Abstract
The ways in which environmental factors participate in the progression of autoimmune diseases are not known. After initiation, it takes years before patients at risk for type 1 diabetes (T1D) develop hyperglycemia. The receptor for advanced glycated endproducts (RAGE) is a scavenger receptor of the immunoglobulin family that binds damage associated molecular patterns (DAMPs) and advanced glycated endproducts (AGEs) and can trigger cell activation. We previously found constitutive intracellular RAGE expression in lymphocytes from patients with T1D. Herein, we show that there is increased RAGE expression in T cells from at-risk euglycemic relatives who progress to T1D compared to healthy control subjects, and in the CD8+ T cells in the at-risk relatives who do vs those who do not progress to T1D. Detectable levels of the RAGE ligand HMGB1 were present in serum from at-risk subjects and patients with T1D. Transcriptome analysis of RAGE+ vs RAGE- T cells from patients with T1D showed differences in signaling pathways associated with increased cell activation and survival‥ Additional markers for effector memory cells and inflammatory function were elevated in the RAGE+ CD8+ cells of T1D patients and at-risk relatives of patients prior to disease onset. These studies suggest that expression of RAGE in T cells of subjects progressing to disease predates dysglycemia. These findings imply that RAGE expression enhances the inflammatory function of T cells and its increased levels observed in T1D patients may account for the chronic autoimmune response when DAMPs are released following cell injury and killing.
Keywords: RAGE, adaptive immunity, Type 1 diabetes, T cell survival
Introduction
RAGE is a pattern recognition receptor belonging to the immunoglobulin superfamily(1, 2). Initially described as the receptor for AGEs, additional ligands belonging to the DAMP superfamily have been identified, including: high mobility group box 1 (HMGB1), S100 calcium binding proteins, amyloid fibrils and nucleic acid backbones (3). RAGE is expressed on a wide range of cells and is thought to be involved in several pathologic conditions ranging from vascular inflammation and adaptive immune responses to Alzheimer’s disease (4–8). RAGE activation triggers downstream signaling through several inflammatory pathways, driving nuclear localization of NF-κB to promote cytokine production, cellular migration, proliferation and survival(4, 9). However, the molecular function for RAGE in adaptive immune cells and in particular its pathological contributions are not known.
Type 1 diabetes mellitus (T1D) is a complex autoimmune disorder resulting from failed tolerance in self-recognition. Disease progression is determined through a combination of genetic and environmental factors. Among the genetic factors, the major histocompatibility complex (MHC) human leukocyte antigen (HLA) genomic region is the most important modifier of disease risk(10). However, the concordance rate for diabetes among even identical twins is low, suggesting that additional environmental factors play an important role in disease progression. The link between the environmental factors and precipitation of disease has not been fully explained. Identifying these environmental factors has been challenging because the kinetics of disease are protracted and generally occur over 3 or more years (11).
T lymphocyte-mediated destruction of insulin producing pancreatic cells is thought to be the main determinant of disease progression. (10). Interestingly, RAGE blockade prevents diabetes transfer by diabetogenic T cells in NOD/scid mice and delays disease recurrence in syngeneic islet transplants (12–14). We have previously shown that T cells from T1D patients express elevated levels of intracellular RAGE compared to patient T cells from other autoimmune diseases and healthy controls (HC)(15). However, it was unclear in those studies whether increased RAGE expression was due to hyperglycemia- induced AGE accumulation or increased RAGE ligands since we also found increased levels of intracellular RAGE in T cells from patients with Type 2 diabetes.
The immunologic significance of RAGE expression on T cells has not been defined. In this study, we report that intracellular RAGE expression precedes dysglycemia in at-risk relatives of patients who later progress to overt diabetes. The ligand HMGB1 augments cytokine production in activated T1D T cells. T cells from patients with T1D express elevated RAGE (Ager) mRNA levels and the RAGE+ T cell population displays a pro- inflammatory gene expression profile. Interestingly, RAGE+ CD8+ T cells show enhanced survival during nutrient deprivation, correlating with higher NF-B nuclear localization. Consistent with these molecular findings, RAGE+CD8+ T cells from patients with and at risk for T1D show higher levels of EOMES and KLRG1 compared to RAGE-CD8+ T cells. Based on these findings, we suggest a mechanism whereby RAGE may link environmental events with progression of autoimmune diabetes by activating T cells that mediate cell killing.
Materials and Methods
Study Design
PBMC were obtained from 22 participants in the TrialNet TN-01, Pathway to Prevention (PTP) study (Clinical Trials.gov NCT00097292). This observational study follows subjects at risk for T1D for possible enrollment in diabetes prevention trials. These subjects were designated “at-risk” because they were relatives of a patient with T1D and found to have a positive autoantibody (anti-GAD65, anti-insulin, anti- ICA512, anti-ZnT8, or ICA). They were studied approximately every 6 months. Of the 22 subjects, 9 did and 13 did not develop T1D over the observation period (Table 1). All of the subjects had a normal hemoglobin A1c (HbA1c) levels and did not have symptoms of diabetes. The samples were obtained between 6/12/2005 and 8/3/2010. Of the 22 subjects, paired samples from 4 who did (“progressors”) and 4 who did not (“non-progressors”) were available for flow cytometric analysis described below. The at-risk population was studied over a period of 501±40 days (mean±SEM). In addition, we collected PBMC from 23 individuals with T1D of > 2 yrs duration and 18 control subjects without a personal or family history of T1D. For subjects shown in figure 6, paired samples were collected from 4 at-risk progressors and 4 at-risk non-progressors 397±9.6 days apart. After collection, the PBMC were frozen (in liquid nitrogen) in our or a central laboratory. Other PBMC’s were obtained from individuals with established T1D and used fresh or stored frozen in our laboratory. IRB approval was obtained at all of the study sites and all subjects or registered legal guardians gave written informed consent for the use of the samples.
Table 1.
Demographics of subjects
| A. At-risk subjects | ||||
|---|---|---|---|---|
| Group | N (Males) | Age (median, range) at 1st collection |
Number of positive autoAbs* |
Interval followed** |
| Progressors | 9 (3) | 17.0 (7.6–41.2) yrs | 3 | 383 days (146–672 days) |
| Non-progressors | 13 (7) | 15.9 (6.5–39.2) yrs | 1 | 585 days (343–742 days) |
| B. Subjects with diabetes | ||||
| N | Age (median, range) |
Disease duration (median, range) |
||
| Female | 9 | 21.1 (13.7–46) | 5.92 (2.67–38)yrs | |
| Male | 14 | 19.5 (13.4–34) | 10.1 (3.6–27) | |
Median
Median (range)
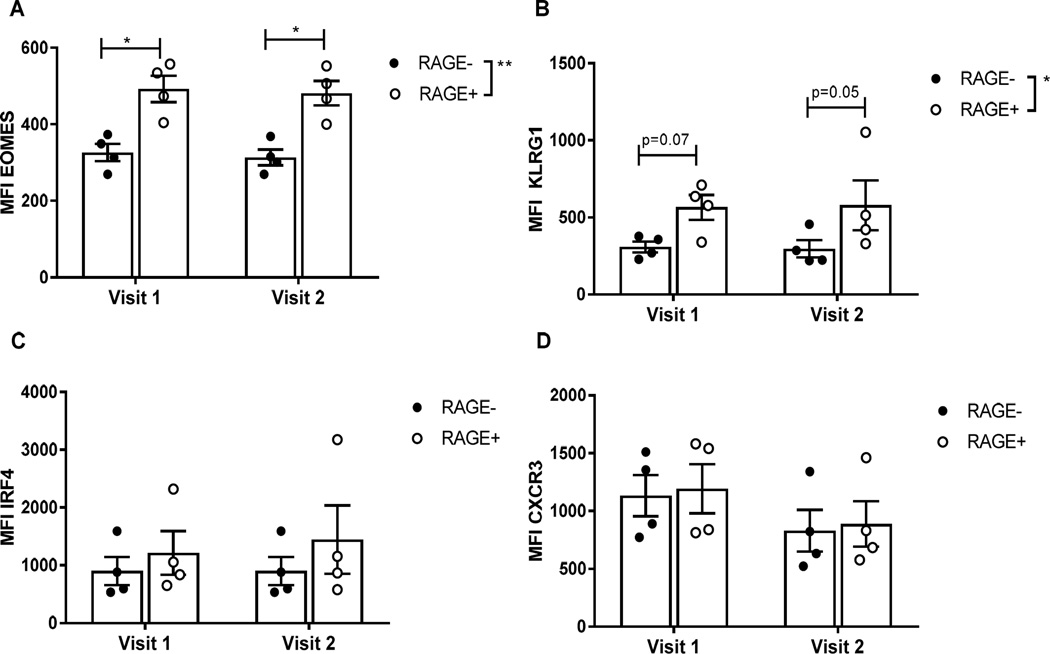
Fig. 6. RAGE+ CD8+ T cells express higher levels of EOMES and KLRG1 from individuals at-risk for T1D prior to the onset of hyperglycemia.
Samples were obtained from at-risk individuals who progressed to T1D on two occasions an average of 397±9.6 days apart. All of the individuals had normal glucose and HbA1c levels at the time that samples were taken. There was increased expression of EOMES (p<0.0016)(A) and KLRG1(p=0.03)(B) in RAGE+ vs RAGE- T cells. We did not detect differences in the expression of RAGE or these markers on the RAGE+ T cells between the two study visits.
Flow Cytometry
T cells were enriched from PBMCs using the Pan T Isolation Kit (Miltenyi Biotec) and stained as outlined in manufacturers protocol (BD Bioscience). Briefly, cells were incubated with LIVE/DEAD Cell Viability Dye (Invitrogen) in 1X PBS, followed by fluorescently labeled cell surface antibodies: anti-human CD4, anti-human CD8, anti-human CXCR3, anti-human KLRG1 (all from BD/Pharmingen) in FACS buffer (1X PBS, 3% FBS, 0.02% sodium azide). For intracellular staining, cells were fixed and permeabilzed with Cytofix/Cytoperm (BD/Pharmingen) and probed with fluorescently labeled intracellular antibodies: anti-human RAGE mAb (FITC labeled in house, Abcam) or AF488-labeled anti-AGER pAb (Bioss), anti-human EOMES, anti-human STAT3, anti- human IFN, IL-17a (all BD/Pharmingen) and anti-human IRF-4 (Miltenyi). Cells were visualized on a BD LSRFortessa (BD Bioscience) and analyzed with FlowJo software (Flowjo).
Cell Cultures
Enriched T cells were cultured in complete RPMI 1640 media (2.05 mM L-glutamine, 1 mM sodium-pyruvate, 10 mM HEPES, 55M -ME, 100 g/mL penicillin/streptomycin) with 5% FBS in the presence or absence of the following stimulants: -anti-CD3 (OKT3, 0.01 µg/mL, a gift from Ortho Pharmaceuticals) + anti-CD28 (0.01 µg/mL, eBioscience), HMGB1 (100 ng/mL, preparation previously described (16)). Following 72h incubation at 37°C, 5% CO2 supernatants were harvested and stored at - 80°C. For intracellular cytokine staining, GolgiStop (BD) was added into stimulation conditions and incubated for 6 hr prior to staining. Cytokines were measured in the supernatants with the Cytokine Human Magnetic 11-plex Panel (Millipore) and quantified with Luminex xMAP Technology (Luminex). The effects of serum deprivation were studied in serum-free media. T cells were washed 2X in 1x PBS and incubated in complete RPMI without serum for 24h at 37°C, 5% CO2. Cells were stained for viability, cell surface and intracellular markers and analyzed by flow cytometry. In siRNA-transfected samples, cells were incubated in complete RPMI without serum for 48h, stained for cell death using the LIVE/DEAD cell viability dye and analyzed by flow cytometry.
Proximity Ligation Assay
Enriched T1D T cells were cultured for 4 hrs in 100 ng/mL of His- tagged HMGB1 or His-tagged human serum albumin (HAS) (Abcam), cytospun onto microscope slides and analyzed for RAGE/HMGB1 protein interaction using Duolink in Situ Proximity Ligation Assay (Sigma) as outlined in manufacturers protocol (17). Both the his-tagged HMGB1 and HSA had < 1EU/mg of detectable LPS. Briefly, mouse -RAGE (Millipore) and rabbit −6X His-tag (Sigma) directed primary antibodies were added to slides and incubated in humidified chamber at RT for 1 hr. Slides were stained with PLA probes recognizing the primary antibody species for 1hr, incubated with ligation-ligase solution for 30 min and amplification-polymerase solution for 100 min. All incubations were performed at 37°C. CD8+ T cells were identified by staining with FITC-conjugated anti-CD3 mAb Hit8a (Biolegend). Slides were mounted with cover slips using Duolink In Situ Mounting Medium with DAPI and analyzed using a multi-channel fluorescent compound microscope (Leico DM500).
RT-PCR
CD4+ and CD8+ T cell populations were isolated using magnetically labeled Dynabeads (Invitrogen) followed by on column cell lysis (RLT Plus + 1% -ME, Qiagen). Total RNA was extracted from lysate using Mini RNA Lysis Kit (Qiagen) and used for cDNA amplification (R&D Systems). cDNA was mixed with SYBR Green master mix (Bio-Rad) and either human Ager (Forward: 5’ CTGGTGCTGAAGTGTAAGGG 3’, Reverse: 5’ GAAGAGGGAGCCGTTGG 3’) or human Gapdh (Forward: 5’ ACCCACTCCTCCACCTTTGAC 3’, Reverse: 5’ TGTTGCTGTAGCCAAATTCGTT 3’) primers for quantification (Bio-Rad iQ5 Cycler).
Nanostring Gene Expression
T cells were isolated from PBMCs from freshly collected blood using the Pan T cell Isolation Kit (Miltenyi Biotec). RAGE+/− CD4+ and CD8+ T cells were sorted into complete RPMI 1640 media using a BD FACSAria Ilu (BD Bioscience). Cells were pelleted by centrifugation and lysed in PDK buffer (Qiagen) supplemented with 10 µL Proteinase K (Qiagen). Samples were sequentially heated at 56°C and 80°C for 12 min each, snap frozen on LN2 and stored at −80°C. Samples were thawed on ice and the nCounter Human Immunology v2 Codeset was used for gene expression analysis as outlined in manufacturer’s protocol (Nanostring Technologies).
Nuclear Localization
Enriched CD8+ T cells were fixed with 3% formalin for 10 min, washed and permeabilized with 0.1% Triton X-100 + 2% FBS in PBS (no Ca/Mg). p65 NF-κB was stained with anti–p65 NF-B (Santa Cruz Biotechnology) and RAGE with anti-AGER mAb (Abnova). PE–labeled donkey Fab2 anti-rabbit IgG (Jackson ImmunoResearch) and AF488-labeled goat anti-mouse IgG (H+L) (Life Technologies) were used to stain corresponding species epitopes. Cells were stained with DAPI (Sigma- Aldrich) nuclear stain for 10 min and washed twice with PBS. Nuclear localization was performed on an Amnis Imagestream-X Mark II at ×40 magnification. Nuclear localization was determined using Amnis IDEAS software (Amnis) by Pearson coefficient colocalization of DAPI and p65 NF-κB.
siRNA Transfection
PBMC were quickly thawed in water bath and incubated overnight in complete RPMI 1640 media at 37°C, 5% CO2. Enriched T cells were transfected with either human Ager (s1168, Invitrogen) or negative control (Negative Control #1, Invitrogen) Silencer Select siRNA using the Amaxa 4D Nucleofector unstimulated primary human T cell, high functionality protocol (Lonza). Immediately after transfection, cells were incubated in complete RPMI 1640 for 48h before use in HMGB1 stimulation, cell death or Western blot assays.
Statistical analysis
The median value for the frequency of RAGE+ CD4+ and CD8+ T cells from the at-risk subjects, was calculated for each individual, using the data from all of the individual time points. Non-parametric tests (Mann-Whitney) were used for group and cell subset comparisons. Comparisons between RAGE+ and RAGE- measurements within each individual were made with a Wilcoxon signed-rank test. In the nanostring analysis, genes that failed to display >20 counts (LN>3) in at least 20% of analyzed samples were determined to be below background and excluded from analysis. For each experiment, the number of individuals providing samples is indicated. All analyses were performed with GraphPad (version 6).
Results
RAGE expression in T cells is increased in at-risk individuals who develop T1D
We analyzed RAGE expression in T cells from 22 at-risk relatives of patients with T1D who were participating in the TrialNet Pathway to Prevention Study (Table 1). Multiple samples (2–5) were obtained over time from participants that did (progressors) or did not (non-progressors) develop T1D over a similar observation period. All of the participants had normal HbA1c and glucose levels at the times samples were obtained.
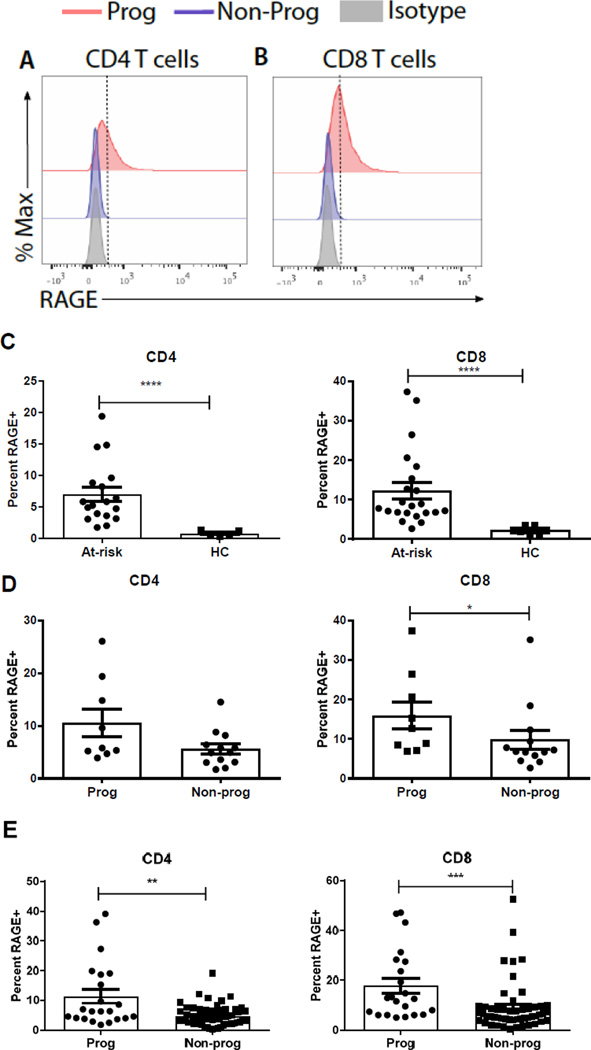
Representative intracellular RAGE staining of CD4+ and CD8+ T cells are shown in Fig 1A. For each individual, the median values describing the frequency of RAGE+ T cells among CD4+ or CD8+ T cells across all of the time points were determined and compared to the values from healthy control (HC) subjects. The median levels of RAGE expression in CD4+ and CD8+ T cells from at-risk individuals were higher than in healthy control subjects (Figure 1C, p<0.0001 for both). In the at-risk subjects, the median levels of RAGE expression were generally higher in progressors compared to non-progressors to T1D in CD4+ (10.5±2.62 v 5.6±0.96, p=0.11) and significantly higher in CD8+ T cells (16.0±3.47 vs 9.81±2.38, p=0.036, Mann Whitney test)(Figure 1D). When all of the individual values were compared, the levels of RAGE expression was significantly higher in both CD4+ and CD8+ T cells in the progressors compared to the non-progressors (p=0.004 and 0.0004 respectively, Mann Whitney test) (Figure 1E).
Fig. 1. At-risk individuals progressing to T1D express significantly higher levels of intracellular RAGE in T cell populations.
(A–B) Representative intracellular RAGE staining in CD4 (A) and CD8 (B) T cells from a single visit in progressors (red; Prog) or non-progressors (blue; NP). Isotype control staining is in grey. (C) The median percentage of RAGE+ CD4 (circles) and CD8 (triangles) T cells was determined for each at-risk subject and for healthy control subjects. There was an increase in the frequency of RAGE+ CD4+ and CD8+ T cells in the at-risk subjects compared to healthy controls (HC) (mean±SEM is shown, p<0.0001 for both, Mann-Whitney test)(D) The frequency of the median of RAGE+ T cells for each individual was compared in the at-risk progressors and non-progressors. There was a significantly greater frequency of RAGE+ CD8+ T cells in the at-risk progressors compared to those who did not develop T1D (p=0.036, mean±SEM, Mann-Whitney test) (E) The frequency of RAGE expression in CD4+ and CD8+ T cells at each study visit is shown. (**p=0.0037***p=0.0004, mean±SEM, Mann-Whitney test).
We also compared the changes in RAGE expression over time in the at-risk subjects. We did not detect a significant relationship between time prior to diagnosis of T1D and the expression of RAGE on CD4+ or CD8+ T cells (not shown).
HMGB1/RAGE stimulation augments inflammatory cytokine release from T1D T cells
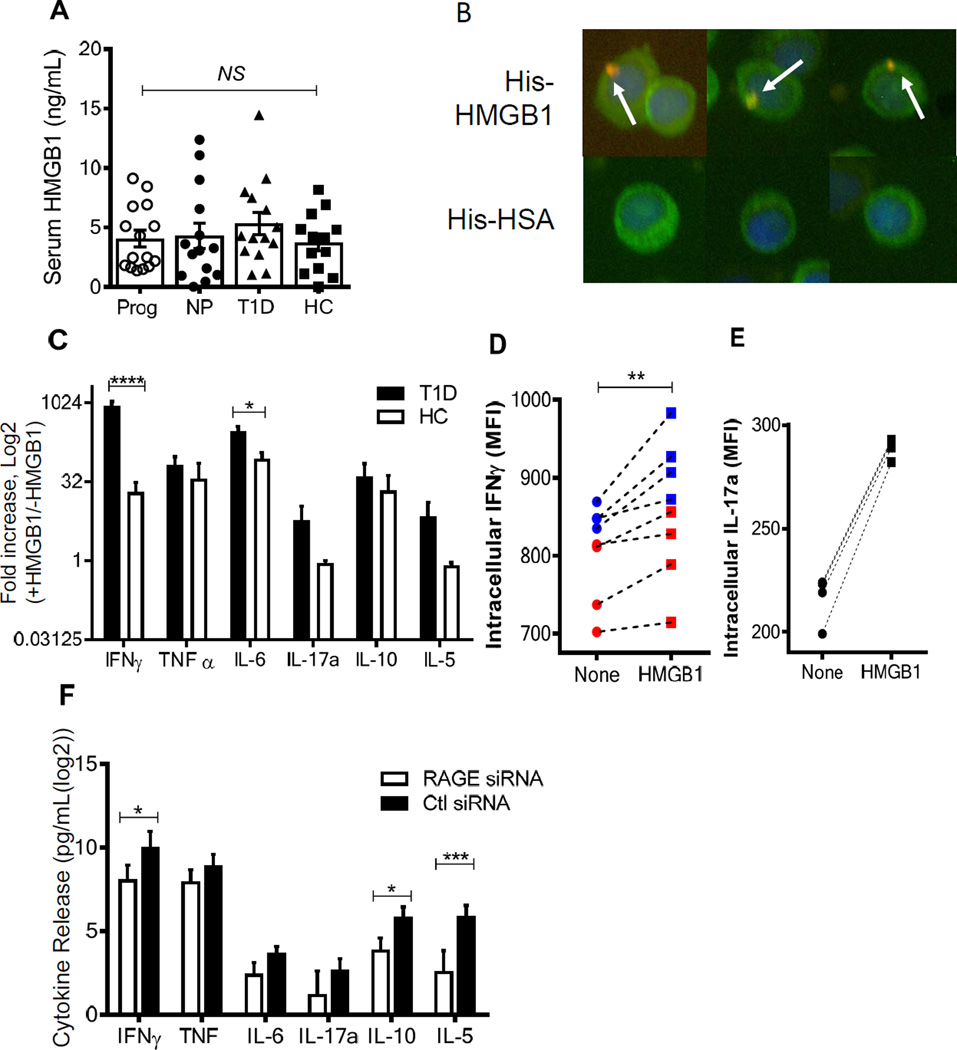
The levels of intracellular RAGE expression in T cells from the at-risk subjects were similar to levels that we had found in patients with T1D (15). Because of limited sample availability from the at-risk subjects, we carried out additional functional studies with T cells from patients after diagnosis (Table 1). Advanced glycation end products, produced with ambient hyperglycemia are not abundant in at-risk subjects in whom glucose levels are normal. We therefore tested whether other known RAGE ligands were available in the serum of patients with and at-risk for T1D and found that HMGB1 was present at similar levels in the serum of at-risk progressors (4.11 ± 0.84%), non-progressors (4.28 ± 1.07%) as in patients with established T1D (5.33 ± 0.96%, Fig 2A).
Fig. 2. Intracellular RAGE binding augments pro-inflammatory cytokine production in activated T1D T cells.
(A) Serum HMGB1 levels were similar in at-risk progressors (open circles), at-risk non-progressors (filled circles), established T1D (triangles) patients and HC (squares) individuals (mean±SEM). (B) Enriched T1D T cells were analyzed for RAGE-HMGB1 interaction using a proximity ligation assay (PLA) in CD8+ T cells from a patient with T1D. After 4 hours, in culture with his-tagged HMGB1 or HSA proximity of the his-tagged protein and RAGE is detected by PLA expression (red, arrows), nuclear staining by DAPI (blue) in CD8+ T cells (green). Data from a single experiment representative of 3 is shown. (C) Cytokine secretion from activated T1D (black bars, n = 3) and HC (white bars, n = 4) T cells were compared as fold induction in the presence of HMGB1 (100 ng/mL) after 72h stimulation. There was greater release of cytokines from cells from patients with T1D vs HC (p<0.0001). Statistical significance was determined using a two-way ANOVA. (**** < 0.0001, *<0.05, mean±SEM). (D-E) Intracellular IFNγ (D, red=CD4, blue=CD8) and IL-17a (E, CD8 only) in activated patient cells with or without HMGB1 stimulation (**p=0.0078 Wilcoxon signed-rank test). (F) HMGB1 induced cytokine secretion was also measured in enriched T cells from T1D patients transfected with RAGE (black) or Ctl (white) siRNA as above (n=6). There was significant reduction in cytokine release in cells that had received RAGE vs Ctl siRNA (p<0.0001)(*p<0.05 ***p<0.001, mean±SEM).
In previous studies we found that RAGE was expressed intracellularly but not on the surfaces of T cells from patients with T1D, whereas RAGE expression on the cell surface of monocytes is abundant (3, 15). Since HMGB1 was found in the serum, it was unclear whether this or other RAGE ligands could interact with RAGE internally localized in T cells. To determine whether extracellular RAGE ligands could interact with intracellular RAGE, we studied RAGE/HMGB1 interactions by proximity ligation (17). Enriched T cells from T1D patients were cultured with or without his-tagged recombinant HMGB1 or human serum albumin (HSA) and stained with PLA probes detecting anti-RAGE and anti-6X his-tag. DAPI was used for nuclear visualization and CD4+ (not shown) or CD8+ T cells were identified by counterstaining. Proximity amplification signal was observed in T1D T cells incubated overnight with 100ng/mL recombinant HMGB1, indicating intracellular interaction between RAGE and HMGB1 (Fig. 2B).
We next tested whether HMGB1 affected functional responses of T cells from patients with T1D and HC. Proliferation of polyclonal T cells was not affected by the addition of HMGB1 to cultures activated with anti-CD3/anti-CD28 mAbs (Fig. S1), but there was increased cytokine production and release when HMGB1 was added to cultures from T1D and HC subjects (Fig. 2C, p<0.0001). Specifically, there was significantly higher release of interferon-γ (p<0.001) and IL-6 (p<0.05) in cultures with cells from patients with T1D vs HC in the presence of HMGB1. Intracellular cytokine staining supported these findings as evident by the increased expression of intracellular IFNγ in CD4+ and CD8+ T cells when HMGB1 was added to low dose anti-CD3+CD28 mAbs (Fig. 2D, p<0.01). While not significant across multiple donors, adding HMGB1 to the cultures induced higher fold increase in interleukin-17a (IL-17a) in patients with T1D versus the HC (Fig. 2C). Interestingly, we found increased levels of IL-17a production in CD8+ T cells in the presence of HMGB1 (Fig. 2E), but not in the CD4+ T cells (data not shown).
HMGB1 may interact with several members of the toll-like receptor (TLR) family (18), which are also present on T cells. To ensure the increasing pro-inflammatory cytokine release from T1D patients was due to RAGE, enriched T cells were transfected with Ager (RAGE) or Ctl siRNA (64 ± 3.68% reduction, p=0.0006) and stimulated as described above. Reducing RAGE expression attenuated global cytokine production in activated T cells stimulated with HMGB1 (p<0.0001). There were statistically significant reductions in IFNγ (p=0.04), IL-10 (p=0.04), and IL-5(p=0.0002)(Fig. 2F).
Inflammatory genes and signaling pathways are constitutively elevated in RAGE+ T cells
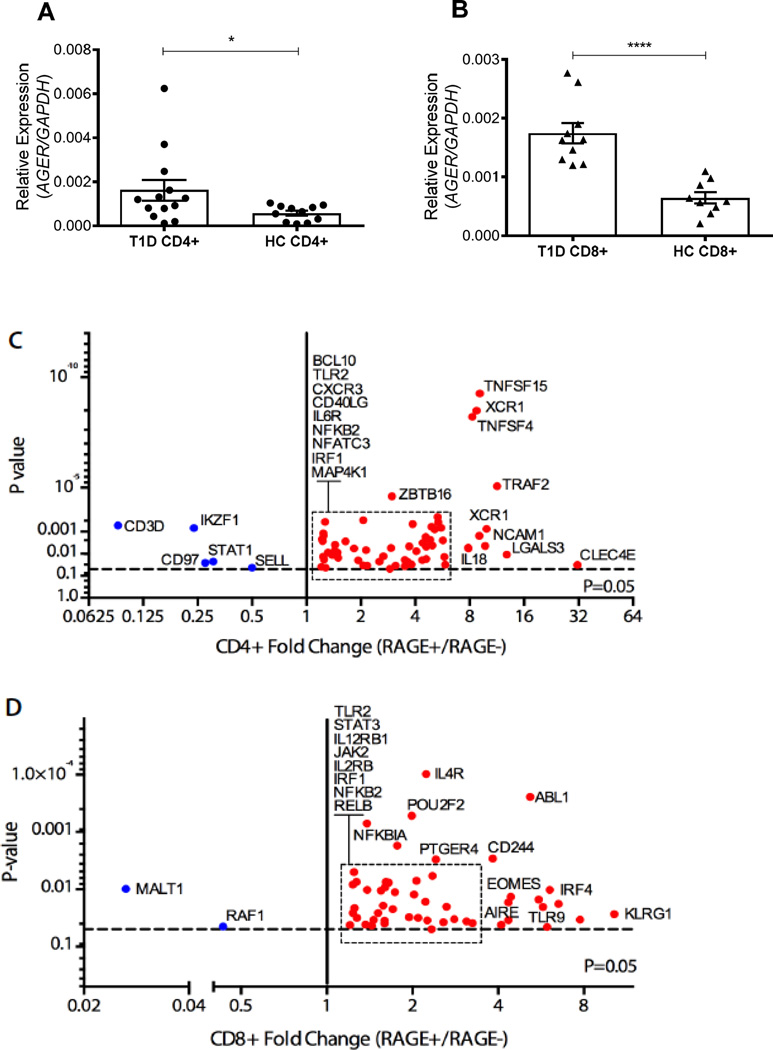
To determine the basis for increased RAGE protein expression we measured Ager gene expression by RT-PCR. Unstimulated CD4+ and CD8+ T cells from T1D patients displayed elevated Ager mRNA levels (2.85-fold and 2.71-fold, respectively) compared to HC cells (p=0.035 and p=0<0.0001 respectively, Fig 3 A–B). We next compared the gene expression profile between FACS- sorted RAGE+ and RAGE- T cells from patients with T1D. We used the Nanostring mRNA probe-based platform (human immunology panel) to identify differently expressed genes following fixing and permeabilization that was needed to identify RAGE by intracellular staining. RAGE+/− cells were compared within the same individual. For this exploratory analysis, gene expression was considered significantly altered if it was changed by > 1.5 fold between RAGE+ and RAGE- populations with a p<0.05. Genes whose expression met these criteria in CD4+ and CD8+ T cells are shown in Figs 3C–D.
Fig. 3. Ager mRNA is elevated in T1D patient T cells and RAGE+ T cells exhibit a pro-inflammatory phenotype.
(A–B) RT-PCR of RAGE (Ager) steady state mRNA levels in unstimulated CD4+ (A, T1D n = 13, HC n = 11) and CD8+ (B, T1D n = 10, HC n = 9) T cells. Values were normalized to GAPDH and statistical significance was determined using an unpaired, Mann-Whitney test (**** < 0.0001, * < 0.05, mean±SEM). (C–D) Genes displaying altered expression levels (≥ 1.5-fold) between RAGE+ and RAGE- unstimulated CD4+ (C, n = 5) and CD8+ (D, n = 6) T1D patient T cells. Statistical significance was determined using a paired Student’s t test between RAGE+ and RAGE- samples from same individual.
We used the data on gene expression to identify differences in pathway expression between RAGE+ and RAGE- T cells with MetaCORE (Table 2). Several pro-inflammatory signaling pathways were different in RAGE+ CD4+ and CD8+ cells, including: NF-κB, IL-6, IL-12 and IL-4. In addition, pathways of cell survival, and T lymphocyte proliferation/differentiation were also increased in RAGE+ T cells.
Table 2.
Pathway analysis determination of biological pathways elevated in RAGE+ versus RAGE- T1D T cells.
| Enriched Biological Pathways – RAGE+ T1D T cells | P-value |
|---|---|
| Apoptosis and survival_Anti-apoptotic TNFs/NF-κB/Bcl-2 pathway | 1.967E-06 |
| Immune response_Naive CD4+ T cell differentiation | 3.846E-06 |
| Immune response_TNF-R2 signaling pathways | 6.334E-05 |
| Signal transduction_NF-κB activation pathways | 1.033E-04 |
| Immune response_IL-6 signaling pathway | 3.699E-04 |
| Immune response_IL-12-induced IFN-gamma production | 5.757E-04 |
| Immune response_IL-4 signaling pathway | 1.034E-03 |
| Immune response_IL-12 signaling pathway | 3.866E-03 |
| Development_PDGF signaling via STATs and NF-κB | 7.332E-03 |
| Immune response_Th17 cell differentiation | 8.698E-03 |
Differences in gene expression in RAGE+ and RAGE- T cells, within the same individual, were used to identify biologic pathways that differ with MetaCORE (Thompson Reuters). The ten most altered T lymphocyte pathways in T1D T cells are ordered based on statistical significance (P-value).
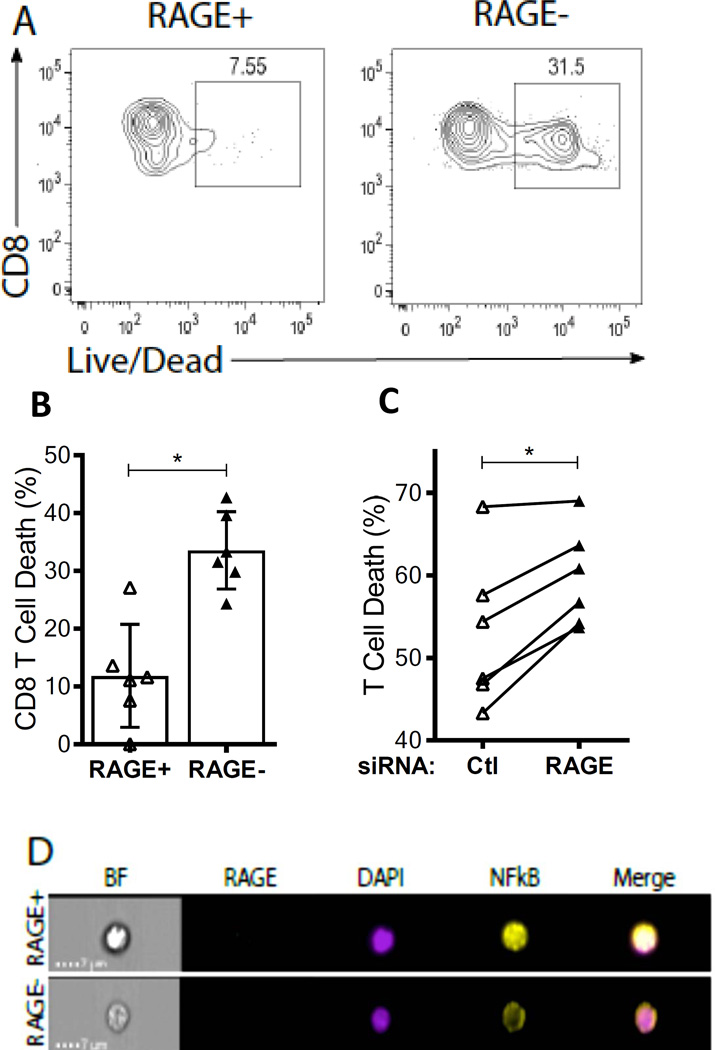
RAGE promotes CD8+ T cell survival under conditions of nutrient deprivation
One of the most highly increased pathways in the RAGE+ CD8+ T cells was pro- survival. We therefore compared the survival of RAGE+ and RAGE- T cells, from patients with T1D, under conditions of serum deprivation. When total T cells, enriched from whole PBMCs, were placed in serum-free media for 24h, markers of cell death were reduced on RAGE+ vs RAGE- CD8+ T cell populations (11.83 ± 3.63% vs 33.50 ± 2.73%, p<0.05, Fig 4A–B). A similar trend was observed in the RAGE+ CD4+ T cells but did not reach statistical significance across multiple donors (20.42 ± 4.62% vs. 35.20 ± 6.06%, p=0.09, Fig S2).
Fig. 4. RAGE protects CD8 T cells from serum starvation-induced cell death in T1D patients.
(A) Representative cell viability staining in T1D CD8+ T cells following 24h serum starvation. RAGE+ T cells are displayed on the left, while RAGE- T cells are on the right. (B) Percent cell death was measured in RAGE+ and RAGE- CD8 T cell populations following 24h serum starvation (n = 6, mean±SEM). (C) Ctl or Ager siRNA transfected T cells from T1D patients were analyzed for percent cell death following 48h serum starvation (n = 5). Statistical significance was determined using a Wilcoxon signed-rank test in each experiment (* < 0.05). (D) CD8+ T cells from T1D patients were stained for intracellular RAGE, NF-κB and the nuclear marker DAPI. Representative single cell images for brightfield (BF), single colors and NF-κB/DAPI colocalization (merge) are displayed from RAGE+ (top) and RAGE- (bottom) populations. Percent NF-κB nuclear localization with DAPI was determined by Pearson coefficient calculations.
To confirm the effects of RAGE on CD8+ T cell survival, RAGE expression was reduced with siRNA (38% reduction ± 4.13%, p=0.0003) and cells were placed in serum- free media. After 48 hrs, the levels of cell death, detected by flow cytometry with a live/dead stain was increased in cells transfected with Ager vs Ctl siRNA (on average by 12 ± 2.73%, p<0.05, Fig. 4C).
Several intrinsic pathways have been proposed to determine T cell death or survival, including NF-κB that was suggested in the Nanostring analysis of the RAGE+ vs RAGE- cells. We analyzed NF-κB localization in RAGE+ and RAGE- CD8+ T cells by confocal flow cytometry. NF-κB nuclear expression was increased in 5/5 individuals with T1D (19 ± 2.88% higher in RAGE+ versus RAGE- cells)(Fig. 4D, p=0.008).
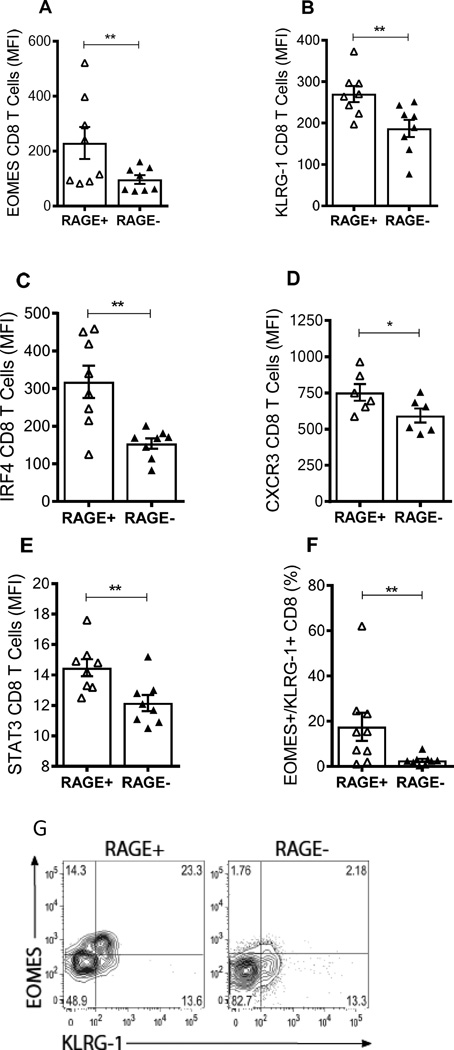
RAGE+ T cells express increased markers of a pro-inflammatory phenotype
Consistent with our nanostring findings, in patients with T1D, there was a higher proportion of RAGE+ CD8+ T cells that were positive for eomesodermin (EOMES) and killer cell lectin-like receptor subfamily G, member 1 (KLRG1) (Fig, 5A,B) and dual+ cells compared to the RAGE- CD8+ T cells (p<0.01. Fig. 5E,F), and both (Fig 5F, G) suggesting a higher proportion of terminally differentiated effector memory cells. The expression of interferon regulatory factor-4 (IRF-4)(p<0.01), CXCR3 (p<0.05), and signal transducer and activator of transcription 3 (STAT3) genes (p<0.01), found on IL-17 producing cells, were also elevated in RAGE+ CD8+ T cells (20). A similar expression patterns were seen in RAGE+ vs RAGE- CD4+ T cell populations (Fig. S3A).
Fig. 5. RAGE+ CD8+ T cells express higher amounts of inflammatory and effector memory markers in T1D patients.
(A) Expression of EOMES and KLRG1 in RAGE+ and RAGE- CD8+ T cells from a patient with T1D. Data from a single patient representative of 8 is shown. (A–F) EOMES (A), KLRG1 (B), IRF4 (C), and CXCR3 (D), STAT3 (E) levels in RAGE+ (open symbols) and RAGE- (closed symbols) CD8+ T cells from patients with T1D. Each symbol represents a single patient. The graphs display mean fluorescence intensity (MFI) of each marker. Statistical significance was calculated with a nonparametric Wilcoxon signed-rank test for all experiments (**<0.01, *<0.05, mean±SEM). (F,G) The frequency of EOMES/KLRG1 dual+ cells is also shown (**p<0.01).
We also analyzed the phenotypes of T cells from at-risk subjects who progressed to T1D using samples from 2 separate study visits approximately 1 year apart (Figure 6). Similar to our findings in patients with T1D, we found that the RAGE+ cells expressed higher levels of EOMES and KLRG1 in CD8+ T cells (p=0.0016 and 0.03 respectively, Fig. 6A, B). The expression of IRF4 was also increased on the RAGE+ vs RAGE- CD8+ T cells (p=0.09, by two way ANOVA), but we did not detect a significant difference in the level of CXCR3 expression (p=0.16, Figs. 6C, 6D). There were similar findings among CD4+ T cells (Fig. S4). We did not detect changes in the frequency of the EOMES, KLRG1, or dual+ cells between the two study visits prior to diabetes onset (Fig 6).
Discussion
After the initiation of autoimmunity, coinciding with the appearance of autoantibodies, it takes years for T1D to occur. Studies from our group and others have suggested that the pathologic process is relatively quiet until the peri-diagnosis period when there is a climactic increase in cell killing and failure in insulin secretion(21). Our studies of RAGE expression and function in T cells from at-risk subjects and patients with T1D suggest a mechanism that may account for the persistence of chronic inflammation and the accelerated pathology in the peri-diagnosis period. We found high levels of RAGE expression in CD4+ and CD8+ T cells of autoantibody positive, at-risk subjects who later progressed to disease prior to the development of dysglycemia or overt diabetes compared to healthy control subjects or to at-risk non-progressors. Gene expression in RAGE+ cells suggested activated inflammatory pathways, confirmed by higher levels of pathologic cytokine production when T cells from patients with T1D were activated with the RAGE ligand HMGB1 compared to T cells from control subjects. We found that the RAGE ligand, HMGB1, is able to gain entry into T cells and bind with the internally expressed receptor. RAGE expression was associated with increased T cell survival. Thus, the expression of RAGE in T1D patient T cells and its functional activity may account for the persistence of underlying inflammation and RAGE+ T cell pathologic function when DAMPs are released following cell injury and killing, or when glycated endproducts are available by transient elevations of glucose levels. Clinical data showing a rapid decline in cell function in the peridiagnosis period compared to earlier periods are consistent with this pathologic mechanism (21, 22).
RAGE expression was higher in T cells from individuals who progressed to overt T1D compared to non-progressors. We do not know the basis for the persistence of RAGE expression on these T cells. RAGE ligands can increase RAGE expression but these subjects did not have overt hyperglycemia that would increase glycated endproducts. We cannot exclude the possibility that there are periodic excursions of glucose levels that generate AGEs, but in addition to normal oral glucose tolerance tests, another measure of hyperglycemia, HbA1c, was also normal in our at risk subjects. Likewise, continuous exposure to antigens may be involved in the maintained expression, but the relatively high frequency of RAGE+ cells is unlikely to be accounted for exclusively by cells that are specific for diabetes associated antigens. We did not identify a change in the frequency of RAGE+ CD4+ or CD8+ T cells over time in the at-risk subjects followed to overt T1D. However, the number of subjects that we studied was limited and the sampling interval was large. A more detailed time course of the kinetics of RAGE expression would allow us to address this question further.
Functional and phenotypic features of RAGE+ T cells suggest a role of these cells in diabetogenesis where RAGE ligands are available. RAGE+ CD8+ T cells expressed CXCR3 and EOMES and a high level of EOMES/KLRG1 dual positivity in patients with T1D suggesting differentiated memory Th1 or Tc1 cells. The gene expression and NF-κB localization findings, indicate that RAGE+ T cells exhibit a predisposed inflammatory phenotype, and when activated in the presence of HMGB1, the cells produced higher levels of IFNγ, an important cytokine released by pathologic T cells in T1D (23). Furthermore, as a result of high levels of CXCR3 expression, the RAGE+ cells may traffic to the islets in response to IP-10 (CXCL10) since IP-10 is abundantly expressed in the pancreatic islets and is found in the serum of newly diagnosed diabetics and at risk individuals (24). We also found increased expression of the Th17/Tc17 cell lineage markers IRF4 and STAT3 in the RAGE+ CD8+ T cells in patients with T1D. We did not find a difference in either IRF4 or CXCR3 expression in the RAGE+ vs RAGE- CD8+ T cells from the at-risk progressors that may suggest that RAGE expression occurs prior to terminal differentiation of the T cells. However, we did find increased frequency of EOMES and KLRG1+ cells in the RAGE+ vs RAGE- CD8+ T cells from the at-risk progressors at both study visits. Further studies of the relationships between RAGE expression and activation and T cell differentiation are needed to resolve the differences in the at-risk subjects and patients. In our previous studies we found increased IL-17A production by T cells following activation with PMA/ionomycin (15), but we did not identify statistically significant differences upon HMGB1 stimulation. There are several possible explanations for this discrepancy including differences in the cell populations studied and methods of activation.
In the endosomes, RAGE may remove DAMPs that are generated within T cells by cellular damage. In this way, intracellular RAGE expression may improve cell survival since in serum-deprived media RAGE+ cells showed improved survival that was lessened by RAGE siRNA. Previous work demonstrates that the NF-κB family of transcription factors is critical for T cell survival and RAGE+ T cells constitutively express high levels of nuclear NF-κB (25). Additional genes (Relb) and signaling pathways (p53) that were activated in RAGE+ T cells have been shown to protect various types of cancer cells from nutrient-deprived apoptosis (26, 27). A role for these mechanisms in terms of RAGE mediated survival of pathologic T cells in T1D requires further evaluation.
Our study has limitations that are important to recognize. Our sample size of at- risk subjects was relatively small but they were studied on multiple occasions. Furthermore, we only followed the non-progressors for the same period of time as the progressors and therefore we cannot conclude that the non-progressors are “protected” from disease. Our studies utilized HMGB1 as the model RAGE ligand but several others have been described (1). HMGB1 is a well characterized ligand that we show is present in the serum of patients with diabetes, those at risk, and even in healthy control subjects (Fig 2A). While other ligands (e.g. glycosylated proteins) may be important in the disease mechanism, our studies with this widely found ligand enabled us to focus on the significance of RAGE expression on T cells. We did not study the effects of RAGE ligands on other immune and non-immune cells that may indirectly affect T cells. Although we have taken measures to limit non-T cell contributions within our experiments, we cannot rule out the indirect effects of other immune cells in vivo. Finally, our data showing increased Ager expression in the T cells from patients with T1D as well as our siRNA studies suggest that RAGE is made by the T cells themselves. However, we cannot exclude the possibility that RAGE is taken up into endosomes from T cells where it may still have effects on the T cells that we describe.
In summary, these studies suggest that intracellular RAGE expression may identify at-risk individuals who progress to T1D. They also suggest mechanisms whereby environmental factors may accelerate T1D progression by activating RAGE+ T cells. Even non-specific mediators may trigger the cells that cause disease explaining the clinical course and highlighting the difficulties in identifying single environmental factors that cause the disease.
Supplementary Material
Acknowledgments
We thank the following individuals for their conrtibutions: L. Devine and C. Wang (Yale Department of Laboratory Medicine) for help in FACS and review of the manuscript. O.I. Henegariu (Yale School of Medicine) for anti-human RAGE fluorophore conjugation, J. Esko (University of California, San Diego) for supplying endogenous HMGB1.
Funding: Support was from NIH grants R01 DK057846, U01 AI102011-04, U01 DK085466 from the NIH and grant SRA 2014-158 from the Juvenile Diabetes Research Foundation. Type 1 Diabetes TrialNet Pathway to Prevention Study Group is a clinical trials netwo,rk funded by the National Institutes of Health (NIH) through the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Allergy and Infectious Diseases, and The Eunice Kennedy Shriver National Institute of Child Health and Human Development, through cooperative agreements listed in the Appendix.
Appendix
The members of the TrialNet Study Group are as follows: Steering Committee: C. Greenbaum (Benaroya Research Institute), M. Anderson (University of California, San Francisco), P. Antinozzi (Wake Forest University), M. Atkinson (University of Florida), M. Battaglia (San Raffaele University), D. Becker (University of Pittsburgh), P. Bingley (University of Bristol), E. Bosi (San Raffaele University), J. Buckner (Benaroya Research Institute), Mark Clements (Children’s Mercy Hospital), P. Colman (Walter & Eliza Hall Institute of Medical Research), L. DiMeglio (Indiana University), S. Gitelman, (University of California, San Francisco), R. Goland (Columbia University), P. Gottlieb (Barbara Davis Center for Childhood Diabetes), K. Herold (Yale University), R. Insel (Juvenile Diabetes Research Foundation), T. Kay (St Vincent’s Institute of Medical Research), M. Knip (University of Helsinki), J. Krischer (University of South Florida), A. Lernmark (Skane University), J.B. Marks (University of Miami), W. Moore (Children’s Mercy Hospital), A. Moran (University of Minnesota), Andrew Muir (Emory University), J. Palmer (University of Washington), M. Peakman (King’s College), L. Philipson (University of Chicago), A. Pugliese (University of Miami), P. Raskin (University of Texas Southwestern), M. Redondo (Baylor University), H. Rodriguez (University of South Florida), B. Roep (Leiden University Medical Center), W. Russell (Vanderbilt University), L. Spain (National Institute of Diabetes and Digestive and Kidney Diseases [NIDDK]), D.A. Schatz (University of Florida), J. Sosenko (University of Miami), D. Wherrett (University of Toronto), D. Wilson (Stanford University), W. Winter (University of Florida), A. Ziegler (Forschergruppe Diabetes); Previous Members: J.S. Skyler (University of Miami, Chair); C. Benoist (Joslin Diabetes Center), J. Blum (Indiana University), K. Bourcier, P. Chase (Barbara Davis Center for Childhood Diabetes), M. Clare-Salzler (University of Florida), R. Clynes (Columbia University), G. Eisenbarth (Barbara Davis Center for Childhood Diabetes), C. G. Fathman (Stanford University), G. Grave (National Institute of Child Health and Human Development), B. Hering (University of Minnesota), F. Kaufman (Children’s Hospital Los Angeles), E. Leschek (NIDDK), J. Mahon (University of Western Ontario), K. Nanto-Salonen University of Turku), G. Nepom (Benaroya Research Institute), T. Orban (Joslin Diabetes Center), R. Parkman (Children’s Hospital Los Angeles), M. Pescovitz (Indiana University), J. Peyman (National Institute of Allergy and Infectious Disease), M. Roncarolo (San Raffaele University), P. Savage (NIDDK), O. Simell (University of Turku), R. Sherwin (Yale University), M. Siegelman (University of Texas Southwestern), A. Steck (Barbara Davis Center for Childhood Diabetes), J. Thomas (Vanderbilt University), M. Trucco (University of Pittsburgh), J. Wagner (University of Minnesota).
References
- 1.Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367–388. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- 2.Bucciarelli LG, Wendt T, Rong L, Lalla E, Hofmann MA, Goova MT, Taguchi A, Yan SF, Yan SD, Stern DM, Schmidt AM. RAGE is a multiligand receptor of the immunoglobulin superfamily: implications for homeostasis and chronic disease. Cell Mol Life Sci. 2002;59:1117–1128. doi: 10.1007/s00018-002-8491-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sirois CM, Jin T, Miller AL, Bertheloot D, Nakamura H, Horvath GL, Mian A, Jiang J, Schrum J, Bossaller L, Pelka K, Garbi N, Brewah Y, Tian J, Chang C, Chowdhury PS, Sims GP, Kolbeck R, Coyle AJ, Humbles AA, Xiao TS, Latz E. RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA. J Exp Med. 2013;210:2447–2463. doi: 10.1084/jem.20120201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hofmann MA, Drury S, Fu C, Qu W, Taguchi A, Lu Y, Avila C, Kambham N, Bierhaus A, Nawroth P, Neurath MF, Slattery T, Beach D, McClary J, Nagashima M, Morser J, Stern D, Schmidt AM. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97:889–901. doi: 10.1016/s0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- 5.Hofmann MA, Drury S, Hudson BI, Gleason MR, Qu W, Lu Y, Lalla E, Chitnis S, Monteiro J, Stickland MH, Bucciarelli LG, Moser B, Moxley G, Itescu S, Grant PJ, Gregersen PK, Stern DM, Schmidt AM. RAGE and arthritis: the G82S polymorphism amplifies the inflammatory response. Genes Immun. 2002;3:123–135. doi: 10.1038/sj.gene.6363861. [DOI] [PubMed] [Google Scholar]
- 6.Hofmann MA, Yang Q, Harja E, Kedia P, Gregersen PK, Cupples LA, Schmidt AM, Hudson BI. The RAGE Gly82Ser polymorphism is not associated with cardiovascular disease in the Framingham offspring study. Atherosclerosis. 2005;182:301–305. doi: 10.1016/j.atherosclerosis.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Yan SD, Chen X, Fu J, Chen M, Zhu H, Roher A, Slattery T, Zhao L, Nagashima M, Morser J, Migheli A, Nawroth P, Stern D, Schmidt AM. RAGE and amyloid-beta peptide neurotoxicity in Alzheimer’s disease. Nature. 1996;382:685–691. doi: 10.1038/382685a0. [DOI] [PubMed] [Google Scholar]
- 8.Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, Parroche P, Drabic S, Golenbock D, Sirois C, Hua J, An LL, Audoly L, La Rosa G, Bierhaus A, Naworth P, Marshak-Rothstein A, Crow MK, Fitzgerald KA, Latz E, Kiener PA, Coyle AJ. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 9.Arumugam T, Simeone DM, Schmidt AM, Logsdon CD. S100P stimulates cell proliferation and survival via receptor for activated glycation end products (RAGE) J Biol Chem. 2004;279:5059–5065. doi: 10.1074/jbc.M310124200. [DOI] [PubMed] [Google Scholar]
- 10.Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N Engl J Med. 2009;360:1646–1654. doi: 10.1056/NEJMra0808284. [DOI] [PubMed] [Google Scholar]
- 11.Herold K, Vignali DA, Cooke A, Bluestone J. Type 1 diabetes: Translating mechanistic observations into effective clinical outcomes. Nat Rev Immunol. 2013:13. doi: 10.1038/nri3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Akirav EM, Chen W, Henegariu O, Moser B, Desai D, Shen JM, Webster JC, Andrews RC, Mjalli AM, Rothlein R, Schmidt AM, Clynes R, Herold KC. RAGE ligation affects T cell activation and controls T cell differentiation. J Immunol. 2008;181:4272–4278. doi: 10.4049/jimmunol.181.6.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Yan SS, Colgan J, Zhang HP, Luban J, Schmidt AM, Stern D, Herold KC. Blockade of late stages of autoimmune diabetes by inhibition of the receptor for advanced glycation end products. J Immunol. 2004;173:1399–1405. doi: 10.4049/jimmunol.173.2.1399. [DOI] [PubMed] [Google Scholar]
- 14.Moser B, Desai DD, Downie MP, Chen Y, Yan SF, Herold K, Schmidt AM, Clynes R. Receptor for advanced glycation end products expression on T cells contributes to antigen-specific cellular expansion in vivo. J Immunol. 2007;179:8051–8058. doi: 10.4049/jimmunol.179.12.8051. [DOI] [PubMed] [Google Scholar]
- 15.Akirav EM, Preston-Hurlburt P, Garyu J, Henegariu O, Clynes R, Schmidt AM, Herold KC. RAGE expression in human T cells: a link between environmental factors and adaptive immune responses. PLoS One. 2012;7:e34698. doi: 10.1371/journal.pone.0034698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu D, Young J, Song D, Esko JD. Heparan sulfate is essential for high mobility group protein 1 (HMGB1) signaling by the receptor for advanced glycation end products (RAGE) J Biol Chem. 2011;286:41736–41744. doi: 10.1074/jbc.M111.299685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, Ostman A, Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 18.Park JS, Svetkauskaite D, He Q, Kim JY, Strassheim D, Ishizaka A, Abraham E. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J Biol Chem. 2004;279:7370–7377. doi: 10.1074/jbc.M306793200. [DOI] [PubMed] [Google Scholar]
- 19.Frigerio S, Junt T, Lu B, Gerard C, Zumsteg U, Hollander GA, Piali L. Beta cells are responsible for CXCR3-mediated T-cell infiltration in insulitis. Nat Med. 2002;8:1414–1420. doi: 10.1038/nm1202-792. [DOI] [PubMed] [Google Scholar]
- 20.Huber M, Brustle A, Reinhard K, Guralnik A, Walter G, Mahiny A, von Low E, Lohoff M. IRF4 is essential for IL-21-mediated induction, amplification, and stabilization of the Th17 phenotype. Proc Natl Acad Sci U S A. 2008;105:20846–20851. doi: 10.1073/pnas.0809077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herold KC, Usmani-Brown S, Ghazi T, Lebastchi J, Beam CA, Bellin MD, Ledizet M, Sosenko JM, Krischer JP, Palmer JP. beta Cell death and dysfunction during type 1 diabetes development in at-risk individuals. J Clin Invest. 2015;125:1163–1173. doi: 10.1172/JCI78142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenbaum CJ, Beam CA, Boulware D, Gitelman SE, Gottlieb PA, Herold KC, Lachin JM, McGee P, Palmer JP, Pescovitz MD, Krause-Steinrauf H, Skyler JS, Sosenko JM. Fall in C-peptide During First 2 Years From Diagnosis: Evidence of at Least Two Distinct Phases From Composite TrialNet Data. Diabetes. 2012 doi: 10.2337/db11-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cardozo AK, Proost P, Gysemans C, Chen MC, Mathieu C, Eizirik DL. IL-1beta and IFN-gamma induce the expression of diverse chemokines and IL-15 in human and rat pancreatic islet cells, and in islets from pre-diabetic NOD mice. Diabetologia. 2003;46:255–266. doi: 10.1007/s00125-002-1017-0. [DOI] [PubMed] [Google Scholar]
- 24.Nicoletti F, Conget I, Di Mauro M, Di Marco R, Mazzarino MC, Bendtzen K, Messina A, Gomis R. Serum concentrations of the interferon-gamma-inducible chemokine IP-10/CXCL10 are augmented in both newly diagnosed Type I diabetes mellitus patients and subjects at risk of developing the disease. Diabetologia. 2002;45:1107–1110. doi: 10.1007/s00125-002-0879-5. [DOI] [PubMed] [Google Scholar]
- 25.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 26.Hamidi T, Algul H, Cano CE, Sandi MJ, Molejon MI, Riemann M, Calvo EL, Lomberk G, Dagorn JC, Weih F, Urrutia R, Schmid RM, Iovanna JL. Nuclear protein 1 promotes pancreatic cancer development and protects cells from stress by inhibiting apoptosis. J Clin Invest. 2012;122:2092–2103. doi: 10.1172/JCI60144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lassus P, Ferlin M, Piette J, Hibner U. Anti-apoptotic activity of low levels of wild-type p53. EMBO J. 1996;15:4566–4573. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.