Abstract
Cell-matrix adhesion complexes are multi-protein structures linking the extracellular matrix (ECM) to the cytoskeleton. They are essential to both cell motility and function by bidirectionally sensing and transmitting mechanical and biochemical stimulations. Several types of cell-matrix adhesions have been identified and they share many key molecular components, such as integrins and actin-integrin linkers. Mechanochemical coupling between ECM molecules and the actin cytoskeleton has been observed from the single cell to the single molecule level and from immune cells to neuronal cells. However, the mechanisms underlying force regulation of integrin-mediated mechanotransduction still need to be elucidated. In this review article, we focus on integrin-mediated adhesions and discuss force regulation of cell-matrix adhesions and key adaptor molecules, three different force-dependent behaviors, and molecular mechanisms for mechanochemical coupling in force regulation.
Keywords: Mechanotransduction, Cell-matrix adhesion, Integrin, Talin, Vinculin, Actin, Force regulation
1. Introduction
Cell–matrix adhesions are discretely distributed on the cell surface and mediate cell interactions with the ECM. They are essential for cellular functions, such as cell rolling, migration, differentiation, tissue remodeling and homeostasis [1]. The adhesions can either be quickly disassembled or progressively matured from focal complexes to fibrillar adhesions in changing the shapes and molecular compositions of the adhesion complexes. Mechanical force plays a key role in adhesion maturation [2]. During the past two decades researchers have focused on force regulation of molecular interactions including integrins and adaptor molecules involved in cell-matrix interactions. Along with the development of sophisticated single molecule experimental techniques, molecular and structural insights into the mechanical regulation of cell-matrix adhesions have been provided [3].
Before we explore the details of force regulation and transmission at the adhesion sites, we first describe characteristics of different integrin-mediated adhesions. Four different cell-matrix adhesion structures have been defined in cultured cells: focal complexes (nascent adhesions), focal adhesions (FA), fibrillar adhesions, and three-dimensional (3D) matrix adhesions [4, 5]. Nascent adhesions are force-independent and are transient adhesion structures, which are formed at the periphery of spreading or migrating cells in early stages of cell attachment. They can mature into a force-dependent FA when proper linkages are established with the actin cytoskeleton, and the typical size of nascent adhesions is less than 0.2 μm2 [6]. Compared to nascent adhesions, FAs are relatively stable and highly regulated structures by modulating association and dissociation of focal adhesion proteins. FAs can further develop into fibrillar adhesions, which are more stable structures. Recent studies have shown that cells in a 3D matrix interact with the environment with distinguished properties and behaviors compared to that of the adhesions formed in the two-dimensional (2D) matrix. This type of cell-matrix adhesion is called 3D matrix adhesion [4, 7].
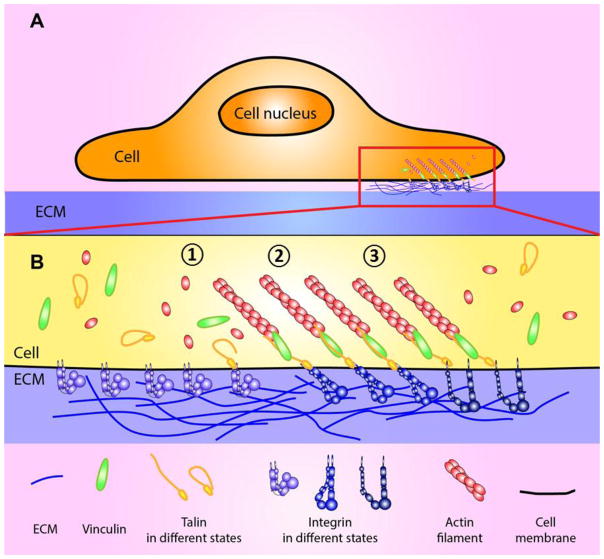
In different stages of cell-matrix interactions, the composition of adhesions and adaptor molecules varies (Fig. 1). Focal complexes are multi-molecular structures that include integrin, talin, paxillin, vinculin, and actin [8–12]. Focal complexes develop into FA by recruiting zyxin [9, 13, 14]; and tyrosine phosphorylation is also required for the recruitment and further maturation of FAs [15, 16]. FAs typically disassemble within 10–20 minutes, however when α5β1-integrin adheres to a fibronectin matrix, FAs further mature into fibrillar adhesions as they elongate the shape of adhesion sites and change the molecular compositions. Stress-fibers are anchored on adhesion sites and the sites move toward to the center of the cell. Fibrillar adhesions specifically contain α5β1 integrin and tensin to form a very stable α5β1-tensin complex, which is scarcely observed in the migrating cell [17, 18]. Throughout the maturation of cell-matrix adhesions, integrins are key players. Integrins are a family of transmembrane proteins: the ectodomain of integrin binds to its ECM ligands and the cytoplasmic tail links to the cytoskeleton via adapter molecules including talin, α-actinin, vinculin, and tensin, thereby connecting the intracellular cytoskeletons to the ECM through the adhesion sites and transmitting bidirectional signals (Table 1).
Figure 1. Schematic of the integrated mechanotransduction at the integrin-mediated adhesion site.
(A) Interaction of cell with the ECM, which is mediated by a multi-protein complex, FA. The red rectangle is enlarged in B. (B) A widely accepted model for force-mediated FA maturation with key adaptor molecules. Integrin interacts with its ECM ligands (e.g. fibronectin) and it triggers signal transduction of followings: (1) Integrin in inactivated state. Talin is recruited to the FA site and starts interacting with integrin β3-tail. Vinculin and other adaptor molecules are also recruited and concentration of free G-actin is accumulated at the adhesion site. (2) Integrin engages with its ligand, and transition to intermediate state. F-actin polymerization is accelerated and force is loaded on the linkage of integrin-adaptor molecule to the actin cytoskeleton. Force-activatable molecules (e.g. vinculin and talin) in the adhesion complex sense the externally applied force and modulate their activation and affinity states for their ligands to transduce the mechanical signaling. (3) Conformational activation of integrin, an extended-open high-affinity state, is fully induced by talin-β-integrin tail binding, integrin ligand binding to integrin head piece, or force applied on integrin. The activation results in modulation of interaction kinetics for its ECM ligands to further develop FAs. FA is matured and stabilized at this stage.
Table 1. Adaptor molecules involved in integrin-mediated mechanotransduction at FAs.
Integrins transmit mechanical outside-in and inside-out signaling via adaptor molecules, which are recruited to the adhesion sites under mechanical force loading conditions.
| Protein | Functions | Force-dependent behavior | Ref |
|---|---|---|---|
| Integrins | Transmembrane receptor Linkage of the actin cytoskeleton to the ECM |
Clustering at FAs Conformational activation with high-affinity for its ligands |
|
| Zyxin | LIM protein Localized at FAs and along the actin cytoskeleton |
Recruiting Ena/VASP and α-actinin Enhancing actin polymerization in cooperation with Ena/VASP |
1*,3* |
| Tensin | Linkage of integrins to the actin cytoskeleton during FA maturation | Maturing FAs into fibrillar adhesion by inducing a5b1 integrin translocation | |
| Talin | Linkage of integrins to F-actin at FAs | Involving in integrin activation by binding to the integrin β-tail Conformational activation with high-affinity for its ligands (e.g. exposing cryptic VBSs) |
39, 40, 41, 2* |
| Paxillin | Interacting with tyrosine kinases (e.g. Src and FAK) and adaptor molecules (e.g. vinculin and talin) | Stabilizing FAs as force increases Recruiting FAK | 4* |
| Vinculin | Linkage of integrin-talin complex to F-actin Binding alternately to tlain or α-actinin |
Conformational activation with regulation of affinity for the ligands (e.g. talin, α-actinin and F-actin) | |
| α-actinin | Linkage of integrin to the actin cytoskeleton via interaction with vinculin, zyxin and F-actin | Stabilizing FAs as tensile force increases | 3*, 4* |
| filamin | Crosslinking two F-actins into an orthogonal structure Interacting with integrin |
Conformational activation with regulation of affinity for the ligands (e.g. integrin) | 5* |
H. Hirata, Zyxin emerges as a key player in the mechanotransduction at cell adhesive structures, 2008, 1(2): 192–5
M. Yao, Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation, Scientific reports, 2014, 4:4610
Reinhard M, An alpha-actinin binding site of zyxin is essential for subcellular zyxin localization and alpha-actinin recruitment, JBC, 1999, 274(19): 13410-
Ye N, Direct observation of α-actinin tension and recruitment at focal adhesions during contact growth, 2014, Exp Cell Res, 327(1):57-
L. Rognoni, Dynamic force sensing of filamin revealed in single-molecule experiments, PNSA, 2012, 109(48):19679–19684.
There are other types of integrin-mediated structure: podosome and invadopodia. Briefly, these are actin-rich dynamic protrusions on the periphery of the plasma membrane [19]. They are found in many cell types, including invasive cancer cells, endothelial cells and immune cells and act as sites of ECM degradation and attachment for cell migration. These two structures share many characteristics in structure and molecular composition (e.g. biomarkers such as WASP, Tks4, Tks5 and MT MMP) [20]. Typically podosomes imply normal cell and invadopodia is cancer cells [21]. Podosomes are highly regulated by many actin regulators, so actin turnover occurs within seconds and the structure lasts only minutes. Compare to that, FAs are more stabilized structures and can last hours.
In this review article, we have focused on molecular mechanisms underlying force regulation of the integrin-mediated adhesion complex. The first part of this review is about the key molecules involved in mechanotransduction and the second part covers characteristics and mechanisms of force regulation.
2. Key adaptor molecules in the integrin-mediated adhesion
1) Integrin
Integrins are the most intensively studied and best characterized among cell-matrix adhesion proteins. In 1986, Hynes and colleagues had revealed structural features of a transmembrane glycoprotein and named the complex ‘integrin’ to denote the role in transmembrane association between ECM and the cytoskeleton [22]. At the same year, Ruoslahti lab also had independently discovered a family of RGD-specific adhesion receptors, a integrin family [23].
Integrins are a heterodimeric transmembrane protein of non-covalently associated α and β subunits. There are 18 α- and 8 β-subunits known, forming at least 24 distinct αβ combinations. Integrins are classified according to their ligands. Collagen-binding integrins include α1β1, α2β1, α10β1, and α11β1 [24–27]; laminin-binding integrins include α3β1, α6β1, α6β4 and α7β1 [28, 29]; and fibronectin-binding integrins include α5β1, α8β1, αIIbβ3 and the αvβ3 [30, 31]. Integrins consist of a relatively large ectodomain, membrane-spanning helixes, and relatively short cytoplasmic tails. The molecular weight of integrins varies from 90 to 160 kDa, and the molecular weight of the ectodomain is between 80 to 150 kDa. Integrins can be classified into two main groups, according to the presence or absence of an αI-domain in the α-subunit. In αI-less integrins, the binding sites to the ligands reside at the interface between the β-propeller domain of α subunits and βI domain of β subunits. In αI integrins, the binding sites are located in the αI domain [32].
The integrin ectodomains bind to ECM glycoproteins (e.g. fibronectins, collagens, and laminins), other cellular receptors (e.g. vascular cell adhesion molecule-1) and the intercellular cell adhesion molecule family (e.g. ICAM-1 and ICAM-3) [24, 33]. The cytoplasmic tails interact with adaptor molecules, such as integrin activators (e.g. talin and kindlin) [34–37] and inhibitors (e.g. filamin) [38, 39]. Competitive binding between these molecules regulates integrin-mediated signal transduction [38, 39]. Focal adhesion kinase (FAK), a cytoplasmic tyrosine kinase, is thought to interact with integrin through a FERM domain [40, 41], which is also found in the talin head and interacts with the integrin β leg, or interact indirectly through vinculin or paxillin [41]. The direct and indirect integrin interactions with FAK and other tyrosine kinases including ILK and TAP20 may trigger downstream signal transduction [42].
Electron microscopy studies suggest that the ectodomains are constrained in either a bent conformation with a height of ~11 nm (Fig. 1B-1) or an extended conformation with a closed headpiece with a height of ~20 nm (Fig. 1B-2). The bent conformation is found to be in the physiological low affinity state for its ligands [43]. The extended-closed headpiece integrin may undergo further changes to a conformation with an open headpiece by swinging the hybrid domain away from the α-subunit during signal transduction (Fig. 1B-3) [43–46]. The extended-open conformation exhibits a high affinity for its ligands compared to the bent conformation [47, 48]. This conformation change is termed integrin activation. The intermediate conformation, the extended- closed headpiece (Fig. 1B-2), is predicted to possess an intermediate affinity to its ligands [44, 45]. The conformational change from a bent to an extended-open conformation with separation of the α and β leg domains consists of three major domain motions: extracellular integrin extension; headpiece opening and hybrid domain swing-out; and separation of the cytoplasmic α and β legs. The order of these motions may depend on the stimulations applied on the integrin [44].
Dynamic regulation of integrin activation is induced by ECM ligand to the ectodomain as well as association of intracellular adaptor molecules (e.g. kindlin and talin) to the cytoplasmic tail. The bidirectional activation processes are so-called outside-in and inside-out signaling: (outside-in) ligand binding to the integrin stimulates the head piece opening followed by integrin extension and the leg separation; (inside-out) the integrin cytoplasmic tail binding to cytoskeleton proteins leads to leg separation and the conformational change propagates to the headpiece.
The inside-out and outside-in signaling processes are not independent or mutually exclusive. In physiological conditions, a small fraction of integrins reside in the extended-open conformation. Integrin binding to specific ECM ligands (e.g. αIIbβ3/αVβ3 to fibronectin) induces and stabilizes the extended-open high-affinity conformation [43]. This conformational change recruits integrin-binding adaptor molecules to the integrin tail, and subsequently triggers further outside-in downstream signaling, such as FAK phosphorylation and recruitment of other signaling proteins. These recruited adaptor proteins can lead inside-out signaling, and vice versa [46, 53, 62, 63].
In the perspective of conformation, both experimental [53] and computational studies [54] showed that the binding of a cytoskeleton protein, talin, to the integrin β cytoplasmic tail is able to induce separation of the α and β legs. Steered molecular dynamics (SMD) stimulation suggested that ligand binding also may induce integrin activation. Soaking αIIbβ3 crystals with linear RGD peptide [57] and soaking α5β1 crystals with cyclic RGD peptide [56] leads to headpiece opening and hybrid domain swing-out. Integrin conformation is also regulated by different divalent cation conditions. Mn2+ activates integrin by inducing the extended conformation, while Ca2+ stabilizes an inactive, bent conformation of integrin [64]. In addition, mechanical force is a key player of integrin activation. The first observed outside-in force transmission by integrins was integrin β1-induced FA formation and cytoskeletal stiffening [49]. External forces enhance integrin activity and clustering, and induce cell-matrix adhesion and cytoskeletal rearrangement. The inside-out force signaling has been observed as actomyosin contraction regulates FA structure and dynamics by strengthening integrin-ECM binding [50–52]. SMD study suggested that mechanical force may induce hybrid domain swing-out and integrin extension [58, 59]. This force-induced extension has been observed in single-molecule experiments as well [60, 61]. More details about force-mediated activation will be discussed in section-3.
Integrin clustering and spacing between the molecules are another important in integrin adhesion and activation. Ligand binding recruits cytoplasmic proteins, such as actin-binding, signaling and adaptor proteins linking integrin to cytoskeletons [65, 66]. The recruited proteins, like talin and kindlin, promote integrin clustering followed by additional recruitment of signaling proteins to FAs [67–72], and leads to reinforcement actin cytoskeleton link to the integrin-mediated adhesion site [73, 74]. As many integrin ligands are multivalent, integrin clustering significantly increases adhesion through avidity modulation [75, 76]. How close is clustered integrin? On the patterned substrates adhesion reinforcement was observed when the distance between integrins was <70nm [77]. Difference integrins play different roles in establishing adhesion. Integrin α5β1 clustered within 40 nm reinforced adhesion strength to the fibronectin under force; however αvβ3 or talin connect to the cytoskeleton and reinforce the connection in response to the force [78].
To measure forces on a single cell or molecule level, different techniques from traction force microscopy to molecular tension sensors have been developed. Forces applied at single adhesion sites were first measured using elastic micropatterned substrates and the stress is found to be 5.5 ± 2 nN/μm2 [79]. Forces generated by single integrin has measured by FRET-based molecular tension sensor [80] and DNA-based sensor in living cells [81] in the range of 1 – 40 pN. Mostly forces exerted at single molecules have read in a range of force, 2 – 40 pN [82–84]. Therefore, single molecular experiments using force microscopies are typically conducted in the range of force.
2) Talin
Talin is the most highlighted among adaptor proteins due to the direct interaction with integrins. Talin is the critical linking component between integrin receptors and the actin cytoskeleton. It was discovered in the focal adhesion in 1983 [85] and in the following year it was found that talin interacts with vinculin [86], another linking component between the cell membrane and actin cytoskeleton. Talin consists of an N-terminal globular head region (~47 kDa) and a large flexible C- terminal rod domain (~220 kDa). The head region is composed of four subdomains: the F1, F2 and F3 domains, which form a FERM (protein 4.1/ezrin/radixin/moesin) domain, and an F0 domain. The F3 subdomain resembles a phosphotyrosine binding-like domain, which binds to the membrane-proximal NPxY motif of the β-integrin cytoplasmic tails [35, 87, 88]. The F1 and F2 domains anchor to the cell membrane. The talin rod (TR) is composed of 62 periodic amphipathic α-helices (H1–H62), which form into a series of 13 helical bundles (R1–R13), followed by a helical dimerization domain (DD) at the C-terminal forming an anti-parallel dimer. In the default physiological native state, talin forms an autoinhibited conformation by intracellular head-to-rod binding. In the inactive autoinhibited conformation, many binding sites, including (at least) three actin, several vinculin, and two integrin binding sites, are cryptic [89–91]. Cell-matrix adhesion stabilization requires talin conformational activation, however the detailed process still remains poorly understood.
Recently, molecular dynamics (MD) simulations have been proposed force-induced activation of talin and its binding sites [92, 93]. The TR contains 11 vinculin binding sites (VBSs) that bind to a hydrophobic pocket in the vinculin head (Vh) [94]. Crystal structure studies revealed that most TR VBSs are buried within the 5-helix bundles stably packed by hydrophobic interactions and result in low binding affinity for vinculin in the inactive state. Magnetic tweezers experiments have demonstrated that mechanical activation by stretching a single talin rod promoted vinculin binding, suggesting that force loading exposes the cryptic VBSs [95]. The R1–R3 domains unfolded under tensile force and vinculin binding to VBSs in those regions inhibited talin refolding after force was removed [58]. These findings suggest that talin acts as a mechanosensor.
Talin also plays a critical role in integrin-mediated inside-out signaling. Talin-induced integrin activation was observed for the first time in CHO cells [87]. Talin-integrin binding is the final step of integrin activation [96] and thought to induce integrin activation by disrupting the association between two α- and β-integrin tails [97]. A structural study showed that the F3 subdomain of the talin head forms a salt bridge (i.e. a strong ionic interaction) with the β-integrin tail. A positively charged patch in the F2 subdomain binds to the negatively charged cell membrane anchoring talin to the membrane [98]. It was found that talin binding alters the integrin β-transmembrane domain topology and disrupts the salt bridge interactions between the α- and β-tail in the transmembrane domain, resulting in conformational activation of the extracellular domain of integrin [53].
3) Vinculin
Vinculin (117 kDa) was first extracted from chicken gizzards and localized at the ends of microfilaments bundles coinciding with cell-matrix adhesion sites in 1979 [99]. Later, vinculin was found to bind to actin filaments [100] and Arp2/3 with high affinity, recruiting Arp2/3 to integrin clusters [101]. In fibroblasts, vinculin localization was correlated with fibronectin distribution on the cell surface [102]. These findings suggest that vinculin is a key player in linking the cytoskeleton to the cell-matrix adhesion site. Electron microscopy studies revealed that vinculin comprises a N-terminal globular head (Vh), a C-terminal rod tail (Vt), and a short flexible proline-rich linker in between them [103–106]. Crystal structure studies revealed that eight antiparallel α-helical bundles organized into five domains in vinculin: the first three domains form the globular head and the last two form the rod-like tail [107, 108].
Vinculin interacts with other structural adaptor molecules: the head domain contains binding sites for talin, α-catenin, α-actinin, and IpaA [109–112]; and the tail domain contains binding sites for actin, paxillin, and PIP2 [113–116]. In physiological conditions, vinculin adopts an autoinhibited conformation [117, 118] formed by the Vh-Vt intracellular interactions through oppositely charged binding sites [119]. The autoinhibition masks the talin and F-actin binding sites on the Vh and Vt respectively and promotes binding to α-actinin and VASP [120, 121]. Vinculin only gets activated at the adhesion site, dependent on actin and talin recruitment [107, 122, 123]. Crystal structure studies have shown that talin-Vh interactions induce a conformational change in the Vh resulting in Vt displacement from the head to open the structure [124]. The open conformation activates vinculin and recruits actin bundles. As mentioned in the talin section, vinculin recruitment to the adhesion site is regulated by force-induced talin activation.
Recently, the development of molecule-based tension sensors allows researchers to measure intracellular molecular tension in vivo. For example, a vinculin-tension sensor was prepared by inserting an elastic protein domain and intermolecular FRET to visualize tension-induced conformational changes. In vivo experiments using the tension sensor have demonstrated that vinculin was required to stabilize FA under force, and tension across vinculin was about 2.5 pN in stable FAs [82].
4) Actin
Actin is a highly conserved protein and found in most eukaryotic cells as an essential element of the cytoskeleton. Actin exits in a freely monomeric state (G-actin; ~42 kDa) or linear filamentous state (F-actin) within the cytoplasm and nucleus [125]. Vertebrates express three main actin isoforms, α, β and ν isoforms. G-actin consists of 375 amino acids and is organized into four subdomains, subdomain 1–4, with an ATP binding site in a cleft between subdomains 2 and 4. Variations between isoforms are differences in few amino acids localized to the proximal N-terminal [126]. The molecules of G-actin spontaneously polymerize to form F-actin, a double-stranded right-hand helical filament. F-actin is a kinetically polarized structure with a fast-growing barbed end and a slow-growing pointed end. The transition between these two states, G-actin and F-actin, is regulated by local concentration of G-actin, different ionic conditions, and a large number of actin-binding proteins. Recently, it has been shown that mechanical forces also affect depolymerization kinetics of G-actin dimer and F-actin [127].
F-actin polymerization is usually powered by ATP hydrolysis, and it occurs over three sequential phases: nucleation, elongation, and steady static phase. During the nucleation phase, few actin subunits assemble spontaneously. Due to a kinetic barrier, the initial aggregate is very slow and unstable, therefore actin nucleators (formin, Arp2/3 and spire) are often required to promote the formation of a stable nucleus [128]. In the elongation phase, the G-actin monomer rapidly associates with the barbed end of F-actin. Filament polymerization reaches an equilibrium state such that association and dissociation of G-actin monomer at both the barbed and the pointed end are balanced in the steady static phase. A group of actin-binding proteins (profilin, formin, Arp2/3, and cofilin) are directly involved in nucleation and polymerization/depolymerization of F-actin and regulate the equilibrium [129, 130]. By regulating actin-binding proteins and adaptor molecules, F-actins form a larger-scale network known as the actin cytoskeleton, which plays critical roles in maintaining the cell shape, providing mechanical support, enabling cell motility, and serving as tracks for transportation. For example, fast polymerization at the barbed end of F-actin pushes the cell membrane and mechanically links it to the adhesion site to protrude forward.
Actin acts as both a force-bearing and force-sensing structure. Tension changes mechanical properties. MD studies have demonstrated that tension on F-actin decreased its twist angle and the conformational change caused an increase of mechanical stiffness [131]. Force loading using optical tweezers exhibited force-dependent distortion of single F-actins [36]. The tension-induced structural distortion affects the affinities for actin-binding proteins, such as cofilin and myosin: F-actin severing was delayed compared to unloaded F-actin, indicating cofilin binding to F-actin was inhibited [37]; and the stretched F-actin showed high affinity for the myosin II domain [34]. These data suggest that mechano-chemical coupling is essential in regulating the actin cytoskeleton.
3. Force regulation of cell-matrix adhesions
Intuitively force would break molecular interactions. However, interestingly, cell-matrix adhesions are strengthened and matured under mechanical force-loading conditions. Two explanations have been proposed for interpretation of the force-induced strengthening and maturation: force-dependent recruitment of adaptor molecules and force-induced stabilization/activation of molecular interactions. Intensive studies have been done in the past decade and provided significant evidence supporting these hypotheses. In many cases, the two hypotheses come together rather than mutually exclude each other. In this section, we will review how force regulates cell-matrix adhesion.
1) Force recruits cell-matrix adhesion molecules
a. Force-dependent recruitment
Studies have shown that force exerted on the cell leads to recruitment of adhesion-related molecules. When force was applied on cells using collagen-coated magnetic beads, filamin A, one of the actin-binding proteins, was recruited to the cell membrane and F-actin polymerization was accelerated [132]. Force loading by an optical trap promoted vinculin and paxillin recruitment in a talin1-dependent manner [133, 134]. Also, internal force generated by actomyosin contraction regulates recruitment of adaptor molecules. Adding myosin kinase inhibitor to block contractile forces resulted in the significant decrease of vinculin accumulation around the adhesion site [133].
Two molecular mechanisms have been proposed to illustrate the force-dependent recruitment: thermodynamics-based and structural mechanism. In thermodynamics-based mechanism, mechanical force leads to a chemical potential shift or molecular density variation in the complex of anchored proteins and results in molecule recruitment to the adhesion site. In structural mechanism, mechanical force induces conformational changes of molecules in the adhesion complex, and results in exposing cryptic binding sites for their ligands, thereby recruiting adaptor molecules to the adhesion site.
i. Thermodynamics-based mechanism
Two thermodynamic mechanisms for molecular recruitment under pulling forces have been proposed [135–137] by Shemesh et al. and Safran et al. In these models, one or two-dimensional aggregate of identical molecules or molecule clusters is considered. The recruited molecules are anchored to the substrate (e.g. cell membrane) and pulling force is applied to the aggregates. The molecules in the anchored aggregate can be exchanged with those freely in the surrounding medium. Shemesh et al. proposed the exchange rate is governed by their chemical potential. If the chemical potential in the anchored aggregate is lower than that in the surrounding medium, the free molecule will be added into the anchored aggregate. In an equilibrium condition, the chemical potentials of the anchored and free molecules are equal and thereby, there is no recruitment. When the anchored molecule is pulled, the chemical potential decreases and leads to recruitment of free molecules from the medium to the anchored site (e.g. adhesion site). This model is largely consistent with experimental observations of FA behaviors under internal and/or external force loading [138, 139]. Safran and his co-workers assumed that the presence of mechanosensors in FA, which cause molecular interactions and are modified by the local density of the protein. Mechanical force deforms FA and changes local density, which in turn affects the molecular interaction. It further induces the downstream biochemical cascade, and recruitment of new molecules to FA [136, 137]. These two different mechanisms could lead to different protein recruitment patterns. A carefully designed fluorescence-recovery-after-photobleaching experiment was suggested by Safran et al. to test these two models [137].
ii. Structural model
The interaction of talin and vinculin is a typical example of force-dependent molecular interactions induced by conformational changes. The talin tail contains at least one cryptic VBS in its native state and when mechanical force is applied to cells, vinculin is recruited to the adhesion site [140]. Therefore, it was proposed that pulling force would induce the conformational change of the talin tail and exposes cryptic VBSs based on MD simulation [92]. In the simulation, VBS1 in the TR rotated ~62.0 degrees and released buried VBSs from the hydrophobic core. This hypothesis has been experimentally validated and it demonstrated that there are more than one cryptic VBS in the tail using magnetic tweezers [95]. As pulling force increased, the number of Vhs bound to the TR increased. Without applying force, only one Vh was observed to bind to the TR; with 2 pN of pulling force, the second and even the third Vh bound to the TR; and when pulling force increased to 12 pN, most TRs were bound with three Vhs. Both the MD simulation and single molecule experiment revealed the possibility that force-induced conformational change, such as exposure of cryptic binding sites, could be one of the molecular mechanisms underlying force-induced recruitment of adaptor molecules.
In thermodynamics-based model, the recruitment of molecules is a thermodynamically favorable consequence of the chemical potential shift or local density variation of FAs, without considering individual molecules in FAs. The structural model focuses on the specific molecules and their binding partners in FAs. However these mechanisms are not exclusive to each other. Both mechanisms may take place concurrently or independently in the recruitment of molecules. The structural mechanism proposed by MD simulation [92] has been validated by single-molecule experiments [95]. The thermodynamics-based mechanisms were proposed based on theoretical considerations. The possible experiments have been proposed to validate the mechanism [135, 137] and further studies are needed to investigate these models.
b. Force induces stabilization of molecular interactions
Another possible explanation for force-induced strengthening of cell-matrix adhesions is that a specific molecular interaction in the adhesion complex is strengthened by the loaded force. There are two possible molecular mechanisms: 1) in case of autoinhibited molecules, mechanical force disrupts the autoinhibited conformation and it prevents competition between autoinhibitory domains and its ligands thereby stabilizing the interaction; and 2) mechanical force directly enhances the bond lifetime of the molecular interaction.
Besides cryptic binding sites, autoinhibition is another mechanism to keep molecules inactive. NMR spectroscopy of talin showed that the C-terminal domain of the talin tail bound to the N-terminal domain of the talin head in its native state, thereby masking the integrin β-tail binding sites on the talin head. Therefore, the integrin β-tail has to compete with the talin tail to bind the talin head for further activation [141]. It suggests that modulation of the talin head-to-tail interaction affects talin affinity for the ligands and talin recruitment to the adhesion site. Similarly, vinculin also consists of head and tail domains. Study with truncation mutants of the Vh and Vt showed that the head and tail directly bind to each other. Pre-incubation of the Vh with talin significantly reduced the intramolecular head-to-tail interaction [117]. This intramolecular interaction also inhibited binding to actin [123]. These studies suggest that talin and the Vt compete to bind to the Vh, and actin and the Vh compete to bind to the Vt. Therefore, vinculin-ligand interactions and vinculin recruitment to the adhesion site can be modulated by affinity changes of the head-to-tail binding. Mechanical force is one of the critical factors which modulates the intramolecular interactions of vinculin and talin. Force applied on the serial molecular bonds in the integrin-mediated adhesion complex induces the open-conformation of talin and vinculin by disrupting the head to tail binding, thereby enhancing the affinity of talin and vinculin for their ligands, such as integrin β-tail, vinculin and actin.
In addition, force directly affects molecular interaction kinetics.
2) Force regulates molecular interaction kinetics
a. Slip, ideal, and catch bond
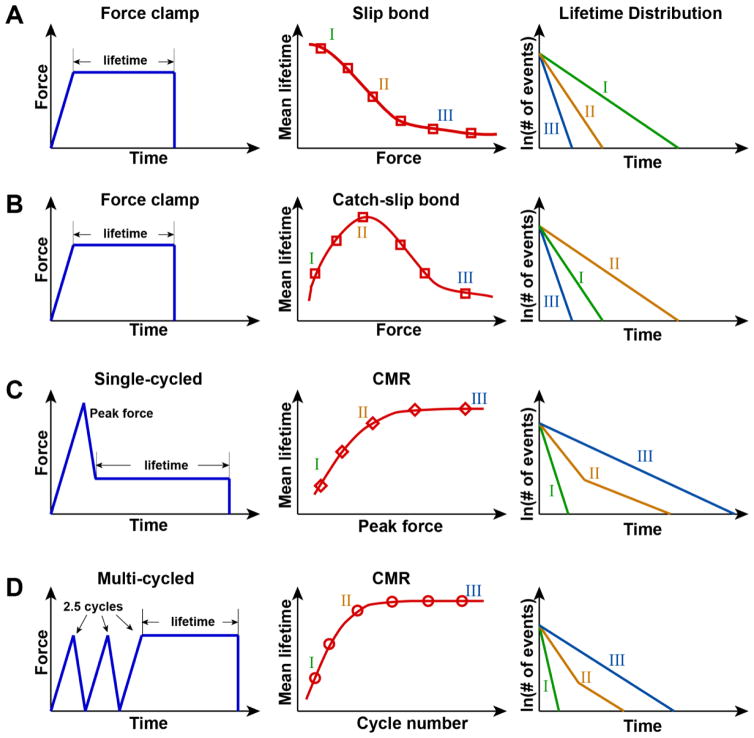
Force directly affects individual molecular interaction kinetics. In 1978, Bell proposed a theoretical model coined in his name. The bell model predicts that force on the bond exponentially increases off-rates of the bond, meaning force weakens molecular bonds, a phenomenon called slip bond (Fig. 2A). A slip bond behavior is intuitive and most commonly observed in biomolecular interactions, such as CD2 and CD48 [48], streptavidin and biotin [55], E-selectin and its ligands [57], integrins and their ligands [142], and antibodies and their antigens [59]. Besides a slip bond, force-independent interaction has been observed, such as the interaction between E-cadherins under certain conditions [47]. The force-independent behavior was theoretically predicted and termed an ideal bond by Dembo et al [143]. They proposed another force-dependent behavior, a catch bond (Fig. 2B). Catch bonds exhibit force-induced bond strengthening distinguished from slip bonds.
Figure 2. Force-dependent interaction kinetics under dynamic tensile force loading conditions: slip and catch bond and cyclic mechanical reinforcement.
The left column is a representative force-time trace loaded on the bond of each force-dependent behavior; the middle column is the curve of mean bond lifetime (MBL) [sec] versus loaded tensile force [pN] in A and B, MBL versus value peak force of single-cycled loading [pN] in C and MBL versus number of cycles of multi-cycled loading in D; and the right is the distribution of bond lifetimes in the middle column. The notation of I, II and III in the middle and right column represents different force ranges, relatively low, intermediate and high, respectively. A typical force range for biomolecular bonds is between 0 and 100 pN. (A) In a slip bond, when tensile force is linearly ramped and clamped at a given force (left), intuitively, mean bond lifetime decreases as force increases (middle). Distributions of bond lifetimes typically exhibit single-state dissociation (right). The lifetime distribution of longer mean lifetime shifts toward to the right, meaning lower off-rate. The off-rate is calculated from the distribution and a reciprocal of the off-rate is a theoretical mean lifetime of the interaction. (B) Typically, a catch bond follows by a slip bond, exhibiting a biphasic behavior, in which counterintuitively mean bond lifetime increases as force increases until to reach a peak and deceases as force further increases (middle) under the same path of force loading with A (left). A typical lifetime distribution is single decay dissociation (right). The distribution of bond lifetime in II force range exhibits the lowest off-rate. (C) CMR induced by mechanical preconditioning with single-cycled tensile force loading. A relatively high peak force for the molecular bond is loaded, reduced to a low force and then clamped to measure bond lifetimes (left). Interestingly, after releasing the peak force, bond lifetime does not immediately revert to the low state, which is measured under linear force-clamping (e.g. A and B), but is significantly enhanced. The degree of reinforcement is much more effective than that by a catch bond behavior. As the peak force increases, mean lifetime increases and reaches a plateau as force further increases, meaning force-history dependence (middle). Under the dynamic force loading condition, a multi-state dissociation is often observed (right). The fraction of bond population in each state changes with different cyclic mechanical preconditionings. (D) CMR induced by multi-cycled tensile loading. A combination of linear ramping to a given peak force and unloading to zero force is repetitively applied (left). As a number of cycle increases, mean lifetime increases and reaches a plateau (middle). The lifetime distribution exhibits the similar feature to the single-cycled reinforcement.
b. Possible mechanisms for catch bonds
A catch bond behavior is widely accepted as a mechanism of mechanical strengthening in biomolecular interactions, in which force prolongs bond lifetimes (i.e. decrease of off-rates). Before any experimental observations of catch bonds, Dembo et al. had proposed theoretical models of catch bonds and predicted its existence in biomolecular interactions [143]. Development of sophisticated single molecule experimental techniques, such as atomic force microscopy (AFM), biomembrane force probe (BFP), an optical and magnetic traps, allow researchers to investigate biomolecular interactions under force loading conditions at the single molecule level. Integrins, such as α5β1, αLβ2, α4β1, and αMβ2, exhibit catch bond behaviors in interaction with their ligands [142, 144–146]. In the last decade, more cell membrane proteins, such as cadherins, selectins, and T-cell receptors, were also found to form a catch bond with their ligands [147–152]. Recently, it was observed that the intracellular protein actin also exhibits a catch bond behavior [127]. Catch bonds are observed only over certain force and time ranges, and followed by a slip bond phase beyond that range. These catch-bond observations attract great attention due to their counterintuitive behavior of possible physiological importance. Theoreticians have proposed many models from different perspectives to explain mechanisms underlying catch bonds. In this review, we will focus on molecular and conformational mechanisms: sliding and rebinding, allosteric regulation, and deformation of energy landscape.
i. Sliding and rebinding
Selectin and PSGL-1 interaction was the first experimentally observed catch bond by single molecule experiment [153, 154]. MD simulation, Monte Carlo simulation and adhesion studies in a flow chamber system showed that force increases the flexibility of L-selectin by breaking a hydrogen bond in an interdomain hinge. The forced opening of the hinge may cause ligands to slide on the binding interface and increase the opportunity of rebinding, thereby enhancing bond lifetimes. G-actin/G-actin and G-actin/F-actin interactions also have shown catch bond behaviors. Steered MD simulation showed that pulling the end unit of F-actin or G-actin dimer resulted in formation of salt bridges, which were induced by sliding and rebinding between the interaction surface of actin subunits [127].
ii. Allosteric regulation
Springer and his co-workers proposed an allosteric regulation model to illustrate catch bond of selectin-ligand interactions. Tensile force exerted on a P-selectin-ligand complex favored an extended conformation and this allosteric change was transmitted to the binding pocket in the lectin-EGF hinge domain, thereby forming high affinity conformation and increasing bond lifetimes [155, 156]. Allosteric and sliding-rebinding models are not necessarily mutually exclusive. Another example of allosteric regulation is interactions between integrins and their ligands. As a major protein in the adhesion complex, characteristics and mechanisms of the integrin catch bond have been extensively studied. During the interaction of integrin and their ligands, force-induced conformational changes, bending and unbending, were observed by BFP experiments [60]. Also, integrin-ligand interactions exhibited multi-state dissociations consisting of short-, intermediate-, and long-lived states, which have fast, intermediate, and slow off-rates, respectively. Tensile force loading switched the short-lived state to the intermediate- or long-lived state, thereby prolonging average bond lifetimes [142].
iii. Deformation of energy landscapes
Besides the structural models, researchers have proposed energy landscape-based models of catch bonds. Evans and his colleagues employed a two-state model based on force spectrum measurements of P-selectin-PSGL-1 interaction [157]. In this model, switching between two dissociation pathways is regulated by force. The first pathway follows Bell’s rule, meaning that the off-rate of this pathway increases with increasing force, like a slip bond. The off-rate of the second pathway is force-independent and slower than that of the first. The first pathway has a deeper energy well under force-free condition and force loading tilts the energy landscape of the bond to shift from the first to the second pathway, resulting in increase of average bond lifetimes. The switching between two dissociation pathways provides a mechanism for the catch bond behavior. In a following study, a simpler, one-state two-pathway model was proposed [158]. In this model, all bonds reside in a single state, but can dissociate along one of two pathways, which have a low and high off-rate respectively. Force tilts the energy landscape and favors bond dissociation from the pathway with a low off-rate, resulting in longer bond lifetime.
c. Effects of force-loading history: cyclic mechanical reinforcement
Most single molecular studies using force microscopies have been conducted under linearly ramped, constant force conditions. In physiological conditions, cells are subject to dynamic force loading rather than static loading, and dynamic force loading over time might act as mechanical preconditioning.
Recent research has demonstrated that mechanical preconditioning by loading dynamic forces on biomolecular bonds significantly strengthened bond interactions. This mechanical reinforcement has been observed in different scales of cellular structure: at the single cell level, cyclic stretching and compression on fibroblasts using AFM induced mechanical stiffening [38]; cyclic shearing on actin-α-actinin meshes mechanically hardened the structure by accelerating α-actinin-mediated actin bundling [39]; and at the single molecule level, applying cyclic tensile forces yielded bond strengthening of both integrin/fibronectin and FimH/mannose interactions using BFP and AFM respectively [61, 159].
The left column of figure 2C and 2D are representative force-time traces of mechanical preconditioning by single-cycle or multi-cycle regimens respectively. In single molecule mechanical preconditioning, the experimental setup typically consists of a combination of bond loading and unloading with a peak force in intermediate to high range, followed by clamping at a given force for lifetime measurement. The middle and right column of figure 2C and 2D represent typical reinforcement after cyclic preconditioning: the stability of tensile force-induced high-affinity states is not only retained, but reinforced; multi-state dissociation is often observed; and different force-loading paths yield different dissociation kinetics.
Interactions between FimH and mannose formed catch bonds in linear force-clamping mode, and the bond strengths were enhanced upon addition of preloaded cycles. All bonds ruptured at significantly higher force compared to those without preloading. Allosterically stabilizing a high affinity conformation by mutations or antibody treatment exhibited the same strengthening effect, suggesting the mechanical reinforcement is due to allosteric regulation [61]. Integrin α5β1 formed catch bonds binding to fibronectin, when the bonds were clamped at a given force. Mechanical preconditioning by cyclic tensile loading further prolonged the bond lifetimes by switching the bond states from a short-lived to long-lived. The fraction of the long-lived population increased as peak force or number of loading cycles increased [159]. Actin also exhibited significant enhancement of bond lifetimes after cyclic mechanical preconditioning compared to catch bond lifetimes induced by constant clamping (data not published). This phenomenon is termed cyclic mechanical reinforcement (CMR) or mechano-memory.
CMR is shown to be significantly more effective in strengthening molecular interactions than the traditional catch bond. In addition to slip and catch bonds, CMR and force-history dependence are possible critical mechanotransduction mechanisms underlying force regulation of integrin-mediated adhesion. [40] [41] [71] [72]
4. Conclusions and future perspectives
We have discussed the key molecules involved in the integrin-mediated cell-matrix interaction and different molecular mechanisms underlying force regulation of integrin and key adaptor molecules. In addition to the traditional force-dependent behaviors, such as a catch, slip and ideal bond, we highlighted CMR, which can mimic physiological conditions more closely. CMR has shown significant effective bond strengthening in some systems and independent of catch or slip behaviors. Therefore, other adhesion molecules also need to be investigated under dynamic force loading to elucidate mechanisms underlying mechanochemical coupling of CMR.
As briefly mentioned in the introduction, recently, cell-matrix adhesions studies have often been conducted on 3D substrates. This has been accompanied by the development of microfluidic-based cell culture systems [70] and 3D ECM-mimicking scaffolds [69]. Although 3D matrix adhesions are comprised of many of the same molecules of 2D cell-matrix adhesions, cell adhesions and behaviors are distinguished from the interactions on the 2D substrates due to inherent complexity of 3D matrices. 2D cell-matrix interactions mostly depend on substrate stiffness and coated ECM molecules, yet in 3D scaffolds, cells sense porosity (pore size and density), topography of ECM ligands or scaffold matrix materials, and isotropicity of material compositions and mechanical properties [67, 68, 160, 161]. Therefore, mechanosensing and mechanotransduction of 3D adhesion differ from traditional 2D adhesion, and in vitro and in vivo studies to elucidate the different characteristics and mechanisms are just beginning.
Force regulation of transmembrane receptors and their ligands has been intensively studied at the single-cell and single–molecule level using AFM, BFP, optical tweezers, and flow chamber systems. However, studies of intracellular force regulation are relatively limited due to the technical hurdles of live cell experiments. A full understanding of the bidirectional signaling cascades requires more attention to intracellular adaptor proteins and cytoskeletons. Single molecule force spectroscopy combined with fluorescence imaging technique or optical microscopy has shown great promise in investigating individual biomolecules both in vivo and in vitro. Cutting edge imaging techniques (e.g. super-resolution-PALM and TIRF-SIM) [162, 163], molecule-based force sensors (e.g. FRET tension sensors) [82, 164] and combinations of single-molecule force spectroscopy and imaging systems [165] are now providing opportunities to further explore the challenging questions regarding mechanosensing and mechanotransduction.
Highlights.
General review of integrin-mediated cell-matrix adhesion
Force regulation of integrin activation and key adaptor proteins
Molecular mechanisms for mechanochemical coupling in force regulation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berrier AL, Yamada KM. Cell–matrix adhesion. Journal of cellular physiology. 2007;213(3):565–573. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 2.Gardel ML, et al. Mechanical integration of actin and adhesion dynamics in cell migration. Annual review of cell and developmental biology. 2010;26:315. doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods. 2008;5(6):491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cukierman E, et al. Taking cell-matrix adhesions to the third dimension. Science. 2001;294(5547):1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 5.Zaman MH, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proceedings of the National Academy of Sciences. 2006;103(29):10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berginski ME, et al. High-resolution quantification of focal adhesion spatiotemporal dynamics in living cells. PloS one. 2011;6(7):e22025. doi: 10.1371/journal.pone.0022025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wozniak MA, et al. Focal adhesion regulation of cell behavior. Biochimica Et Biophysica Acta-Molecular Cell Research. 2004;1692(2–3):103–119. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Geiger B, Yamada KM. Molecular architecture and function of matrix adhesions. Cold Spring Harbor perspectives in biology. 2011;3(5):a005033. doi: 10.1101/cshperspect.a005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaidel-Bar R, et al. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. Journal of cell science. 2003;116(22):4605–4613. doi: 10.1242/jcs.00792. [DOI] [PubMed] [Google Scholar]
- 10.Giannone G, et al. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 2004;116(3):431–443. doi: 10.1016/s0092-8674(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 11.Alexandrova AY, et al. Comparative dynamics of retrograde actin flow and focal adhesions: formation of nascent adhesions triggers transition from fast to slow flow. PloS one. 2008;3(9):e3234–e3234. doi: 10.1371/journal.pone.0003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giannone G, et al. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 2007;128(3):561–575. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beningo KA, et al. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. The Journal of cell biology. 2001;153(4):881–888. doi: 10.1083/jcb.153.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimerman B, Volberg T, Geiger B. Early molecular events in the assembly of the focal adhesion-stress fiber complex during fibroblast spreading. Cell Motil Cytoskeleton. 2004;58(3):143–59. doi: 10.1002/cm.20005. [DOI] [PubMed] [Google Scholar]
- 15.Zaidel-Bar R, et al. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. Journal of cell science. 2007;120(1):137–148. doi: 10.1242/jcs.03314. [DOI] [PubMed] [Google Scholar]
- 16.Ballestrem C, et al. Molecular mapping of tyrosine-phosphorylated proteins in focal adhesions using fluorescence resonance energy transfer. Journal of cell science. 2006;119(5):866–875. doi: 10.1242/jcs.02794. [DOI] [PubMed] [Google Scholar]
- 17.Pankov R, et al. Integrin Dynamics and Matrix Assembly Tensin-Dependent Translocation of α5β1 Integrins Promotes Early Fibronectin Fibrillogenesis. The Journal of cell biology. 2000;148(5):1075–1090. doi: 10.1083/jcb.148.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zamir E, et al. Dynamics and segregation of cell–matrix adhesions in cultured fibroblasts. Nature cell biology. 2000;2(4):191–196. doi: 10.1038/35008607. [DOI] [PubMed] [Google Scholar]
- 19.Linder S, Kopp P. Podosomes at a glance. J Cell Sci. 2005;118(Pt 10):2079–82. doi: 10.1242/jcs.02390. [DOI] [PubMed] [Google Scholar]
- 20.Gimona M, et al. Assembly and biological role of podosomes and invadopodia. Curr Opin Cell Biol. 2008;20(2):235–41. doi: 10.1016/j.ceb.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Flynn DC, et al. Podosomes and Invadopodia: Related structures with Common Protein Components that May Promote Breast Cancer Cellular Invasion. Breast Cancer (Auckl) 2008;2:17–29. doi: 10.4137/bcbcr.s789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamkun JW, et al. Structure of integrin, a glycoprotein involved in the transmembrane linkage between fibronectin and actin. Cell. 1986;46(2):271–82. doi: 10.1016/0092-8674(86)90744-0. [DOI] [PubMed] [Google Scholar]
- 23.Pytela R, et al. Platelet membrane glycoprotein IIb/IIIa: member of a family of Arg-Gly-Asp--specific adhesion receptors. Science. 1986;231(4745):1559–62. doi: 10.1126/science.2420006. [DOI] [PubMed] [Google Scholar]
- 24.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110(6):673–87. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 25.Ruggiero F, et al. Structural requirements for alpha 1 beta 1 and alpha 2 beta 1 integrin mediated cell adhesion to collagen V. J Cell Sci. 1996;109( Pt 7):1865–74. doi: 10.1242/jcs.109.7.1865. [DOI] [PubMed] [Google Scholar]
- 26.Velling T, et al. cDNA cloning and chromosomal localization of human alpha(11) integrin. A collagen-binding, I domain-containing, beta(1)-associated integrin alpha-chain present in muscle tissues. J Biol Chem. 1999;274(36):25735–42. doi: 10.1074/jbc.274.36.25735. [DOI] [PubMed] [Google Scholar]
- 27.Camper L, Hellman U, Lundgren-Akerlund E. Isolation, cloning, and sequence analysis of the integrin subunit alpha10, a beta1-associated collagen binding integrin expressed on chondrocytes. J Biol Chem. 1998;273(32):20383–9. doi: 10.1074/jbc.273.32.20383. [DOI] [PubMed] [Google Scholar]
- 28.Tashiro K, et al. An IKLLI-containing peptide derived from the laminin alpha1 chain mediating heparin-binding, cell adhesion, neurite outgrowth and proliferation, represents a binding site for integrin alpha3beta1 and heparan sulphate proteoglycan. Biochem J. 1999;340( Pt 1):119–26. [PMC free article] [PubMed] [Google Scholar]
- 29.Nishiuchi R, et al. Ligand-binding specificities of laminin-binding integrins: A comprehensive survey of laminin-integrin interactions using recombinant alpha 3 beta 1, alpha 6 beta 1, alpha 7 beta 1 and alpha 6 beta 4 integrins. Matrix Biology. 2006;25(3):189–197. doi: 10.1016/j.matbio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 30.van der Flier A, Sonnenberg A. Function and interactions of integrins. Cell Tissue Res. 2001;305(3):285–98. doi: 10.1007/s004410100417. [DOI] [PubMed] [Google Scholar]
- 31.Singh P, Carraher C, Schwarzbauer JE. Assembly of fibronectin extracellular matrix. Annu Rev Cell Dev Biol. 2010;26:397–419. doi: 10.1146/annurev-cellbio-100109-104020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie C, et al. Structure of an integrin with an alphaI domain, complement receptor type 4. EMBO J. 2010;29(3):666–79. doi: 10.1038/emboj.2009.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plow EF, et al. Ligand binding to integrins. J Biol Chem. 2000;275(29):21785–8. doi: 10.1074/jbc.R000003200. [DOI] [PubMed] [Google Scholar]
- 34.Uyeda TQ, et al. Stretching actin filaments within cells enhances their affinity for the myosin II motor domain. PLoS One. 2011;6(10):e26200. doi: 10.1371/journal.pone.0026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moser M, et al. The tail of integrins, talin, and kindlins. Science. 2009;324(5929):895–899. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- 36.Shimozawa T, Ishiwata S. Mechanical distortion of single actin filaments induced by external force: detection by fluorescence imaging. Biophys J. 2009;96(3):1036–44. doi: 10.1016/j.bpj.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayakawa K, Tatsumi H, Sokabe M. Actin filaments function as a tension sensor by tension-dependent binding of cofilin to the filament. J Cell Biol. 2011;195(5):721–7. doi: 10.1083/jcb.201102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe-Nakayama T, et al. Direct detection of cellular adaptation to local cyclic stretching at the single cell level by atomic force microscopy. Biophys J. 2011;100(3):564–72. doi: 10.1016/j.bpj.2010.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmoller KM, et al. Cyclic hardening in bundled actin networks. Nat Commun. 2010;1:134. doi: 10.1038/ncomms1134. [DOI] [PubMed] [Google Scholar]
- 40.Hirata H, Tatsumi H, Sokabe M. Zyxin emerges as a key player in the mechanotransduction at cell adhesive structures. Commun Integr Biol. 2008;1(2):192–5. doi: 10.4161/cib.1.2.7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinhard M, et al. An alpha-actinin binding site of zyxin is essential for subcellular zyxin localization and alpha-actinin recruitment. J Biol Chem. 1999;274(19):13410–8. doi: 10.1074/jbc.274.19.13410. [DOI] [PubMed] [Google Scholar]
- 42.Liu S, Calderwood DA, Ginsberg MH. Integrin cytoplasmic domain-binding proteins. Journal of cell science. 2000;113(20):3563–3571. doi: 10.1242/jcs.113.20.3563. [DOI] [PubMed] [Google Scholar]
- 43.Takagi J, et al. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110(5):599–611. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 44.Xiao T, et al. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 2004;432(7013):59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo B-H, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annual review of immunology. 2007;25:619. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Springer TA, Dustin ML. Integrin inside-out signaling and the immunological synapse. Current opinion in cell biology. 2012;24(1):107–115. doi: 10.1016/j.ceb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rakshit S, et al. Ideal, catch, and slip bonds in cadherin adhesion. Proc Natl Acad Sci U S A. 2012;109(46):18815–20. doi: 10.1073/pnas.1208349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pierres A, et al. Determination of the lifetime and force dependence of interactions of single bonds between surface-attached CD2 and CD48 adhesion molecules. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(26):15114–15118. doi: 10.1073/pnas.93.26.15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang N, Butler JP, Ingber DE. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260(5111):1124–7. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 50.Choquet D, Felsenfeld DP, Sheetz MP. Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell. 1997;88(1):39–48. doi: 10.1016/s0092-8674(00)81856-5. [DOI] [PubMed] [Google Scholar]
- 51.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94(25):13661–5. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guilluy C, et al. The Rho GEFs LARG and GEF-H1 regulate the mechanical response to force on integrins. Nat Cell Biol. 2011;13(6):722–7. doi: 10.1038/ncb2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim C, et al. Talin activates integrins by altering the topology of the β transmembrane domain. The Journal of cell biology. 2012;197(5):605–611. doi: 10.1083/jcb.201112141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Isralewitz B, Izrailev S, Schulten K. Binding pathway of retinal to bacterio-opsin: a prediction by molecular dynamics simulations. Biophys J. 1997;73(6):2972–9. doi: 10.1016/S0006-3495(97)78326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Izrailev S, et al. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys J. 1997;72(4):1568–81. doi: 10.1016/S0006-3495(97)78804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Humphrey W, Bamberg E, Schulten K. Photoproducts of bacteriorhodopsin mutants: a molecular dynamics study. Biophys J. 1997;72(3):1347–56. doi: 10.1016/S0006-3495(97)78781-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wayman AM, et al. Triphasic force dependence of E-selectin/ligand dissociation governs cell rolling under flow. Biophys J. 2010;99(4):1166–74. doi: 10.1016/j.bpj.2010.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yao M, et al. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci Rep. 2014;4:4610. doi: 10.1038/srep04610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shang H, Lee GU. Magnetic tweezers measurement of the bond lifetime-force behavior of the IgG-protein A specific molecular interaction. J Am Chem Soc. 2007;129(20):6640–6. doi: 10.1021/ja071215c. [DOI] [PubMed] [Google Scholar]
- 60.Chen W, et al. Observing force-regulated conformational changes and ligand dissociation from a single integrin on cells. The Journal of cell biology. 2012;199(3):497–512. doi: 10.1083/jcb.201201091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yakovenko O, et al. FimH forms catch bonds that are enhanced by mechanical force due to allosteric regulation. J Biol Chem. 2008;283(17):11596–605. doi: 10.1074/jbc.M707815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ye F, et al. Recreation of the terminal events in physiological integrin activation. The Journal of cell biology. 2010;188(1):157–173. doi: 10.1083/jcb.200908045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu P, Luo BH. Integrin bi-directional signaling across the plasma membrane. Journal of cellular physiology. 2013;228(2):306–312. doi: 10.1002/jcp.24154. [DOI] [PubMed] [Google Scholar]
- 64.Campbell ID, Humphries MJ. Integrin structure, activation, and interactions. Cold Spring Harb Perspect Biol. 2011;3(3) doi: 10.1101/cshperspect.a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Calderwood DA, Campbell ID, Critchley DR. Talins and kindlins: partners in integrin-mediated adhesion. Nat Rev Mol Cell Biol. 2013;14(8):503–17. doi: 10.1038/nrm3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ye F, Snider AK, Ginsberg MH. Talin and kindlin: the one-two punch in integrin activation. Front Med. 2014;8(1):6–16. doi: 10.1007/s11684-014-0317-3. [DOI] [PubMed] [Google Scholar]
- 67.Hollister SJ. Porous scaffold design for tissue engineering. Nat Mater. 2005;4(7):518–24. doi: 10.1038/nmat1421. [DOI] [PubMed] [Google Scholar]
- 68.Bruzauskaite I, et al. Scaffolds and cells for tissue regeneration: different scaffold pore sizes-different cell effects. Cytotechnology. 2015 doi: 10.1007/s10616-015-9895-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Doyle AD, Yamada KM. Mechanosensing via cell-matrix adhesions in 3D microenvironments. Exp Cell Res. 2015 doi: 10.1016/j.yexcr.2015.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Velve-Casquillas G, et al. Microfluidic tools for cell biological research. Nano Today. 2010;5(1):28–47. doi: 10.1016/j.nantod.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ye N, et al. Direct observation of alpha-actinin tension and recruitment at focal adhesions during contact growth. Exp Cell Res. 2014;327(1):57–67. doi: 10.1016/j.yexcr.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rognoni L, et al. Dynamic force sensing of filamin revealed in single-molecule experiments. Proc Natl Acad Sci U S A. 2012;109(48):19679–84. doi: 10.1073/pnas.1211274109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saltel F, et al. New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control beta3-integrin clustering. J Cell Biol. 2009;187(5):715–31. doi: 10.1083/jcb.200908134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Helsten TL, et al. Differences in regulation of Drosophila and vertebrate integrin affinity by talin. Mol Biol Cell. 2008;19(8):3589–98. doi: 10.1091/mbc.E08-01-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bunch TA. Integrin alphaIIbbeta3 activation in Chinese hamster ovary cells and platelets increases clustering rather than affinity. J Biol Chem. 2010;285(3):1841–9. doi: 10.1074/jbc.M109.057349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shattil SJ, Kim C, Ginsberg MH. The final steps of integrin activation: the end game. Nat Rev Mol Cell Biol. 2010;11(4):288–300. doi: 10.1038/nrm2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Selhuber-Unkel C, et al. Cooperativity in adhesion cluster formation during initial cell adhesion. Biophys J. 2008;95(11):5424–31. doi: 10.1529/biophysj.108.139584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roca-Cusachs P, et al. Clustering of alpha(5)beta(1) integrins determines adhesion strength whereas alpha(v)beta(3) and talin enable mechanotransduction. Proc Natl Acad Sci U S A. 2009;106(38):16245–50. doi: 10.1073/pnas.0902818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Balaban NQ, et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3(5):466–72. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 80.Morimatsu M, et al. Molecular tension sensors report forces generated by single integrin molecules in living cells. Nano Lett. 2013;13(9):3985–9. doi: 10.1021/nl4005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang X, Ha T. Defining single molecular forces required to activate integrin and notch signaling. Science. 2013;340(6135):991–4. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grashoff C, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466(7303):263–6. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiang G, et al. Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature. 2003;424(6946):334–7. doi: 10.1038/nature01805. [DOI] [PubMed] [Google Scholar]
- 84.Thoumine O, et al. Short-term binding of fibroblasts to fibronectin: optical tweezers experiments and probabilistic analysis. Eur Biophys J. 2000;29(6):398–408. doi: 10.1007/s002490000087. [DOI] [PubMed] [Google Scholar]
- 85.Burridge K, Connell L. A new protein of adhesion plaques and ruffling membranes. The Journal of cell biology. 1983;97(2):359–367. doi: 10.1083/jcb.97.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burridge K, Mangeat P. An Interaction between Vinculin and Talin. Nature. 1984;308(5961):744–746. doi: 10.1038/308744a0. [DOI] [PubMed] [Google Scholar]
- 87.Calderwood DA, et al. The talin head domain binds to integrin β subunit cytoplasmic tails and regulates integrin activation. Journal of Biological Chemistry. 1999;274(40):28071–28074. doi: 10.1074/jbc.274.40.28071. [DOI] [PubMed] [Google Scholar]
- 88.Calderwood DA, et al. The phosphotyrosine binding-like domain of talin activates integrins. Journal of Biological Chemistry. 2002;277(24):21749–21758. doi: 10.1074/jbc.M111996200. [DOI] [PubMed] [Google Scholar]
- 89.Papagrigoriou E, et al. Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle. The EMBO journal. 2004;23(15):2942–2951. doi: 10.1038/sj.emboj.7600285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fillingham I, et al. A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head. Structure. 2005;13(1):65–74. doi: 10.1016/j.str.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 91.Himmel M, et al. Control of high affinity interactions in the talin C terminus: how talin domains coordinate protein dynamics in cell adhesions. J Biol Chem. 2009;284(20):13832–42. doi: 10.1074/jbc.M900266200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee SE, Kamm RD, Mofrad MR. Force-induced activation of talin and its possible role in focal adhesion mechanotransduction. Journal of biomechanics. 2007;40(9):2096–2106. doi: 10.1016/j.jbiomech.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 93.Hytonen VP, Vogel V. How force might activate talin’s vinculin binding sites: SMD reveals a structural mechanism. PLoS Comput Biol. 2008;4(2):e24. doi: 10.1371/journal.pcbi.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gingras AR, et al. Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod. Journal of Biological Chemistry. 2005;280(44):37217–37224. doi: 10.1074/jbc.M508060200. [DOI] [PubMed] [Google Scholar]
- 95.del Rio A, et al. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323(5914):638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tadokoro S, et al. Talin binding to integrin beta tails: a final common step in integrin activation. Science. 2003;302(5642):103–6. doi: 10.1126/science.1086652. [DOI] [PubMed] [Google Scholar]
- 97.Vinogradova O, et al. A structural mechanism of integrin alpha(IIb)beta(3) “inside-out” activation as regulated by its cytoplasmic face. Cell. 2002;110(5):587–97. doi: 10.1016/s0092-8674(02)00906-6. [DOI] [PubMed] [Google Scholar]
- 98.Anthis NJ, et al. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. The EMBO journal. 2009;28(22):3623–3632. doi: 10.1038/emboj.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Geiger B. A 130K protein from chicken gizzard: its localization at the termini of microfilament bundles in cultured chicken cells. Cell. 1979;18(1):193–205. doi: 10.1016/0092-8674(79)90368-4. [DOI] [PubMed] [Google Scholar]
- 100.Wilkins JA, Lin S. High-affinity interaction of vinculin with actin filaments in vitro. Cell. 1982;28(1):83–90. doi: 10.1016/0092-8674(82)90377-4. [DOI] [PubMed] [Google Scholar]
- 101.DeMali KA, Barlow CA, Burridge K. Recruitment of the Arp2/3 complex to vinculin coupling membrane protrusion to matrix adhesion. The Journal of cell biology. 2002;159(5):881–891. doi: 10.1083/jcb.200206043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Burridge K, Feramisco JR. Microinjection and localization of a 130K protein in living fibroblasts: a relationship to actin and fibronectin. Cell. 1980;19(3):587–595. doi: 10.1016/s0092-8674(80)80035-3. [DOI] [PubMed] [Google Scholar]
- 103.Isenberg G, Leonard K, Jockusch BM. Structural aspects of vinculin-actin interactions. Journal of molecular biology. 1982;158(2):231–249. doi: 10.1016/0022-2836(82)90431-4. [DOI] [PubMed] [Google Scholar]
- 104.Coutu MD, Craig SW. cDNA-derived sequence of chicken embryo vinculin. Proceedings of the National Academy of Sciences. 1988;85(22):8535–8539. doi: 10.1073/pnas.85.22.8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Eimer W, et al. Molecular shape of vinculin in aqueous solution. Journal of molecular biology. 1993;229(1):146–152. doi: 10.1006/jmbi.1993.1014. [DOI] [PubMed] [Google Scholar]
- 106.Milam LM. Electron microscopy of rotary shadowed vinculin and vinculin complexes. Journal of molecular biology. 1985;184(3):543–545. doi: 10.1016/0022-2836(85)90301-8. [DOI] [PubMed] [Google Scholar]
- 107.Bakolitsa C, et al. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430(6999):583–586. doi: 10.1038/nature02610. [DOI] [PubMed] [Google Scholar]
- 108.Borgon RA, et al. Crystal structure of human vinculin. Structure. 2004;12(7):1189–1197. doi: 10.1016/j.str.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 109.Burridge K, Mangeat P. An interaction between vinculin and talin. 1984 doi: 10.1038/308744a0. [DOI] [PubMed] [Google Scholar]
- 110.Hazan RB, et al. Vinculin is associated with the E-cadherin adhesion complex. Journal of Biological Chemistry. 1997;272(51):32448–32453. doi: 10.1074/jbc.272.51.32448. [DOI] [PubMed] [Google Scholar]
- 111.Van Nhieu GT, Ben-Ze’ev A, Sansonetti P. Modulation of bacterial entry into epithelial cells by association between vinculin and the Shigella IpaA invasin. The EMBO Journal. 1997;16(10):2717–2729. doi: 10.1093/emboj/16.10.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Weiss EE, et al. Vinculin is part of the cadherin–catenin junctional complex: complex formation between α-catenin and vinculin. The Journal of cell biology. 1998;141(3):755–764. doi: 10.1083/jcb.141.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Turner CE, Glenney JR, Burridge K. Paxillin: a new vinculin-binding protein present in focal adhesions. The Journal of Cell Biology. 1990;111(3):1059–1068. doi: 10.1083/jcb.111.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wood C, et al. Characterisation of the paxillin-binding site and the C-terminal focal adhesion targeting sequence in vinculin. Journal of cell science. 1994;107(2):709–717. [PubMed] [Google Scholar]
- 115.Johnson RP, Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. 1995 doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- 116.Johnson RP, Craig SW. The carboxy-terminal tail domain of vinculin contains a cryptic binding site for acidic phospholipids. Biochemical and biophysical research communications. 1995;210(1):159–164. doi: 10.1006/bbrc.1995.1641. [DOI] [PubMed] [Google Scholar]
- 117.Johnson RP, Craig SW. An intramolecular association between the head and tail domains of vinculin modulates talin binding. Journal of Biological Chemistry. 1994;269(17):12611–12619. [PubMed] [Google Scholar]
- 118.Johnson RP, Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 1995;373:261–264. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- 119.Miller GJ, Dunn SD, Ball EH. Interaction of the N-and C-terminal Domains of Vinculin CHARACTERIZATION AND MAPPING STUDIES. Journal of Biological Chemistry. 2001;276(15):11729–11734. doi: 10.1074/jbc.M008646200. [DOI] [PubMed] [Google Scholar]
- 120.Kroemker M, et al. Intramolecular interactions in vinculin control α-actinin binding to the vinculin head. FEBS letters. 1994;355(3):259–262. doi: 10.1016/0014-5793(94)01216-4. [DOI] [PubMed] [Google Scholar]
- 121.Hüttelmaier S, et al. The interaction of the cell-contact proteins VASP and vinculin is regulated by phosphatidylinositol-4, 5-bisphosphate. Current biology. 1998;8(9):479–488. doi: 10.1016/s0960-9822(98)70199-x. [DOI] [PubMed] [Google Scholar]
- 122.Chen H, Choudhury DM, Craig SW. Coincidence of actin filaments and talin is required to activate vinculin. Journal of Biological Chemistry. 2006;281(52):40389–40398. doi: 10.1074/jbc.M607324200. [DOI] [PubMed] [Google Scholar]
- 123.Cohen DM, et al. Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. Journal of Biological Chemistry. 2005;280(17):17109–17117. doi: 10.1074/jbc.M414704200. [DOI] [PubMed] [Google Scholar]
- 124.Izard T, et al. Vinculin activation by talin through helical bundle conversion. Nature. 2004;427(6970):171–175. doi: 10.1038/nature02281. [DOI] [PubMed] [Google Scholar]
- 125.Grummt I. Actin and myosin as transcription factors. Current opinion in genetics & development. 2006;16(2):191–196. doi: 10.1016/j.gde.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 126.Dominguez R, Holmes KC. Actin structure and function. Annual review of biophysics. 2011;40:169. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lee C-y, et al. Actin depolymerization under force is governed by lysine 113: glutamic acid 195-mediated catch-slip bonds. Proceedings of the National Academy of Sciences. 2013;110(13):5022–5027. doi: 10.1073/pnas.1218407110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001;81(2):667–74. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326(5957):1208–12. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–76. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 131.Matsushita S, et al. Effect of tensile force on the mechanical behavior of actin filaments. J Biomech. 2011;44(9):1776–81. doi: 10.1016/j.jbiomech.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 132.Glogauer M, et al. The role of actin-binding protein 280 in integrin-dependent mechanoprotection. Journal of Biological Chemistry. 1998;273(3):1689–1698. doi: 10.1074/jbc.273.3.1689. [DOI] [PubMed] [Google Scholar]
- 133.Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. The Journal of cell biology. 2002;159(4):695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Giannone G, et al. Talin1 is critical for force-dependent reinforcement of initial integrin–cytoskeleton bonds but not tyrosine kinase activation. The Journal of cell biology. 2003;163(2):409–419. doi: 10.1083/jcb.200302001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shemesh T, et al. Focal adhesions as mechanosensors: a physical mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(35):12383–12388. doi: 10.1073/pnas.0500254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nicolas A, Geiger B, Safran SA. Cell mechanosensitivity controls the anisotropy of focal adhesions. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(34):12520–12525. doi: 10.1073/pnas.0403539101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Besser A, Safran SA. Force-induced adsorption and anisotropic growth of focal adhesions. Biophysical journal. 2006;90(10):3469–3484. doi: 10.1529/biophysj.105.074377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Riveline D, et al. Focal contacts as mechanosensors externally applied local mechanical force induces growth of focal contacts by an mdia1-dependent and rock-independent mechanism. The Journal of cell biology. 2001;153(6):1175–1186. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Balaban NQ, et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nature cell biology. 2001;3(5):466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 140.Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. Journal of Cell Biology. 2002;159(4):695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Goksoy E, et al. Structural basis for the autoinhibition of talin in regulating integrin activation. Molecular cell. 2008;31(1):124–133. doi: 10.1016/j.molcel.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen W, Lou J, Zhu C. Forcing switch from short-to intermediate-and long-lived states of the αA domain generates LFA-1/ICAM-1 catch bonds. Journal of Biological Chemistry. 2010;285(46):35967–35978. doi: 10.1074/jbc.M110.155770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dembo M, et al. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proceedings of the Royal Society of London B: Biological Sciences. 1988;234(1274):55–83. doi: 10.1098/rspb.1988.0038. [DOI] [PubMed] [Google Scholar]
- 144.Kong F, et al. Demonstration of catch bonds between an integrin and its ligand. The Journal of cell biology. 2009;185(7):1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fiore VF, et al. Dynamic catch of a Thy-1–α5β1+ syndecan-4 trimolecular complex. Nature communications. 2014:5. doi: 10.1038/ncomms5886. [DOI] [PubMed] [Google Scholar]
- 146.Rosetti F, et al. A Lupus-Associated Mac-1 Variant Has Defects in Integrin Allostery and Interaction with Ligands under Force. Cell reports. 2015;10(10):1655–1664. doi: 10.1016/j.celrep.2015.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Marshall BT, et al. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423(6936):190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 148.Sarangapani KK, et al. Low force decelerates L-selectin dissociation from P-selectin glycoprotein ligand-1 and endoglycan. Journal of Biological Chemistry. 2004;279(3):2291–2298. doi: 10.1074/jbc.M310396200. [DOI] [PubMed] [Google Scholar]
- 149.Wayman AM, et al. Triphasic force dependence of E-selectin/ligand dissociation governs cell rolling under flow. Biophysical journal. 2010;99(4):1166–1174. doi: 10.1016/j.bpj.2010.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rakshit S, et al. Ideal, catch, and slip bonds in cadherin adhesion. Proceedings of the National Academy of Sciences. 2012;109(46):18815–18820. doi: 10.1073/pnas.1208349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Buckley CD, et al. The minimal cadherin-catenin complex binds to actin filaments under force. Science. 2014;346(6209):1254211. doi: 10.1126/science.1254211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Manibog K, et al. Resolving the molecular mechanism of cadherin catch bond formation. Nature communications. 2014:5. doi: 10.1038/ncomms4941. [DOI] [PubMed] [Google Scholar]
- 153.Lou J, et al. Flow-enhanced adhesion regulated by a selectin interdomain hinge. The Journal of cell biology. 2006;174(7):1107–1117. doi: 10.1083/jcb.200606056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lou J, Zhu C. A structure-based sliding-rebinding mechanism for catch bonds. Biophysical journal. 2007;92(5):1471–1485. doi: 10.1529/biophysj.106.097048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Springer TA. Structural basis for selectin mechanochemistry. Proceedings of the National Academy of Sciences. 2009;106(1):91–96. doi: 10.1073/pnas.0810784105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Waldron TT, Springer TA. Transmission of allostery through the lectin domain in selectin-mediated cell adhesion. Proceedings of the National Academy of Sciences. 2009;106(1):85–90. doi: 10.1073/pnas.0810620105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Evans E, et al. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(31):11281–11286. doi: 10.1073/pnas.0401870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pereverzev YV, et al. The two-pathway model for the catch-slip transition in biological adhesion. Biophysical journal. 2005;89(3):1446–1454. doi: 10.1529/biophysj.105.062158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kong F, et al. Cyclic mechanical reinforcement of integrin-ligand interactions. Mol Cell. 2013;49(6):1060–8. doi: 10.1016/j.molcel.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sun H, Tawa G, Wallqvist A. Classification of scaffold-hopping approaches. Drug Discov Today. 2012;17(7–8):310–24. doi: 10.1016/j.drudis.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hakkinen KM, et al. Direct comparisons of the morphology, migration, cell adhesions, and actin cytoskeleton of fibroblasts in four different three-dimensional extracellular matrices. Tissue Eng Part A. 2011;17(5–6):713–24. doi: 10.1089/ten.tea.2010.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kner P, et al. Super-resolution video microscopy of live cells by structured illumination. Nat Methods. 2009;6(5):339–42. doi: 10.1038/nmeth.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Greenfield D, et al. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7(6):e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.LaCroix AS, et al. Construction, imaging, and analysis of FRET-based tension sensors in living cells. Methods Cell Biol. 2015;125:161–86. doi: 10.1016/bs.mcb.2014.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cordova JC, et al. Combining single-molecule manipulation and single-molecule detection. Curr Opin Struct Biol. 2014;28:142–8. doi: 10.1016/j.sbi.2014.08.010. [DOI] [PubMed] [Google Scholar]