Abstract
Prognosis and survival for malignant melanoma is highly dependent on early diagnosis and treatment. While the American Joint Committee on Cancer (AJCC) criterion provides a means of staging melanomas and guiding treatment approaches, it is unable to identify the risk of disease progression of early stage tumors or provide reliable stratification for novel adjuvant therapies. The demand for credible prognostic/companion biomarkers able to identify high-risk melanoma subgroups as well as guide more effective personalized/precision-based therapy is therefore of paramount importance. Autophagy, the principle lysosomal-mediated process for the degradation/recycling of cellular debris, is a hot topic in cancer medicine, and observations of its deregulation in melanoma have brought its potential as a prognostic biomarker to the forefront of current research. Key regulatory proteins, including Atg8/microtubule-associated light chain 3 (LC3) and BECN1 (Beclin 1), have been proposed as potential prognostic biomarkers. However, given the dynamic nature of autophagy, their expression in vitro does not translate to their use as a prognostic biomarker for melanoma in vivo. We have recently identified the expression levels of Sequestosome1/SQSTM1 (p62) and activating molecule in Beclin 1-regulated autophagy protein 1 (AMBRA1) as novel independent prognostic biomarkers for early stage melanomas. While increasing followed by subsequent decreasing levels of p62 expression reflects the paradoxical role of autophagy in melanoma, expression levels additionally define a novel prognostic biomarker for AJCC stage II tumors. Conversely, loss of AMBRA1 in the epidermis overlying primary melanomas defines a novel prognostic biomarker for AJCC stage I tumors. Collectively, the definition of AMBRA1 and p62 as prognostic biomarkers for early stage melanomas provides novel and accurate means through which to identify tumors at risk of disease progression, facilitating earlier patient therapeutic intervention and stratification tools for novel personalized therapeutic approaches to improve clinical outcome.
Keywords: autophagy biomarkers, malignant melanoma, Ambra1, p62, prognostic biomarkers
Malignant melanoma, the most aggressive form of skin cancer arising from the malignant transformation of melanocytes, is an increasing public health concern worldwide with incidence rates doubling every 10–20 years (1), which now renders this malignancy accountable for 75% of all skin cancer deaths and the most common cause of cancer-related mortality in young individuals between 20 and 35 years of age (2).
As with many cancers, prognosis and survival for melanoma is highly dependent on early detection, diagnosis, and treatment. In line with this need and coupled with the emergence of novel targeted and immunotherapies, current interest is focused on the discovery of predictive and prognostic biomarkers. A biomarker refers to any measurable diagnostic indicator that is used to assess the risk or presence of disease (3). While predictive biomarkers are able to indicate which patient subgroups are likely to benefit from certain treatments (4), prognostic markers enable stratification of patients at initial diagnosis according to eventual outcome, which can be used clinically to guide patient management including the earlier initiation of adjuvant therapies in patients at high risk of disease progression, potentially preventing the development of untreatable metastatic disease (5).
Some of the best established prognostic biomarkers for melanoma are incorporated into the current American Joint Committee on Cancer (AJCC) 2009 staging criterion, the most comprehensive staging system for melanoma to date, which remains the international standard for disease staging and as a guide for treatment approaches. AJCC staging combines several prognostic factors for melanoma, including the depth of invasion (Breslow depth), rate of mitoses, presence of ulceration (loss of the epidermis overlying the tumor), evidence of metastatic spread, and changes in serum lactate dehydrogenase (LDH) to allow risk stratification of morbidity and mortality at the initial diagnosis (6). In general, this divides malignant melanoma into four stages: stages I–II comprising primary tumors of distinct thickness (defined as early stage disease), stage III where locoregional spread of disease (mainly to local lymph node basins) is present, and stage IV where there is presence of distant metastasis.
While early stage melanoma is largely curative by surgical excision, metastatic disease represents the cause of death from melanoma in the vast majority of cases due to a lack of consistently beneficial treatment regimens for late stage disease. Furthermore, despite its comprehensiveness, AJCC staging as a prognostic biomarker is limited by the inability of its criteria to accurately identify high-risk melanoma subgroups that will go on to progress; a particular problem in seemingly “low risk” AJCC stage I melanomas where up to 10% of tumors subsequently metastasize. This emphasizes the urgent need for novel credible biomarkers to identify high-risk tumors as well as the stratification of such patients for more efficacious and earlier therapeutic approaches (6).
Observations of deregulated autophagy in many cancers, including melanoma, have brought this key signaling mechanism to the forefront of much research (7, 8), including its potential capacity as a prognostic biomarker. Autophagy, the principle catabolic process for lysosomal-mediated degradation of intracellular components to sustain cellular energy and survival, is regulated by a complex signaling cascade involving ubiquitin-like conjugation systems, autophagy regulatory proteins [BECN1/Beclin 1 (Beclin-1), activating molecule in Beclin 1-regulated autophagy protein 1 (AMBRA1), Atg8/microtubule-associated light chain 3 LC3 (LC3), and Sequestosome1/SQSTM1 (p62)], and the inactivation of mammalian target of rapamycin (mTOR) to induce activation. Cellular debris within the cytoplasm is sequestered into double-membrane autophagosomes, which are then delivered to lysosomes for degradation and recycling (9). In cancer, however, autophagy plays a paradoxical role; on the one hand, preventing build-up of toxic cellular components that result in genomic stress and instability, thereby promoting tumorigenesis, while, on the other hand, promoting tumor survival of advanced stage solid tumors such as melanoma in a nutrient-deprived hypoxic environment (10). Consistent with the paradoxical role of autophagy and in contrast to observations in benign nevi, electron microscopy studies have shown an increased presence and vacuolization (suggesting degradation) of double-membraned autophagosomes in the cytoplasm of metastatic melanoma cells, thus supporting the notion of increased autophagic activity in advanced stage disease (11, 12).
BRAF is a member of the RAF group of serine/threonine protein kinases, and as such functions to regulate the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) cellular growth pathway (13). Activating mutations in BRAF, present and in up to 70% of all melanomas along with NRas mutations (present in approximately 15–20% of melanomas), result in constitutive activation of MAPK signaling, promoting growth, survival, and chemoresistance (14–17). Interestingly, BRAF mutational status in melanoma has also been shown to variably influence autophagy. Following treatment of BRAF wild-type melanoma cells with endoplasmic reticulum (ER) stress-inducing agents in vitro, autophagy is activated in line with its pro-survival role; however, when autophagy is inhibited exogenously, this leads to increased cell death (12, 18). Conversely, oncogenic BRAF induces a chronic ER stress status, resulting in enhanced basal autophagy [as evidenced by increased Atg8-PE/LC3-II (LC3-II) expression], resistance of melanoma cells to apoptosis, and insensitivity to further autophagy induction. This suggests that although melanomas rely on increased innate autophagic activity, BRAF-mutated tumors are resistant to further mTOR-dependant stimulation of autophagy and that while combined inhibition of autophagy with chemotherapy might be a viable therapeutic avenue for BRAF wild-type melanomas, targeted therapies that attenuate ER stress may prove a more effective treatment strategy for BRAF mutant melanomas (12, 18, 19).
To date, several markers of autophagy, including LC3 and Beclin 1, have been identified as potential prognostic biomarkers for melanoma. Under normal homeostatic conditions, exogenous LC3 is cytoplasmic but upon autophagy induction becomes conjugated to phosphatidylethanolamine (PE) to form the membrane bound form LC3-II, thus acting as a marker of autophagy induction (20). Increased immunohistochemical expression of LC3 has been shown in malignant melanomas compared to benign nevi (21) and is associated with the development of metastatic disease and poorer outcomes (22). In addition, studies of Beclin 1 expression suggest its’ downregulation parallels melanoma disease stage progression, further supporting a role for autophagy in tumor invasion and metastasis (23, 24). However, although the expression of LC3 provides an indication of autophagy status in melanoma, it is important to note that conversion of LC3-I to LC3-II is a dynamic process, thus limiting the capacity of endogenous LC3 expression as an accurate biomarker of autophagy status and importantly, the reflection of autophagic flux (15). Moreover, although reported to be downregulated in melanoma, there is also conflicting evidence of Beclin 1 overexpression in advanced melanoma, thus questioning its expression as a reliable prognostic biomarker (25).
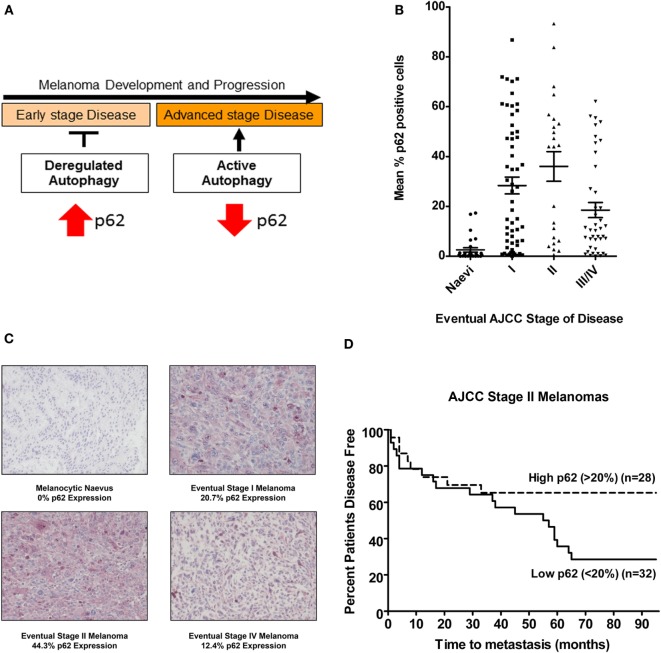
Sequestosome1/SQSTM1 is a scaffold protein that shuttles ubiquitinated proteins into the autophagosome, later degraded along with other autophagosomal contents upon fusion with a lysosome. Impairment of autophagy is therefore reflected by an associated accumulation of p62, a process reported to be a key to the onset of tumorigenesis (17). Conversely, decreased levels of p62 reflect active autophagy, as observed in advanced stage melanomas where autophagy is commonly reactivated to enhance tumor survival (Figure 1A). Data from our lab have further defined p62 expression as a prognostic biomarker for melanoma where a stepwise increase in expression is observed in early AJCC stage melanomas (increased above basal levels in benign nevi and reflecting deregulated autophagy) but which is subsequently decreased in advanced metastatic tumors, consistent with the reactivation of autophagy and its paradoxical role in cancer [Figures 1B,C; (26)]. Furthermore, univariate analysis showed a significantly increased risk of metastasis in AJCC stage II tumors with low p62 expression (<20% median p62 expression) compared to those with high expression (>20% p62 expression) [Figure 1D; (26)]. Moreover, since there was no association with Breslow depth or tumor ulceration, p62 expression defines an independent stratifying variable from AJCC staging prognosticators. Collectively, these data highlight p62 as a novel independent prognostic biomarker for AJCC stage II melanomas, providing a powerful tool for refining the risk of disease progression and enabling earlier patient therapeutic intervention. In addition, p62 expression may represent a companion biomarker of response to autophagy modulation in vivo, an important concept in view of emerging autophagy modulator therapies (27) and their potential to improve overall clinical outcome for patients with metastatic melanoma.
Figure 1.
p62 expression is a prognostic biomarker for AJCC stage II melanomas. (A) Schematic of the paradoxical role of autophagy in melanoma in the context of p62 expression; impairment of autophagy drives tumorigenesis of early stage melanomas reflected by p62 accumulation, whereas decreased levels of p62 seen in advanced disease reflect autophagy reactivation. (B) Mean % p62 expression in a cohort of eventual AJCC stage I, II, III, and IV melanomas or benign nevi after a minimum 5-year follow-up. Each point represents the mean % of p62 positive cells. Horizontal lines representing median p62 expression levels indicate an increase in median p62 expression levels between benign nevi and AJCC stage I or II melanomas and a relative decrease in expression in advanced AJCC stages III and IV tumors (Kruskal–Wallis P < 0.0001) (26). (C) Photomicrographs of immunohistochemical p62 expression and mean % in a melanocytic nevus or an eventual AJCC stage I, II, or IV melanoma. Scale bar = 100 μm. (D) Univariate analysis of mean p62 expression in AJCC stage II primary tumors demonstrating an increased risk of metastasis in tumors expressing >20% p62 [Log-Rank (Mantel–Cox) P = 0.031, HR 2.29 (95% CI 1.08–4.86)], and highlighting the potential of p62 as a prognostic biomarker (26).
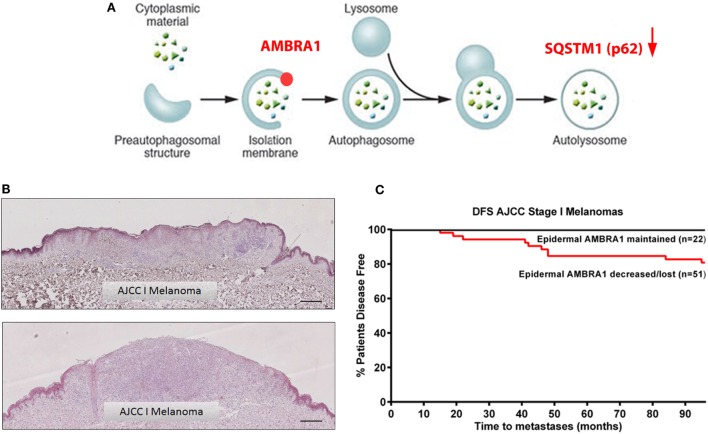
Activating molecule in Beclin 1-regulated protein 1 (AMBRA1) is a component of the Beclin 1/VPS34 complex and involved in the formation of PI3K rich membranes during the nucleation phase of autophagy [Figure 2A; (28)]. As a key autophagy initiating regulatory protein, AMBRA1 represents a potential marker of autophagy induction as well as a possible therapeutic target for autophagy inhibition. However, in addition to its functional role in autophagy, a growing body of evidence supports a role for AMBRA1 in cellular differentiation (29, 30) including in the early differentiation of neuronal stem cells in which autophagy is activated to fulfill the high energy demands of this process (28, 31). In line with these findings, we have recently demonstrated the role of AMBRA1 in epidermal differentiation with the expression in vivo increasing in line with keratinocyte differentiation from the basal layer of the epidermis to the uppermost layer, the stratum corneum (32). Unlike p62, however, the expression of AMBRA1 in primary melanomas is variable and as such its value as a tumoral biomarker remains undefined. Strikingly, however, our recent data demonstrate the decreased or even complete loss of AMBRA1 expression in the epidermis overlying many AJCC stage I melanomas (Figure 2B), which did not correlate with the degree of epidermal invasion and was not observed in benign nevi (32). These data suggest that the expression of AMBRA1 in the melanoma microenvironment may have prognostic potential. Univariate analysis of an initial cohort of 129 all AJCC stage melanomas further revealed decreased or loss of epidermal AMBRA1 expression was significantly associated with decreased disease-free survival, with stratification for AJCC stage I disease, additionally revealing epidermal AMBRA1 expression as a putative biomarker of disease progression [Figure 2C; (32)]. Again, there was no correlation with Breslow depth suggesting, such as p62, that epidermal AMBRA1 expression is also a biologically distinct marker for AJCC stage I melanomas. This is a striking finding considering that these tumors are normally regarded as low risk, with currently no alternative means of identifying specific individuals whose tumors are likely to progress.
Figure 2.
Loss of epidermal AMBRA1 identifies a high-risk AJCC stage I melanoma subgroup. (A) Schematic representation of the autophagy pathway highlighting the role of AMBRA1 in the nucleation phase of autophagy and indicating interplay of p62. (B) Representative immunohistochemistry images of epidermal AMBRA1 expression depicting maintained (top image) or loss of AMBRA1 expression (bottom image) in the epidermis overlying AJCC stage I melanomas. Loss of epidermal AMBRA1 expression overlying the tumor tissue creates a “watershed” area, where the epidermis distant to the tumor reveals a normal pattern of AMBRA1 expression. Scale bar = 100 μm. (C) Kaplan–Meier curve showing decreased 7-year disease-free survival in 51 AJCC stage I tumors where epidermal AMBRA1 was decreased or lost as compared with 22 tumors where AMBRA1 expression was maintained [Log-Rank (Mantel–Cox) test P < 0.03, HR 4.3 (95% CI 1.14–16.51)]. Epidermal AMBRA1 expression of each tumor was recorded as being either maintained, decreased, or lost based on the perceived degree of loss of epidermal AMBRA 1 expression overlying the tumor bulk compared to normal epidermis within the same section.
Since the current and universally adopted AJCC staging system is unable to identify the risk of disease progression in seemingly “low risk” early stage melanomas, such tumors are only identified after the onset of metastatic disease progression, at which point treatment options are limited and frequently ineffective. Critically, identifying, refining, and validating prognostic biomarkers for early stage melanomas such as the proposed biomarkers of autophagy will thus enable the identification of high-risk tumor subgroups. Both p62 and AMBRA1 expression exemplify how autophagy can be harnessed as prognostic biomarkers for melanoma, each providing clinically relevant information over and above AJCC staging, which in particular will be useful for refining the risk of melanoma progression in patients with AJCC stage I or II melanomas. Ultimately, further validation of these biomarkers will allow application in a clinical context, facilitating both earlier therapeutic intervention and the refinement of personalized therapies for malignant melanoma to improve clinical outcome and the prevention of premature loss of life.
Author Contributions
DT: main author of the paper and corresponding author involved in all aspects of authorship. RE: substantial contributions to the acquisition, analysis, and interpretation of data for the work including production of figures for the paper. PL: senior author, contribution to the analysis of data and the writing of the manuscript; accountable for all aspects of the work. All the authors have undertaken final approval of the final manuscript version to be published.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
DT is sponsored by a fellowship from The British Skin Foundation, RE and PL are supported by research grants from Melanoma Focus, North Eastern Skin Research Fund, and The Newcastle Healthcare Charity.
References
- 1.Lens MB, Dawes M. Global perspectives of contemporary epidemiological trends of cutaneous malignant melanoma. Br J Dermatol (2004) 150(2):179–85. 10.1111/j.1365-2133.2004.05708.x [DOI] [PubMed] [Google Scholar]
- 2.Cichorek M, Wachulska M, Stasiewicz A, Tyminska A. Skin melanocytes: biology and development. Postepy Dermatol Alergol (2013) 30(1):30–41. 10.5114/pdia.2013.33376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gogas H, Eggermont AM, Hauschild A, Hersey P, Mohr P, Schadendorf D, et al. Biomarkers in melanoma. Ann Oncol (2009) 20(Suppl 6):vi8–13. 10.1093/annonc/mdp251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. REporting recommendations for tumor MARKer prognostic studies (REMARK). Nat Clin Pract Urol (2005) 2(8):416–22. 10.1038/ncponc0252 [DOI] [PubMed] [Google Scholar]
- 5.Gould Rothberg BE, Bracken MB, Rimm DL. Tissue biomarkers for prognosis in cutaneous melanoma: a systematic review and meta-analysis. J Natl Cancer Inst (2009) 101(7):452–74. 10.1093/jnci/djp038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol (2009) 27(36):6199–206. 10.1200/JCO.2009.23.4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Checinska A, Soengas MS. The gluttonous side of malignant melanoma: basic and clinical implications of macroautophagy. Pigment Cell Melanoma Res (2011) 24(6):1116–32. 10.1111/j.1755-148X.2011.00927.x [DOI] [PubMed] [Google Scholar]
- 8.Corazzari M, Fimia GM, Lovat P, Piacentini M. Why is autophagy important for melanoma? Molecular mechanisms and therapeutic implications. Semin Cancer Biol (2013) 23(5):337–43. 10.1016/j.semcancer.2013.07.001 [DOI] [PubMed] [Google Scholar]
- 9.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol (2009) 335:1–32. 10.1007/978-3-642-00302-8_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathew R, White E. Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr Opin Genet Dev (2011) 21(1):113–9. 10.1016/j.gde.2010.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma XH, Piao S, Wang D, McAfee QW, Nathanson KL, Lum JJ, et al. Measurements of tumor cell autophagy predict invasiveness, resistance to chemotherapy, and survival in melanoma. Clin Cancer Res (2011) 17(10):3478–89. 10.1158/1078-0432.CCR-10-2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armstrong JL, Corazzari M, Martin S, Pagliarini V, Falasca L, Hill DS, et al. Oncogenic B-RAF signaling in melanoma impairs the therapeutic advantage of autophagy inhibition. Clin Cancer Res (2011) 17(8):2216–26. 10.1158/1078-0432.CCR-10-3003 [DOI] [PubMed] [Google Scholar]
- 13.Cantwell-Dorris ER, O’Leary JJ, Sheils OM. BRAFV600E: implications for carcinogenesis and molecular therapy. Mol Cancer Ther (2011) 10(3):385–94. 10.1158/1535-7163.MCT-10-0799 [DOI] [PubMed] [Google Scholar]
- 14.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature (2002) 417(6892):949–54. 10.1038/nature00766 [DOI] [PubMed] [Google Scholar]
- 15.Soengas MS, Lowe SW. Apoptosis and melanoma chemoresistance. Oncogene (2003) 22(20):3138–51. 10.1038/sj.onc.1206454 [DOI] [PubMed] [Google Scholar]
- 16.Gray-Schopfer VC, da Rocha Dias S, Marais R. The role of B-RAF in melanoma. Cancer Metastasis Rev (2005) 24(1):165–83. 10.1007/s10555-005-5865-1 [DOI] [PubMed] [Google Scholar]
- 17.Lee JH, Choi JW, Kim YS. Frequencies of BRAF and NRAS mutations are different in histological types and sites of origin of cutaneous melanoma: a meta-analysis. Br J Dermatol (2011) 164(4):776–84. 10.1111/j.1365-2133.2010.10185.x [DOI] [PubMed] [Google Scholar]
- 18.Maddodi N, Huang W, Havighurst T, Kim K, Longley BJ, Setaluri V. Induction of autophagy and inhibition of melanoma growth in vitro and in vivo by hyperactivation of oncogenic BRAF. J Invest Dermatol (2010) 130(6):1657–67. 10.1038/jid.2010.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corazzari M, Rapino F, Ciccosanti F, Giglio P, Antonioli M, Conti B, et al. Oncogenic BRAF induces chronic ER stress condition resulting in increased basal autophagy and apoptotic resistance of cutaneous melanoma. Cell Death Differ (2015) 22(6):946–58. 10.1038/cdd.2014.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanida I, Ueno T, Kominami E. LC3 and autophagy. Methods Mol Biol (2008) 445:77–88. 10.1007/978-1-59745-157-4_4 [DOI] [PubMed] [Google Scholar]
- 21.Lazova R, Klump V, Pawelek J. Autophagy in cutaneous malignant melanoma. J Cutan Pathol (2010) 37(2):256–68. 10.1111/j.1600-0560.2009.01359.x [DOI] [PubMed] [Google Scholar]
- 22.Lazova R, Camp RL, Klump V, Siddiqui SF, Amaravadi RK, Pawelek JM. Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin Cancer Res (2012) 18(2):370–9. 10.1158/1078-0432.CCR-11-1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miracco C, Cevenini G, Franchi A, Luzi P, Cosci E, Mourmouras V, et al. Beclin 1 and LC3 autophagic gene expression in cutaneous melanocytic lesions. Hum Pathol (2010) 41(4):503–12. 10.1016/j.humpath.2009.09.004 [DOI] [PubMed] [Google Scholar]
- 24.Sivridis E, Koukourakis MI, Mendrinos SE, Karpouzis A, Fiska A, Kouskoukis C, et al. Beclin-1 and LC3A expression in cutaneous malignant melanomas: a biphasic survival pattern for beclin-1. Melanoma Res (2011) 21(3):188–95. 10.1097/CMR.0b013e328346612c [DOI] [PubMed] [Google Scholar]
- 25.Hara Y, Nakamura M. Overexpression of autophagy-related beclin-1 in advanced malignant melanoma and its low expression in melanoma-in-situ. Eur J Dermatol (2012) 22(1):128–9. 10.1684/ejd.2011.1562 [DOI] [PubMed] [Google Scholar]
- 26.Ellis RA, Horswell S, Ness T, Lumsdon J, Tooze SA, Kirkham N, et al. Prognostic impact of p62 expression in cutaneous malignant melanoma. J Invest Dermatol (2014) 134(5):1476–8. 10.1038/jid.2013.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Armstrong JL, Hill DS, McKee CS, Hernandez-Tiedra S, Lorente M, Lopez-Valero I, et al. Exploiting cannabinoid-induced cytotoxic autophagy to drive melanoma cell death. J Invest Dermatol (2015) 135(6):1629–37. 10.1038/jid.2015.45 [DOI] [PubMed] [Google Scholar]
- 28.Fimia GM, Di Bartolomeo S, Piacentini M, Cecconi F. Unleashing the Ambra1-Beclin 1 complex from dynein chains: Ulk1 sets Ambra1 free to induce autophagy. Autophagy (2011) 7(1):115–7. 10.4161/auto.7.1.14071 [DOI] [PubMed] [Google Scholar]
- 29.Hamanaka RB, Chandel NS. Mitochondrial metabolism as a regulator of keratinocyte differentiation. Cell Logist (2013) 3(1):e25456. 10.4161/cl.25456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamanaka RB, Glasauer A, Hoover P, Yang S, Blatt H, Mullen AR, et al. Mitochondrial reactive oxygen species promote epidermal differentiation and hair follicle development. Sci Signal (2013) 6(261):ra8. 10.1126/scisignal.2003638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vazquez P, Arroba AI, Cecconi F, de la Rosa EJ, Boya P, de Pablo F. Atg5 and Ambra1 differentially modulate neurogenesis in neural stem cells. Autophagy (2012) 8(2):187–99. 10.4161/auto.8.2.18535 [DOI] [PubMed] [Google Scholar]
- 32.Tang D, Greenwood A, Carling E, Horswell S, Lovat P, Ellis RA. Epidermal AMBRA1 and Loricrin as Novel Prognostic Biomarkers for Melanoma. BSID Annual Meeting 2016. Dundee, Scotland: Wiley-Blackwell; (2016). [Google Scholar]