Abstract
Foxp3+RORγt+ T cells have recently been characterized as an immunoregulatory population highly enriched in the colon lamina propria. However, their developmental origin and relation to RORγt− Treg and TH17 cells remains unclear. Here, we use a fixed TCRβ system to show that the TCR repertoire of the Foxp3+RORγt+ population is mostly distinct compared to other colonic T cell subsets. However, a fraction of these TCRs are also found in the TH17 subset, suggesting that TCR repertoire overlap may contribute to the reported ability of Foxp3+RORγt+ cells to regulate TH17 immunity. Naïve transgenic T cells expressing a Foxp3+RORγt+ restricted TCR first acquire a Foxp3+RORγt− phenotype before co-expressing RORγt, suggesting that Foxp3+RORγt+ cell development can occur via an RORγt− Treg intermediate.
Keywords: T cells, T cell Receptors, Cell Differentiation, Repertoire Development, Mucosa
Introduction
The maintenance of tolerance towards commensal bacteria represents a unique challenge for the host immune system. In order to protect against infection from pathogenic microbes, the host must be capable of clearing harmful bacteria, while simultaneously limiting responses that target commensal, though no less foreign, species. This balance is achieved in part by the induction of effector and tolerogenic CD4+ T cell responses by specific commensal species. For example, segmented filamentous bacteria is capable of promoting the differentiation of retinoid orphan receptor (ROR)γt-expressing T helper (TH)17 cells, which are able to promote anti-bacterial responses (1). Conversely, Clostridial species (2) are capable of promoting the differentiation of forkhead box P3 (Foxp3)-expressing regulatory T (Treg) cells, which can limit inflammation (3). Dysregulation between these subsets and subsequent loss of immune homeostasis is thought to represent an important component of inflammatory bowel disease (IBD) pathogenesis (4).
The classical model of CD4+ T cell differentiation involves the generation of several unambiguous subsets, distinguished by expression of lineage-defining transcription factors. However, this paradigm has been challenged in the case of Treg cells as several subsets of Foxp3+ cells have been identified that co-express transcription factors normally associated with effector lineages. This has led to the hypothesis that effector transcription factor co-expression allows Treg cells to specifically target corresponding effector subsets for suppression (5). For example, T-bet+ Treg cells are involved in restraining TH1-mediated gut inflammation (6). However, the precise developmental origin of such transcription factor co-expressing Treg cells is incompletely understood.
Recently, a subset of intestinal CD4+ T cells co-expressing both Foxp3 and RORγt was identified (7–9). While small numbers of these cells have been found in other peripheral lymphoid tissues (10, 11), the lamina propria appears uniquely enriched for this population. Foxp3+RORγt+ cells are dependent on the presence of commensal bacteria and are capable of producing IL-10. Moreover, the absence of these cells exacerbates pathogenesis of several models of mucosal autoimmunity, suggesting that these Foxp3+RORγt+ T cells represent another Treg cell subset.
Yet, the unique shared developmental requirements of Treg and TH17 cells suggests an alternative interpretation for the role of Foxp3+RORγt+ T cells. TGFβ can promote the peripheral development of both Treg and TH17 cells from naïve CD4+ T cells (12). This raises the possibility that, instead of a stable Treg subset, Foxp3+RORγt+ T cells could represent a common precursor of both Treg and Th17 cells, as previously proposed (12). Thus, the relationship between Treg, Th17, and Foxp3+RORγt+ T cells remains poorly defined.
Here, we use TCR sequencing and a TCR transgenic (TCRtg) system to elucidate the relationships between these Foxp3+RORγt+ cells and conventional Treg and TH17 cells. We found that the TCR repertoire of Foxp3+RORγt+ cells is largely unique compared to other colonic T cell subsets suggesting that TCR specificity is sufficient to mediate Foxp3+RORγt+ cell development. Yet, we also observe a subset of Foxp3+RORγt+ TCRs that are shared with TH17 cells and may contribute to the ability of this population to suppress TH17 inflammation. Naïve T cells expressing a TCR restricted to the Foxp3+RORγt+ subset likely develop via an RORγt− Treg intermediate without significant TH17 differentiation. In addition, we show that, similar to TH17 cells, Foxp3+ cells are partially dependent on CX3CR1+ antigen presenting cells (APCs) for subsequent RORγt expression. Together, our data suggest that the dominant portion of Foxp3+RORγt+ T cells develop as a subset of Treg cells.
Experimental Procedures
Mice
TCli TCRβ Tcrα+/− mice, used as previously described (13), were bred to Foxp3IRES-Thy1.1 (14), a gift from A. Rudensky (MSK); and RORγtgfp, obtained from Jackson Laboratory (stock# 007572). CT2 transgenic mice were generated as described (15) with microinjection into B6 × 129 fertilized eggs and backcrossed 5+ generations to B6 background. CD11cCre Notch2fl/fl (16) were a gift from Ken Murphy (WashU). CX3CR1DTR (17) were purchased from Jackson Laboratory (stock# 025629) and bred to CD11cCre. CD45.1 mice are housed together and interbred to maintain microbial integrity as assessed by CT2 transfer experiments. Animal experiments were performed in a specific pathogen-free facility under the guidelines of the Institutional Animal Care and Use Committee at Washington University.
Cell isolation and flow cytometry
Lamina propria tissue was isolated and digested in RPMI media containing 3% FBS, 20mM Hepes, 1mM DTT, and 50mM EDTA for 20min at 37° C. Additional washes in RPMI + 22.5mM EDTA were done to completely remove IELs. Remaining tissue was minced and digested in digested in 28.3µg/mL liberase TL (Roche) and 200µg/mL RNase 1 (Roche) for 30min at 37° C to release lamina propria lymphocytes. Suspended cells were filtered through a 40µm filter prior to use. Distal mLN was isolated due to its lymphatic drainage of the colon (18). Fluorescently conjugated antibodies were purchased from Biolegend and eBioscience. Samples were sorted and analyzed with a FACSAria IIu (BD) and data was processed in FlowJo (Treestar).
TCR sequencing
CD4+Vα2+ cells were sorted from TCli TCRβ Tcrα+/− Foxp3IRES-Thy1.1Rorγtgfp/+ mice at ≥8wks old (mean age 18 weeks). TCRα cDNA was prepared using a Cα-specific primer for reverse transcription. TRAV14 libraries were generated using a nested PCR protocol with primers indicated in Supplemental Table II. Paired end reads were generated from 250 cycle sequencing using Illumina MiSeq at the Washington University Center for Genome Sciences. V, J, and CDR3 regions were then determined with a custom BLAST application using sequence data from IMGT (19).
Adoptive transfer experiments
Naïve (CD4+ CD44−lo CD62L−hi CXCR3− Foxp3−RORγt−) T cells were FACS purified from peripheral LNs and spleen of Foxp3IRES-Thy1.1Rorγtgfp/+Rag1−/− TCRtg mice. CD45.2 TCRtg mice were used for transfer experiments into wild type CD45.1 hosts. CD45.1 TCRtg mice were used in experiments with CD45.2 DC-deficient hosts. 5×104 cells were injected retro-orbitally into 3 week old hosts and analyzed 3 weeks post-transfer, unless otherwise indicated.
Statistical analysis
Diversity profiles were generated using Renyi entropy values with alpha/order values ranging from 0 (natural logarithm of species richness) through 2 (natural logarithm of the inverse Simpson index) (20, 21). This includes alpha = 1, which represents the commonly used Shannon entropy. Evenness was calculated from the ratio of each point on the diversity profile to Renyi entropy at alpha = 0, resulting in an evenness profile of relative evenness indices (RLE0,alpha) (22). Pielou’s evenness represents the special case of RLE0,alpha=1. Euclidean distance was used to generate hierarchical clusters from these diversity profiles and Pearson correlation was used for clustering of Evenness profiles (23). Coverage was calculated as described (24). For multiple comparisons of individual TCR enrichment between samples, Benjamini-Hochberg false discovery rate adjusted p-values were used. To generate TCR perturbation scores, CDR3 sequences within a sample were represented by their amino acid length to generate a spectratype distribution (25). These spectratype distributions were then compared using the Morisita-Horn index and hierarchically clustered. All statistical analysis was performed in R (v3.3.0) with the use of the vegan (v2.3-5, diversity and similarity analysis), DESeq2 (v1.12.0, differential TCR usage), and pvclust (v2.0-0, bootstrapped dendrograms) packages. Mann-Whitney U or Kruskal-Wallis with post-hoc Dunn’s tests were used for between group analysis. Our analytical code can be found at https://github.com/BenSolomon/Solomon-Hsieh-JI-2016.
Results
Sequencing of the Foxp3+RORγt+ TCR repertoire
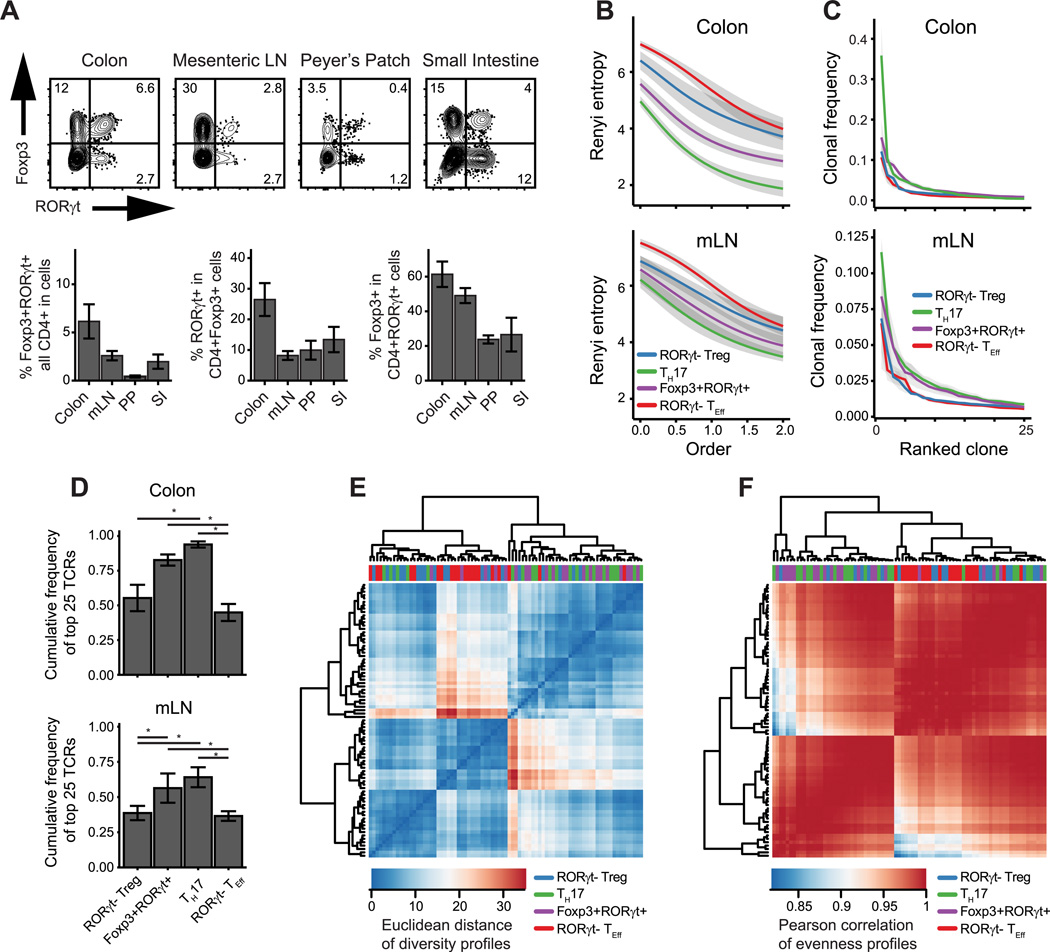
Similar to data reported in recent publications, we found enrichment of Foxp3+RORγt+ T cells in mucosal tissue, particularly the colon lamina propria (Fig. 1A). To address how this population relates to other colonic T cell subsets, we asked whether Foxp3+RORγt+ T cells utilized the same TCR repertoire as other Foxp3+ or RORγt+ cells. In order to circumvent analytical problem posed by the diversity of the fully polyclonal TCR repertoire, we analyzed TRAV14 TCRα transcripts from a TCRβ-tg system in which TCR diversity is restricted to the TCRα chain (13). From mucosal tissues we sorted and sequenced mature CD4+ T cells (CD44−hi CD62L−lo) into RORγt− Treg (Foxp3+RORγt−) and Foxp3+RORγt+ populations, as well as TH17 (Foxp3− RORγt+) and remaining RORγt− T effector (TEff) cell (Foxp3− RORγt-) populations (Supplemental Fig. 1). In order to limit overlap with TH1 cells, all populations except the RORγt− TEff population were further gated to exclude CXCR3+ cells (26) (Supplemental Fig 1C).
Figure 1. TCR repertoire diversity of mucosal Foxp3+ and RORγt+ cells.
(A) Phenotype of CD4+ CD44−hi CD62L−lo CXCR3− cells in indicated tissues. (B–F) TCR repertoire analysis of indicated populations sorted from TCli TCRβ Tcrα+/− mice. (B) Mean Renyi diversity. Increasing order indicates increasing weight on high frequency clones. (C) Mean clonal frequency of top 25 clones in each population. (D) Oligoclonality indicated by average cumulative frequency of top 25 clones (E–F) Hierarchical clustering of (E) diversity and (F) evenness profiles calculated from TCR sequence counts. n=6, each of 2 pooled mice. Error bars/ribbon = ± 1 s.e.m. (*) adjusted p-value < 0.05.
We first quantified the diversity of the Foxp3+RORγt+ TCR repertoire using Renyi entropies, which depict population diversity across a spectrum of orders that increasingly weight unique TCR sequences according to their abundance (20). Higher Renyi entropy values indicate greater sequence diversity and a steeper decrease in entropy with increasing order reflects a TCR repertoire more dominated by high frequency clones. Compared to the RORγt− TEff and RORγt− Treg populations, the Foxp3+RORγt+ and TH17 T cell populations appear to have lower overall diversity (Fig. 1B), with a larger proportion of the repertoire composed of high frequency clones (Fig. 1C, 1D).
The relationship between the diversity patterns of Foxp3+RORγt+ and TH17 cell repertoires can be further quantified using hierarchical clustering. A distance metric can be calculated between two samples’ Renyi diversity profiles (23), as defined by each sample’s series of entropy values across a range of orders. Similarly, a correlation metric can be applied to evenness, a quantitative measure of clonal dominance reflected in the steepness of a curve of Renyi entropies (22). An evenness profile can be calculated from a series of Renyi entropies relativized to their order 0 entropy. Correlation and clustering of population diversity (Fig. 1E) and evenness profiles (Fig. 1F; Supplemental Fig. 1F) across all tissues groups samples into two broad sets, one composed mainly of TH17 and Foxp3+RORγt+ cell samples and the other composed mostly of RORγt− TEff and RORγt− Treg cell samples. Thus, these data suggest that the TCR repertoire diversity of Foxp3+RORγt+ cells is closer to that of the TH17 cell population and that these cells may be driven by a more restricted set of antigens than other Foxp3+ cells.
Foxp3+RORγt+ cells share limited sequence similarity with TH17 cells
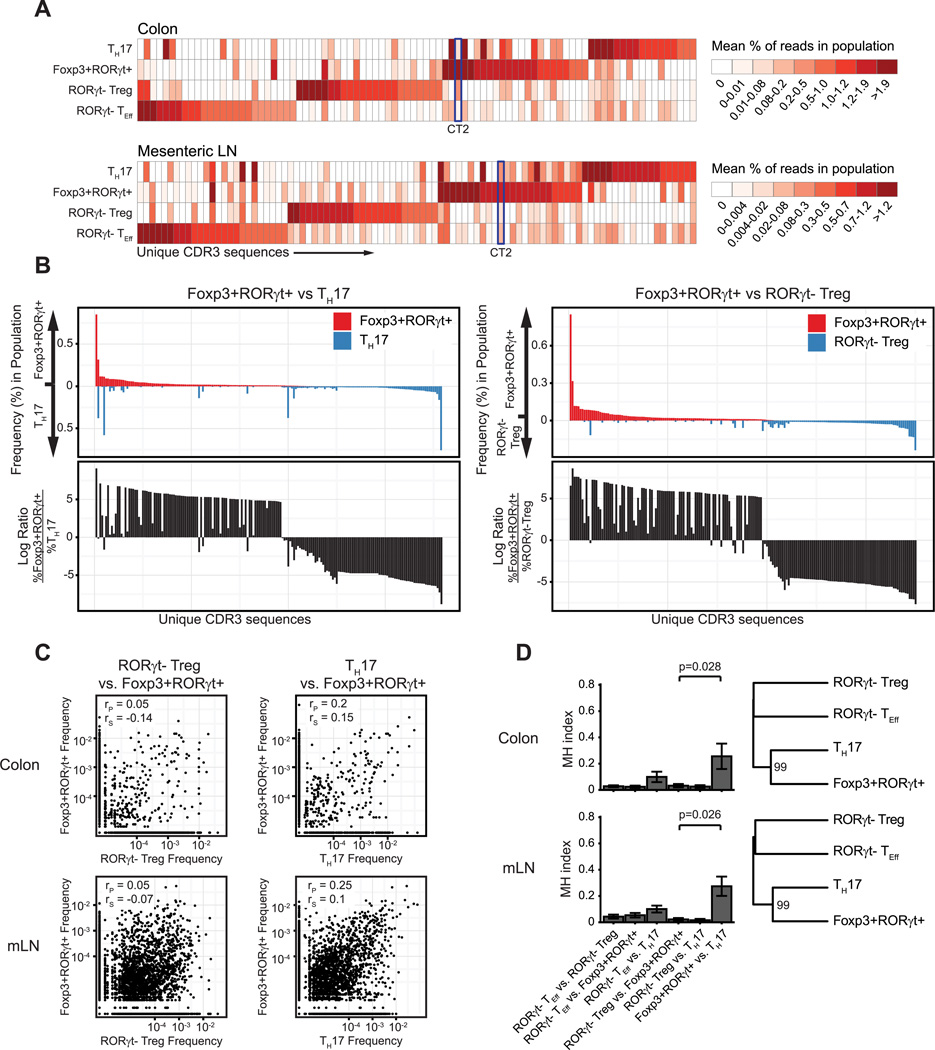
To assess similarity of the Foxp3+RORγt+ and TH17 cell repertories at the level of antigen specificity, we compared the usage of individual TCR sequences across each of the sequenced populations. For most of the sequenced subsets, the majority of top clones for each population are largely unshared by any of the other populations (Fig. 2A, 2B). However, the Foxp3+RORγt+ T cell population shares several of its more common TCR specificities with the TH17 population (Fig. 2C).
Figure 2. Foxp3+RORγt+ T cell population shares limited number of TCR specificities with TH17 cell population.
(A) Top 25 TCR sequences from each population plotted by mean relative frequency in each population. The TCR CT2 is indicated. (B) Mean clonal frequencies (top) or ratio (bottom) of all TCRs with ≥0.01% clonal frequency in either of indicated populations in the colon. (C) Pearson (rP) and Spearman (rS) correlation of TCR frequencies between indicated populations. n=6, each of 2 pooled mice. (D) Mean Morisita-Horn (MH) similarity index (0=complete dissimilarity, 1=identical) between indicated populations. Dendrogram of mean MH-based distance. Value indicates bootstrap reproducibility of branch point. Error bars/ribbon = ± 1 s.e.m.
We further quantified this repertoire overlap using the Morisita-Horn (MH) index. This index represents the probability that the sequence drawn from each of the two populations being compared will represent the same TCR, relative to the probability that two sequences drawn from all possible sequences, regardless of population, will represent the same TCR. This metric results in a continuum of values from 0, indicating complete dissimilarity, to 1, indicating complete similarity. Similar to previous observations of effector T cells in our lab, the repertoires of each of the four sorted populations were largely unique. However, the Foxp3+RORγt+ and TH17 repertoires demonstrated some degree of similarity that was significantly greater than that between the Foxp3+ RORγt+ population and any other sequenced subset (Fig. 2D, Supplemental Fig. 2A). We also utilized TCR repertoire perturbation scores, which applies the MH-similarity index to TCR’s according to their amino acid lengths (i.e. CDR3 spectratype), rather than the TCR CDR3 amino acid sequences (25), to compare the general structure of TCRs between samples. This also showed that the Foxp3+RORγt+ repertoire frequently cluster with the TH17 (Supplemental Fig. 2B, 2C).
However, analysis of the repertoire at the level of individual TCRs revealed that only a limited number of shared high frequency clones accounts for the repertoire similarity seen between these two populations (Fig. 2B). This is further supported by the difference in Pearson and Spearman correlation values for comparisons of TCR frequencies between populations. Pearson correlation is more affected by outliers, like the high frequency TCRs shared by the Foxp3+RORγt+ and TH17 populations, while Spearman correlation is less affected. We observe that, while a modest correlation between the Foxp3+RORγt+ and TH17 populations is suggested by Pearson correlation, this relationship is reduced or absent when considering Spearman correlation. These data suggest that the Foxp3+RORγt+ population possesses a largely unique TCR repertoire, yet shares a limited subset of high frequency antigen receptor sequences with the TH17 population.
TCR specificity can mediate acquisition of the Foxp3+RORγt+ phenotype
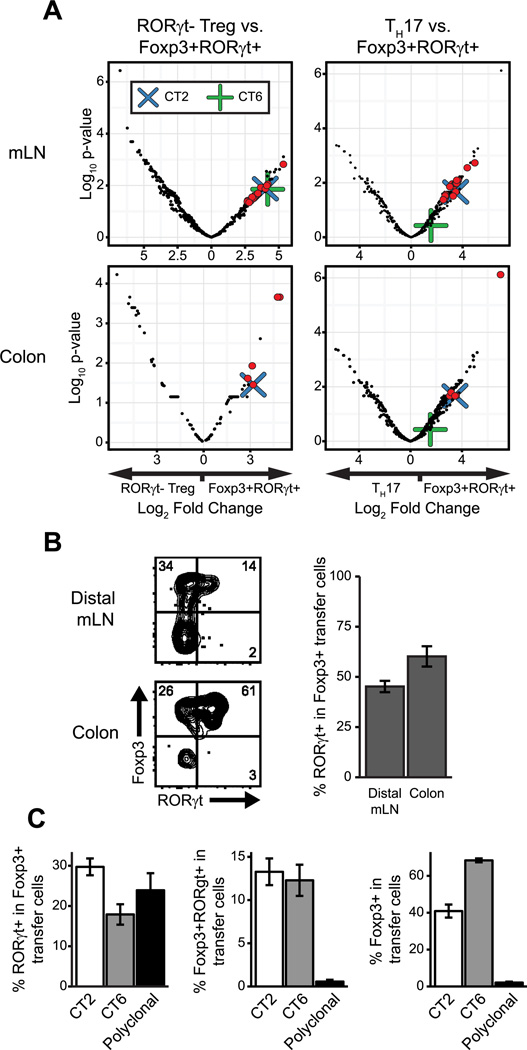
Though limited to a subset of clones, the similarity seen between the Foxp3+RORγt+ TCR repertoire and TH17 TCR repertoire raises the possibility that Foxp3+RORγt+ cells could represent a developmental subset of the TH17 population. However, as previous work has characterized Foxp3+RORγt+ cells as immunoregulatory, this population could represent a developmental subset of Treg cells instead. In order to distinguish between these alternatives, we utilized a monoclonal T cell-based approach. From our sequencing data, we identified several TCRs highly enriched in the Foxp3+RORγt+ T cell population. Moreover, as the majority of Foxp3+RORγt+ TCRs are restricted to this population, we narrowed our search to TCRs with high relative enrichment compared to all other sorted populations (Fig. 3A, Supplemental Table I). Notably, the TCR CT2, previously identified by our lab as a colonic Treg TCR specific to gut bacteria (27), was the single most highly represented TCR in the Foxp3+RORγt+ T cell population.
Figure 3. Antigen specificity can mediate development of Foxp3+RORγt+ T cells.
(A) Enrichment of TCRs in Foxp3+RORγt+ population relative to indicated population vs. FDR adjusted p-value. Red dots are TCRs > 2-fold enriched in the Foxp+RORγt+ population over both Treg and TH17 populations with an adjusted p-value < 0.05. Blue X indicates TCR CT2. Green cross indicates TCR CT6. n=6, each of 2 pooled mice. (B) Phenotype of CT2-TCRtg CD4+ cells 3 weeks after transfer of 5×104 naïve cells into WT hosts. (C) Frequencies of indicated populations from distal mLN of WT host mice 3 weeks after transfer of 5×104 naïve cells. n≥5 for each group. Data representative of ≥2 independent experiments. Error bars/ribbon = ±1 s.e.m
We first sought to test whether this TCR could specifically mediate Foxp3+RORγt+ T cell development using TCR transgenic (TCRtg) mice expressing CT2. We adoptively transferred sorted naive Foxp3−RORγt− TCRtg cells into wild type host mice. By 3 weeks post-transfer, these cells showed significant co-expression of Foxp3 and RORγt, with greater than 50% of all colonic Treg cells expressing RORγt (Fig. 3B). While the proportion of Foxp3+ cells expressing RORγt is similar for transferred polyclonal cells, this conversion is much less efficient than CT2 TCRtg cells. With polyclonal cells, only ~1% of all cells transferred become Foxp3+ RORγt+, compare to ~13% of all CT2 TCRtg cells at 3 weeks in the distal mesenteric LN (Fig. 3C). This suggests that different TCRs have varying ability to mediate Foxp3+RORγt+ T cell development and, thus, that antigen specificity is an important determinant for differentiation into this phenotype. This is further supported by the observation that an alternative mucosal Treg TCR, CT6, also mediates Foxp3+RORγt+ conversion, but to a lesser degree than CT2 (Fig. 3C).
CT2 TCRtg T cells develop into Foxp3+RORγt+ T cells via a Treg cell intermediate
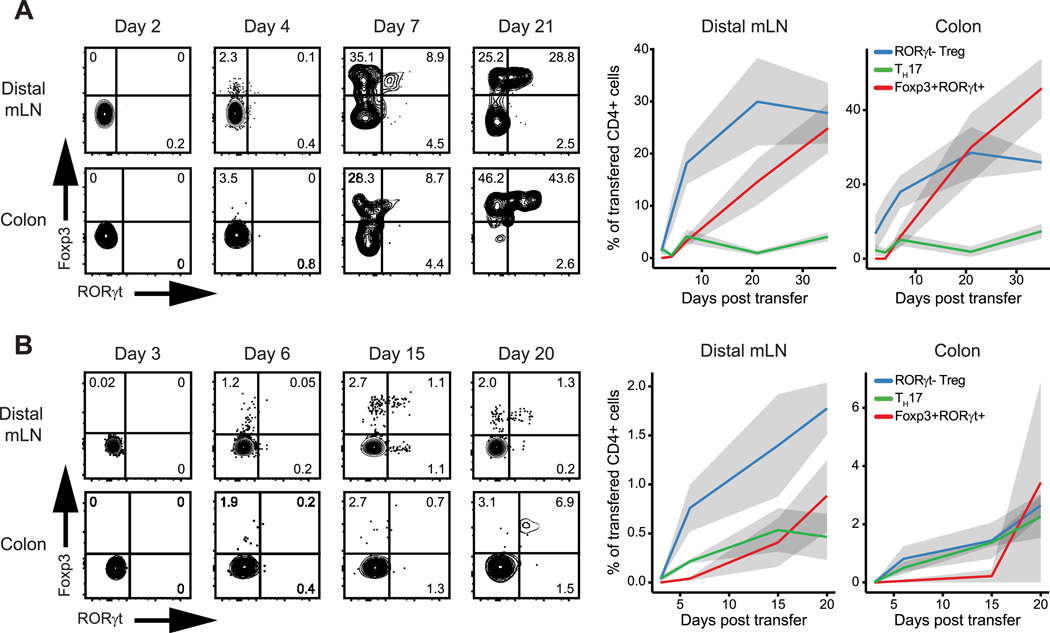
To more precisely delineate the developmental relationships between Foxp3+RORγt+, Treg, and TH17 cells, we next performed kinetic experiments using this monoclonal TCR system. After transfer of naïve CT2 cells into wild type hosts, we analyzed Foxp3 and RORγt expression in the transfer population at multiple time points. Foxp3 expression was first observed as early as day 4 post-transfer. RORγt expression followed by day 7 post-transfer and was mostly restricted to the Foxp3+ population (Fig. 4A). By 3 weeks post-transfer, RORγt expression is substantial and entirely restricted to the Foxp3+ population in both the colon and distal mLN.
Figure 4. Foxp3+RORγt+ T cells develop from RORγt− Treg cells.
Phenotype of transferred CD4+ cells at indicated time points after transfer of 5×104 naïve (A) CT2 TCRtg or (B) polyclonal cells into WT hosts. n≥5 for each group/time point. Data representative of ≥2 independent experiments. Error bars/ribbon = ±1 s.e.m
We also validated these results in a polyclonal setting. We obtained similar results as seen with CT2 TCRtg cells, although the kinetics and extent of differentiation were somewhat diminished in the polyclonal population (Fig. 4B). In addition, the TH17 population was more prevalent in the polyclonal setting. Together, these data suggest that the majority of Foxp3+RORγt+ T cells begin as naïve CD4+ T cells in the periphery and pass through a RORγt− Treg intermediate before co-expressing RORγt.
CX3CR1+ APCs can provide signals for Foxp3+RORγt+ differentiation
There are several phenotypically distinct APC subsets found in the gut, several of which support the differentiation of particular CD4+ T cells subsets. For example, monocyte-derived MHC2−hi APCs (28) contribute to the development of mucosal TH17 cells, while Treg cells in the gut broadly require CD103+ cDCs (29). Recent work from our lab suggests that mucosal Treg development is also partially dependent on the CD11b+ fraction of CD103+ DCs in a monoclonal setting (30). Previous work has demonstrated that CD11c+ DCs are necessary for Foxp3+RORγt+ T cell development (8), but the role of specific APC subsets remains unclear.
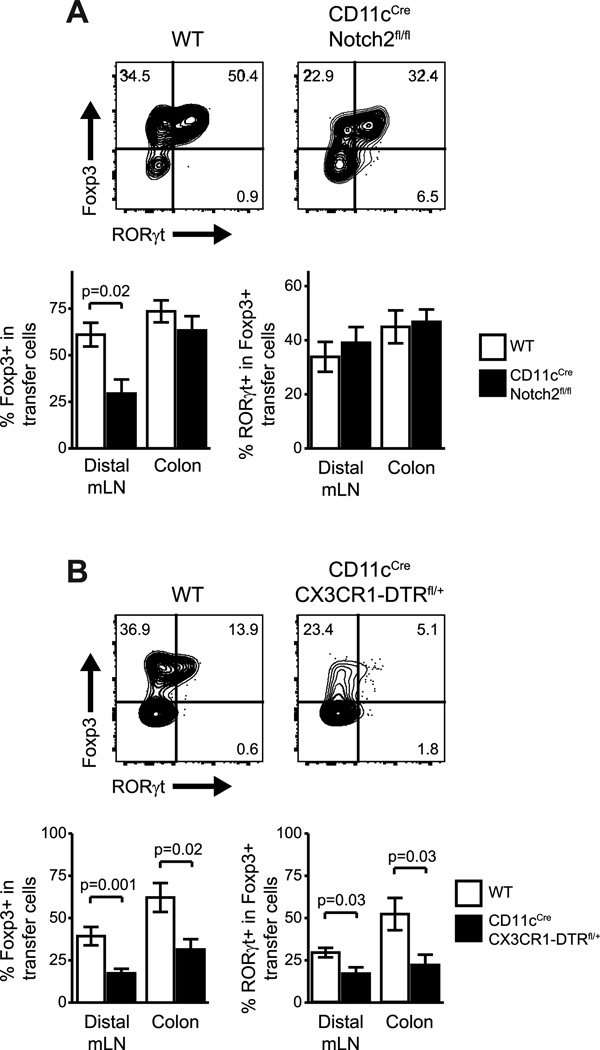
To test whether there are similar APC requirements for Foxp3 and RORγt expression in RORγt+ Treg cells, we adoptively transferred naïve CT2 TCRtg T cells into hosts that lack specific subsets of APCs. Since we have previously demonstrated a role for Notch2-dependent CD103+CD11b+ DC in CT2 Treg development, we first tested if RORγt+ Treg cells are similarly dependent on this DC. While we confirmed a general defect in Foxp3+ cells development, DC-specific Notch2 deficiency in host mice (31) did not specifically affect RORγt+ Treg cells (Fig. 5A).
Figure 5. The development of Foxp3+RORγt+ T cells is not affected by the loss of individual APC subsets.
(A) Phenotype of CT2 TCRtg CD4+ cells in Notch2+ APC-deficient host mice 3 weeks after transfer of 5×104 naïve cells. (B) Phenotype of CT2 TCRtg CD4+ cells in CXC3R1-APC deficient host mice 10 days after transfer of 5×104 naïve cells. For deletion of CX3CR1+ APCs, DT was administered (20ng/g) for two days prior to transfer and (5ng/g) every other day after transfer until analysis. n≥7 for each group. Data representative of ≥2 independent experiments. Error bars = ±1 s.e.m
Because of their reported contribution to SFB-mediated TH17 induction, we also tested the role of CX3CR1+ APCs in Foxp3+RORγt+ T cell development using CX3CR1-DTR mice. These mice express DTR upon CD11c-Cre mediated excision of a Loxp-Stop cassette, resulting in loss of the of the CX3CR1+ APC population after DT treatment. Indeed, we observed a specific defect in the generation of RORγt-expressing Treg cells (Fig. 5B), similar to the impaired development of RORγ+ TH17 cells reporter previously. Interestingly, we also observed a deficit in overall Foxp3+ cell frequencies in absence of CX3CR1+ APCs. This shows that the CX3CR1+ APC may be involved in the differentiation of multiple T cell subsets and are important for the specific development of RORγ+ Treg cells.
Discussion
The expression of Foxp3 or RORγt in T cells leads to the acquisition of generally antagonistic cellular phenotypes. Yet, a significant proportion of gut lamina propria T cells co-express these two transcription factors. Despite evidence suggesting that these Foxp3+RORγt+ T cells play a regulatory, rather than inflammatory role, it is still unclear how these cells develop in relation to classical Treg and TH17 cells. Here, we present data which shows that Foxp3+RORγt+ T cells are a developmental subset of regulatory T cells.
Among CD4+ T cell subsets, Treg and TH17 cells possess a unique relationship. Peripheral development of both subsets can be induced by TGFβ, raising the possibility that Treg and TH17 cells share a developmental intermediate (12). The identification of Foxp3+RORγt+ T cells represents an appealing candidate for such an intermediate. However, using our TCRtg system, we demonstrate that the first cells to arise from naïve precursors are RORγt− Treg cells, preceding the appearance of Foxp3+RORγt+ T cells by several days. These TCRtg cells infrequently develop into TH17 cells, despite reaching a Foxp3+RORγt+ state, suggesting that this is a mature rather than intermediate phenotype. This conclusion is supported by data showing the stability of the Foxp3+RORγt+ T cell gene expression profile (9).
If Foxp3+RORγt+ T cells do represent a subset of Treg cells, one might expect to see more similarity between the TCR repertoires of these two populations. However, as previously reported, >70% of all Foxp3+RORγt+ T cell development occurs by 9 weeks of age (8). As the mice in our experiments were sequenced at a mean 18 weeks of age, the majority of cells in our data set capable of differentiating into Foxp3+RORγt+ cells likely had already done so. Therefore, even if the Foxp3+RORγt+ population is a subset of Treg cells, these two groups could still appear distinct by TCR sequencing.
In fact, the repertoire of Foxp3+RORγt+ cells appears largely distinct from all sequenced populations. The simplest explanation for this observation is that Foxp3+RORγt+ cells recognize a distinct set of antigens. However, it remains possible that they may recognize the same antigens, but that different affinities to these or other antigens results the differential expression of Foxp3 or RORγt, akin to that seen with Treg cell selecting ligands in the thymus (32, 33). Future studies will be required to determine the precise features of antigen specificity that dictate Foxp3+RORγt+ development.
Yet, the Foxp3+RORγt+ population does share some TCR sequences with the TH17 population and, though they represent a limited subset of the overall repertoire, this raises the question of how these clones develop. One possibility is that cells with these shared TCRs sequentially develop from TH17 to Foxp3+RORγt+ cells, in contrast to the CT2 TCR studied. Alternatively, the Foxp3+RORγt+ and TH17 phenotypes may develop via parallel pathways due to stochastic expression of cytokines or other signals associated with the cognate antigen. Further studies with additional TCRs are required to determine the extent that CT2 represents Foxp3+RORγt+ development, as well as establish the progression of transcription factor expression in cells expressing TCR shared by Foxp3+RORγt+ and TH17 repertoires.
Notably, our sequencing data differs from that of a previous study which found no repertoire similarity between Foxp3+RORγt+ T cells and any other subset (9). However, these experiments used samples pooled from the spleen and peripheral lymph nodes, two tissues in which the population of RORγt+ Treg cells is dramatically smaller than that seen in the lamina propria. Given the relationship between Foxp3+RORγt+ T cells and the gut microbiota, we believe our analysis, which includes the lamina propria, more accurately reflects the physiology of these cells. Moreover, the use of fully polyclonal mice in the former study can make identification of shared clones difficult due the unknown TCRβ pairing, as well as the greater diversity in the TCR repertoire. Our use of a fixed TCRβ system restricts the TCR repertoire to a size capable of revealing clonal relationships, such as that seen between the Foxp3+RORγt+ and TH17 cell populations.
The developmental path of Foxp3+RORγt+ cells is reflected in their requirements for specific APC subsets. As we have shown previously (30) and in this report, Foxp3+ T cell development is influenced by Notch2-dependent, CD103+CD11b+ DCs. Moreover, we demonstrate that development of the RORγt+ fraction of Treg cells is specifically affected by the loss of CX3CR1+ APCs. Although future studies are required to determine the specific role of these APC subsets, one hypothetical model consistent with the current data is that naïve T cells first upregulate Foxp3 in response to signals from CD103+CD11b+ DCs and then express RORγt in response to CX3CR1+ APC-derived factors.
Several groups have demonstrated that co-expression of effector T cell transcription factors by a Treg cell confers upon it the ability to specifically regulate the corresponding effector T cell subset. In the case of T-bet+ Treg cells, this is achieved, in part, by the T-bet-mediated expression of the chemokine receptor CXCR3, which allows these cells to traffic to the same locations as CXCR3-expressing TH1 cells (6). However, it remains to be shown whether these Treg subsets recognize antigens similar to those of their corresponding effector subsets and if such an overlap in TCR specificity contributes to the function of these Treg cells.
Here, we show that a noticeable fraction of high frequency Foxp3+RORγt+ TCR clones are shared with the TH17 population. As previous work demonstrated that TH17 inflammation is enhanced in the absence of RORγt+ Treg cells (7, 9), our observation suggests effector-specific Treg populations can share antigen specificities with their target effector populations. TCR-dependent activation of these cells would be expected to occur at the same sites of corresponding TH17 activation, thus providing TH17-specific regulation. Loss of these clones in Foxp3CreRORγtflx could account for the reported TH17-regulatory phenotype. One caveat to this model comes from an additional study, which instead reported that RORγt+ Treg cells are involved in regulating type-2 immunity (8). However, as we did not specifically sequence the Th2 subset, we cannot rule out the possibility of a similar set of Foxp3+RORγt+ TCRs that overlap with this population. Future work is needed to determine the role of overlap in antigen specificity for regulatory T cell control of specific effector T cell subsets.
Supplementary Material
Acknowledgments
We thank T. Egawa, T. Ai, and J. Chai (WashU) for critical reading of the manuscript.
C.S.H. is supported by NIH R01 DK094995, R21 AI097535, the CCFA, and the Burroughs Wellcome Fund. B.D.S is additionally supported by NIH F30 DK102214 and the Hu & Zheng Immunology Fellowship.
B.D.S and C.S.H. conceived the project, designed the experiments, and wrote the manuscript. B.D.S performed the experiments.
Footnotes
Raw sequence data can be found in the European Nucleotide Archive, accession # PRJEB15258 (http://www.ebi.ac.uk/ena/data/view/PRJEB15258).
References
- 1.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee Ca, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 3.Josefowicz SZ, Lu L-F, Rudensky AY. Regulatory T Cells: Mechanisms of Differentiation and Function. Annu. Rev. Immunol. 2012:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng Y, Chaudhry A, Kas A, DeRoos P, Kim JM, Chu T-T, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koch Ma, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 2009;10:213–215. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, Ghosh S, Earl A, Snapper SB, Jupp R, Kasper D, Mathis D, Benoist C. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science. 2015;349:993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohnmacht C, Park J-HJ, Cording S, Wing JB, Atarashi K, Obata Y, Gaboriau-Routhiau V, Marques R, Dulauroy S, Fedoseeva M, Busslinger M, Cerf-Bensussan N, Boneca IG, Voehringer D, Hase K, Honda K, Sakaguchi S, Eberl G. The microbiota regulates type 2 immunity through RORγt+ T cells. Science. 2015;349:989–993. doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]
- 9.Yang B-H, Hagemann S, Mamareli P, Lauer U, Hoffmann U, Beckstette M, Föhse L, Prinz I, Pezoldt J, Suerbaum S, Sparwasser T, Hamann A, Floess S, Huehn J, Lochner M. Foxp3+ T cells expressing RORγt represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol. 2015;205:1381–1393. doi: 10.1038/mi.2015.74. [DOI] [PubMed] [Google Scholar]
- 10.Lochner M, Bérard M, Sawa S, Hauer S, Gaboriau-Routhiau V, Fernandez TD, Snel J, Bousso P, Cerf-Bensussan N, Eberl G. Restricted microbiota and absence of cognate TCR antigen leads to an unbalanced generation of Th17 cells. J. Immunol. 2011;186:1531–1537. doi: 10.4049/jimmunol.1001723. [DOI] [PubMed] [Google Scholar]
- 11.Lochner M, Peduto L, Cherrier M, Sawa S, Langa F, Varona R, Riethmacher D, Si-Tahar M, Di Santo JP, Eberl G. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J. Exp. Med. 2008;205:1381–1393. doi: 10.1084/jem.20080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou L, Lopes JE, Chong MMW, Ivanov II, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, Ziegler SF, Littman DR. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsieh C-S, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat. Immunol. 2006;7:401–410. doi: 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
- 14.Liston A, Nutsch KM, Farr AG, Lund JM, Rasmussen JP, Koni Pa, Rudensky AY. Differentiation of regulatory Foxp3+ T cells in the thymic cortex. Proc. Natl. Acad. Sci. U. S. A. 2008;105:11903–11908. doi: 10.1073/pnas.0801506105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bautista JL, Lio C-WJ, Lathrop SK, Forbush K, Liang Y, Luo J, Rudensky AY, Hsieh C-S. Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat. Immunol. 2009;10:610–617. doi: 10.1038/ni.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCright B, Lozier J, Gridley T. Generation of new Notch2 mutant alleles. Genesis. 2006;44:29–33. doi: 10.1002/gene.20181. [DOI] [PubMed] [Google Scholar]
- 17.Diehl GE, Longman RS, Zhang J-X, Breart B, Galan C, Cuesta A, Schwab SR, Littman DR. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494:116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 19.Lefranc MP. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 2003;31:307–310. doi: 10.1093/nar/gkg085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jost L. Entropy and diversity. Oikos. 2006;113:363–375. [Google Scholar]
- 21.Pacholczyk R, Kern J, Singh N, Iwashima M, Kraj P, Ignatowicz L. Nonself-antigens are the cognate specificities of Foxp3+ regulatory T cells. Immunity. 2007;27:493–504. doi: 10.1016/j.immuni.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jost L. The relation between evenness and diversity. Diversity. 2010;2:207–232. [Google Scholar]
- 23.Greiff V, Bhat P, Cook SC, Menzel U, Kang W, Reddy ST. A bioinformatic framework for immune repertoire diversity profiling enables detection of immunological status. Genome Med. 2015;7:49. doi: 10.1186/s13073-015-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao A, Jost L. Coverage-based rarefaction and extrapolation: Standardizing samples by completeness rather than size. Ecology. 2012;93:2533–2547. doi: 10.1890/11-1952.1. [DOI] [PubMed] [Google Scholar]
- 25.Bergot AS, Chaara W, Ruggiero E, Mariotti-Ferrandiz E, Dulauroy S, Schmidt M, von Kalle C, Six A, Klatzmann D. TCR sequences and tissue distribution discriminate the subsets of naïve and activated/memory Treg cells in mice. Eur. J. Immunol. 2015;45:1524–1534. doi: 10.1002/eji.201445269. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto J, Adachi Y, Onoue Y, Adachi YS, Okabe Y, Itazawa T, Toyoda M, Seki T, Morohashi M, Matsushima K, Miyawaki T. Differential expression of the chemokine receptors by the Th1- and Th2-type effector populations within circulating CD4+ T cells. J. Leukoc. Biol. 2000;68:568–574. [PubMed] [Google Scholar]
- 27.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio C-W, Santacruz N, Peterson Da, Stappenbeck TS, Hsieh C-S. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panea C, Farkas AM, Goto Y, Abdollahi-Roodsaz S, Lee C, Koscsó B, Gowda K, Hohl TM, Bogunovic M, Ivanov II. Intestinal Monocyte-Derived Macrophages Control Commensal-Specific Th17 Responses. Cell Rep. 2015;12:1314–1324. doi: 10.1016/j.celrep.2015.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coombes JL, Siddiqui KRR, Arancibia-Cárcamo CV, Hall J, Sun C-M, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J. Exp. Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nutsch K, Chai JN, Ai TL, Russler-Germain E, Feehley T, Nagler CR, Hsieh CS. Rapid and efficient generation of regulatory T cells to commensal antigens in the periphery. Cell Reports. doi: 10.1016/j.celrep.2016.08.092. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satpathy AT, Briseño CG, Lee JS, Ng D, Manieri NA, Kc W, Wu X, Thomas SR, Lee W-L, Turkoz M, McDonald KG, Meredith MM, Song C, Guidos CJ, Newberry RD, Ouyang W, Murphy TL, Stappenbeck TS, Gommerman JL, Nussenzweig MC, Colonna M, Kopan R, Murphy KM. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 2013;14:937–948. doi: 10.1038/ni.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shafiani S, Dinh C, Ertelt JM, Moguche AO, Siddiqui I, Smigiel KS, Sharma P, Campbell DJ, Way SS, Urdahl KB. Pathogen-Specific Treg Cells Expand Early during Mycobacterium tuberculosis Infection but Are Later Eliminated in Response to Interleukin-12. Immunity. 2013;38:1261–1270. doi: 10.1016/j.immuni.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee HM, Bautista JL, Scott-Browne J, Mohan JF, Hsieh CS. A Broad Range of Self-Reactivity Drives Thymic Regulatory T Cell Selection to Limit Responses to Self. Immunity. 2012;37:475–486. doi: 10.1016/j.immuni.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.