Abstract
The mammalian target of rapamycin (mTOR) is essential for helper T cell proliferation and effector differentiation, making the mTOR signaling network an attractive immunomodulatory target for autoimmune related diseases. While direct targeting of mTOR-complex-1 (mTORC1) with rapamycin can provide clinical benefit, targeting downstream enzymes has potential to offer more selective immunosuppression. Here we evaluated p70 ribosomal protein S6 Kinase 2 (S6K2), a downstream effector of mTORC1, for its role in T cell function and autoimmunity. S6K2 is a direct substrate of mTORC1, with a potential role in Th17 differentiation suggested by biochemical studies. Using a genetic approach with S6K2 knockout mice, we found that S6K2 loss reduces Th17 skewing and increases Treg differentiation in vitro when cultured in RPMI media. However, S6K2 was dispensable for Th17 differentiation in IMDM media. In an in vivo EAE model in which rapamycin suppresses disease, S6K2 knockout mice did not exhibit differences in clinical score or Th17 differentiation. These results suggest that S6K2 is dispensable for Th17-driven autoimmunity and highlight how distinct experimental conditions can produce significantly different results in T cell differentiation.
Introduction
The mTOR protein is a serine-threonine kinase that forms two separate complexes, mTORC1 and mTORC2. Each complex has distinct regulation and downstream target substrates (1, 2). mTOR signaling in both naïve CD4+ and CD8+ T cells can be activated by engagement of a recognized antigen through the PI3K-Akt-mTOR pathway (3, 4). The complex mTORC1, defined by the regulatory-associated protein of mTOR (Raptor) subunit, regulates both bioenergetic and biosynthetic processes that are crucial for T cell growth and division (5, 6). These include protein translation, lipid biogenesis, and suppression of autophagy. Downstream of mTORC1, two directly phosphorylated substrates are ribosomal S6 Kinases (S6Ks) and eukaryotic initiation factor 4E (eIF4E) binding proteins (4E-BPs), both of which have evolutionarily conserved roles in cell growth and metabolism (1, 7, 8).
Newly activated T-lymphocytes utilize mTOR signaling to initiate transcription of metabolic regulatory genes (9), a process crucial for driving proliferation and differentiation. The ability to produce a sustainable effector program selectively requires mTORC1 activity to integrate both immune and nutrient cues. In the context of an autoimmune response, proinflammatory Th1 and Th17 subsets are highly dependent upon glucose consumption, whereas suppressive Tregs favor increased mitochondrial respiration (10–12). Interestingly, while mTORC1 activity is required for differentiation and effector cytokine production of both Th1 and Th17 subsets (13, 14), the production of induced Tregs increases when mTORC1 is suppressed (13, 15, 16). The function of established Tregs is impaired when mTORC1 activity is either reduced (12) or increased (17).
Rapamycin, a potent inhibitor of mTORC1 activity, suppresses pathology in several animal models of T cell-driven autoimmunity (18–22) and at low doses extends lifespan of mice (23). Rapamycin has been tested in clinical trials of human autoimmune diseases and is often prescribed to such patients off label (24–26). However, sustained rapamycin treatment has some detrimental side effects (27, 28). Targeting mTORC1 substrate pathways is a reasonable alternative that might provide a better therapeutic window.
There are two S6K isoforms, S6K1 and S6K2, both of which are expressed in T cells (29, 30). Activated mTORC1 phosphorylates S6Ks, which in turn phosphorylate the ribosomal protein S6 and other substrates involved in protein and lipid biosynthesis (7). When mTORC1 phosphorylates 4E-BPs, these proteins are released from eIF4E to promote cap-dependent translation (31). Thus, S6K and 4E-BP signaling pathways converge to regulate mRNA translation. The inhibition of mTORC1 by rapamycin results in profound suppression of S6K phosphorylation and activity (32, 33), but only partial inhibition of 4E-BP1 phosphorylation (34). However, it is unclear which mTOR functions are mediated by S6Ks during T helper (Th) cell differentiation.
Previous studies have provided limited and sometimes conflicting information about the functions of S6Ks in T cell growth, proliferation and differentiation. S6K1−/− mice have a small size phenotype but apparently normal lymphocyte function, associated with massive compensatory upregulation of S6K2 in T cells (30). One study reported that the S6K1 inhibitor PF-4708671 suppresses CD4 T cell growth and proliferation (35). However, we found that PF-4708671 has non-specific effects at the 10μM concentration used (36). Mice with a germline deletion of both S6K1 and S6K2 exhibit perinatal lethality, making it challenging to analyze mature immune cells (37). In the few adult double knockout mice that survive, T cell growth and proliferation is surprisingly unimpaired (36). Koyasu and colleagues used gain-of-function approaches to suggest unique roles for S6K1 and S6K2 in Th17 differentiation (14). A later study by Sasaki et al. (38) utilized S6K1−/− mice to show a role for S6K1 in differentiation of Th17 cells, but not other CD4+ T cell subsets. S6K1 was found to promote expression of Th17 associated genes (e.g. IL17a, IL17f, and IL23R); however, it did not affect expression of the Th17 master transcription factor retinoic acid receptor-related orphan receptor gamma (RORγ). In addition, mice lacking S6K1 did not show improvement of clinical disease scores in experimental autoimmune encephalomyelitis (EAE), a mouse autoimmunity model.
In this study, we investigated the specific role(s) of S6K2 in mouse T cell activation, development, and proliferation. S6K2 germline knockout mice are viable, fertile and of normal size; moreover, S6K1 expression is unchanged in S6K2-deficient T cells. We found that S6K2 knockout (S6K2−/−) mice had no defects in T cell development and early CD4 T cell activation, despite a measurable decrease in overall S6K activity as determined by S6 protein phosphorylation. Interestingly, S6K2−/− cells exhibited a context-specific Th differentiation phenotype. In vitro experiments using RPMI culture media revealed decreased Th17 and increased Treg differentiation in S6K2−/− cells. However, in an in vivo EAE model of autoimmunity, S6K2−/− mice showed no difference in clinical outcome or Th17 differentiation. Re-evaluation of the in vitro differentiation assay conditions surprisingly showed that the Th17 differentiation defect depends on the culture medium used. IMDM medium, known to induce more robust Th17 differentiation (39), led to Th17 skewing and robust proliferation that was indistinguishable between S6K2−/− and wildtype T cells. This unexpected result indicates that S6K2 is dispensable for Th17 differentiation under optimal activation conditions. In addition, our findings suggest that the use of IMDM media for in vitro Th17 studies better predicts the in vivo response in EAE models.
Materials and Methods
Animals
All mice were kept in specific pathogen–free animal facilities and in accordance with guidelines of the University of California Institutional Animal Care and Use Committee. C57BL/6 mice were obtained from Jackson Laboratories. S6K2−/− mice were a gift from Sara Kozma.
Media and cell culture
Primary cell culture was performed in complete media consisting of RPMI or IMDM media (Gibco) supplemented with 10% FCS (Gibco), L-glutamine (Corning), βME and antibiotics (Gibco).
Antibodies and reagents
Stimulatory anti-CD3 (2C11) and anti-CD28 (37.51), as well as neutralizing anti-IFN-γ (XMG1.2), anti-IL-2 (JES6-1A12), anti-IL-12p40 (C17.8), and anti-IL-4 (11B11) were from eBioscience. For differentiation, recombinant cytokines were used: TGF-beta (R&D systems), mouse IL-6 and IL-12 (eBioscience). For flow cytometry analysis, surface and intracellular staining antibodies were purchased from eBioscience and Cell Signaling Technologies. For immunoblotting, rabbit antibodies specific for total and phosphorylated forms were obtained from Cell Signaling Technologies: AKT (#2965, #9267, #4058, #4685), rS6 (#4838, #2215, #2211), S6K (#9205, #2708), pCAD (#12662). For imaging, anti-Mouse RORγt (B2D) from eBioscience was used.
Cell purification
Splenic CD4+ T cells depleted of natural Tregs were purified by negative selection using the EasySep Mouse CD4+ T Cell Isolation Kit from Stemcell Technologies with biotinylated anti-CD25 (0.5 μg/ml) added.
Cell cycle analysis
T cells were stimulated in T-conventional (Tconv), Th17, or Treg skewing conditions for 40h and fixed with 100% ethanol. Cells were incubated with RNAse A and propidium iodine (PI) diluted in cell cycle buffer (5mM EDTA in PBS) before cell cycle analysis was performed using FACS.
Cell stimulation and differentiation
Unless otherwise stated, T cells were stimulated as Tconv, with plate-bound anti-CD3 (5 μg/ml) and soluble anti-CD28 (2 μg/ml) from eBioscience. 10 nM of Rapamycin (LC Laboratories) and 500 nM of LY2584702 (S6K1i) (Eli Lilly & Company) were also used when indicated. When proliferation is measured, isolated T cells were prelabeled with CFSE (eBioscience). Cell stimulation cocktail (500×) from eBioscience containing PMA (phorbol 12-myristate 13-acetate), ionomycin, Brefeldin A, and monensin were used for restimulation (detection of intracellular cytokines). For Th polarization assays, isolated naïve T cells were skewed with anti-CD3 and anti-CD28 for 4 days (unless otherwise noted), restimulated for 4 hours, and harvested for flow cytometry analysis. Skewing conditions were as follows: TH1-IL-12 (20 ng/ml) and anti-IL-4 (10 μg/ml); TH17, TGF-β (5 ng/ml), IL-6 (20 ng/ml), anti-IFN-γ (10 μg/ml), anti-IL-4 (10 μg/ml) and IL-2 (1 ng/ml); Treg, TGF-β (5 ng/ml).
Intracellular staining for flow cytometry analysis
After restimulation with cell stimulation cocktail, cells were fixed and made permeable with BD Cytofix/Cytoperm and then stained for cell surface markers and cytokines. Intracellular staining of transcription factors (i.e. FoxP3) was done without restimulation. Gates were set appropriately with unstimulated controls and voltages were set on the basis of isotype-matched control antibodies. Data were acquired using a BD FACSCalibur flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star).
ELISA
Cells were harvested after indicated stimulation periods and supernatants were collected to measure cytokine production according to manufacturer’s instructions. IL-2 and IL-10 ELISA kits were obtained from eBioscience and IL-17A ELISA kit was obtained from R&D systems.
Experimental Autoimmune Encephalomyelitis
10–12 week-old female wildtype and S6K2−/− mice were immunized with 100μg of MOG35-55 antigen in 200μg of CFA per mouse. Pertussis was administered i.v. on day 0 and 1 at 400ng per injection. Mice were scored on a scale of 0–5: 0-no clinical symptoms; 1-limp tail; 2-limp tail and partial hind limb paralysis; 3-limp tail and complete hind limb paralysis; 3.5-partial front or one-sided paralysis; 4-moribund; 5-death. Mice scoring 3.5 or more for two consecutive days were euthanized. Spinal cords were harvested and centrifuged through a Percoll gradient. Isolated infiltrating lymphocytes were restimulated for 4h before staining intracellularly for IFNγ and IL-17A. To analyze the phenotype of activated CD4+ T cells during disease induction, wildtype and S6K2−/− mice were immunized with MOG peptide without pertussis and sacrificed after 10 days. Lymph nodes were harvested for analysis of the percentage of pS6+ among CD4+CD44hi cells. Splenocytes were restimulated with MOG (10 and 50μg/mL), IL-23 (20ng/mL), and anti-IFNγ antibody (10μg/mL) for 3 days and then harvested for cytokine measurement in the supernatant, as well as stained intracellularly for IL-17A and FoxP3 expressing CD4+ T cells.
Immunofluorescence Microscopy
CD4+ T cells were harvested after 40 hours of stimulation, washed, and centrifuged. The pellets were fixed with 4% formaldehyde for 10 min at 37°C, permeabilized with 0.1% Saponin (Acros Organics) in PBS for 10 min at room temperature, blocked with 5% anti-rat serum in 1X PBS / 0.1% Saponin and stained with anti-mouse RORγT (eBioscience) for 2 hr at 37°C in PBS containing 1% BSA / 0.1% Saponin. F(ab′)2 Anti-Rat IgG PE (eBioscience) were used as a secondary antibody. Specimens were further stained with DAPI (Thermo Fisher Scientific) and Rhodamine-phalloidin (Thermo Fisher Scientific). Single cell suspensions are spun onto a microscope slide by use of a cytocentrifuge Images were obtained using a Leica TCS SP8 confocal scanning microscope and LAS X core microscopy software. For statistical analysis, the Manders’ Overlap Coefficient was used to measure RORγ localization in the nucleus (as determined by RORγ and DAPI staining overlap) in at least three representative fields of view for each condition.
Metabolic studies
Metabolic flux analysis was performed in complete Seahorse media (Seahorse Bioscience, 102365) supplemented with 25mM D-Glucose, 1mM Na-Pyruvate and 2mM L-glutamine.
Treg suppression assays
nTregs were sorted by CD4+CD25+ expression from combined spleen and lymph nodes of aged matched mice. nTregs were then seeded with CFSE labeled naïve WT CD4+ T cells at a 1:2 ratio. The wells were pre-coated with 2μg/mL of CD3 and 1μg/mL of soluble CD28 was added. After 3 days of stimulation, the cells were washed and analyzed by FACS.
Statistical Analysis
Mean and SEM values were calculated from at least three independent experiments. p values were calculated using a two-tailed unpaired Student’s t-test when determining differences between groups, unless the Wilcoxon signed-rank test was used to determine pairwise differences (specifically indicated in figure legends).
Results
S6K2−/− mice have normal T cell development and proliferation
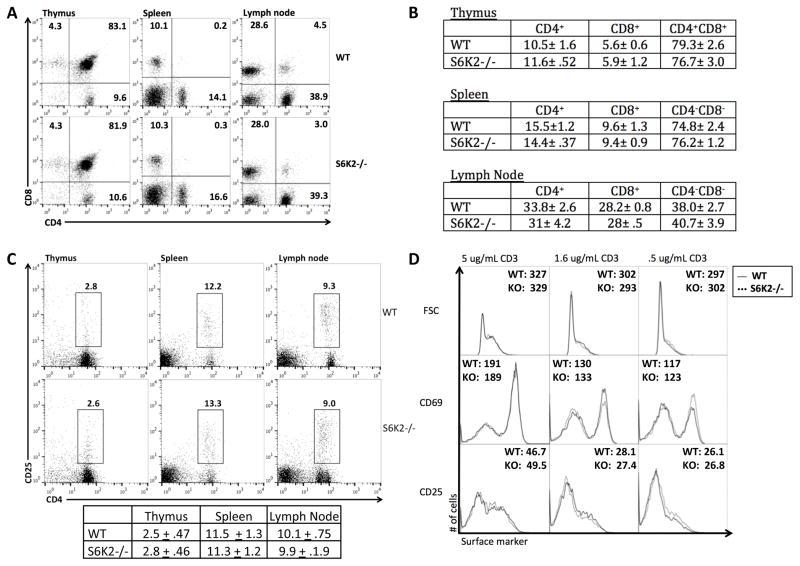
S6K2−/− mice are viable, fertile, and exhibit normal development. They exhibit no evidence of autoimmunity or inflammatory conditions. Using these mice, we sought to determine the role of S6K2 in T cell development and activation. Analysis of peripheral lymphoid organs in S6K2−/− mice revealed normal percentages and ratios of CD4+ and CD8+ populations (Fig. 1A and 1B), and developing T cell subpopulations in the thymus were indistinguishable (Fig. 1A and 1B). Natural Treg (nTreg) populations (CD4+CD25+) were also comparable in percentage in the peripheral lymphoid organs (Fig. 1C). The suppressive ability of nTregs was comparable when comparing CD4+CD25+ cells sorted from WT versus S6K2−/− mice (Supplemental Fig. 1A).
Figure 1.
S6K2−/− mice have normal T cell compartments and CD4+ T cells display normal activation marker upregulation. (A) Flow cytometry analysis of CD4+ and CD8+ populations in the thymus, spleen, and lymph nodes from a representative experiment in (B). (B) Quantitative analysis of CD4+, CD8+, and CD4+CD8+ populations in the thymus, spleen, and lymph nodes (numbers indicate millions, mean +/− SEM, n = 3 independent experiments). (C) nTreg populations were analyzed by CD4+CD25+ expression in the thymus, spleen, and lymph nodes. Average percentage mean (from total CD4+ populations) +/− SEM is shown below (n = 3 independent experiments). (D) Flow cytometry analysis of activation markers and cell size (forward scatter; FSC) of WT and S6K2−/− T cells after 24 hr of stimulation using varying concentrations of anti-CD3 under neutral conditions (n = 3 independent experiments).
We then isolated naïve conventional T cells (CD4+CD25−) and stimulated them with anti-CD3 and anti-CD28 for 24h to assess the role of S6K2 in early T cell activation. Expression of the activation markers CD25 and CD69, and the induction of blasting as measured by forward scatter, were similar between wildtype (WT) and S6K2−/− T cells (Fig. 1D). Production of IL-2 after 40hr stimulation was also unimpaired (Supplemental Fig. 1B).
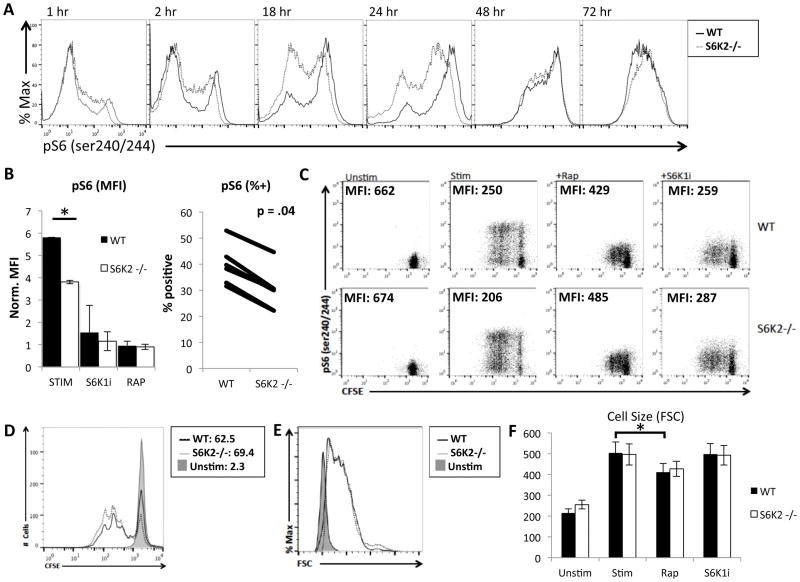
Upon TCR engagement, the enzymes PI3K and Akt are essential for the induction of numerous signaling cascades (4). mTORC1 is a main effector downstream of PI3K and Akt, yet little is known about the roles of S6Ks in these processes. First we assessed the contribution of S6K2 to total S6K activity. Naïve T cells were stimulated for different times and analyzed by flow cytometry to measure phosphorylation of the S6 ribosomal protein (pS6). Phosphorylation of S6 at the Ser 240 and 244 residues (pS6) is S6K-dependent and a robust readout of mTORC1 and S6K activity (37, 40). This analysis revealed that S6K2−/− T cells have a reduction in the fraction of cells with high pS6 at early time points, and a delayed increase in pS6 positive cells when compared to WT T cells (Fig. 2A). Focusing on the 24hr time point, we observed a significant reduction in both the MFI and fraction of the population positive for pS6 in S6K2−/− cells (Fig. 2B). Rapamycin was used as a positive control to show that the pS6 measurements were completely mTORC1-dependent. We also assessed the contribution of S6K1 to this response using the selective inhibitor LY2584702 (here termed S6K1i) (41). We used S6K1i instead of S6K1−/− mice because of the reported compensatory upregulation of S6K2 in such mice (30). The results showed that S6K1 inhibition strongly suppressed the activation-induced increase in pS6 in both WT and S6K2−/− T cells.
Figure 2.
S6K2−/− CD4 T cells have reduced S6 phosphorylation during the first 24 hours of stimulation compared to WT, but have comparable proliferation and cell size under neutral stimulation conditions. (A) Histogram overlays of p-S6 comparing WT and S6K2−/− CD4 T cells during a time course of neutral stimulation, representative of 3 independent experiments. (B) Left, Graph of normalized average mean fluorescence intensities (MFIs) of p-S6 in 24hr-activated CD4 T cells, * p < 0.05 (n = 5 independent experiments). Error bars indicate SEM. Right, The percentage of p-S6-positive T cells was graphed for 6 independent experiments comparing WT and S6K2−/− CD4 T cells after 24hr stimulation. The Wilcoxon signed-rank test was used to assess p value. (C) CFSE and p-S6 staining of WT and S6K2−/− naïve CD4+ T cells unstimulated or stimulated for 3 days, representative of 3 independent experiments. (D) CFSE histogram overlay of WT and S6K2−/− T cells from (C). (E) Cell size histogram overlay WT and S6K2−/− naïve CD4+ T cells stimulated for 24 hours. (F) Average FSC MFI plotted from (E) is shown, * p < 0.05 (n = 3 independent experiements). Error bars indicate SEM.
We recently demonstrated that S6Ks are surprisingly dispensable for both growth and division in activated T cells (36). Confirming these findings, S6K2−/− T cells showed comparable proliferation to WT T cells at 72hr (Fig. 2C and D). At an earlier time point of 40hr, there was no difference in cell cycle distribution between WT and S6K2−/− T cells (Supplemental Fig. 1C). Addition of S6K1i to both S6K2−/− and WT T cells also did not impair their proliferation despite fully suppressing pS6 induction (Fig. 2C). Similarly, there was no difference in growth (cell size measured by forward scatter (FSC)) in WT and S6K2−/− T cells treated with or without the S6K1i inhibitor (Fig. 2E and 2F). We also tested allogeneic stimulation by mixed lymphocyte reaction and also observed no difference in proliferation or cell death (data not shown). These results show that in the absence of S6K activity, T cells are still able to grow and proliferate under neutral stimulation conditions.
Distinct signaling roles for S6K1 and S6K2
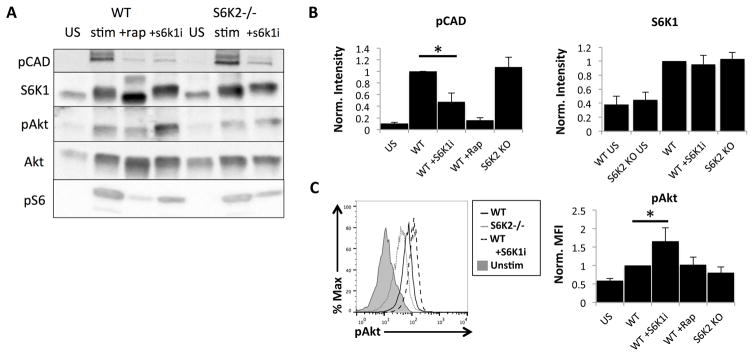
The first study of S6K1−/− mice reported that S6K1-deficient lymphocytes have a compensatory upregulation of S6K2, even under resting conditions (30). To determine whether S6K2−/− T cells have altered expression of S6K1, we used Western blots to measure total S6K1 in resting and 24h stimulated T cells. S6K1 expression in the S6K2−/− was similar to WT T cells under basal and activated conditions (Fig. 3A and 3B). This confirms that S6K1 is not upregulated to compensate for loss of S6K2 in the S6K2−/− T cells.
Figure 3.
S6K2 and S6K1 have distinct substrates in activated T cells and have different roles in feedback to Akt. (A) Immunoblot analysis of lysates from WT and S6K2−/− naïve CD4+ T cells unstimulated or stimulated for 24 hours, representative of 3 independent experiments. Rapamycin (+rap) or S6K1 inhibitor (S6Ki) was included during stimulation in the samples indicated. Antibodies used to probe the blots are shown on the left. (B) Analysis of phospho CAD and S6K1 expression from immunoblots, mean +/− SEM, n = 3 independent experiments. *p < 0.05. (C) Intracellular phosphoflow staining of Akt and analysis by normalized MFI, mean +/− SEM, n = 3 independent experiments. *p < 0.05.
In many cells, S6K1 mediates strong negative feedback on growth factor receptor signaling and the activation of PI3K and AKT (42). Consequently, inhibition of mTORC1 or S6K1 often leads to increased phosphorylation of Akt (pAkt). In accord, WT naïve T cells treated with S6K1i had a statistically significant increase in pAkt following TCR stimulation (Fig. 3C). This increase was not observed in S6K2−/− T cells, which instead showed a variable reduction in pAkt. These results are in accord with a study of breast cancer cells, which reported reduced pAkt when S6K2 was suppressed (43). We also measured phosphorylated CAD, reported to be a selective substrate of S6K1 (44, 45). CAD is essential for the de novo synthesis of pyrimidine nucleotides and is directly phosphorylated by S6K1 at Ser1859. Though S6K1i significantly reduced pCAD in activated WT T cells, loss of S6K2 had no apparent effect (Fig. 3A and 3B). These signaling studies support the conclusion that S6K1 and S6K2 have distinct substrates in activated T cells and contribute differently to feedback pathways.
S6K2−/− mice generate more Tregs and fewer Th17s in vitro
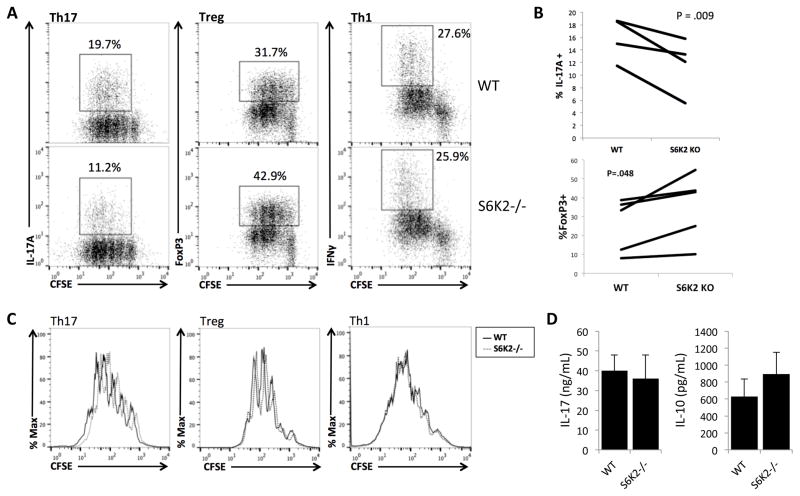
Although S6K2−/− T cells proliferated normally under neutral stimulation conditions, we considered the possibility that S6K2 deficiency impairs proliferation and differentiation under T helper skewing conditions. For these in vitro differentiation assays, highly purified naïve CD4+CD25− T cells were labeled with CFSE and activated with under conditions that polarize cells towards Th1, Th17, or Treg differentiation. On day 4, the cells were harvested and stained for intracellular cytokines or transcription factors indicative of each subset. Though S6K2 loss did not affect skewing to the Th1 subset, the percentage of IL-17-producing cells was reduced under Th17 culture conditions when compared to WT T cells (Fig. 4A and 4B). Conversely, S6K2−/− T cells showed an increased percentage of FoxP3+ cells when stimulated under Treg skewing conditions (Fig. 4 and 4B). Importantly, S6K2−/− T cells showed unimpaired IL-2 production (Supplemental Fig. 1B) and cell cycle distribution at 40hr (Supplemental Fig. 1D), and proliferation was comparable to WT T cells at 72hr for both Th17 and Treg subsets (Fig. 4C), indicating that altered differentiation was not linked to changes in cell division.
Figure 4.
Naïve S6K2−/− CD4+ T cells generate fewer Treg and more Th17 cells, with little effect on Th1, Th17, or Treg proliferation. (A) Intracellular staining of IL-17A, FoxP3, and IFNγ in WT and S6K2−/− T cells stained with CFSE for cell division tracking. Naïve WT or S6K2−/− CD4+ T cells were cultured in RPMI media under indicated conditions for 4 days. Shown plots are representative of data analyzed in (B). (B) Paired analysis of IL-17A and IFNγ positive cells from multiple experiments performed as in (A) (Th17: n = 4 independent experiments, Treg: n = 5 independent experiments). The Wilcoxon signed-rank test was used to assess p value. (C) Histogram overlays of CFSE-labeled T cells from (A). (D) IL-17 and IL-10 cytokine measurements by ELISA from the supernatant of WT and S6K2−/− T cells cultured in Th17 or Treg skewing conditions for 4 days (mean +/− SEM, n = 3 independent experiments).
Next we measured IL-10 production under Treg skewing conditions and IL-17A production under Th17 conditions, 4 days after stimulation. WT and S6K2−/− T cells produced comparable amounts of each cytokine, though there was a trend towards increased IL-10 production by S6K2−/− T cells (Fig. 4D).
S6K2 deficiency does not ameliorate EAE clinical symptoms
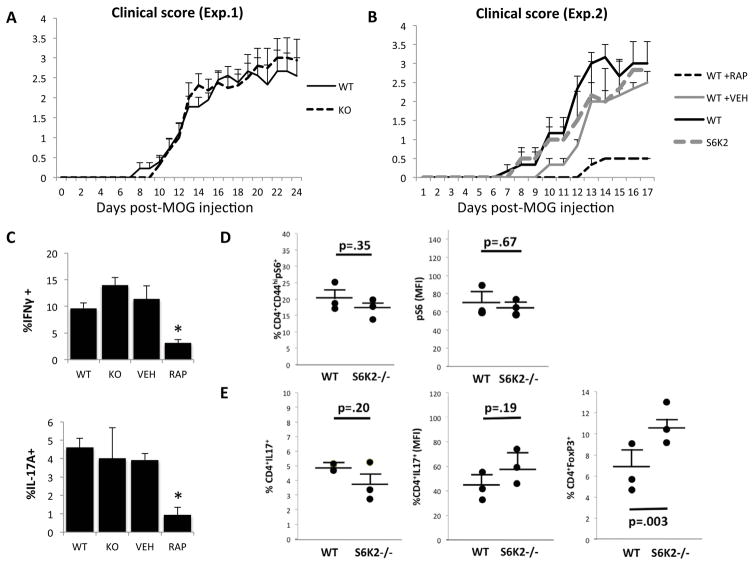
Drawing upon our findings of reduced Th17 and increased Treg generation in vitro, we tested whether S6K2−/− mice were protected from autoimmune disease in a mouse model. We chose MOG35-55 induced experimental autoimmune encephalomyelitis (EAE), a classical mouse model of multiple sclerosis (MS) in which Th17 cells are required and mTORC1 activity plays a prominent role (13, 14, 46, 47). The pathogenesis of EAE is strongly reinforced by a Th17/Treg imbalance (48). Surprisingly, disease onset and severity was comparable in S6K2−/− and WT mice (Fig. 5A). We also quantitated cytokine production of infiltrating lymphocytes in the spinal cord and found similar percentages of IFNγ and IL-17A producing CD4+ T cells (Supplemental Fig. 1E). To confirm the role of mTORC1 in our hands, we conducted a second EAE experiment comparing WT mice treated with rapamycin or vehicle. As reported (13, 19), rapamycin treatment significantly lowered EAE scores (Fig. 5B) and reduced the percentage of CD4 T cells producing IFNγ or IL-17A in the spinal cord (Fig. 5C). Concurrently, we analyzed cohorts of WT and S6K2−/− mice that were not injected with rapamycin or vehicle. Again the absence of S6K2 did not affect EAE onset or severity (Fig. 5B), and did not affect inflammatory cytokine production in CD4+ T cells isolated from spinal cord infiltrates (Fig. 5C).
Figure 5.
S6K2−/− and WT mice exhibit comparable EAE development. (A) Clinical scores of EAE-induced WT and S6K2−/− mice (mean +/− SEM, n = 8 for each group). (B) Clinical scores of EAE-induced WT, WT treated with Rap or vehicle (Veh), and S6K2−/− mice (mean +/− SEM, n = 3 for each group). (C) Graphed analysis of IFNγ and IL-17A positive CD4+ T cells in the spinal cord of EAE-induced mice from (B), * p < 0.05 (mean +/− SEM, n = 3). (D) pS6 (MFI and % positive) in CD4+ T cells from freshly harvested lymph nodes from 10 day MOG immunized WT and S6K2−/− mice (mean +/− SEM, n = 3). (E) CD4+IL17A+ (% positive and MFI) and CD4+FoxP3+ (% positive) populations in 3 day MOG restimulated splenocytes harvested from the same mice as used in (D) (mean +/− SEM, n = 3).
Because we did not detect any differences in proinflammatory responses at the peak of disease, MOG immunization was also used to characterize T effector and Treg responses in vivo during immune response development. Cohorts of WT and S6K2−/− mice were immunized with MOG35-55 without pertussis to characterize T cell differentiation. This model allows assessment of antigen-specific immune responses to complement polyclonal in vitro differentiation assays. After 10 days, the mice were sacrificed and lymph nodes were harvested to measure pS6 amounts in CD4+CD44hi T cells. WT and S6K2−/− cells had similar pS6 as measured by both MFI and % positive (Fig. 5D). Splenocytes were restimulated with MOG peptide in cultures that included IL-23 and anti-IFNγ antibody for 3 days, to specifically measure frequencies of MOG-specific Th17 and Treg cells. Though the percentage and MFI of CD4+IL17A+ T cells were no different, CD4+FoxP3+ populations was significantly increased in cultures from S6K2−/− mice (Fig. 5E). IL-17 production measurement by ELISA also showed no difference between WT and S6K2−/− mice (Supplemental Fig. 1F). In summary, our data suggest that S6K2 is not required for in vivo Th17 differentiation, but does seems to suppress Treg generation in this model.
The role of S6K2 in Th17 differentiation is context-specific
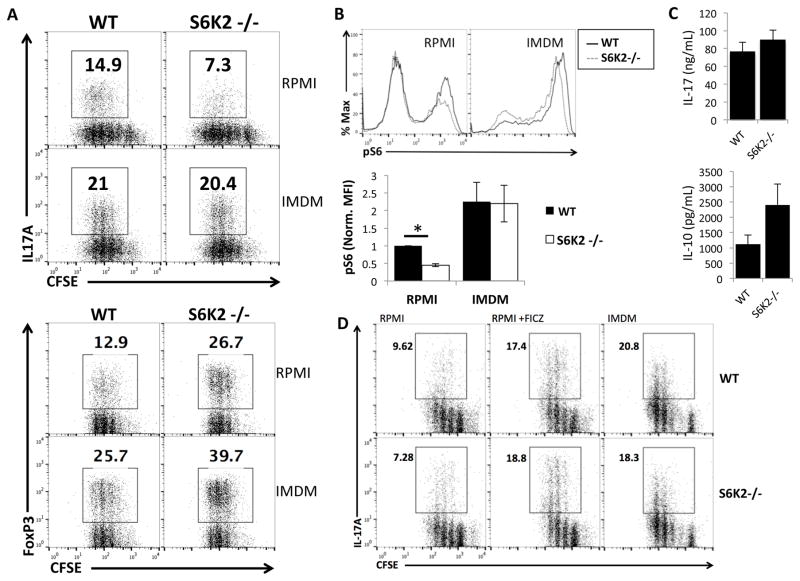
To understand why significant in vitro differences between S6K2−/− and WT T cells did not translate into altered Th17 differentiation in vivo, we reassessed the cell culture media used for T cell differentiation. We initially used RPMI media, which is commonly used for primary lymphocyte culture but supports very low levels of Th17 differentiation (39). Thus, we compared RPMI to IMDM, the latter a medium rich in aromatic amino acids that was shown to support robust Th17 differentiation (39). Not only did IMDM media enhance Th17 skewing in WT T cells, it also restored Th17 differentiation of S6K2−/− cells to levels equivalent to WT (Fig. 6A and Supplemental Fig. 2A). In contrast to the rescue of Th17 differentiation, S6K2−/− T cells in IMDM still generated a higher percentage of FoxP3+ cells under Treg skewing conditions compared to WT (Fig. 6A). IL-17A and IL-10 cytokine production under Th17 and Treg skewing conditions, respectively, were comparable between WT and S6K2−/− T cells (Fig. 6C).
Figure 6.
IMDM rescues Th17 differentiation in S6K2−/− T cells. (A) Intracellular staining of IL-17A and FoxP3 in WT and S6K2−/− T cells stained with CFSE. Naïve WT or S6K2−/− CD4+ T cells were cultured under indicated conditions for 4 days in RPMI or IMDM. Plots are representative of 3 independent experiments. (B) Comparative flow cytometric analysis and quantitative analysis for normalized MFI of p-S6 signaling between WT and S6K2−/− T cells 24 hr of neutral stimulation, *p < 0.05 (mean +/− SEM, n = 3 independent experiments). (C) IL-17 and IL-10 cytokine measurements by ELISA from the supernatant of WT and S6K2−/− T cells cultured in Th17 or Treg skewing conditions for 4 days (mean +/− SEM, n = 3 independent experiments). (D) Intracellular staining of IL-17A in CFSE-labeled WT and S6K2−/− T cells cultured in Th17 skewing conditions for 4 days in RPMI, RPMI +FICZ, or IMDM. Plots are representative of 3 independent experiments.
The tryptophan metabolite 6-formylindolo[3,2-b] carbazole (FICZ) found in IMDM can enhance differentiation of Th17 cells in RPMI media (39). Therefore, we supplemented RPMI with 300 nm FICZ to see if this could account for restored Th17 differentiation in S6K2−/− T cells. Indeed, the proportion of IL-17+ cells was comparable between WT and S6K2−/− T cells when cultured in RPMI plus FICZ (Fig. 6C and Supplemental Fig. 2B). Proliferation, however, was not restored to the robust response obtained using IMDM (Supplemental Fig. 2C). To further explore these media-based phenotypic differences, we measured mTORC1 signaling output as assessed by pS6 intracellular staining. 24hr after T cell stimulation under neutral conditions, cells cultured in IMDM exhibited significantly higher mTORC1 activity compared to cells cultured in RPMI, as assessed by the MFI of pS6 as well as the proportion of pS6-high cells (Fig. 6B). This difference was observed in both S6K2−/− and WT populations.
A previous study by Kurebayashi et al. reported that S6K2 functions downstream of mTORC1 to shuttle the Th17 transcription factor RORγ to the nucleus (14). This study did not assess the role of RORγ localization in Th17 differentiation in S6K2−/− T cells. Therefore, we measured nuclear localization of RORγ in WT and S6K2−/− T cells after 40hr of stimulation in Th17 conditions, comparing both RPMI and IMDM media. In RPMI, S6K2−/− T cells indeed showed significantly reduced RORγ nuclear localization when compared to WT T cells (Supplemental Fig. 3A). Culturing cells in IMDM media resulted in significantly increased nuclear localization for S6K2−/− T cells, but did not rescue this to WT levels (Supplemental Fig. 3B). WT cells cultured in IMDM did not exhibit an increase in RORγ nuclear localization compared to RPMI conditions.
Because mTORC1 activity is strongly tied to cell metabolism, we used a Seahorse metabolic flux analyzer to analyze metabolism in both WT and S6K2−/− cells activated in Th17, Treg, and neutral (Tneu) differentiation conditions. In these experiments, initial stimulation was done in IMDM media for Th17 conditions and RPMI for Tneu and Treg conditions. Interestingly, basal glycolysis (extracellular acidification rate (ECAR)) and oxidative phosphorylation (oxygen consumption rate (OCR)) of all three subsets were not significantly different (Supplemental Fig. 4A and 4B). Rapamycin treated WT cells were used as a positive control in reducing both ECAR and OCR of Tneu stimulated cells (49).
Discussion
S6K1 and S6K2 are canonical mTORC1 substrates whose inhibition has potential to modulate T helper differentiation and treat T-cell mediated autoimmune diseases. Indeed, S6K1 knockdown reduces Th17 differentiation in both WT cells and in cells with hyperactive mTORC1 resulting from TSC1 deficiency (17). Although S6K2 is also prominently expressed in lymphocytes (29), the role of this isoform in T cells has not been investigated. Here we used S6K2−/− mice to assess the impact of S6K2 deficiency on T cell development, proliferation and differentiation. Previous genetic studies of T cells lacking mTOR pathway components (i.e. mTOR, raptor, and Rheb) have generally reported no changes in early T cell development and homeostasis. Our data show similarly that S6K2−/− T cells develop normally and that CD4+ T cells have no defects in early activation. This allowed us to ensure that functional studies were unaffected by preexisting cell-intrinsic defects. Proliferation of S6K2−/− T cells was unimpaired under neutral stimulation conditions or when polarized to Th1, Th17 or Treg subsets. Analysis of T helper differentiation in vitro showed a modest but significant decrease in Th17 differentiation using RPMI media, but not in IMDM. Using MOG35-55-induced EAE as an in vivo model of Th17-driven autoimmunity, we compared WT and S6K2−/− mice and observed comparable clinical scores and Th17 cell percentages in the spinal cord. Utilizing the same model to study Th17 and Treg responses to a specific antigen in vivo, we gated on CD4+CD44hi infiltrating T cells in the cervical lymph nodes and found no change in pS6 signaling, which corresponds with the in vitro signaling data in cells cultured in IMDM. We also restimulated splenocytes with MOG and detected no difference between WT and S6K2−/− mice in IL-17A expression. However, there was a significant increase in CD4+FoxP3+ populations. These findings suggest that S6K2 may play a negative role in Treg generation, but is not a key driver of Th17 differentiation or autoimmune pathology in vivo, at least in this EAE model. Moreover, the discordant findings using RPMI media compared to IMDM and to the EAE model emphasize the importance of using optimal in vitro activation conditions for assessing Th17 differentiation.
Signaling measurements indicate a lesser role for S6K2 compared to S6K1 in phosphorylation events triggered by TCR/CD3 engagement. The phosphorylation of S6 protein was modestly reduced within 24hr of activation, indicating that S6K2 contributes significantly to the overall S6K signaling output. However, S6K1 blockade with the selective inhibitor LY2584702 caused a stronger inhibition of pS6 at all time points assessed. Furthermore, S6K1 inhibition but not S6K2 loss reduced phosphorylation of CAD, a key enzyme in de novo pyrimidine synthesis (44, 45). mTORC1 and S6K1 mediate negative feedback loops that suppress upstream phosphorylation of Akt in many cellular systems (42, 50, 51). In accord, S6K1i treatment of WT T cells significantly increased pAkt, indicating a removal of feedback signaling. In contrast, pAkt in activated S6K2−/− T cells was similar or slightly reduced compared to WT cells. We also found that S6K1 is not upregulated in the absence of S6K2, whereas a study of S6K1−/− mice reported massive compensatory upregulation of S6K2 in T cells (30). These distinctions further support the conclusion that S6K1 loss has greater impact on T cell biology compared to S6K2 loss. Nevertheless, the fact that S6K2 is expressed in T cells suggests that this kinase has a functional role, prompting us to assess T cell proliferation and differentiation.
S6K2−/− T cells became activated (based on expression of CD25 and CD69 surface markers and production of IL-2) and proliferated (assessed by CFSE dilution at 72hr and cell cycle at 40hr) to a comparable extent as WT cells. In fact, when we fully suppressed S6K function by treating S6K2−/− T cells with S6K1i, proliferation was unimpaired. These data agree with our recent finding that S6Ks are dispensable for lymphocyte proliferation downstream of mTORC1; instead, the 4E-BP/eIF4E axis is the main driver of lymphocyte clonal expansion (36). It is reasonable to propose that the primary role of S6Ks in T cells is to control differentiation rather than proliferation. A role for S6K1 in Th17 differentiation is supported by both gain- and loss-of-function studies (14, 17). S6K2 can promote nuclear localization of RORγT, the Th17 cell hallmark transcription factor (14), but whether this function is required for Th17 differentiation has not been determined. Therefore, we tested S6K2−/− CD4 T cells for the ability to differentiate into Th17 and other Th cell subsets.
Our results show that S6K2 has a context-specific role in Th17 differentiation in vitro. When CD4 T cells were cultured in RPMI media under Th17 polarizing conditions, S6K2−/− cells generated a significantly lower percentage of IL-17A+ cells. This correlated with reduced nuclear localization of RORγT. This finding is consistent with the model that S6K2 and not S6K1 shuttles RORγT to the nucleus as proposed by Kurebayashi et al. (14). Importantly, when cells were cultured in IMDM to support more robust Th17 differentiation (39), both WT and S6K2−/− CD4 T cells generated higher percentages of IL-17A+ cells and there was no difference between the genotypes. The IMDM condition also fully rescued the S6 phosphorylation defect in S6K2−/− T cells, and partially restored nuclear localization of RORγT. It is not clear how the distinct composition of RPMI and IMDM media can affect the wiring of signaling pathways, such as the S6K2-dependence of S6 phosphorylation and RORγT localization. Compared to RPMI, IMDM has increased D-glucose and L-glutamine amounts in addition to higher concentrations of inorganic salts, vitamins, and amino acids. A key difference in IMDM is the greater abundance of aromatic amino acids that are precursors of endogenous agonists for the arylhydrocarbon receptor (AhR) that drives Th17 differentiation (39). It is possible that AhR activity alters gene expression in ways that reprogram T cells to be less dependent on S6K2. In support, addition of an exogenous AhR ligand FICZ corrected the Th17 differentiation defect in S6K2−/− T cells.
We chose to assess Th17 differentiation in vivo using the MOG35-55-induced model of EAE, since chemical and genetic approaches have established a role for mTORC1 in this model (13, 14, 19, 20). As expected, rapamycin treatment muted clinically apparent EAE symptoms and reduced the percentage of IL-17A+ as well as IFNγ+ cells in the spinal cord. However, S6K2−/− mice showed no difference in clinical scores or inflammatory T cells. This result suggests that selective targeting of S6K2 has limited potential for the treatment of T cell-mediated autoimmunity. Another implication of this result is that in vitro Th17 differentiation in IMDM, compared to RPMI, provided a better prediction of in vivo outcomes. Differences in media composition might underlie some of the differences reported among various studies of specific mTORC1 pathway components in T helper differentiation (9, 13, 14).
mTOR inhibition enhances the formation of induced Tregs (13, 15), yet the relevant downstream substrates involved in this effect are not clear. Interestingly, naïve S6K2−/− T cells had an increased propensity to differentiate into FoxP3+ cells under Treg biasing conditions in vitro, regardless of the media used. The increase was consistent across experiments but quantitatively modest, with the percent FoxP3+ cells averaging 26% for WT and 35% for S6K2−/− T cells. In addition, the significant increase seen in Treg generation in 10 days MOG immunized S6K2−/− mice suggests a milder inflammatory model may benefit from inhibition of S6K2. Further studies are needed to determine whether this increase in induced Treg generation influences the severity of other autoimmune disease models and anti-tumor immunity. Notably, S6K2−/− mice had normal numbers of nTregs with similar suppressive potential as WT nTregs.
Both S6K1 and S6K2 are expressed in T cells and contribute to robust phosphorylation of S6 and other substrates following TCR stimulation. However, it remains unclear how these kinases contribute to T cell activation and function. Though Sasaki and colleagues have shown S6K1 knockout mice to have decreased Th17 differentiation in vitro, their EAE studies showed no improvement in clinical scores (38). Taken in consideration with our own in vivo studies suggest S6Ks to not be a main mTORC1 downstream effector in driving EAE pathogenesis. Therefore, the efficacy of rapamycin in the EAE model might be the result of blocking T cell clonal expansion and/or a specific effect on T helper differentiation. We initiated treatment several days after immunization to minimize effects on proliferation. This suggests that rapamycin targets other mTORC1 downstream substrates to produce significantly delayed and improved clinical scores in EAE. It is possible that S6Ks have a more prominent function in other autoimmune conditions or in certain T cell responses to infection.
There is interest in targeting S6K2 for cancer treatment, as over 88% of tumors express higher levels than corresponding healthy tissues (52–56). Selective chemical inhibitors of S6K2 have not yet been described. Should such compounds become available, the data presented here suggest that blockade of S6K2 will not be broadly immunosuppressive compared to rapamycin. Selective inhibition or conditional deletion of S6K1 and S6K2 in various cell types will be necessary to achieve a more detailed understanding of the unique and shared functions of these kinases in different aspects of the immune response.
Supplementary Material
Acknowledgments
The authors thank Yumay Chen and Ping H. Wang (UC Irvine) for use and assistance of the Seahorse XF24 analyzer, and Lindsey Araujo, Adam K. Idica, and Sara E. Lutz for technical advice. We are grateful to Sara Kozma (IDIBELL) for providing the S6K2−/− mouse strain, originally generated at the Friedrich Miescher Institute.
Footnotes
This work was supported by NIH grant R21-AI099656 (to D.A.F.) and National Multiple Sclerosis Society CA1058-A8 (to C.M.W.).
References
- 1.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cybulski N, Hall MN. TOR complex 2: a signaling pathway of its own. Trends Biochem Sci. 2009;34:620–627. doi: 10.1016/j.tibs.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Finlay D, Cantrell DA. Metabolism, migration and memory in cytotoxic T cells. Nat Rev Immunol. 2011;11:109–117. doi: 10.1038/nri2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okkenhaug K. Signaling by the phosphoinositide 3-kinase family in immune cells. Annu Rev Immunol. 2013;31:675–704. doi: 10.1146/annurev-immunol-032712-095946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng H, Chi H. mTOR and lymphocyte metabolism. Curr Opin Immunol. 2013;25:347–355. doi: 10.1016/j.coi.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annu Rev Immunol. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 8.Lawrence JC, Abraham RT. PHAS/4E-BPs as regulators of mRNA translation and cell proliferation. Trends Biochem Sci. 1997;22:345–349. doi: 10.1016/s0968-0004(97)01101-8. [DOI] [PubMed] [Google Scholar]
- 9.Yang K, Shrestha S, Zeng H, Karmaus PW, Neale G, Vogel P, Guertin DA, Lamb RF, Chi H. T cell exit from quiescence and differentiation into Th2 cells depend on Raptor-mTORC1-mediated metabolic reprogramming. Immunity. 2013;39:1043–1056. doi: 10.1016/j.immuni.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michalek RD, V, Gerriets A, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499:485–490. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, Powell JD. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurebayashi Y, Nagai S, Ikejiri A, Ohtani M, Ichiyama K, Baba Y, Yamada T, Egami S, Hoshii T, Hirao A, Matsuda S, Koyasu S. PI3K-Akt-mTORC1-S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORγ. Cell Rep. 2012;1:360–373. doi: 10.1016/j.celrep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Procaccini C, De Rosa V, Galgani M, Abanni L, Calì G, Porcellini A, Carbone F, Fontana S, Horvath TL, La Cava A, Matarese G. An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity. 2010;33:929–941. doi: 10.1016/j.immuni.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park Y, Jin HS, Lopez J, Elly C, Kim G, Murai M, Kronenberg M, Liu YC. TSC1 regulates the balance between effector and regulatory T cells. J Clin Invest. 2013;123:5165–5178. doi: 10.1172/JCI69751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Branisteanu DD, Mathieu C, Bouillon R. Synergism between sirolimus and 1,25-dihydroxyvitamin D3 in vitro and in vivo. J Neuroimmunol. 1997;79:138–147. doi: 10.1016/s0165-5728(97)00116-1. [DOI] [PubMed] [Google Scholar]
- 19.Esposito M, Ruffini F, Bellone M, Gagliani N, Battaglia M, Martino G, Furlan R. Rapamycin inhibits relapsing experimental autoimmune encephalomyelitis by both effector and regulatory T cells modulation. J Neuroimmunol. 2010;220:52–63. doi: 10.1016/j.jneuroim.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Donia M, Mangano K, Amoroso A, Mazzarino MC, Imbesi R, Castrogiovanni P, Coco M, Meroni P, Nicoletti F. Treatment with rapamycin ameliorates clinical and histological signs of protracted relapsing experimental allergic encephalomyelitis in Dark Agouti rats and induces expansion of peripheral CD4+CD25+Foxp3+ regulatory T cells. J Autoimmun. 2009;33:135–140. doi: 10.1016/j.jaut.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Lisi L, Navarra P, Cirocchi R, Sharp A, Stigliano E, Feinstein DL, Dello Russo C. Rapamycin reduces clinical signs and neuropathic pain in a chronic model of experimental autoimmune encephalomyelitis. J Neuroimmunol. 2012;243:43–51. doi: 10.1016/j.jneuroim.2011.12.018. [DOI] [PubMed] [Google Scholar]
- 22.Carlson RP, Baeder WL, Caccese RG, Warner LM, Sehgal SN. Effects of orally administered rapamycin in animal models of arthritis and other autoimmune diseases. Ann N Y Acad Sci. 1993;685:86–113. doi: 10.1111/j.1749-6632.1993.tb35855.x. [DOI] [PubMed] [Google Scholar]
- 23.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perl A. mTOR activation is a biomarker and a central pathway to autoimmune disorders, cancer, obesity, and aging. Ann N Y Acad Sci. 2015;1346:33–44. doi: 10.1111/nyas.12756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perl A. Activation of mTOR (mechanistic target of rapamycin) in rheumatic diseases. Nat Rev Rheumatol. 2016;12:169–182. doi: 10.1038/nrrheum.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai ZW, Borsuk R, Shadakshari A, Yu J, Dawood M, Garcia R, Francis L, Tily H, Bartos A, Faraone SV, Phillips P, Perl A. Mechanistic target of rapamycin activation triggers IL-4 production and necrotic death of double-negative T cells in patients with systemic lupus erythematosus. J Immunol. 2013;191:2236–2246. doi: 10.4049/jimmunol.1301005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer KE, Gelfond JA, Soto VY, Han C, Someya S, Richardson A, Austad SN. Health Effects of Long-Term Rapamycin Treatment: The Impact on Mouse Health of Enteric Rapamycin Treatment from Four Months of Age throughout Life. PLoS ONE. 2015;10:e0126644. doi: 10.1371/journal.pone.0126644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pallet N, Legendre C. Adverse events associated with mTOR inhibitors. Expert Opin Drug Saf. 2013;12:177–186. doi: 10.1517/14740338.2013.752814. [DOI] [PubMed] [Google Scholar]
- 29.Nardella C, Lunardi A, Fedele G, Clohessy JG, Alimonti A, Kozma SC, Thomas G, Loda M, Pandolfi PP. Differential expression of S6K2 dictates tissue-specific requirement for S6K1 in mediating aberrant mTORC1 signaling and tumorigenesis. Cancer Res. 2011;71:3669–3675. doi: 10.1158/0008-5472.CAN-10-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma SC. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 1998;17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelletier J, Graff J, Ruggero D, Sonenberg N. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res. 2015;75:250–263. doi: 10.1158/0008-5472.CAN-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung J, Kuo CJ, Crabtree GR, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 33.Kuo CJ, Chung J, Fiorentino DF, Flanagan WM, Blenis J, Crabtree GR. Rapamycin selectively inhibits interleukin-2 activation of p70 S6 kinase. Nature. 1992;358:70–73. doi: 10.1038/358070a0. [DOI] [PubMed] [Google Scholar]
- 34.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salmond RJ, Brownlie RJ, Meyuhas O, Zamoyska R. Mechanistic Target of Rapamycin Complex 1/S6 Kinase 1 Signals Influence T Cell Activation Independently of Ribosomal Protein S6 Phosphorylation. J Immunol. 2015;195:4615–4622. doi: 10.4049/jimmunol.1501473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.So L, Lee J, Palafox M, Mallya S, Woxland C, Arguello M, Truitt M, Sonenberg N, Ruggero D, Fruman DA. The 4E-BP/eIF4E axis controls rapamycin-sensitive growth and proliferation in lymphocytes. 2016 doi: 10.1126/scisignal.aad8463. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. S6K1(−/−)/S6K2(−/−) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasaki CY, Chen G, Munk R, Eitan E, Martindale J, Longo DL, Ghosh P. p(70S6K1) in the TORC1 pathway is essential for the differentiation of Th17 Cells, but not Th1, Th2, or Treg cells in mice. Eur J Immunol. 2016;46:212–222. doi: 10.1002/eji.201445422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veldhoen M, Hirota K, Christensen J, O’Garra A, Stockinger B. Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J Exp Med. 2009;206:43–49. doi: 10.1084/jem.20081438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez OD, Nolan GP. Phospho-proteomic immune analysis by flow cytometry: from mechanism to translational medicine at the single-cell level. Immunol Rev. 2006;210:208–228. doi: 10.1111/j.0105-2896.2006.00364.x. [DOI] [PubMed] [Google Scholar]
- 41.Tolcher A, Goldman J, Patnaik A, Papadopoulos KP, Westwood P, Kelly CS, Bumgardner W, Sams L, Geeganage S, Wang T, Capen AR, Huang J, Joseph S, Miller J, Benhadji KA, Brail LH, Rosen LS. A phase I trial of LY2584702 tosylate, a p70 S6 kinase inhibitor, in patients with advanced solid tumours. Eur J Cancer. 2014;50:867–875. doi: 10.1016/j.ejca.2013.11.039. [DOI] [PubMed] [Google Scholar]
- 42.Wan X, Harkavy B, Shen N, Grohar P, Helman LJ. Rapamycin induces feedback activation of Akt signaling through an IGF-1R-dependent mechanism. Oncogene. 2007;26:1932–1940. doi: 10.1038/sj.onc.1209990. [DOI] [PubMed] [Google Scholar]
- 43.Sridharan S, Basu A. S6 kinase 2 promotes breast cancer cell survival via Akt. Cancer Res. 2011;71:2590–2599. doi: 10.1158/0008-5472.CAN-10-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- 45.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eisenstein EM, Williams CB. The T(reg)/Th17 cell balance: a new paradigm for autoimmunity. Pediatr Res. 2009;65:26R–31R. doi: 10.1203/PDR.0b013e31819e76c7. [DOI] [PubMed] [Google Scholar]
- 49.Pollizzi KN, Waickman AT, Patel CH, Sun IH, Powell JD. Cellular size as a means of tracking mTOR activity and cell fate of CD4+ T cells upon antigen recognition. PLoS ONE. 2015;10:e0121710. doi: 10.1371/journal.pone.0121710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, Marto JA, Sabatini DM. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villén J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pérez-Tenorio G, Karlsson E, Waltersson MA, Olsson B, Holmlund B, Nordenskjöld B, Fornander T, Skoog L, Stål O. Clinical potential of the mTOR targets S6K1 and S6K2 in breast cancer. Breast Cancer Res Treat. 2011;128:713–723. doi: 10.1007/s10549-010-1058-x. [DOI] [PubMed] [Google Scholar]
- 53.Filonenko VV, Tytarenko R, Azatjan SK, Savinska LO, Gaydar YA, Gout IT, Usenko VS, Lyzogubov VV. Immunohistochemical analysis of S6K1 and S6K2 localization in human breast tumors. Exp Oncol. 2004;26:294–299. [PubMed] [Google Scholar]
- 54.Lyzogubov VV, Lytvyn DI, Dudchenko TM, Lubchenko NV, Pogrybniy PV, Nespryadko SV, Vinnitska AB, Usenko VS, Gout IT, Filonenko VV. Immunohistochemical analysis of S6K1 and S6K2 expression in endometrial adenocarcinomas. Exp Oncol. 2004;26:287–293. [PubMed] [Google Scholar]
- 55.Pardo OE, Wellbrock C, Khanzada UK, Aubert M, Arozarena I, Davidson S, Bowen F, Parker PJ, Filonenko VV, Gout IT, Sebire N, Marais R, Downward J, Seckl MJ. FGF-2 protects small cell lung cancer cells from apoptosis through a complex involving PKCepsilon, B-Raf and S6K2. EMBO J. 2006;25:3078–3088. doi: 10.1038/sj.emboj.7601198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pardo OE, Seckl MJ. S6K2: The Neglected S6 Kinase Family Member. Front Oncol. 2013;3:191. doi: 10.3389/fonc.2013.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.