Abstract
Purpose of Review
Long-term culture of adult progenitor cells in 3D is a recently emerging technology that inhabits the space between 2D cell lines and organ slice culture.
Recent Findings
Adaptations to defined media components in the wake of advances in ES and iPS cell culture has led to the identification of conditions that maintained intestinal cell progenitors in culture. These conditions retain cellular heterogeneity of the normal or tumour tissue, and the cultures have been shown to be genetically stable, such that substantial biobanks are being created from patient derived material. This coupled with advances in analytical tools has generated a field, characterized by the term “organoid culture”, that has huge potential for advancing drug discovery, regenerative medicine, and furthering the understanding of fundamental intestinal biology.
Summary
In this review, we describe the approaches available for the long-term culture of intestinal cells from normal and diseased tissue, the current challenges, and how the technology is likely to develop further.
Keywords: Intestinal, Organoid culture, Intestinal cell progenitors, Cancer stem cell, Disease models, Colorectal cancer
Introduction
In 2007, after decades of research, the intestinal cell progenitors (ICPs) that are responsible for regenerating the surface of the normal intestine every 2–7 days were identified and shown to reside within crypt bases of the intestinal epithelium [1]. Up to this point, a variety of approaches had been used in an attempt to grow this tissue in vitro, including 2D immortalized cell lines often derived from either benign or malignant tumours, short-lived primary tissue isolates, as animal xenografts, and tissue pieces/slices. Each system not only had strengths but also shortcomings such as limited population doublings (primary isolates), ethical concerns (xenografts), or short-term viability (tissue slices). Following the identification of the ICPs, there was a period that saw major advances in the understanding of basic stem cell biology and refinements of progenitor cell culture. In 2007, it was demonstrated that 3D spheroids derived from CD133+ intestinal cancer stem cells (CSCs) could be maintained by utilizing culture medium refinements from neurosphere culture methods [2]. This formed part of a progression of developments, mainly pioneered by researchers then based within the laboratory of Hans Clevers, that in 2009 led to the identification of Lgr5 as an ICP marker and publication of a 3D culture technique which allowed single murine intestinal stem cells to be grown into organoids that contained protruding crypt structures with all the cell lineages that comprise the small intestinal crypt in vivo [3, 4, 5•]. These cultures were grown in a mesenchyme-free environment comprised Matrigel (a reconstituted basement membrane gel [6]) in a medium with three organoid supporting supplements: epidermal growth factor (EGF); Noggin, which is a BMP signalling inhibitor that maintains an undifferentiated state; and R-Spondin, a modulator of the Wnt pathway and potent stimulator of adult stem cell proliferation [7]. The generation of mice harbouring an Lgr5 driven GFP reporter [8] has enabled work that further characterized the crypt niche [9] along with identifying other ICP markers, notably Bmi1 [10], and indeed proved crucial to the identification of R-Spondin as a key modulator of Wnt signalling. It was later observed that cultures of mouse colonic epithelium required the addition of Wnt3A to enable their indefinite expansion, suggesting that the organoid Wnt ligand production is insufficient to maintain colonic stem cells [11]. This work was then successfully translated into patient-derived ICP containing organoids utilizing similar media, although human intestinal and colonic organoids required both p38 and TGF-β inhibition (to suppress differentiation), with human colon culture additionally requiring Wnt3A, Prostaglandin E2 (that promoted organoid integrity through blocking anoikis and promoting proliferation), and Nicotinamide (a vitamin shown to inhibit differentiation) [11, 12].
This review discusses the progress made over the last 3 years in using organoid culture of tissue-derived ICPs. Related developments in which intestinal cultures are generated by the directed differentiation of embryonic or induced pluripotent stem cells are described and reviewed elsewhere [13–16]. Within this review, we will introduce the areas in which long-term tissue-derived ICP cultures are finding utility; (1) their application in studying disease processes (particularly CSC biology), (2) the prospective clinical applications of long-term ICP culture models, (3) the ongoing cell culture refinements and elaborations of ex vivo ICP models, and (4) an overview of the analytical technologies around the use of ICP organoids that will lead to the proliferation of ICP organoid platforms.
Study of ICPs in Disease
ICP-generated 3D organoids retain in vivo cell-to-cell contacts, mass transport properties, mechanical properties, and metabolic profiles, whilst incorporating many cell types, modelling cell proliferation/differentiation, combined with long-term genomic stability [17•] and gene expression patterns. Thus, the organoids maintain their integrity, unlike classical 2D cell culture with its inherent loss of heterogeneity and the genomic rearrangements associated with the culture ‘crisis’/cellular senescence events that occur during cellular adaption. This maintenance of cell identity and genetic integrity within ICP containing organoid cultures makes them the current gold standard tool for interrogating basic and diseased intestinal biology ex vivo and the protocols for isolation of human intestinal progenitor cells from resected surgical samples and biopsies are now well established [18, 19]. Indeed the derivation of ICP organoid cultures from normal tissue and tumour material is carried out in such a way that cells are never grown directly upon culture plastic, as opposed to spheroid or tumoursphere culture models that are generated from established 2D cell lines. These organoid cultures have been particularly used in the study of colorectal cancer (CRC), and are being applied to translational settings such as regenerative medicine, diagnostic tests, and disease modelling [20–24].
CRC Modelling Using Ex Vivo ICP Culture
Ex vivo ICP culture has been highly relevant in CRC, where the ISC has been identified as the cell of origin [25]. Previously, cancer research has relied heavily on the use of genetically modified mouse models (GEMMs) to explore the genes and pathways associated with the disease. However, these models predominantly develop tumours in the small intestine and not the large intestine, the reverse of the human situation. Indeed, for the time being ex vivo ICP-based culture systems do not yet fully recapitulate in vivo 3D architecture, nor the contributions of the stroma, endothelial cells, oxygen tension, blood supply, immune system, or innervations that are afforded by CRC GEMMs. Further, the herbivorous mouse small intestine microbiome bears little resemblance to the omnivorous human large intestine microbiome and does not model the significant role that the environment plays in CRC risk. Despite these current drawbacks, the potential major benefit of long-term ICP culture will be the reduction in the current reliance on GEMMs and a shift towards using human and mouse organoid cultures and systems that will increasingly reflect the in vivo CRC environment. Indeed, establishing reliable sources of this material is now crucial for researchers to understand normal and malignant ICPs. The following section will summarize the recent developments in using diseased ICP cells that have either come from patients, GEMMs, or that have been ‘engineered’/gene edited from one of the first two sources.
Patient-Derived ICPs
There are a number of paths to modelling CRC based upon the culture of ICPs, perhaps the most direct involving the collection of patient tumour material and the preparation of stable long-term cultures. This has led to the development of CRC organoid collections (or biobanks) which have the aim of representing the diversity of CRC disease, infection, and drug responses that exists within the patient population [17•, 26, 27]. An issue with such an approach is the need to develop a biobank that truly reflects the main disease subtypes. There are currently four clinical subtypes of CRC that have been recently assigned by the CRC Subtyping Consortium, they are microsatellite instability immune (CMS1; characterized by hyper-mutated, microsatellite unstable and strong immune activation), canonical (CMS2; epithelial, with marked Wnt and Myc signalling activation), metabolic (CMS3; epithelial and evident metabolic dysregulation), and mesenchymal (CMS4; prominent TGF-β activation, stromal invasion, and angiogenesis) [28]. The interpretation of drug response studies is notably more complex in the CMS1 subtype because whilst the genetic changes will classify the cultures the impact of immune cell components is missing from the assay readouts [17•]. Such collections require not only enough patients per subtype to power future analysis, but also culture conditions that address concerns regarding any selective bias favouring one CRC subtype over another. The key advantages of patient-derived ICP cultures are the capture of genetic combinations that are known drivers of human disease, the maintenance of disease associated epigenetic history, and the ability to recapitulate the histology of primary disease.
GEMM-Derived ICPs
The second major source of malignant ICP cultures is from genetically characterized GEMMs. Despite the drawbacks previously mentioned, they are still invaluable tools for research and can be combined with organoid culture to great effect. This was epitomized by the work from the group of Doug Winton, who used ICPs from GEMMs to generate organoids that could contain a singly mutated ICP to understand the altered crypt dynamics and clonal advantage induced by the most common genetic alterations pervading CRC biology (Apc loss, Kras activation, and P53 mutation) [29]. Indeed over the past 3 years, GEMMs and organoids have been used to investigate the myriad of ISC- and CRC-associated genes and their relevant pathways to identify which are driving the tumour and which are passengers, e.g. Troy [30], Brg1 [31], Cdx2 [32], Kcnq [33], Prox1 [34], Fzd7 [35], Yap [36]. As well as the continuing exploration of intestinal biology, the GEMM-derived organoids are also being deployed as platforms to screen how normal gut and primary tumours with defined mutations respond to potential cancer therapeutics and determine the mechanisms of action. For example, Lorenzi et al. have used this approach to demonstrate that the resistance of FBXW7-mutated CRC cells to certain types of chemotherapy (e.g. Fluorouracil (5-FU)) is due to an inhibition of terminal differentiation indicating the that they could be overcome by using differentiating therapies [37]. ICP culture has been used to investigate the p300-CREB-MYB protein interactions and its role in Oxaliplatin resistance [38]. It has been demonstrated that the changes in gene expression pattern in a malignant intestinal stem cell are also closely tied to radiation resistance; Ladang et al. identified that the expression of Elp3 plays a key role in the radio-resistance of the Lgr5+Dclk1+ malignant ISC due to its promotion of Sox9 translation [39]. There are of course limitations to GEMM work that are driving the development of the human CRC models described above, such as the costs associated with their creation, their maintenance, and their inability to accurately reflect the biology of the human large intestine, particularly as the majority of GEMM studies on intestinal tumourigenesis have performed in the mouse small intestine. A key advantage going forwards will be the ability to reduce the cost of GEMM studies through combining defined genetic backgrounds with the ability to establish them as long-term organoid cultures and in doing so obtain much more data per animal.
Genome Editing of ICPs
A third approach capable of utilizing both of the above CRC culture derivations is through the exploitation of current DNA manipulation technologies to manipulate normal mouse and human ICPs into cancer ICPs that reflect human CRC. Using shRNA to target APC, P53, and PTEN, Onuma et al. have used lentiviral vectors in normal ICPs that has enabled them to generate recapitulated intestinal tumour organoids without generating gene-modified mice [40]. Similarly, Wang et al. [41] using adenovirus vectors and Ju et al. using food derived exosome delivery of nanoparticles [42] have provided proof of principle studies to demonstrate these techniques as effective gene delivery vehicles for genetic manipulation in 3D organoid cultures. However, future work is likely to use genome editing technology to a greater extent. The groups of Toshiro Sato and Hans Clevers have pioneered the use of CRISPR-Cas9 in ICP culture to demonstrate that common CRC mutations (often termed as ‘driver mutations’) confer niche-independent stem cell maintenance but not to metastatic progression, with data indicating that additional molecular lesions are also necessary for invasive tumour behaviour [43•]. A key feature of these techniques is the ability to modify genes in a stepwise fashion enabling the immediate analysis of the effect each gene has on an ICP [40, 44]. ICP culture has demonstrated that oncogenic alterations activating the MAPK and Wnt/β-catenin pathways must be consecutively and coordinately selected to assure stem cell maintenance during colon cancer initiation and progression [45]. Germann et al. [46] used organoids generated from Apc min/+ mice noting an aberrant cyst-like sphere morphology induced by a constitutively activated Wnt pathway that was responsible for increasing both self-renewal and growth while reducing differentiation. This observation combined with deletion studies elegantly described the engagement of the Wnt, Notch, and Myb transcriptional pathways in intestinal tumourigenesis and further highlighted the Wnt pathway as a therapeutic target in CRC. It seems likely that these last techniques will slowly lead us away from our current reliance on GEMMs.
Clinical Applications of Long-Term ICP Culture Models
This section summarizes in turn, personalized medicine, drug discovery, and regenerative medicine, the three main areas of clinical application for long-term ICP cultures.
Personalised Medicine
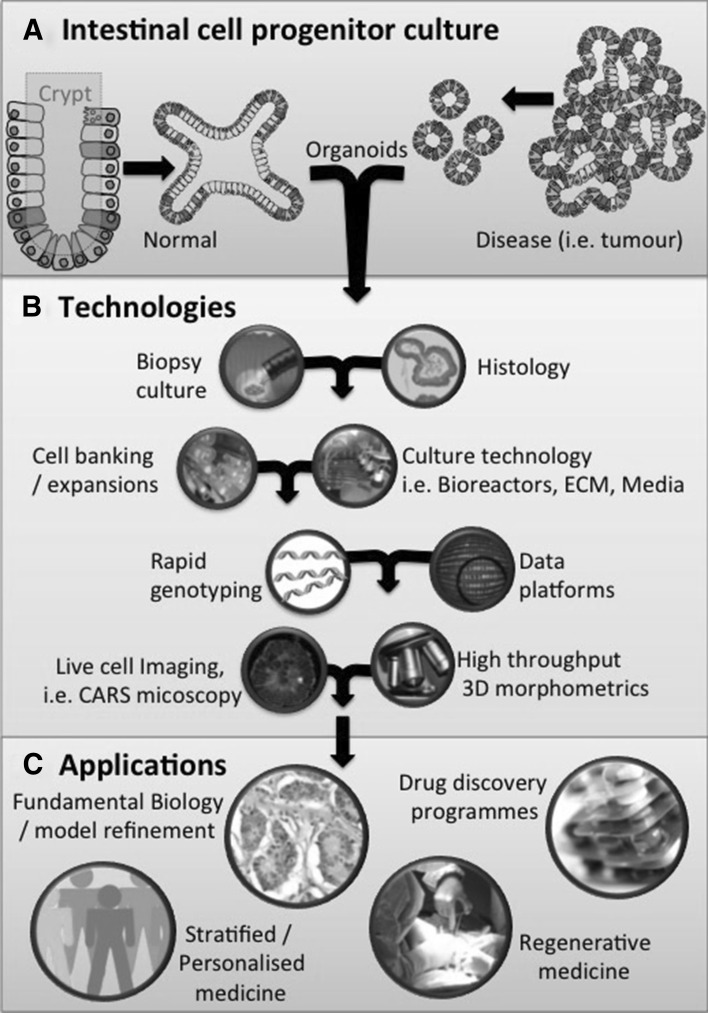
One of the greatest potential benefits of ICP cultures is to deliver a personalized medicine paradigm, where much like a biomedical service such as microbiology or virobiology a small sample is supplied and then cultured for rapid analysis. Where cultures act as ‘avatars’ of patients in the dish, an emerging application recently summarized by [47] allows clinical feedback of sample response to available therapeutics (Fig. 1). Ideally, these samples could also seed the biobanks described in the previous section, increasing the potential to break the existing 2D cell line paradigm that is prevalent in commercial drug/toxicity testing [48].
Fig. 1.
Schema showing the scope of tissue-derived ICP culture. a Intestinal cell progenitor culture, the origin of different organoids (normal and disease). b A collection of techniques and technologies that need to be in place in order to fully exploit the potential of ICP long-term cultures, for example the need for rapid genotyping to ensure the integrity of cultures from passage to passage. c The main applications and areas of ongoing research for potential deployment long-term ICP cultures in biomedical and clinical settings
Drug Discovery
Diverse collections of patient samples reflecting population disease profiles may power future drug discovery programmes, wherein organoids representative of specific disease subtypes would be tested against panels of compounds in drug titration assays in order to determine their potential efficacy. Key to shifting the current 2D culture led paradigm will be the generation of defined batches of organoids through scalable processes for commercial drug discovery programmes. Already the use of ICP culture is currently expanding knowledge of the role of individual genes in response to injury [49], and chemical-induced injury, e.g. ID1 [50].
Regenerative Medicine
Increasing our understanding of the influence of diet on the ISC using ex vivo culture is identifying the mechanisms for understanding the cause of disease resistance and simultaneously opening up potential avenues to be exploited for regenerative medicine. Potentially, the relationship between a high fat diet (which has been shown to increase the self-renewal potential of intestinal organoids), and susceptibility to CRC could be exploited to aid regeneration following intestinal injury [51]. Further, it has recently been demonstrated that transplantation of ICP organoids can potentially be used to increase the absorptive area in patients with short bowel syndrome [52] or alternatively tissue reconstruction, i.e. bowel reconstruction after disease, could be achieved using de-cellularized scaffolds for growing functional epithelium. The tools of organoid culture have also been explored in single-gene hereditary defects affecting the intestine, notably in studies of the cystic fibrosis transmembrane conductor receptor where organoids derived from the ISCs of cystic fibrosis patients have facilitated functional studies, drug development, personalized medicine, and gene repair approaches to treating the disease [53–55]. Gene manipulation in vivo and ex vivo has led to the conversion of ICPs into insulin producing “neo β-cell islets”, providing a potentially abundant and accessible source of functional insulin producing cells [56].
However, for organoids to achieve their clinical potential, biobanks of intestinal disease (i.e. CRC) would ideally (1) contain the tumour, blood (germline DNA), and early passage organoid set for DNA/expression analysis, which would allow the checking for a faithful recapitulation of the tumour by the organoid culture, alongside the identification of mutations from polymorphisms, (2) the ability to supply/maintain organoids in sufficient quantities and under standardized conditions to facilitate drug titration assays, which will require the development of bioreactors capable of standardizing growth and assay material, and (3) a sample collection reflecting a broad genetic diversity enabling toxicity studies where the range of likely toxic responses can be monitored.
Advances in Ex Vivo ICP Culture
Increasingly, the literature demonstrates a proliferation of the use of, and the number of, applications of ICP models (Table 1). Indeed, there have been some attempts to define and standardize conditions through publications of detailed protocols [57], and commercial production of specific media. Further streamlining of key components of the culture platform will be required to generate fully defined and reproducible growth conditions, a key example is the 3D support/extracellular matrix. The most commonly used support matrix, Matrigel, is a biological derived product that has a protein matrix composition of laminin, entactin, collagen, and heparan sulphate proteoglycans with batch variations compounded by varying concentrations of growth factors, such as bFGF, EGF, IGF-1, PDGF, NGF, and TGF-β [58]. Despite the work of some groups to date a defined hydrogel suitable for long-term ICP culture has not been reported, matrix biologists from the field of directed stem cell differentiation are now engaging with organoid culture [59].
Table 1.
Overview of intestinal cell progenitor culture model applications ranging across many areas of intestinal biology
| Subjects | Area of investigation | Tools/technology | Culture | Species (tissue) | Publication/s |
|---|---|---|---|---|---|
| Tumour biology | Modelling colorectal cancer progression | CRISPR-Cas9 introduction of APC, KRAS, SMAD4, TP53 and PIK3CA | Wnt, R-spondin, epidermal growth factor (EGF), noggin, transforming growth factor (TGF)-β inhibitors | Human | Matano et al. [43•] |
| Oncogenic BRAF induced loss of intestinal stem cells is antagonized by β-catenin activity | GEMM BRAFV637E knock-in mice |
Varied | Mouse | Riemer et al. [45] | |
| Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration | GEMM LGR5-GFP reporter mice Lgr5-LacZ reporter mice |
Co-culture with innate lymphoid cells Defined EGF; Noggin, R-spondin organoid media |
Human and mouse | Lindemans et al. [76] | |
| Organoid metabolism, particularly aspects of the Warburg shift in tumorigenesis | Lgr5-EGFP-IRES-creERT2 mice Measuring energy metabolism |
Defined BMP antagonist LDN-193189, EGF, R-spondin, Wnt conditioned medium | Mouse | Fan et al. [74] | |
| Regenerative medicine | Short bowel syndrome Small intestine reconstruction |
Culture onto a polyglycolic acid scaffold | Organoids cultured on scaffolds DMEM, 10 % serum |
Human and mouse cultures into NOD/SCID mice | Grant et al. [52] |
| Colonic mucosal injury | EGFP labelled donor mice | Defined EGF, Noggin, R-spondin organoid media | Mouse into mouse model | Fukuda et al. [49] | |
| Intestinal stem cell biology | A high-throughput platform for stem cell niche co-cultures and downstream gene expression analysis | Lgr5EGFP-CreERT2 mice Sox9EGFP:CAGDsRed Single cell microfluidics platform to interrogate stemness, transcriptional characterization of niche lineages |
Single cell culture AdvDMEM, N2, B27, Nac, 5 % FBS |
Mouse | Gracz et al. [67] |
| Studying cdx2 and its role in intestinal progenitor cell lineage Transformation of intestinal stem cells into gastric stem cells on loss of Cdx2 |
Lgr5-EGFP-Ires-CreERT2 and Cdx2+/− mice | Intestinal organoids EGF, Noggin, R-spondin Stomach organoids EGF, Noggin, R-spondin, Wnt, Fgf, Gastrin |
Mouse | Simmini et al. [32] | |
| Metabolism | Mapping early fate determination in Lgr5+ crypt stem cells using a novel Ki67-RFP allele | LGR5 cell cycling and Wnt signalling, ki67 labelled mouse model | Mouse | Basak et al. [9] | |
| Infection models | Impact of bacteria on intestinal epithelial cell biology, i.e. infection and irritable bowel disease | Salmonella (co-culture) | EGF, Noggin, R-spondin based with study specific additions | Mouse Human |
Zhang et al. [50] Rouch et al. [77] |
|
Helicobacter pylori
Organoid microinjection |
EGF, Noggin, R-spondin, Wnt, Fgf, Gastrin | Human (normal and tumour) | Bartfeld et al. [79] | ||
| Escherichia coli (various pathogenic strains) | Wnt, R-spondin, Noggin | Human | VanDussen et al. [26] | ||
| Tissue engineering/co-culture models | Gastric epithelium stem cell maintenance | Co-culture Collagen gels |
Myofibroblast cell line Collagen gel in an air–liquid interface environment, Ham’s F12, with 20 % FBS |
Mouse (gastric) | Katano et al. [63] |
| Intestinal crypt fission | Atomic force microscopy Lgr5-EGFP-ires-CreERT2 mice |
EGF, Noggin, R-Spondin | Mouse (glandular stomach culture) | Pin et al. [62] |
Aside from standardization of existing models, ongoing elaborations of ICP niche culture are being pursued in efforts to draw models still closer to recapitulating the organ in a dish (Table 1). There are a growing number of co-culturing methods to elaborate organoids with other cell types from the intestinal niche: (1) nerve and fibroblast cells replacing the need for exogenous Wnt signalling [60], myofibroblasts [61–63], gut nerve cells [64], (2) microorganisms of the gut microbiome modulating nutrient availability [65, 66], and (3) the use of microfluidics to explore niche dynamics [67, 68] (Table 1). Many of the above uses of these cultures are also discussed by Fatehullah et al. [69]. Again, the key is to refine culture techniques that accurately replicate the in vivo environment whilst addressing the challenge of establishing reproducible systems suitable for their application in drug discovery programmes.
Analysis of ICP Organoids
The use of ICP culture and technology to alter gene behaviour can elicit a myriad of cellular biology; thus, there is a requirement for techniques that can be used to analyse and quantify relevant parameters within complex 3D cellular environments [19]. This section sets out the cell culture analysis tools that can be used with 3D organoid cultures to quantify the complexity of organoid systems that display a wider range of biology compared with ‘classical’ 2D culture models. In some cases, tools have been modified to better work in 3D culture conditions.
3D Organoid Visualization
A change from 2D to 3D biology represents a sizable image analysis challenge. Standard light microscopy image analysis (often achieved using whole well or plate scanning imaging apparatus) is limited to 2D images, offering relatively few extra parameters (i.e. organoid diameter, organoid area, and number of organoids), although they do facilitate non-destructive serial measurements. In order to exploit the readouts of commonly used immunohistochemistry and immunofluorescence methods (for example live/dead cell detection, cell polarity, cell lineages, and organoid architecture), microscopic imaging platforms, such as wide field and confocal high content microscopes, are being used to analyse organoid cultures in 3D. Software analysis pipelines that are capable of analysing hundreds of 3D morphometric parameters and readouts can be used to identify subtle biological effects of media conditions and drug treatments [70]. These workflows are being designed to be compatible with the medium- to high-throughput drug screening demands of the pharmaceutical industry, and have led the development of morphometric analysis tools able to handle and interpret the volume of image stacks generated. The power and continued evolution of these screening platforms will be needed if 3D ICP-containing tumour models are to replace 2D cell lines in the preclinical drug discovery process. Image analysis of organoids may also be aided by the development of label-free imaging approaches such as coherence anti-Stokes–Raman spectroscopy (CARS) since cell type identification within complex 3D cellular environments currently requires organoid fixation and permeabilization [71, 72]. With the enhanced microscopy platforms, such as CARS, it will very likely be the timely analysis and interpretation of massive volumes of data created that represent the rate-limiting step to their application in drug screening and discovery programmes.
Single Cell Analysis
The ability to isolate and examine single cells within an organoid is an important feature of ICP culture. As the next-generation single-cell gene profiling becomes increasingly available in research labs, further progress will be made in understanding intestinal biology and disease. Existing next-generation sequencing technology has already been adapted and exploited to identify new intestinal cell types within organoids based on single cell messenger RNA sequencing [73]. This will allow lineage tracing experiments and a greater understanding of the dynamics of normal and diseased cells within the crypts. Although the readouts are still relatively limited, standard biochemical assays (live/dead ATP assays) have also been adapted for use in 3D culture, and other bioassay techniques are also coming on line. Techniques such as those reported by Fan et al. [74] wherein they adapted existing tools to create a platform capable of tracking dynamic energy metabolism in organoids and demonstrated a Warburg-like metabolic profile associated with colon tumourigenesis.
For the current rate of progress in the use of organoids to be maintained, it will be important that the development of technology, for obtaining and interpreting the vast quantities of information that can be gained from long-term ICP cultures, does not lag far behind.
Bringing Ex Vivo Closer to In Vivo: The Next Challenge
The complexity of the intestinal niche necessitates further elaboration of culture model systems that will likely include gut microbiota and diet research. Current models do not include stroma, elements of the immune system, a disease-specific ECM, or gut bacteria (the latter applicable to drug development for infection models). The ability of ICPs to faithfully maintain physiological relevance over time is vitally important for their use as research tools. However, replicating the environment in the human large intestine is crucial to our understanding how CRC develops. Culture conditions that manipulate the signalling pathways essential for ISC function are continually being identified and refined to better replicate and understand the ISC niche [75, 76]. Recent research has taken a reductionist approach to begin understanding the enormous complexity of this system. Reports investigating the role of single components of the diet [51], microbiome [26, 77–81], metabolome [66], immune system [81], and stroma [63, 82] on ICP using ex vivo culture are starting to emerge. These have demonstrated an increased understanding of how the environment elicits cellular and epigenetic alterations [83, 84] that are relevant to human health and intestinal diseases. Beyaz et al., using an ex vivo model, recapitulated ex vivo the environment associated with a high fat diet and established a PPARδ-dependent link to an increase in stemness within the intestinal niche that predisposes to CRC. Although these reductionist approaches are yielding greater insight into ICP, ultimately there will need to be greater efforts made to bring ex vivo culture techniques closer to the in vivo environment. The challenge for the future is to develop the tools for ICP culture to the point where human intestine can be recapitulated in the laboratory, such as elaborate co-culturing systems involving microfluidic linked culture vessels mimicking multi-compartment and even multi-tissue interactions. This would enable the exploration of the interactions between the full range of factors (diet, microbiome, metabolome, stroma, and immune system) that impact on the ISC and the roles they play in promoting, preventing, initiating, and driving intestinal diseases.
Acknowledgments
The authors thank Professor Trevor C. Dale (School of Biosciences, Cardiff University) for his discussion and advice in preparing this review. Andrew J. Hollins is currently supported by the Cancer Research UK (CR-UK) Centre Cardiff, having been previously supported by the Cardiff Experimental Cancer Medicines Centre (ECMC). The Wales Cancer Bank (including the affiliated Histopathology Team), as well as Clinical Teams with the Colorectal Surgical Multi-disciplinary Group within the University Hospital of Wales, Cardiff, supports Andrew’s research upon a patient-derived organoid platform. Lee Parry is supported by a Fellowship from the European Cancer Stem Cell Research Institute.
Compliance with Ethical Guidelines
Conflict of Interest
Andrew J. Hollins and Lee Parry declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
This article is part of the Topical Collection on Tissue Pathobiology: Stem Cells, Reprogramming, Regenerative Medicine, Tissue Engineering.
Contributor Information
Andrew J. Hollins, Email: HollinsAJ@cardiff.ac.uk
Lee Parry, Email: ParryL3@cardiff.ac.uk.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance
- 1.Brittan M, Wright NA. Gastrointestinal stem cells. J Pathol. 2002;197:492–509. doi: 10.1002/path.1155. [DOI] [PubMed] [Google Scholar]
- 2.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 3.Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 4.Ootani A, Li X, Sangiorgi E, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.• Ohta Y, Sato T (2014) Intestinal tumor in a dish. Front Med 1:1–4. doi:10.3389/fmed.2014.00014. An excellent review of the history of ICP organoid culture methodology, and potential future applications [DOI] [PMC free article] [PubMed]
- 6.Taub M, Wang Y, Szczesny TM, Kleinman HK. Epidermal growth factor or transforming growth factor alpha is required for kidney tubulogenesis in matrigel cultures in serum-free medium. Proc Natl Acad Sci USA. 1990;87:4002–4006. doi: 10.1073/pnas.87.10.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Lau WB, Snel B, Clevers HC. The R-spondin protein family. Genome Biol. 2012;13(3):242–252. doi: 10.1186/gb-2012-13-3-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 9.Basak O, van de Born M, Korving J, et al. Mapping early fate determination in Lgr5+ crypt stem cells using a novel Ki67-RFP allele. EMBO J. 2014;33:2057–2068. doi: 10.15252/embj.201488017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato T, Stange DE, Ferrante M, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 12.Jung P, Sato T, Merlos-suárez A, et al. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 13.Spence JR, Mayhew CN, Rankin SA, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCracken KW, Catá EM, Crawford CM, et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wells JM, Spence JR. How to make an intestine. Development. 2014;141:752–760. doi: 10.1242/dev.097386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huch M, Koo B-KB, Ahlgren U, et al. Modeling mouse and human development using organoid cultures. Development. 2015;142:3113–3125. doi: 10.1242/dev.118570. [DOI] [PubMed] [Google Scholar]
- 17.• van de Wetering M, Francies HE, Francis JM et al (2015) Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161:933–945. doi:10.1016/j.cell.2015.03.053. First to produce and comprehensively characterise a significant collection of patient derived (normal and tumour) organoids, the paper shows the long term genetic stability of the cultures, sets out the requirements of a cancer organoid biobank, and their future utility [DOI] [PMC free article] [PubMed]
- 18.Mahe MM, Sundaram N, Watson CL, et al. Establishment of human epithelial enteroids and colonoids from whole tissue and biopsy. J Vis Exp. 2015 doi: 10.3791/52483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parris A, Williams MR. A human colonic crypt culture system to study regulation of stem cell-driven tissue renewal and physiological function. Methods Mol Biol. 2015;1212:141–161. doi: 10.1007/7651_2015_197. [DOI] [PubMed] [Google Scholar]
- 20.Kuratnik A, Giardina C. Intestinal organoids as tissue surrogates for toxicological and pharmacological studies. Biochem Pharmacol. 2013;85:1721–1726. doi: 10.1016/j.bcp.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 21.Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 22.Krausova M, Korinek V. Wnt signaling in adult intestinal stem cells and cancer. Cell Signal. 2014;26:570–579. doi: 10.1016/j.cellsig.2013.11.032. [DOI] [PubMed] [Google Scholar]
- 23.Date S, Sato T. Mini-gut organoids: reconstitution of the stem cell niche. Annu Rev Cell Dev Biol. 2015;31:269–289. doi: 10.1146/annurev-cellbio-100814-125218. [DOI] [PubMed] [Google Scholar]
- 24.Kondo J, Endo H, Okuyama H, et al. Retaining cell–cell contact enables preparation and culture of spheroids composed of pure primary cancer cells from colorectal cancer. Proc Natl Acad Sci USA. 2011;108:6235–6240. doi: 10.1073/pnas.1015938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barker N, Ridgway RA, van Es JH, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 26.VanDussen KL, Marinshaw JM, Shaikh N, et al. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut. 2014 doi: 10.1136/gutjnl-2013-306651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujii M, Shimokawa M, Date S, et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vermeulen L, Morrissey E, van der Heijden M, et al. Defining stem cell dynamics in models of intestinal tumor initiation. Science. 2013;342:995–998. doi: 10.1126/science.1243148. [DOI] [PubMed] [Google Scholar]
- 30.Fafilek B, Krausova M, Vojtechova M, et al. Troy, a tumor necrosis factor receptor family member, interacts with Lgr5 to inhibit Wnt signaling in intestinal stem cells. Gastroenterology. 2013;144:381–391. doi: 10.1053/j.gastro.2012.10.048. [DOI] [PubMed] [Google Scholar]
- 31.Holik AZ, Krzystyniak J, Young M, et al. Brg1 is required for stem cell maintenance in the murine intestinal epithelium in a tissue-specific manner. Stem Cells. 2013;31:2457–2466. doi: 10.1002/stem.1498. [DOI] [PubMed] [Google Scholar]
- 32.Simmini S, Bialecka M, Huch M, et al. Transformation of intestinal stem cells into gastric stem cells on loss of transcription factor Cdx2. Nat Commun. 2014;5:5728. doi: 10.1038/ncomms6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Than BLN, Goos JACM, Sarver AL, et al. The role of KCNQ1 in mouse and human gastrointestinal cancers. Oncogene. 2014;33:3861–3868. doi: 10.1038/onc.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiener Z, Högström J, Hyvönen V, et al. Prox1 promotes expansion of the colorectal cancer stem cell population to fuel tumor growth and ischemia resistance. Cell Rep. 2014;8:1943–1956. doi: 10.1016/j.celrep.2014.08.034. [DOI] [PubMed] [Google Scholar]
- 35.Flanagan DJ, Phesse TJ, Barker N, et al. Frizzled7 functions as a Wnt receptor in intestinal epithelial Lgr5+ stem cells. Stem Cell Rep. 2015;4:1–9. doi: 10.1016/j.stemcr.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gregorieff A, Liu Y, Inanlou MR, et al. Yap-dependent reprogramming of Lgr5+ stem cells drives intestinal regeneration and cancer. Nature. 2015;526:715–718. doi: 10.1038/nature15382. [DOI] [PubMed] [Google Scholar]
- 37.Lorenzi F, Babaei-Jadidi R, Sheard J, et al. Fbxw7-associated drug resistance is reversed by induction of terminal differentiation in murine intestinal organoid culture. Mol Ther Methods Clin Dev. 2016;3:16024. doi: 10.1038/mtm.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sampurno S, Bijenhof A, Cheasley D, et al. The Myb-p300-CREB axis modulates intestine homeostasis, radiosensitivity and tumorigenesis. Cell Death Dis. 2013;4:e605. doi: 10.1038/cddis.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ladang A, Rapino F, Heukamp LC, et al. Elp3 drives Wnt-dependent tumor initiation and regeneration in the intestine. J Exp Med. 2015;212:2057–2075. doi: 10.1084/jem.20142288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Onuma K, Ochiai M, Orihashi K, et al. Genetic reconstitution of tumorigenesis in primary intestinal cells. Proc Natl Acad Sci USA. 2013;110:11127–11132. doi: 10.1073/pnas.1221926110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang N, Zhang H, Zhang BQ, et al. Adenovirus-mediated efficient gene transfer into cultured three-dimensional organoids. PLoS One. 2014 doi: 10.1371/journal.pone.0093608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ju S, Mu J, Dokland T, et al. Grape exosome-like nanoparticles induce intestinal stem cells and protect mice from DSS-induced colitis. Mol Ther. 2013;21:1345–1357. doi: 10.1038/mt.2013.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.• Matano M, Date S, Shimokawa M et al (2015) Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med 21:256–262. doi:10.1038/nm.3802. Demonstrated the effects of the addition of 5 common colorectal cancer driver mutations (APC, KRAS, SMAD4, TP53 and PIK3CA) to human normal colorectal organoids, using a systemicatic approach and cutting edge CRISPR-Cas9 tools [DOI] [PubMed]
- 44.Drost J, van Jaarsveld RH, Ponsioen B, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 45.Riemer P, Sreekumar A, Reinke S, et al. Transgenic expression of oncogenic BRAF induces loss of stem cells in the mouse intestine, which is antagonized by β-catenin activity. Oncogene. 2015;34:3164–3175. doi: 10.1038/onc.2014.247. [DOI] [PubMed] [Google Scholar]
- 46.Germann M, Xu H, Malaterre J, et al. Tripartite interactions between Wnt signaling, Notch and Myb for stem/progenitor cell functions during intestinal tumorigenesis. Stem Cell Res. 2014;13:355–366. doi: 10.1016/j.scr.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Shroyer NF. Tumor organoids fill the niche. Cell Stem Cell. 2016;18:686–687. doi: 10.1016/j.stem.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 48.Cox MC, Reese LM, Bickford LR, Verbridge SS. Toward the broad adoption of 3D tumor models in the cancer drug pipeline. ACS Biomater Sci Eng. 2015;1:877–894. doi: 10.1021/acsbiomaterials.5b00172. [DOI] [PubMed] [Google Scholar]
- 49.Fukuda M, Mizutani T, Mochizuki W, et al. Small intestinal stem cell identity is maintained with functional Paneth cells in heterotopically grafted epithelium onto the colon. Genes Dev. 2014;28:1752–1757. doi: 10.1101/gad.245233.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang N, Yantiss RK, Nam H, et al. ID1 is a functional marker for intestinal stem and progenitor cells required for normal response to injury. Stem Cell Rep. 2014;3:716–724. doi: 10.1016/j.stemcr.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beyaz S, Mana MD, Roper J, et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature. 2016;531:53–58. doi: 10.1038/nature17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grant CN, Salvador GM, Sala FG, et al. Human and mouse tissue-engineered small intestine both demonstrate digestive and absorptive function. Am J Physiol Gastrointest Liver Physiol. 2015 doi: 10.1152/ajpgi.00111.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dekkers JF, Wiegerinck CL, de Jonge HR, et al. WS14.5 A functional CFTR assay using primary cystic fibrosis intestinal organoids. J Cyst Fibros. 2012;11:S32. doi: 10.1016/S1569-1993(12)60101-5. [DOI] [PubMed] [Google Scholar]
- 54.Dekkers JF, van der Ent CK, Beekman JM. Novel opportunities for CFTR-targeting drug development using organoids. Rare Dis (Austin Tex) 2013;1:e27112. doi: 10.4161/rdis.27112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dekkers JF, Van Mourik P, Vonk AM, et al. Potentiator synergy in rectal organoids carrying S1251N, G551D, or F508del CFTR mutations. J Cyst Fibros. 2016 doi: 10.1016/j.jcf.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 56.Chen YJ, Finkbeiner SR, Weinblatt D, et al. De novo formation of insulin-producing “Neo-ß Cell Islets” from intestinal crypts. Cell Rep. 2014;6:1046–1058. doi: 10.1016/j.celrep.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Es JH, Clevers H. Generation and analysis of mouse intestinal tumors and organoids harboring APC and K-Ras mutations. Methods Mol Biol. 2015;1267:125–144. doi: 10.1007/978-1-4939-2297-0_6. [DOI] [PubMed] [Google Scholar]
- 58.Hughes CS, Postovit LM, Lajoie GA. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–1890. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- 59.Gjorevski N, Ranga A, Lutolf MP. Bioengineering approaches to guide stem cell-based organogenesis. Development. 2014;141:1794–1804. doi: 10.1242/dev.101048. [DOI] [PubMed] [Google Scholar]
- 60.Pastuła A, Middelhoff M, Brandtner A, et al. Three-dimensional gastrointestinal organoid culture in combination with nerves or fibroblasts: a method to characterize the gastrointestinal stem cell niche. Stem Cells Int. 2016 doi: 10.1155/2016/3710836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katano T, Ootani A, Mizoshita T, et al. Establishment of a long-term three-dimensional primary culture of mouse glandular stomach epithelial cells within the stem cell niche. Biochem Biophys Res Commun. 2013;432:558–563. doi: 10.1016/j.bbrc.2013.02.051. [DOI] [PubMed] [Google Scholar]
- 62.Pin C, Parker A, Gunning AP, et al. An individual based computational model of intestinal crypt fission and its application to predicting unrestrictive growth of the intestinal epithelium. Integr Biol (Camb) 2015;7:213–228. doi: 10.1039/C4IB00236A. [DOI] [PubMed] [Google Scholar]
- 63.Katano T, Ootani A, Mizoshita T, et al. Gastric mesenchymal myofibroblasts maintain stem cell activity and proliferation of murine gastric epithelium in vitro. Am J Pathol. 2015;185:798–807. doi: 10.1016/j.ajpath.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 64.Schrenk S, Schuster A, Klotz M, et al. Vascular and neural stem cells in the gut: Do they need each other? Histochem Cell Biol. 2015;143:397–410. doi: 10.1007/s00418-014-1288-9. [DOI] [PubMed] [Google Scholar]
- 65.Kaiko GE, Ryu SH, Koues OI, et al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell. 2016;165:1708–1720. doi: 10.1016/j.cell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Donohoe DR, Holley D, Collins LB, et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014;4:1387–1397. doi: 10.1158/2159-8290.CD-14-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gracz AD, Williamson IA, Roche KC, et al. A high-throughput platform for stem cell niche co-cultures and downstream gene expression analysis. Nat Cell Biol. 2015;17:340–349. doi: 10.1038/ncb3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reardon S. “Organs-on-chips” go mainstream. Nature. 2015;7560:266. doi: 10.1038/523266a. [DOI] [PubMed] [Google Scholar]
- 69.Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 70.Sandercock AM, Rust S, Guillard S, et al. Identification of anti-tumour biologics using primary tumour models, 3-D phenotypic screening and image-based multi-parametric profiling. Mol Cancer. 2015;14:147. doi: 10.1186/s12943-015-0415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4:359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bradley J, Pope I, Masia F, et al. Quantitative imaging of lipids in live mouse oocytes and early embryos using CARS microscopy. Development. 2016 doi: 10.1242/dev.129908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grün D, Lyubimova A, Kester L, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 74.Fan Y-Y, Davidson LA, Callaway ES, et al. A bioassay to measure energy metabolism in mouse colonic crypts, organoids and sorted stem cells. Am J Physiol Gastrointest Liver Physiol. 2015 doi: 10.1152/ajpgi.00052.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yin X, Farin HF, van Es JH, et al. Niche-independent high-purity cultures of Lgr5(+) intestinal stem cells and their progeny. Nat Methods. 2014;11:106–112. doi: 10.1038/nmeth.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lindemans CA, Calafiore M, Mertelsmann AM, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528:560–564. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rouch JD, Scott A, Lei NY, et al. Development of functional microfold (M) cells from intestinal stem cells in primary human enteroids. PLoS One. 2016;11:e0148216. doi: 10.1371/journal.pone.0148216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y-G, Wu S, Xia Y, Sun J. Salmonella-infected crypt-derived intestinal organoid culture system for host–bacterial interactions. Physiol Rep. 2014;2:e12147. doi: 10.14814/phy2.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bartfeld S, Bayram T, Van De Wetering M, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148(126–136):e6. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chandrakesan P, Roy B, Jakkula LUMR, et al. Utility of a bacterial infection model to study epithelial–mesenchymal transition, mesenchymal–epithelial transition or tumorigenesis. Oncogene. 2014;33:2639–2654. doi: 10.1038/onc.2013.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Farin HF, Karthaus WR, Kujala P, et al. Paneth cell extrusion and release of antimicrobial products is directly controlled by immune cell-derived IFN-γ. J Exp Med. 2014;211:1393–1405. doi: 10.1084/jem.20130753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hirokawa Y, Yip KHY, Tan CW, Burgess AW. Colonic myofibroblast cell line stimulates colonoid formation. Am J Physiol Gastrointest Liver Physiol. 2014;306:G547–G556. doi: 10.1152/ajpgi.00267.2013. [DOI] [PubMed] [Google Scholar]
- 83.Cao L, Kuratnik A, Xu W, et al. Development of intestinal organoids as tissue surrogates: cell composition and the epigenetic control of differentiation. Mol Carcinog. 2015;54:189–202. doi: 10.1002/mc.22089. [DOI] [PubMed] [Google Scholar]
- 84.Zimberlin CD, Lancini C, Sno R, et al. HDAC1 and HDAC2 collectively regulate intestinal stem cell homeostasis. FASEB J. 2015;29:2070–2080. doi: 10.1096/fj.14-257931. [DOI] [PubMed] [Google Scholar]