Summary
Expression changes of competing endogenous RNAs (ceRNAs) have been proposed to influence microRNA (miRNA) activity and thereby regulate other transcripts containing miRNA-binding sites. Here, we find that although miRNA levels define the extent of repression, they have little effect on the magnitude of the ceRNA expression change required to observe derepression. Canonical 6-nt sites, which typically mediate modest repression, can nonetheless compete for miRNA binding, with potency ∼20% of that observed for canonical 8-nt sites. In aggregate, low-affinity/background sites also contribute to competition. Sites with extensive additional complementarity can appear as more potent, but only because they induce miRNA degradation. Cooperative binding of proximal sites for the same or different miRNAs does increase potency. These results provide quantitative insights into the stoichiometric relationship between miRNAs and target abundance, target-site spacing, and affinity requirements for ceRNA-mediated gene regulation, and the unusual circumstances in which ceRNA-mediated gene regulation might be observed.
Keywords: competing endogenous RNA, miRNA, target abundance, cooperatively, gene regulation, base pair complementarity, miRNA degradation
Graphical Abstract
Highlights
-
•
ceRNA-mediated derepression is typically insensitive to reduced miRNA activity
-
•
Extensively paired sites can reduce derepression thresholds by triggering miRNA decay
-
•
Weak sites can contribute to target-site competition without imparting repression
-
•
Closely spaced sites of the same or different miRNAs cooperatively sequester miRNAs
Denzler et al. show that effects of competing miRNA sites are insensitive to reduced miRNA activity, low-affinity/background miRNA sites contribute to competition, and adjacent miRNA sites can cooperatively sequester miRNAs. Overall, their results reduce the prospects of observing an effect from a ceRNA.
Introduction
MicroRNA (miRNA) levels have long been known to influence the magnitude of target-gene repression (Bartel, 2009). More recent studies point out that the number of predicted binding sites present in the transcriptome also affects the activity of miRNAs (Arvey et al., 2010, Garcia et al., 2011). Consistent with this concept, strong overexpression of natural or artificial RNAs that contain miRNA sites can titrate miRNAs away from natural targets, thereby reducing the repression of these transcripts (Ebert et al., 2007, Franco-Zorrilla et al., 2007, Mukherji et al., 2011, Hansen et al., 2013, Memczak et al., 2013). These observations are extended by the notion that a site-containing transcript found naturally within cells can act as competing endogenous RNA (ceRNA) and regulate other site-containing transcripts by increasing or decreasing the miRNA activity (Poliseno et al., 2010, Cesana et al., 2011, Salmena et al., 2011, Karreth et al., 2015).
The ceRNA hypothesis remains controversial due to the lack of a plausible explanation for how modulating the expression of a single endogenous gene could perceptibly influence miRNA activity across all of its target sites. Two recent studies have empirically assessed the ceRNA hypothesis by quantifying the number of miRNA response elements (MREs) that must be added to detect ceRNA-mediated gene regulation (Bosson et al., 2014, Denzler et al., 2014). Both studies agree that determining the number of transcriptomic miRNA-binding sites is crucial for evaluating the potential for ceRNA regulation and that miRNA-binding sites are generally higher than the number of miRNA molecules. However, they differ in two aspects: (1) the experimental approaches used to determine the number of “effective” transcriptomic miRNA-binding sites and (2) the impact miRNA concentrations have on the number of binding sites that must be added to detect target gene derepression (derepression threshold [DRT]).
The discrepancies between these studies lead to different conclusions with respect to the likelihood of observing ceRNA effects in natural settings. The first study concluded that changes in ceRNAs must approach a miRNA’s target abundance before they can exert a detectable effect on gene regulation (Denzler et al., 2014). Furthermore, because target abundance for a typical miRNA is very high, regulation of gene expression by ceRNAs is unlikely to occur in differentiated cells under physiological settings or most disease settings (Denzler et al., 2014). In addition, the study shows that the DRT remains constant when miRNA activity is reduced. A subsequent review presents a mathematical model that assesses binding-site occupancy and competition at different assumed target abundances (Jens and Rajewsky, 2015). This in silico model predicts that only global and collective changes in binding sites can produce an effect on target abundance large enough to detectably derepress target genes, which concurs with the results and conclusions of Denzler et al. (2014).
The second study presents a “hierarchical affinity model,” in which the miRNA abundance is proposed to determine the respective susceptibility to ceRNA-mediated regulation (Bosson et al., 2014). In this model, the suggestion is that, as miRNA concentration increases and Ago-miRNA complexes spread to weaker and weaker sites (with affinity inferred from the site hierarchy of 8-nt > 7-nt > 6-nt site), the effective target-site abundance grows too large for physiological ranges of ceRNA expression to influence repression. By this reasoning, physiological ceRNA changes can nevertheless influence repression by a more modestly expressed miRNA, with its correspondingly lower effective target-site abundance. Moreover, the use of high-throughput cross-linking to detect targets leads to lower target-abundance estimates, which further increases the plausibility of ceRNA regulation (Bosson et al., 2014). However, experimental support for the proposed influence of miRNA concentration is correlative and lacks direct experimental evidence, such as manipulation of miRNA activity and measurement of resulting DRT changes.
Denzler et al. (2014) propose that sites of all different affinities contribute to the effective target abundance, regardless of the miRNA concentration. Here, we call the model of Denzler et al. (2014) the “mixed-affinity model” to distinguish it from the hierarchical affinity model. The mixed-affinity model recognizes that a high-affinity site will contribute more to effective target-site abundance than a low-affinity site (Denzler et al., 2014). However, in aggregate, low-affinity sites, because of their high numbers within the transcriptome, still make a substantial contribution to the effective target-site abundance for each miRNA—even for more modestly expressed miRNAs.
Other studies suggest that the ceRNA crosstalk of two transcripts is stronger and more specific when they share a large number of sites to different miRNA seed families. This hypothesis emerged from observations in cancer models, in which the expression of a particular oncogene correlates with its pseudogene, and both transcripts share a high sequence homology in their 3′ UTRs and are reported to co-regulate each other through a ceRNA mechanism (Poliseno et al., 2010, Karreth et al., 2015). Even if transcripts containing multiple sites can exert an additive effect of independently acting binding sites, sites for each miRNA family would still have to individually reach the high thresholds necessary to observe target-gene derepression. Therefore the simple presence of multiple binding sites alone would not be expected to be sufficient to increase the likelihood of a ceRNA effect, unless the sites acted through a cooperative mechanism. Although the effect of cooperativity has been studied in the context of target-gene repression (Doench et al., 2003, Grimson et al., 2007, Saetrom et al., 2007, Broderick et al., 2011), it is unclear whether closely spaced miRNA-binding sites can sequester miRNA in a non-independent manner and hence increase the prospects of a ceRNA effect.
In this study, we examine the impact that miRNA levels have on the DRT and thereby address a key difference between the hierarchical affinity and mixed-affinity models. We then analyze the influence of target-site complementarity on ceRNA-mediated gene regulation and examine the extent to which closely spaced miRNA-binding sites can cooperatively influence the potency of target-gene derepression. Finally, we develop a mathematical model, which incorporates both the mixed-affinity binding and the repressive activities of miRNAs to recapitulate our results.
Results
miR-294 Is Susceptible to Competition Despite High Expression Levels
A powerful tool for studying competition among MREs is a single-cell reporter assay that transcribes mCherry mRNA (with or without MREs in its 3′ UTR) and enhanced yellow fluorescent protein (eYFP) mRNA as an internal measure of reporter transcription (Mukherji et al., 2011, Bosson et al., 2014). Using analytical flow cytometry, mCherry reporter readout can be assessed over a broad range of added MREs. At high expression levels, MREs can compete with each other for miRNA binding, thereby causing derepression. Using this assay in embryonic stem cells (ESCs), some miRNAs need fewer competing MREs to mediate reporter derepression and are therefore more susceptible to ceRNAs than other miRNAs (Bosson et al., 2014).
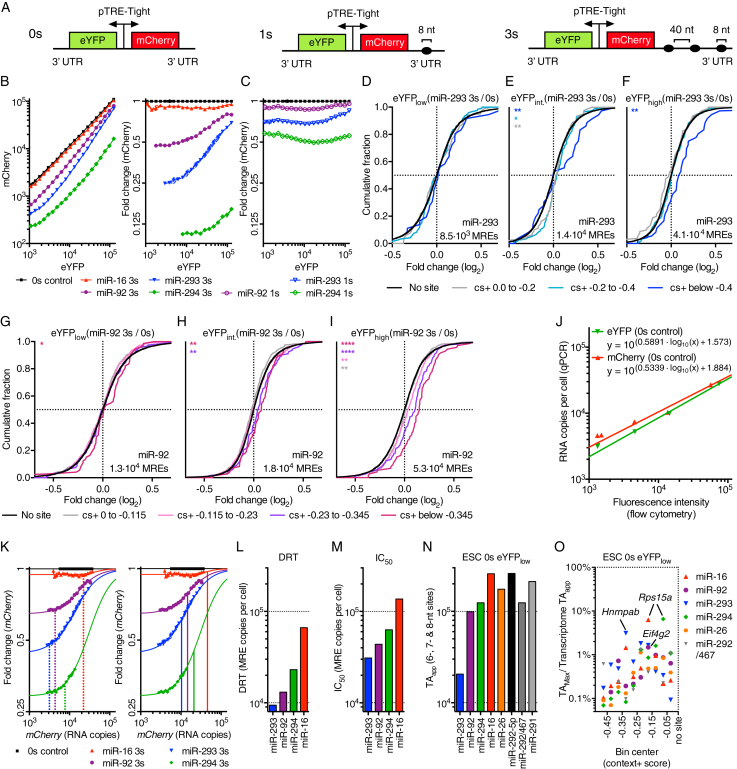
To explore these different susceptibilities, we created reporter constructs for six highly expressed ESC miRNAs (miR-294, -293, -92, -16, -26, and -292-5p) (Bosson et al., 2014), containing zero (0s), or three (3s) 8-nt miRNA sites in the 3′ UTR of mCherry (Figure 1A). For the miRNA families miR-294, -293, and -92, reporters containing a single (1s) miRNA-binding site were also created. Sites for miR-294, -293, -92, and -16 (Figures 1B and 1C), but not those for miR-26 and -292-5p (data not shown) caused detectable miRNA-mediated repression of mCherry. The extent of repression of reporters for miR-294, -293, and -92 resembled that observed previously, as did the derepression of mCherry constructs harboring sites for miR-293 or miR-92 (Bosson et al., 2014). However, the 3s reporter construct for miR-294, a miRNA reported to be insensitive to competitor perturbations (Bosson et al., 2014), and the reporter for miR-16 were derepressed when eYFP fluorescence exceeded 2.2 × 104 or 2.8 × 104, respectively (Figure 1B). The ability to observe derepression of the miR-294 reporter presumably resulted from improvements to the equipment and protocol that enabled more precise measurements, as indicated by the improved SEM values, although differences between the ESCs might have also played a role. These results showing derepression of the mCherry reporter at similar competitor levels for both miR-294 and miR-16, two miRNAs present at very different levels in ESCs, and with very different miRNA:target ratios estimated by Bosson et al. (2014), support the mixed-affinity model.
Figure 1.
Derepression of Target mRNAs Occurs at a High Threshold of Added Target Sites
(A) Dual-color fluorescent reporter constructs containing zero (0s), one (1s), or three (3s) 8-nt miRNA site(s) in the 3′ UTR of mCherry.
(B and C) ESCs transfected with either a 3s (B), 1s (C), or 0s reporter construct (n = 3) with miRNA-binding sites for miR-294, -293, -92, or -16. Mean mCherry fluorescence (B, left), and mCherry fluorescence normalized to the 0s control (B, right and C) across 20 bins of eYFP.
(D–I) RNA-seq results (n = 2) of sorted ESCs shown in Figure S1A. ESCs were transfected with a 3s reporter for miR-293 (D–F) or miR-92 (G–I), or a 0s control, and gated for cells with low (eYFPlow) (D and G), intermediate (eYFPint.) (E and H), or high eYFP (eYFPhigh) (F and I) expression. Cumulative distribution function (CDF) of mRNA changes for predicted target genes with the indicated context+ score (cs+) bins (color) or for genes with no miRNA site (black). mCherry MREs per cell evaluated by qPCR are shown on each graph. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, one-sided Kolmogorov-Smirnov (K-S) test. Also see Figures S1B and S1C.
(J) Relationship between reporter protein fluorescence measured by flow cytometry and RNA copies per cell evaluated by qPCR of ESCs transfected with the 0s reporter and sorted into four different bins of eYFP-expressing cells. Line represents non-linear regression of data points; respective equations are shown.
(K) Protein fluorescence values shown in (B) transformed to RNA copies per cell using the equations shown in (J). Vertical lines represent the DRT (dotted lines) or IC50 (solid lines).
(L and M) Bar plot of DRT (L) and IC50 (M) shown in (K).
(N) Transcriptome TAapp of ESCs transfected with the 0s reporter and sorted for low eYFP-expressing cells (ESC 0 s eYFPlow).
(O) Fractional contribution of the largest potential contributors to transcriptome TAapp of ESC 0s eYFPlow. Potential contributors were binned by their context+ score, and the top potential contributors are plotted within each bin.
Data represent mean ± SEM for (B), (C), and (K).
Derepression of Target mRNAs Occurs at a High Threshold of Added Target Sites
The competition among MREs for miRNA binding is expected to occur not only between the added MREs within the mCherry mRNA but also between the added MREs and those of the endogenous targets. To examine the effect on endogenous targets, ESCs transfected with the 0s or a 3s reporter for either miR-293 or miR-92 were sorted into three bins based on their eYFP expression (Figure S1A, available online). RNA sequencing (RNA-seq) of each bin revealed the number of MREs added per cell as well as differences in endogenous mRNA levels for cells with the 3s reporter compared to those with the 0s reporter. Endogenous mRNAs with predicted MREs were grouped based on the strength of their predicted response to the miRNA, as scored by the context+ model of TargetScan 6.2 (Garcia et al., 2011). For the middle, but not the lower, bin (1.4 × 104 and 0.85 × 104 added miR-293 MREs per cell, respectively), endogenous miR-293 targets were derepressed, as indicated by the significant shift in the distribution of mRNA fold-change values of the top predicted miR-293 targets (Figures 1D–1F and S1D–S1F; Table S1). Likewise, convincing miR-92 target derepression was not observed until exceeding 1.3 × 104 added miR-92 MREs (Figures 1G–1I and S1G–S1I).
Comparison of mCherry and eYFP fluorescence with the corresponding transcript copy numbers, as measured by qRT-PCR (qPCR), revealed that fluorescence and mRNA abundance were highly correlated, although the relationship was not one-to-one (Figure 1J). Because protein fluorescence intensity is an indirect readout that is not directly relevant to the competition that occurs on the level of mRNA and miRNA, we transformed the fluorescence values measured by flow cytometry in Figure 1B to transcript copies per cell by employing the standard curves of Figure 1J (Figures 1K and S1J). Strikingly, the DRT observed for miR-293 and miR-92 reporters (0.9 × 104 and 1.3 × 104 sites per cell, respectively; Figures 1K and 1L) resembled those observed by RNA-seq for endogenous targets, thereby validating the reporter output (after transforming fluorescence to transcript copy number) for endogenous target derepression.
We next calculated the number of MREs that must be added per cell to observe half-maximal derepression (termed half-maximal inhibitory concentration, or IC50) of the different reporter constructs (Figures 1K and 1M). The number of miRNA molecules per ESC is reported to be 5.7 × 104 for miR-294, 2.6 × 103 for miR-293, 1.7 × 103 for miR-92, and 1.8 × 103 for miR-16 (Bosson et al., 2014), which was consistent with the relative levels of these miRNAs in our ESCs, as determined by small-RNA-seq (Figure S1L; Table S2). Thus, as observed for miR-122 in hepatocytes (Denzler et al., 2014), the IC50 values exceeded the number of miRNA molecules per ESC. In such a regime, the IC50 provides an empirical measure of the effective endogenous target-site abundance, as half-maximal derepression should be achieved when the competing sites reach an effective concentration matching that of the endogenous sites (Denzler et al., 2014).
In hepatocytes, the miR-122 IC50 (4.5 × 105 sites per cell) happens to correspond to the sum of all 3′ UTR 6-, 7-, and 8-nt sites of the transcriptome, leading to the idea that this sum, defined as the TAapp, can provide an estimate of the effective target-site abundance for other miRNAs (Denzler et al., 2014). To test this idea, we examined the correspondence between the newly determined IC50 values and the TAapp values for the ESC transcriptome. When comparing RNA-seq data with absolute copy numbers of mCherry, eYFP, and three differently expressed genes, a linear association was observed (Figure S1K), which provided a standard curve to transform RNA-seq data to absolute mRNA copies per cell, enabling TAapp values for eight active ESC miRNAs to be determined (Figure 1N). For all four miRNAs with IC50 values, the TAapp approached the IC50, ranging from ∼2-fold above the IC50 (miR-16, -92, and -294), to 1.5-fold below the IC50 (miR-293). Because TA values estimated from cross-linking (Bosson et al., 2014) strongly correlated with TAapp values (Figures S1M and S1N), but were ∼7-fold lower, the cross-linking immunoprecipitation (CLIP)-estimated TA values were not more informative for the purposes of estimating the effective target-site abundance. We conclude that summing of 3′ UTR 6-, 7-, and 8-nt sites in the transcriptome provides a reasonable approximation of effective abundance of endogenous target sites.
The DRTs ranged between 12% (miR-92) and 30% (miR-293) of TAapp. Importantly, no endogenous transcript contributed such a large percentage to transcriptome TAapp of the ESC miRNAs examined. The largest contributor was ribosomal protein S15A (Rps15a) mRNA, which contributed 6.5% of the TAapp for both miR-294 and miR-16 (Figure 1O). Thus in ESCs, as in hepatocytes (Denzler et al., 2014), ceRNA-regulated gene expression through upregulation or downregulation of a single transcript is unlikely. Similar results have been reported in HEK293 cells (Yuan et al., 2015).
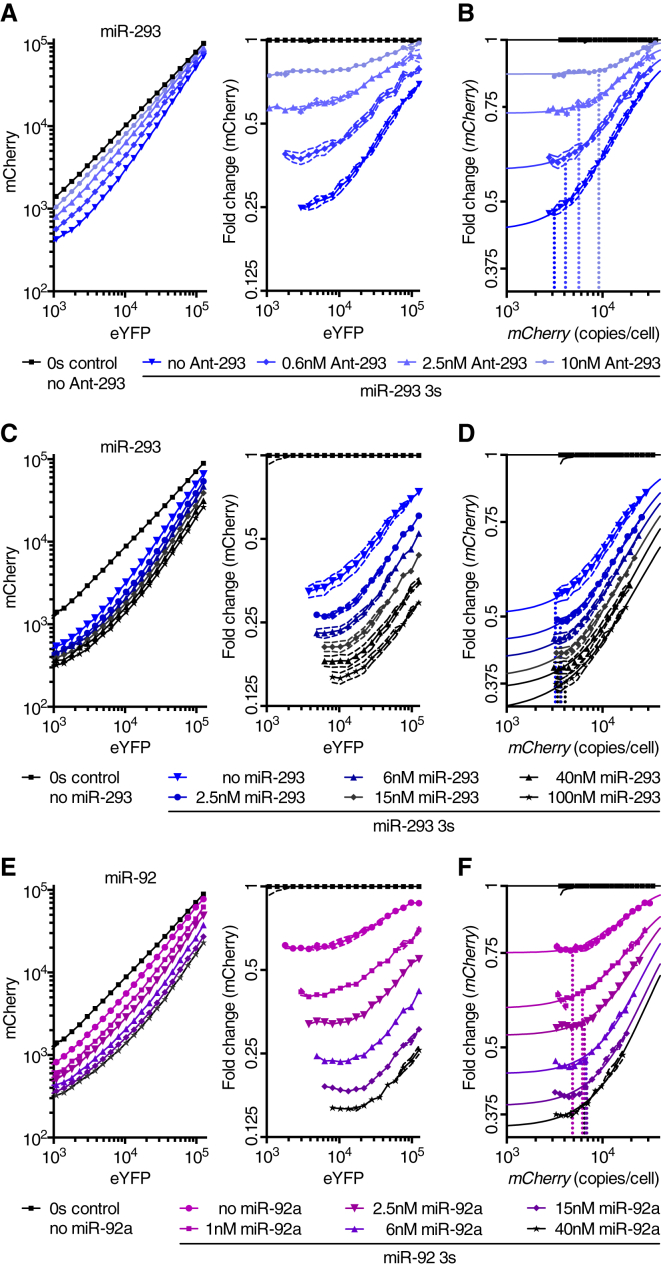
Derepression Threshold Values Are Insensitive to Changes in miRNA Activity
A key difference between the mixed-affinity and the hierarchical affinity models is the impact that miRNA levels have on the threshold required to detect derepression of target genes (Bosson et al., 2014, Denzler et al., 2014). To investigate this issue, we examined the influence that reduced miRNA activity has on the DRT in the single-cell assay. ESCs were transfected with either 0s or 3s miR-293 reporters, in addition to different concentrations of Antagomir-293 (Ant-293). Reduction of mCherry repression correlated with increasing Ant-293 concentrations, confirming that miR-293 activity was reduced in Antagomir-treated ESCs (Figure 2A). As observed for miR-122 in hepatocytes (Denzler et al., 2014), the DRTs and IC50 values did not decrease as miR-293 activity was reduced in ESCs (Figures 2B and S2A–S2C).
Figure 2.
Derepression Threshold Values Are Insensitive to Changes in miRNA Activity
(A–F) ESCs co-transfected with a 3s reporter for miR-293 (A–D), miR-92 (E and F), or respective 0s reporter control, and different concentrations of Ant-293 (n = 3) (A and B), miR-293 (n = 6) (C and D), or miR-92 (n = 6) (E and F).
(A, C, and E) Mean mCherry fluorescence (left), and mCherry fluorescence normalized to the 0s control (right) across 20 bins of eYFP.
(B, D, and F) Protein fluorescence values shown in (A), (C), and (E) were transformed to RNA copies per cell using the equations shown in Figure 1J. Vertical, dotted lines denote the DRT.
Data represent mean ± SEM for all panels.
We next increased miRNA activity and examined the effect on the DRT. ESCs were transfected with the dual-fluorescent reporter and different concentrations of miRNA duplex. When quantified with respect to eYFP fluorescence or eYFP mRNA copies, we detected an increase in the DRT as more miRNA was transfected (Figures 2C, 2E, S2D, S2E, S2G, and S2H). However, eYFP, unlike mCherry, is not a good measure for MRE induction as it is not repressed by the miRNA and hence does not inform how many MREs are actually expressed in a cell. Indeed, when quantified with respect to mCherry transcript abundance, the DRT of competing transcripts remained constant as more miRNA was transfected (Figures 2D, 2F, S2F, and S2I). These results monitoring DRTs after decreasing or increasing miRNA activities supported the mixed-affinity model, in which less abundant miRNAs should be no more susceptible to ceRNA effects than are more abundant miRNAs (Denzler et al., 2014).
Extensively Paired Sites Are More Potent Than 8-nt Sites and Trigger miRNA Decay
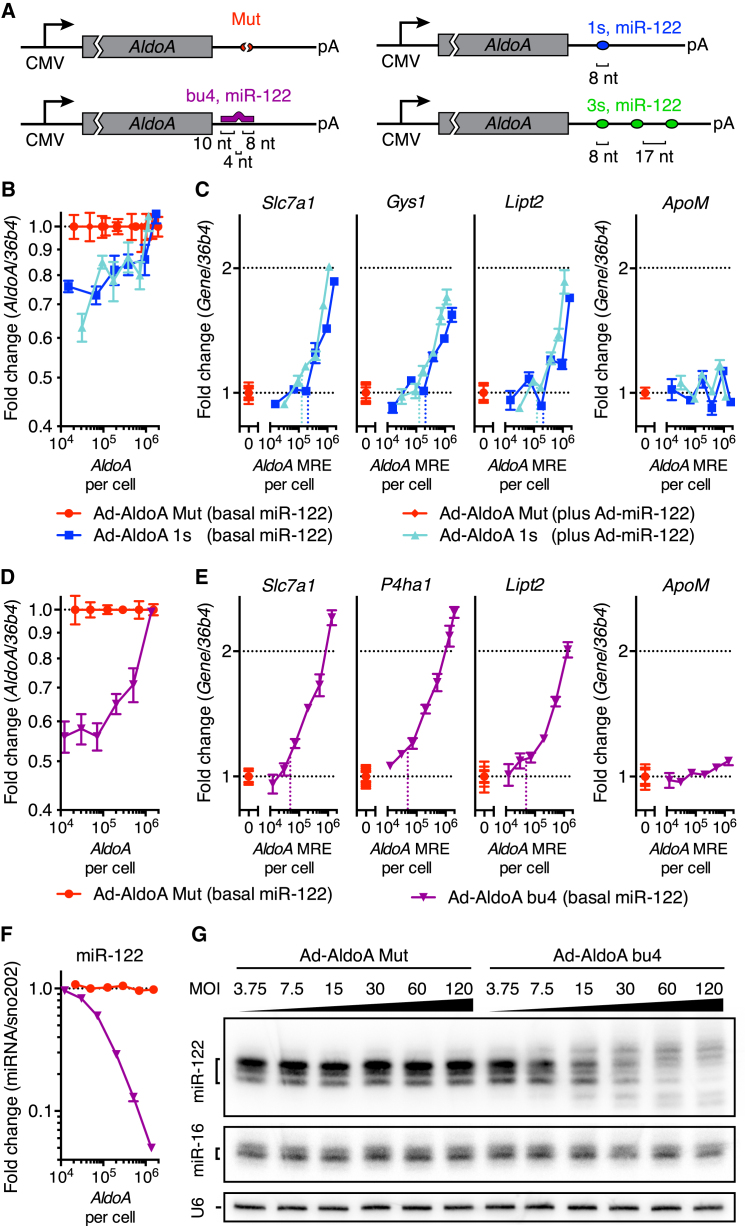
We investigated whether the DRT was also insensitive to increased miRNA levels in primary hepatocytes. A 4-fold increase in miR-122, attained by infecting hepatocytes with a recombinant adenovirus expressing the miR-122 precursor (Ad-miR-122), resulted in decreased levels of endogenous miR-122 target mRNAs (Figures S3A–S3C). To manipulate miR-122 MREs and measure the subsequent effects on miR-122 target genes, we increased the levels of the miR-122 target AldolaseA (AldoA) mRNA using an adenovirus (Ad-AldoA) that carried either a mutated site (Mut), one (1s), or three sites (3s) to miR-122 (Figure 3A). Hepatocytes were infected at different multiplicities of infection (MOIs), at either basal or elevated miR-122 levels (Figures 3B and S3D–S3G). At endogenous miR-122 levels, we began to observe miR-122 target derepression when more than 2.1 × 105 miR-122 MREs were introduced (Figure 3C). The DRT did not increase when endogenous miR-122 levels were raised 4-fold (Figure 3C), which is in agreement with our observations in ESCs. Of note, the higher DRT observed in hepatocytes compared to ESCs is expected based on the larger cytoplasm and number of mRNAs per cell in hepatocytes.
Figure 3.
Extensively Paired Sites Are More Potent Than 8-nt Sites and Trigger miRNA Decay
(A) Schematic overview of the different AldoA-expressing adenovirus constructs.
(B–G) Primary hepatocytes (n = 4) infected with different MOIs of Ad-AldoA 1s (B and C), bu4 (D–G), or respective Ad-AldoA Mut controls at either basal miR-122 levels (B–G) or with co-infected Ad-miR-122 (B and C). Relative levels of AldoA (B and D), miR-122 target genes and control non-target gene (ApoM) (C and E), or miR-122 (F). Vertical, dotted lines denote the DRT. miRNA levels are relative to the lowest MOI of Ad-AldoA Mut at basal miR-122 levels. (G) Northern blot analysis of miR-122, miR-16, and U6 at basal miR-122 levels.
Data represent mean ± SEM for all panels. Also see Figure S7.
Our finding that derepression occurred at only high thresholds of added target sites seemed to disagree with a study in HeLa cells that used “bulged” binding sites with near-perfect complementarity (Mukherji et al., 2011). We sought to test the possibility that sites with perfect complementarity to the miRNA 3′ region might yield different results because they mediate miRNA degradation. Hepatocytes were infected with Ad-AldoA containing either a mutated or a bulged (bu4) binding site (Figure 3A). Interestingly, derepression was already observed when exceeding only 5 × 104 bulged miR-122 MREs per cell (Figures 3D, 3E, S3H, and S3I), confirming that bulged sites are more efficient than 8-nt sites in influencing miRNA activity. The efficiency of target-mRNA derepression mediated by bulged sites correlated well with a decrease of miR-122, but not miR-16, levels (Figures 3F and S3J), suggesting that derepression was induced by enhanced miRNA degradation rather than direct competition between miRNA-binding sites.
Target-mediated miRNA decay is associated with tailing and trimming of the miRNA (Ameres et al., 2010). Indeed, we observed reduced miR-122 signal with evidence of tailing and trimming when bulged, but not 8-nt, seed matches caused target-gene derepression (Figures 3G and S3K). These results confirmed that bulged sites with perfect complementarity to the miR-122 3′ region reduce miRNA activity primarily through miRNA degradation rather than competition with other binding sites. Therefore, to be effective, these bulged sites need not approach the effective abundance of the miRNA target sites, but need only to be sufficiently abundant that the amount of target-mediated RNA decay substantially decreases the miRNA abundance.
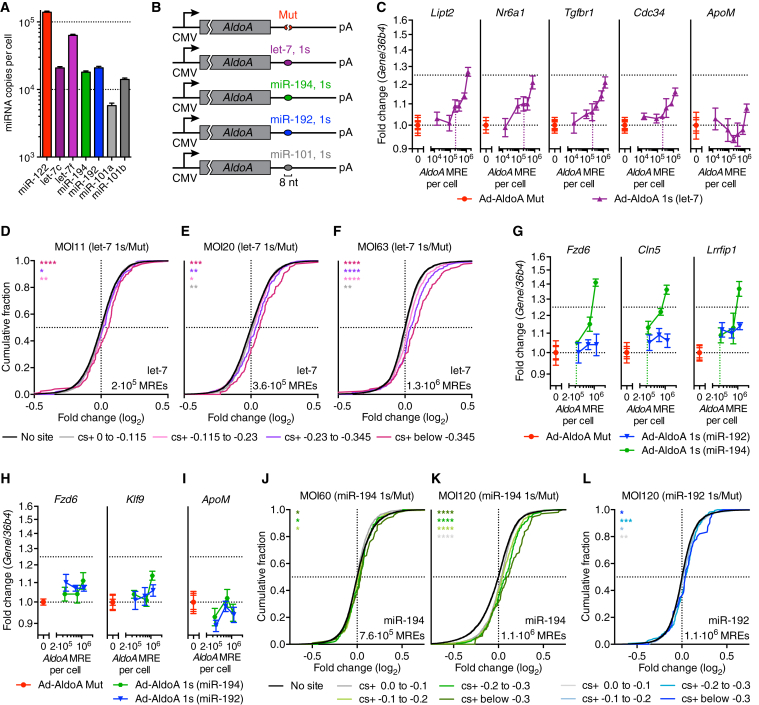
miRNA Target Derepression for let-7, miR-194, and miR-192 Also Occurs at a High Threshold of Added MREs
To consider the susceptibility of other hepatocyte miRNAs to ceRNA-mediated gene regulation, we first measured absolute levels of miR-122 and six other miRNA seed families highly expressed in liver (Denzler et al., 2014). These levels ranged from 3.8 × 103 to 1.4 × 105 copies per cell (Figures 4A and S4A) and correlated well with small-RNA-seq data (Figure S4B; Table S2). We selected four families (let-7, miR-194, -192, and -101) that were not influenced by control virus expression (Figure S4C) and were expressed above 1.8 × 104 copies per cell. To study the sensitivity of these four miRNA families to competing RNA perturbations, Ad-AldoA constructs were generated in which the miR-122 site was replaced with a single 8-nt site (1s) for the respective miRNA (Figure 4B). We first infected hepatocytes with different MOIs of Ad-AldoA Mut or 1 s (let-7). Derepression of let-7 targets, which were validated by transfection of let-7f mimics (Figures S4D–S4G), was observed when >2.1 × 105 let-7 MREs were expressed per cell (Figures 4C, S4H, and S4I). This DRT was consistent with RNA-seq results (Figures 4D–4F and S4J–S4L; Table S3). In contrast, addition of up to 106 MREs of either miR-192, miR-194, or miR-101 through respective Ad-AldoA infections did not result in detectable derepression of validated targets (Figures S4M–S4P; data not shown), suggesting that the endogenous level of 1.8 × 105 miRNA molecules per cell did not impart sufficient repression upon which derepression could act. We therefore performed the analogous experiment under conditions of elevated miR-192 and miR-194 levels using recombinant adenovirus expression (Ad-miR-192/194) (Figure S4M). Increasing miR-194 by 4.5-fold increased repression to a level at which target derepression could be observed, with a DRT of >3.2 × 105 added miR-194 MREs per cell (Figures 4G, 4I, S4Q, and S4R). RNA-seq analysis confirmed a similar DRT for predicted miR-194 targets (Figures 4J, 4K, S4S, and S4T). Because repression of predicted targets was not readily observed when increasing miR-192 levels by 3.7-fold (Figure S4O), derepression was also difficult to measure (Figures 4H, 4I, S4Q, and S4R), although some signal for derepression was detected when 1.1 × 106 miR-192 MREs were added (Figures 4L and S4U). Together, these results indicated that derepression of let-7, miR-192, and miR-194 targets in hepatocytes occurred at similar or higher DRTs than previously observed for miR-122 targets.
Figure 4.
miRNA Target Derepression for let-7, miR-194, and miR-192 Also Occurs at a High Threshold of Added MREs
(A) Absolute copies per cell of hepatocyte miRNAs.
(B) Schematic overview of Ad-AldoA constructs harboring a mutated site (Mut), or one (1s) 8-nt binding site for let-7, miR-194, -192, or -101.
(C–F) Primary hepatocytes infected with different MOIs of Ad-AldoA Mut or 1s (let-7).
(C) Relative expression of let-7 target genes and control non-target gene (ApoM).
(D–F) CDF of RNA-seq data (n = 2) showing mRNA changes for predicted target genes of let-7 with the indicated cs+ bins (color) or for transcripts with no miRNA site (black).
(G–L) Hepatocytes infected with different MOIs of Ad-AldoA Mut, 1s (miR-192), or 1s (miR-194), in addition to MOI 15 Ad-miR-192/194. Relative levels of miR-194 (G) or miR-192 (H) target genes, and control non-target gene (ApoM) (I). CDF of RNA-seq data (n = 2) showing mRNA changes for predicted target genes of miR-194 (J and K) or miR-192 (L) with the indicated cs+ bins (color) or for genes with no miRNA site (black).
AldoA MREs per cell evaluated by qPCR are shown on each graph. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, one-sided K-S test. Vertical, dotted lines denote the DRT.
Data represent mean ± SEM (n = 4) for all panels.
The 6-, 7-, and 8-nt Sites Contribute Comparably to Target Abundance
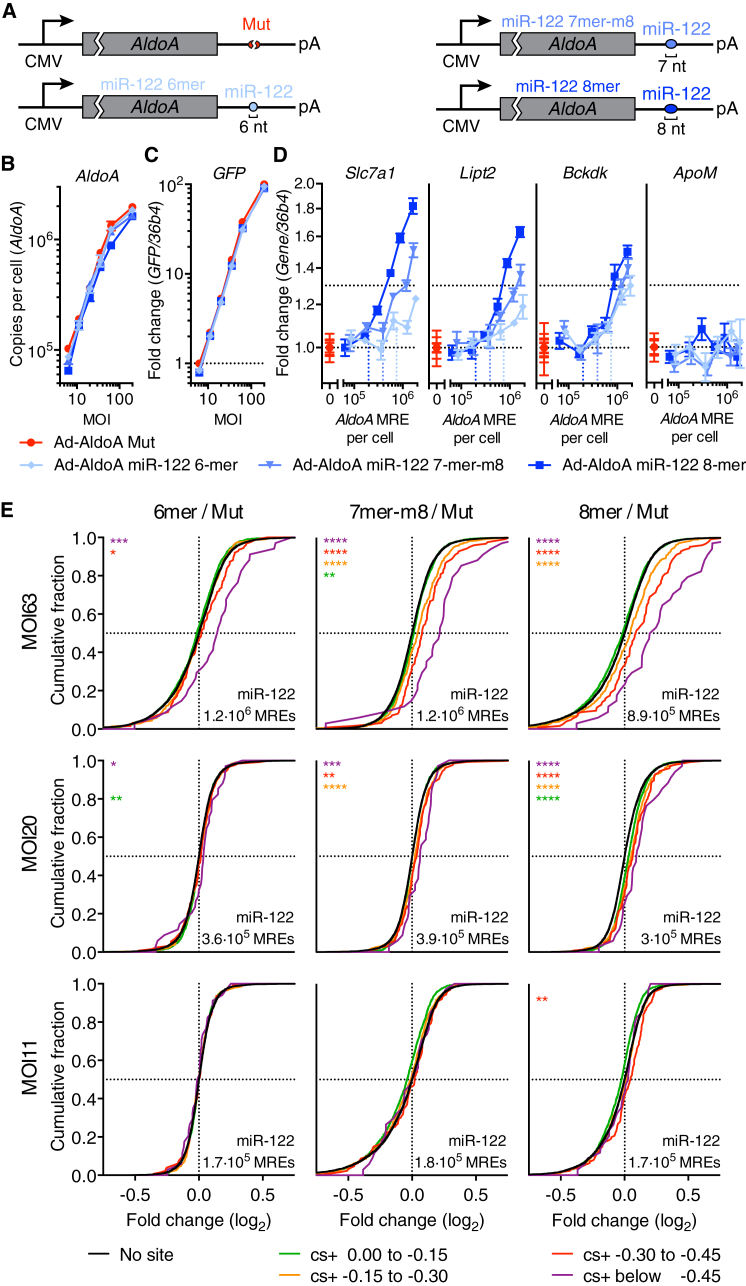
The different levels of repression efficacy and preferential conservation observed for 6-, 7-, and 8-nt site types (Bartel, 2009) raised the question as to the extent to which these site types differ in their efficacy as competitors. Accordingly, we infected hepatocytes with different MOIs of Ad-AldoA constructs harboring either a mutated or one 6-, 7-, or 8-nt site to miR-122 (Figure 5A). Derepression of miR-122 targets was observed when adding each of the three site types, with a clear relationship between competitor site type and DRT (Figures 5B–5D). This relationship, in which DRT increased as site size decreased, was also observed when extending our analysis to the transcriptome (Figures 5E and S5A). For example, the derepression observed for the 6-nt site at MOI 63 was between that observed for the 8-nt site at MOI 11 and 20, suggesting that as a competitor it was about 20% as effective as the 8-nt site. With respect to the 7-nt site, derepression at MOI 63 exceeded that observed for the 8-nt site at MOI 20, and derepression at MOI 20 surpassed that measured for the 8-nt site at MOI 11, suggesting that as a competitor the 7-nt site was about 50% as effective as the 8-nt site. Employing these factors to calculate a weighted TAapp only decreased the TAapp, without affecting the relative ranking of the respective miRNA TAapp (Figure S5B). These results indicate that, in aggregate, 7-nt sites, which are 3- to 8-fold more abundant than 8-nt sites, contribute more to effective target-site abundance than do 8-nt sites, and that 6-nt sites contribute more to effective target-site abundance than might have been expected from their marginal efficacy in target repression.
Figure 5.
The 6-, 7-, and 8-nt Sites Contribute Comparably to Target Abundance of miR-122
(A) Schematic overview of Ad-AldoA constructs used in this figure.
(B–E) Primary hepatocytes infected with different MOIs of Ad-AldoA miR-122 8-mer, miR-122 7-mer-m8, miR-122 6-mer, or Mut. Absolute copy numbers per cell of AldoA (B), relative gene expression of GFP (C), and of miR-122 target genes or control non-target gene (ApoM) (D). (E) CDF of RNA-seq data (n = 2) showing mRNA changes for predicted target genes of miR-122 with the indicated cs+ bins (color) or for genes with no respective miRNA site (black). AldoA MREs per cell evaluated by qPCR are shown on each graph. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, one-sided K-S test. See also Figures S5C and S5D.
Vertical, dotted lines denote the DRT. Data represent mean ± SEM (n = 4) for all panels.
Derepression Is Enhanced When Mediated by Closely Spaced MREs
Although the cooperative effect of closely spaced miRNA-binding sites has been studied in the context of mRNA repression (Doench et al., 2003, Grimson et al., 2007, Saetrom et al., 2007, Broderick et al., 2011), the role of cooperatively spaced miRNA-binding sites has not been investigated in the setting of site competition. We therefore analyzed whether closely spaced miRNA-binding sites can cooperatively sequester miRNA molecules and hence reduce the number of sites required for derepression.
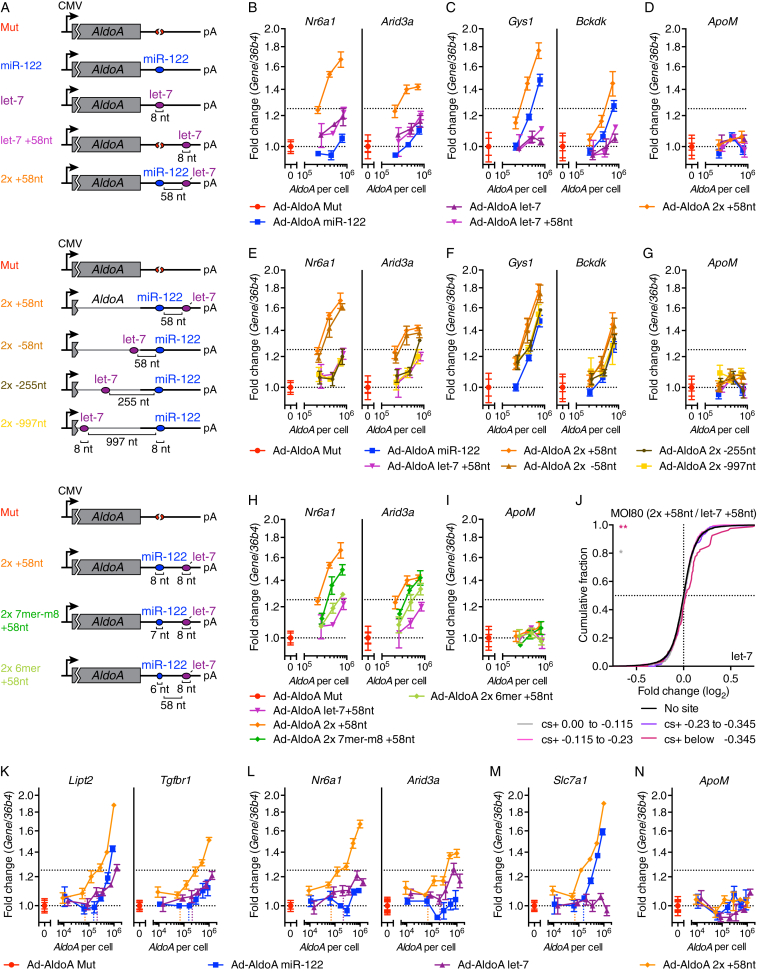
Cooperatively acting MREs within endogenous 3′ UTRs tend to be between 8 and ∼60 nt apart (Grimson et al., 2007, Saetrom et al., 2007). We thus generated Ad-AldoA constructs harboring one 8-nt site for miR-122 and one for let-7, separated by 58 nt (Ad-AldoA 2x +58nt), or respective single-site controls (Figure 6A), and infected hepatocytes at different MOIs. Interestingly, predicted let-7 targets that lacked miR-122 sites showed stronger derepression when let-7 MREs were added through constructs harboring a nearby miR-122 site (Figures 6B–6D, S6A, and S6B). Analogous results were obtained for the derepression of miR-122 targets by miR-122 MREs that had an adjacent let-7 site, showing that competition for binding to one miRNA family can be influenced by a nearby site of a different family. To achieve the same level of derepression conferred by isolated sites, the sites with nearby cooperative sites required only 20%–50% as many molecules per cell (Figure 6B).
Figure 6.
Derepression Is Enhanced When Mediated by Closely Spaced MREs
(A) Schematic overview of Ad-AldoA constructs used in this figure.
(B–J) Primary hepatocytes infected with different MOIs of Ad-AldoA constructs shown in (A). Relative gene expression of let-7 target genes (B, E, and H), miR-122 target genes (C and F), or control non-target gene (ApoM) (D, G, and I). (J) CDF of RNA-seq results (n = 2) showing mRNA changes from hepatocytes infected with MOI 80 of Ad-AldoA let-7 +58nt or 2x +58nt for predicted target genes of let-7 (with no predicted target sites for miR-122) with the indicated cs+ bins (color) or for genes with no let-7 or miR-122 miRNA sites (black). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, one-sided K-S test.
(K–N) Hepatocytes infected with different MOIs of Ad-AldoA Mut, miR-122, let-7, or 2x +58nt. Relative gene expression of predicted target genes for both let-7 and miR-122 (K), let-7 target genes (L), a miR-122 target gene (M), or a control non-target gene (ApoM) (N). Vertical, dotted lines denote the DRT.
Data represent mean ± SEM (n = 4) for all panels.
To study the influence that the spacing of the miR-122 and let-7 sites has on the ability to cause cooperative competition, we infected hepatocytes with various MOIs of differently spaced Ad-AldoA 2x constructs (Figure 6A). Although the cooperative effect of Ad-AldoA-2x-mediated gene regulation persisted independently of whether the let-7–binding site was 58 nt upstream or downstream of the miR-122 site—indicating that a specific intervening sequence or structure was not required—no cooperative effect was observed when the two sites were 255 or 997 nt apart (Figures 6E–6G, S6C, and S6D). When changing the 8-nt miR-122 site on Ad-AldoA 2x +58nt to a 7- or 6-nt site (Figure 6A), a strong relationship was observed between site type and the magnitude of the cooperative effect (Figures 6H, 6I, and S6E–S6G). Moreover, at the transcriptome level, predicted let-7 targets that lacked predicted miR-122 sites were significantly more derepressed if the competing let-7 site had an adjacent miR-122 site (Figures 6J and S6H), thereby confirming that closely spaced binding sites of co-expressed miRNAs can boost the efficacy of competing sites.
We then investigated whether the DRT was lower in conditions in which cooperativity was present by infecting hepatocytes with different MOIs using Ad-AldoA miR-122, let-7, and 2x +58nt. Derepression of miR-122 and let-7 target genes was detected when 7 × 104 AldoA copies of the 2x +58nt construct were exceeded (Figures 6K–6N, S6I, and S6J). Depending on whether binding sites of either miR-122 or let-7 alone, or both together are included in the cooperative DRT, the 7 × 104 AldoA copies would correspond to a DRT of either 7 or 14 × 104 MREs, which is either 3.1- or 1.5-fold lower (let-7), or 2.2- or 1.1-fold lower (miR-122), respectively, than the previously determined DRTs. Regardless of the DRT interpretation, these results indicate that the cooperative action of sites can detectibly boost the prospects of ceRNA-mediated gene derepression.
Mathematical Framework for the Mixed-Affinity Model
Mathematical simulations of miRNA-target interactions have been used to evaluate the potential effects of competing MREs (Mukherji et al., 2011, Ala et al., 2013, Bosson et al., 2014, Jens and Rajewsky, 2015, Schmiedel et al., 2015). However, these simulations either model only the extent to which different site types are occupied by a miRNA without modeling repressive effects that are needed for comparison to experimental results (Bosson et al., 2014, Jens and Rajewsky, 2015), or they model the repression of mRNA from one or two genes without modeling competition of sites from other expressed transcripts (Mukherji et al., 2011, Ala et al., 2013, Schmiedel et al., 2015). These latter simulations also omit the bound form of the mRNA from its simulated abundance. Most importantly, previous simulations also ignore the influence of the large number of low-affinity, non-canonical/background sites.
We therefore built a mathematical framework that incorporates site-type occupancy, mRNA destabilization, and the range of binding-site affinities intrinsic to the mixed-affinity model. This framework was used to predict the influence of both target-site abundance and miRNA level on target derepression. As with previous simulations (Mukherji et al., 2011, Bosson et al., 2014, Jens and Rajewsky, 2015), we assumed that (1) molecular species are well mixed within the cytosol and their concentrations are not influenced by cell growth and division; (2) each mRNA and miRNA is produced at a constant rate, and unbound mRNAs and miRNAs undergo constant first-order decay; (3) upon association with a miRNA, the mRNA degradation rate increases, regardless of the site type; and (4) miRNA binding is reversible, and upon miRNA dissociation the mRNA degradation rate reverts to its original value. We also assumed that the Michaelis constant (KM) describing mRNA degradation with respect to the miRNA-mRNA complex is well approximated by the complex dissociation constant (KD), and that both bound and unbound mRNA are translated.
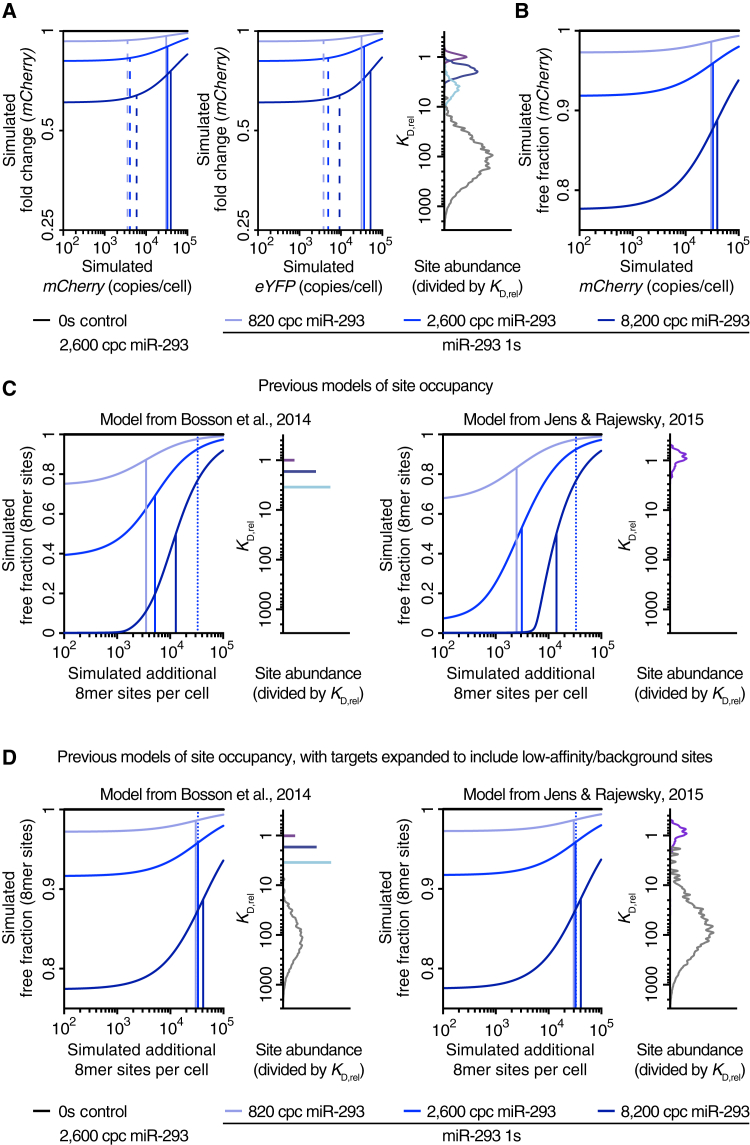
We first simulated the results of adding the 1s reporter for miR-293 to ESCs, as done in Figure 1C, setting levels of miR-293 and its canonical 3′ UTR sites to those measured by sequencing. Binding affinities of 6-, 7-, and 8-nt sites were modeled with distributions centering on their measured affinities, and a distribution of low-affinity sites was added such that the simulated IC50 reflected the experimentally determined value of ∼3 × 104 copies per cell (Figure 7A). With this target-site distribution (Figure 7A, right), the simulation recapitulated other features of our results. For example, DRT values were only marginally sensitive to 10-fold changes in miRNA (Figure 7A, left), and this sensitivity seemed greater when plotted as a function of eYFP, the co-expressed mRNA lacking a miR-293 site (Figure 7A, middle). Moreover, the mCherry IC50 values were even less sensitive to miRNA changes (Figure 7A, left) and corresponded to the half-maximal occupancy values (Figure 7B).
Figure 7.
Mathematical Simulation of the Mixed-Affinity Model
(A) Simulated effects of changing miR-293 concentrations in ESCs on 8-nt target site repression (as performed in Figure 2), using the mathematical framework of the mixed-affinity model. Simulated mCherry fold-changes of the 1s reporter normalized to the 0s control are either plotted against mCherry (left) or eYFP (middle), indicating the IC50 (solid lines) and DRT (dashed lines), for each of the three simulated miR-293 levels (in copies per cell [cpc]). Also plotted is the binding affinity distribution of all simulated target sites (right), with the KD of each site normalized to that of an 8-nt site and the abundance of each site scaled by its normalized KD. Abundance of 8-, 7-, and 6-nt, and low-affinity sites for miR-293 are plotted separately (purple, blue, cyan, and gray, respectively). The abundance of the canonical sites was determined by sequencing and that of the low-affinity sites was set such that the IC50 matched that observed in Figure 1C.
(B) Site occupancy for the simulations in (A). Plotted is the simulated free fraction of mCherry 1s reporter as a function of its expression, otherwise as in (A).
(C) Simulated effects of changing miR-293 concentrations in ESCs on 8-nt target site occupancy using the mathematical models of site competition from Bosson et al. (2014) (left) or Jens and Rajewsky (2015) (right). Simulated free fraction of an added 8-nt target site is plotted as a function of its expression as in (B), using the binding-affinity distributions of the simulated target sites of the original studies, plotted as in (A). The IC50 inferred from Figure 1C is indicated (dotted lines).
(D) Simulations using the models of (C) but adding low-affinity sites to alleviate the discrepancy between the simulated and experimental results, otherwise as in (C).
Plotting the competition in terms of site occupancy (Figure 7B) allowed comparison to previous simulations that do not consider mRNA repression. Reconstructing the simulation of miR-293 binding in ESCs from Bosson et al. (2014), using their values for site affinity and abundance, showed that sensitivity to additional 8-nt sites was much greater than that observed in our experiments, as was the influence of miRNA levels on half-occupancy values (Figure 7C, left). Similar results were observed when applying the model of Jens and Rajewsky (2015), which uses the same mathematical framework as Bosson et al. (2014) but a continuous distribution of canonical site affinities (Figure 7C, right). Remarkably, after adding low-affinity sites such that the half-maximal occupancy value matched that inferred from our experimental results, both of the previous frameworks behaved indistinguishably from ours (Figures 7B and 7D). Thus, the fundamental difference between the mixed-affinity model and the other models, which enables our simulation to better match the experimental results, is the greater effective target abundance that results from consideration of many low-affinity sites.
Discussion
Our results support the mixed-affinity model for miRNA site competition. In agreement with this model, we found that DRTs did not correlate with endogenous miRNA abundance and changed only modestly with experimental manipulations that increased or decreased miRNA levels. Because reducing miRNA levels does not substantially reduce the very high number of added MREs that are necessary to impart detectable derepression, changes in ceRNAs are not more likely to influence targets of miRNAs expressed at lower levels. Thus, the previous conclusion that a ceRNA effect on miR-122 targets in hepatocytes is unlikely to occur in normal physiological or disease conditions (Denzler et al., 2014) can now be more confidently extended to targets of other miRNAs in other cell types. Indeed, using two different cell types, testing several different miRNA families, and employing complementary single-transcript and high-throughput methods, we found that competing sites must approach ∼10%–40% of a miRNA’s TAapp in order to detectably influence miRNA activity. As nearly all transcripts each contribute <5% to TAapp, ceRNA-mediated gene regulation is very unlikely to occur under normal homeostatic conditions.
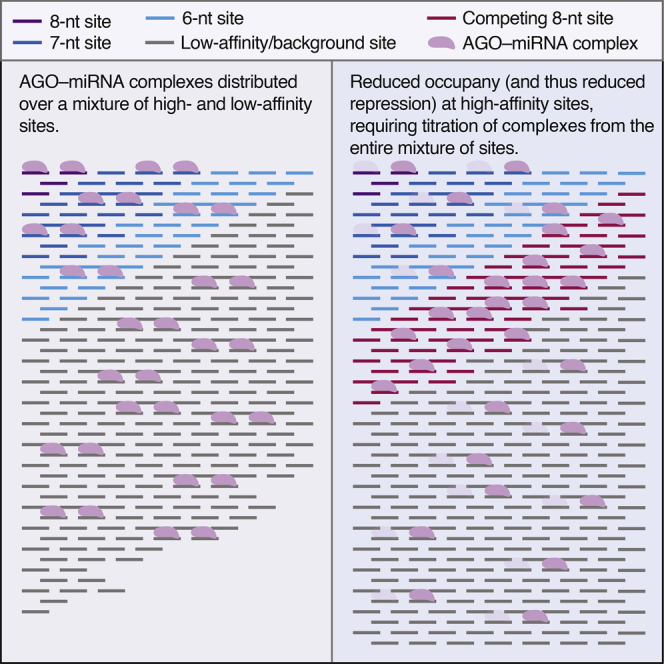
In disfavoring the hierarchical affinity model for site competition, we are not questioning the biochemical fact that some sites have more affinity than others, and thus low- and high-affinity sites exhibit differential occupancy. Indeed, although we disfavor the hierarchical affinity model with respect to site competition, it is nonetheless useful for explaining miRNA-mediated repression: when a miRNA is lowly expressed, only the highest-affinity sites are sufficiently occupied to mediate repression, but as miRNA expression increases, more and more intermediate- and low-affinity sites have occupancies sufficient to mediate repression. This model for repression is consistent with conclusions from cross-linking studies as well as those from mRNA-profiling studies showing a strong signal for derepression at 6-nt sites after loss of very highly expressed miRNAs (Giraldez et al., 2006, Bosson et al., 2014). The difference between modeling repression and modeling competition is that weak sites (including 6-nt, non-canonical, and background sites) all compete for binding even if they impart marginal or negligible repression and, importantly, this competition occurs regardless of the miRNA level. Although occupancy at any individual weak site is low, it cannot be discounted when modeling competition because weak, low-occupancy sites are in vast excess over high-affinity sites. The idea that these weak sites make a substantial contribution to effective target abundance is supported by our mathematical modeling showing that experimental results cannot be accurately simulated without considering the aggregate contribution of low-affinity sites. Also supporting this idea are single-molecule results showing that 6-nt sites and even some sites with only partial seed matches associate with the miRNA silencing complex at rates resembling those of the higher-affinity sites (Chandradoss et al., 2015, Salomon et al., 2015). Thus, even a miRNA expressed at a very low level, such as one molecule per cell, is expected to sample very many weak sites before (and after) occupying a high-affinity site.
Although miRNA levels do not affect the DRT, miRNA levels are important insomuch as they define the magnitude at which targets are initially repressed and hence the magnitude of effect that could theoretically be observed upon changes in ceRNA expression. Thus, ceRNA-regulated gene expression is expected to be more easily observed and more biologically relevant when miRNA levels are high.
When we conclude that miRNA levels do not substantially influence the DRT, we refer to the DRT as the number of sites that were measured in steady-state conditions in the presence of miRNA-mediated repression, such as those represented by the mCherry transcripts in the dual-fluorescence reporter system. In this system, it was important to account for the miRNA-mediated degradation of the competitor, as our conclusions would have differed if we determined the competitor concentration in the absence of miRNA repression, represented by the output of the co-transcribed eYFP reporter.
Bulged and fully complementary sites were the first site types to be investigated in the context of regulating miRNA activity through competition (Ebert et al., 2007, Franco-Zorrilla et al., 2007), and they have been widely used to inhibit or measure miRNA activity (Doench et al., 2003, Broderick et al., 2011, Mukherji et al., 2011, Mullokandov et al., 2012, Xie et al., 2012). However, these sites with extensive complementarity to the 3′ region of the miRNA can trigger degradation of the miRNA (Ameres et al., 2010). Indeed, we observed target-directed miRNA degradation in hepatocytes when adding bulged sites of miR-122. Hence, bulged sites can reduce miRNA activity predominantly through triggering miRNA degradation rather than by competing with other miRNA-binding sites. Likewise, endogenous transcripts with highly complementary binding sites might affect miRNA activity through degradation rather than competition, especially in situations of low or intermediate miRNA levels, with this degradation mechanism requiring much lower expression levels to be consequential. For example, potent target-directed degradation has been described in primary neurons (de la Mata et al., 2015), and a highly complementary binding site has been identified in the linc-MD1 long non-coding RNA and implicated in muscle differentiation through a ceRNA mechanism (Cesana et al., 2011). Whether this complementary site can induce miRNA degradation or whether other such sites exist remains to be shown.
We found that 7-nt sites were 50% as effective as 8-nt sites in contributing to target abundance, and 6-nt sites were 20% as effective. This 20% efficacy compared to 8-nt sites was much greater than might have been expected from the marginal repression typically imparted by 6-nt sites, again illustrating how competition efficacy imperfectly mirrors repression efficacy. Because miRNA association rates (kon values) of 6-, 7-, or 8-nt sites are similar (Chandradoss et al., 2015, Salomon et al., 2015), the difference between competition and repression presumably relates to the different dissociation rates (koff values) of these site types. Perhaps, before any repression can begin, some time is required to remodel the target transcript, assembling TNRC6 (trinucleotide repeat containing 6) and the deadenylation complexes, such that the dwell time of the miRNA on 6-nt sites only rarely exceeds this lag time. Similar models have been proposed to explain the poor repression efficacy of sites in the path of the ribosome (Grimson et al., 2007) and inefficacy of non-canonical sites in 3′ UTRs, despite the compelling CLIP evidence for binding to the ineffective sites (Agarwal et al., 2015). In this way, site types that are marginal or ineffective with respect to repression can nonetheless contribute meaningfully to effective target-site abundance. Indeed, our mathematical simulations illustrate that, in aggregate, low-affinity, non-canonical/background sites contribute more to the effective target-site abundance than do the canonical sites.
As the sum of 6-, 7-, and 8-nt sites in transcriptome 3′ UTRs, TAapp is a crude approximation of the effective target site abundance, in that it overcounts effects of 6- and 7-nt 3′ UTR sites and misses both the sites outside of 3′ UTRs and the weak but highly abundant non-canonical sites in 3′ UTRs. Nonetheless, summing up all 6-, 7-, or 8-nt 3′ UTR sites equally without weighting approximated IC50 within a few fold, presumably because overcounting the effects of some sites largely offset the failure to count other sites.
Our competition results provided mechanistic insight into the cooperative effect sometimes observed for adjacent sites. Whereas two distantly spaced 3′ UTR sites typically confer the repression expected from their independent action, two more closely spaced sites often confer more repression than expected from independent action (Grimson et al., 2007, Saetrom et al., 2007). Previous studies of this phenomenon using repression as the output do not distinguish between cooperative binding of the two sites or some other type of cooperative function in repression. Our use of competition as the output, with the observation that transcripts containing two miRNA-binding sites spaced 58 nt apart cooperatively sequester miRNAs corresponding to each site, uniquely shows that cooperative binding occurs.
Although the mechanism of this cooperative binding is unknown, an attractive hypothesis is that nearby Argonaute proteins might be tethered to each other through binding of the same TNRC6 molecule, also known as glycine-tryptophan protein of 182 kDa (GW182) in flies (Huntzinger and Izaurralde, 2011, Fabian and Sonenberg, 2012). TNRC6 contains multiple Argonaute-binding sites that might simultaneously interact with multiple miRNA-loaded Argonaute proteins (Schirle and MacRae, 2012, Pfaff et al., 2013), thereby enabling adjacent transcript-bound Argonaute proteins to prolong the dwell times of each other.
In the previous study of miR-122 site competition in hepatocytes, the three-site construct appears only 3-fold more effective than the one-site construct, as would be expected for non-cooperative, independent action of the three sites (Denzler et al., 2014). Suspecting that cooperativity was not observed in this context because the number of different MOIs examined was insufficient to detect subtle differences, we revisited potential cooperative binding of sites within the Ad-AldoA 3s construct at more MOIs. Derepression started to occur at 1.1 × 105 MREs (Figure S7), a DRT about 50% lower than that observed for the Ad-AldoA 1s construct. Thus, as expected, cooperativity can be observed for miRNAs of the same family as well as for miRNAs of different families.
Among the features that we analyzed, cooperative binding of miRNAs was the only one that increased the feasibility of regulation through changes in ceRNA levels, lowering the number of competing sites needed to detect derepression by ∼50%. However, this 50% difference does not seem large enough to substantially improve the prospects of observing a ceRNA effect in a physiological setting. Perhaps in unusual cases cooperativity provides more than a 50% difference, a good candidate for unusually strong cooperativity being the circular RNA CDR1as/ciRS-7 with >60 closely spaced sites to miR-7 (Hansen et al., 2013, Memczak et al., 2013). However, very few other circRNAs have more miRNA-binding sites than expected by chance (Guo et al., 2014, Rybak-Wolf et al., 2015), which brings the focus back to linear transcripts as a more abundant source of potential ceRNA candidates. For cooperativity to be a factor, such a transcript would need to be very highly expressed and have multiple sites that fall in a cooperative sequence context, and sites would need to correspond to miRNA families that are each expressed at levels sufficient to actively repress target genes. If or how frequently such conditions occur in vivo is currently unknown, but if such a candidate is found, recently developed gene-editing methods offer the opportunity to introduce precise mutations of the sites within their genomic context (without induced overexpression) and thereby provide the first convincing evidence of ceRNA regulation in vivo.
Experimental Procedures
See Supplemental Experimental Procedures for details.
Single-Cell Reporter Assay
The fluorescent reporter plasmids are based on the pTRE-Tight-BI (Clontech) system, in which a bidirectional Tet promoter expresses eYFP and mCherry (Mukherji et al., 2011). The 3′ UTR of mCherry contains either zero (0s), one (1s), or three consecutive (3s) 48-nt-long sequence stretches, which are comprised of one 8-mer MRE and ±20-bp flanking regions (Bosson et al., 2014). ESC line E14 was transfected with reporter and rtTA plasmids, induced with doxycycline 6 hr post-transfection, and harvested 18 hr later. Samples were analyzed using a FACSAria IIIu flow cytometer and eYFP and mCherry fluorescent values were corrected for autofluorescence as described in Bosson et al. (2014).
Hepatocyte Isolation and Viral Infections
Animal experiments were approved by the Kantonale Veterinäramt Zürich. Hepatocytes of 8- to 12-week-old male C57BL/6N mice (Janvier) were counted and plated at 300,000 cells per well in surface-treated six-well plates (Corning) in low-glucose media. Four to 6 hr after plating, cells were infected with adenovirus constructs in Hepatozyme media (Life Technologies) and harvested 24 hr post-infection.
Adenoviruses
Recombinant adenoviruses generated in this study are based on the AldoA constructs described in Denzler et al. (2014) and express GFP from an independent promoter. See Tables S5 and S6 for the nucleotide sequences of all Ad-AldoA constructs.
miRNA and Gene Expression Analysis
qPCRs were performed using TaqMan MicroRNA Assays (Life Technologies) for miRNA or gene-specific primer pairs (Table S4) for gene expression, respectively. Relative expression values were calculated using the ddCT method employing snoRNA202 for miRNA or mouse 36b4 (Rplp0) for gene expression normalization.
Author Contributions
R.D. designed and performed experiments, analyzed and interpreted data, and drafted the manuscript. S.E.M. developed the mathematical model and performed associated analyses. A.C.T. performed RNA blots and helped with experiments shown in Figure 1. V.A. processed RNA-seq raw data. M.S. and D.P.B. designed experiments, interpreted data, and revised the manuscript. R.D., S.E.M., A.C.T., V.A., D.P.B., and M.S. reviewed the results and contributed to writing the manuscript.
Acknowledgments
We would like to thank J. Zamudio, C. JnBaptise, and P. Sharp for technical advice and helpful discussions; B. Kleaveland for small-RNA-seq; and C. Ciaudo for providing ESCs. This study was supported in part by the National Science Foundation Graduate Research Fellowship (V.A.), an ERC grant “Metabolomirs” and NCCR “RNA and Biology” (M.S.), and NIH grant GM067031 (D.P.B.). D.P.B. is a Howard Hughes Medical Institute Investigator.
Published: October 27, 2016
Footnotes
Supplemental Information contains Supplemental Experimental Procedures, seven figures, and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2016.09.027.
Contributor Information
David P. Bartel, Email: dbartel@wi.mit.edu.
Markus Stoffel, Email: stoffel@biol.ethz.ch.
Accession Numbers
The accession number for the data reported in this paper is GEO: GSE76288.
Supplemental Information
Related to Figures 1 and S1. Table of gene expression levels, measured in fragments per kilobase per million (FPKM), log2 fold-changes, predicted target-site efficacies (context + scores), and site types of nine ESC samples sequenced, corresponding to all 9,547 detectably expressed (FPKM above 2.0) protein-coding genes annotated in Ensembl. More comprehensive gene expression data can be found as raw or processed data deposited at GEO (GEO: GSE76288).
Related to Figures S1 and S4. Table of miRNA expression levels, measured in counts and reads per million (RPM) of one ESC and one primary hepatocyte sample sequenced; data can be found as raw or processed data deposited at GEO (GEO: GSE76288).
Related to Figures 4, 5, 6, and S4–S7. Table of gene expression levels, measured in fragments per kilobase per million (FPKM), log2 fold changes, predicted target site efficacies (context + scores), and site types of all 23 primary hepatocyte samples sequenced, corresponding to all 9,205 detectably expressed (FPKM above 1.0) protein-coding genes annotated in Ensembl. More comprehensive gene expression data can be found as raw or processed data deposited at GEO (GEO: GSE76288).
References
- Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:4. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ala U., Karreth F.A., Bosia C., Pagnani A., Taulli R., Léopold V., Tay Y., Provero P., Zecchina R., Pandolfi P.P. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc. Natl. Acad. Sci. USA. 2013;110:7154–7159. doi: 10.1073/pnas.1222509110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres S.L., Horwich M.D., Hung J.H., Xu J., Ghildiyal M., Weng Z., Zamore P.D. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvey A., Larsson E., Sander C., Leslie C.S., Marks D.S. Target mRNA abundance dilutes microRNA and siRNA activity. Mol. Syst. Biol. 2010;6:363. doi: 10.1038/msb.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosson A.D., Zamudio J.R., Sharp P.A. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell. 2014;56:347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick J.A., Salomon W.E., Ryder S.P., Aronin N., Zamore P.D. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA. 2011;17:1858–1869. doi: 10.1261/rna.2778911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandradoss S.D., Schirle N.T., Szczepaniak M., MacRae I.J., Joo C. A dynamic search process underlies microRNA targeting. Cell. 2015;162:96–107. doi: 10.1016/j.cell.2015.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M., Gaidatzis D., Vitanescu M., Stadler M.B., Wentzel C., Scheiffele P., Filipowicz W., Großhans H. Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep. 2015;16:500–511. doi: 10.15252/embr.201540078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzler R., Agarwal V., Stefano J., Bartel D.P., Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Petersen C.P., Sharp P.A. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert M.S., Neilson J.R., Sharp P.A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian M.R., Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- Garcia D.M., Baek D., Shin C., Bell G.W., Grimson A., Bartel D.P. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Huntzinger E., Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Jens M., Rajewsky N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat. Rev. Genet. 2015;16:113–126. doi: 10.1038/nrg3853. [DOI] [PubMed] [Google Scholar]
- Karreth F.A., Reschke M., Ruocco A., Ng C., Chapuy B., Léopold V., Sjoberg M., Keane T.M., Verma A., Ala U. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161:319–332. doi: 10.1016/j.cell.2015.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Mukherji S., Ebert M.S., Zheng G.X., Tsang J.S., Sharp P.A., van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullokandov G., Baccarini A., Ruzo A., Jayaprakash A.D., Tung N., Israelow B., Evans M.J., Sachidanandam R., Brown B.D. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods. 2012;9:840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff J., Hennig J., Herzog F., Aebersold R., Sattler M., Niessing D., Meister G. Structural features of Argonaute-GW182 protein interactions. Proc. Natl. Acad. Sci. USA. 2013;110:E3770–E3779. doi: 10.1073/pnas.1308510110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J., Pandolfi P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A., Stottmeister C., Glažar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- Saetrom P., Heale B.S., Snøve O., Jr., Aagaard L., Alluin J., Rossi J.J. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007;35:2333–2342. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon W.E., Jolly S.M., Moore M.J., Zamore P.D., Serebrov V. Single-molecule imaging reveals that Argonaute reshapes the binding properties of its nucleic acid guides. Cell. 2015;162:84–95. doi: 10.1016/j.cell.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle N.T., MacRae I.J. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedel J.M., Klemm S.L., Zheng Y., Sahay A., Blüthgen N., Marks D.S., van Oudenaarden A. Gene expression. MicroRNA control of protein expression noise. Science. 2015;348:128–132. doi: 10.1126/science.aaa1738. [DOI] [PubMed] [Google Scholar]
- Xie J., Ameres S.L., Friedline R., Hung J.H., Zhang Y., Xie Q., Zhong L., Su Q., He R., Li M. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat. Methods. 2012;9:403–409. doi: 10.1038/nmeth.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Liu B., Xie P., Zhang M.Q., Li Y., Xie Z., Wang X. Model-guided quantitative analysis of microRNA-mediated regulation on competing endogenous RNAs using a synthetic gene circuit. Proc. Natl. Acad. Sci. USA. 2015;112:3158–3163. doi: 10.1073/pnas.1413896112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figures 1 and S1. Table of gene expression levels, measured in fragments per kilobase per million (FPKM), log2 fold-changes, predicted target-site efficacies (context + scores), and site types of nine ESC samples sequenced, corresponding to all 9,547 detectably expressed (FPKM above 2.0) protein-coding genes annotated in Ensembl. More comprehensive gene expression data can be found as raw or processed data deposited at GEO (GEO: GSE76288).
Related to Figures S1 and S4. Table of miRNA expression levels, measured in counts and reads per million (RPM) of one ESC and one primary hepatocyte sample sequenced; data can be found as raw or processed data deposited at GEO (GEO: GSE76288).
Related to Figures 4, 5, 6, and S4–S7. Table of gene expression levels, measured in fragments per kilobase per million (FPKM), log2 fold changes, predicted target site efficacies (context + scores), and site types of all 23 primary hepatocyte samples sequenced, corresponding to all 9,205 detectably expressed (FPKM above 1.0) protein-coding genes annotated in Ensembl. More comprehensive gene expression data can be found as raw or processed data deposited at GEO (GEO: GSE76288).