia 10,11,29,30, with both TMEM119 and P2RY12 displaying a broad membrane and punctate vesicular expression pattern, consistently with their roles as part of the microglial sensome (Fig. 3e, f). The majority of pMGLs at this stage were positive for TMEM119, IBA1 and CD45 (Fig. 3g).
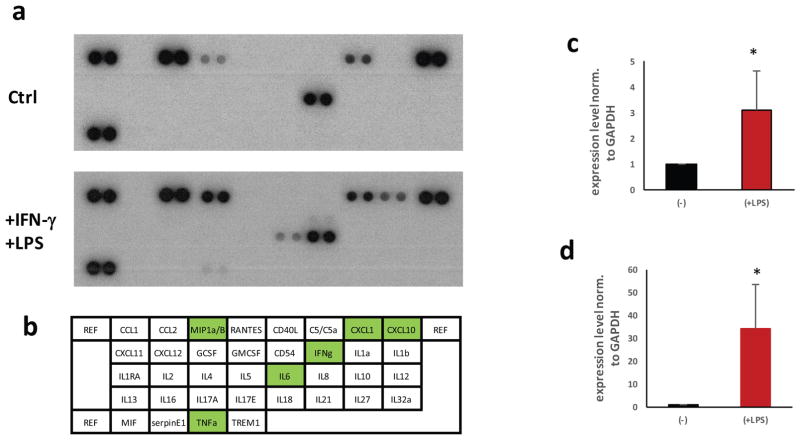
We assessed cytokine and chemokine profiles released in the supernatant of pMGL cultures. Unstimulated pMGLs secreted detectable levels of various cytokines and chemokines, including IL-8, CXCL1 or CCL2 (Fig. 4a, b). Furthermore, the addition of IFN-γ and endotoxin (LPS) triggered the release of chemokine and cytokines above baseline (Fig. 4a–d). In particular, CXCL10, MIP1a, IL-6 and TNFα were increased more than 2-fold within the dynamic range of this assay. We verified that this effect was detectable at the transcriptional level, with an increase in mRNA of IL-6 and TNFα (Fig. 4c, d).
Figure 4. pMGLs cytokine profiles in response to endotoxin challenge.
(a) Baseline cytokine profiler assay from 105 pMGLs conditioning 2mL of NGD in 24 hours. Top panel: 10′ exposure, showing baseline secretion of detectable CCL2, MIP1α/β, CXCL1 and IL8. Lower Panel: profile after stimulation for 24hours with 100ng/mL LPS and 20ng/mL IFN-γ. IL6, TNFα, MIP1α/β and CXCL10 display a dramatic increase. (b) map, to scale, of the spots blotted in panel a. Green highlights cytokines showing a significant upregulation. (c, d) Quantitative qPCR of TNFα (c) and IL6 (d) transcription in pMGLs at baseline (−) and after LPS stimulation (+). Data are presented as mean ± s.e.m. from 2 biological replicates, t-test significance is reported.
These results indicate that monocultures of pMGLs can be a substrate to study the functional and morphological changes in microglia, derived from isogenic parental cells, or to screen for modulators of inflammation.
pMGLs resemble primary human microglia, and differ from peripheral macrophages
We used the molecular signature specific of microglia, which is distinct from other macrophages 11,29,31, to compare the transcriptome of pMGLs with bona fide primary human microglia. Isolated human fetal microglia and pMGLs were grown in the same maintenance medium for a week (MGM). This comparison is relevant since pMGLs are directly isolated from differentiating pluripotent stem cell cultures and grown in serum-free neuroglial-compatible conditions. We found that pMGLs display a signature strikingly resembling that of fetal human microglia kept in the same conditions. Supplementary Table 3 shows that the top GO categories coincide between both cell types. pMGLs differ in the enrichment of gene categories related to the establishment and modulation of the extracellular matrix (ECM, Suppl. Tables 4 and 5). The differentially expressed genes between primary cells and in vitro derived pMGLs did not include any of the canonical myeloid ontology terms, supporting their use as surrogate for human microglia. Several individual genes were highly expressed in primary human fetal microglia and pMGLs, consistent with their identity (Suppl. Fig. 2a). Among those genes were CD11b (ITGAM), ITGB2, CSF1R, CD45, IBA1, ADORA3 or LGMN. In addition, pMGLs highly expressed genes relevant to nervous system disorders such as APOE, CD33 and TREM2 (Supplementary Fig. 2b). Likewise, HEXB, a causal gene for the lysosomal storage Sandhoff disease, was highly expressed in pMGLs, supporting the interest in microglial catabolism of GM2 ganglioside 32. Neural progenitors derived from the same pluripotent stem cells and further differentiated into complex cultures of neurons and glia (Diff. NPCs, see Fig. 6), express these genes to much lower levels (Fig. 5 and Supplementary Fig. 2), highlighting the lack of overlap between these mesodermal and neuro-ectodermal lineages, despite their side-by-side residence in the brain, in vivo.
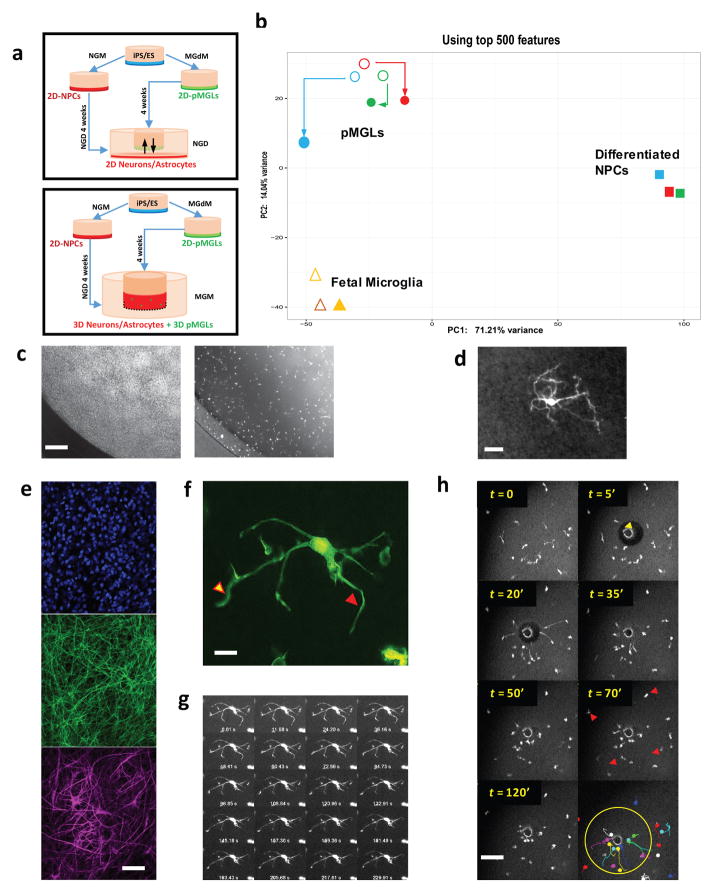
Figure 6. neural co-cultures enhance the microglial signature of pMGLs.
(a) top: schematic representation of transwell culture system exposing pMGLs to conditioned medium from differentiating neuro-glial cultures. Bottom: schematic representation of direct re-aggregation after GFP transduction, as free-floating spheroids or 3D stacks in transwells. (b) Principal component analysis comparing RNAseq profiles from differentiated neural cultures (full squares, NPC1 in red, NPC2 in blue, NPC3 in green) to those of primary fetal microglia (fMG1 and fMG2, open triangles. fMG1+ NCM, closed triangle), and pMGLs (circles, pMGL1 in red, pMGL2 in blue, and pMGL3 in green) in the absence (open circles) or presence (full circles) of neural progenitor conditioned medium (+NCM). N=1 for each sequencing data point. (c) phase contrast (left) and fluorescence (right) images depicting the relative tiling position of GFP-labeled pMGLs in 3d culture on transwell. Scale bar: 200 μm. (d) representative image of a GFP-positive pMGL (grey scale) after 4 weeks in 3D neural stacks (live observation). Scale bar: 10μm. (e): Optical section through a fixed 3D neuroglial culture (without embedded pMGLs), stained for DAPI (blue, top), neuronal MAP2 (green, middle) and astrocytic GFAP (magenta, bottom). Scale bar: 50μm. (f) Maximum projection of supplementary movie 5 (scale bar: 2μm) pointing to rapidly extending (yellow arrowhead) and retracting (red arrowhead) protrusions in the 3D neuroglial cultures. (g) 1/5 frames of supplementary movies S5 and S6, showing branch movements over 300s. (h) Example montage of time-lapse images depicting GFP-labeled pMGLs (grayscale) response to localized cellular damage in 3D culture. Yellow arrowhead indicates the site of two-photon laser ablation after 5 minutes of acquisition. Red arrowheads point to microglia-like cells further away from injury, not reacting to the damage. Bottom right panel represent the color-coded individual trajectories of pMGLs during acquisition, highlighting radial migration towards the injury. Scale bar = 100μm.
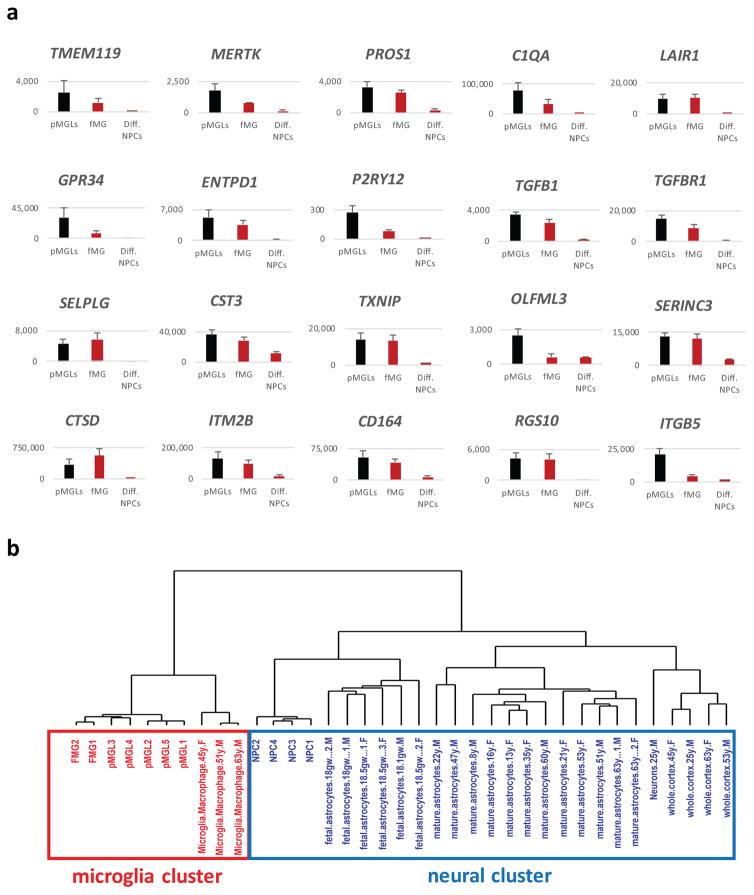
Figure 5. pMGLs recapitulate the consensus signature distinguishing primary microglia from other macrophages.
(a) Quantification of normalized expression (counts from RNAseq data) of the indicated genes for pMGLs (black bars, N=5), primary human fetal microglia (red bars, hFMG, N=2), and differentiated human neural progenitors (red bars, Diff. NPCs, N=4). Data are presented as mean ± s.e.m. (b) Dendrogram depicting the results of unbiased hierarchical clustering of fMGs (N=2), pMGLs (N=5) and NPCs (N=4) derived in this study, compared to a published dataset (GSE73721) 31 for adult primary microglia (N=3), fetal astrocytes (N=6), mature astrocytes (N=12), neurons (N=1) and whole cortex (N=4), using genes in Suppl. Table 8.
To characterize pMGLs we used a consensus subset of genes that is highly expressed in microglia but poorly expressed in other macrophages, combining the most relevant genes of the microglial sensome 11, the unique TGFβ-dependent signature of microglia 29, and the microglial cassette found to be upregulated in TMEM119+ microglia during postnatal mouse development 10 (see Suppl. Table 8 for microglial genes compounded from Bennett et al. 2016). These include genes coding for the transmembrane protein TMEM119, the proto-oncogene MERTK, the phospholipid binding protein PROS1, the putative synaptic-tagging complement factor C1QA, the inhibitory receptor LAIR1, the GPCR GPR34, the ectonucleotidase ENTPD1, the purinergic sensors P2Y12/13, and the main TGFβ signaling partners TGFβ1 and TGFβR1. The expression of these markers appears to be positively correlated with maturation time in vivo, increasing in adult animals as compared to newborns 29. Conversely, their expression seems to be negatively correlated with time in culture, being highest in freshly isolated cells and lowest in maintained primary cells and transformed lines. Most of these genes are in fact expressed at higher levels in pMGLs than in primary human fetal microglia (Fig. 5A, supplementary Fig. 2), supporting their identity as in vitro generated microglia. Notably, these genes are expressed at low levels in cultured macrophage cell lines as well as in previously described mouse ES-derived microglia 33. Conversely, genes such SLPI, SAA1/2, PRG4, CFP, CD5l, CRIP1 are expected to be highly expressed in peripheral macrophages, and low in microglia 11: these genes are in the lower expression quartile of our dataset. Genes such as GPR56 and BIN1 are expressed in microglia and in other CNS cell types, but have not been described in peripheral macrophages. Supplementary Figure 2c shows their expression in pMGLs, fMG, as well as differentiated NPCs. We performed unbiased hierarchical clustering of our data against previously published sequencing for primary and induced macrophages. As expected, all myeloid cells cluster away from parental iPS cells and NPCs. Strikingly, we find that pMGLs and primary fetal microglia cluster together, separately from other macrophages (Suppl. Figure 3b).
In the mouse, several microglia-specific genes appear to be upregulated in brain-resident microglia through early postnatal development 10. For example, SELPLG, CST3, TXNIP, P2RY13, OLFML3 are upregulated (respectively 16, 11, 9, 5 and 5-fold) between embryonic day 17 and postnatal day 60. In our cultures, we find that SELPLG, CST3 and TXNIP are already expressed at high levels in both FMGs and pMGLs, while OLFML3 and P2RY13 are more than 4-fold higher in pMGLs compared to FMGs. We also note that pMGLs display high expression of both PU.1 and IRF8, but very low levels of MYB (Suppl. Fig 3a) consistent with microglia emerging from PU.1 and IRF8-dependent, but MYB-independent cells 34–37.
We further performed unbiased hierarchical clustering of our data against previously published data on primary human brain cells isolated by immuno-panning, including neurons, fetal and mature astrocytes, and adult microglia 31. We find that pMGLs form a cluster with both fetal and mature primary microglia, while iPS-derived neural cells cluster with primary neurons and glia, close to fetal astrocytes (Figure 5b).
The pMGL phenotype is affected by co-cultivation with neurons
We observed that exposure to undifferentiated neural progenitor conditioned medium maintains the rounded phenotype of immature microglia and promotes their proliferation. We investigated whether maturation of pMGLs in the presence of human pluripotent stem cell-derived mature neural cells would further refine their molecular signature (Fig. 6a, top panel). Principal component analysis of the transcriptome revealed that primary fetal microglia and pMGLs cluster tightly together along PC1, and segregate away from differentiated neural progenitors cultured in the same conditions (Fig. 6b). The difference between NPCs and pMGLs along PC1 may reflect the main myeloid vs neural identity, accounting for ~77% of sample variance, while the variation along PC2 (~12% of sample variance) may be refined by tissue residency, ECM and cell/cell interactions. To directly test this notion, we grew pMGLs in conditioned medium from differentiating neural cultures (defined before conditioning, with no other variation in recombinant growth factor concentrations). Fig. 6b shows that, after 2 weeks, the pMGLs partially shifted their signature towards that of primary microglia along the PC2 axis.
To test whether physical embedding and maturation of pMGLs in a 3D organotypic neuroglial environment would accentuate their branching patterns and allow us to observe their surveying behavior, we transduced immature pMGLs with a GFP lentivirus and re-aggregated the GFP-positive pMGLs with pre-differentiated neural cultures, at a stage where neurons have already become post-mitotic and gliogenesis is ongoing (>4weeks). The re-aggregates were either kept in spinning suspension cultures to maximize viability, or as 200μm-thick cultures in transwells, so as to avoid triggering pMGL activation from hypoxia of the surrounding tissue and allow live observation. In this context, an entire spheroid or cellular stack is self-assembling, laying down its own extracellular matrix without need for additional scaffold (Fig. 6a, lower panel). The neural progenitors progressively differentiated into neurons and macro-glia (astrocytes, oligodendrocytes), and would be devoid of any microglia without the exogenous addition of pMGLs. GFP-labeled pMGLs, which had adopted rounded to first-order ramified morphologies when cultured on plastic, integrated into the three-dimensional cultures, tiled the volume (Fig. 6c), and projected highly branched ramifications (Fig. 6d). The 3D culture matrix is a cortex-like mesh of MAP2 positive neurites and GFAP positive astrocytic branches (Fig. 6e). Live Imaging of the GFP positive pMGL branches in situ showed rapid extension and retraction of filopodial arbor termini (Fig. 6f, g and Supplementary Movies 6, 7). These results suggest that pMGLs can integrate into organotypic neural cultures, and mature into what is currently defined as resting yet dynamically motile microglia. We note that this magnitude of extension and retraction is not observed in 2D cultures on Primaria or glass coverslips, where non-amoeboid pMGLs only display terminal ruffle movement, on a much smaller scale.
Embedded pMGLs rapidly respond to cellular damage
One of the characteristics of microglia in vivo is their ability to survey the parenchyma from their static tiling positions, yet revert to an amoeboid and actively migrating state in response to injury. Migration towards a site of cellular damage is driven by purinergic receptors such as P2RY12/13, in response to ATP/ADP release from dying cells. We took advantage of our 3D culture system to model such localized damage and observe microglial behavior in real-time. Figure 6h and Supplementary Movies 8, 9 display the time-lapse reaction of embedded GFP+ microglia to a focal laser injury (yellow arrowhead at 5′). Microglia reacted within minutes by extending a single long process towards the injury center, making contact with the damaged zone (t =20′). They then rapidly migrated their cell body to surround the damaged area. In contrast, microglia away from the injury site stayed in place.
DISCUSSION
In this work we have established a robust protocol that allows the derivation of microglia-like cells from human pluripotent stem cells. These cells are derived in a defined medium that mimics the serum-free environment of the CNS interstitial milieu, supporting electrophysiological maturation of neurons, proliferation and maturation of astrocytes, and differentiation and maintenance of oligodendrocytes. We demonstrated that pMGLs are highly phagocytic and, based on their phenotypic and gene expression profiles, resemble primary fetal and adult human microglia. In particular, they bypass a colony-forming monocytic stage, are initially amoeboid and capable of proliferation, extensive migration and phagocytosis. They eventually settle into a highly ramified morphology, enhanced by co-culture with differentiated neurons and glia. These cells express many of the markers expected to be characteristic of microglial expression phenotypes, such as P2RY12/13, HEXB, GPR34 and TMEM119. Transcriptome sequence analysis shows that they resemble human primary fetal and adult microglia, unlike other macrophages.
Primary microglia and cell lines have been maintained for short periods on canonical polymer linkers such as poly-D-Lysine (PDL) + Laminin, but this is also a substrate for neural progenitors, neurons and glia. To exclude the growth of neuro-ectodermal derivatives we used Primaria plastic to positively select pMGLs. It is notable that coatings such as PDL or matrigel can represent another layer of variability between cultures, and their exclusion contributes to the reproducibility of this protocol.
Macrophage colony stimulating factor (CSF1) is a primary actor of proliferation, differentiation and function of myeloid cells, but microglia develop normally in CSF1 mutant mice, though their numbers are drastically decreased in CSF1R mutant animals 16. Therefore, it was suggested that an additional cognate ligand of CSF1R is crucial for microglia development. Addition of the recently identified alternative ligand for CSF1R, Interleukin-34 (IL-34) to the base medium improved the morphological phenotype of microglia-like cells. Immature pMGLs delaminate and adopt directly a macrophage identity, with no colony-forming intermediate. This is consistent with our current understanding of microglial development, which supports the notion that microglia derive from non-monocytic primitive myeloid cells acting as precursors, unlike adult bone marrow-derived macrophages, but similarly to some other tissue-resident macrophages 37. As pMGLs populate the substrate, they clearly isolate from their nearest neighbors, an in vitro behavior which may correspond to the ability to tile the CNS by their in vivo counterparts. Unlike other tissue-resident macrophages, microglia derive from PU.1+/IRF8+/MYB− precursors 34,35. Expression of the same markers in pMGLs supports this ontogeny. pMGLs kept in mono-culture express high levels of SELPLG, TXNIP, CST3 and OLFML3, suggesting they adopt a relatively more mature profile than primary fetal cells.
Embryonic microglia, when first colonizing the nervous system, do not display the extensively ramified morphology observed in the adult CNS, instead adopting an amoeboid morphology as they migrate and spread through the parenchyma. Likewise, pMGLs progressively adopt a ramified morphology when cultured in isolation, as they lose their proliferation potential. In isolation, their morphology and function can be analyzed for cell-autonomous phenotypes. We showed that MECP2 mutant microglia are smaller than their isogenic counterpart: this may contribute to functional differences in Rett microglia worthy of further investigation.
pMGLs further adopt canonical high-order branching patterns once embedded in a differentiated neuro-glial environment. Their surveying behavior is reminiscent of that observed in vivo in the landmark two-photon imaging studies of the dynamic process motility of microglia in intact mouse brains 38. Furthermore, they respond to localized injury by migrating and encapsulating the damaged area. pMGLs are capable of major and rapid morphological changes, as well as secretion of highly bioactive cytokines and chemokines. It will be particularly interesting to study their secretomes in the context of disease models to which we applied this paradigm.
It is well established that microglia perform key functions in the development of the brain such as in circuit refinement, neurogenesis, and neuronal growth 39. Microglia mediated inflammation can have deleterious consequences for the brain and much evidence indicates that microglial activation contributes to neurodegenerative disease such as AD, PD and ALD 40,41. In ALD, phagocytes derived from autologous bone marrow transplants likely elicit their beneficial effects by replacing mutant microglia, as they stall terminal neuro-inflammation and demyelination 42. In addition, other brain dysfunctions including depression 43, schizophrenia and autism have been suggested to involve microglial dysfunction. The availability of a robust protocol to generate and maintain microglia from patients suffering from these conditions allows studying the interaction of neurons and microglia and will facilitate the investigation of these diseases in defined culture conditions.
MATERIALS AND METHODS
Study design
no statistical tests were used to pre-determine sample size. Experimenters were not blinded to the genotypes or culture conditions, except for transcriptomic where line genotype was not explicit for analysis. 2-tailed Student’s t-tests were performed where appropriate, p<0.05 was considered significant.
Statement of compliance with IRBs
all experiments involving cells from human donors and animals were performed in compliance with established IRB protocols at the Whitehead institute.
Human ES and iPS cell cultures
Human ES cell line WIBR1/2/3 and hES-Rett were previously described 26,44 and cultured in 5% O2 on mitomycin C-inactivated mouse embryonic fibroblasts (MEFs) in hESC medium, containing DMEM/F12 (Thermo), 15% fetal bovine serum (Hyclone), 5% knockout serum replacement (Thermo), 1% non-essential amino acids (Invitrogen), 1mM glutamine (Thermo), 0.1mM β-mercaptoethanol (Sigma) and 4ng/ml bFGF (Thermo). Cultures were passaged manually or with 1mg/ml collagenase type IV (Thermo) every 5–7 days. iPS-wt1/2/3, iPS-fAD2, iPS-AMN1/2/3 and iPS-ALD1/2/3/4 were reprogrammed from patient fibroblasts using the constitutive excisable STEMCCA lentivirus (OSKM) and maintained in the same conditions as WIBR1/2/3. iPS-wt4/5, iPS-fAD1/3 were reprogrammed using non-integrative Sendai virus (OSKML), and maintained on inactivated mouse embryonic fibroblasts in serum-free hES medium containing DMEM/F12 (Thermo), 20% knockout serum replacement (Thermo), 1% non-essential amino acids (Thermo), 1mM Glutamax (Thermo), 0.1mM β-mercaptoethanol (Sigma) and 12ng/ml bFGF (Thermo). hES-WIBR3 is a female line, all other lines are male. All parental lines were maintained for over 50 passages, verified for stable expression of Oct3/4, Nanog and TRA1-60, and routinely tested for mycoplasma negativity.
NGD formulation justification
We adjusted the component concentrations to match those of human cerebrospinal fluid, providing metabolic substrates necessary for individual cell types growing in the absence of others. Ionic concentrations were chosen to match human cerebrospinal fluid osmolality 45, thus providing extracellular sodium concentrations necessary for proper electrophysiological function. We followed a rationale matched by recent efforts to improve electrophysiological activity in iPS derived neurons46. Iron salts were excluded, instead using iron-loaded transferrin in Neurobasal, in order to avoid dangerous redox cycling through Fenton chemistry. Glutamate was omitted to avoid the excitotoxicity of this amino-acid in dissociated cultures 47. We included additional pyruvate48, and lactic acid 49 as energy-providing and neuroprotective substrates in addition to the more canonical oxidative substrates (glucose, galactose and glutamine/Glutamax). Biotin and lipid-loaded albumin (Albumax) were added to support de novo synthesis lipid bilayers (e.g. axons, myelin sheaths).
Differentiation to neural progenitors and maintenance
Differentiation of human ES and iPS cells to neural progenitors in 2-D adherent culture was performed as follows: 2 million human ES or iPS were passaged onto matrigel-coated dishes using PBS w/o Ca2+/Mg2+, filtered through a 40μm mesh to remove mEFS, and cultured directly in NGD medium containing dorsomorphin (2.5mM, Stemgent), bFGF (10ng/mL, Thermo) and Insulin (additional 10ng/mL) for 3 days until super-confluent. bFGF and Insulin were subsequently removed, and NGD + dorsomorphin was replaced every day for 10 days. Cells were subsequently passaged 1:1 with PBS−/− when rosette lawns were observed throughout the culture. Rho-associated protein kinase (ROCK) inhibitor Y27632 (10mM, Stemgent) was added to the medium during each of the first 3 passages. Initial passaging at no more than 1:2 ratio, followed by 1:3 to 1:6 every 5 days. Neural progenitors were expanded and maintained in NGD medium with 10ng/ml bFGF, and additional 10ng/mL Insulin (NGM medium).
Differentiation towards neurons and glia
NPCs were differentiated into neurons and glia by removing FGF and culturing in NGD base levels of insulin (5ug/mL) without retinoic acid addition (NGD contains no additional retinoid). To initiate differentiation, NPCs were plated in a 35mm dish on 1% matrigel at 5*105/cm2 and fed 5mL of NGD every 2–3 days. At 4 weeks, neurons appeared in the culture and a final dissociation was performed. The culture was dissociated in the presence of 0.05% DNase I by incubation with Accutase (Stem Cell Technologies) for 30′ at 37degC with gentle agitation (bacterial rotator), re-suspended in chilled HBSS/0.1% BSA and filtered through a 40μM mesh before being centrifuged through a cushion of 4% BSA to remove cellular debris. For 2D cultures and for medium conditioning, cells were re-plated at 5*105/cm2 on 0.1% PEI-coated plastic or glass. For re-aggregation with pMGLs, 3D cultures were initiated by plating 1.5×106cells in a 0.3cm2 PET transwell with 0.4mm pores coated with 0.1% PEI. Cells were mixed with transduced pMGLs in a 1:10 ratio. Spheroid formation was initiated by re-aggregating 3*104 NPCs per well in 96-well ultra-low attachment plate (corning), mixed with pMGLs at 1:10 ratio.
Differentiation towards pMGLs
Colonies were treated with collagenase IV (1.5mg/mL) and mildly triturated to form a suspension of uniform clumps, transferred directly to 5mL MGdM (NGD + 10ng/mL IL-34 + 10ng/mL CSF1), in ultra-low attachment 6-well plates (corning). 6 confluent wells (~3×106 cells) were pooled into one suspension well. Embryoid bodies were monitored for appearance of two main identifiable types. The first group was composed of compact phase-bright neuralized spheroids, the second group were large, expanding cystic bodies, YS-EBs. Every 5 days, EBs were gently triturated to shear off loose cells of interest, settled, and the supernatant placed in a single well of a Primaria 6-well plate. Unattached cells and small EBs were washed with fresh MGdM. Attached cells were monitored for morphological characteristics of microglia/microglial precursors (compact nucleus, vacuoles, membrane ruffles, motility), and wells from 6 consecutive productions (one month) were pooled to constitute one synchronized population. Further maintenance was performed in MGM (NGD + 100ng/mL IL-34 + 5ng/mL CSF1). Cells are sensitive to passaging, but can be lifted with Accutase, or preferentially with ice-cold PBS with 5ng/mL CSF1. Assays and imaging were performed 4–5 days after passaging or feeding. pMGL staining displays cells derived from iPS-wt5.
Lentivirus production and transduction
FU-GFP-IRES-PURO-W lentivirus constructs were used for pMGL transduction. VSVG-coated lentiviruses were generated in HEK293 cells as previously described 50. Sub-confluent HEK293 cells were transfected using X-tremeGENE 9 (Roche), with a mixture of lentiviral construct and second-generation packaging plasmids. Culture medium was changed 12 hours after transfection and collected 96 hours later. Virus-containing medium was filtered through 0.45μm filter and concentrated by ultracentrifugation (23krpm, 90′, 4°C). pMGLs cultured in MGdM were transduced at an MOI of 10 (titer assessed by p24 ELISA). Medium was replaced after 12 hours, and cells were left to recover. Expression was evaluated by fluorescence microscopy without additional selection.
EdU click-it assay
EdU (10μM, Life Technologies) was added to MGdM medium for 24 hours, after which cells or tissues were fixed using ice-cold methanol. EdU click-it assay was performed on fixed cells per the manufacturer’s instruction (Life Technologies), followed by fluorescent immuno-staining and imaging. Counting was performed by automated particle counting with Fiji. Data is represented as a ratio of EDU positive to DAPI positive nuclei. 6 random fields were chosen at 100x magnification, and averaged between two biological replicates to generate s.e.m. and t-test values.
Cell size comparison
At least 200 cells were imaged in phase contrast in each of 3 replicate wells of 2 isogenic lines of pMGLs (Rett and control). FiJi Gaussian blur was applied to the images, followed by background subtraction and thresholding. The particle counting function was used to measure average pixel counts (surface) of cell masks, and average perimeters, in each replicate. Data is given as mean ± s.e.m., with significance below 0.05 for the t-test.
Imaging
Cells and tissues were fixed with ice-cold methanol or 4% paraformaldehyde in PBS. After PFA fixation, permeabilization was effected with PBS containing 0.3% triton. Fixed and permeabilized cells were blocked with 3% normal donkey serum. Primary antibodies were against TREM2 (Abcam, 1:500), CD11b (Abcam,1:500), TMEM119 (Sigma/Atlas, 1:50), P2RY12 (Sigma/Atlas, 1:50), PU.1 (cell signaling, 1:500) and visualized by ad hoc secondary antibodies conjugated with Alexa 488, 568, 594, 647 (Life Technologies, 1:1000), followed by counter-staining with DAPI. Phase contrast and Fluorescent images of immuno-staining, as well as time lapse movies, were captured on a wide-field Nikon Ti2000 mounted with a SPOT RT monochrome camera. For 3D embedded microglia, observation was performed in the culture transwell, placed on a 1.5 coverslip Matek glass-bottom 35mm dish. Optical sectioning through 3D neural cultures was performed on a Zeiss LSM 700 at 20X magnification. Live imaging and wound assay in 3D culture of cells derived from iPS-wt5 was performed on Zeiss LSM710, photo-ablation was performed with 2-photon 780nm laser at 20% power, acquiring a 25um optical slice at 10X.
Flow Cytometry
Single cell suspensions of pMGLs were blocked with FcR blocking reagent, 1% BSA, to avoid non-specific antibody binding, in NGD medium without phenol red. 10^5 cells were labeled in 100μL with pre-conjugated rat anti-CD11B-FITC (1:100, StemCells Tech.), mouse anti-CD45-AF647 (1:100, Biolegend), and unconjugated rabbit anti-IBA1 (1:100, Abcam) followed by AF647 conjugated secondary. Sorting was performed on a customized FACS Aria (BD).
Phagocytosis assay
For qualitative assessment of phagocytosis, 1μm polystyrene red-orange fluospheres (Thermo) were left to settle at a density of 107/cm2 on PEI coated plastic. pMGLs were added the next day at a density of 1000/cm2. Cells were observed after 6 hours, displaying their migration on the lawn and the cytoplasmic bead uptake. Alternatively, beads were added directly to a pMGL culture medium at a concentration of 108 beads/mL. Phagocytosis was observed in real time, highlighting the rapid active uptake.
Primary mouse microglia preparation
All culture conditions were initially tested with mouse primary microglia. C57Bl/6 neonates were cold/CO2 anesthetized in a dry ice chamber following approved protocols and whole brains were harvested. After meningeal removal, cortical cups were isolated, minced on ice, and crudely triturated with a wide-bore fire-polished pipette in a solution of Accutase (Life). The preparation was incubated at 37°C with rotation for 10′, followed by further trituration with a narrowed pipette. The suspension was further incubated for 10′ at 37°C with DNase I 0.1%. The final trituration was left to settle for 1 minute, and the supernatant was filtered through a 70μm mesh, and spun through a 4% BSA cushion to remove cell debris. This preparation was plated directly on matrigel coated plate, and grown in complete Neurobasal with 5% serum. After a week, loosely adherent microglia were harvested and further used on different surfaces. Alternatively, MACS sorting (Miltenyi) for CD11b was applied to the cortical suspension, following the manufacturer’s instructions. Flow-through was discarded while retained cells were eluted directly onto test surfaces and used accordingly.
Primary fetal human microglia
Cells were obtained from first trimester aborted fetuses from ScienCell (#1900), and cultured on Primaria in MGdM and MGM. ScienCell strictly adheres to the guidelines for human tissue collection and distribution according to established protocols. Human tissue used for the isolation of primary cells is derived from donors who have signed informed consent by the donor themselves or an authorized agent acting on the donor’s behalf.
RNA extraction, reverse transcription and quantitative PCR
Cells and tissues were homogenized and total RNA extracted using the RNeasy Micro kit (Qiagen) following the manufacturer’s instructions. Total RNA concentrations were measured using NanoDrop ND-1000 spectrophotometer. For RNAseq, RNA was directly analyzed and quality checked before sequencing. RNA was reverse transcribed into cDNA using Superscript III reverse transcriptase (Invitrogen) with random hexamer primers. Transcript abundance was determined by quantitative PCR using SYBR Green PCR mix (Applied Biosystems), with primer pairs against IL6, TNFα and GAPDH.
RNAseq and Analysis
Samples were prepared using the SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Clontech), according to manufacturer’s protocols and using 10 ng of input. Final libraries were evaluated for size using a Fragment Analyzer (Advanced Analytical Technologies) and quantified using both Qubit (Thermo Fisher) and qPCR (Roche LightCycler 480, KAPA Illumina Library Quantification Kit) before sequencing. RNA-seq 75 bp paired-end reads from Illumina were checked with FastQC and fastq_screen. Reads were mapped to human genome hg19 using TopHat v 2.0.13 with Ensembl annotation (GRCh37.75) in gtf format. FeatureCounts was used to obtain gene counts with default option. The gene counts were normalized with DESeq, and PCA plot was created with plotPCA function. Bar charts represent the average normalized raw reads for different PSC lines and primary samples. Gene ontology differently selected top features for FMG, pMGLs and differentiated NPCs, ran on the online Gene Ontology Consortium tool (geneontology.org). Hierarchical clustering was performed on quantile normalized FPKMs from the different GEO datasets. For HMDM and iPSDM, we used GSE55536. For the comparison to adult human microglia, and other brain cells, we used GSE73721. Biological replicates of pMGLs, FMGs and differentiated NPCs were used for analyses. The values for pMGL3 were the average of two technical replicates.
Gene ontology
For supplementary table 3, average expression values for fMGs and pMGLs were ranked, the gene list associated with the top 300 values in each set was submitted to the panther algorithm at geneontology.org. For Suppl. Table 4–7, the ratios of expression were ranked, and the gene lists corresponding to the top 200 values were submitted.
Cytokine profiler
2mL of NGD was added to 100k pMGLs in a 35mm well and allowed to condition for 24 hours. For IFN-γ/LPS stimulation, LPS was added at 100ng/mL along with 20ng/mL IFN-γ at the onset. 400uL of this supernatant were added to a prepared cytokine antibody panel membrane, per the manufacturer’s instructions (RnD Systems, kits ARY005 and replacement ARY005B). After incubation, the membranes were revealed with ECL reagent.
Supplementary Material
Acknowledgments
The authors thank Laure Freland-Gobaille, Dongdong Fu, Raaji Alagappan, Tenzin Lungjangwa, and Sarah Elmsaouri, for technical support, and all members of the Jaenisch lab for helpful discussions. We thank Prathapan Thiru and George Bell at WIBR’s BARC for help and advice with sequencing data analysis. Confocal microscopy and wound assays were performed at the Keck Facility, with the precious help of Wendy Salmon. We thank P. Wisniewski and C. Zollo for help with cell sorting. We thank T. Volkert, J. Love and S. Gupta at the WIGTC for help with library preparation and sequencing. J.M. received funding from the European Leukodystrophy Association, and a NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation. Y.L. received funding from a Simons Postdoctoral Fellowship, an International Rett Syndrome Foundation (IRSF) Postdoctoral Fellowship, and a NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation. S.C. was the recipient of a Simons undergraduate scholarship and an Amgen Scholarship. G.B. was supported by H.H.M.I. Work for this project was supported by a grant from the Simons Foundation (SFARI 204106 R.J.), NIH grants HD 045022, R37-CA084198, the ELA foundation, the Emerald foundation, and Biogen (to R.J.) and NIH 1RF1 AG042978 to L-H. T.
Footnotes
Accession
Raw data from RNA-seq analyses have been submitted to the NCBI Gene Expression Omnibus under accession number GSE85839.
Author Contributions
J. Muffat, Y. Li and R. Jaenisch conceived the project, designed and supervised the experiments, interpreted results and wrote the paper with input from R. Ransohoff, and all other authors. J. Muffat, Y. Li and R. Jaenisch designed the differentiation method and growth conditions for pMGLs. B. Yuan performed transcriptome profile analyses and comparisons. M. Mitalipova provided additional hES and hiPS lines for the study and performed their pMGL differentiation. J. Muffat and Y. Li performed and analyzed all other experiments. A. Omer, G. Bakiasi and S. Corcoran assisted with cell culture, sample preparation and data analysis.
Competing Financial Interests
R. Jaenisch is an adviser to Stemgent, a cofounder of Fate Therapeutics and Fulcrum Therapeutics. R. Ransohoff is Senior Research Fellow with Biogen. P. Aubourg is cofounder of GTDesign.
Primary Sources
- 1.Tremblay ME, et al. The role of microglia in the healthy brain. J Neurosci. 2011;31:16064–16069. doi: 10.1523/JNEUROSCI.4158-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alliot F, Godin I, Pessac B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res Dev Brain Res. 1999;117:145–152. doi: 10.1016/s0165-3806(99)00113-3. [DOI] [PubMed] [Google Scholar]
- 3.Vitry S, Bertrand JY, Cumano A, Dubois-Dalcq M. Primordial hematopoietic stem cells generate microglia but not myelin-forming cells in a neural environment. J Neurosci. 2003;23:10724–10731. doi: 10.1523/JNEUROSCI.23-33-10724.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong S, Dissing-Olesen L, Stevens B. New insights on the role of microglia in synaptic pruning in health and disease. Curr Opin Neurobiol. 2016;36:128–134. doi: 10.1016/j.conb.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith AM, Gibbons HM, Lill C, Faull RL, Dragunow M. Isolation and culture of adult human microglia within mixed glial cultures for functional experimentation and high-content analysis. Methods Mol Biol. 2013;1041:41–51. doi: 10.1007/978-1-62703-520-0_6. [DOI] [PubMed] [Google Scholar]
- 7.Hu BY, Du ZW, Li XJ, Ayala M, Zhang SC. Human oligodendrocytes from embryonic stem cells: conserved SHH signaling networks and divergent FGF effects. Development. 2009;136:1443–1452. doi: 10.1242/dev.029447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu BY, Zhang SC. Differentiation of spinal motor neurons from pluripotent human stem cells. Nat Protoc. 2009;4:1295–1304. doi: 10.1038/nprot.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grigoriadis AE, et al. Directed differentiation of hematopoietic precursors and functional osteoclasts from human ES and iPS cells. Blood. 2010;115:2769–2776. doi: 10.1182/blood-2009-07-234690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett ML, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hickman SE, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ginhoux F, Prinz M. Origin of microglia: current concepts and past controversies. Cold Spring Harb Perspect Biol. 2015;7:a020537. doi: 10.1101/cshperspect.a020537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butovsky O, Bukshpan S, Kunis G, Jung S, Schwartz M. Microglia can be induced by IFN-gamma or IL-4 to express neural or dendritic-like markers. Mol Cell Neurosci. 2007;35:490–500. doi: 10.1016/j.mcn.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, et al. NS21: re-defined and modified supplement B27 for neuronal cultures. J Neurosci Methods. 2008;171:239–247. doi: 10.1016/j.jneumeth.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, et al. High-efficiency induction of neural conversion in human ESCs and human induced pluripotent stem cells with a single chemical inhibitor of transforming growth factor beta superfamily receptors. Stem Cells. 2010;28:1741–1750. doi: 10.1002/stem.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei S, et al. Functional overlap but differential expression of CSF-1 and IL-34 in their CSF-1 receptor-mediated regulation of myeloid cells. J Leukoc Biol. 2010;88:495–505. doi: 10.1189/jlb.1209822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol. 2012;13:753–760. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palis J, McGrath KE, Kingsley PD. Initiation of hematopoiesis and vasculogenesis in murine yolk sac explants. Blood. 1995;86:156–163. [PubMed] [Google Scholar]
- 19.Hoeffel G, Ginhoux F. Ontogeny of Tissue-Resident Macrophages. Front Immunol. 2015;6:486. doi: 10.3389/fimmu.2015.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sturgeon CM, Ditadi A, Awong G, Kennedy M, Keller G. Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nat Biotechnol. 2014;32:554–561. doi: 10.1038/nbt.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKercher SR, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 22.Smith AM, et al. The transcription factor PU.1 is critical for viability and function of human brain microglia. Glia. 2013;61:929–942. doi: 10.1002/glia.22486. [DOI] [PubMed] [Google Scholar]
- 23.Feng R, et al. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci U S A. 2008;105:6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito D, et al. Microglia-specific localisation of a novel calcium binding protein, Iba1. Brain Res Mol Brain Res. 1998;57:1–9. doi: 10.1016/s0169-328x(98)00040-0. [DOI] [PubMed] [Google Scholar]
- 25.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, et al. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell. 2013;13:446–458. doi: 10.1016/j.stem.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derecki NC, Cronk JC, Kipnis J. The role of microglia in brain maintenance: implications for Rett syndrome. Trends Immunol. 2013;34:144–150. doi: 10.1016/j.it.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derecki NC, et al. Wild-type microglia arrest pathology in a mouse model of Rett syndrome. Nature. 2012;484:105–109. doi: 10.1038/nature10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butovsky O, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haynes SE, et al. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat Neurosci. 2006;9:1512–1519. doi: 10.1038/nn1805. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vitner EB, Futerman AH, Platt N. Innate immune responses in the brain of sphingolipid lysosomal storage diseases. Biol Chem. 2015;396:659–667. doi: 10.1515/hsz-2014-0301. [DOI] [PubMed] [Google Scholar]
- 33.Beutner C, Roy K, Linnartz B, Napoli I, Neumann H. Generation of microglial cells from mouse embryonic stem cells. Nat Protoc. 2010;5:1481–1494. doi: 10.1038/nprot.2010.90. [DOI] [PubMed] [Google Scholar]
- 34.Kierdorf K, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 35.Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 36.Hoeffel G, et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity. 2015;42:665–678. doi: 10.1016/j.immuni.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ginhoux F, Guilliams M. Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity. 2016;44:439–449. doi: 10.1016/j.immuni.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 38.Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 39.Wu Y, Dissing-Olesen L, MacVicar BA, Stevens B. Microglia: Dynamic Mediators of Synapse Development and Plasticity. Trends Immunol. 2015;36:605–613. doi: 10.1016/j.it.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aubourg P. Cerebral adrenoleukodystrophy: a demyelinating disease that leaves the door wide open. Brain. 2015;138:3133–3136. doi: 10.1093/brain/awv271. [DOI] [PubMed] [Google Scholar]
- 41.Ransohoff RM, El Khoury J. Microglia in Health and Disease. Cold Spring Harb Perspect Biol. 2015;8 doi: 10.1101/cshperspect.a020560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cartier N, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 43.Yirmiya R, Rimmerman N, Reshef R. Depression as a microglial disease. Trends Neurosci. 2015;38:637–658. doi: 10.1016/j.tins.2015.08.001. [DOI] [PubMed] [Google Scholar]
Secondary Sources
- 44.Lengner CJ, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 45.Harrington MG, et al. Cerebrospinal fluid sodium rhythms. Cerebrospinal Fluid Res. 2010;7:3. doi: 10.1186/1743-8454-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bardy C, et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc Natl Acad Sci U S A. 2015;112:E2725–2734. doi: 10.1073/pnas.1504393112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hogins J, Crawford DC, Zorumski CF, Mennerick S. Excitotoxicity triggered by Neurobasal culture medium. PLoS One. 2011;6:e25633. doi: 10.1371/journal.pone.0025633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakamichi N, et al. Protection by exogenous pyruvate through a mechanism related to monocarboxylate transporters against cell death induced by hydrogen peroxide in cultured rat cortical neurons. J Neurochem. 2005;93:84–93. doi: 10.1111/j.1471-4159.2005.02999.x. [DOI] [PubMed] [Google Scholar]
- 49.Jourdain P, et al. L-Lactate protects neurons against excitotoxicity: implication of an ATP-mediated signaling cascade. Sci Rep. 2016;6:21250. doi: 10.1038/srep21250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soldner F, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.