Abstract
HIV-1 infection is associated with substantial damage to the gastrointestinal (GI) tract resulting in structural impairment of the epithelial barrier and a disruption of intestinal homeostasis. The accompanying translocation of microbial products and potentially microbes themselves from the lumen into systemic circulation has been linked to immune activation, inflammation, and HIV-1 disease progression. The importance of microbial translocation in the setting of HIV-1 infection has led to a recent focus on understanding how the communities of microbes that make up the intestinal microbiome are altered during HIV-1 infection and how they interact with mucosal immune cells to contribute to inflammation. This review details the dysbiotic intestinal communities associated with HIV-1 infection and their potential link to HIV-1 pathogenesis. We detail studies that begin to address the mechanisms driving microbiota-associated immune activation and inflammation and the various treatment strategies aimed at correcting dysbiosis and improving the overall health of HIV-1 infected individuals. Finally, we discuss how this relatively new field of research can advance to provide a more comprehensive understanding of the contribution of the gut microbiome to HIV-1 pathogenesis.
Introduction
HIV-1 infection dramatically alters the intestinal landscape resulting in pronounced changes in the structural and immunological properties of this critical organ system[1-11]. HIV-1 replicates in gut-associated lymphoid tissue in the early stages of infection, resulting in a massive depletion of mucosal effector CD4+ T cells, including T helper (Th) 17 and Th22 cells which are critical in maintaining the integrity of the gut. HIV-1 infection is also associated with enteropathy, mucosal inflammation and aberrant cytokine production, and with substantial intestinal epithelial cell damage. Disruption of the epithelial barrier is a major factor associated with local and systemic inflammation during HIV infection by allowing the movement of bacterial products, and perhaps even viable bacteria, through the lamina propria (LP) and into the systemic circulation (microbial translocation; MT)[12]. MT has been associated with systemic immune activation[13] and predicts disease progression in untreated HIV-1 infected individuals[14]. Its persistence during effective anti-retroviral therapy (ART) predicts mortality and is associated with a number of co-morbidities (e.g. cardiovascular disease, neurocognitive dementia) linked to chronic immune activation and inflammation[15-17].
The intestinal microbiome harbors trillions of bacteria, many of which play critical roles in maintaining intestinal immune homeostasis[18-20]. In recent years, numerous studies have linked alterations in the intestinal microbiota (dysbiosis)[21] to diseases and metabolic disorders such as inflammatory bowel diseases (IBD)[22-24], obesity[25, 26], type 1 diabetes[27-29], periodontal disease[30], rheumatic diseases[31], atherosclerosis and cardiovascular disease (CVD)[32-34]. Numerous studies now suggest that HIV-1 infection is also associated with changes in the enteric microbiome. The relationship between HIV-1-associated mucosal pathogenesis and the microbiome is likely a two-way street with changes in mucosa leading to dysbiosis and with dysbiosis subsequently playing a critical role in sustaining the disruption in intestinal homeostasis and further contributing to HIV-1 associated immune activation and inflammation. Because of this important interplay between the microbiome and features of HIV-1 pathogenesis, it is crucial to thoroughly understand 1) the mechanisms by which HIV-1 infection impacts the composition and diversity of the intestinal microbiome; 2) the downstream pathologic and clinical consequences of dysbiosis; 3) the impact of viral suppression with effective ART on dysbiosis and gut homeostasis; and 4) whether we can therapeutically modulate the intestinal microbiome to restore intestinal function, limit MT-mediated inflammation, and thereby greatly improve the overall health of HIV-infected individuals.
Microbiome Analysis
The advent of high-throughput sequencing technologies which allows for the comprehensive, rapid, and cost-effective evaluation of the microbial communities that exist in, on, and around us[35, 36] has dramatically advanced our understanding of host-microbe interactions. Studies investigating the intestinal microbiome in the setting of HIV-1 infection have used a number of different sequence-based technologies to comprehensively identify and enumerate the constituents of complex, host-associated bacterial communities[37-53]. Although most recent studies used the highly conserved gene encoding the small-subunit ribosomal RNA gene (16S in bacteria/archaea or 18S rRNA in eucaryotes) to infer the types of microorganisms present in a community, the particular hypervariable (V) sub-regions of the 16S gene that were targeted (due to the fact that most next-generation sequencing platforms cannot generate full-length 16S sequences) differed; a number of studies amplified and sequenced the ~250 bp V4 region[37, 40, 42, 43, 53], whereas other studies amplified V1V3[49], V3V4[46, 48, 52], V3V5[39, 50] or V6[47] regions (~530 bp, ~460 bp, ~570 bp, and ~120 bp, respectively). The choice of hypervariable region and particular broad-range primers used for PCR amplification undoubtedly affect the resulting sequence dataset(s) since V regions differ intrinsically in their abilities to resolve phylogeny[24, 54, 55].
Other methodological variations include the type and anatomical site of samples collected, which included intestinal tissue biopsies[37, 44, 51, 53], stool samples[37, 39-41, 43-47, 49], as well as rectal mucosal samples collected using a sponge-based sampling technique[42] and duodenum samples acquired by brushing the mucosa[52]. Given that bacterial communities differ not only between GI-tract mucosa versus fecal samples, but also between mucosa at different sites along the GI tract itself [56-58], it is likely that the sampling site is a contributing factor to study-specific differences. HIV disease status (acute/recent versus chronic infection versus elite controllers versus AIDS designation), treatment status (untreated versus ART), sex, race, and geographical background also differed among studies. The majority of studies have had small sample sizes of HIV-infected study participants and various uninfected control populations and thus were hypothesis-generating rather than being powered to address microbiome endpoints. Finally, a recent study highlighted the importance of appropriately matching control study participants by sexual preference[45] (discussed in more detail below). Thus clinical as well as technical variables may help to explain the inconsistencies observed among studies investigating the enteric microbiome in the setting of HIV-1 infection.
Alterations in the Diversity of Intestinal Bacterial Communities During HIV-1 Infection
The microbial diversity in a sample, which can be inferred from both the number of taxa present in the sample and the relative abundances of these taxa, is a useful means of comparing microbiota between individuals; reduced diversity has been observed across a wide range of disease states. Indeed, reductions in diversity have been observed in both untreated and treated HIV-infected subjects compared to uninfected control subjects in mucosal[42, 44] and fecal samples[43, 44, 46, 48]. Surprisingly, bacterial diversity further decreased in stool samples of HIV-1 infected study participants who were followed after the introduction of ART[46] suggesting antiretroviral drugs themselves may impact the intestinal microbiota. Men who have sex with men (MSM) were found to have greater fecal microbial diversity than non-MSM, but HIV-associated reductions in diversity persisted even when HIV-1 infected subjects were stratified for MSM vs. non-MSM[45]. Mucosal samples exhibited greater differences in diversity than did fecal samples[44]. Decreased diversity in fecal samples was associated with systemic indicators of MT and immune activation and was an independent predicator of blood CD4+ T cell counts in untreated HIV-1 infected study participants[46]. In contrast, Lozupone et al. observed significantly higher diversity in fecal samples from individuals with chronic untreated HIV-1 infection compared to uninfected controls[40]. This finding may be attributed in part to differences in sexual practices between infected and control participants. A number of other studies observed no differences in diversity in either mucosal samples from untreated[38, 51, 52] or treated[51] HIV-1 infected individuals or in fecal samples from treated HIV-1 infected study participants[39, 50].
Alterations in the Composition of Intestinal Bacterial Communities During HIV-1 infection
In 2008, Gori and colleagues published the first study suggesting that HIV-1 infection altered the gut microbiome[59]. Applying fluorescence in situ hybridization and quantitative PCR (qPCR) techniques to fecal samples, they observed increased levels of the opportunistic pathogens Pseudomonas aeruginosa and Candida albicans in conjunction with decreased levels of Bifidobacteria spp. and Lactobacilli spp., in untreated HIV-1 infected study participants compared to historical controls. Further evidence that HIV-1 infection was associated with dysbiosis came from a pilot study conducted by Ellis et al., who were the first to use qPCR to quantify overall 16S rRNA gene expression and the bacterial orders Enterobacteriales, Bacteroidales and Clostridiales in fecal samples[60]. In that study, lower levels of total fecal bacterial loads correlated with increased levels of T cell activation in duodenal tissue of HIV-1 infected study participants. Moreover, HIV-1 infection was associated with a trend towards increased Enterobacteriales, which correlated with lower frequencies of duodenal CD4+ T cells.
In 2013, the first publications using next generation sequencing (NGS) technology emerged and so began the intensive investigations into the role(s) of the microbiome in HIV-1 pathogenesis (Table 1). Alterations in bacterial community composition were typically observed in the 3 most dominant phyla found in the distal adult GI tract: Proteobacteria, Bacteroidetes and Firmicutes (Table 2). In general, dysbiosis was characterized by increased relative abundances of commensal Gram-negative bacteria, encompassing many bacterial species with reported pathogenic potential (termed pathobionts[61]), in conjunction with decreased abundances of both Gram-positive and other Gram-negative bacteria, including those with known immune-regulatory properties[62]. Furthermore, several cross-sectional studies found dysbiosis in HIV-infected individuals on effective ART with viral suppression, and in the few studies in which HIV-1-infected study participants were followed longitudinally after beginning ART, shifts in the fecal microbiome of a number of individuals persisted[40, 46]. Moreover, administration of ART appeared to further alter these microbial communities[46].
Table 1.
Features of various studies investigating the gut microbiome during HIV-1 infection.
| Vujkovic-Cvijin et al.[51] | Lozupone et al.[40] | Perez-Santiago et al[47]. | McHardy et al.[42] | |
| HIV-1 infected (M/F) | 23 (23/0) | 25 (22/3) | 13 (13/0) | 40 (40/0) |
| HIV-1 status | ||||
| Chronic | 22 | 22 | - | 40 |
| Early | - | 3 | 13 | - |
| AIDS | - | - | - | - |
| Other | - | LTNP N=1 | - | - |
| ART | 16 (+2)1 | 14 (8, 6)2 | 133 | 20 |
| Controls (M/F) | 9 (9/0) | 13 (8/5) | - | 20 (20/0) |
| Geographical Location | USA | USA | USA | USA |
| Sample site | Rectosigmoid biopsies | Fecal | Fecal | Rectal mucosa (sponge) |
| Mutlu et al.[44] | Dillon et al.[37, 38] | Lozupone et al.[41] | Vazquez-Castellanos et al.[49] | |
| HIV-1 infected (M/F) | 21 (16/5) | 18 (13/5) | 40 (M/F not reported) | 15 (12/3) |
| HIV-1 status | ||||
| Chronic | 21 | 18 | 40 | 15 |
| Early | - | - | - | - |
| AIDS | - | - | - | - |
| Other | - | - | - | - |
| ART | 19 | - | 28 (17/11)5 | 15 |
| Controls (M/F) | 22 (17/5) | 14 (9/5) | 15 (M/F not reported) | 15 (8/7) |
| Geographical Location | USA | USA | USA | Spain |
| Sample site | Multiple biopsy sites4, fecal | Colon biopsies, fecal | Fecal | Fecal |
| Volpe et al.[50] | Dinh et al.[39] | Nowak et al.[46] | Yang et al.[52] | |
| HIV-1 infected (M/F) | 15 (13/2) | 21 (17/4) | 31 (16/15) | 8 (5/3) |
| HIV-1 status | ||||
| Chronic | 15 | 21 | 31 | 87 |
| Early | - | - | - | - |
| AIDS | - | - | - | - |
| Other | - | - | Elite controller N=3 | - |
| ART | 15 | 21 | 196 | - |
| Controls (M/F) | 17 (13/4) | 16 (12/4) | 9 (5/4) | 8 (4/4) |
| Geographical Location | USA | USA | Sweden | USA |
| Sample site | Fecal | Fecal | Fecal | Duodenum (mucosal brushes)8 |
| Noguera-Julian et al.9[45] | Monaco et al.[43] | Sun et al.[48] | ||
| HIV-1 infected (M/F) | 129 (101/28) | 82 (31/51) | 13 (9/4) | |
| HIV-1 status | ||||
| Chronic | 129 | 8211 | 13 | |
| Early | - | - | - | |
| AIDS | 11 | 25 | - | |
| Other | Elite controller N=8 | - | - | |
| ART | 84 | 40 | 11 | |
| Controls (M/F) | 27 (23/3/110) | 40 (20/20) | 7 (5/2) | |
| Geographical Location | Spain | Uganda | China | |
| Sample site | Fecal | Fecal | Fecal | |
LTNP: Long-term non-progressor.
Two viremic untreated study participants returned after 9 months of effective ART.
Of the 14 chronically HIV-1 infected study participants on ART, 8 study participants were on long-term (≥ 12 months) ART and 6 study participants on short-term (≤12 months) ART with 3 of the short-term ART sampled before and after ART initiation.
All HIV-1 infected study participants started ART within 1 week of enrollment and followed for longitudinally for 48 weeks.
Samples collected from terminal ileum, right colon, left colon.
Of the 28 chronically HIV-1 infected study participants on ART, 17 were on long-term (≥ 12 months) ART and 11 study participants were on short-term (≤12 months) ART.
Of the 31 HIV-1 infected subjects, 19 were followed for a median 10months after initiating ART.
Years since first HIV-seropositive test reported as median 2.1yrs with a range of 0.16-7yrs.
Samples were also collected from esophagus and stomach.
Study included an external validation cohort; data reported for test cohort.
Of uninfected study participants, one study participant reported as Transgender.
Time since diagnosis not reported.
Table 2.
Major findings characterizing dysbiosis in HIV-1 infection compared to uninfected individuals: grouped by taxonomic hierarchy and sample site.
| Mucosal tissue | References | Fecal samples | References | |
|---|---|---|---|---|
| Phylum Proteobacteria | ↑ | [37, 51, 52] | ↑ | [39, 48, 50] |
| Family | ||||
| Enterobacteriaceae | ↑ | [44, 51] | ↑ | [39] |
| Brucellaceae | ↑ | [37] | ||
| Xanthomonadaceae | ↑ | [37] | ||
| Rhodspirillaceae | ↓ | [37] | ||
| Genus | ||||
| Escherichia | ↑ | [44, 51] | ||
| Serratia | ↑ | [51] | ||
| Shigella | ↑ | [51] | ||
| Klebsiella | ↑ | [51] | ||
| Ralstonia | ↑ | [44] | ||
| Actinobacter | ↑ | [37] | ||
| Burkholderia | ↑ | [52] | ||
| Desulfovibrio | ↑ | [40] | ||
| Thalossospira | ↓ | [37] | ||
| Phylum Bacteroidetes | ||||
| Family | ||||
| Rikenellaceae | ↓ | [42, 44] | ↓ | [39, 40] |
| Bacteroidaceae | ↓ | [37] | ↓ | [37, 40, 48] |
| Prevotellaceae | ↑ | [37] | ↑ | [40] |
| Genus | ||||
| Bacteroides | ↓ | [37, 44, 51] | ↓ | [37, 40, 48, 49] |
| Alistipes | ↓ | [37, 42, 51] | ↓ | [37, 39, 40, 50] |
| Barnesiella | ↓ | [37] | ↓ | [37] |
| ↑ | [39] | |||
| Prevotella | ↑ | [37, 44] | ↑ | [40, 49] |
| Phylum Firmicutes | ↓ | [37, 42, 52] | ||
| Family | ||||
| Lachnospiraceae | ↓ | [37, 42, 44] | ||
| Ruminococcaceae | ↓ | [37, 42, 44] | ↓ | [37] |
| Christensenellaceae | ↓ | [37] | ||
| Erysipelotrichaceae | ↑ | [51] | ↑ | [39, 40] |
| Veillonellaceae | ↑ | [40] | ||
| Genus | ||||
| Coprococcus | ↓ | [37, 42, 44] | ||
| Faecalibacterium | ↓ | [44] | ↓ | [46, 48, 49] |
| Blautia | ↓ | [37, 44] | ||
| Rumminococcus | ↓ | [42, 44] | ||
| Dorea | ↓ | [44] | ||
| Oscillospira | ↓ | [44] | ||
| Lachnospira | ↓ | [42, 44] | ↓ | [48] |
| Roseburia | ↓ | [42, 44] | ↓ | [48, 49] |
| Dialister | ↓ | [44] | ↑ | [40] |
| Eubacterium | ↓ | [42, 44] | ||
| Lactobacillus | ↓ | [52] | ↑ | [46] |
| Mitsuokella | ↑ | [40] | ||
| Catenibacterium | ↑ | [44] | ↑ | [40] |
| Mogibacterium | ↑ | [44] | ||
| Bulleidia | ↑ | [40] |
An overall enrichment of diverse commensal bacteria of the phylum Proteobacteria, a phylum that includes not only pathogenic bacteria (e.g. Salmonella, Yersinia, Escherichia), but also pathobionts (e.g. Acinetobacter), was observed in the mucosal tissue of both untreated[37, 51, 52] and treated[44] HIV-1 infected individuals. One study observed HIV-associated increases in Proteobacteria only in the colonic mucosa but not in subject-matched stool samples of untreated individuals[37], whereas other studies have shown increased Proteobacteria abundance in the stool of untreated[40] and treated [39, 48, 50] HIV-1 infected individuals. In recent studies in which bacteria were evaluated to the species level, enrichment for Burkholderia fungorium, Bradyrhizobium pachyrhizi[52], Acinetobacter junii and Schlegella thermodepolymerans[38] in the colonic mucosa and Desulfovibrio piger in the stool[40] during untreated HIV-1 infection were observed.
In contrast to the increase in Proteobacteria, numerous studies have noted decreased abundances of members of the phylum Firmicutes, which comprises a rather diverse group of Gram-positive species (Staphylococci, Streptococci, Lactobacilli, Clostridia), including many groups with immune-regulatory properties and even some used as probiotics. Reductions in Firmicutes abundance were shown to be mucosa-specific in some studies but also observed in stool samples in others, in the setting of both untreated and treated HIV-1 infection[37, 42-44, 46-49, 51, 52]. As an example, Lactobacilli were decreased in duodenal tissue from untreated HIV-1 infected subjects[52], and lower Lactobacillus abundance was also detected in stool samples of HIV-1 infected subjects with lower blood CD4+ T cell count and higher viral loads compared to those individuals with higher CD4+ T cell counts and lower viral load[47]. Surprisingly, a number of genera belonging to the Firmicutes phylum (e.g. Lachnospira, Oribacterium) were reduced after the introduction of ART[46] suggesting a direct drug effect on the gut microbiota.
The Firmicutes also include several species that produce the short chain fatty acid (SCFA) butyrate as a byproduct of fiber fermentation[63-65]. Butyrate is important in regulating intestinal homeostasis as both an energy source for epithelial cells and as a signaling molecule that modulates intestinal immune cell responses[63]. Of note, a number of the genera known to contain these butyrate producing bacteria (e.g. Roseburia, Faecalibacterium, Coprococcus, Eubacterium) were decreased in both untreated[37, 42, 46] and treated[44, 48, 49] HIV-1 infected subjects. However, not all Firmicutes species are reduced during HIV-1 infection. For instance, the family Erysipelotrichaceae, reported to be linked to other inflammatory disorders[66], was increased in the mucosa and stool of untreated and treated HIV-1 infected study participants in several studies[39, 40, 51].
One of the most consistent dysbiotic profiles observed in HIV-1 infected subjects has been a decrease in Bacteroides abundance which, in many studies, occurred with a concomitant increase in Prevotella spp. abundance. Both of these are genera of the phylum Bacteroidetes[37, 40, 42-44, 48, 49, 51, 53]. These shifts were observed irrespective of anatomical site or treatment status. Prevotella spp. are reported to have pro-inflammatory properties[38, 67-70], whereas some Bacteroides spp. are inducers of regulatory T cell function[71]. A recent study suggested that the Prevotella-rich/Bacteroides-poor community structure was more prevalent among men who have sex with men (MSM), rather than with HIV-1 infection status per se[45]. This finding highlighted the importance of appropriately accounting for the myriad of possible factors that might confound HIV-1 microbiome studies. However, decreased abundances of Bacteroides were still observed in studies in which controls were risk-matched by sexual practice and/or histories of exposure to HIV from interactions with HIV-1-infected partners[51]. Moreover, Prevotella abundances decreased after the introduction of ART[46], suggesting the potential for at least an indirect involvement of HIV-1 infection in these microbiome changes. Moreover, Prevotella spp. have been linked to HIV-1-associated mucosal immune activation in vivo and in vitro (see below) suggesting that although the increase in Prevotella spp. may not be due solely to HIV-1 infection, greater abundances of Prevotella in the enteric microbiomes of MSM may be an important contributing factor to HIV-1-associated gut pathology.
Role of Diet in HIV-associated Dysbiosis
Diet has a profound influence on the relationship between the intestinal microbiota and the host[72]. To date, few studies have investigated the impact of diet on the intestinal microbiome in the setting of HIV-1 infection. Using a limited dietary questionnaire, our group found a positive association between the number of servings of red meat and the relative abundance of Bacteroides in uninfected individuals, a relationship which was disrupted in persons with untreated HIV-1 infection [37]. Furthermore, meta-analysis revealed that the stool microbiota of HIV-1 infected subjects in the USA resembled that of the Prevotella-rich/Bacteroides-poor profiles associated with agrarian cultures[40]. Indeed, a failure to see relatively greater Prevotella abundance in HIV-1 infected individuals in Uganda was suggested to be related, at least in part, to the high abundance of Prevotella typically observed among healthy uninfected study participants in this population[43]. However, given the recent evidence that the Prevotella-rich profile observed in HIV-1 infection may be more closely linked to sexual practice than to diet or HIV-1 infection status[45], future studies will need to include extensive dietary questionnaires that are validated within the target populations with appropriately matched control participants. In this way, the impact of diet on changes in the microbiome during HIV-1 infection can be better addressed.
Linking Dysbiosis to HIV-1 Pathogenesis
In an effort to elucidate the relationship between gut dysbiosis and features of HIV-1 pathogenesis, several groups performed analyses to assess the associations between indicators of dysbiosis and various HIV-related clinical, virological and immunological parameters (Table 3). In one study, lower bacterial diversity in the stool was associated with higher levels of markers indicative of systemic MT (lipopolysaccharide (LPS), LPS binding protein (LBP)) and monocyte activation (sCD14, sCD63)[46]. Further, bacterial diversity independently associated with blood CD4+ T cell count which suggested a close relationship between the number of bacterial species and the extent of immune dysfunction[46]. Overall changes in stool microbial community composition of ART-treated HIV-1 infected study subjects associated with blood T cell activation and with high sensitivity C-reactive Protein (hs-CRP), an systemic indicator of inflammation[49]. In several other studies, overall changes in mucosa-associated bacterial communities associated with blood T cell activation[37, 51] and with various indicators of systemic inflammation[51] and MT[37, 44]. Moreover, changes in the mucosal bacterial community as a whole also positively correlated with colonic mucosal T cell activation[37, 51] as well as with levels of activated colonic myeloid dendritic cells (mDC)[37]. The latter are antigen presenting cells critical in initiating inflammatory responses and driving adaptive immunity[73]. Conversely, HIV-associated mucosal microbiome changes were inversely associated with numbers of mucosal Th22 cells[37].
Table 3.
Reported associations between features of dysbiosis and HIV-1 pathogenesis.
| HIV-associated Dysbiosis | Clinical, Virological, Immunological Parameter | Ref. | |
|---|---|---|---|
| Positive association: | Negative association: | ||
| Bacterial diversity | |||
| Stool diversity | Blood CD4+ T cell count | Systemic LPS, LBP, sCD14, sCD1631 | [46] |
| Blood CD4+ T cell count2 | [45] | ||
| Overall microbial community composition | |||
| Mucosa-associated | Blood CD4 and CD8 T cell activation | [37, 51] | |
| Systemic LPS | [37] | ||
| Systemic LTA1 | [44] | ||
| Systemic IP-101, kynurenine/tryptophan ratio | [51] | ||
| Colonic T cell activation | [37, 51] | ||
| Colonic mDC activation | [37] | ||
| Colonic IFNγ-producing CD8 T cells | Colonic Th22 cells | [37] | |
| Stool-associated | Blood CD4 and CD8 T cell activation | [49] | |
| Systemic hs-CRP1 | [49] | ||
| Individual Taxa (mucosa-associated) | |||
| Bacteroidetes Phylum: | |||
| Rikenellaeae | Colonic IL-17 production | [51] | |
| Bacteroides | Colonic iNKT cell frequencies | [53] | |
| Colonic IL-4 producing iNKT cells | [53] | ||
| Systemic LTA | Systemic IL-61 | [44] | |
| Prevotella | Colonic mDC activation | [37, 38] | |
| Colonic T cell activation | [37] | ||
| Colonic iNKT cell frequencies | [53] | ||
| Proteobacteria Phylum | |||
| Enterobacteriaceae | Colonic T cell activation | [51] | |
| Ralstonia, B. fungorum, B. pachyrhizi | Blood CD4+ T cell counts | [52] | |
| Firmicutes Phylum | |||
| Clostridia, Lachnospiraceae | Systemic TNFα1 | [44] | |
| Fecalibacterium | Systemic sCD14 | [44] | |
| Lachnospira | Systemic sCD14, plasma viral load | [37] | |
| Ruminococcus | Systemic LTA | [44] | |
| Roseburia | Systemic LPS | [37] | |
| Individual Taxa (Stool) | |||
| Bacteroidetes Phylum: | |||
| Barnesiella | Systemic TNFα | [39] | |
| Prevotella | Systemic sCD14 | [49] | |
| Proteobacteria Phylum: | |||
| Enterobacteriaceae | Systemic sCD14, IFNγ, IL-1β1 | [39] | |
| Firmicutes Phylum: | |||
| Erysipelotrichi | Systemic IL-1β | Systemic EndoCAb1 | [39] |
| Lactobacillales3 | Blood CD4+ T cells | Plasma viral load, systemic sCD14 | [47] |
| Blautia, Rumminococcus | Systemic TNFα | [44] | |
| Fecalibacterium | Blood CD8+ T cells expressing CD57 | [49] | |
| Eubacterium | Blood CD8+ T cells expressing CD57 | [49] | |
| Coprococcus | Blood CD8 T cell activation | [49] | |
| R. bromii, R. callidus | Blood CD4+ T cell count4 | [43] | |
Systemic indicators of Microbial Translocation: LPS, LBP, LTA, EndoCAb; Monocyte Activation: sCD14, sCD163; Inflammation: high-sensitivity C-reactive protein (hs-CRP), IP-10, TNFα, IFNγ, IL-1β, IL-6.
Immune discordant (CD4 counts <300 cells/mm3) individuals had lowest microbiome richness, immune concordant (CD4+ T cell count>500cells/mm3) had lower microbiome richness than HIV-1 negative study participants, but not as low as immune discordant individuals.
Detailed for HIV-1 infected subjects pre-ART; Higher levels of Lactobacillales associated with higher percent blood CD4+ T cells, less microbial translocation (sCD14), less systemic immune activation (CD8 T cell activation), less gut CD4 T cell proliferation and higher percent of gut CD4 T cells post-ART (≤48weeks).
Abundance of R. bromii and R. callidus were higher in HIV-infected individuals with CD4+ T cell counts >200cells/μl compared to HIV-infected individuals with CD4+ T cell counts <200cells/μl. 13 bacterial families within Proteobacteria (n=3), Firmicutes (n=7), Actinobacteria (n=2) and Fusobacteria (n=1) phylum associated with HIV-infected individuals with CD4+ T cell counts <200cells/μl.
Expanding on analyses that focused on changes in the microbiota as a whole, investigations into the potential relationship between individual bacteria taxa and various indicators of HIV-1 pathogenesis found that increasing levels of mucosa-associated Prevotella spp. correlated positively with levels of activated colonic mucosal mDC and T cells[37, 38] and inversely with frequencies of gut invariant natural killer T cells (iNKT) [53], innate-like T cells that respond to lipid antigens[74]. Conversely, levels of Bacteroides spp. were positively associated with iNKT frequencies[53]. Relative abundance of taxa belonging to the phylum Proteobacteria (e.g. Enterobacteriaceae) positively associated with markers of monocyte activation (sCD14), inflammation (IFNγ, IL-1β)[39] and colonic T cell activation[51] and inversely correlated with blood CD4+ T cell count[52]. Decreased levels of bacterial taxa known to exhibit immune-regulatory properties (e.g., Bacteroides, Fecalibacterium) appeared to associate with increasing levels of inflammation and monocyte activation (e.g. systemic levels of IL-6, sCD14)[39, 44]. Lactobacillales, an order of bacteria that contains probiotic species (see below) positively associated with blood CD4+ T cell count and inversely correlated with HIV-1 viral load and sCD14[47]. Relative abundance of Roseburia, a genus known to produce butyrate, an important energy source for epithelial cells[65], was inversely associated with systemic levels of LPS[37], a marker for MT. Taken together, results of these studies (Table 3) suggest a link between dysbiosis and numerous indicators of HIV-1 pathogenesis and disease progression. However, they do not demonstrate causality and do not directly address whether changes in the microbiota are causative or a rather a result of mucosal or systemic HIV-1-associated immune activation and inflammation. Moreover, not all studies corrected for multiple comparisons or other potentially confounding variables (e.g. diet, sexual practices). Therefore, caution should be taken in concluding that these individual strains of bacteria directly cause immune activation, inflammation, and/or MT.
Linking Alterations in Microbe-associated Metabolites to HIV Pathogenesis
In addition to evaluating the composition of the intestinal microbiome at the taxonomic level, a number of technologies now allow assessment of microbial community function. Understanding key bacterial functional pathways that are altered during HIV-1 infection will provide a better understanding of the relationship between the host microbiota and immune system and potentially provide new and novel treatments to improve the health of HIV-1 infected individuals.
Enrichment of mucosa-adherent bacteria capable of catabolizing tryptophan through the kynurenine pathway was observed in HIV-1 infected subjects[51]. When taken in conjunction with previous studies linking kynurenine derivatives to mucosal immune disruption and HIV-1 disease pathogenesis[75], these observations suggest that outgrowth of tryptophan-catabolizing bacteria may directly contribute to mucosal inflammation in HIV-1 infection. The metabolite Trimethylamine-N-oxide (TMAO), a derivative of Trimethylamine (TMA) that is produced as a waste product of carnitine and choline metabolism by intestinal bacteria, is strongly linked to increased risk for CVD[76-78]. Given that HIV-1 infection is associated with both microbiome changes and with increased CVD risk, a number of recent studies have investigated potential links between TMAO and various aspects of CVD in HIV-1 infected individuals[79-82]. In these pilot studies with relatively small sample sizes, serum and plasma levels of TMAO were not higher in HIV-1 infected study participants compared to uninfected controls and limited evidence for an important role of TMAO in the pathogenesis of coronary heart disease in HIV-1 infected individuals was observed. However, levels of TMA, the precursor to TMAO, in ART-treated HIV-1 infected study participants positively associated with a number of coronary plaque features[82].
Metagenomic functional capacity of rectal microbiota, imputed from 16S data, differed between untreated HIV-1 infected subjects and healthy controls[42]. Specifically, the microbiota of HIV-1 infected subjects was enriched in genes and pathways involved in glutathione metabolism, selenocompound metabolism, folate biosynthesis, and siderophore biosynthetic genes. In contrast, genes involved in amino acid production and metabolism, fructose/mannose metabolism and CoA biosynthesis were depleted. These functions did not appear to be fully restored with ART. Shotgun metagenomic sequencing of stool samples from ART-treated, virally suppressed HIV-1 infected individuals elucidated an altered functional profile compared to uninfected study participants. The former was characterized by enrichment of genes involved in LPS biosynthesis, bacterial translocation and inflammation, with depletion of genes encoding factors contributing to amino acid metabolism and energy processes[49]. A recent study suggested that HIV-1 infection results in metabolic alterations associated with changes in gut microbiota that differ from those induced in other diseases[83]. This “disease-dependent” impact on gut microbial activity was most notably characterized by a bacterial community with a reduced ability to synthesize a set of specific amino acids whereas these same amino acids are metabolized by bacteria from patients with systemic lupus erythaematosus (SLE) or Clostridium difficile infection[83].
Modeling the Impact of HIV-1-assoicated Dysbiosis on Immune Cell Function
A number of groups have begun to use in vitro models to better understand the potential mechanisms that drive microbiota-associated immune activation and inflammation in the setting of HIV-1 infection. For example, exposure of peripheral blood mononuclear cells (PBMC) from HIV-1 infected subjects to bacterial lysates from Bacteroides spp, Prevotella spp., and Erysipelotrichaceae elicited greater production of TNFα and IL-10 compared to similarly stimulated PBMC from uninfected subjects[40]. However, proliferative CD4+ T cell responses to Bacteroides spp. were reduced in HIV-1 infected subjects. Using a model of primary human intestinal lamina propria mononuclear cells (LPMC), exposure of LPMC to enteric E. coli enhanced HIV-1-infection of LP CD4+ T cells, particularly Th17 cells[84] and promoted apoptotic T cell death in vitro[85].
More recent studies have utilized this in vitro LPMC model to investigate the impact on intestinal immune cell function following exposure to bacterial species altered in the colonic mucosa of HIV-1 infected individuals[38, 86]. LPMC were exposed to a panel of bacteria that represented species in each of the 3 major phyla (Bacteroidetes, Proteobacteria, and Firmicutes) that were increased or decreased in relative abundances in HIV-1 infected subjects and that were identified in the majority of HIV-1 infected and uninfected study participants[86]. Although all commensal bacteria enhanced HIV-1 infection in LP CD4+ T cells to some extent, Gram-negative commensal bacteria (including Prevotella spp) increased productive infection and CD4+ T cell death to a greater degree than Gram-positive bacteria. Enhanced infection was primarily mediated indirectly through increased expression of CCR5 on LP CD4+ T cells following bacterial exposure. Ex vivo modeling studies also revealed a requirement for LP mDC in the activation and expansion of bacteria-reactive T cells and in the commensal bacteria-associated enhancement of LP CD4+ T cell infection[84, 87]. Moreover, commensal Prevotella spp. induced inflammatory cytokine production by gut mDC in vitro, highlighting the pro-inflammatory potential of these bacteria[38].
The Microbiome in Simian Immunodeficiency Virus (SIV) Infection of Nonhuman Primates
Major advances in our understanding of HIV-1 pathogenesis have been possible by the extensive use of the nonhuman primate models of SIV infection[88, 89]. Pathogenic SIV infection of macaques (e.g. Rhesus, Pigtail) leads to simian AIDS with disease progression bearing many similarities to HIV/AIDS including phases of acute and chronic viremia, chronic immune activation, MT, mucosal dysfunction and ineffective innate and adaptive immunity[88]. Moreover, virus replication can be suppressed by various ART regimens, providing opportunities to address pathogenesis in the setting of viral suppression. In the context of microbiome studies, these animal models provide the opportunity to further our understanding of possible mechanisms driving dysbiosis and are able to control for potential confounding factors associated with human studies such as diet and lifestyle. However, these same factors may limit our ability to directly transfer observations made in the macaque to humans.
McKenna et al. first characterized the macaque intestinal microbiome using next generation sequencing technologies [90]. Similar to humans, the macaque microbiome was dominated by the phyla Firmicutes and Bacteroidetes with lower abundances of Proteobacteria and Actinobacteria. Unlike the human intestinal microbiome, the phylum Spirochaetes was well represented but Verrucomicrobia was not, and Prevotella relative abundance was more common than Bacteroides. Similar to humans[56-58], the microbiome differed across anatomical sites.
In contrast to human studies, investigations into the impact of SIV infection have generally found only modest differences in the fecal bacterial composition between chronic SIV-infected macaques relative to uninfected macaques[90-94]. However, decreased diversity, depletion of Firmicutes, and a transient increase in Proteobacteria have been observed in fecal [93, 94] and jejunal mucosa[95] samples during acute SIV infection. Moreover, decreased abundances of Firmicutes in jejunum tissue samples[95] and decreased fecal abundance of Lactobacillus spp. persisted into chronic SIV infection[93, 94]. Alterations in the mucosa-associated microbiome during acute infection correlated with changes in cytokine gene expression and with expression of genes important in innate recognition of microbes in both the mucosa and mesenteric lymph node (MLN)[95, 96]. Depletion of stool Lactobacillus was associated with increased indoleamine 2,3-dioxygenase1 (IDO1) enzyme activity and loss of blood Th17 cells suggesting a role for Lactobacillus in the control of the IDO pathway and SIV pathogenesis[94]. Increased levels of Proteobacteria in the mesenteric lymph nodes (MLNs) of chronic SIV-infected macaques was associated with MLN CD4+ T cell activation, leading to the suggestion that bacterial species in this phylum may preferentially translocate and contribute to immune activation[93].
Similar to HIV studies[46], administration of ART altered the stool microbiome by reducing the abundance of Bacteroidetes and Firmicutes and increasing Proteobacteria, although compositional changes were transient and resembled pre-therapy abundances within 2 weeks despite continued use of ART[93].
Treatments to Restore the Intestinal Microbiome
Although not definitive, the preponderance of evidence indicates that changes in gut microbiota secondary to HIV-1 infection contribute to the severity and quality of HIV-1 related chronic inflammation. Consequently, a number of studies have attempted to modify the microbiome in an effort to reduce HIV-1-associated inflammation, immune activation, MT, and poor clinical outcomes (Table 4). To date, modification of the gut microbiome is typically achieved through the administration of probiotics which introduce “good” bacteria into the host, through the administration of prebiotics which are non-digestible fiber compounds that stimulate the growth and/or activity of beneficial bacteria in the host[97], or via ingestion of products that contain a combination of both prebiotics and probiotics (synbiotics). Probiotic yogurt administration improved blood CD4+ T cell count in HIV-1 infected women in Nigeria[98] and in men and women living in Tanzania[99]. However, other studies reported minimal beneficial impact on blood CD4+ T cell outcomes with probiotic supplementation (reviewed in [100]). Administration of specific probiotics, both in single and multi-strain formats, decreased systemic indicators of MT or inflammation in several studies of virally suppressed, ART-treated HIV-1 infected subjects[101-103], but in other studies, probiotics had minimal impact[104]. A reduced CD4+ T cell decline concurrent with lower blood CD4+ T cell activation was observed in untreated HIV-1 infected subjects taking a prebiotic nutritional supplement [105]. However, no change in CD8+ T cell activation was observed. In ART-treated, HIV-1 infected women, levels of MT as well as T cell and monocyte activation remained largely unchanged after 4 weeks of synbiotic treatment[106].
Table 4.
Treatments to modify the intestinal microbiome and improve clinical outcomes.
| Probiotics |
| • Yogurt supplemented with single (e.g. Lactobacillus rhamnosus) and multiple strains of bacteria (e.g. L. rhamnosus, L. reuteri). |
| • Fermented milk supplemented with single (e.g. L. casei) and multiple strains of bacteria (e.g. Lactobacillus spp., Bifidobacteria spp.). |
| • Capsules with single (e.g. Saccharomyces boulardii; Bacillus coagulans) and multiple strains of bacteria (e.g. Streptococcus spp., Bifidobacteria spp., Lactobacillus spp.). |
| Prebiotics |
| • Single compounds (e.g. oligosaccharides: short-chain galacto-oligosaccharides, long chain fructo-oligosaccharides (inulin), pectin hydrolysate-derived acidic - oligosaccharides). |
| • Multiple compounds (e.g. oligosaccharides, bovine colstrum protein, cysteine, lipids, lactose, glucose). |
| Synbiotics |
| • Multiple strains of bacteria (e.g. Pediococcus pentosaceus, Leuconostoc mesenteroides, L. paracasei, L. plantarum) + nondigestible, fermentable dietary fibers (e.g. betaglucan, inulin, pectin and resistant starch). |
| Fecal microbiota transplantation |
Few studies have directly addressed the effects of dietary supplements on the intestinal microbiome in HIV-1 infection. Specific prebiotic oligosaccharide mixtures altered the stool microbiota of untreated HIV-1 infected study participants by increasing bifidobacteria and decreasing abundances of pathogenic bacteria[107]. Moreover, prebiotic supplementation reduced blood T cell activation and sCD14 levels concurrent with those microbiome changes. Administration of probiotics (fermented milk supplemented with Lactobacillus and Bifidobacterium spp) to virally suppressed, ART-treated HIV-1 infected subjects increased stool abundances of Firmicutes and Actinobacteria, primarily through increased Lactobacillus and Bifidobacterium respectively, while decreasing RA of Bacteroidetes, mainly due to decreased Bacteroides[108]. However, despite these changes in the microbiome and significant reductions in both D-dimer, a marker of coagulation, as well reduced levels of soluble markers of inflammation (e.g. IL-6), levels of MT (LPS) were similar between the probiotic treated and untreated groups.
To date, the majority of studies investigating the impact of probiotics during SIV infection have focused on VSL#3, a probiotic mixture of 7 bacterial strains including 4 that are Lactobacillus spp. Administration of VSL#3 to chronically SIV-infected macaques decreased plasma IDO1 activity which had been associated with a loss of Lactobacillus and blood Th17 cells in animals without probiotic treatment (detailed above) [94]. Synbiotics (VSL#3/prebiotic inulin) administration to ART-treated SIV-infected macaques improved mucosal immunity characterized by decreased levels of colonic T cell activation and fibrosis and increased frequencies of colonic CD4+ T cells and antigen-presenting cells[109]. Further, administration of VSL#3 in conjunction with IL-21, a cytokine important in Th17 homeostasis[11, 110], to ART-treated SIV-infected macaques enhanced the expansion of polyfunctional Th17 cells, increased relative abundances of Bifidobacterium and reduced markers of MT over ART-only treated animals. Treatment of healthy macaques with VSL#3 increased frequencies of protective mucosal immune cells (IgA-expressing B cells, innate immune cells) and suggested that probiotic treatment would also enhance mucosal vaccination strategies and protection from mucosal infections[111]. Additional human clinical studies evaluating the ability of Visbiome, a multi-strain probiotic similar to VSL#3, to reduce systemic immune activation and inflammation and to restore normal intestinal immune function are currently enrolling in the USA (AIDS Clinical Trials Group A5350; ClinicalTrials.gov NCT02706717) and Canada[112].
Restoring a healthy microbiome through Fecal Microbiota Transplantation (FMT) has proven to be an effective therapeutic strategy for Clostridium difficile infection[113]. In SIV-infected macaques pre-clinical evaluation of the safety and efficacy of FMT as a therapy for HIV-1 infection demonstrated that it was well tolerated with no negative side effects[114]. A clinical study to examine the safety and durability of FMT in HIV-1 infected individuals on ART is currently underway (ClinicalTrials.gov NCT02256592).
Alterations of the Enteric Virome in HIV-1 Infection
In addition to bacteria, the enteric microbiome also encompasses viruses, fungi and archaea. In 2012, Handley and colleagues used next-generation sequencing of fecal samples to be the first to demonstrate that pathogenic SIV infection is associated with the expansion of the enteric virome[92]. In a follow up longitudinal study of SIV-infected monkeys, progression of SIV infection to AIDS was associated with expansion of gastrointestinal adenoviruses, adeno-associated viruses and picornaviruses[91]. Moreover, effective vaccination and specific vaccine-elicited immune responses were associated with a prevention of expansion of the fecal virome[91]. Findings in HIV-1 infection paralleled those of SIV infection with expansion of enteric adenoviruses associated with AIDS whereas, in the absence of immunodeficiency, the enteric virome remained largely unchanged[43]. Taken together, these observations more strongly suggest a potential role for the enteric virome in contributing to progression to AIDS rather than driving immune activation and inflammation during chronic HIV-1 infection.
Future Directions and Unanswered Questions
In the past decade, significant advances have been made in our knowledge of how HIV-1 infection disrupts intestinal function and the downstream consequences of the breakdown in intestinal homeostasis on clinical outcomes. Using recent technical advancements that more easily allow assessment of the intestinal microbiome, the HIV research community has begun to unravel the complex interactions between the intestinal microbial community and the host in the setting of HIV-1 infection. These initial studies overwhelming suggest that, like in many other inflammatory disorders, alterations in the microbial community are present during HIV-1 infection and that HIV-1-associated dysbiosis is linked, in some way, to inflammation and disease pathogenesis (Figure 1). However, these studies have also highlighted that addressing the microbiome in the setting of HIV-1 infection comes with its own repertoire of confounding factors that must be addressed for a more comprehensive understanding of the true nature as well as consequences of HIV-associated microbiome changes. This has been most keenly demonstrated with the recent study suggesting that the fecal dysbiotic profiles observed in HIV-1 infected individuals may be reflective of MSM status rather than of HIV-1 infection itself[45]. Furthermore, HIV-1 infected individuals vary by gender, mode of transmission, length of infection, ART treatment status, response to ART (e.g. immunological non-responders), and HIV-1 disease status (e.g elite controllers, long-term non-progressors (LTNP), viremics). Furthermore, dietary habits differ among HIV-infected individuals living within as well as between countries and cultures.
Figure 1.
HIV-1 infection is associated with alterations in the intestinal microbiome including changes in both diversity and composition (1). Dysbiosis is typically characterized by decreased abundances of bacteria important in maintaining epithelial barrier health and in immuneregulation in combination with increased abundance of bacteria with pro-inflammatory potential (pathobionts). Translocation of pathobionts through the disrupted epithelial barrier (2) leads to activation of innate immune cells (3). Activated innate immune cells not only contribute to local inflammation (4), but activated myeloid dendritic cells (mDC) drive increased T cell activation and expansion leading to increased infection and death and a loss of protective Th17 and Th22 cells (5). Increased T cell activation, in conjunction with a lack of protective Th cells further contributes to the inflammatory environment (6) which potentiates epithelial barrier breakdown (leading to increased microbial translocation) and may promote dysbiosis (7). In addition to mucosal inflammation and the breakdown in intestinal homeostasis, other factors such as diet, sexual behaviour and anti-retroviral therapy (ART) may also contribute to alterations in the intestinal microbiome (8). Ultimately, the combined effect of dysbiosis, microbial translocation and mucosal inflammation lead to chronic generalized immune activation, systemic inflammation and associated co-morbidities.
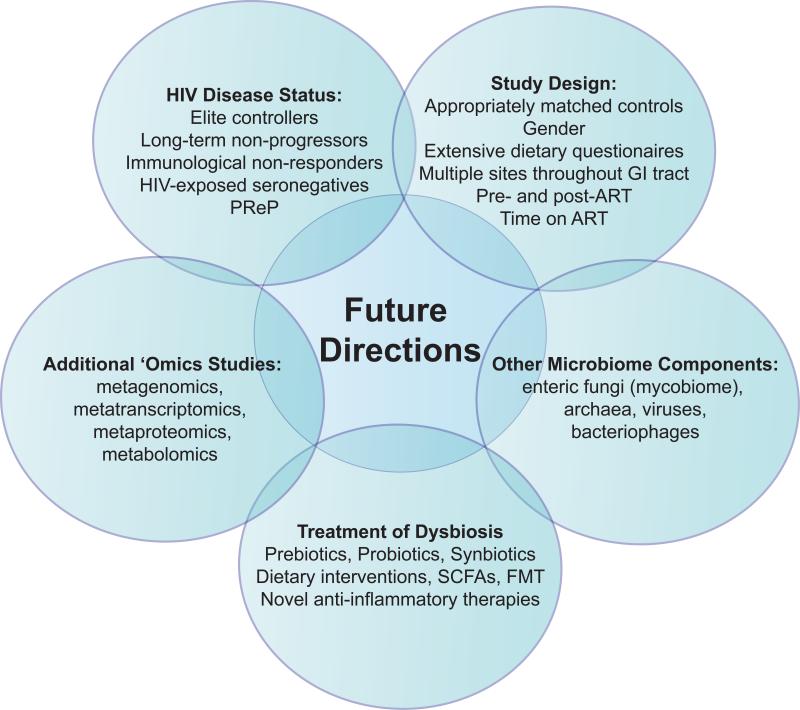
Consequently, to better understand the impact of HIV-1 on the intestinal microbiome, future studies (Figure 2) must more thoroughly account for these various aspects through the use of appropriately matched controls, undertaking longitudinal studies into the impact of ART on the microbiome and analyzing microbiomes in immunological non-responders, elite controllers, LTNP and in HIV-exposed seronegative individuals (HESN). Furthermore, future studies should investigate how MSM-associated dysbiosis impacts HIV-1 transmission, as well as whether anti-retroviral pre-exposure prophylaxis (PrEP) induces a dysbiotic profile that modifies the risk of HIV transmission in certain individuals. The impact of diet must also be addressed through the use of extensive dietary questionnaires. Finally, although information can be gained from the more easily accessible fecal samples, to best understand the direct effect of the microbiome on gut biology, evaluation of mucosa-associated microbial communities, as would be achieved by direct assessment of intestinal tissue samples, is necessary. Although a challenge, sampling GI sites other than the distal colon, such as the ileum, may provide further insight into how dysbiosis arises and, in turn, influences immune homeostasis and gut barrier function.
Figure 2.
Future Research Directions to Investigate the Relationship Between the Gut Microbiome and HIV-1 Pathogenesis.
An additional imperative is to further explore whether differences in microbial communities translate into alterations in key bacterial functional pathways that drive mucosal dysfunction rather than to simply focus on differences based on taxonomy. Moreover, currently published studies have been biased towards assessing bacterial communities; however, the microbiome also includes enteric fungi (mycobiome), archaea, viruses and bacteriophage, and microbial eukaryotes. Delineating the contribution of these under-appreciated components of the gut microbial ecosystem to enteric health undoubtedly will lead to a richer understanding of HIV-1-associated gut pathogenesis and perhaps suggest new treatment paradigms.
HIV microbiome studies are truly in their infancy and, as with other studies investigating human dysbiosis, determination of the biological and clinical consequences of dysbiosis remain[21]. If imbalances in gut microbial communities exacerbate HIV-1 disease pathogenesis, then restoring “normal” community structure could reduce pathogenesis. Alternatively, if dysbiosis arises solely as a consequence of HIV-1 infection, but does not contribute to pathogenesis, then treating dysbiosis would be ineffective. Conversely, HIV-1 associated mucosal pathogenesis, or indeed current anti-viral therapies, could promote dysbiosis, in which case improving mucosal immunity and treatment strategies could induce changes in the microbiome. Understanding the host mechanisms, especially inflammatory factors that influence microbial community structures could lead to novel approaches to durably restore intestinal homeostasis. Finally, disruption of the microbiome unrelated to HIV-1 infection itself (e.g. MSM), could contribute to both HIV-1 transmission and severity of disease. Therefore, determining the etiology of dysbiosis[21] through carefully designed studies and with more extensive use of in vitro modeling and animal studies will have a fundamental impact on the type of clinical interventions required to restore balance to the microbe/host relationship in the setting of HIV-1 infection.
REFERENCES
- 1.Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chege D, Sheth PM, Kain T, Kim CJ, Kovacs C, Loutfy M, et al. Sigmoid Th17 populations, the HIV latent reservoir, and microbial translocation in men on long-term antiretroviral therapy. AIDS. 2011;25:741–749. doi: 10.1097/QAD.0b013e328344cefb. [DOI] [PubMed] [Google Scholar]
- 3.Guadalupe M, Reay E, Sankaran S, Prindiville T, Flamm J, McNeil A, et al. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol. 2003;77:11708–11717. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mehandru S, Poles MA, Tenner-Racz K, Manuelli V, Jean-Pierre P, Lopez P, et al. Mechanisms of gastrointestinal CD4+ T-cell depletion during acute and early human immunodeficiency virus type 1 infection. J Virol. 2007;81:599–612. doi: 10.1128/JVI.01739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sankaran S, George MD, Reay E, Guadalupe M, Flamm J, Prindiville T, et al. Rapid onset of intestinal epithelial barrier dysfunction in primary human immunodeficiency virus infection is driven by an imbalance between immune response and mucosal repair and regeneration. J Virol. 2008;82:538–545. doi: 10.1128/JVI.01449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epple HJ, Schneider T, Troeger H, Kunkel D, Allers K, Moos V, et al. Impairment of the intestinal barrier is evident in untreated but absent in suppressively treated HIV-infected patients. Gut. 2009;58:220–227. doi: 10.1136/gut.2008.150425. [DOI] [PubMed] [Google Scholar]
- 7.Keating J, Bjarnason I, Somasundaram S, Macpherson A, Francis N, Price AB, et al. Intestinal absorptive capacity, intestinal permeability and jejunal histology in HIV and their relation to diarrhoea. Gut. 1995;37:623–629. doi: 10.1136/gut.37.5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ullrich R, Zeitz M, Riecken EO. Enteric immunologic abnormalities in human immunodeficiency virus infection. Semin Liver Dis. 1992;12:167–174. doi: 10.1055/s-2007-1007388. [DOI] [PubMed] [Google Scholar]
- 9.Kim CJ, Nazli A, Rojas OL, Chege D, Alidina Z, Huibner S, et al. A role for mucosal IL-22 production and Th22 cells in HIV-associated mucosal immunopathogenesis. Mucosal Immunol. 2012;5:670–680. doi: 10.1038/mi.2012.72. [DOI] [PubMed] [Google Scholar]
- 10.Kok A, Hocqueloux L, Hocini H, Carriere M, Lefrou L, Guguin A, et al. Early initiation of combined antiretroviral therapy preserves immune function in the gut of HIV-infected patients. Mucosal Immunol. 2014 doi: 10.1038/mi.2014.50. [DOI] [PubMed] [Google Scholar]
- 11.Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, Asher TE, et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood. 2008;112:2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annu Rev Immunol. 2012;30:149–173. doi: 10.1146/annurev-immunol-020711-075001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 14.Marchetti G, Cozzi-Lepri A, Merlini E, Bellistri GM, Castagna A, Galli M, et al. Microbial translocation predicts disease progression of HIV-infected antiretroviral-naive patients with high CD4+ cell count. AIDS. 2011;25:1385–1394. doi: 10.1097/QAD.0b013e3283471d10. [DOI] [PubMed] [Google Scholar]
- 15.Marchetti G, Tincati C, Silvestri G. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin Microbiol Rev. 2013;26:2–18. doi: 10.1128/CMR.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zevin AS, McKinnon L, Burgener A, Klatt NR. Microbial translocation and microbiome dysbiosis in HIV-associated immune activation. Curr Opin HIV AIDS. 2016;11:182–190. doi: 10.1097/COH.0000000000000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu DC, Sereti I. Serious Non-AIDS Events: Therapeutic Targets of Immune Activation and Chronic Inflammation in HIV Infection. Drugs. 2016;76:533–549. doi: 10.1007/s40265-016-0546-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geuking MB, Koller Y, Rupp S, McCoy KD. The interplay between the gut microbiota and the immune system. Gut Microbes. 2014;5:411–418. doi: 10.4161/gmic.29330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDermott AJ, Huffnagle GB. The microbiome and regulation of mucosal immunity. Immunology. 2014;142:24–31. doi: 10.1111/imm.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank DN, Zhu W, Sartor RB, Li E. Investigating the biological and clinical significance of human dysbioses. Trends Microbiol. 2011;19:427–434. doi: 10.1016/j.tim.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:179–184. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li E, Hamm CM, Gulati AS, Sartor RB, Chen H, Wu X, et al. Inflammatory bowel diseases phenotype, C. difficile and NOD2 genotype are associated with shifts in human ileum associated microbial composition. PLoS One. 2012;7:e26284. doi: 10.1371/journal.pone.0026284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown K, DeCoffe D, Molcan E, Gibson DL. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients. 2012;4:1095–1119. doi: 10.3390/nu4081095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.John GK, Mullin GE. The Gut Microbiome and Obesity. Curr Oncol Rep. 2016;18:45. doi: 10.1007/s11912-016-0528-7. [DOI] [PubMed] [Google Scholar]
- 27.Alkanani AK, Hara N, Gottlieb PA, Ir D, Robertson CE, Wagner BD, et al. Alterations in Intestinal Microbiota Correlate With Susceptibility to Type 1 Diabetes. Diabetes. 2015;64:3510–3520. doi: 10.2337/db14-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 29.Mathis D, Benoist C. The influence of the microbiota on type-1 diabetes: on the threshold of a leap forward in our understanding. Immunol Rev. 2012;245:239–249. doi: 10.1111/j.1600-065X.2011.01084.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Qi J, Zhao H, He S, Zhang Y, Wei S, et al. Metagenomic sequencing reveals microbiota and its functional potential associated with periodontal disease. Sci Rep. 2013;3:1843. doi: 10.1038/srep01843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeoh N, Burton JP, Suppiah P, Reid G, Stebbings S. The role of the microbiome in rheumatic diseases. Curr Rheumatol Rep. 2013;15:314. doi: 10.1007/s11926-012-0314-y. [DOI] [PubMed] [Google Scholar]
- 32.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh V, Yeoh BS, Vijay-Kumar M. Gut microbiome as a novel cardiovascular therapeutic target. Curr Opin Pharmacol. 2016;27:8–12. doi: 10.1016/j.coph.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Bella JM, Bao Y, Gloor GB, Burton JP, Reid G. High throughput sequencing methods and analysis for microbiome research. J Microbiol Methods. 2013;95:401–414. doi: 10.1016/j.mimet.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 36.Frank DN, Pace NR. Gastrointestinal microbiology enters the metagenomics era. Curr Opin Gastroenterol. 2008;24:4–10. doi: 10.1097/MOG.0b013e3282f2b0e8. [DOI] [PubMed] [Google Scholar]
- 37.Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7:983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dillon SM, Lee EJ, Kotter CV, Austin GL, Gianella S, Siewe B, et al. Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T-cell activation in untreated HIV-1 infection. Mucosal Immunol. 2016;9:24–37. doi: 10.1038/mi.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dinh DM, Volpe GE, Duffalo C, Bhalchandra S, Tai AK, Kane AV, et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis. 2015;211:19–27. doi: 10.1093/infdis/jiu409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14:329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lozupone CA, Rhodes ME, Neff CP, Fontenot AP, Campbell TB, Palmer BE. HIV-induced alteration in gut microbiota: driving factors, consequences, and effects of antiretroviral therapy. Gut Microbes. 2014;5:562–570. doi: 10.4161/gmic.32132. [DOI] [PubMed] [Google Scholar]
- 42.McHardy IH, Li X, Tong M, Ruegger P, Jacobs J, Borneman J, et al. HIV infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. 2013;1:26. doi: 10.1186/2049-2618-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monaco CL, Gootenberg DB, Zhao G, Handley SA, Ghebremichael MS, Lim ES, et al. Altered Virome and Bacterial Microbiome in Human Immunodeficiency Virus-Associated Acquired Immunodeficiency Syndrome. Cell Host Microbe. 2016;19:311–322. doi: 10.1016/j.chom.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mutlu EA, Keshavarzian A, Losurdo J, Swanson G, Siewe B, Forsyth C, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog. 2014;10:e1003829. doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noguera-Julian M, Rocafort M, Guillén Y, Rivera J, Casadellà M, Nowak P, et al. Gut Microbiota Linked to Sexual Preference and HIV Infection. EBioMedicine. 2016 doi: 10.1016/j.ebiom.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nowak P, Troseid M, Avershina E, Barqasho B, Neogi U, Holm K, et al. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS. 2015;29:2409–2418. doi: 10.1097/QAD.0000000000000869. [DOI] [PubMed] [Google Scholar]
- 47.Perez-Santiago J, Gianella S, Massanella M, Spina CA, Karris MY, Var SR, et al. Gut Lactobacillales are associated with higher CD4 and less microbial translocation during HIV infection. AIDS. 2013;27:1921–1931. doi: 10.1097/qad.0b013e3283611816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Y, Ma Y, Lin P, Tang YW, Yang L, Shen Y, et al. Fecal bacterial microbiome diversity in chronic HIV-infected patients in China. Emerg Microbes Infect. 2016;5:e31. doi: 10.1038/emi.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vazquez-Castellanos JF, Serrano-Villar S, Latorre A, Artacho A, Ferrus ML, Madrid N, et al. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol. 2014 doi: 10.1038/mi.2014.107. [DOI] [PubMed] [Google Scholar]
- 50.Volpe GE, Ward H, Mwamburi M, Dinh D, Bhalchandra S, Wanke C, et al. Associations of cocaine use and HIV infection with the intestinal microbiota, microbial translocation, and inflammation. J Stud Alcohol Drugs. 2014;75:347–357. doi: 10.15288/jsad.2014.75.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, et al. Dysbiosis of the Gut Microbiota Is Associated with HIV Disease Progression and Tryptophan Catabolism. Sci Transl Med. 2013;5:193ra191. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang L, Poles MA, Fisch GS, Ma Y, Nossa C, Phelan JA, et al. HIV-induced immunosuppression is associated with colonization of the proximal gut by environmental bacteria. AIDS. 2016;30:19–29. doi: 10.1097/QAD.0000000000000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paquin-Proulx D, Ching C, Vujkovic-Cvijin I, Fadrosh D, Loh L, Huang Y, et al. Bacteroides are associated with GALT iNKT cell function and reduction of microbial translocation in HIV-1 infection. Mucosal Immunol. 2016 doi: 10.1038/mi.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fettweis JM, Serrano MG, Sheth NU, Mayer CM, Glascock AL, Brooks JP, et al. Species-level classification of the vaginal microbiome. BMC Genomics. 2012;13(Suppl 8):S17. doi: 10.1186/1471-2164-13-S8-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008;74:2461–2470. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Durban A, Abellan JJ, Jimenez-Hernandez N, Ponce M, Ponce J, Sala T, et al. Assessing gut microbial diversity from feces and rectal mucosa. Microb Ecol. 2011;61:123–133. doi: 10.1007/s00248-010-9738-y. [DOI] [PubMed] [Google Scholar]
- 57.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gori A, Tincati C, Rizzardini G, Torti C, Quirino T, Haarman M, et al. Early impairment of gut function and gut flora supporting a role for alteration of gastrointestinal mucosa in human immunodeficiency virus pathogenesis. J Clin Microbiol. 2008;46:757–758. doi: 10.1128/JCM.01729-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ellis CL, Ma ZM, Mann SK, Li CS, Wu J, Knight TH, et al. Molecular characterization of stool microbiota in HIV-infected subjects by panbacterial and order-level 16S ribosomal DNA (rDNA) quantification and correlations with immune activation. J Acquir Immune Defic Syndr. 2011;57:363–370. doi: 10.1097/QAI.0b013e31821a603c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011;23:473–480. doi: 10.1016/j.coi.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duerkop BA, Vaishnava S, Hooper LV. Immune responses to the microbiota at the intestinal mucosal surface. Immunity. 2009;31:368–376. doi: 10.1016/j.immuni.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 63.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 64.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- 65.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 66.Kaakoush NO. Insights into the Role of Erysipelotrichaceae in the Human Host. Front Cell Infect Microbiol. 2015;5:84. doi: 10.3389/fcimb.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82:338–344. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 68.Lucke K, Miehlke S, Jacobs E, Schuppler M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J Med Microbiol. 2006;55:617–624. doi: 10.1099/jmm.0.46198-0. [DOI] [PubMed] [Google Scholar]
- 69.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Troy EB, Kasper DL. Beneficial effects of Bacteroides fragilis polysaccharides on the immune system. Front Biosci (Landmark Ed) 2010;15:25–34. doi: 10.2741/3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee HK, Iwasaki A. Innate control of adaptive immunity: dendritic cells and beyond. Semin Immunol. 2007;19:48–55. doi: 10.1016/j.smim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 74.Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol. 2010;11:197–206. doi: 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- 75.Favre D, Mold J, Hunt PW, Kanwar B, Loke P, Seu L, et al. Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci Transl Med. 2010;2:32ra36. doi: 10.1126/scitranslmed.3000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brown JM, Hazen SL. The gut microbial endocrine organ: bacterially derived signals driving cardiometabolic diseases. Annu Rev Med. 2015;66:343–359. doi: 10.1146/annurev-med-060513-093205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Romano KA, Vivas EI, Amador-Noguez D, Rey FE. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. MBio. 2015;6:e02481. doi: 10.1128/mBio.02481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tang WH, Hazen SL. The contributory role of gut microbiota in cardiovascular disease. J Clin Invest. 2014;124:4204–4211. doi: 10.1172/JCI72331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haissman JM, Knudsen A, Hoel H, Kjaer A, Kristoffersen US, Berge RK, et al. Microbiota-Dependent Marker TMAO Is Elevated in Silent Ischemia but Is Not Associated With First-Time Myocardial Infarction in HIV Infection. J Acquir Immune Defic Syndr. 2016;71:130–136. doi: 10.1097/QAI.0000000000000843. [DOI] [PubMed] [Google Scholar]
- 80.Knudsen A, Christensen TE, Thorsteinsson K, Ghotbi AA, Hasbak P, Lebech AM, et al. Microbiota-Dependent Marker TMAO is Not Associated With Decreased Myocardial Perfusion in Well-Treated HIV-Infected Patients as Assessed by 82Rubidium PET/CT. J Acquir Immune Defic Syndr. 2016;72:e83–85. doi: 10.1097/QAI.0000000000001044. [DOI] [PubMed] [Google Scholar]
- 81.Miller PE, Haberlen SA, Brown TT, Margolick JB, DiDonato JA, Hazen SL, et al. Brief Report: Intestinal Microbiota-Produced Trimethylamine-N-Oxide and Its Association With Coronary Stenosis and HIV Serostatus. J Acquir Immune Defic Syndr. 2016;72:114–118. doi: 10.1097/QAI.0000000000000937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Srinivasa S, Fitch KV, Lo J, Kadar H, Knight R, Wong K, et al. Plaque burden in HIV-infected patients is associated with serum intestinal microbiota-generated trimethylamine. AIDS. 2015;29:443–452. doi: 10.1097/QAD.0000000000000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Serrano-Villar S, Rojo D, Martinez-Martinez M, Deusch S, Vazquez-Castellanos JF, Sainz T, et al. HIV infection results in metabolic alterations in the gut microbiota different from those induced by other diseases. Sci Rep. 2016;6:26192. doi: 10.1038/srep26192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dillon SM, Manuzak JA, Leone AK, Lee EJ, Rogers LM, McCarter MD, et al. HIV-1 infection of human intestinal lamina propria CD4+ T cells in vitro is enhanced by exposure to commensal Escherichia coli. J Immunol. 2012;189:885–896. doi: 10.4049/jimmunol.1200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Steele AK, Lee EJ, Manuzak JA, Dillon SM, Beckham JD, McCarter MD, et al. Microbial exposure alters HIV-1-induced mucosal CD4+ T cell death pathways Ex vivo. Retrovirology. 2014;11:14. doi: 10.1186/1742-4690-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dillon SM, Lee EJ, Donovan AM, Guo K, Harper MS, Frank DN, et al. Enhancement of HIV-1 infection and intestinal CD4+ T cell depletion ex vivo by gut microbes altered during chronic HIV-1 infection. Retrovirology. 2016;13:5. doi: 10.1186/s12977-016-0237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Howe R, Dillon S, Rogers L, McCarter M, Kelly C, Gonzalez R, et al. Evidence for dendritic cell-dependent CD4(+) T helper-1 type responses to commensal bacteria in normal human intestinal lamina propria. Clin Immunol. 2009;131:317–332. doi: 10.1016/j.clim.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Evans DT, Silvestri G. Nonhuman primate models in AIDS research. Curr Opin HIV AIDS. 2013;8:255–261. doi: 10.1097/COH.0b013e328361cee8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Klatt NR, Silvestri G, Hirsch V. Nonpathogenic simian immunodeficiency virus infections. Cold Spring Harb Perspect Med. 2012;2:a007153. doi: 10.1101/cshperspect.a007153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McKenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, Liu Z, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Handley SA, Desai C, Zhao G, Droit L, Monaco CL, Schroeder AC, et al. SIV Infection-Mediated Changes in Gastrointestinal Bacterial Microbiome and Virome Are Associated with Immunodeficiency and Prevented by Vaccination. Cell Host Microbe. 2016;19:323–335. doi: 10.1016/j.chom.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klase Z, Ortiz A, Deleage C, Mudd JC, Quinones M, Schwartzman E, et al. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol. 2015 doi: 10.1038/mi.2014.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vujkovic-Cvijin I, Swainson LA, Chu SN, Ortiz AM, Santee CA, Petriello A, et al. Gut-Resident Lactobacillus Abundance Associates with IDO1 Inhibition and Th17 Dynamics in SIV-Infected Macaques. Cell Rep. 2015;13:1589–1597. doi: 10.1016/j.celrep.2015.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Glavan TW, Gaulke CA, Santos Rocha C, Sankaran-Walters S, Hirao LA, Raffatellu M, et al. Gut immune dysfunction through impaired innate pattern recognition receptor expression and gut microbiota dysbiosis in chronic SIV infection. Mucosal Immunol. 2016;9:677–688. doi: 10.1038/mi.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Glavan TW, Gaulke CA, Hirao LA, Sankaran-Walters S, Dandekar S. SIV-infection-driven changes of pattern recognition receptor expression in mesenteric lymph nodes and gut microbiota dysbiosis. J Med Primatol. 2015;44:241–252. doi: 10.1111/jmp.12187. [DOI] [PubMed] [Google Scholar]
- 97.Slavin J. Fiber and prebiotics: mechanisms and health benefits. Nutrients. 2013;5:1417–1435. doi: 10.3390/nu5041417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Anukam KC, Osazuwa EO, Osadolor HB, Bruce AW, Reid G. Yogurt containing probiotic Lactobacillus rhamnosus GR-1 and L. reuteri RC-14 helps resolve moderate diarrhea and increases CD4 count in HIV/AIDS patients. J Clin Gastroenterol. 2008;42:239–243. doi: 10.1097/MCG.0b013e31802c7465. [DOI] [PubMed] [Google Scholar]
- 99.Irvine SL, Hummelen R, Hekmat S, Looman CW, Habbema JD, Reid G. Probiotic yogurt consumption is associated with an increase of CD4 count among people living with HIV/AIDS. J Clin Gastroenterol. 2010;44:e201–205. doi: 10.1097/MCG.0b013e3181d8fba8. [DOI] [PubMed] [Google Scholar]
- 100.Miller H, Ferris R, Phelps BR. The effect of probiotics on CD4 counts among people living with HIV: a systematic review. Benef Microbes. 2016;7:345–351. doi: 10.3920/BM2015.0163. [DOI] [PubMed] [Google Scholar]
- 101.d'Ettorre G, Ceccarelli G, Giustini N, Serafino S, Calantone N, De Girolamo G, et al. Probiotics Reduce Inflammation in Antiretroviral Treated, HIV-Infected Individuals: Results of the “Probio-HIV” Clinical Trial. PLoS One. 2015;10:e0137200. doi: 10.1371/journal.pone.0137200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Falasca K, Vecchiet J, Ucciferri C, Di Nicola M, D'Angelo C, Reale M. Effect of Probiotic Supplement on Cytokine Levels in HIV-Infected Individuals: A Preliminary Study. Nutrients. 2015;7:8335–8347. doi: 10.3390/nu7105396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Villar-Garcia J, Hernandez JJ, Guerri-Fernandez R, Gonzalez A, Lerma E, Guelar A, et al. Effect of probiotics (Saccharomyces boulardii) on microbial translocation and inflammation in HIV-treated patients: a double-blind, randomized, placebo-controlled trial. J Acquir Immune Defic Syndr. 2015;68:256–263. doi: 10.1097/QAI.0000000000000468. [DOI] [PubMed] [Google Scholar]
- 104.Yang OO, Kelesidis T, Cordova R, Khanlou H. Immunomodulation of antiretroviral drug-suppressed chronic HIV-1 infection in an oral probiotic double-blind placebo-controlled trial. AIDS Res Hum Retroviruses. 2014;30:988–995. doi: 10.1089/aid.2014.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cahn P, Ruxrungtham K, Gazzard B, Diaz RS, Gori A, Kotler DP, et al. The immunomodulatory nutritional intervention NR100157 reduced CD4+ T-cell decline and immune activation: a 1-year multicenter randomized controlled double-blind trial in HIV-infected persons not receiving antiretroviral therapy (The BITE Study). Clin Infect Dis. 2013;57:139–146. doi: 10.1093/cid/cit171. [DOI] [PubMed] [Google Scholar]
- 106.Schunter M, Chu H, Hayes TL, McConnell D, Crawford SS, Luciw PA, et al. Randomized pilot trial of a synbiotic dietary supplement in chronic HIV-1 infection. BMC Complement Altern Med. 2012;12:84. doi: 10.1186/1472-6882-12-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gori A, Rizzardini G, Van't Land B, Amor KB, van Schaik J, Torti C, et al. Specific prebiotics modulate gut microbiota and immune activation in HAART-naive HIV-infected adults: results of the “COPA” pilot randomized trial. Mucosal Immunol. 2011;4:554–563. doi: 10.1038/mi.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stiksrud B, Nowak P, Nwosu FC, Kvale D, Thalme A, Sonnerborg A, et al. Reduced Levels of D-dimer and Changes in Gut Microbiota Composition After Probiotic Intervention in HIV-Infected Individuals on Stable ART. J Acquir Immune Defic Syndr. 2015;70:329–337. doi: 10.1097/QAI.0000000000000784. [DOI] [PubMed] [Google Scholar]
- 109.Klatt NR, Canary LA, Sun X, Vinton CL, Funderburg NT, Morcock DR, et al. Probiotic/prebiotic supplementation of antiretrovirals improves gastrointestinal immunity in SIV-infected macaques. J Clin Invest. 2013;123:903–907. doi: 10.1172/JCI66227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Micci L, Cervasi B, Ende ZS, Iriele RI, Reyes-Aviles E, Vinton C, et al. Paucity of IL-21-producing CD4(+) T cells is associated with Th17 cell depletion in SIV infection of rhesus macaques. Blood. 2012;120:3925–3935. doi: 10.1182/blood-2012-04-420240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Manuzak JA, Hensley-McBain T, Zevin AS, Miller C, Cubas R, Agricola B, et al. Enhancement of Microbiota in Healthy Macaques Results in Beneficial Modulation of Mucosal and Systemic Immune Function. J Immunol. 2016;196:2401–2409. doi: 10.4049/jimmunol.1502470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim CJ, Walmsley SL, Raboud JM, Kovacs C, Coburn B, Rousseau R, et al. Can Probiotics Reduce Inflammation and Enhance Gut Immune Health in People Living with HIV: Study Designs for the Probiotic Visbiome for Inflammation and Translocation (PROOV IT) Pilot Trials. HIV Clin Trials. 2016;17:147–157. doi: 10.1080/15284336.2016.1184827. [DOI] [PubMed] [Google Scholar]
- 113.Choi HH, Cho YS. Fecal Microbiota Transplantation: Current Applications, Effectiveness, and Future Perspectives. Clin Endosc. 2016;49:257–265. doi: 10.5946/ce.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hensley-McBain T, Zevin AS, Manuzak J, Smith E, Gile J, Miller C, et al. The effects of fecal microbial transplantation on microbiome and immunity in SIV-infected macaques. J Virol. 2016 doi: 10.1128/JVI.00099-16. [DOI] [PMC free article] [PubMed] [Google Scholar]