ABSTRACT
Bdellovibrio bacteriovorus and Micavibrio aeruginosavorus are predatory bacteria that naturally—and obligately—prey on other Gram-negative bacteria, and their use has been proposed as a potential new approach to control microbial infection. The ability of predatory bacteria to prey on Gram-negative human pathogens in vitro is well documented; however, the in vivo safety and efficacy of predatory bacteria have yet to be fully assessed. In this study, we examined whether predatory bacteria can reduce bacterial burden in the lungs in an in vivo mammalian system. Initial safety studies were performed by intranasal inoculation of rats with predatory bacteria. No adverse effects or lung pathology were observed in rats exposed to high concentrations of predatory bacteria at up to 10 days postinoculation. Enzyme-linked immunosorbent assay (ELISA) of the immune response revealed a slight increase in inflammatory cytokine levels at 1 h postinoculation that was not sustained by 48 h. Additionally, dissemination experiments showed that predators were efficiently cleared from the host by 10 days postinoculation. To measure the ability of predatory bacteria to reduce microbial burden in vivo, we introduced sublethal concentrations of Klebsiella pneumoniae into the lungs of rats via intranasal inoculation and followed with multiple doses of predatory bacteria over 24 h. Predatory bacteria were able to reduce K. pneumoniae bacterial burden, on average, by more than 3.0 log10 in the lungs of most rats as measured by CFU plating. The work presented here provides further support for the idea of developing predatory bacteria as a novel biocontrol agent.
IMPORTANCE
A widely held notion is that antibiotics are the greatest medical advance of the last 50 years. However, the rise of multidrug-resistant (MDR) bacterial infections has become a global health crisis over the last decade. As we enter the postantibiotic era, it is crucial that we begin to develop new strategies to combat bacterial infection. Here, we report one such new approach: the use of predatory bacteria (Bdellovibrio bacteriovorus and Micavibrio aeruginosavorus) that naturally—and obligately—prey on other Gram-negative bacteria. To our knowledge, this is the first study that demonstrated the ability of predatory bacteria to attenuate the bacterial burden of a key human pathogen in an in vivo mammalian system. As the prevalence of MDR infections continues to rise each year, our results may represent a shift in how we approach treating microbial infections in the future.
INTRODUCTION
The identification of penicillin by Alexander Fleming in 1928 ushered in the golden age of antibacterial treatment and is widely regarded as the greatest medical advance of the last 50 years (1). However, due to the overuse of our antibiotic supplies, among other reasons, the rise of multidrug-resistant (MDR) bacterial infections has become a global health crisis over the last decade (2). The issue of MDR infections and the lack of antibiotics in the development pipeline have spurred researchers to consider new ways to combat bacterial infection in the coming postantibiotic era. One such approach is the use of naturally occurring predatory bacteria.
Bdellovibrio bacteriovorus and Micavibrio aeruginosavorus are small Gram-negative proteobacteria that are obligate predators of other Gram-negative bacteria (3, 4). B. bacteriovorus randomly moves through the environment via the use of a single, polar flagellum and invades across the prey outer cell membrane into the periplasmic space of the Gram-negative prey, establishing a bdelloplast. The mechanism by which B. bacteriovorus bacteria invade their prey is not completely elucidated, but type IV pili have been implicated as essential in the process (5–7). Once inside, B. bacteriovorus grows in a filamentous manner by digesting the prey cell from within, divides into a number of progeny, and then lyses the bdelloplast to continue looking for more prey to invade. M. aeruginosavorus are epibiotic predators that do not invade their prey. Instead, they attach themselves to the outer membrane of a prey cell and digest the contents in a “vampire”-like fashion (4, 8, 9).
Multiple studies have already demonstrated the effectiveness with which both B. bacteriovorus and M. aeruginosavorus are able to control key human pathogens, including in MDR infections (10), in vitro (11–14). Predatory bacteria are unable to invade and prey on mammalian cells (15), mitigating potentially harmful off-target effects. Additionally, predatory bacteria are nonpathogenic in a variety of animal models, including mice, rabbits, guinea pigs, and chicks (14, 16–19). Furthermore, development of genetically stable resistance to predation has yet to be confirmed (20). However, most studies examining the ability of predatory bacteria to control human pathogens have been performed in vitro; thus, the efficacy with which predatory bacteria can control a bacterial infection in a live host is still not known.
In this study, first, the safety of intranasal inoculation of predatory bacteria in Sprague-Dawley (SD) rats was assessed. Then, lungs of rats were exposed to Klebsiella pneumoniae and were treated with multiple doses of predatory bacteria to determine their ability to reduce bacterial burden within the lungs. To our knowledge, this is the first study that demonstrated the ability of predatory bacteria to attenuate the bacterial burden of a key human pathogen in an in vivo mammalian system. The work presented here further supports the potential development of predatory bacteria into a biocontrol agent.
RESULTS
Host morbidity.
While the safety of administering predatory bacteria into the lungs of mice has already been demonstrated (19), we began by investigating if the predators are compatible with the animal model being used in this study, Sprague-Dawley (SD) rats. To examine the effect on host morbidity of introducing predatory bacteria into the respiratory tract of SD rats, we performed intranasal inoculations of 6.0 × 108 PFU/rat of B. bacteriovorus 109J, 1.1 × 109 PFU/rat of B. bacteriovorus HD100, or 5.0 × 107 PFU/rat of M. aeruginosavorus ARL-13 into rats in three groups containing six rats each. Another group of six rats was inoculated as a control with the vehicle, phosphate-buffered saline (PBS).
The animals were housed and monitored for up to 10 days postinoculation for any signs of discomfort, illness, or infection. At 10 days, all 18 rats inoculated with PBS or either strain of B. bacteriovorus were visually healthy with no apparent signs of illness (Table 1). One rat inoculated with M. aeruginosavorus died immediately after inoculation; however, veterinary consultation proposed asphyxiation as the probable cause of death. Blood analysis of the specific rat confirmed no elevated concentrations of white and red blood cell counts, and red blood cells were found with normal morphology (data not shown), suggesting that death was not caused by an acute infection resulting from M. aeruginosavorus inoculation. The remaining five rats inoculated with M. aeruginosavorus were found to be healthy at 10 days postinoculation (Table 1).
TABLE 1 .
Numbers of rats showing visual signs of morbidity after intranasal inoculation with predatory bacteria
| Treatment | No. of rats showing visual signs of morbidity at indicated time after inoculation/total no. of rats |
|||
|---|---|---|---|---|
| 1 h | 24 h | 48 h | 10 days | |
| Control (PBS) | 0/12 | 0/12 | 0/12 | 0/6 |
| B. bacteriovorus 109J | 0/12 | 0/12 | 0/12 | 0/6 |
| B. bacteriovorus HD100 | 0/12 | 0/12 | 0/12 | 0/6 |
| M. aeruginosavorus ARL-13 | 0/12 | 0/12 | 0/12 | 1/6a |
| K. pneumoniaeb | 0/8 | 0/8 | 0/8 | |
One rat died immediately after inoculation with predatory bacteria; however, asphyxiation was suggested as the cause of death.
With animal well-being in mind, rats inoculated with K. pneumoniae were not kept past 48 h postinoculation.
Histological examination of lung tissue from the 10-day exposure experiment revealed no pathology due to inoculation with predatory bacteria in any treatment group (Fig. 1). All specimens were characterized by lungs showing partial collapse of alveolar sacs without signs of inflammation or other abnormalities. Together, the data suggest that when inhaled, predatory bacteria have no adverse effects on rat morbidity.
FIG 1 .
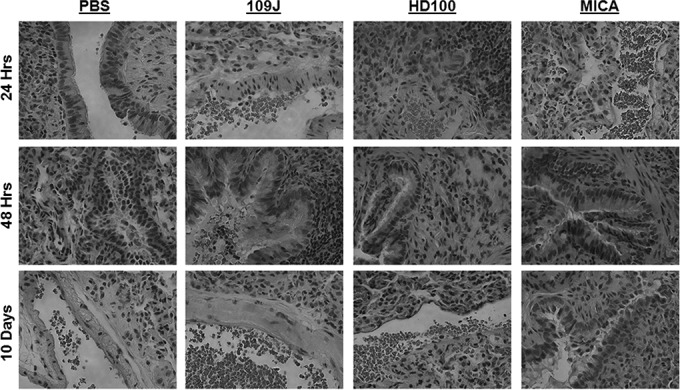
Histological examination of rat lungs after intranasal introduction of predatory bacteria. B. bacteriovorus 109J or HD100 bacteria or M. aeruginosavorus ARL-13 (MICA) bacteria were introduced into the lungs of SD rats via intranasal inoculation. Histological examination of rat lungs revealed no pathological abnormalities compared to rats inoculated with the control (PBS). All images are representative micrographs taken at 24, 48 h, and 10 days postintranasal inoculation and at ×40 total magnification.
Inflammatory response and histopathology.
To examine the effect of predatory bacteria on the host immune response within the lungs of SD rats, we introduced PBS, 3.8 × 108 PFU/rat of B. bacteriovorus 109J, 2.5 × 108 PFU/rat of B. bacteriovorus HD100, or 2.0 × 108 PFU/rat of M. aeruginosavorus ARL-13 into four groups of 36 rats each. An additional 28 rats were inoculated with a sublethal dose of 2.8 × 107 CFU/rat of K. pneumoniae, a known respiratory pathogen. Animals were again visually monitored for any signs of illness for up to 48 h. All 108 animals inoculated with any strain of predatory bacteria were found to be healthy for the duration of the experiment (Table 1). Twelve animals from each group (eight for the K. pneumoniae group) were sacrificed at 1, 24, and 48 h postinoculation, and the lung samples were harvested for histological examination and to assess inflammatory cytokine levels.
All rats that were inoculated with predatory bacteria did not show any visual signs of illness or discomfort. Histological examination of lung tissue revealed no abnormal pathology due to B. bacteriovorus 109J or M. aeruginosavorus at 24 h postinoculation (Fig. 1). At 48 h, examined lungs from rats inoculated with B. bacteriovorus 109J or M. aeruginosavorus showed only mild acute inflammation with stromal eosinophil infiltration and bronchioles containing a greater than normal amount of a proteinaceous substance consistent with mucus (Fig. 1). Lungs inoculated with B. bacteriovorus HD100 exhibited reactive lymphoid hyperplasia and acute inflammation at 24 h postinoculation; however, no pathological abnormalities were seen at 48 h (Fig. 1). Lungs inoculated with K. pneumoniae showed no abnormalities at 24 h postinfection; in contrast, at 48 h, lungs exhibited acute inflammation, stromal infiltration of eosinophils, germinal center formation within the lymphoid component of the inflammatory infiltrate, and bronchioles with a greater than normal amount of proteinaceous substance consistent with mucus. Lungs inoculated with the vehicle, PBS, exhibited no histological abnormalities at any time point examined.
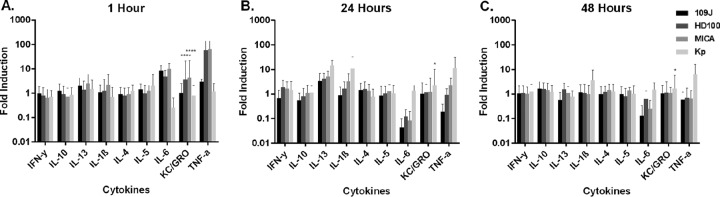
Enzyme-linked immunosorbent assay (ELISA) analysis of inflammatory proteins revealed 59.3- and 63.9-fold increases of tumor necrosis factor alpha (TNF-α) levels and 3.7- and 4.4-fold increases of KC/GRO (CXCL1) levels at 1 h postinoculation in rats inoculated with B. bacteriovorus HD100 and M. aeruginosavorus, respectively (Fig. 2A). However, none of the increases were sustained, as the levels of TNF-α and KC/GRO returned to baseline by 24 h postinoculation (Fig. 2B). We also observed 8.6-, 4.8-, and 10.0-fold increases in levels of interleukin-6 (IL-6) in rats inoculated with B. bacteriovorus 109J, B. bacteriovorus HD100, and M. aeruginosavorus, respectively, at 1 h postinoculation (Fig. 2A), but again, levels returned to baseline by 24 h (Fig. 2B). IL-13 levels increased 3.5-, 4.1-, and 5.2-fold in rats treated with B. bacteriovorus 109J, B. bacteriovorus HD100, and M. aeruginosavorus, respectively, at 24 h postinoculation (Fig. 2B) before reverting to physiological levels at 48 h (Fig. 2C). Rats inoculated with M. aeruginosavorus also demonstrated a 3.3-fold increase in IL-1β levels at 24 h (Fig. 2B). No levels of other inflammatory cytokines were found to be increased more than 3.0-fold at any time point examined in any group treated with predatory bacteria (Fig. 2). In contrast, rats inoculated with K. pneumoniae still exhibited 14.4-, 11.0-, and 11.4-fold increases in the levels of IL-13, IL-1β, and TNF-α, respectively, at 24 h postinoculation (Fig. 2B). At 48 h postinoculation, levels of IL-1β and TNF-α were still found to be substantially increased (3.6- and 6.3-fold, respectively) (Fig. 2C). Collectively, the data indicate that administering predatory bacteria at high concentrations into the lungs of rats does not result in adverse tissue pathology or provoke a sustained inflammatory response.
FIG 2 .
Inflammatory protein profile within rat lungs in response to intranasal inoculation of predatory bacteria. ELISA analysis of responses of IL-1β, IL-4, IL-5, IL-8, IL-10, IL-13, CXCL-1/KC, gamma interferon (IFN-γ), and TNF to intranasal inoculation of predatory bacteria relative to those seen with a PBS control was performed. B. bacteriovorus 109J or HD100 bacteria or M. aeruginosavorus ARL-13 bacteria (and also K. pneumoniae [Kp] bacteria as a control) were introduced into the lungs of SD rats via intranasal inoculation. Inflammatory proteins were assessed within the lungs at (A) 1, (B) 24, and (C) 48 h postinoculation. Twelve rats per treatment group (eight for K. pneumoniae) were used at each time point. Data are combined from two independent experiments. Data represent means ± standard errors of the means. Significant differences between treatment groups and respective control were determined using ANOVA (*, P < 0.05; ****, P < 0.0001).
Predatory bacterial dissemination.
To determine the predatory bacterial load within the lungs and other organs at 1, 24, and 48 h and 10 days post-intranasal inoculation, lung, liver, kidney, and spleen samples were also harvested during the previously described experiments and analyzed for the presence of predatory bacteria 16S rRNA using quantitative PCR (qPCR). Rats sacrificed at 1, 24, or 48 h were inoculated as described for the inflammatory response and histopathology experiment, while rats sacrificed at 10 days were treated as described above (see “Host morbidity”).
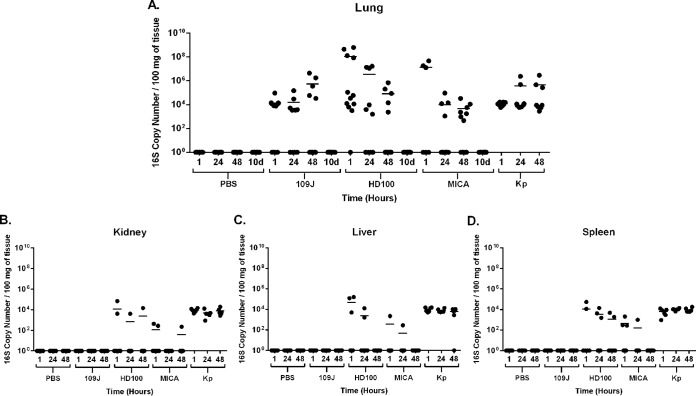
At 1 h postinoculation, B. bacteriovorus 109J was detected in the lungs of 7/12 rats (at levels ranging from 7.5 × 103 to 9.2 × 104 copy numbers), B. bacteriovorus HD100 in 11/12 rats (3.1 × 103 to 6.1 × 108), and M. aeruginosavorus in 3/6 rats (1.3 × 107 to 4.6 × 107) (Fig. 3A). Samples were available for only 6 (of 12) of the rats inoculated with M. aeruginosavorus at the 1 h time point. Detectable levels of B. bacteriovorus HD100 and M. aeruginosavorus in animals decreased with time. At 48 h, B. bacteriovorus HD100 was detected in only 5/12 rats (at levels ranging from 2.4 × 103 to 2.0 × 105 copy numbers) and M. aeruginosavorus in 7/12 rats (4.6 × 102 to 3.2 × 104) (Fig. 3A). By 10 days postinoculation, no B. bacteriovorus HD100 or M. aeruginosavorus bacteria were detected in any rats inoculated (Fig. 3A). Interestingly, while B. bacteriovorus 109J was detected in the lungs of only 5/12 rats at 48 h postinoculation, the average level of B. bacteriovorus 109J actually increased to 5.4 × 105 copy numbers (Fig. 3A). However, this increase was not sustained, as no B. bacteriovorus 109J was detected in the lungs of rats at 10 days postinoculation (Fig. 3A). In comparison, K. pneumoniae 16S rRNA was detected in the lungs of all rats at every time point examined and at higher average levels (1.2 × 104 to 4.6 × 105 copy numbers; Fig. 3A).
FIG 3 .
Predatory bacterial dissemination within hosts. qPCR detection of predatory bacteria within the host was performed. The (A) lungs, (B) kidneys, (C) livers, and (D) spleens were probed for B. bacteriovorus 109J and HD100, M. aeruginosavorus (MICA), and K. pneumoniae at 1, 24, and 48 h and 10 days (d) (lung only) postinoculation. Twelve rats per treatment group (eight for K. pneumoniae) were analyzed at each time point for the lungs, with the exception of the M. aeruginosavorus 1-h treatment group, which had six rats. Only six rats per treatment group were analyzed for the kidney, liver, and spleen. Each data point represents a single rat’s respective bacterial load. A line represents the mean of the results from each treatment set. Data are combined from the results of two independent experiments.
Predatory bacteria 16S rRNA was detected in only a limited number of the kidneys, livers, and spleens of rats inoculated with either B. bacteriovorus HD100 or M. aeruginosavorus at 1 h postinoculation (Fig. 3B to D). The kidneys, livers, and spleens of only six rats per treatment group at each time point were assessed. The trend exhibited a decrease in the concentration of predatory bacteria, as well as a decrease in the number of animals with predators that disseminated from the lungs to other organs examined. By 48 h postinoculation, B. bacteriovorus HD100 was detected in the kidneys of only 1/6 rats, in the livers of 0/6 rats, and the kidneys of only 2/6 rats (Fig. 3B to D). Similarly, M. aeruginosavorus was detected in the kidneys of only 1/6 rats and in the livers and kidneys of none of the rats inoculated at 48 h (Fig. 3B to D). No B. bacteriovorus 109J was detected in the kidneys, liver, or spleen of any rat inoculated at any time point assessed (Fig. 3B to D). In contrast, K. pneumoniae disseminated at high levels to the kidney, liver, and spleen in all rats inoculated by 1 h postinfection and continued to be detectable at similarly high levels at 48 h (Fig. 3B to D). In conclusion, the data suggest that predatory bacterial dissemination to other organs after respiratory inoculation is limited, while predators are quickly and efficiently cleared from the host by 10 days postinoculation.
Pathogen inoculation and treatment.
To determine whether predatory bacteria are able to reduce bacterial burden in the lungs, 36 rats were introduced with 3.3 × 107 CFU/rat of K. pneumoniae via intranasal inoculation (“experimental group”). Thirty-six more rats were inoculated with PBS (“control group”). Twelve rats each from the control and experimental groups were dosed four times with PBS, 4.6 × 108 PFU/rat of B. bacteriovorus 109J, or 6.6 × 107 PFU/rat of M. aeruginosavorus ARL-13 over a 24-h period postinfection. Dosages of PBS or predatory bacteria were administered through intranasal inoculation at 30 min and 6, 12, and 18 h before animals were sacrificed and organs harvested at 24 h postinoculation.
As before, examination of lung tissue revealed no significant histological abnormalities due to predatory bacteria at 24 h postinoculation. Tissue from rats inoculated with K. pneumoniae and treated with either B. bacteriovorus 109J or M. aeruginosavorus was similar to the tissue from the PBS-inoculated control group and generally showed peribronchiolar aggregates of eosinophils and macrophages, though a few samples showed partial collapse of the alveolar sacs (Fig. 4). Rats inoculated with K. pneumoniae and not treated with predatory bacteria exhibited bronchiole-associated lymphoid tissue with reactive lymphoid hyperplasia, as well as peribronchiolar infiltration of eosinophils (Fig. 4).
FIG 4 .
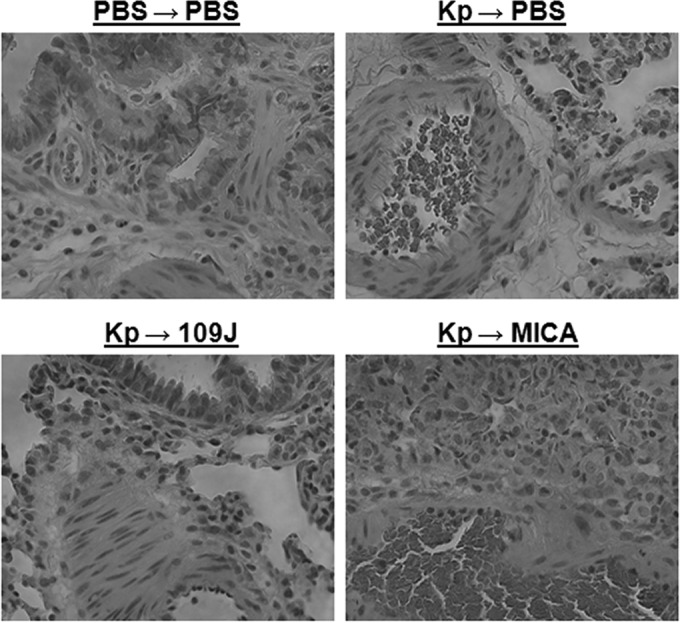
Histological examination of rat lungs after treatment of K. pneumoniae inoculation with predatory bacteria. K. pneumoniae (or PBS for control groups) was initially introduced into the lungs of rats via intranasal inoculation. Animals were then treated with PBS, B. bacteriovorus 109J, or M. aeruginosavorus (MICA) at 30 min and 6, 12, and 18 h postinoculation. All images are representative micrographs taken at 24 h postinoculation and at ×40 total magnification.
Lung samples were homogenized and plated on MacConkey agar. As expected, no colonies of K. pneumoniae from the lungs of rats in the PBS-inoculated control group were isolated on agar plates (Fig. 5). In the experimental group, a median of 1.6 × 104 CFU/ml (mean = 4.7 × 105 CFU/ml) of K. pneumoniae was isolated from the lungs of 75.0% of rats initially inoculated with K. pneumoniae and treated with PBS (Fig. 5). In contrast, we recovered colonies of K. pneumoniae from the lungs of only 58.3% of rats inoculated with K. pneumoniae and treated with B. bacteriovorus 109J, with a median of 1.9 × 102 CFU/ml (mean = 1.8 × 104 CFU/ml) (Fig. 5). K. pneumoniae was also recovered from the lungs of 66.6% of rats treated with M. aeruginosavorus, with a median of 1.2 × 102 CFU/ml (mean = 1.7 × 105 CFU/ml). However, for both the B. bacteriovorus 109J and M. aeruginosavorus treatments, levels of K. pneumoniae CFU showed high variability. Strikingly, 83.3% of B. bacteriovorus 109J-treated rats and 66.6% of M. aeruginosavorus-treated rats exhibited greater than 3.0 log10 reductions in copy numbers of K. pneumoniae recovered compared to the mean of the results from PBS-treated rats (Fig. 5). Furthermore, we recovered no K. pneumoniae from 41.6% of B. bacteriovorus 109J-treated rats and 33.3% of M. aeruginosavorus-treated rats. Taken together, the data indicate that B. bacteriovorus 109J and M. aeruginosavorus are able to efficiently reduce bacterial burden in the lungs of rats.
FIG 5 .
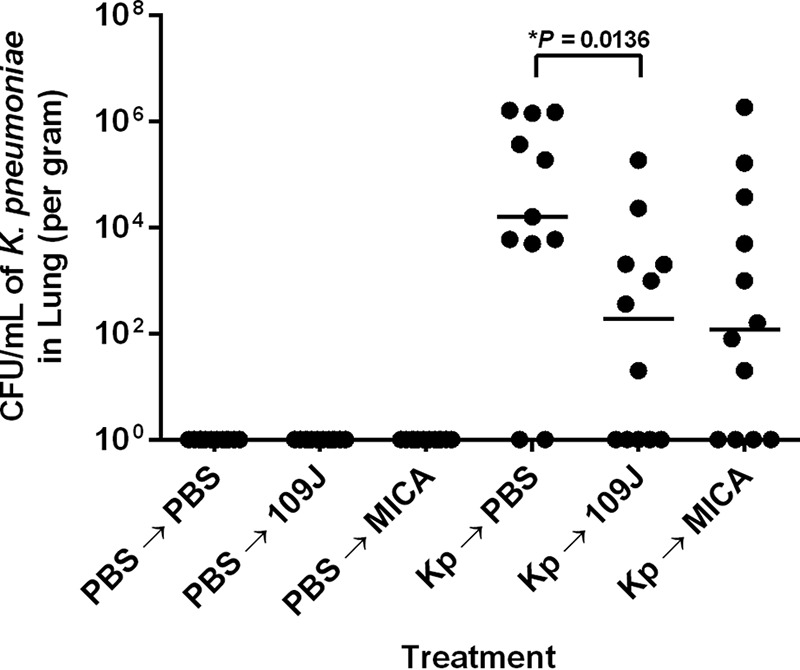
K. pneumoniae bacterial burden within lungs of rats after treatment with predatory bacteria. K. pneumoniae (or PBS for control groups) was initially introduced into the lungs of rats via intranasal inoculation. Animals were then treated with PBS, B. bacteriovorus 109J, or M. aeruginosavorus (MICA) at 30 min and 6, 12, and 18 h postinoculation. At 24 h, lungs were harvested, homogenized, and plated on MacConkey agar plates to recover K. pneumoniae CFU. Twelve rats per treatment group were used at each time point. Each data point represents a single rat’s respective bacterial load. Horizontal lines represent the median of the results from each treatment set. Data are combined from the results from two independent experiments. Analyses of significant differences between treatment groups and respective controls were performed using the Mann-Whitney test (*, P < 0.05).
DISCUSSION
The prevalence of antibiotic-resistant infections has climbed to frightening levels over the last decade (2). Compounding this problem is the fact that since the development of linezolid in the 1970s, virtually no new class of antibiotics, particularly those that can target Gram-negative bacterial pathogens, has been discovered through traditional drug screening techniques. Recently, the first MDR infection caused by a member of the Enterobacteriaceae harboring the mcr-1 plasmid-borne colistin resistance gene was detected in the United States, potentially signaling the emergence of truly pan-drug-resistant bacteria (21). For these reasons, among others, it is crucial that we begin to develop new treatments to combat bacterial infection. Researchers have already begun investigating potential new antimicrobial strategies (22), such as the use of antimicrobial peptides (23, 24), phage therapy (25, 26), and gene-editing enzymes (27, 28). Here, we report another promising novel approach: the use of predatory bacteria to control Gram-negative human pathogens.
In this study, we first determined the safety of intranasal inoculation of predatory bacteria in rats. High doses (>107 PFU/ml) of B. bacteriovorus 109J or HD100 or of M. aeruginosavorus ARL-13 were administered via intranasal inoculation into the lungs of SD rats. Two different B. bacteriovorus strains were examined to ensure that the results were not strain specific; unfortunately, we were unable to obtain additional M. aeruginosavorus strains. Of the total of 126 rats inoculated with predatory bacteria during the safety experiments, 125 rats were healthy, with no visual signs of adverse effects. Histological examination of tissue also revealed no adverse pathology associated with respiratory inoculation of predatory bacteria.
The lack of toxicity due to inoculation with predatory bacteria is in agreement with previous studies demonstrating the safety of introducing predatory bacteria in a variety of animal models (16–19). In particular, we observed similar results in our previous study, in which C57BL/6 mice were intranasally inoculated with high concentrations of viable or heat-killed B. bacteriovorus or M. aeruginosavorus (19). In that study, no mouse exhibited either morbidity or any histological abnormalities at up to 50 days postinoculation, signaling that predatory bacteria are nontoxic when inhaled.
We next looked to determine the immune response to predatory bacteria within the lungs of rats. A slight increase in levels of inflammatory cytokines (IL-1β, IL-6, IL-13, TNF-α, KC/GRO) after inoculation with predatory bacteria was observed at the earlier time points of 1 and 24 h; however, inflammatory protein levels returned to baseline levels by 48 h postinoculation. The cytokine response in rats inoculated with the positive control, K. pneumoniae, was not as highly elevated as the response reported in other studies (29, 30). An explanation for this is the fact we used a sublethal dose to ensure that the animals would not succumb to the infection before administration of predatory bacteria in the lung infection model. Furthermore, we demonstrated that predatory bacteria do not disseminate efficiently to other organs and are quickly cleared from the lungs of the host by 10 days postinoculation. As the aforementioned inflammatory cytokines are key players in the primary immune response, we suspect that the predatory bacteria are promptly and efficiently cleared through innate immunity mechanisms.
Our results seen with the rat model align with our previous study, where intranasal or intravenous inoculation of high doses of either B. bacteriovorus or M. aeruginosavorus in mice caused no tissue pathology and induced only a modest inflammatory response that returned to baseline levels within 24 h postinoculation (19). Furthermore, predatory bacteria were completely cleared from the animals by an innate immune response (possibly via neutrophils) within 48 h postinoculation (19). The consistency in our results provides further evidence that B. bacteriovorus and M. aeruginosavorus are inherently nonpathogenic in mammalian models. The lack of a productive and sustained immune response to predatory bacteria may be partially a result of the presence of an altered lipopolysaccharide (LPS) (31). The negatively charged phosphate residues of the LPS on the surface of Gram-negative bacteria are primarily responsible for antagonizing an immune response by strongly binding to Toll-like receptors on immune cells. B. bacteriovorus HD100 is known to express a neutrally charged LPS which has been shown to provoke only a weak inflammatory response in vitro (31), which may explain the lack of immunogenicity in our in vivo models. It is still unknown if M. aeruginosavorus expresses an altered LPS, as well.
The main objective of our study was to determine whether predatory bacteria are able to reduce the bacterial burden in the lungs in an in vivo mammalian system. We intranasally inoculated rats with K. pneumoniae and treated them with PBS, B. bacteriovorus 109J, or M. aeruginosavorus at 30 min and 6, 12, and 18 h postinoculation. In order to limit the number of animals being sacrificed, only B. bacteriovorus 109J was used in efficacy experiments. It was predicted that B. bacteriovorus 109J would provide a greater chance of reducing the bacterial burden than strain HD100 due to its weak innate immune response profile and its ability to remain longer in the lungs of the rats, as measured in dissemination experiments. Histological examination of lung tissue exhibited no significant pathological abnormalities due to treatment with predatory bacteria at 24 h postinoculation. Lung samples were homogenized and plated on MacConkey agar, a selective medium used to isolate Gram-negative and enteric bacteria, in order to determine K. pneumoniae concentrations (32). While the average reduction of K. pneumoniae bacterial burden due to treatment with B. bacteriovorus 109J or M. aeruginosavorus was approximately ≤1 log10, 83.3% of B. bacteriovorus 109J-treated rats and 66.6% of M. aeruginosavorus-treated rats exhibited greater than 3.0 log10 reductions in the levels of K. pneumoniae recovered compared to the mean of the results seen with PBS-treated rats. Furthermore, 5/12 B. bacteriovorus 109J-treated rats and 4/12 of M. aeruginosavorus-treated rats had no detectable K. pneumoniae, suggesting clearance of the pathogen from the host. The observed amount of K. pneumoniae reduced by predatory bacteria in vivo is similar to the reported change in vitro. One such study demonstrated the ability of predatory bacteria to reduce levels of five different strains of K. pneumoniae (10). B. bacteriovorus 109J and M. aeruginosavorus were able to reduce K. pneumoniae levels in vitro by averages of 3.4 and 3.0 log10 CFU/ml, respectively (10). Although active predation could be the sole contributor to the reduction of the K. pneumoniae load in the animals treated with predatory bacteria, one might suggest that additional immune response elements, elicited by the presence of the predatory bacteria, also played a role in reducing the microbial burden. Thus, the potential synergistic effects of predatory bacteria and the immune system should be a basis of future studies.
We are aware of only one other study that has tested the in vivo efficacy of treatments using predatory bacteria. That previous study determined whether oral administration of B. bacteriovorus could reduce the level of colonizing Salmonella in the guts of young chicks (18). The authors observed an average of 0.64 to 1.09 log10 reduction of levels of Salmonella (18). While the mean reductions in the chick study were similar to the mean values that we obtained, a major difference in our study was that the majority of rats treated with predatory bacteria showed a greater than 3.0 log10 reduction in K. pneumoniae levels. Note that, in the previous study, chicks were dosed once with predatory bacteria, whereas we dosed the rats four times over 24 h, modeling the dosing regimen in a typical antibiotic dosing schedule. Another major difference between the studies was in the physiological conditions of the animals, as birds have a physiological body temperature of 42°C. Predatory bacteria may prey upon the host bacteria and grow more efficiently in a mammal whose physiological temperature is 37°C, which is closer to the optimum predation conditions for predatory bacteria at 30°C. Furthermore, it is difficult for predatory bacteria to survive passage through the acidic environment of the stomach, and while an antacid was administered concurrently with predatory bacteria to increase the pH in the chick study, it is possible that the overall conditions of the chick gut are not optimal for predation.
It is important to emphasize the limitations of our model for extrapolation to the treatment of humans. We acknowledge that treatment with predatory bacteria at 30 min post-K. pneumoniae inoculation does not represent abrogation of an established infection. Rather, this study was an experimental exercise designed to determine whether predatory bacteria have the ability to reduce bacterial burden in an in vivo mammalian system. Our treatment scheme is similar to that of a recent study examining the efficacy of structurally nanoengineered antimicrobial peptide polymers (SNAPPs) in reducing MDR infections in mice (33). Mice were “infected” with a pathogen (Acinetobacter baumannii) and, similarly to our study method, were treated with SNAPPs at 30 min and 4 and 8 h postinoculation before being euthanized at 24 h.
In conclusion, our results indicate that predatory bacteria are safe to administer intranasally to a mammalian host, are able to attenuate pathogen burden in the lungs of rats, and may provide a novel way to combat infection caused by Gram-negative pathogens. Future studies will focus on using established in vivo models of infection to further determine whether the use of predatory bacteria is a viable treatment for Gram-negative infections, including MDR infections.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Predatory bacteria examined in this study were Bdellovibrio bacteriovorus strain 109J (ATCC 43826), Bdellovibrio bacteriovorus strain HD100 (ATCC 15356) (34), and Micavibrio aeruginosavorus strain ARL-13 (9). As the pathogen, Klebsiella pneumoniae ATCC 43816 was used and grown in Luria-Bertani (LB) medium (35). Predatory bacteria were cultured and processed as previously described (11, 15). Escherichia coli WM3064, a diaminipimelic acid (DAP) auxotroph, was used as prey and grown overnight in LB medium supplemented with 0.3 mM DAP. Predator stock lysates were made by coculturing the predators with prey cells in HEPES buffer (25 mM) supplemented with 3 mM MgCl2 and 2 mM CaCl2. Cocultures were incubated at 30°C until the culture cleared (stock lysates). In order to obtain high concentrations of B. bacteriovorus for inoculation experiments, 10 ml of a washed overnight culture of E. coli WM3064 cells (~1 × 109 CFU/ml) was resuspended in 80 ml of HEPES medium containing 10 ml of predatory bacteria from the stock lysates and incubated for 24 h on a rotary shaker. To obtain higher M. aeruginosavorus concentrations, M. aeruginosavorus cocultures were prepared in 200 ml of HEPES medium containing 25 ml of prey and 25 ml of M. aeruginosavorus stock lysates and incubated on a rotary shaker for 72 h. Cocultures were passed two times through a 0.45-µm-pore-size Millex filter (Millipore) to remove residual prey and cell debris (filtered lysate). Filtered lysates were pelleted three times by centrifugation at 29,000 × g for 45 min using a Sorvall LYNX 4000 centrifuge (Thermo Fisher Scientific Inc.) to further purify and concentrate predator samples. Each time, the pellet was washed and resuspended in 50 ml of phosphate-buffered saline (PBS). For the final wash, the predator pellet was resuspended in 1 to 2 ml of PBS solution to reach final optical densities at 600 nm (OD600) of 0.2 ± 0.02 for B. bacteriovorus and 0.1 ± 0.02 for M. aeruginosavorus, which corresponded to PFU values of between ~5 × 109 and 5 × 1010 PFU/ml and between ~5 × 108 and 5 × 109 PFU/ml, respectively. The standard double-layered agar method was used to determine predator cell concentrations (36). Fifty microliters of the predator samples was plated on DAP-supplemented LB agar and tryptic soy broth (TSB)-blood plates to confirm that the samples were free of prey cells and contaminants. Since the predatory bacteria were used directly after isolation, the actual viable predator dose was known only a few days after each experiment, as the PFU appeared. Therefore, in some experiments, mainly involving M. aeruginosavorus, the inoculation sizes differed somewhat. The actual predator inoculation doses are specified for each experiment.
Rats.
Wild-type male Sprague-Dawley (SD) rats (4 to 6 weeks old) were purchased from Charles River Laboratories (Wilmington, MA). All rats were housed under pathogen-free conditions at the Rutgers New Jersey Medical School animal facility. Guidelines from the Rutgers New Jersey Medical School Institutional Animal Care and Use Committee (protocol 15012) and the Animal Care and Use Review Office of the U.S. Army Medical Research and Material Command were followed in handling the animals.
Intranasal inoculation of bacteria.
Predatory bacteria were introduced into the lungs of SD rats by intranasal inoculation to model a respiratory infection. After animals were anesthetized with 4% isoflurane–oxygen for 5 min using an isoflurane vaporizer, 50 µl of purified bacterial suspension was applied at both nostrils using a pipette. Rats were inoculated with PBS, B. bacteriovorus strain 109J, B. bacteriovorus strain HD100, M. aeruginosavorus strain ARL-13, or K. pneumoniae. To avoid cross contamination, animals were caged according to treatment group and time point to be sacrificed. After every set of inoculations was performed in each experiment, animals were visually assessed for signs of infection, illness, and discomfort. At 1, 24, and 48 h and 10 days postinoculation, lung, liver, spleen, and kidney samples were harvested for histological examination, inflammatory protein analysis, and bacterial dissemination experiments.
Histological examination.
Lung samples designated for histology were stored in formalin at 4°C before examination. All histopathological examinations were performed by a pathologist blind to each specimen’s treatment group. Formalin-fixed organ segments from infected mice were paraffin embedded and stained with hematoxylin and eosin (H&E) for analysis of cellular composition as previously described. Stained sections were analyzed and photographed using an EVOS FL cell imaging system (Life Technologies, Carlsbad, CA).
Inflammatory protein analysis (ELISA).
Lung samples were harvested in Lysing Matrix D tubes (MP Biomedicals) containing 1.0 ml of PBS with protease inhibitor. Samples were homogenized at 5.0 m/s for 30 s on a FastPrep-24 instrument (MP Biomedicals) before being stored at −80°C. At the time of analysis, homogenized tissues were thawed and centrifuged at >13,000 × g relative centrifugal force (RCF) for 10 min at 4°C. The resulting supernatant was filtered through a 0.22 µm-pore-size filter at 12 × g RCF for 4 min. Inflammatory proteins were measured using a V-Plex proinflammatory Panel 2 (rat) kit (K15059D-1; Meso Scale Discovery) according to the manufacturer’s instructions and read on a Sector Imager 2400 instrument (Meso Scale Discovery).
Nucleic acid extraction.
Samples were prepared as previously described (37). Lung, liver, spleen, and kidney samples designated for nucleic acid extraction were harvested in Lysing Matrix D tubes containing 1.0 ml of TRIzol (Invitrogen). Samples were homogenized at 5.0 m/s for 30 s on a FastPrep-24 instrument before being stored at −80°C. Total RNA was extracted as previously described. Briefly, liquefied samples were spun down at >13,000 × g RCF for 20 min at 4°C to remove tissue debris. Two hundred microliters of chloroform was added to the supernatants, and the reaction mixture was centrifuged again at >13,000 × g RCF for 15 min at 4°C. An equal volume of isopropanol was added to the aqueous phase, and then the reaction mixture was centrifuged at >13,000 × g RCF for 15 min to pellet the precipitated RNA. After removal of the remaining isopropanol, pellets were washed twice with 500 ml of ice-cold 70% ethanol and then resuspended in 30 µl of nuclease-free water. Total RNA was then purified using the “RNA Cleanup” protocol described in the instructions provided for the RNeasy minikit (Qiagen) and stored at −80°C.
Bacterial dissemination.
As organs were originally stored in TRIzol for gene expression analysis studies to be performed in the future, only RNA was available for dissemination analysis. cDNA synthesis was performed on total RNA isolated using iScript Reverse Transcription Supermix (Bio-Rad Laboratories) according to manufacturer’s instructions. The following primers specifically targeting the 16S rRNA gene of each predatory bacterial strain were synthesized: for B. bacteriovorus 109J and HD100 (38), (Forward) 5′-GGAGGCAGCAGTAGGGAATA-3′ and (Reverse) 5′-GCTAGGATCCCTCGTCTTACC-3′; for M. aeruginosavorus ARL-13, (Forward) 5′-GGCTTCACTTTGTCCAGAGC-3′ and (Reverse) 5′ CAGAAAAACGCGAAATCCTC 3′; for K. pneumoniae (39), (Forward) 5′ AGCACAGAGAGCTTGC 3′ and (Reverse) 5′ ACTTTGGTCTTGCGAC 3′. qPCR was performed on the samples in triplicate, with each reaction mixture containing the following components: template (1.0 µl of cDNA synthesized as described above), SsoAdvanced Universal SYBR green Supermix (Bio-Rad Laboratories), and a 500 nM (for 109J and Micavibrio) or 900 nM (for HD100) concentration of each primer (synthesized at the Rutgers New Jersey Medical School Molecular Resource Facility). A CFX384 Touch real-time PCR detection system (Bio-Rad Laboratories) was used with the following protocol: 50°C for 2 min (1 cycle), 95°C for 10 min (1 cycle), 95°C for 15 s and 60°C for 1 min (40 cycles), and 95°C for 15 s, 60°C for 15 s, and 95°C for 15 s (1 cycle). For each qPCR run, a 10-fold dilution series of the standard (purified DNA from each predatory strain) was assessed in triplicate to validate qPCR performance and facilitate quantification (E = 97%, R2 = 0.970, slope = −3.397). In addition, each qPCR run included negative controls (no template). 16S rRNA copy numbers were calculated using “Calculator for determining the number of copies of a template” (URI Genomics and Sequencing Center; http://cels.uri.edu/gsc/cndna.html) (40).
Pathogen inoculation and treatment.
Animals were anesthetized and inoculated intranasally with 50 µl of K. pneumoniae as previously described. A 50-µl volume of predatory bacteria was administered to rats at 30 min and 6, 12, and 18 h postinfection for a total of four doses over a 24-h period. At 24 h postinoculation, animals were euthanized and lung samples harvested for histological examination and CFU plating. Lung samples designated for CFU were stored in Lysing Matrix D tubes containing 1.0 ml of PBS and placed on ice. Samples were immediately homogenized at 6.0 m/s for 1 min on a FastPrep-24 instrument. Liquefied samples were then serially diluted and plated on MacConkey agar in order to determine K. pneumoniae concentrations. Histological examination was performed as described above.
Statistical analysis.
ELISA data are presented as means ± standard errors of the means; significant differences between the data from the treated samples and the data from the respective controls were determined using analysis of variance (ANOVA). K. pneumoniae reduction data are presented as medians; significant differences between treatment groups and respective controls were analyzed using the Mann-Whitney test. A P value of <0.05 was considered significant. Graphs were prepared and statistical analyses were performed using GraphPad Prism 6.05.
ACKNOWLEDGMENTS
We are grateful to Juli-Anne Royes Russo (Connell laboratory, Rutgers New Jersey Medical School) for administrative help in managing different aspects of the study. We also are appreciative to Nadeem Obaydou (Connell laboratory, Rutgers New Jersey Medical School) and Suresh Bhatt (Comparative Medicine Resources, Rutgers New Jersey Medical School) for their support in working with the animals.
Research was sponsored by the U.S. Army Research Office and the Defense Advanced Research Projects Agency (DARPA) and was accomplished under cooperative agreement number W911NF-15-2-0036.
The views and conclusions contained in this document are ours and should not be interpreted as representing the official policies, either expressed or implied, of the Army Research Office, DARPA, or the United States Government. The United States Government is authorized to reproduce and distribute reprints for government purposes notwithstanding any copyright notation hereon.
K.S., N.D.C., and D.E.K. conceived and designed the experiments. K.S., E.S., C.T., M.Z., S.S., S.G., S.D., and O.O. performed the experiments. K.S., C.T., and J.R. analyzed the data. K.S. drafted the manuscript. K.S., N.D.C., and D.E.K. supervised the study and revised the manuscript. All of us reviewed the manuscript.
Funding Statement
Research was sponsored by the U.S. Army Research Office and the Defense Advanced Research Projects Agency and was accomplished under Cooperative Agreement Number W911NF-15-2-0036. The views and conclusions contained in this document are those of the authors and should not be interpreted as representing the official policies, either expressed or implied, of the Army Research Office, DARPA, or the U.S. Government. The U.S. Government is authorized to reproduce and distribute reprints for Government purposes notwithstanding any copyright notation hereon.
Footnotes
Citation Shatzkes K, Singleton E, Tang C, Zuena M, Shukla S, Gupta S, Dharani S, Onyile O, Rinaggio J, Connell ND, Kadouri DE. 2016. Predatory bacteria attenuate Klebsiella pneumoniae burden in rat lungs. mBio 7(6):e01847-16. doi:10.1128/mBio.01847-16.
REFERENCES
- 1.Fleming A. 1929. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br J Exp Pathol 10:226–236. [Google Scholar]
- 2.WHO 2014. Antimicrobial resistance: global report on surveillance 2014. WHO, Geneva Switzerland. [Google Scholar]
- 3.Stolp H, Starr MP. 1963. Bdellovibrio bacteriovorus gen. et sp. n., a predatory, ectoparasitic, and bacteriolytic microorganism. Antonie Van Leeuwenhoek 29:217–248. doi: 10.1007/BF02046064. [DOI] [PubMed] [Google Scholar]
- 4.Lambina VA, Afinogenova AV, Romay Penobad Z, Konovalova SM, Andreev LV. 1983. New species of exoparasitic bacteria of the genus Micavibrio infecting gram-positive bacteria. Mikrobiologiia 52:777–780. [PubMed] [Google Scholar]
- 5.Chanyi RM, Koval SF. 2014. Role of type IV pili in predation by Bdellovibrio bacteriovorus. PLoS One 9:e113404. doi: 10.1371/journal.pone.0113404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans KJ, Lambert C, Sockett RE. 2007. Predation by Bdellovibrio bacteriovorus HD100 requires type IV pili. J Bacteriol 189:4850–4859. doi: 10.1128/JB.01942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahmoud KK, Koval SF. 2010. Characterization of type IV pili in the life cycle of the predator bacterium Bdellovibrio. Microbiology 156:1040–1051. doi: 10.1099/mic.0.036137-0. [DOI] [PubMed] [Google Scholar]
- 8.Lambina VA, Afinogenova AV, Romai Penabad S, Konovalova SM, Pushkareva AP. 1982. Micavibrio admirandus gen. et sp. nov. Mikrobiologiia 51:114–117. [PubMed] [Google Scholar]
- 9.Wang Z, Kadouri DE, Wu M. 2011. Genomic insights into an obligate epibiotic bacterial predator: Micavibrio aeruginosavorus ARL-13. BMC Genomics 12:453. doi: 10.1186/1471-2164-12-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadouri DE, To K, Shanks RM, Doi Y. 2013. Predatory bacteria: a potential ally against multidrug-resistant gram-negative pathogens. PLoS One 8:e63397. doi: 10.1371/journal.pone.0063397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dashiff A, Junka RA, Libera M, Kadouri DE. 2011. Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J Appl Microbiol 110:431–444. doi: 10.1111/j.1365-2672.2010.04900.x. [DOI] [PubMed] [Google Scholar]
- 12.Kadouri D, O’Toole GA. 2005. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl Environ Microbiol 71:4044–4051. doi: 10.1128/AEM.71.7.4044-4051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadouri D, Venzon NC, O’Toole GA. 2007. Vulnerability of pathogenic biofilms to Micavibrio aeruginosavorus. Appl Environ Microbiol 73:605–614. doi: 10.1128/AEM.01893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dwidar M, Monnappa AK, Mitchell RJ. 2012. The dual probiotic and antibiotic nature of Bdellovibrio bacteriovorus. BMB Rep 45:71–78. doi: 10.5483/BMBRep.2012.45.2.71. [DOI] [PubMed] [Google Scholar]
- 15.Shanks RM, Davra VR, Romanowski EG, Brothers KM, Stella NA, Godboley D, Kadouri DE. 2013. An eye to a kill: using predatory bacteria to control gram-negative pathogens associated with ocular infections. PLoS One 8:e66723. doi: 10.1371/journal.pone.0066723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verklova ZS. 1973. Study of the virulence, toxicity and immunogenicity of different strains of Bdellovibrio bacteriovorus. Gig Sanit 38:10–13. (In Russian.) [PubMed] [Google Scholar]
- 17.Westergaard JM, Kramer TT. 1977. Bdellovibrio and the intestinal flora of vertebrates. Appl Environ Microbiol 34:506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atterbury RJ, Hobley L, Till R, Lambert C, Capeness MJ, Lerner TR, Fenton AK, Barrow P, Sockett RE. 2011. Effects of orally administered Bdellovibrio bacteriovorus on the well-being and Salmonella colonization of young chicks. Appl Environ Microbiol 77:5794–5803. doi: 10.1128/AEM.00426-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shatzkes K, Chae R, Tang C, Ramirez GC, Mukherjee S, Tsenova L, Connell ND, Kadouri DE. 2015. Examining the safety of respiratory and intravenous inoculation of Bdellovibrio bacteriovorus and Micavibrio aeruginosavorus in a mouse model. Sci Rep 5:12899. doi: 10.1038/srep12899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shemesh Y, Jurkevitch E. 2004. Plastic phenotypic resistance to predation by Bdellovibrio and like organisms in bacterial prey. Environ Microbiol 6:12–18. doi: 10.1046/j.1462-2920.2003.00530.x. [DOI] [PubMed] [Google Scholar]
- 21.McGann P, Snesrud E, Maybank R, Corey B, Ong AC, Clifford R, Hinkle M, Whitman T, Lesho E, Schaecher KE. 2016. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob Agents Chemother 60:4420–4421. doi: 10.1128/AAC.01103-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Czaplewski L, Bax R, Clokie M, Dawson M, Fairhead H, Fischetti VA, Foster S, Gilmore BF, Hancock RE, Harper D, Henderson IR, Hilpert K, Jones BV, Kadioglu A, Knowles D, Ólafsdóttir S, Payne D, Projan S, Shaunak S, Silverman J, Thomas CM, Trust TJ, Warn P, Rex JH. 2016. Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect Dis 16:239–251. doi: 10.1016/S1473-3099(15)00466-1. [DOI] [PubMed] [Google Scholar]
- 23.Brunetti J, Falciani C, Bracci L, Pini A. 13 July 2016. Models of in-vivo bacterial infections for the development of antimicrobial peptide-based drugs. Curr Top Med Chem https://www.ncbi.nlm.nih.gov/pubmed/27411321. [DOI] [PubMed] [Google Scholar]
- 24.Guralp SA, Murgha YE, Rouillard JM, Gulari E. 2013. From design to screening: a new antimicrobial peptide discovery pipeline. PLoS One 8:e59305. doi: 10.1371/journal.pone.0059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sybesma W, Zbinden R, Chanishvili N, Kutateladze M, Chkhotua A, Ujmajuridze A, Mehnert U, Kessler TM. 2016. Bacteriophages as potential treatment for urinary tract infections. Front Microbiol 7:465. doi: 10.3389/fmicb.2016.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furusawa T, Iwano H, Hiyashimizu Y, Matsubara K, Higuchi H, Nagahata H, Niwa H, Katayama Y, Kinoshita Y, Hagiwara K, Iwasaki T, Tanji Y, Yokota H, Tamura Y. 2016. Phage therapy is effective in a mouse model of bacterial equine keratitis. Appl Environ Microbiol 82:5332–5339 doi: 10.1128/aem.01166-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JS, Cho DH, Park M, Chung WJ, Shin D, Ko KS, Kweon DH. 2016. CRISPR/Cas9-mediated re-sensitization of antibiotic-resistant Escherichia coli harboring extended-spectrum beta-lactamases. J Microbiol Biotechnol 26:394–401. doi: 10.4014/jmb.1508.08080. [DOI] [PubMed] [Google Scholar]
- 28.Shahbazi Dastjerdeh M, Kouhpayeh S, Sabzehei F, Khanahmad H, Salehi M, Mohammadi Z, Shariati L, Hejazi Z, Rabiei P, Manian M. 2016. Zinc finger nuclease: a new approach to overcome beta-lactam antibiotic resistance. Jundishapur J Microbiol 9:e29384. doi: 10.5812/jjm.29384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartlett KH, McCray PB Jr, Thorne PS. 2003. Novispirin G10-induced lung toxicity in a Klebsiella pneumoniae infection model. Antimicrob Agents Chemother 47:3901–3906. doi: 10.1128/AAC.47.12.3901-3906.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenberger MJ, Kunkel SL, Strieter RM, Lukacs NW, Bramson J, Gauldie J, Graham FL, Hitt M, Danforth JM, Standiford TJ. 1996. IL-12 gene therapy protects mice in lethal Klebsiella pneumoniae. J Immunol 157:3006–3012. [PubMed] [Google Scholar]
- 31.Schwudke D, Linscheid M, Strauch E, Appel B, Zahringer U, Moll H, Muller M, Brecker L, Gronow S, Lindner B. 2003. The obligate predatory Bdellovibrio bacteriovorus possesses a neutral lipid A containing alpha-d-mannoses that replace phosphate residues: similarities and differences between the lipid as and the lipopolysaccharides of the wild type strain B. bacteriovorus HD100 and its host-independent derivative HI100. J Biol Chem 278:27502–27512. doi: 10.1074/jbc.M303012200. [DOI] [PubMed] [Google Scholar]
- 32.Mossel DAA, Mengerink WHJ, Scholts HH. 1962. Use of a modified MacConkey agar medium for the selective growth and enumeration of Enterobacteriaceae. J Bacteriol 84:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam SJ, O’Brien-Simpson NM, Pantarat N, Sulistio A, Wong EHH, Chen Y-Y, Lenzo JC, Holden JA, Blencowe A, Reynolds EC, Qiao GG. 2016. Combating multidrug-resistant gram-negative bacteria with structurally nanoengineered antimicrobial peptide polymers. Nat Microbiol 1:16162. [DOI] [PubMed] [Google Scholar]
- 34.Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, Keller H, Lambert C, Evans KJ, Goesmann A, Meyer F, Sockett RE, Schuster SC. 2004. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- 35.Lawlor MS, Hsu J, Rick PD, Miller VL. 2005. Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol Microbiol 58:1054–1073. doi: 10.1111/j.1365-2958.2005.04918.x. [DOI] [PubMed] [Google Scholar]
- 36.Shilo M, Bruff B. 1965. Lysis of gram-negative bacteria by host-independent ectoparasitic Bdellovibrio bacteriovorus isolates. J Gen Microbiol 40:317–328. doi: 10.1099/00221287-40-3-317. [DOI] [PubMed] [Google Scholar]
- 37.Bhatt K, Kim A, Kim A, Mathur S, Salgame P. 2013. Equivalent functions for B7.1 and B7.2 costimulation in mediating host resistance to Mycobacterium tuberculosis. Cell Immunol 285:69–75. doi: 10.1016/j.cellimm.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Van Essche M, Sliepen I, Loozen G, Van Eldere J, Quirynen M, Davidov Y, Jurkevitch E, Boon N, Teughels W. 2009. Development and performance of a quantitative PCR for the enumeration of Bdellovibrionaceae. Environ Microbiol Rep 1:228–233. doi: 10.1111/j.1758-2229.2009.00034.x. [DOI] [PubMed] [Google Scholar]
- 39.Kurupati P, Chow C, Kumarasinghe G, Poh CL. 2004. Rapid detection of Klebsiella pneumoniae from blood culture bottles by real-time PCR. J Clin Microbiol 42:1337–1340. doi: 10.1128/JCM.42.3.1337-1340.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Preiser V, Goetsch D, Sulyok M, Krska R, Mach RL, Farnleitner A, Brunner K. 2015. The development of a multiplex real-time PCR to quantify Fusarium DNA of trichothecene and fumonisin producing strains in maize. Anal Methods 7:1358–1365. doi: 10.1039/C4AY02581D. [DOI] [Google Scholar]