Abstract
Patterning of a multicellular body plan involves a coordinated set of developmental processes that includes cell division, morphogenesis, and cellular differentiation. These processes have been most intensively studied in animals and land plants; however, deep insight can also be gained by studying development in simpler multicellular organisms. The multicellular green alga Volvox carteri (Volvox) is an excellent model for the investigation of developmental mechanisms and their evolutionary origins. Volvox has a streamlined body plan that contains only a few thousand cells and two distinct cell types: reproductive germ cells and terminally differentiated somatic cells. Patterning of the Volvox body plan is achieved through a stereotyped developmental program that includes embryonic cleavage with asymmetric cell division, morphogenesis, and cell-type differentiation. In this review we provide an overview of how these three developmental processes give rise to the adult form in Volvox and how developmental mutants have provided insights into the mechanisms behind these events. We highlight the accessibility and tractability of Volvox and its relatives that provide a unique opportunity for studying development.
Graphical Abstract
Introduction
How a multicellular body plan becomes patterned is a central question in developmental biology. Development from a single progenitor cell or group of cells into a fully formed individual requires a coordinated set of processes that include growth, cell division, morphogenesis and cell differentiation. Eukaryotic multicellularity, and hence developmental mechanisms, have evolved independently over two dozen times (Grosberg and Strathmann, 2007), but beyond animals and land plants the developmental diversity of eukaryotes has not been well explored. The study of developmental mechanisms in other multicellular groups has the potential to broaden our understanding of developmental tool kits and patterning mechanisms that may ultimately lead to new ideas and elucidate common underlying principles governing eukaryotic development (Herron et al., 2013). Green algae are a potentially rich group of organisms in which to investigate developmental biology because several independent occurrences of either multicellular or coenocytic developmental mechanisms evolved just within this clade (Coneva and Chitwood, 2015; Leliaert et al., 2012; Umen, 2014). Furthermore, multicellular green algae exhibit simplified body plans with tractable developmental programs, thereby providing unique opportunities to dissect fundamental mechanisms underlying developmental patterning.
Here we explore development in the multicellular green alga Volvox carteri (Volvox) whose appeal for developmental biology studies is manifold. Volvox has a small and streamlined body plan that is composed of only a few thousand cells and two distinct cell types—germ and somatic (Fig. 1). The Volvox body plan is patterned through a stereotyped developmental program that is characterized by processes similar to those found in animals and land plants such as embryogenesis from a single cell, tissue remodeling, and spatially controlled cell type specification. Volvox is a well-developed model organism that is easy to culture, has relatively few cells and cell types, and possesses fast generation times. A growing arsenal of genetic and molecular genetic tools has also been developed for Volvox, including a reference genome sequence (Prochnik et al., 2010), nuclear transformation and expression of transgenes (Cheng et al., 2003, Geng et al., 2014, Ishida, 2007, Kirk et al., 1999, Miller and Kirk, 1999, Nishii et al., 2003, Pappas and Miller, 2009, Schiedlmeier et al., 1994, Stark et al., 2001 and Ueki and Nishii, 2009), forward genetics through crosses and transposon-tagging (Huskey et al., 1979, Miller et al., 1993 and Ueki and Nishii, 2008), and reverse genetics through RNAi-mediated or antisense knockdown (Cheng et al., 2006 and Geng et al., 2014).
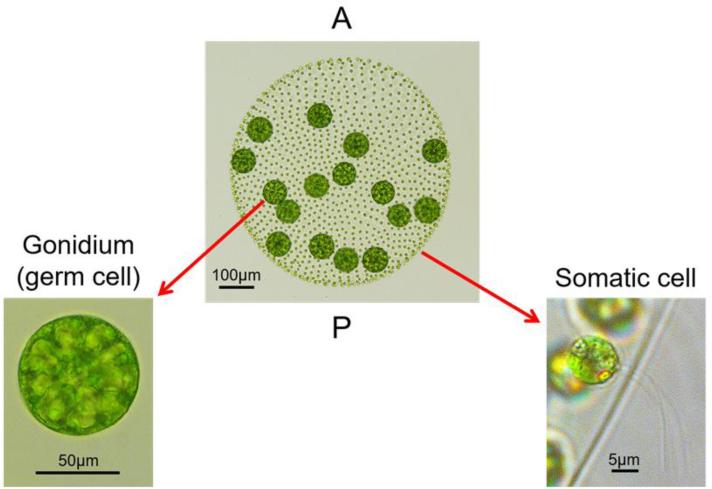
Figure 1. Volvox carteri body plan and cell types.
Center, an adult vegetative Volvox spheroid with two distinct cell types: ~2000 small, flagellated somatic cells (right inset) and ~16 large, aflagellate germ cells called gonidia (left inset). Somatic cells are on the outer surface of the spheroid with flagella oriented towards the exterior. Gonidia are just beneath the somatic cell layer in the posterior hemisphere. All cells are embedded within a clear, compartmentalized extracellular matrix. Anterior (A) and posterior (P) poles of the spheroid are labeled.
An equally important and compelling attribute of Volvox is its position within a larger monophyletic grouping collectively known as volvocine algae comprising multicellular species with different body sizes, cell numbers and degrees of cell-type specialization (Coleman, 2012; Herron et al., 2009)(Fig. 2). Importantly, volvocine algae include the well-studied model unicellular green flagellate, Chlamydomonas reinhardtii, that serves as an outgroup to the multicellular species (Coleman, 1999; Harris, 2001; Merchant et al., 2007; Nozaki, 2003; Nozaki et al., 2000) and provides an excellent reference point for comparing the similarities and differences underlying unicellular and multicellular organization (Miller, 2010; Nishii and Miller, 2010). The shared biology and experimental tractability of the volvocine algae make them an ideal group in which to investigate the origins of developmental patterning mechanisms and the potential constraints on how they evolved.
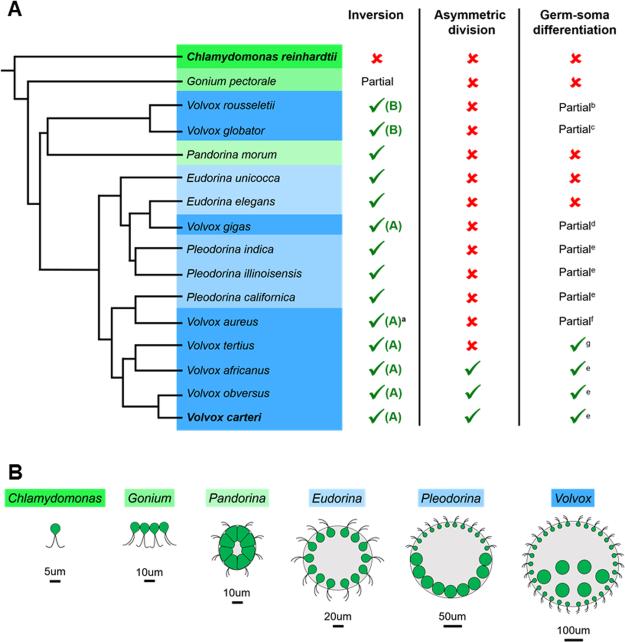
Figure 2. Grades of volvocine algal body plan complexity and polyphyletic origins of the genus Volvox.
(A, left) Abbreviated volvocine phylogeny adapted from Herron and Michod (2008) and Herron et al. (2009). Different genera are highlighted by different colored boxes. Body plan schematics of each genus are shown in (B). The genus Volvox (dark blue highlighted species), which is characterized by spheroid size (typically >500 µm diameter), large cell number (>500), and composition of mostly terminally differentiated somatic cells (Coleman, 2012), is polyphyletic with at least three separate origins. Volvox carteri, the species that is the focus of this review, and Chlamydomonas reinhardtii, a unicellular outgroup for all multicellular volvocine species, are highlighted in bold. (A, right) Table indicating the presence or absence of developmental innovations that are discussed in this review. Complete inversion: inverted embryo forms a closed spheroid; partial inversion: inverted embryo reverses curvature but does not form a closed spheroid (Kirk, 2005). Type of inversion (A or B) refers to the spatial and temporal sequences of tissue and cell shape changes during inversion that differ between species (Hallmann, 2006). Presence/absence of asymmetric cell division as described in (Desnitski, 1995 and Herron et al., 2010). Complete germ-soma differentiation: germ and somatic cells have completely distinct fates; partial germ-soma differentiation: germ cell precursors first proceed through a flagellated somatic phase prior to acquiring a germ fate (Kirk, 2005 and Ransick, 1993). Note that asymmetric cell division and complete germ-soma differentiation are not universal traits of Volvox and are likely derived in V. carteri from a simpler ancestral program with no asymmetric division and partial germ-soma differentiation. (B) Cartoons illustrating volvocine body plan size and organization based on genus. Chlamydomonas, single celled; Gonium, discoidal body plan of 4–16 cells; Pandorina, spheroidal body plan with 16 cells; Eudorina, spheroidal body plan with 32–64 cells and expanded ECM; Pleodorina, spheroidal body plan with 32–128 cells, expanded ECM, and partial germ-soma differentiation; Volvox, spheroidal body plan with 500–50,000 cells, a small proportion of germ cells, and expanded ECM. aV. aureus was reported in Hallmann (2006) to undergo type B inversion; however its inversion appears closer to type A in (Darden, 1966). bPocock (1933a); cKirk et al.(1986); dPocock (1933b); eRansick (1993); fDarden (1966); gRansick (1993) noted that conclusive documentation for complete germ-soma differentiation in V. tertius is lacking, but Pocock (1938) reports that the gonidial initials of V. tertius are differentiated prior to the conclusion of embryogenesis implying that they do not have a transient somatic stage.
Here we focus on three processes that play key roles in Volvox development: embryonic cleavage, morphogenesis, and cellular differentiation. We chose these processes as they illuminate areas where Volvox is particularly well suited to tackle fundamental questions in developmental biology with clear parallels in other taxa. Note that the genus Volvox is polyphyletic with at least two or three separate origins within the volvocine clade (Fig. 2A). Throughout this review we use Volvox to indicate Volvox carteri, and in places where we refer to other species in the genus Volvox we use italicized Latin names.
Like all volvocine algae, Volvox is haploid and spheroids can reproduce both asexually (vegetatively) and sexually. While the sexual cycle of Volvox holds a great deal of interest for investigating evolutionary and developmental questions (Callahan and Huskey, 1980; Geng et al., 2014; Hiraide et al., 2013; Kirk, 2006; Nozaki, 1996; Nozaki et al., 2006; Umen, 2011), the somewhat simpler vegetative developmental cycle where spheroids reproduce clonally is the focus of this review.
Vegetative Volvox spheroids exhibit a streamlined form of cellular differentiation with only two cell types: ~16 large germ cells (or stem cells) called gonidia and ~2,000 small terminally differentiated somatic cells that are precisely positioned and oriented within a clear and highly structured glycoprotein-rich extracellular matrix (ECM) that occupies the majority of the adult spheroid volume (Hallmann, 2003; Kirk et al., 1986)(Fig. 1). The somatic cells are positioned around the surface of the spheroid with approximately even spacing between cells, each with a pair of apical flagella extending outside the ECM boundary (Hoops, 1993)(Fig. 1, right inset). Proper positioning and orientation of somatic cells is critical for their flagella to beat collectively and efficiently propel the phototaxing spheroid through the water (Brumley et al., 2014; Hoops, 1993; Huskey, 1979; Ueki et al., 2010). Somatic cells are very similar in size and structure to a Chlamydomonas cell with highly organized cytoskeletons and stereotyped positioning of intracellular organelles (Johnson and Porter, 1968; Kochert and Olson, 1970). However unlike Chlamydomonas cells that can grow and reproduce mitotically, somatic cells in Volvox are terminally differentiated and fail to divide in the adult. Consequently somatic cells have a limited lifetime and eventually undergo senescence and cell death (Pommerville and Kochert, 1982, 1981). In contrast to somatic cells the ~16 gonidial cells form the asexual germ line of Volvox. Gonidia are positioned just beneath the somatic cell layer within the posterior region of the spheroid (Fig. 1). Gonidial cells are round, aflagellate and capable of growing throughout the Volvox life cycle. At maturity they are over five hundred times larger in volume than somatic cells. Each gonidial cell undergoes a program of embryonic cleavage, morphogenesis and differentiation to produce a new vegetative spheroid. As described in detail below, the Volvox body plan is patterned through developmental processes that are shared by more complex organisms. Volvox embryogenesis has particularly striking similarities to embryogenesis in some animals that begins with rapid cleavage divisions of a large spherical zygote to produce a blastula followed by coordinated cell movements and contortions that shape the embryo with three primary axes and presumptive germ layers (Graham and Morgan, 1966; O'Farrell et al., 2004; Solnica-Krezel and Sepich, 2012).
Overview of the Volvox asexual life cycle
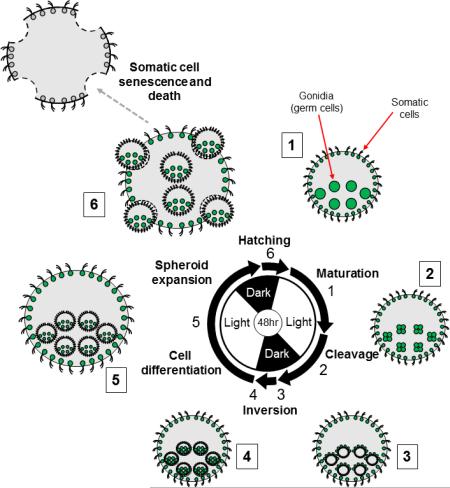
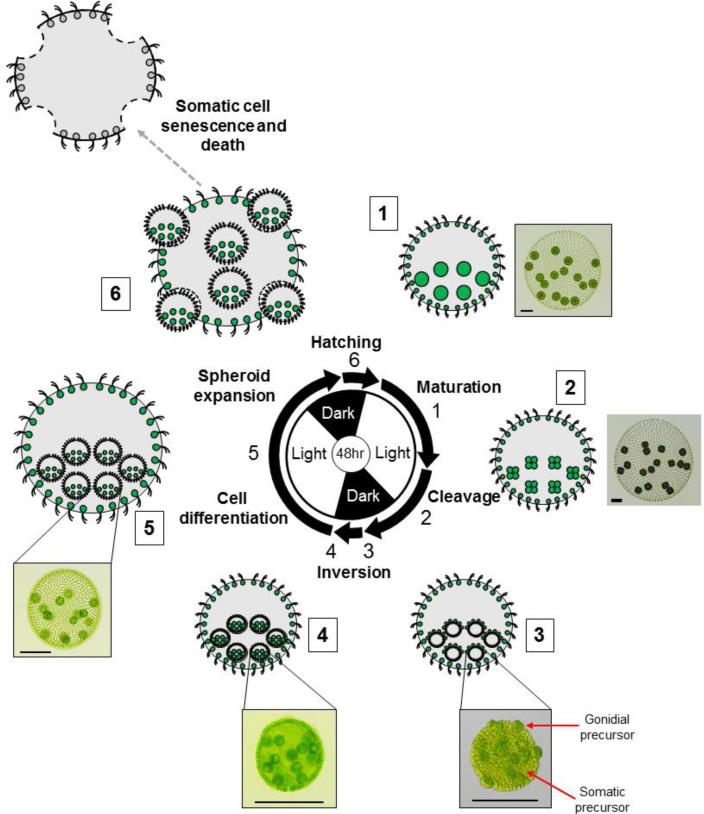
The asexual life cycle of Volvox follows a recursive pattern of reproduction with each gonidial cell undergoing embryogenesis to produce a new miniature juvenile spheroid still residing within its original mother spheroid (Fig. 3). Juveniles grow and mature within the mother until they eventually hatch out, while the somatic cells of the mother spheroid undergo senescence and cell death. The hatched juvenile spheroids continue to grow through expansion of ECM and enlargement of gonidia, and when mature begin the reproductive cycle again. Under laboratory conditions the entire vegetative life cycle can be synchronized over a 48-hour period with two light-dark cycles (Fig. 3).
Figure 3. Asexual life cycle of Volvox.
Stages of vegetative reproduction are depicted proceeding clockwise from upper right with cartoons and micrographs of selected stages. Center, two-day diurnal regime used to synchronize the Volvox life cycle with alternating light and dark periods of 16hrs and 8hrs respectively. Arrows represent the approximate duration of each labeled phase. Numbers next to cartoons and micrographs correspond to numbers next to arrows on diurnal diagram showing the stage depicted. 1, Mature adult spheroid with pre-cleavage gonidia (large green circles) and flagellated somatic cells (small green circles) embedded within extracellular matrix (grey). 2, Adult mother spheroid with four-cell stage embryos derived from gonidia. Note that adult somatic cells do not divide. 3, Adult mother spheroid with post-cleavage embryos prior to inversion. Note the large gonidial precursor cells on the exterior surface. 4, Adult mother spheroid with post-inversion juveniles. 5, Adult mother spheroid with differentiated and expanded juveniles. 6, Late stage juveniles hatching from their mother spheroid. The somatic cells from the mother spheroid undergo senescence and eventual death. Scale bars = 100μm.
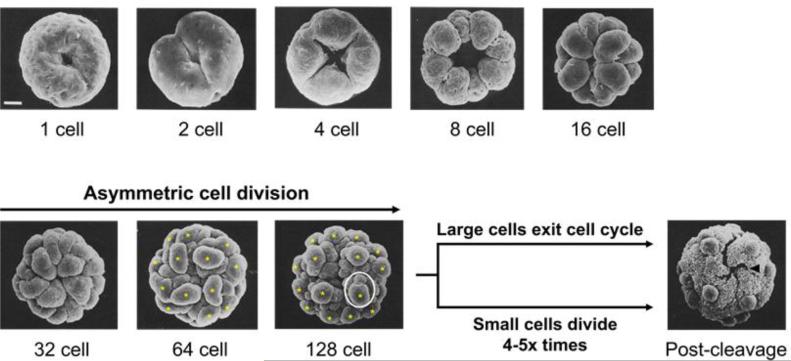
The life cycle starts when each gonidium of a mature adult initiates embryogenesis by undergoing a series of 11 or 12 cleavage divisions over a period of 6–8 h (Fig. 3). At the sixth cleavage division sixteen of the anterior cells in the embryo divide asymmetrically to produce large and small daughters, with the large cells halting division after one or two more asymmetric cleavages and the small cells continuing to divide until the end of embryogenesis (Green and Kirk, 1981 and Starr, 1970Fig. 4). Large post-cleavage embryonic cells are destined to become the gonidia of the next generation whereas small cells will differentiate into somatic cells. At the end of embryonic cleavage each embryo is a hollow sphere with 2000–4000 tightly packed small cells arranged around the surface and sixteen large cells protruding outward (Fig. 3 panel 3; Fig. 4, Post-cleavage) (Green and Kirk, 1981). A distinct feature of the embryonic division mechanism in Volvox and most other volvocine species is that after division the apical ends of cells marked by basal bodies and nuclei are oriented inward toward the interior of the embryo, a configuration that must be reversed in order for flagella to project outward in the adult (Coleman, 2012). Volvox solves this topological problem through a remarkable process of morphogenesis called inversion that is discussed in detail below. During inversion the entire embryo turns itself inside-out allowing the flagella of somatic cells to project outwards and placing the gonidial initials on the inside (Viamontes and Kirk, 1977). Cells within the post-inversion embryo then begin the process of germ-soma differentiation in which presumptive gonidia resorb their flagellar stubs and grow dramatically in size, while presumptive somatic cells elongate their flagella and undergo limited growth (Bell, 1985, Coggin and Kochert, 1986, Kirk et al., 1993 and Nozaki, 1994). Cytodifferentiation is accompanied by expansion of the spheroid, mostly through secretion of ECM that continues throughout the remainder of the life cycle. On the day after embryogenesis and inversion the juveniles hatch out of the mother spheroid while the discarded parental somatic cells undergo senescence and cell death (Pommerville and Kochert, 1982 and Pommerville and Kochert, 1981). After hatching, the now adult-stage spheroids continue growing until their own gonidial cells are large enough to undergo embryogenesis and begin a new round of the vegetative reproductive cycle (Fig. 3).
Figure 4. Volvox embryogenesis.
Scanning electron micrographs showing the anterior hemisphere of a cleaving gonidium at indicated stages. Yellow asterisks mark 16 large daughters produced by asymmetric division at the 6th cleavage cycle (32→64 cell stage) and 7th cleavage cycle (64→128 cell stage). Within the white circle at the 128 cell stage is a large cell undergoing visibly asymmetric cell division. The cross-shaped phialopore opening in the post-cleavage embryo before inversion is indicated by a black arrowhead. Scale bar, 10um. EM micrographs used with permission from (Green and Kirk, 1981) ©1981 Green and Kirk. Journal of Cell Biology. 91:743–755.
Embryogenesis: the early embryonic cell cycle and asymmetric cell division
During embryogenesis each gonidium undergoes multiple rounds of rapid and synchronous cell division with little or no intervening growth to give rise to all the cells that will be present in the adult. This type of cell cycle is termed multiple fission (or palintomy) and is an ancestral feature shared by most volvocine species including unicellular Chlamydomonas. The multiple fission cell cycle has a prolonged G1 phase where cells can enlarge in size by many-fold. At the end of G1 cells undergo a rapid succession of alternating S phases and mitotic divisions to produce 2n daughter cells (Bišová and Zachleder, 2014, Cross and Umen, 2015, Herron et al., 2010, Kirk, 2005, Kirk, 1998, Setlik and Zachleder, 1984 and Umen and Olson, 2012). The variable n has a range for each volvocine species that scales with organismal size and also with the amount of growth required before division can begin. In the case of Volvox, n is 11 or 12 corresponding to a final cell number of ~2000 or ~4000 respectively. Multiple fission cell cycles are used not just in green algae and protists (Cavalier-Smith, 1980) but also during embryogenesis of many animals (O'Farrell et al., 2004), and have even been observed in some of the earliest known metazoan fossil embryos (Butterfield, 2011 and Huldtgren et al., 2011).
The first sign of embryogenesis in Volvox is loss of radial symmetry in gonidia as they begin to adopt a polarized cytology along an anterior-posterior (A-P) axis. The nucleus is re-positioned near the anterior pole just beneath the basal bodies (Green et al., 1981 and Kirk, 1998) which behave as centrioles during division by coordinating the plane of cleavage relative to the position of the mitotic spindle (Coss, 1974 and Ehler et al., 1995). The first five rounds of division are symmetric and give rise to a 32-celled embryo whose cells are all equal-sized (Fig. 4). The first two division planes are offset by 90° and slightly oblique to each other, after which the mitotic apparatus and subsequent cleavage planes rotate clockwise with each division giving rise to a helical twist pattern that is reminiscent of spiralian embryos from certain animal lineages such as annelids, molluscs, and bryozoans (Green and Kirk, 1981 and Lambert, 2010). These spatially patterned divisions during embryogenesis, combined with the fact that from the third division onward the cells elongate in the circumferential direction, results in the production of a spiral-shaped hollow sphere with an opening at one end called the phialopore, which is formed in the region where elongating non-sister cells come into contact (Fig. 4; Green and Kirk, 1981). At the 6th round of division (32→64 cell stage), each of the 16 cells in the anterior half of the embryo divides asymmetrically giving rise to a large and a small daughter, while each of the 16 cells in the posterior half continues to divide symmetrically (Fig. 4). The larger daughters formed in the anterior divide asymmetrically one or two more times, then exit from cell division, while all of the small daughters in both hemispheres continue dividing symmetrically 4 or 5 more times (Green and Kirk, 1981). At the end of cleavage the embryo contains all the cells that will be found in the adult: ~16 large gonidial precursors and ~2000 small somatic precursors. The combination of asymmetric division and premature exit from cell division for the large cells results in them having a ~30-fold larger volume than the remaining small cells (Kirk et al., 1993).
Cytological and genetic control of embryonic patterning
The cell cycle program of embryogenesis in Volvox shares several ancestral features with that of its single-celled relative Chlamydomonas, including rapid division cycles characteristic of multiple fission and rotation of division planes between each cell division. However, there are three modifications of cell division that are derived in Volvox and are key contributors to morphogenesis and cell type differentiation: incomplete cytokinesis, asymmetric cell division, and premature cell cycle exit of large asymmetric daughter cells (Herron and Michod, 2008; Kirk, 2005).
Incomplete cytokinesis during Volvox embryogenesis results in thin cytoplasmic bridges remaining between daughters after each round of division. All bridges form at around the middle of each cell with respect to its anterior-posterior axis, and by the end of embryogenesis the bridges have formed a coherent band that circumscribes the entire embryo, except at its most anterior region where a cross-shaped opening called the phialopore remains (Green et al., 1981; Fig. 4 Post-cleavage, Fig. 5A). The cytoplasmic bridge network serves to maintain the integrity of the multicellular embryo by physically attaching cells to their neighbors and also provides a stable structural framework against which cells can exert force to mediate the morphogenetic process of inversion described below.
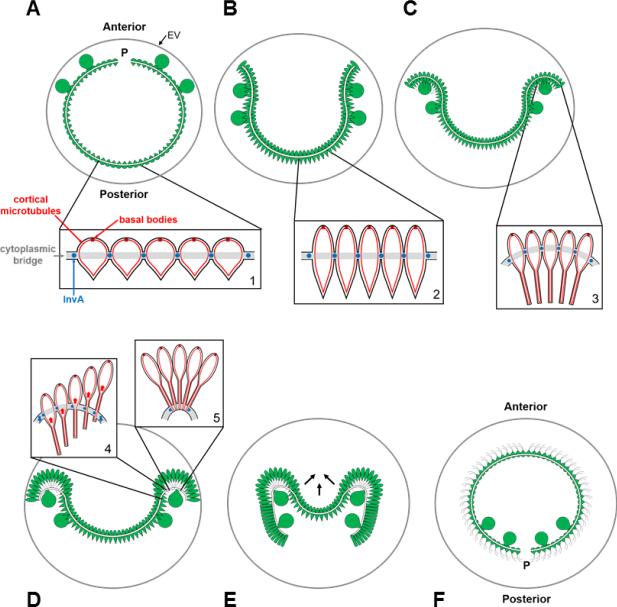
Figure 5. Stages of inversion and associated cell shape changes.
A) Pre-inversion embryo. Presumptive gonidia (large cells) protrude from the exterior surface of the anterior hemisphere and flagellar ends of presumptive somatic cells (pointed ends of small pear-shaped cells) (inset 1) face the interior of the embryo. Cytoplasmic bridges (white band) connect cells at their midpoints and traverse the embryo except at the phialopore (P). A glycoprotein embryonic vesicle (gray circle labeled EV) surrounds the embryo and expands during inversion. Anterior and posterior poles of the embryo are labeled. B) Presumptive somatic cells transition to a spindle shape (inset 2) causing the embryo to shrink and allowing the phialopore lips to begin opening. C) Cells near the phialopore transition to an elongated flask shape with their cortical microtubules extending into their posterior ends (inset 3). D) The kinesin motor protein InvA that is anchored to the cytoplasmic bridges migrates along cortical microtubules in cells near the phialopore driving cell body movement relative to the cytoplasmic bridges (inset 4) to cause increased local curvature and acute bending near the phialopore lips (inset 5). A wave of InvA generated cell movement relative to bridges and local bending proceeds from the anterior region towards the equator. E) The bend reaches the equator and the posterior hemisphere contracts (arrows) allowing it to snap through the already inverted anterior hemisphere. Cells that have passed through the bend region undergo a third shape change to a compact columnar shape. F) Inversion is complete and the gonidia and phialopore are now positioned at the posterior pole of the juvenile spheroid.
The second derived feature of Volvox embryogenesis is asymmetric cell division that is critical for generating embryonic cell-size asymmetry at the 6th division, a process that ultimately dictates post-embryonic cell-type differentiation. Isolation and characterization of mutants with aberrant asymmetric division patterns has shed light on the genetic basis of asymmetric division in Volvox. In the gonidialess mutant, glsA, asymmetric division does not occur and embryos end up with all small cells that differentiate somatically. Without germ cells, gonidialess mutants would normally have a lethal phenotype so they can only be isolated in a strain background that contains another mutation, regA, whose phenotype allows somatic cells to become reproductive (see below) (Kirk et al., 1991). The glsA gene encodes a zuotin-family J domain chaperone that is a member of a large family of J domain proteins (Miller and Kirk, 1999). J domain proteins interact with Hsp70A heat shock proteins and it was initially proposed that GlsA, in partnership with Hsp70A, effects asymmetric division by altering the position of the mitotic spindle and cleavage furrow (Kirk et al., 1991 and Miller and Kirk, 1999). Indeed, it has been shown that interaction of GlsA with the cytoplasmic Hsp70A isoform is essential for asymmetric division in Volvox (Cheng et al., 2005). However, GlsA and Hsp70A co-localize with histones during mitosis (Cheng et al., 2005) and GlsA binds directly to histones (Pappas and Miller, 2009). These localization data suggest that GlsA and Hsp70A may control asymmetric cell division via a nuclear function, possibly as a transcription factor complex. In either case, localization studies of these two proteins provide a potential explanation for why asymmetric cell division is restricted to the anterior hemisphere of the embryo: While GlsA is distributed uniformly throughout the embryo throughout embryogenesis, Hsp70A becomes enriched in the anterior hemisphere of the embryo following the fifth round of division immediately before asymmetric division occurs (Cheng et al., 2005). It is therefore possible that functional GlsA-Hsp70A complexes can only form in the anterior half of the embryo and thus spatially restrict asymmetric division to this hemisphere. multiple gonidia (mul) mutants define a second class of asymmetric division mutants. As the name indicates, the phenotype of mul mutants is supernumerary gonidia which are thought to arise because the timing of asymmetric division is shifted one or two cycles later than normal (Callahan and Huskey, 1980, Huskey et al., 1979 and Kirk et al., 1991). Mutations at several loci are known to generate mul phenotypes, but no mul genes have yet been identified or molecularly characterized. It is possible that mul genes or gene products interact with GlsA-Hsp70A in some manner to control the timing for onset of asymmetric division during embryogenesis. For example, mul gene products may be responsible for restricting Hsp70A localization to the anterior hemisphere at a specific cell division cycle number or activating GlsA-HSP70A when blastomeres reach a specific size. Alternatively, mul genes may be under the control of GlsA-Hsp70A transcription factor complexes as downstream targets. Although not discussed here, it is notable that the timing of asymmetric division in embryos undergoing sexual development is different from that of vegetative embryos and also different between males and females (Starr, 1970, Starr, 1969 and Umen, 2011). These differences underscore the precise genetic control that is exerted over the timing of asymmetric cell division and provide further motivation for understanding its bases. Identification of affected loci in mul mutants and characterization of mul gene products will play an important role in dissecting the control mechanisms that govern asymmetric cell division.
The third derived feature of Volvox embryogenesis is the early withdrawal of large asymmetric daughters from the embryonic cleavage program. In premature cessation of division (pcd) mutants all of the embryonic cells exit from cleavage earlier than in wild-type embryos resulting in a collection of abnormally large post-embryonic cells (Pall, 1975). As will be discussed in detail below, cell size determines cell fate in Volvox, so pcdmutants differentiate after cleavage with mostly reproductive cells and few or no somatic cells (Pall, 1975). Identification of pcd loci and characterization of PCD gene products may help elucidate the mechanisms that enable different cell cycle programs to be executed in large versus small embryonic cells after asymmetric cell divisions.
Less well characterized but fascinating mutants that alter the anterior-posterior (A-P) axis of polarity have previously been described (Sessoms and Huskey, 1973). One such mutant led to a “donut” phenotype with partially duplicated anterior hemispheres including gonidia and phialopore openings produced in both halves of the embryo. Another class of mutant altered a subtler patterning feature of somatic cells that normally show a graded decrease in eyespot size along the A-P axis, but in the mutant were changed to an all posterior pattern without affecting other aspects of A-P morphogenesis. Understanding the wild-type functions of the patterning genes discussed above awaits their cloning and further characterization that should be facilitated by the Volvox genome sequence (Prochnik et al., 2010) and other new tools that are being developed such as genetic markers (Harper et al., 1987, Huskey et al., 1979 and Kirk, 1998) and RNAi-mediated knockdown for functional characterization of gene products (Cheng et al., 2006 and Geng et al., 2014).
Inversion: green algal “gastrulation”
The relative simplicity of Volvox inversion makes it a well-suited model in which to investigate how locally coordinated changes in cell shape and movements can lead to global changes in tissue configuration. While plant morphogenesis is constrained by the rigid wall surrounding each cell, this is not true in Volvox and other multicellular volvocine algae where dynamic cell shape changes play a key role in driving inversion, just as they do in animal gastrulation (Keller and Shook, 2011). Moreover, compared with animal gastrulation Volvox inversion is simpler and can be broken down into defined stages with mutants that block specific steps. However unlike the case in animal embryos where cells can move with respect to their neighbors (Nishimura and Takeichi, 2009; Solnica-Krezel and Sepich, 2012), postembryonic cells of Volvox are connected to their neighbors via cytoplasmic bridges that are formed by incomplete cytokinesis. As a result, nearly all Volvox cells maintain fixed spatial relationships with their nearest neighbors throughout development. The cytoplasmic bridges are a key contributor to the process of inversion as they provide a stable structural framework against which cells exert force to effect inversion (Green et al., 1981; Nishii and Ogihara, 1999; Nishii et al., 2003). Additionally, the cytoplasmic bridges are flexible, which allows the cellular sheet of Volvox to bend backwards during inversion and eventually flip inside-out. Remarkably, the process of inversion is not identical in all species of Volvox, with two patterns termed type A and type B that are used in distantly related clades (Hallmann, 2006; Höhn and Hallmann, 2011)(Fig. 2A). This review focuses on type A inversion that is used by Volvox carteri and which is better understood in terms of molecular mechanisms. It is also worth noting that nearly all multicellular volvocine species undergo some form of inversion after cell division (see Hoops and Floyd (1982) and Arakaki et al., (2013) for two interesting exceptions). Gonium is the only genus in which inversion is “incomplete” meaning that a curved sheet of post-mitotic embryonic cells reverses its curvature so that the initially concave face where flagella will form becomes the convex face; but the post-inversion cell sheet never closes into a ball (Iida et al., 2013). In the larger-sized genera with complete inversion—Pandorina, Eudorina, and Pleodorina—embryos start out bowl-shaped and also reverse their curvature, but then undergo a process of closure where the free edges of the embryo join together to form an enclosed ball shape (Hallmann, 2006; Kikuchi, 1978; Marchant, 1977). Volvox embryos also undergo complete inversion as they start out sphere-shaped and are able to flip completely inside out to make an inverted sphere-shaped embryo. Despite their differences, complete and incomplete inversion accomplish similar topological reversals in cell curvature to place the flagellar ends of the embryo on the convex face of the adult organism. It is likely that complete inversion is derived from modification of incomplete inversion, but testing this idea will require more detailed understanding of the molecular and cytological details of inversion in additional volvocine species.
Early inversion: embryonic contraction and opening of the phialopore
Inversion is initiated when the embryo contracts slightly, separating itself from the glycoprotein vesicle wall that encloses it. This vesicle is present throughout embryogenesis and the rest of development where it enlarges along with the embryo and juvenile spheroid until hatching when the vesicle is partially disrupted to allow adult spheroids to exit their mother spheroid (Fig. 5). The initial contraction of the embryo is driven by elongation of presumptive somatic cells into spindle-shapes resulting in a 40% reduction in their diameters and consequently a ~10% reduction in the diameter of the embryo as a whole (Viamontes and Kirk, 1977; Viamontes et al., 1979). As mentioned above, the only embryonic region of Volvox that lacks cytoplasmic bridges is the phialopore, a cross shaped pair of slits at the anterior pole that provides the opening through which the embryo passes itself during inversion (Fig. 4, Post-cleavage). Contraction of the embryo leads to separation of the embryo from its surrounding vesicle and a release of tensile stress at the phialopore opening that allows the lips of the phialopore to begin curling back (Fig. 5B).
Mid-inversion: tissue bending and bend propagation
The next stage of inversion begins with a shape change in cells near the phialopore from spindle to flask morphology. The flask shape is the result of constriction of the outward-facing chloroplast ends of cells into a long stalk-like structure (Viamontes and Kirk, 1977 and Viamontes et al., 1979), and this localized shape change causes additional backward bending of the phialopore lips (Nishii et al., 2003; Fig. 5C). The flask-shaped cells in Volvox inversion are analogous in structure and function to bottle-shaped epithelial cells that form during metazoan gastrulation. Bottle cells in animal embryos have highly constricted apical ends and enlarged basolateral ends, and their formation is coupled to an inward fold of an initially flat cell sheet at the beginning of gastrulation (Sawyer et al., 2010 and Solnica-Krezel and Sepich, 2012). Thus similar types of shape changes in embryonic cells evolved independently in the Volvox lineage and metazoans to help drive similar morphogenetic processes.
At the time that the first flask-shaped cells are forming in Volvox, the cytoplasmic bridges that were initially located near the cellular midpoints, relocate to the outermost ends of the cells near the tips of the stalks (Green et al., 1981; Viamontes and Kirk, 1977; Viamontes et al., 1979)(Fig. 5D). The combination of the cell shape change to an elongated flask morphology and the relocation of the cytoplasmic bridges to the narrow distal ends of the flask cells generates tensile stress that leads to a more acute localized backward bend of the cell sheet (Fig. 5D). Several possible mechanisms by which cytoplasmic bridges could be relocated have been proposed: (1) Bridges at the cellular midpoints could be broken and new bridges formed at the chloroplast ends; (2) bridges could be actively moved from the cell midpoints to the tips; or (3) the cells could move relative to the bridges (Green et al., 1981). Through observation of small clusters of cells isolated from inverting embryos it was shown that the latter mechanism where cells move with respect to fixed bridges is what occurs during inversion (Green et al., 1981). This observation suggests that the lengths of cytoplasmic bridges between cells remain fixed throughout inversion, and the tension that builds up in the bridges as a result of cellular movement is released through a bending of the bridge network.
The transition to flask shape is temporary and after assuming this conformation the cells end up in a third shape called columnar in which the cytoplasmic bridges remain at their basal ends but the overall cell shape becomes more compact (Viamontes and Kirk, 1977)(Fig. 5E). Inversion proceeds as a wave of spindle-to-flask-to-columnar shape changes that propagates from the anterior towards the equator until a half-inverted embryo is formed (Fig. 5E). Viewed from the anterior pole this wave would appear as a ring propagating from pole to equator.
Late inversion: snapping through the posterior hemisphere
The first half of inversion is driven by a concerted wave of cell sheet bending that propagates to the equator. Once the bend region reaches the equator considerable tensile stress has accumulated within the half-inverted embryo where the bend region forms an equatorial ring that surrounds the uninverted posterior hemisphere. It was hypothesized that this geometrical constraint would cause the posterior hemisphere of the embryo to undergo an elastic “snap-through” motion, such that it would flip inside-out much more rapidly compared to the inversion of the anterior hemisphere (Viamontes et al., 1979). Careful observations confirmed that inversion of the posterior hemisphere proceeds around seven times faster than anterior hemisphere inversion, and outpaces the wave of cell shape changes that drive the first half of the process (Fig. 5E; Nishii and Ogihara, 1999 and Viamontes et al., 1979). Thus inversion of the posterior hemisphere is not driven by the same cell shape changes that underlie inversion of the anterior hemisphere, but rather by mechanical forces that act to relieve accumulated stress at the equator. However, as described below, the second half of inversion also requires a general shrinkage of the posterior hemisphere mediated by the actin cytoskeleton that allows it to squeeze through the opening at the equator (Nishii and Ogihara, 1999).
Upon completion of inversion the embryo is in its adult configuration with presumptive gonidial cells on the inside and presumptive somatic cells oriented with their anterior ends containing basal bodies and flagella facing outward (Fig. 5F). The process of cytodifferentiation then begins, starting with loss of cytoplasmic bridges, elongation of flagella on somatic cells, and deposition of extracellular matrix material that “glues” back together the phialopore opening and begins to push apart neighboring somatic cells as the juvenile spheroid enlarges.
Mutant and inhibitor studies reveal critical steps of inversion
The role of the cytoskeleton in driving cell shape changes during inversion has been revealed through inhibitor studies and by mutants that arrest inversion at different stages. Blocking microtubule polymerization arrests the inversion process at an early stage, but does not affect the formation of spindle-shaped cells in the early embryo or the subsequent opening of the phialopore (Viamontes et al., 1979). The absence of microtubules allows the phialopore to widen, yet the phialopore lips are unable to curl backward (Viamontes et al., 1979). Closer examination showed that cells from inhibitor-treated embryos were unable to transition from spindle shape to flask shape, thus demonstrating the importance of these shape changes for causing tissue bending during inversion (Viamontes et al., 1979).
An additional role of the microtubule cytoskeleton was revealed by the isolation and characterization of an inversionless mutant, invA. invA− mutants begin the process of inversion normally with the formation of flask-shaped cells in the correct location and a slight curving back of the phialopore lips; however, the acute backward bend in the cell sheet adjacent to the phialopore does not form, thus precluding further progress of inversion (Nishii et al., 2003). Closer examination of invA− cells revealed that cell shape changes were normal, but that the cytoplasmic bridges in flask-shaped cells failed to relocate to the narrow stalk-ends (Nishii et al., 2003). These observations demonstrated that invA is required for cellular movement relative to the cytoplasmic bridges. Further strengthening this idea was the finding that the invA locus encodes a predicted microtubule motor protein from the kinesin family. InvA is transcribed and translated during inversion with InvA protein localizing specifically to cytoplasmic bridges (Nishii et al., 2003). Based on these data a model was proposed where InvA is anchored to the cytoplasmic bridges and uses its kinesin domain to migrate along cortical microtubules that extend into the tips of the stalks, thereby moving the cell bodies relative to the cytoplasmic bridges to ultimately cause bending of the cell sheet (Nishii et al., 2003; Fig. 5D, inset 3). InvA defines a volvocine-algal-specific sub-family of kinesins with a Chlamydomonas ortholog, IAR1 (invA-related), that can substitute for invA in complementing the inversion defect of an invA− mutant (Nishii and Miller, 2010). The function of IAR1 in Chlamydomonas is unknown, but it is intriguing that IAR1 can mediate cellular changes during Volvox morphogenesis when the unicellular Chlamydomonas has no documented cytoplasmic bridges and is not known to undergo any shape changes during its life cycle that in any way resemble those of Volvox cells during inversion. This cross-species complementation result highlights the potential for evolutionary developmental studies in Volvox and Chlamydomonas to inform about the functions carried out by genes in a unicellular ancestor that may have been co-opted for morphogenetic functions in a multicellular descendant. It also underscores the value of having access to model organisms representing two ends of the spectrum of complexity and multicellular organization in the volvocine lineage.
The actin cytoskeleton also plays a role in inversion, but is required specifically during the second half of the process for inversion of the posterior hemisphere. Inhibitors of actin polymerization or of the actin-associated motor protein myosin arrest the inversion process at the midway point, after the anterior hemisphere has inverted but before the posterior hemisphere has snapped-through (Nishii and Ogihara, 1999). Consistent with this finding, normal cell shape changes and tissue bending occurred in the anterior hemisphere when actomyosin activity was blocked (Nishii and Ogihara, 1999). When observing the posterior hemisphere, it was found that the diameter of the posterior hemisphere was larger in embryos treated with actomyosin inhibitors compared to that in untreated embryos, demonstrating that actomyosin activity is necessary for contraction of the posterior hemisphere during inversion and suggesting that an uncontracted posterior hemisphere is too wide to fit through the phialopore. When the anterior hemisphere was microsurgically removed from an embryo whose inversion was arrested with an actomyosin inhibitor, the posterior hemisphere was able to complete inversion without contracting (Nishii and Ogihara, 1999). This finding confirms that the width of the phialopore is the limiting factor for inversion of the posterior hemisphere and that during the second half of inversion actomyosin activity must drive the contraction of the posterior hemisphere in order for the posterior hemisphere to squeeze through the phialopore opening that has narrowed as a result of inversion of the anterior hemisphere. The actomyosin-driven cell shape or cell size changes that cause posterior hemisphere contraction during inversion have not been fully characterized, but as mentioned above they are not related to either the spindle-to-flask shape transition or to cytoplasmic bridge migrations that both occur normally in embryos treated with actin or myosin inhibitors (Nishii and Ogihara, 1999). Mutants that phenocopy the action of actin and myosin inhibitors have not been described, but could be informative about how the actomyosin cytoskeleton drives contraction of the posterior hemisphere.
Two additional inversionless mutants highlight the importance of the extra-embryonic vesicle in allowing morphogenesis to occur. invB and invC mutants appear to arrest the process of inversion completely without even the first stage of embryo contraction or phialopore opening occurring. Subsequent analyses revealed that embryos in both mutants are enclosed by an abnormally small glycoprotein vesicle that constrains phialopore opening and normal backward bending of the cell sheet (Ueki and Nishii, 2009, 2008). invB encodes a GDP-mannose transporter and invC encodes a predicted Golgi-localized glycosyltransferase, both of which are potentially implicated in secretory processes. While in theory either of these secretory functions might be required in some direct manner within the embryonic cells for proper morphogenesis, in the case of InvB this idea was ruled out by mechanically releasing invB mutant embryos from their vesicles whereupon normal inversion was able to occur (Ueki and Nishii, 2009). Based on their predicted functions and vesicle expansion phenotypes it is thought that the primary role of both InvB and InvC is in secretion of extra-embryonic ECM material that allows the embryonic vesicle to expand appropriately around the embryo, giving it space to invert.
Cell-type differentiation
Cell size as a determinant of cell fate
Upon completion of embryogenesis, a wild-type juvenile spheroid contains ~16 large cells and ~2,000 small cells. The large cells will become gonidia; whereas, all the small cells will differentiate as somatic cells. The qualitative contents within a cell's cytoplasm or the position of a cell relative to other tissues or cell types following asymmetric cell division(s) have been repeatedly shown to play a role in specifying cell fate in animals (Davidson, 1986; Lécuyer et al., 2007; Palacios, 2007) and plants (Benfey, 2016; Fisher and Sozzani, 2016; Moreno-Risueno et al., 2015). Yet the possibility that the size of a cell may have an effect on its subsequent fate has not been extensively examined. The strictly anterior location of asymmetric cell division in Volvox indicates that an anterior-posterior axis of polarity exists within cleaving embryos, and this axis could in principle be involved in later cell fate decisions. For example, the presumptive germ cells in Volvox might receive a specialized subset of anterior cytoplasm that determines their fate as do primordial germ cells of some animals such as pole cells in Drosophila embryos and the P4 cell in C. elegans embryos (Strome and Lehmann, 2007). Despite the many precedents and suggestive evidence for spatially dictated mechanisms of cell fate specification in Volvox, an elegant and thorough study by Kirk and colleagues convincingly ruled out such mechanisms (Kirk et al., 1993). Instead the authors found that regardless of its origin within the cleaving embryo, final cell size and not cell position was responsible for cell fate determination. To rule out other mechanisms, Kirk and colleagues made use of a wide range of genetic, physiological and mechanical manipulations that altered the timing or location of asymmetric cell division and/or the sizes of cells at the end of division. In each case they found that large cells (>8μm in diameter), regardless of location, adopted a gonidial fate while the opposite was true for small cells. Similar findings were made by Pall and colleagues using pcd (premature cessation of division) mutants, mentioned above, whose abnormally large cells in both hemispheres of the embryo all went on to adopt a germ cell fate (Pall, 1975). All data gathered to date point to cell size, not cytoplasmic content, as the key determinant of cell fate in Volvox carteri.
Interestingly, as with inversion, not all species of Volvox use the same mechanisms for germ-soma differentiation. For example, Volvox obversus has asymmetric embryonic cell divisions like V. carteri, but in this species both position within the embryo and cell size influence gonidial cell fate specification (Ransick, 1991). Contrasting with the above two species are more distantly related species of Volvox that specify germ cells without using any asymmetric embryonic cell divisions (e.g. Volvox aureus, Volvox gigas, Volvox rousseletii and Volvoxglobator) (Fig. 2A). In these species, germ-soma differentiation is not complete as all cells in the embryo initially adopt a somatic fate, where they develop fully functional flagella, and then acquire a gonidial fate by resorbing their flagella and growing in size (Darden, 1966; Kirk, 1998; Pocock, 1933a, 1933b; Ransick, 1993). In these cases, germ-soma differentiation is controlled in a temporal manner that is reminiscent of the transition from a flagellated motile phase to an aflagellated reproductive phase during the life cycle of the unicellular Chlamydomonas. Thus, this temporal program of germ-soma specification is ancestral to the Volvocine algae and was likely co-opted by V. carteri and V. obversus and regulated in a spatial context following asymmetric cell division. An exciting avenue for future work will be to elucidate the molecular determinants of germ-soma specification in different clades of Volvox (Fig. 2) and determine whether a coherent cellular differentiation program is conserved in each sub-lineage but subject to different regulatory inputs including spatial and temporal cues, or whether germ-soma differentiation is independently derived in the separate clades.
Converting cell size to a cell fate decision
The mechanism by which cell size triggers differential cell fates in Volvox remains an intriguing puzzle. Cell-type differentiation in Volvox is remarkably sensitive to light and normally only initiates at the onset of the light period following completion of embryogenesis (Kirk and Kirk, 1983)(Fig. 3, Cell differentiation). It is known that numerous changes in protein synthetic patterns occur at dark-to-light transitions in Volvox and these changes occur independently of transcription or photosynthetic activity, suggesting that transcripts are selectively translated in Volvox in response to light signaling (Kirk and Kirk, 1983, 1985). In addition, some differential polypeptide labeling was observed between early gonidial and somatic cells exposed to light and became more pronounced later (Kirk and Kirk, 1983), yet connections between translation of specific mRNAs and the process of cytodifferentiation have not been examined further. Although photosynthetic activity was not directly tied to control of translation in differentiating juvenile spheroids (Kirk and Kirk, 1985), it is still possible that it could serve as a means of coupling physiological differences in large versus small cells to the activity of differentiation factors. Both large and small postembryonic cells have a single nucleus and the same DNA content, but vastly different surface to volume ratios, and by extension, different amounts of cytoplasm, chloroplast and mitochondria. Consequently, rates of photosynthesis and accumulation of metabolites and metabolic by-products such as reactive oxygen species (ROS) are almost certainly different in large versus small cells. Regulation of gene expression can be highly sensitive to cellular metabolic status (Goodenough et al., 2014; Görke and Stülke, 2008; Hua et al., 2004; McLaughlin and Smith, 1994; Wang et al., 2010; Zubay et al., 1970), so it is possible that in Volvox one or more transcription or translation factors is altered in response to the different metabolic outputs of large and small cells and thereby triggers a cascade of events that leads to cells locking in a germ or somatic fate. Stress in Volvox, including exposure to ROS, has been shown to be a potent cue for sexual differentiation (Kirk and Kirk, 1986; Nedelcu and Michod, 2003; Nedelcu et al., 2004), but the relationship between metabolic stress or other physiological activities and vegetative germ-soma differentiation has not been examined.
Another possibility for conversion of cell size to cell fate decisions that is not mutually exclusive with the metabolic control scenario described above is through cooption of the size control mechanism that operates during multiple fission. In Chlamydomonas, the retinoblastoma (RB) tumor suppressor pathway governs the relationship between cell size and cell cycle entry or exit. The RB pathway influences the critical size that cells must reach before they can commit to divide, and it is required to ensure that mother cells of different sizes divide the appropriate number of times to produce 2n uniform-sized daughters (Fang et al., 2006; Umen and Goodenough, 2001). Moreover, a recently characterized protein in Chlamydomonas, CDKG1, has been proposed to act as a “sizer” whose abundance is coupled to mother cell division number (Li et al., 2016). The proteins of the Chlamydomonas RB pathway, including CDKG1, are conserved across the volvocine algae (Ferris et al., 2010; Hanschen et al., 2016; Hiraide et al., 2013; Prochnik et al., 2010) meaning that the machinery needed to sense cell size and convert it to a binary fate decision (i.e. divide or not) was already in place when Volvox evolved. It is therefore possible that the cell-size-dependent decision-making mechanism used by the RB pathway in volvocine algae may have been partially rewired and coopted to couple cell size to a cell fate decision (reproduction or terminal differentiation) in Volvox. The RB pathway is also likely to be involved in other aspects of embryogenesis where, for example, its activity is predicted to be modified in the large daughters formed by asymmetric cell division in order for them to stop dividing at a different cell size and cycle number compared to the somatic cell precursors (Kirk, 2005). The pcd mutants described above (Kirk et al., 1993; Pall, 1975) might be particularly informative on the topic of differential cell division control. One interpretation of pcd phenotypes is that all blastomeres in a pcd embryo adopt the division pattern of large asymmetric daughters that exit division five or six cycles earlier than the remaining cells. Further progress and testing of this idea awaits cloning of pcd genes and characterization pcd gene products.
Cytodifferentiation
Visible signs of cytodifferentiation appear upon exposure to light after embryogenesis is completed. Cytoplasmic bridges disappear and secretion of ECM begins (Dauwalder et al., 1980; Schmitt et al., 1992; Sessoms and Huskey, 1973; Wenzl et al., 1984). Gonidial initials develop radial symmetry, resorb their flagellar stubs that are present at the end of embryonic cell divisions, and begin to grow—eventually increasing by 140-fold in volume; whereas somatic initials exhibit limited growth with around a 9-fold volume increase and develop a highly polarized cytology including elongation of two apical flagella (Coggin and Kochert, 1986; Hoops, 1993; Kirk, 1998; Kirk et al., 1993; Koufopanou, 1994; Nozaki, 1994).
As described above, gonidial- and somatic-initials exhibit progressively diverging patterns of protein synthesis during the light period following completion of embryogenesis (Kirk and Kirk, 1983). Subsequent work identified a few dozen mRNAs that accumulate specifically in either gonidial or somatic cells during differentiation (Tam and Kirk, 1991; Tam et al., 1991). Sequencing of gonidial-specific genes showed that 15 of the 19 encoded proteins were predicted to be involved in photosynthesis-related functions, suggesting that photosynthetic activity may be suppressed in somatic cells (Meissner et al., 1999). This finding has led to the idea that suppression of photosynthesis may be the primary mechanism for inducing a senescent fate in somatic cells, such that somatic cells are unable to grow sufficiently large or rapidly enough to reproduce (Kirk, 2001; Meissner et al., 1999; Michod, 2007; Nedelcu and Michod, 2004; Nedelcu, 2009). It should be noted, however, that information about comparative rates and productivity of photosynthesis in somatic versus gonidial cells is lacking, and that somatic cells have been shown to actively fix CO2 (Pommerville and Kochert, 1982) and must also consume energy to maintain basal metabolism, flagellar motility and biosynthesis and secretion of ECM.
Somatic cell senescence and death
A hallmark of somatic cells is their terminal differentiation. After juvenile spheroids hatch the parental somatic cells undergo senescence and eventually die. Multiple lines of evidence suggest that the progression of somatic cells through senescence and death is part of a genetically controlled developmental program as opposed to being a passive process of necrosis. Rather than dying stochastically, somatic cells experience a synchronized and precipitous loss in viability ~100hr after embryogenesis is completed. Furthermore, this loss in viability is delayed when the population is treated with a protein synthesis inhibitor suggesting that cell death is an active process (Pommerville and Kochert, 1982, 1981). Despite it appearing to be under active control, we refrain here from using the term programmed cell death since it implies a stereotyped set of events as defined by the process of apoptosis in animal cells that have not been examined in Volvox. Senescing Volvox somatic cells appear to enter a starvation-like state with changes in cyto-architecture towards a more disorganized chloroplast thylakoid system and accumulation of lipid bodies (Pommerville and Kochert, 1981). During senescence, chlorophyll and protein levels decline rapidly as do rates of photosynthesis (Pommerville and Kochert, 1982, 1981), just as occurs in senescing plant leaves (Guo and Gan, 2005). During leaf senescence, essential nutrients are released and reused by the rest of the plant to support the growth of young tissues (Guo and Gan, 2005). Whether an analogous process of nutrient recycling occurs in Volvox remains to be determined.
Mutants that affect germ-soma differentiation
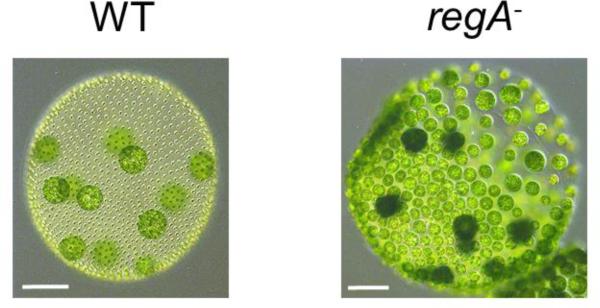
Because the dichotomy between germ and somatic cell identities is so clear in Volvox, abrogation of this dichotomy can be readily observed in culture and has allowed the isolation and genetic characterization of mutants that have defects at specific stages of cell-type specification. The most well-characterized of these mutants is the somatic regenerator (regA) mutant, in which somatic cell identity is not maintained. regA− mutants have a striking phenotype where embryogenesis and cytodifferentiation initially appear normal; however, regA− somatic cells lose their terminally differentiated state as they subsequently resorb their flagella and eyespots, and redifferentiate as gonidial cells that grow and reproduce. Consequently, regA− mutants give rise to dense, misshapen spheroids with motility defects due to absence of functional somatic cells (Huskey and Griffin, 1979; Starr, 1970)(Fig. 6). The regenerator phenotype has been found to arise from mutations at a single locus, regA, that encodes a putative transcription factor (Huskey and Griffin, 1979; Huskey et al., 1979; Kirk et al., 1999). The RegA protein is part of a small family called VARL (volvocine algal regA-like) characterized by a conserved SAND domain that is found in animal and plant transcription factors and has DNA binding activity (Bottomley et al., 2001; Carles and Fletcher, 2009; Duncan et al., 2007). Consistent with its somatic phenotype and predicted function, regA expression is restricted to somatic cells and the RegA protein is nuclear-localized (Kirk et al., 1999). Using a strain with a temperature-sensitive regA mutation RegA function was found to be required during cytodifferentiation (Huskey and Griffin, 1979). Since regA encodes a putative transcription factor it is likely that RegA binds to its target genes during cytodifferentiation and the gene products of these targets carry out their function after cytodifferentiation to maintain somatic cell fate. When regA was ectopically expressed in gonidial precursors a fruitless phenotype was seen where presumptive gonidia had slow or arrested growth, and the strains died after one or two generations due to a lack of functional gonidia (Stark et al., 2001). Therefore, regA is sufficient to suppress a reproductive or stem-cell-like fate. No mutations at loci other than the regA locus have been identified and confirmed to give rise to a regenerator-like phenotype in Volvox despite extensive searching (Huskey and Griffin, 1979; Huskey et al., 1979; Kirk, 1998).
Figure 6. The somatic regenerator (regA) mutant phenotype.
Micrographs of a wild type (WT) adult spheroid (left) and a regA− mutant (right). Note the cells that initially were somatic in the regA− spheroid have re-differentiated as gonidia and enlarged, and will eventually undergo embyrogenesis. Scale bars, 100um.
Gonidial cell specification is also under genetic control. late gonidia (lag) loci suppress the somatic identity in gonidial initials. In lag mutants gonidial initials initially develop some somatic-like features including a smaller than normal size and elongation of full length flagella. These cells will then differentiate as functional gonidial cells by resorbing their flagella and growing further in size prior to division (Kirk, 2001, 1998, 1988). It is unclear whether the reduced size of lag gonidia is caused by supernumerary division cycles during embryogenesis or a failure of gonidial initials to grow during cytodifferentiation; but these two possibilities are testable. In contrast to the reg phenotype, which is caused by mutations at only one locus (regA), mutations at multiple loci have been found that give rise to lag phenotypes (Kirk, 2001; Schmitt, 2003). To date no lag mutants have been cloned so their function in germ-soma differentiation awaits further study. The formal relationships between genes that are required for normal germ-soma differentiation are diagrammed in Figure 7.
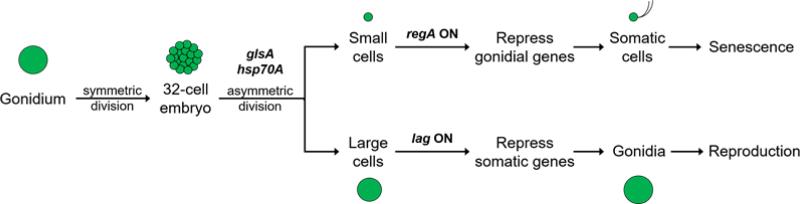
Figure 7. Formal genetic pathway for germ-soma differentiation in V. carteri.
During embryogenesis GlsA and Hsp70A are required for asymmetric divisions that give rise to small and large blastomeres. Cell size determines the fate of postembryonic cells with regA activated in small cells to repress germ cell fate, and lag genes required in large cells to partially suppress somatic fate. Modified from (Kirk, 2001).
Future Perspectives
We conclude by posing some of the key questions that remain unanswered in Volvox developmental biology and by describing some of the new tools and resources that are available to address them.
Key questions for future studies
What are the mechanisms for establishing and interpreting embryonic polarity?
Embryonic polarity plays crucial roles during Volvox development but it is unclear how anterior-posterior asymmetry is established and converted into a signal for downstream processes that include asymmetric cell division, morphogenetic events of inversion that proceed in a defined sequence along the A-P axis, and somatic cell morphological differences that also follow a gradient defined by the A-P axis. Are any developmental regulators asymmetrically localized within a gonidium prior to embryogenesis? How do these factors influence the occurrence of asymmetric division or other morphogenetic events?
How does cell size determine cell fate?
The relationship between cell size and cell fate is at the heart of germ soma differentiation in Volvox, yet it is still unclear how cell size is converted into a differentiation signal. Does cell size influence cell physiology in a way that influences differentiation? What are the factors that detect cell size and how do they influence downstream cell-type master regulators, such as RegA and Lag proteins? Is the cell size “sensing” mechanism of Volvox used in other developmental contexts or found in other species?
What are the differences between germ and somatic cells and what are the regulators that control differentiation?
We currently know only a handful of genes that show germ-soma expression dichotomy (Kianianmomeni and Hallmann, 2014; Nematollahi et al., 2006; Tam and Kirk, 1991). To begin understanding what differences underlie this most fundamental division of labor will require a more systematic investigation. The ease with which the two cell types can be separated and distinguished make this one of the most promising areas for exploration using high throughput sequencing and other genomics and genetics tools as described below. Additionally, elucidating the target genes of master regulators of cell-type differentiation in Volvox, such as RegA and GlsA, will be critical for understanding the molecular-genetic control of germ-soma differentiation.
What are the origins of developmental innovations in Volvox and other volvocine algae?
One of the major advantages of working with Volvox is that developmental mechanisms can be placed within an evolutionary framework that allows their origins to be inferred. Simpler volvocine genera lack one or more of the derived innovations found in V. carteri, such as asymmetric division, complete inversion, or germ-soma differentiation (Fig. 2). By comparing the genetic toolkits of Volvox and other volvocine relatives, we can begin to trace the molecular-genetic origins of developmental traits in this lineage and even to compare independent paths of evolution that led to different clades of Volvox (Fig. 2). For example, the GlsA protein in Volvox that is required for asymmetric cell division has a Chlamydomonas ortholog, GAR1, that can functionally substitute for the Volvox protein in a glsA mutant even though Chlamydomonas does not exhibit asymmetric cell division (Cheng et al., 2003). Similarly, the InvA kinesin from Volvox that is required for relocation of cytoplasmic bridges along the microtubule cytoskeleton during inversion also has a Chlamydomonas ortholog, IAR1, even though Chlamydomonas does not form cytoplasmic bridges or undergo the cell shape changes and cytoskeletal reorganizations that are characteristic of Volvox cells during inversion (Nishii and Miller, 2010). We have much to learn about the biology of Chlamydomonas and other volvocine algae that may be applied towards an understanding of how their conserved toolkits were modified or coopted in Volvox to participate in novel developmental processes.
New tools and resources for Volvox and volvocine algal biology
The Volvox experimental toolkit already contains many methods that are useful for developmental biology including forward and reverse genetics (Geng et al., 2014; Huskey et al., 1979; Miller et al., 1993; Ueki and Nishii, 2008), nuclear transformation and expression of transgenes including fluorescent proteins (Geng et al., 2014; Ishida, 2007; Pappas and Miller, 2009; Schiedlmeier et al., 1994), and regulated promoters (Hallmann and Sumper, 1994; von der Heyde et al., 2015). CRISPR-mediated gene targeting has been reported for Chlamydomonas (Shin et al., 2016) and could probably be adapted for use in Volvox and other volvocine species. Additional tools and resources developed in recent years will make tackling some of the above questions easier (Herron, 2016; Umen and Olson, 2012). Foremost among these are genome sequences that provide a foundation for molecular genetics and genomics approaches that were not available during the heyday of Volvox forward genetics in the 1970s-2000s. Genome sequences are now available for three volvocine species: Chlamydomonas reinhardtii, Volvox carteri and Gonium pectorale (Hanschen et al., 2016; Merchant et al., 2007; Prochnik et al., 2010). As the cost of sequencing goes down, more volvocine genomes will surely follow. Genome sequences enable large-scale systematic examinations of gene content evolution and expression profiling. They also make easier the process of mapping and cloning developmental mutants, many of which have been lost from V. carteri strain collections but could be re-isolated. Rough genetic maps of Volvox have been produced using mutant loci and/or RAPD markers (Harper et al., 1987; Huskey et al., 1979; Kirk, 1998), but with new genome sequencing technology the potential exists to identify many more markers and to make use of new strategies of cloning by re-sequencing that can accelerate the speed with which causative changes in developmental mutants are identified (Blumenstiel et al., 2009; Doitsidou et al., 2010; Dutcher et al., 2012). This re-sequencing approach can bypass the effective but still unreliable and time consuming approach of transposon tagging that has been the mainstay of Volvox forward genetics since the 1990s (Kirk et al., 1999; Miller and Kirk, 1999; Miller et al., 1993; Nishii et al., 2003; Ueki and Nishii, 2008). At the same time, new imaging technologies might be used to speed up the identification and characterization of Volvox developmental mutants. High throughput flow cytometry or digital image processing methods could be employed to help automate the search for new developmental mutants. In addition, Volvox is well suited to methods such as selective plane illumination microscopy (SPIM) for collecting real time high resolution morphological data on live specimens (Haas and Goldstein, 2015; Huisken and Stainier, 2009). Finally, the similarities in gene content (Prochnik et al., 2010) and underlying cell biology (Coleman, 2012; Hoops, 1984; Johnson and Porter, 1968; Kirk, 1998; Kochert and Olson, 1970; Lang, 1963) of Chlamydomonas and Volvox should motivate and enable cross-species studies that can take advantage of tools in both systems to help illuminate the nature and origin of developmental novelties in Volvox, many of which seem to have arisen through modification of an ancestral cell biological process still present in Chlamydomonas (Cheng et al., 2003; Geng et al., 2014; Kirk, 2005; Nedelcu, 2009; Nishii et al., 2003; Prochnik et al., 2010). With so many new tools and resources available now or in the near future, developmental biology in Volvox has never looked more promising or exciting.
Highlights.
Volvox is a simple tractable model for investigating fundamental developmental questions.
Patterned symmetric and asymmetric embryonic cell divisions underlie cell fate specification.
Differences in postembryonic cell size dictate differentiation as germ or soma.
Cell shape changes drive inversion, a morphogenetic process analogous to animal gastrulation.
New technologies will enhance Volvox and its relatives as a model for evolution and development.
Acknowledgements
This work was supported by NIH grant GM078376 to JGU and by a William H. Danforth Plant Sciences Graduate Fellowship to GM. We thank David Kirk for providing us with permissions to use scanning electron microscope images of Volvox embryogenesis. We thank the three anonymous reviewers of this manuscript for their valuable comments and suggestions for improvement.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arakaki Y, Kawai-Toyooka H, Hamamura Y, Higashiyama T, Noga A, Hirono M, Olson BJSC, Nozaki H. The simplest integrated multicellular organism unveiled. PLoS One. 2013;8 doi: 10.1371/journal.pone.0081641. doi:10.1371/journal.pone.0081641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. The origin and early evolution of germ cells as illustrated by the Volvocales. In: Halvorson H, Monroy A, editors. The Origin and Evolution of Sex. Wood's Hole Symposium. Liss, New York: 1985. pp. 221–256. [Google Scholar]
- Benfey PN. Essays on Developmental Biology, Part A. Elsevier Inc.; 2016. Defining the Path from Stem Cells to Differentiated Tissue; pp. 35–43. doi:10.1016/bs.ctdb.2015.12.002. [DOI] [PubMed] [Google Scholar]
- Bišová K, Zachleder V. Cell-cycle regulation in green algae dividing by multiple fission. J. Exp. Bot. 2014;65:2585–602. doi: 10.1093/jxb/ert466. doi:10.1093/jxb/ert466. [DOI] [PubMed] [Google Scholar]
- Blumenstiel JP, Noll AC, Griffiths JA, Perera AG, Walton KN, Gilliland WD, Hawley RS, Staehling-Hampton K. Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics. 2009;182:25–32. doi: 10.1534/genetics.109.101998. doi:10.1534/genetics.109.101998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomley MJ, Collard MW, Huggenvik JI, Liu Z, Gibson TJ, Sattler M. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat. Struct. Biol. 2001;8:626–33. doi: 10.1038/89675. doi:10.1038/89675. [DOI] [PubMed] [Google Scholar]
- Brumley DR, Wan KY, Polin M, Goldstein RE. Flagellar synchronization through direct hydrodynamic interactions. Elife. 2014;3:e02750. doi: 10.7554/eLife.02750. doi:10.7554/eLife.02750.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield NJ. Paleontology. Terminal developments in Ediacaran embryology. Science. 2011;334:1655–6. doi: 10.1126/science.1216125. doi:10.1126/science.1216125. [DOI] [PubMed] [Google Scholar]
- Callahan AM, Huskey RJ. Genetic control of sexual development in Volvox. Dev. Biol. 1980;80:419–35. doi: 10.1016/0012-1606(80)90416-9. [DOI] [PubMed] [Google Scholar]
- Carles CC, Fletcher JC. The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes Dev. 2009;23:2723–8. doi: 10.1101/gad.1812609. doi:10.1101/gad.1812609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Fowler R, Tam L, Edwards L, Miller SM. The role of GlsA in the evolution of asymmetric cell division in the green alga Volvox carteri. Dev. Genes Evol. 2003;213:328–35. doi: 10.1007/s00427-003-0332-x. doi:10.1007/s00427-003-0332-x. [DOI] [PubMed] [Google Scholar]
- Cheng Q, Hallmann A, Edwards L, Miller SM. Characterization of a heat-shock-inducible hsp70 gene of the green alga Volvox carteri. Gene. 2006;371:112–120. doi: 10.1016/j.gene.2005.11.026. doi:10.1016/j.gene.2005.11.026. [DOI] [PubMed] [Google Scholar]
- Cheng Q, Pappas V, Hallmann A, Miller SM. Hsp70A and GlsA interact as partner chaperones to regulate asymmetric division in Volvox. Dev. Biol. 2005;286:537–48. doi: 10.1016/j.ydbio.2005.08.028. doi:10.1016/j.ydbio.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Coggin SJ, Kochert G. Flagellar Development and Regeneration in Volvox carteri (Chlorophyta). J. Phycol. 1986;22:370–381. doi:10.1111/j.1529-8817.1986.tb00038.x. [Google Scholar]
- Coleman AW. A Comparative Analysis of the Volvocaceae (Chlorophyta). J. Phycol. 2012;48:491–513. doi: 10.1111/j.1529-8817.2012.01168.x. doi:10.1111/j.1529-8817.2012.01168.x. [DOI] [PubMed] [Google Scholar]
- Coleman AW. Phylogenetic analysis of “Volvocacae” for comparative genetic studies. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13892–13897. doi: 10.1073/pnas.96.24.13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coneva V, Chitwood DH. Plant architecture without multicellularity: quandaries over patterning and the soma-germline divide in siphonous algae. Front. Plant Sci. 2015;6:1–6. doi: 10.3389/fpls.2015.00287. doi:10.3389/fpls.2015.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coss RA. Mitosis in Chlamydomonas reinhardtii basal bodies and the mitotic apparatus. J. Cell Biol. 1974;63:325–9. doi: 10.1083/jcb.63.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR, Umen JG. The Chlamydomonas cell cycle. Plant J. 2015;82:370–392. doi: 10.1111/tpj.12795. doi:10.1111/tpj.12795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauwalder M, Whaley WG, Starr RC. Differentiation and secretion in Volvox. J. Ultrastruct. Res. 1980;70:318–35. doi: 10.1016/s0022-5320(80)80015-3. [DOI] [PubMed] [Google Scholar]
- Davidson E. Gene Activity in Early Development. 3rd ed. Academic Press, Inc; Orlando, FL.: 1986. [Google Scholar]
- Deason TR, Darden WH, Ely S. The development of sperm packets of the M5 strain of Volvox aureus. J. Ultrastruct. Res. 1969;26:85–94. doi: 10.1016/s0022-5320(69)90037-9. doi:10.1016/S0022-5320(69)90037-9. [DOI] [PubMed] [Google Scholar]
- Desnitski AG. A review on the evolution of development in Volvox — morphological and physiological aspects. Eur. J. Protistol. 1995;31:241–247. doi:10.1016/S0932-4739(11)80087-8. [Google Scholar]
- Doitsidou M, Poole RJ, Sarin S, Bigelow H, Hobert O. C. elegans Mutant Identification with a One-Step Whole-Genome-Sequencing and SNP Mapping Strategy. PLoS One. 2010;5:e15435. doi: 10.1371/journal.pone.0015435. doi:10.1371/journal.pone.0015435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan L, Nishii I, Harryman A, Buckley S, Howard A, Friedman NR, Miller SM. The VARL gene family and the evolutionary origins of the master cell-type regulatory gene, regA, in Volvox carteri. J. Mol. Evol. 2007;65:1–11. doi: 10.1007/s00239-006-0225-5. doi:10.1007/s00239-006-0225-5. [DOI] [PubMed] [Google Scholar]
- Dutcher SK, Li L, Lin H, Meyer L, Giddings TH, Kwan AL, Lewis BL. Whole-Genome Sequencing to Identify Mutants and Polymorphisms in Chlamydomonas reinhardtii. G3 (Bethesda) 2012;2:15–22. doi: 10.1534/g3.111.000919. doi:10.1534/g3.111.000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehler LL, Holmes JA, Dutcher SK. Loss of spatial control of the mitotic spindle apparatus in a Chlamydomonas reinhardtii mutant strain lacking basal bodies. Genetics. 1995;141:945–60. doi: 10.1093/genetics/141.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S-C, de los Reyes C, Umen JG. Cell size checkpoint control by the retinoblastoma tumor suppressor pathway. PLoS Genet. 2006;2:e167. doi: 10.1371/journal.pgen.0020167. doi:10.1371/journal.pgen.0020167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P, Olson BJSC, De Hoff PL, Douglass S, Casero D, Prochnik S, Geng S, Rai R, Grimwood J, Schmutz J, Nishii I, Hamaji T, Nozaki H, Pellegrini M, Umen JG. Evolution of an expanded sex-determining locus in Volvox. Science. 2010;328:351–4. doi: 10.1126/science.1186222. doi:10.1126/science.1186222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AP, Sozzani R. Uncovering the networks involved in stem cell maintenance and asymmetric cell division in the Arabidopsis root. Curr. Opin. Plant Biol. 2016;29:38–43. doi: 10.1016/j.pbi.2015.11.002. doi:10.1016/j.pbi.2015.11.002. [DOI] [PubMed] [Google Scholar]
- Geng S, De Hoff P, Umen JG. Evolution of Sexes from an Ancestral Mating-Type Specification Pathway. PLoS Biol. 2014;12:e1001904. doi: 10.1371/journal.pbio.1001904. doi:10.1371/journal.pbio.1001904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, Blaby I, Casero D, Gallaher SD, Goodson C, Johnson S, Lee JH, Merchant SS, Pellegrini M, Roth R, Rusch J, Singh M, Umen JG, Weiss TL, Wulan T. The path to triacylglyceride obesity in the sta6 strain of Chlamydomonas reinhardtii. Eukaryot. Cell. 2014;13:591–613. doi: 10.1128/EC.00013-14. doi:10.1128/EC.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008;6:613–24. doi: 10.1038/nrmicro1932. doi:10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- Graham C, Morgan R. Changes in the cell cycle during early amphibian development. Dev. Biol. 1966;14:439–460. doi:10.1016/0012-1606(66)90024-8. [Google Scholar]
- Green KJ, Kirk DL. Cleavage patterns, cell lineages, and development of a cytoplasmic bridge system in Volvox embryos. J. Cell Biol. 1981;91:743–55. doi: 10.1083/jcb.91.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KJ, Viamontes GI, Kirk DL. Mechanism of formation, ultrastructure, and function of the cytoplasmic bridge system during morphogenesis in Volvox. J. Cell Biol. 1981;91:756–69. doi: 10.1083/jcb.91.3.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosberg RK, Strathmann RR. The Evolution of Multicellularity: A Minor Major Transition? Annu. Rev. Ecol. Evol. Syst. 2007;38:621–654. doi:10.1146/annurev.ecolsys.36.102403.114735. [Google Scholar]
- Guo Y, Gan S. Leaf Senescence: Signals, Execution, and Regulation. Curr. Top. Dev. Biol. 2005;71:83–112. doi: 10.1016/S0070-2153(05)71003-6. doi:10.1016/S0070-2153(05)71003-6. [DOI] [PubMed] [Google Scholar]
- Haas P. a, Goldstein RE. Elasticity and glocality: initiation of embryonic inversion in Volvox. J. R. Soc. Interface. 2015;12 doi: 10.1098/rsif.2015.0671. doi:10.1098/rsif.2015.0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmann A. Morphogenesis in the Family Volvocaceae: Different Tactics for Turning an Embryo Right-side Out. Protist. 2006;157:445–461. doi: 10.1016/j.protis.2006.05.010. doi:10.1016/j.protis.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Hallmann A. Extracellular matrix and sex-inducing pheromone in Volvox. Int. Rev. Cytol. 2003;227:131–182. doi: 10.1016/s0074-7696(03)01009-x. doi:10.1016/S0074-7696(03)01009-X. [DOI] [PubMed] [Google Scholar]
- Hallmann A, Sumper M. Reporter genes and highly regulated promoters as tools for transformation experiments in Volvox carteri. Proc. Natl. Acad. Sci. U. S. A. 1994;91:11562–6. doi: 10.1073/pnas.91.24.11562. doi:10.1073/pnas.91.24.11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanschen ER, Marriage TN, Ferris PJ, Hamaji T, Toyoda A, Fujiyama A, Neme R, Noguchi H, Minakuchi Y, Suzuki M, Kawai-Toyooka H, Smith DR, Sparks H, Anderson J, Bakarić R, Luria V, Karger A, Kirschner MW, Durand PM, Michod RE, Nozaki H, Olson BJSC. The Gonium pectorale genome demonstrates co-option of cell cycle regulation during the evolution of multicellularity. Nat. Commun. 2016;7:11370. doi: 10.1038/ncomms11370. doi:10.1038/ncomms11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JF, Huson KS, Kirk DL. Use of repetitive sequences to identify DNA polymorphisms linked to regA, a developmentally important locus in Volvox. Genes Dev. 1987;1:573–584. doi: 10.1101/gad.1.6.573. doi:10.1101/gad.1.6.573. [DOI] [PubMed] [Google Scholar]
- Harris EH. Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:363–406. doi: 10.1146/annurev.arplant.52.1.363. doi:10.1146/annurev.arplant.52.1.363. [DOI] [PubMed] [Google Scholar]
- Herron MD. Origins of multicellular complexity: Volvox and the volvocine algae. Mol. Ecol. 2016;25:1213–23. doi: 10.1111/mec.13551. doi:10.1111/mec.13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron MD, Desnitskiy AG, Michod RE. Evolution of Developmental Programs in Volvox (Chlorophyta). J. Phycol. 2010;46:316–324. doi:10.1111/j.1529-8817.2009.00803.x. [Google Scholar]
- Herron MD, Hackett JD, Aylward FO, Michod RE. Triassic origin and early radiation of multicellular volvocine algae. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3254–8. doi: 10.1073/pnas.0811205106. doi:10.1073/pnas.0811205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron MD, Michod RE. Evolution of complexity in the volvocine algae: transitions in individuality through Darwin's eye. Evolution. 2008;62:436–51. doi: 10.1111/j.1558-5646.2007.00304.x. doi:10.1111/j.1558-5646.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- Herron MD, Rashidi A, Shelton DE, Driscoll WW. Cellular differentiation and individuality in the “minor” multicellular taxa. Biol. Rev. Camb. Philos. Soc. 2013;88:844–61. doi: 10.1111/brv.12031. doi:10.1111/brv.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraide R, Kawai-Toyooka H, Hamaji T, Matsuzaki R, Kawafune K, Abe J, Sekimoto H, Umen J, Nozaki H. The evolution of male-female sexual dimorphism predates the gender-based divergence of the mating locus gene MAT3/RB. Mol. Biol. Evol. 2013;30:1038–1040. doi: 10.1093/molbev/mst018. doi:10.1093/molbev/mst018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhn S, Hallmann A. There is more than one way to turn a spherical cellular monolayer inside out: type B embryo inversion in Volvox globator. BMC Biol. 2011;9:89. doi: 10.1186/1741-7007-9-89. doi:10.1186/1741-7007-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoops HJ. Flagellar , cellular and organismal polarity in Volvox carteri. J. Cell Sci. 1993;117:105–117. [Google Scholar]
- Hoops HJ. Somatic cell flagellar apparatuses in two species of Volvox (Chlorophyceae). J. Phycol. 1984;20:20–27. doi:10.1111/j.0022-3646.1984.00020.x. [Google Scholar]
- Hoops HJ, Floyd GL. Mitosis, cytokinesis and colony formation in the colonial green alga Astrephomene gubernaculifera. Br. Phycol. J. 1982;17:297–310. doi:10.1080/00071618200650301. [Google Scholar]
- Hua Q, Yang C, Oshima T, Mori H, Shimizu K. Analysis of gene expression in Escherichia coli in response to changes of growth-limiting nutrient in chemostat cultures. Appl. Environ. Microbiol. 2004;70:2354–66. doi: 10.1128/AEM.70.4.2354-2366.2004. doi:10.1128/AEM.70.4.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisken J, Stainier DYR. Selective plane illumination microscopy techniques in developmental biology. Development. 2009;136:1963–75. doi: 10.1242/dev.022426. doi:10.1242/dev.022426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huldtgren T, Cunningham JA, Yin C, Stampanoni M, Marone F, Donoghue PCJ, Bengtson S. Fossilized nuclei and germination structures identify Ediacaran “animal embryos” as encysting protists. Science. 2011;334:1696–9. doi: 10.1126/science.1209537. doi:10.1126/science.1209537. [DOI] [PubMed] [Google Scholar]
- Huskey RJ. Mutants affecting vegetative cell orientation in Volvox carteri. Dev. Biol. 1979;72:236–43. doi: 10.1016/0012-1606(79)90114-3. [DOI] [PubMed] [Google Scholar]
- Huskey RJ, Griffin BE. Genetic control of somatic cell differentiation in Volvox: Analysis of somatic regenerator mutants. Dev. Biol. 1979;72:226–35. doi: 10.1016/0012-1606(79)90113-1. [DOI] [PubMed] [Google Scholar]
- Huskey RJ, Griffin BE, Cecil PO, Callahan AM. A Preliminary Genetic Investigation of Volvox carteri. Genetics. 1979;91:229–44. doi: 10.1093/genetics/91.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida H, Ota S, Inouye I. Cleavage, incomplete inversion, and cytoplasmic bridges in Gonium pectorale (Volvocales, Chlorophyta). J. Plant Res. 2013;126:699–707. doi: 10.1007/s10265-013-0553-7. doi:10.1007/s10265-013-0553-7. [DOI] [PubMed] [Google Scholar]
- Ishida K. Sexual pheromone induces diffusion of the pheromone-homologous polypeptide in the extracellular matrix of Volvox carteri. Eukaryot. Cell. 2007;6:2157–2162. doi: 10.1128/EC.00304-07. doi:10.1128/EC.00304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson UG, Porter KR. Fine structure of cell division in Chlamydomonas reinhardi. Basal bodies and microtubules. J. Cell Biol. 1968;38:403–25. doi: 10.1083/jcb.38.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R, Shook D. The bending of cell sheets-from folding to rolling. BMC Biol. 2011;9:90. doi: 10.1186/1741-7007-9-90. doi:10.1186/1741-7007-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kianianmomeni A, Hallmann A. Transcriptional analysis of Volvox photoreceptors suggests the existence of different cell-type specific light-signaling pathways. Curr. Genet. 2014:3–18. doi: 10.1007/s00294-014-0440-3. doi:10.1007/s00294-014-0440-3. [DOI] [PubMed] [Google Scholar]
- Kikuchi K. Cellular differentiation in Pleodorina californica. Cytologia (Tokyo) 1978;43:153–160. [Google Scholar]
- Kirk DL. Oogamy: inventing the sexes. Curr. Biol. 2006;16:R1028–30. doi: 10.1016/j.cub.2006.11.015. doi:10.1016/j.cub.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Kirk DL. A twelve-step program for evolving multicellularity and a division of labor. Bioessays. 2005;27:299–310. doi: 10.1002/bies.20197. doi:10.1002/bies.20197. [DOI] [PubMed] [Google Scholar]
- Kirk DL. Germ-Soma Differentiation in Volvox. Dev. Biol. 2001;238:213–23. doi: 10.1006/dbio.2001.0402. doi:10.1006/dbio.2001.0402. [DOI] [PubMed] [Google Scholar]
- Kirk DL. Volvox: Molecular-Genetic Origins of Multicellularity and Cellular Differentiation. Cambridge University Press; Cambridge, UK.: 1998. [Google Scholar]
- Kirk DL. The ontogeny and phylogeny of cellular differentiation in Volvox. Trends Genet. 1988;4:32–6. doi: 10.1016/0168-9525(88)90063-7. [DOI] [PubMed] [Google Scholar]
- Kirk DL, Birchem R, King N. The extracellular matrix of Volvox: a comparative study and proposed system of nomenclature. J. Cell Sci. 1986;80:207–31. doi: 10.1242/jcs.80.1.207. [DOI] [PubMed] [Google Scholar]
- Kirk DL, Kaufman MR, Keeling RM, Stamer K. a. Genetic and cytological control of the asymmetric divisions that pattern the Volvox embryo. Dev. Suppl. 1991;1:67–82. [PubMed] [Google Scholar]
- Kirk DL, Kirk MM. Heat shock elicits production of sexual inducer in Volvox. Science. 1986;231:51–4. doi: 10.1126/science.3941891. [DOI] [PubMed] [Google Scholar]
- Kirk DL, Kirk MM. Protein synthetic patterns during the asexual life cycle of Volvox carteri. Dev. Biol. 1983;96:493–506. doi: 10.1016/0012-1606(83)90186-0. [DOI] [PubMed] [Google Scholar]
- Kirk MM, Kirk DL. Translational regulation of protein synthesis, in response to light, at a critical stage of Volvox development. Cell. 1985;41:419–428. doi: 10.1016/s0092-8674(85)80015-5. [DOI] [PubMed] [Google Scholar]
- Kirk MM, Ransick A, McRae SE, Kirk DL. The relationship between cell size and cell fate in Volvox carteri. J. Cell Biol. 1993;123:191–208. doi: 10.1083/jcb.123.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk MM, Stark K, Miller SM, Müller W, Taillon BE, Gruber H, Schmitt R, Kirk DL. regA, a Volvox gene that plays a central role in germ-soma differentiation, encodes a novel regulatory protein. Development. 1999;126:639–47. doi: 10.1242/dev.126.4.639. [DOI] [PubMed] [Google Scholar]
- Kochert G, Olson LW. Ultrastructure of Volvox carteri. Arch. Mikrobiol. 1970;74:19–30. doi:10.1007/BF00408684. [Google Scholar]
- Koufopanou V. The Evolution of Soma in the Volvocales. Am. Nat. 1994;143:907–931. [Google Scholar]
- Lambert JD. Developmental patterns in spiralian embryos. Curr. Biol. 2010;20:R72–7. doi: 10.1016/j.cub.2009.11.041. doi:10.1016/j.cub.2009.11.041. [DOI] [PubMed] [Google Scholar]
- Lang NJ. Electron Microscopy of the Volvocaceae and Astrephomenaceae. Am. J. Bot. 1963;50:280–300. doi:10.2307/2440022. [Google Scholar]
- Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–87. doi: 10.1016/j.cell.2007.08.003. doi:10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, De Clerck O. Phylogeny and Molecular Evolution of the Green Algae. CRC. Crit. Rev. Plant Sci. 2012;31:1–46. doi:10.1080/07352689.2011.615705. [Google Scholar]
- Li Y, Liu D, López-Paz C, Olson BJ, Umen JG. A new class of cyclin dependent kinase in Chlamydomonas is required for coupling cell size to cell division. Elife. 2016;5:1–28. doi: 10.7554/eLife.10767. doi:10.7554/eLife.10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant HJ. Colony formation and inversion in the green alga Eudorina elegans. Protoplasma. 1977;93:325–339. doi:10.1007/BF01275663. [Google Scholar]
- McLaughlin J, Smith S. Metabolic regulation of glyoxylate-cycle enzyme synthesis in detached cucumber cotyledons and protoplasts. Planta. 1994;195:22–28. doi:10.1007/BF00206287. [Google Scholar]
- Meissner M, Stark K, Cresnar B, Kirk DL, Schmitt R. Volvox germline-specific genes that are putative targets of RegA repression encode chloroplast proteins. Curr. Genet. 1999;36:363–70. doi: 10.1007/s002940050511. [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, Marshall WF, Qu L, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Cardol P, Cerutti H, Chanfreau G, Chen C-L, Cognat V, Croft MT, Dent R, Dutcher S, Fernández E, Fukuzawa H, González-Ballester D, González-Halphen D, Hallmann A, Hanikenne M, Hippler M, Inwood W, Jabbari K, Kalanon M, Kuras R, Lefebvre PA, Lemaire SD, Lobanov AV, Lohr M, Manuell A, Meier I, Mets L, Mittag M, Mittelmeier T, Moroney JV, Moseley J, Napoli C, Nedelcu AM, Niyogi K, Novoselov SV, Paulsen IT, Pazour G, Purton S, Ral J-P, Riaño-Pachón DM, Riekhof W, Rymarquis L, Schroda M, Stern D, Umen J, Willows R, Wilson N, Zimmer SL, Allmer J, Balk J, Bisova K, Chen C-J, Elias M, Gendler K, Hauser C, Lamb MR, Ledford H, Long JC, Minagawa J, Page MD, Pan J, Pootakham W, Roje S, Rose A, Stahlberg E, Terauchi AM, Yang P, Ball S, Bowler C, Dieckmann CL, Gladyshev VN, Green P, Jorgensen R, Mayfield S, Mueller-Roeber B, Rajamani S, Sayre RT, Brokstein P, Dubchak I, Goodstein D, Hornick L, Huang YW, Jhaveri J, Luo Y, Martínez D, Ngau WCA, Otillar B, Poliakov A, Porter A, Szajkowski L, Werner G, Zhou K, Grigoriev IV, Rokhsar DS, Grossman AR. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–50. doi: 10.1126/science.1143609. doi:10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michod RE. Evolution of individuality during the transition from unicellular to multicellular life. Proc. Natl. Acad. Sci. U. S. A. 2007;104(Suppl):8613–8. doi: 10.1073/pnas.0701489104. doi:10.1073/pnas.0701489104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. Volvox, Chlamydomonas, and the evolution of multicellularity. Nat. Educ. 2010;3:65. [Google Scholar]
- Miller SM, Kirk DL. glsA, a Volvox gene required for asymmetric division and germ cell specification, encodes a chaperone-like protein. Development. 1999;126:649–58. doi: 10.1242/dev.126.4.649. [DOI] [PubMed] [Google Scholar]
- Miller SM, Schmitt R, Kirk DL. Jordan, an active Volvox transposable element similar to higher plant transposons. Plant Cell. 1993;5:1125–38. doi: 10.1105/tpc.5.9.1125. doi:10.1105/tpc.5.9.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Sozzani R, Yardımcı GG, Petricka JJ, Vernoux T, Blilou I, Alonso J, Winter CM, Ohler U, Scheres B, Benfey PN. Transcriptional control of tissue formation throughout root development. Science. 2015;350:426–30. doi: 10.1126/science.aad1171. doi:10.1126/science.aad1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelcu AM. Environmentally induced responses co-opted for reproductive altruism. Biol. Lett. 2009;5:805–8. doi: 10.1098/rsbl.2009.0334. doi:10.1098/rsbl.2009.0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelcu AM, Marcu O, Michod RE. Sex as a response to oxidative stress: a twofold increase in cellular reactive oxygen species activates sex genes. Proc. Biol. Sci. 2004;271:1591–6. doi: 10.1098/rspb.2004.2747. doi:10.1098/rspb.2004.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelcu AM, Michod RE. Modularity in Development and Evolution. University of Chicago Press; Chicago and London: 2004. Evolvability , Modularity , and Individuality During the Transition to Multicellularity in Volvocalean Green Algae; pp. 466–489. [Google Scholar]
- Nedelcu AM, Michod RE. Sex as a response to oxidative stress: the effect of antioxidants on sexual induction in a facultatively sexual lineage. Proc. Biol. Sci. 2003;270(Suppl):S136–9. doi: 10.1098/rsbl.2003.0062. doi:10.1098/rsbl.2003.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nematollahi G, Kianianmomeni A, Hallmann A. Quantitative analysis of cell-type specific gene expression in the green alga Volvox carteri. BMC Genomics. 2006;7:321. doi: 10.1186/1471-2164-7-321. doi:10.1186/1471-2164-7-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishii I, Miller SM. Volvox: simple steps to developmental complexity? Curr. Opin. Plant Biol. 2010;13:646–53. doi: 10.1016/j.pbi.2010.10.005. doi:10.1016/j.pbi.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Nishii I, Ogihara S. Actomyosin contraction of the posterior hemisphere is required for inversion of the Volvox embryo. Development. 1999;126:2117–2127. doi: 10.1242/dev.126.10.2117. [DOI] [PubMed] [Google Scholar]
- Nishii I, Ogihara S, Kirk DL. A kinesin, invA, plays an essential role in volvox morphogenesis. Cell. 2003;113:743–53. doi: 10.1016/s0092-8674(03)00431-8. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Takeichi M. Current Topics in Developmental Biology. 1st ed. Elsevier Inc.; 2009. Remodeling of the Adherens Junctions During Morphogenesis. doi:10.1016/S0070-2153(09)89002-9. [DOI] [PubMed] [Google Scholar]
- Nozaki H. Origin and evolution of the genera Pleodorina and Volvox (Volvocales). Biologia (Bratisl) 2003;58:425–431. [Google Scholar]
- Nozaki H. Morphology and evolution of sexual reproduction in the Volvocaceae (Chlorophyta). J. Plant Res. 1996;109:353–361. doi:10.1007/BF02344484. [Google Scholar]
- Nozaki H. Unequal flagellar formation in Volvox. Phycologia. 1994;33:58–61. doi:dx.doi.org/10.2216/i0031-8884-33-1-58.1. [Google Scholar]
- Nozaki H, Misawa K, Kajita T, Kato M, Nohara S, Watanabe MM. Origin and evolution of the colonial volvocales (Chlorophyceae) as inferred from multiple, chloroplast gene sequences. Mol. Phylogenet. Evol. 2000;17:256–68. doi: 10.1006/mpev.2000.0831. doi:10.1006/mpev.2000.0831. [DOI] [PubMed] [Google Scholar]
- Nozaki H, Mori T, Misumi O, Matsunaga S, Kuroiwa T. Males evolved from the dominant isogametic mating type. Curr. Biol. 2006;16:1018–1020. doi: 10.1016/j.cub.2006.11.019. doi:10.1016/j.cub.2006.11.019. [DOI] [PubMed] [Google Scholar]
- O'Farrell PH, Stumpff J, Su TT. Embryonic Cleavage Cycles: How Is a Mouse Like a Fly? Curr. Biol. 2004;14:35–45. doi: 10.1016/j.cub.2003.12.022. doi:10.1016/S0960-9822(03)00933-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios IM. How does an mRNA find its way? Intracellular localisation of transcripts. Semin. Cell Dev. Biol. 2007;18:163–170. doi: 10.1016/j.semcdb.2007.01.008. doi:10.1016/j.semcdb.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall ML. Mutants of Volvox showing premature cessation of division: Evidence for a relationship between cell size and reproductive cell differentiation. In: McMahon D, Fox FC, editors. Developmental Biology: Pattern Formation, Gene Regulation. W.A. Benjamin, Inc.; Menlo Park, CA: 1975. pp. 148–156. [Google Scholar]
- Pappas V, Miller SM. Functional analysis of the Volvox carteri asymmetric division protein GlsA. Mech. Dev. 2009;126:842–51. doi: 10.1016/j.mod.2009.07.007. doi:10.1016/j.mod.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Pocock MA. Volvox in South Africa. Ann. South African Museum. 1933a;16:523–646. [Google Scholar]
- Pocock MA. Volvox and associated algae from Kimberley. Ann. South African Museum. 1933b;16:473–521. [Google Scholar]
- Pommerville J, Kochert G. Effects of senescence on somatic cell physiology in the green alga Volvox carteri. Exp. Cell Res. 1982;140:39–45. doi: 10.1016/0014-4827(82)90153-7. [DOI] [PubMed] [Google Scholar]
- Pommerville JC, Kochert GD. Changes in somatic cell structure during senescence of Volvox carteri. Eur. J. Cell Biol. 1981;24:236–43. [PubMed] [Google Scholar]
- Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK, Hellsten U, Chapman J, Simakov O, Rensing S. a, Terry A, Pangilinan J, Kapitonov V, Jurka J, Salamov A, Shapiro H, Schmutz J, Grimwood J, Lindquist E, Lucas S, Grigoriev IV, Schmitt R, Kirk D, Rokhsar DS. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329:223–6. doi: 10.1126/science.1188800. doi:10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransick A. Evolutionary Conservation of Developtmental Mechanisms. Wiley-Liss; 1993. Specification of Reproductive Cells in Volvox; pp. 55–70. [Google Scholar]
- Ransick A. Reproductive cell specification during Volvox obversus development. Dev. Biol. 1991;143:185–98. doi: 10.1016/0012-1606(91)90065-b. [DOI] [PubMed] [Google Scholar]
- Sawyer JM, Harrell JR, Shemer G, Sullivan-Brown J, Roh-Johnson M, Goldstein B. Apical constriction: A cell shape change that can drive morphogenesis. Dev. Biol. 2010;341:5–19. doi: 10.1016/j.ydbio.2009.09.009. doi:10.1016/j.ydbio.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiedlmeier B, Schmitt R, Müller W, Kirk MM, Gruber H, Mages W, Kirk DL. Nuclear transformation of Volvox carteri. Proc. Natl. Acad. Sci. U. S. A. 1994;91:5080–4. doi: 10.1073/pnas.91.11.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt R. Differentiation of germinal and somatic cells in Volvox carteri. Curr. Opin. Microbiol. 2003;6:608–613. doi: 10.1016/j.mib.2003.10.007. doi:10.1016/j.mib.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Schmitt R, Fabry S, Kirk DL. In search of molecular origins of cellular differentiation in Volvox and its relatives. Int. Rev. Cytol. 1992;139:189–265. doi: 10.1016/s0074-7696(08)61413-8. [DOI] [PubMed] [Google Scholar]
- Sessoms AH, Huskey RJ. Genetic control of development in Volvox: isolation and characterization of morphogenetic mutants. Proc. Natl. Acad. Sci. U. S. A. 1973;70:1335–1338. doi: 10.1073/pnas.70.5.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlik I, Zachleder V. The Microbial Cell Cycle. CRC; Boca Raton, FL: 1984. The multiple fission cell reproductive patterns in algae; pp. 253–279. [Google Scholar]
- Shin S-E, Lim J-M, Koh HG, Kim EK, Kang NK, Jeon S, Kwon S, Shin W-S, Lee B, Hwangbo K, Kim J, Ye SH, Yun J-Y, Seo H, Oh H-M, Kim K-J, Kim J-S, Jeong W-J, Chang YK, Jeong B. CRISPR/Cas9-induced knockout and knock-in mutations in Chlamydomonas reinhardtii. Sci. Rep. 2016;6 doi: 10.1038/srep27810. doi:10.1038/srep27810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnica-Krezel L, Sepich DS. Gastrulation: Making and Shaping Germ Layers. Annu. Rev. Cell Dev. Biol. 2012;28:687–717. doi: 10.1146/annurev-cellbio-092910-154043. doi:10.1146/annurev-cellbio-092910-154043. [DOI] [PubMed] [Google Scholar]
- Stark K, Kirk DL, Schmitt R. Two enhancers and one silencer located in the introns of regA control somatic cell differentiation in Volvox carteri. Genes Dev. 2001;15:1449–60. doi: 10.1101/gad.195101. doi:10.1101/gad.195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr RC. Control of differentiation in Volvox. Symp. Soc. Dev. Biol. 1970;29:59–100. doi: 10.1016/b978-0-12-395534-0.50009-1. [DOI] [PubMed] [Google Scholar]
- Starr RC. Structure, reproduction, and differentiation in Volvox carteri f. nagariensis Iyengar, Strains HK 9 & 10. Arch. Protistenk. 1969;111:204–222. [Google Scholar]
- Strome S, Lehmann R. Germ versus soma decisions: lessons from flies and worms. Science. 2007;316:392–393. doi: 10.1126/science.1140846. doi:10.1126/science.1140846. [DOI] [PubMed] [Google Scholar]
- Tam LW, Kirk DL. Identification of cell-type-specific genes of Volvox carteri and characterization of their expression during the asexual life cycle. Dev. Biol. 1991;145:51–66. doi: 10.1016/0012-1606(91)90212-l. [DOI] [PubMed] [Google Scholar]
- Tam LW, Stamer KA, Kirk DL. Early and late gene expression programs in developing somatic cells of Volvox carteri. Dev. Biol. 1991;145:67–76. doi: 10.1016/0012-1606(91)90213-m. [DOI] [PubMed] [Google Scholar]
- Ueki N, Matsunaga S, Inouye I, Hallmann A. How 5000 independent rowers coordinate their strokes in order to row into the sunlight: phototaxis in the multicellular green alga Volvox. BMC Biol. 2010;8:103. doi: 10.1186/1741-7007-8-103. doi:10.1186/1741-7007-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki N, Nishii I. Controlled enlargement of the glycoprotein vesicle surrounding a volvox embryo requires the InvB nucleotide-sugar transporter and is required for normal morphogenesis. Plant Cell. 2009;21:1166–81. doi: 10.1105/tpc.109.066159. doi:10.1105/tpc.109.066159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki N, Nishii I. Idaten is a new cold-inducible transposon of Volvox carteri that can be used for tagging developmentally important genes. Genetics. 2008;180:1343–1353. doi: 10.1534/genetics.108.094672. doi:10.1534/genetics.108.094672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG. Green Algae and the Origins of Multicellularity in the Plant Kingdom. Cold Spring Harb. Perspect. Biol. 2014 doi: 10.1101/cshperspect.a016170. doi:10.1101/cshperspect.a016170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG. Evolution of sex and mating loci: an expanded view from Volvocine algae. Curr. Opin. Microbiol. 2011;14:634–41. doi: 10.1016/j.mib.2011.10.005. doi:10.1016/j.mib.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Goodenough UW. Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev. 2001;15:1652–61. doi: 10.1101/gad.892101. doi:10.1101/gad.892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Olson BJSC. Genomics of Volvocine Algae. Adv. Bot. Res. 2012;64 doi: 10.1016/B978-0-12-391499-6.00006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viamontes GI, Fochtmann LJ, Kirk DL. Morphogenesis in Volvox: analysis of critical variables. Cell. 1979;17:537–50. doi: 10.1016/0092-8674(79)90262-9. [DOI] [PubMed] [Google Scholar]
- Viamontes GI, Kirk DL. Cell shape changes and the mechanism of inversion in Volvox. J. Cell Biol. 1977;75:719–30. doi: 10.1083/jcb.75.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Heyde EL, Klein B, Abram L, Hallmann A. The inducible nitA promoter provides a powerful molecular switch for transgene expression in Volvox carteri. BMC Biotechnol. 2015:1–13. doi: 10.1186/s12896-015-0122-3. doi:10.1186/s12896-015-0122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Jiang JC, Jazwinski SM. Gene regulatory changes in yeast during life extension by nutrient limitation. Exp. Gerontol. 2010;45:621–631. doi: 10.1016/j.exger.2010.02.008. doi:10.1016/j.exger.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzl S, Thym D, Sumper M. Development-dependent modification of the extracellular matrix by a sulphated glycoprotein in Volvox carteri. EMBO J. 1984;3:739–44. doi: 10.1002/j.1460-2075.1984.tb01877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubay G, Schwartz D, Beckwith J. Mechanism of activation of catabolite-sensitive genes: a positive control system. Proc. Natl. Acad. Sci. U. S. A. 1970;66:104–10. doi: 10.1073/pnas.66.1.104. doi:10.1101/SQB.1970.035.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]