Abstract
A novel ligand bearing a central piperidinyl sulfonamide group, N(SO2pip)dpa, and its corresponding Re tricarbonyl complex, [Re(CO)3(N(SO2pip)dpa)]+, have been synthesized in good yield. The methylene CH2 signal seen as a singlet (4.54 ppm) in a 1H NMR spectrum of the ligand in DMSO-d 6 appears as two doublets (5.39, 5.01 ppm) in a spectrum of the [Re(CO)3(N(SO2pip)dpa)]+ complex and confirms the presence of magnetically nonequivalent protons upon coordination to Re. Structural results revealed that the Re–N bond lengths fall within the normal range establishing coordination of ligand to metal. The presence of intraligand π → π ⁎ and n → π ⁎ transitions is indicated by the absorption peaks around 200–250 nm in UV-visible spectra. Absorption peaks in UV-visible spectra around 300 nm for metal complexes were identified as MLCT transitions. The S–N stretch observed as a strong peak at 923 cm−1 for N(SO2pip)dpa appeared at a shorter frequency, at 830 cm−1 in an FTIR spectrum of the [Re(CO)3(N(SO2pip)dpa)]+. The intense fluorescence displayed by the N(SO2pip)dpa ligand has quenched upon coordination to Re. Relatively low IC50 values given by human breast cancer cells, MCF-7, (N(SO2pip)dpa = 139 μM, [Re(CO)3(N(SO2pip)dpa)]+ = 360 μM) indicate that N(SO2pip)dpa and [Re(CO)3(N(SO2pip)dpa)]+ are promising novel compounds that can be further investigated on their usage as potential anticancer agents.
1. Introduction
Historically, fac-Re(CO)3L complexes have provided a good model system to interpret the nature of its analogous 99mTc agents which are widely used in imaging. The fac-[Re(CO)3(H2O)3]+ precursor may be easily utilized [1] for the synthesis of new organometallic complexes having suitable pharmaceutical properties. Furthermore, the triaquatricarbonyl complex of 99mTc, which is the group seven congener of rhenium, is commercially available as a kit preparation [2], enabling the easy simulation of the nonradioactive rhenium complexes [3]. The use of nonradioactive rhenium facilitates the synthesis of the metal complexes rather than handling radioactive metal tracers.
Metal complexes (M = 99mTc, Re) bearing symmetrical linear tridentate ligands have been reported to possess lower bioaccumulation than complexes bearing bidentate or monodentate ligands [2], resulting in fewer side effects when used on the long term. Rhenium, in its lowest oxidation state, is less sensitive to chemical attack by the reactive species that may be present in cellular environment [2]. Facially arranged carbonyl ligands cover one face of the distorted octahedral complex thus protecting the metal center. Metal carbon bonds are greatly stabilized by strong backdonation from the carbonyl ligand to the metal. As rhenium is a soft metal center, it prefers soft donors such as nitrogen and tridentate metal complexes with nitrogen donors are widely used [2, 4, 5].
Among various types of chronic human diseases, certain types of cancers are widely treated by organometallic complexes [6, 7]. Breast cancer is one of the most abundant types of cancer in recent times. However, only a few metallopharmaceuticals are available for diagnostic and therapeutic purposes for human breast cancer [7, 8].
Sigma receptors are a distinct class of proteins that are mostly found in the central nervous system [9]. Sigma receptors are virtually absent in healthy human breast cells; however, they appear in a high density in breast tumor biopsy tissue [9, 10]. Proliferative human breast cancer cells express approximately ten times more sigma receptors per cell than the quiescent cells [9]. Therefore, high densities of sigma receptors in rapidly proliferating breast cancer cells make them ideal candidates for diagnosis as well as therapy of breast cancer. Due to the multiple binding sites containing lipophilic sterol binding domains on the protein molecule, various types of exogenous ligands, such as several derivatives of piperidine, piperazine [11, 12], benzamides [13], and alkylamine [14], have been reported as potential pharmaceuticals for therapy of human breast cancer, mainly because these types of compounds preferentially bind with sigma receptors [10].
To the best of our knowledge, there are only a few radiotracers in which the piperidinyl group has been utilized towards targeting sigma receptors; Satpati et al. have synthesized 99mTc based cancer diagnostic agents [10], Choi et al. have developed a 99mTc labeled sigma-2 receptor-specific ligand as a potential breast tumor imaging agent [9], and Caveliers et al. have utilized N-[2-(1′-piperidinyl)ethyl]-3-123I-iodo-4-methoxybenzamide [8]. Only the latter has entered clinical trials for the diagnosis of patients with primary breast cancer.
We based our study on the fact that fac-[Re(CO)3(N(R)dpa)]+ complexes have been reported to possess promising biomedical properties where R equals a suitable substituent linked by a N–C bond at the central nitrogen in dipicolylamine [15]. Furthermore, the tertiary sulfonamide linkage has recently been utilized to propose a new approach to radiopharmaceutical bioconjugation [5]. We also envisage that the target specificity may be increased by incorporating a piperidinyl derivative into the novel compounds.
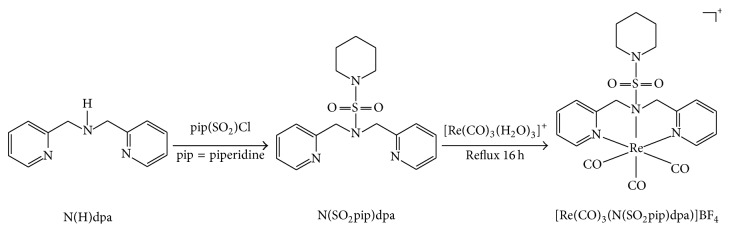
Our goal has been to synthesize a novel ligand and its rhenium tricarbonyl complex having possible therapeutic potential, with the long term goal of extending this study to 99mTc complexes towards the eventual diagnosis of breast cancer. Thus, in this study we report the synthesis and characterization of a novel piperidinyl appended dipicolylamine ligand and its corresponding Re tricarbonyl complex and go one step further to report their potential applicability in biological systems as anticancer agents (Scheme 1).
Scheme 1.
Synthetic routes for the preparation of N(SO2pip)dpa and [Re(CO)3(N(SO2pip)dpa)]BF4.
2. Experimental
2.1. Materials and Methods
Re2(CO)10, AgOTf, piperidine-1-sulfonyl chloride, di(2-picolyl)amine, NaBF4, anhydrous sodium sulphate, acetone, methanol, dichloromethane, chromasolv water, and dioxane were obtained from Sigma Aldrich, USA. Human breast cancer cell line MCF-7 was obtained from American Type Culture Collection. All the solvents and chemicals were of analytical grade and were used as received, without further purification.
2.2. NMR Measurements
1H NMR spectra were recorded in DMSO-d 6 on a Bruker 400 MHz spectrometer. Peak positions are relative to trimethylsilane (TMS) as reference. All NMR data were processed with TopSpin 3.2 and MestReC software.
2.3. X-Ray Data Collection and Structure Determination
Single crystals were placed in a cooled nitrogen gas stream at 90 K on a Bruker Kappa Apex-II DUO diffractometer equipped with Cu Kα (λ = 1.54178 Ǻ) radiation (N(SO2pip)dpa ligand) or Mo Kα radiation (λ = 0.71073 Å) ([Re(CO)3(N(SO2pip)dpa)]BF4 complex). Refinement was performed by full-matrix least squares methods using SHELXL (Sheldrick (2008)) [16], with H atoms in idealized positions. Molecular graphics are drawn using ORTEP-3 for windows [17].
2.4. UV-Visible Spectroscopy
Electronic spectra for ligand and metal complex were obtained on Spectro UV-VIS autoversion 3.10, UV-2602 spectrophotometer. The spectral range was 190 nm–1100 nm. Spectra were obtained in methanol with base line correction. Spectral data were processed with UV WIN software.
2.5. FTIR Analysis
FTIR spectra were recorded on a Thermo Scientific Nicolet iS10 spectrophotometer. ATR spectra were obtained within the 4000–600 cm−1 spectral range. Spectral data were processed with OMNIC software.
2.6. Fluorometric Analysis
Excitation and emission spectra for ligand and metal complex were obtained in methanol and acetonitrile on a Thermo Scientific Lumina spectrophotometer. A 150 W Xenon lamp was used as the excitation source. Spectral data were processed with Luminous software.
2.7. Synthesis
In order to synthesize metal complexes, [Re(CO)3(H2O)3]OTf precursor was prepared using Re2(CO)10 as the starting material according to a known procedure [1].
2.7.1. N(SO2pip)dpa Ligand
A solution of piperidine-1-sulfonyl chloride (0.034 g, 5 mmol) in 25 mL of dioxane was added dropwise over a period of 2 hours to a solution of N(H)dpa (0.057 g, 10 mmol) in 100 mL of dioxane at 20°C. The reaction mixture was stirred at room temperature for 24 hours and then filtered to remove any precipitate. Thereafter, the dioxane was completely removed by rotary evaporation. Slightly acidic water (30 mL, pH ~ 5) was added to the resulting compound, and the product was extracted into CH2Cl2 (2 × 25 mL). The CH2Cl2 extracts were combined, washed with water (2 × 25 mL), and taken to dryness. Grey color, plate-like crystals were obtained. 1H NMR signals (ppm) in DMSO-d 6 are 8.49(d, 2H, H6/H6′), 7.75(t, 2H, H4/H4′), 7.35(d, 2H, H3/H3′), 7.27(t, 2H, H5/H5′), 4.53(s, 2CH2), 3.09(s, 2H, Ha), and 1.40(s, 2H, Hb).
2.7.2. [Re(CO)3(N(SO2pip)dpa)]BF4 Complex
A solution of N(SO2pip)dpa (0.0346 g, 0.1 mmol) in 3 mL methanol was treated with aqueous [Re(CO)3(H2O)3]OTf (0.1 mmol, 4 mL). Methanol (3 mL) was added to dissolve the precipitate which formed. Acidity of the solution was measured (pH ~ 6) and the clear solution was heated at reflux for 16 h. A slight excess of NaBF4 was added to the clear, brownish yellow color solution. The solution was then allowed to develop crystals over three days at room temperature after which block-like, colorless crystals were obtained. 1H NMR signals (ppm) in DMSO-d 6 are 8.84(d, 2H, H6/H6′), 8.06(t, 2H, H4/H4′), 7.61(d, 2H, H3/H3′), 7.47(t, 2H, H5/H5′), 5.39(d, endo-H), 5.01(d, exo-H), 3.68(s, 2H, Ha), and 1.68(s, 2H, Hb).
2.8. Biological Assays
2.8.1. Cytotoxicity Assessment
The novel ligand and its metal complex were investigated for their cytotoxicity against MCF-7 (breast cancer) cells. Cells were cultured in 96-well culture plates and exposed to 25 μg/mL, 50 μg/mL, 100 μg/mL, 200 μg/mL, and 400 μg/mL concentrations of ligand and complex for 24 h, 48 h, and 72 h, respectively, and cytotoxicity was assessed by Sulforhodamine B assay [18]. All exposures were carried out in triplicate. Briefly, the cell supernatant was completely removed and washed with phosphate buffer solution. Trichloroacetic acid (50%, 25 μL) was added on top of fetal bovine serum-free fresh medium (200 μL) to make final concentration of 10% trichloroacetic acid and was incubated at 4°C for one hour former to the SRB assay. The plate was then washed with five washing cycles with water and dried completely. An aliquot of 100 μL of 0.4% Sulforhodamine B dissolved in 1% trichloroacetic acid, was added to each well, and was allowed to stain for 15 minutes. The plate was again washed with five washing cycles to remove unbound dye using 1% (vol/vol) acetic acid after removing the stain. The protein bound dye was solubilized with trisbase (10 mM, pH 7.5, 200 μL), after air-drying. The plates were then shaken for 60 minutes to homogenize the dye solution. The absorbance was then measured at 540 nm using Synergy HTBioTek microplate reader. The percentage viability was calculated by the equation given below:
| (1) |
3. Results and Discussion
3.1. Structural Results
Crystal data and details of the structural refinement for N(SO2pip)dpa and [Re(CO)3(N(SO2pip)dpa)]+ are summarized in Table 1. Crystallographic data are deposited with the Cambridge Crystallographic Data Centre under deposition numbers CCDC 1495607 and 1495608.
Table 1.
Crystal data and structure refinement for N(SO2pip)dpa and [Re(CO)3(N(SO2pip)dpa)]+.
| Crystal data | N(SO2pip)dpa | [Re(CO)3(N(SO2pip)dpa)]+ |
|---|---|---|
| Empirical formula | C17H22N4O2S | C20H22N4O5ReS·BF40.682(CH3OH)·0.318(H2O) |
| Mr | 346.44 | 731.03 |
| Crystal system | Triclinic | Monoclinic |
| Space group | P21/c | |
| Unit cell dimensions | ||
| a (Å) | 8.6557 (3) | 10.8932 (4) |
| b (Ǻ) | 10.1132 (3) | 11.7517 (5) |
| c (Ǻ) | 10.4022 (3) | 20.7973 (8) |
| α (deg) | 71.979 (1) | — |
| β (deg) | 81.262 (1) | 102.260 (2) |
| γ (deg) | 74.382 (1) | — |
| V (Å3) | 831.59 (5) | 2601.62 (18) |
| T (K) | 90 | 90 |
| Z | 2 | 4 |
| ρ calc (g/cm3) | 1.384 | 1.866 |
| abs coeff (mm−1) | 1.88 | 4.82 |
| 2θ max (deg) | 135 | 66.2 |
| R[F 2 > 2σ(F 2)] | 0.030 | 0.027 |
| wR(F2) | 0.086 | 0.058 |
| Res density | 0.29, −0.41 | 1.50, −1.65 |
| Data/parameters | 2819/217 | 9917/354 |
| Radiation type | Cu Kα | Mo Kα |
| Radiation wavelength/(Å) | 1.54178 | 0.71073 |
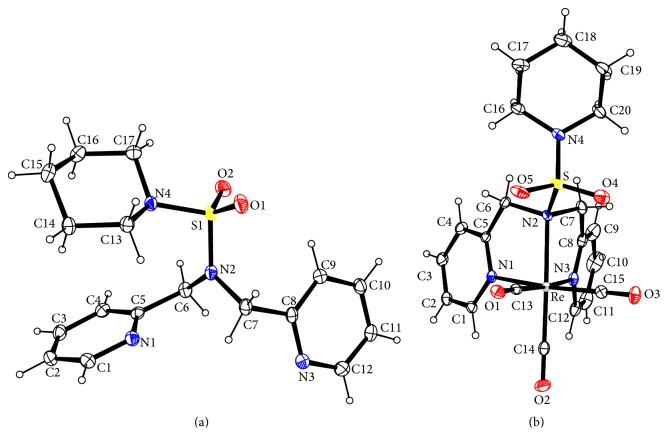
The uncoordinated ligand (Figure 1) possesses S–N(2) bond length of 1.6194 (11) Å (Table 2). However, upon forming the metal complex, the S–N(2) bond length has increased (1.777 (2) Å). This may be attributed to the donation of lone electron pair on sulfonamide nitrogen to Re, which results in lowering of the strength of the S–N bond. Previous studies have reported that the S–N distance is ~1.73–1.76 Å [5] when Re–N(sulfonamide) bond is strong [19]. The above observation provides evidence that the complexation of the ligand with Re has occurred successfully. Weakly coordinated or noncoordinated sulfonamide groups have considerable double character. This is because of the conjugation of lone electron pair of the sulfonamide nitrogen with S=O across the S–N bond [19].
Figure 1.
ORTEP plot of N(SO2pip)dpa (a) and the cation in [Re(CO)3(N(SO2pip)dpa)]+ (b). Thermal ellipsoids are drawn with 50% probability.
Table 2.
Selected bond distances (Å) for N(SO2pip)dpa.
| Bond length (Å) | |
|---|---|
| S1–O1 | 1.4363 (10) |
| S1–O2 | 1.4369 (10) |
| S1–N2 | 1.6194 (11) |
| S1–N4 | 1.6329 (11) |
| N1–C5 | 1.3391 (18) |
| N1–C1 | 1.3422 (18) |
| N3–C8 | 1.3373 (18) |
| N3–C12 | 1.3459 (18) |
| N2–C7 | 1.4657 (16) |
| N2–C6 | 1.4615 (16) |
| N4–C13 | 1.4848 (16) |
| N4–C17 | 1.4838 (17) |
In the N(SO2pip)dpa ligand, planar geometry is observed at N2 nitrogen which is sp2 hybridized. Therefore, C6, C7, and S species are oriented in an angle of ~120° to the central N2 (Table 3). All the other bond angles of the ligand lie within the normal values depending on their hybridization.
Table 3.
Selected bond angles (deg) for N(SO2pip)dpa.
| Bond angle (deg) | |
|---|---|
| O1–S1–O2 | 116.88 (6) |
| O1–S1–N2 | 106.19 (6) |
| O2–S1–N2 | 111.30 (6) |
| O1–S1–N4 | 113.83 (6) |
| O2–S1–N4 | 106.15 (6) |
| N2–S1–N4 | 101.48 (5) |
| N2–C6–C5 | 113.65 (10) |
| N1–C5–C4 | 122.65 (12) |
| N1–C5–C6 | 116.86 (11) |
| C4–C5–C6 | 120.47 (12) |
| C5–N1–C1 | 117.42 (11) |
| C8–N3–C12 | 117.28 (11) |
| C6–N2–C7 | 118.01 (10) |
| C6–N2–S1 | 118.33 (9) |
| C7–N2–S1 | 122.54 (9) |
| C17–N4–C13 | 113.03 (10) |
In the [Re(CO)3(N(SO2pip)dpa)]+ complex, the coordination geometry at Re exhibits a pseudo octahedral structure (Figure 1). This deviation may be due to the facial arrangement of tridentate chelating unit which leads to forming two strained, five-membered rings at Re resulting in a N–Re–N bond angle < 90°. The remaining three coordination sites of the opposite face of the octahedron are occupied by three carbonyl ligands.
The distance of new bonds formed between Re and nitrogen atoms in dipicolylamine lies within the range of normal values. Generally, the distance between Re and sp2 hybridized nitrogen in a Re complex with prototypical NNN donor ligand is 2.14–2.18 Å [4, 20]. The coordination bond between Re and sp2 hybridized nitrogen in the pyridyl rings which are denoted as N1 and N3 (Figure 1) exhibits a bond distance close to that of the prototypical bond (Table 4). Similarly, the coordination bond between Re and sp3 hybridized tertiary nitrogen (2.251 (2) Å), though exceeding the normal range (2.23–2.29 Å) [4, 20], is less than the Re–N2 distance of 2.2826 (16) Å of [Re(CO)3(N(SO2Me)dpa]PF6 [5] and confirms rehybridization of sulfonamide nitrogen from sp2 to sp3 when bound to Re. Further evidence for the rehybridization of sulfonamide nitrogen from sp2 to sp3 upon binding to Re is provided by the fact that the bond angles of this tertiary sulfonamide nitrogen are closer to 109.5° (Table 5). All the other bond angles lie within the normal range of reported compounds [5].
Table 4.
Selected bond distances (Ǻ) for [Re(CO)3(N(SO2pip)dpa)]+.
| Bond length (Ǻ) | |
|---|---|
| Re–N1 | 2.1746 (19) |
| Re–N2 | 2.251 (2) |
| Re–N3 | 2.179 (2) |
| Re–C13 | 1.919 (3) |
| Re–C14 | 1.904 (3) |
| Re–C15 | 1.934 (2) |
| S–O4 | 1.425 (2) |
| S–O5 | 1.422 (2) |
| S–N2 | 1.777 (2) |
| S–N4 | 1.588 (2) |
Table 5.
Selected bond angles (deg) for [Re(CO)3(N(SO2pip)dpa)]+.
| Bond angle (deg) | |
|---|---|
| N1–Re–N3 | 77.55 (7) |
| N1–Re–N2 | 77.98 (8) |
| N3–Re–N2 | 76.58 (8) |
| C13–Re–C15 | 88.25 (10) |
| C13–Re–N1 | 98.09 (9) |
| N2–Re–C15 | 97.00 (9) |
| N2–Re–C14 | 171.70 (9) |
| N2–Re–C13 | 98.43 (9) |
| C14–Re–N3 | 96.37 (10) |
| C15–Re–N3 | 95.81 (9) |
| C6–N2–Re | 108.74 (14) |
| C7–N2–Re | 107.06 (15) |
| S–N2–Re | 113.31 (9) |
| N1–Re–C15 | 172.39 (9) |
| N1–Re–C14 | 96.36 (9) |
3.2. 1H NMR Analysis
N(SO2pip)dpa and [Re(CO)3(N(SO2pip)dpa)]+ were characterized using 1H NMR spectroscopy in DMSO-d 6. Peaks related to residual solvents were also identified [21, 22]. Signals related to the dipicolylamine were assigned according to previously reported data of related compounds [5, 23]. Protons attached to the pyridyl nitrogen (H6/6′) are more deshielded and thereby signals are located downfield in the spectrum (Figure 2). Protons in the piperidinyl ring were assigned with the aid of reported spectra of related compounds [10]. The signal for protons in the methylene groups of the N(SO2pip)dpa appears as a singlet at (4.53 ppm) because the protons attached to the two methylene groups are magnetically equivalent due to free rotation. Singlet peaks at 3.3, 2.5, 5.8, and 3.5 ppm are due to residual solvents including water, DMSO, dichloromethane, and dioxane, respectively [21, 22].
Figure 2.
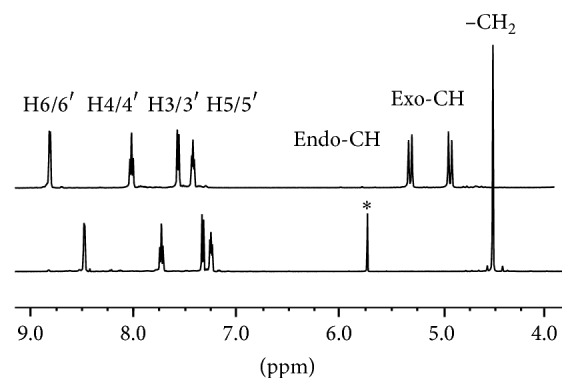
1H NMR spectra in DMSO-d 6 of N(SO2pip)dpa (lower) and [Re(CO)3(N(SO2pip)dpa)]+ (upper). ∗Peak due to trace amounts of CH2Cl2 solvent.
According to the spatial arrangement of the atoms in [Re(CO)3(N(SO2pip)dpa)]+, the methylene protons are projected towards and away from the carbonyl ligands attached to rhenium and may be designated as endo-H and exo-H, respectively. Therefore, those methylene protons are not magnetically equivalent. Hence, the singlet at 4.53 ppm related to the CH2 group of the free ligand appears as two doublets further downfield upon binding to the metal (Figure 2). All the signals related to protons attached to pyridyl nitrogen have shifted downfield upon binding to the metal (Table 6). These shifts of peaks may be attributed to the electron withdrawing inductive effect as a result of the formation of direct Re–N bond and provide strong evidence of the formation of the metal complex.
Table 6.
Comparison of the 1H NMR shifts of N(SO2pip)dpa ligand and [Re(CO)3(N(SO2pip)dpa)]+ complex in DMSO-d 6.
| H6/6′ | H5/5′ | H4/4′ | H3/3′ | −CH2 | |
|---|---|---|---|---|---|
| N(SO2pip)dpa | 8.49 (d) | 7.27 (t) | 7.75 (t) | 7.35 (d) | 4.53 (s) |
| [Re(CO)3(N(SO2pip)dpa)]+ | 8.84 (d) | 7.47 (t) | 8.06 (t) | 7.61 (d) | 5.39 (d), 5.01 (d) |
3.3. UV-Visible Analysis
The absorption peak at 307 nm in [Re(CO)3(N(SO2pip)dpa)]+ may be attributed to MLCT transition (Figure 3). Rhenium tricarbonyl complexes with suitable ligands display MLCT transitions [24]. Other high energy absorption peaks may be due to intraligand π → π ∗ and n → π ∗ transitions [24, 25].
Figure 3.
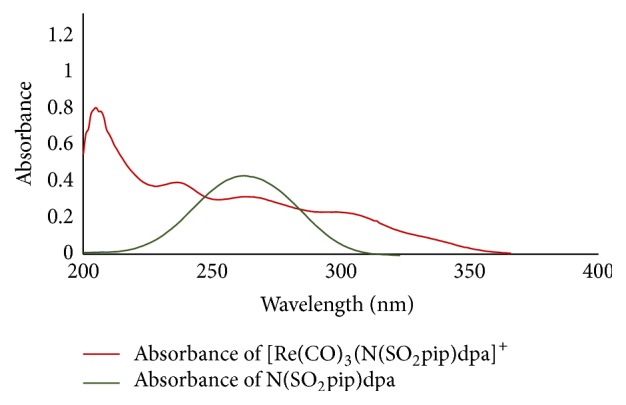
UV-visible spectra of N(SO2pip)dpa and [Re(CO)3(N(SO2pip)dpa)]+.
3.4. FTIR Analysis
Literature has greatly aided in assigning FTIR spectra of novel ligand and complex. In an FTIR spectrum of N(SO2pip)dpa, a short absorption band appears at 3077 cm−1 due to the asymmetric stretching vibration of aliphatic systems [26]. A narrow and sharp absorption band at 2942 cm−1 is attributed to the C–H asymmetric stretching vibration of aliphatic systems [26]. A strong absorption band at 1136 cm−1 may be due to the symmetric stretching vibration of S=O bond of the sulfonamide group [26]. A collection of absorption peaks in the range between 1294 cm−1 and 1590 cm−1 is due to the symmetric and asymmetric stretching vibrations of C=C bonds [26] in aromatic rings and C=N stretching vibration mode in the ligand [26]. Stretching vibration due to S–N bond is identified as the strong absorption peak at 923 cm−1 [27].
Most of the peaks attributed to ligand also appear in a spectrum of the [Re(CO)3(N(SO2pip)dpa)]+ complex and, in addition, two strong and intense absorption peaks at 2035 cm−1 and 1916 cm−1 are attributed to the stretching vibrations of CO ligands in the Re(CO)3 core [28]. C–N stretching vibrations in the pyridyl rings have shifted towards lower frequencies. S–N stretching vibration has shifted to a lower frequency (836 cm−1) due to lowering of initial bond energy resulting from the sigma donation of lone electron pairs on sp3 hybridized orbital in sulfonamide nitrogen.
3.5. Fluorometric Analysis
Fluorescence spectra were obtained for N(SO2pip)dpa and [Re(CO)3(N(SO2pip)dpa)]+ in methanol and acetonitrile. The concentrations of the test samples were approximately 0.01 mol/dm3. 3D scans were done for the analysis and the relevant excitation and emission details are summarized in Table 7.
Table 7.
Excitation and emission wave lengths of N(SO2pip)dpa and [Re(CO)3(N(SO2pip)dpa)]+ in methanol and acetonitrile.
| Test sample | Solvent | Excitation wave length/nm | Emission wave length/nm |
|---|---|---|---|
| N(SO2pip)dpa | Methanol | 320 | 468 |
| Acetonitrile | 325 | 475 | |
|
| |||
| [Re(CO)3(N(SO2pip)dpa)]+ | Methanol | 320 | 430 |
| Acetonitrile | 325 | 435 | |
N(SO2pip)dpa displays high fluorescence emission in methanol and the intensity was enhanced even more in acetonitrile (Figure 4). These emissions may occur due to the intraligand π → π ∗ and n → π ∗ transitions. However, fluorescence intensity of [Re(CO)3(N(SO2pip)dpa)]+ has lowered possibly due to the quenching of fluorescence upon direct binding of sulfonamide nitrogen to the metal. Nevertheless, weak fluorescence of [Re(CO)3(N(SO2pip)dpa)]+ may arise due to the MLCT transition.
Figure 4.
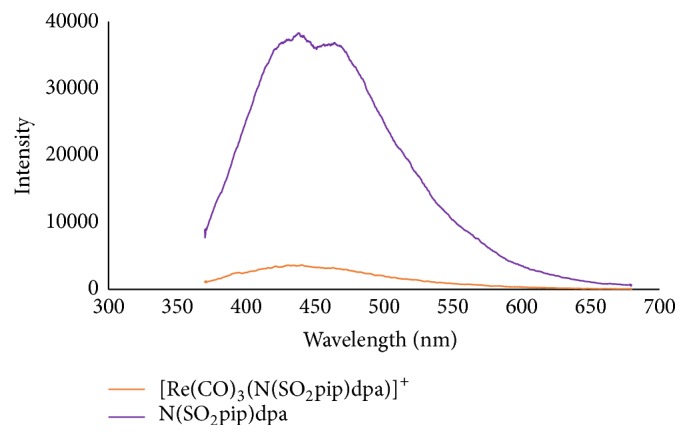
Fluorescence spectra of N(SO2pip)dpa and [Re(CO)3(N(SO2pip)dpa)]+ in acetonitrile.
3.6. Antiproliferative Activity
Cytotoxicity in human breast cancer cells (MCF-7) was induced upon addition of ligand and complex at IC50 of 139 μM and 360 μM, respectively, after 24 hr (Figure 5). This shows that, comparatively, human breast cancer cells show higher sensitivity to the ligand than its metal complex. These findings emphasize that N(SO2pip)dpa and [Re(CO)3(N(SO2pip)dpa)]+ are promising novel compounds that can be further investigated on their usage as anticancer agents and cancer cell imaging agents.
Figure 5.
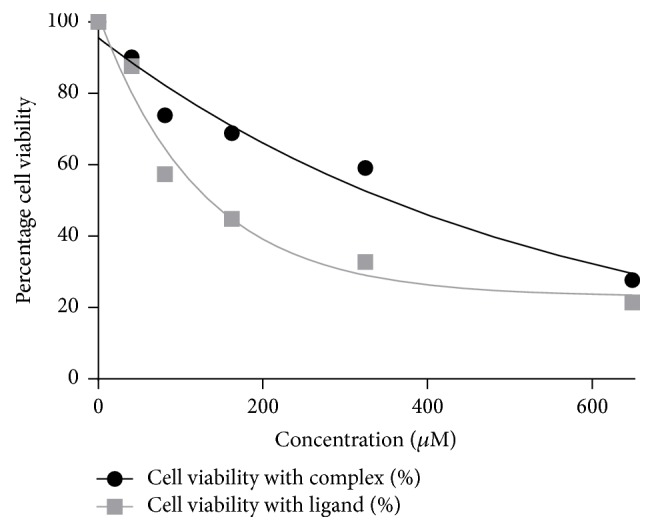
The plot of percentage cell viability versus concentration of the test compounds (μM) obtained by Sulforhodamine B assay.
Comparisons of the IC50 values obtained for ligand and the complex at 24, 48, and 72 hr have shown that the minimum test compound concentration needed to affect cell viability increased with increasing incubation time (Table 8) for the complex whereas it has decreased with that of the ligand. This emphasizes that, comparatively, ligand cytotoxicity is much higher than the metal complex toxicity. Decreasing cytotoxicity may be an indicator of activation of repair mechanisms after the initial cellular damage.
Table 8.
IC50 values reported for ligand and the complex at 24, 48, and 72 hr incubation period.
| Test compound | IC50 values/µM | ||
|---|---|---|---|
| 24 hr | 48 hr | 72 hr | |
| Ligand | 262 | 209 | 95 |
| Complex | 283 | 391 | 915 |
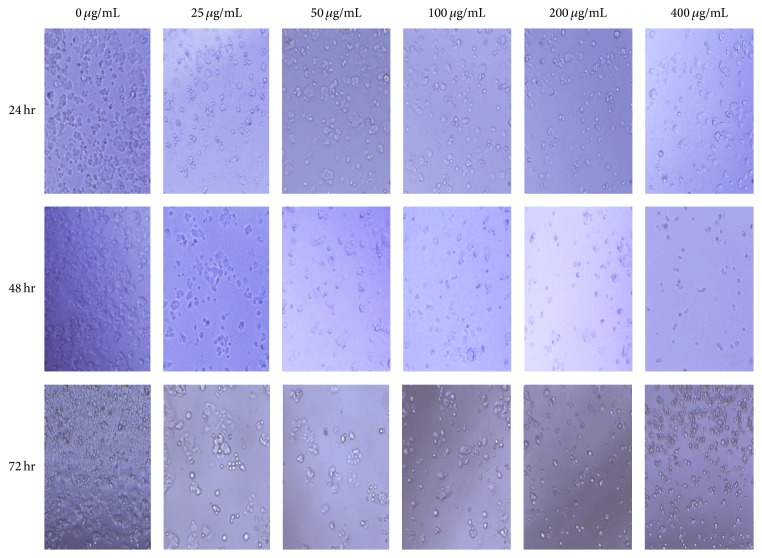
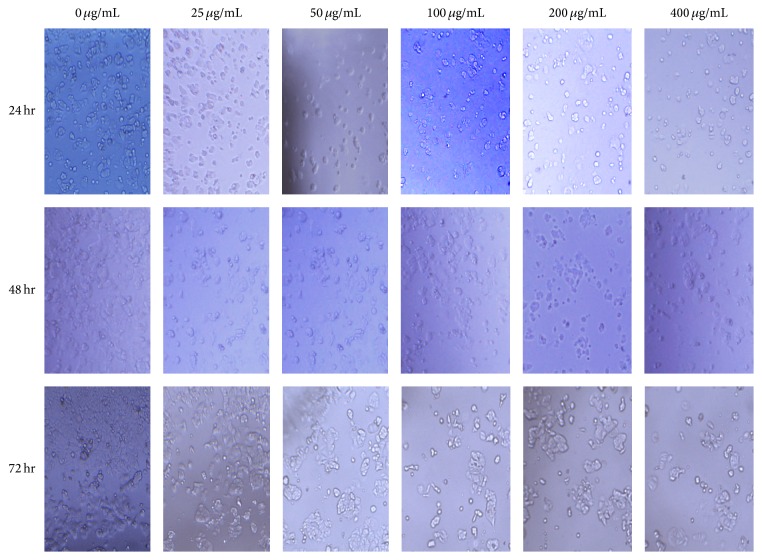
Morphology of human breast cancer cells upon treating with test compounds at 24, 48, and 72 hr time periods was micrographed and images were obtained as in Figures 6 and 7.
Figure 6.
Morphology of human breast cancer cells (MCF-7), after 24, 48, and 72 hr incubating with the N(SO2pip)dpa ligand at increasing concentrations.
Figure 7.
Morphology of human breast cancer cells (MCF-7), after 24, 48, and 72 hr incubating with the [Re(CO)3(N(SO2pip)dpa)]+ complex at increasing concentrations.
Morphology of the cells does not show significant change under light microscopy indicative of antiproliferative activity. However, the cell numbers in wells have an apparent change. Beside cytotoxic effects, the ligand shows an acute effect by 48 hours but numbers seem to be increasing by 72 hours. Cells may have resorted to a repair mechanism against the cytotoxic effect of the ligand. However, when complexed with Re, the decline of cell population seems to be continuing beyond 72 hr, which shows a persistent effect and continued antiproliferative activity.
4. Conclusions
A novel ligand and its corresponding rhenium complex were synthesized in good yield and purity and characterized using single crystal X-ray diffraction, NMR, UV-visible, and FTIR spectroscopy. The near-normal Re–N bond lengths as well as the relatively long N–S bond demonstrate that tertiary sulfonamide serves as a good donor in the [Re(CO)3(N(SO2pip)dpa)]+ complex. The sp2 to sp3 rehybridization of sulfonamide nitrogen has facilitated the binding of tertiary sulfonamide nitrogen in the [Re(CO)3(N(SO2pip)dpa)]+ complex. The fluorescence of the free N(SO2pip)dpa ligand was quenched upon direct binding of the sulfonamide nitrogen to Re in the metal complex resulting in weak fluorescence for [Re(CO)3(N(SO2pip)dpa)]+.
Cytotoxicity for both N(SO2pip)dpa and [Re(CO)3(N(SO2pip)dpa)]+ was determined against human breast cancer cells (MCF-7) by Sulforhodamine B assay. Effects of complex on breast cancer cells seem to decrease with time but the ligand continues to exert the cytotoxic effect where both compounds show promising antiproliferative activity.
In conclusion, utilizing the conjugation approach described by Perera and coworkers [5], we demonstrate that a piperidinyl group may be successfully introduced to develop therapeutic and diagnostic agents because we envisage that the synthesis reported here for nonradioactive rhenium complexes may be extended to the 99mTc core.
Acknowledgments
Financial assistance by Grant no. ASP/06/RE/SCI/2013/08 of the University of Sri Jayewardenepura is gratefully acknowledged.
Competing Interests
The authors declare no competing interests regarding the publication of this paper.
References
- 1.He H., Lipowska M., Xu X., Taylor A. T., Carlone M., Marzilli L. G. Re(CO)3 complexes synthesized via an improved preparation of aqueous fac-[Re(CO)3(H2O)3]+ as an Aid in Assessing 99mTc imaging agents. structural characterization and solution behavior of complexes with thioether-bearing amino acids as tridentate ligands. Chemistry. 2005;44(15):5437–5446. doi: 10.1021/ic0501869. [DOI] [PubMed] [Google Scholar]
- 2.Schibli R., Schubiger A. P. Current use and future potential of organometallic radiopharmaceuticals. European Journal of Nuclear Medicine. 2002;29(11):1529–1542. doi: 10.1007/s00259-002-0900-8. [DOI] [PubMed] [Google Scholar]
- 3.Stephenson K. A., Banerjee S. R., Besanger T., et al. Bridging the gap between in vitro and in vivo imaging: isostructural Re and 99mTc complexes for correlating fluorescence and radioimaging studies. Journal of the American Chemical Society. 2004;126(28):8598–8599. doi: 10.1021/ja047751b. [DOI] [PubMed] [Google Scholar]
- 4.Christoforou A. M., Marzilli P. A., Fronczek F. R., Marzilli L. G. Fac-[Re(CO)3L]+ complexes with N-CH2-CH2-X-CH2-CH2-N tridentate ligands. Synthetic, X-ray crystallographic, and NMR spectroscopic investigations. Inorganic Chemistry. 2007;46(26):11173–11182. doi: 10.1021/ic701576u. [DOI] [PubMed] [Google Scholar]
- 5.Perera T., Abhayawardhana P., Marzilli P. A., Fronczek F. R., Marzilli L. G. Formation of a metal-to-nitrogen bond of normal length by a neutral sufonamide group within a tridentate ligand. A new approach to radiopharmaceutical bioconjugation. Inorganic Chemistry. 2013;52(5):2412–2421. doi: 10.1021/ic302180t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aird R. E., Cummings J., Ritchie A. A., et al. In vitro and in vivo activity and cross resistance profiles of novel ruthenium (II) organometallic arene complexes in human ovarian cancer. British Journal of Cancer. 2002;86(10):1652–1657. doi: 10.1038/sj.bjc.6600290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergamo A., Masi A., Dyson P. J., Sava G. Modulation of the metastatic progression of breast cancer with an organometallic ruthenium compound. International Journal of Oncology. 2008;33(6):1281–1289. doi: 10.3892/ijo_00000119. [DOI] [PubMed] [Google Scholar]
- 8.Caveliers V., Everaert H., John C. S., Lahoutte T., Bossuyt A. Sigma receptor scintigraphy with N-[2-(1′-piperidinyl) ethyl]-3-123I-iodo-4-methoxybenzamide of patients with suspected primary breast cancer: first clinical results. Journal of Nuclear Medicine. 2002;43:1647–1649. [PubMed] [Google Scholar]
- 9.Choi S.-R., Yang B., Plössl K., et al. Development of a Tc-99m labeled sigma-2 receptor-specific ligand as a potential breast tumor imaging agent. Nuclear Medicine and Biology. 2001;28(6):657–666. doi: 10.1016/s0969-8051(01)00234-7. [DOI] [PubMed] [Google Scholar]
- 10.Satpati D., Bapat K., Sarma H. D., Venkatesh M., Banerjee S. Synthesis, characterization and biological activity of 99mTc- labeled piperidine analogues targeting sigma receptors. Journal of Labelled Compounds and Radiopharmaceuticals. 2010;53(4):198–204. doi: 10.1002/jlcr.1750. [DOI] [Google Scholar]
- 11.Quaglia W., Giannella M., Piergentili A., et al. 1′-benzyl-3, 4-dihydrospiro [2 H-1-benzothiopyran-2, 4′-piperidine](Spipethiane), a potent and highly selective σ1 ligand. Journal of Medicinal Chemistry. 1998;41:1557–1560. doi: 10.1021/jm970740r. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y., Williams W., Torrence-Campbell C., Bowen W. D., Rice K. C. Characterization of novel N,N'-disubstituted piperazines as σ receptor ligands. Journal of Medicinal Chemistry. 1998;41(25):4950–4957. doi: 10.1021/jm980143k. [DOI] [PubMed] [Google Scholar]
- 13.Dence C. S., John C. S., Bowen W. D., Welch M. J. Synthesis and evaluation of [18F] labeled benzamides: high affinity sigma receptor ligands for PET imaging. Nuclear Medicine and Biology. 1997;24(4):333–340. doi: 10.1016/s0969-8051(97)00001-2. [DOI] [PubMed] [Google Scholar]
- 14.Berardi F., Colabufo N. A., Giudice G., et al. New σ and 5-HT1A receptor ligands: ω-(tetralin-1-yl)-n-alkylamine derivatives. Journal of Medicinal Chemistry. 1996;39:176–182. doi: 10.1021/jm950409c. [DOI] [PubMed] [Google Scholar]
- 15.Storr T., Sugai Y., Barta C. A., et al. Carbohydrate-appended 2,2′-dipicolylamine metal complexes as potential imaging agents. Inorganic Chemistry. 2005;44(8):2698–2705. doi: 10.1021/ic048528i. [DOI] [PubMed] [Google Scholar]
- 16.Sheldrick G. M. A short history of SHELX. Acta Crystallographica Section A. 2008;64(1):112–122. doi: 10.1107/s0108767307043930. [DOI] [PubMed] [Google Scholar]
- 17.Farrugia L. J. WinGX and ORTEP for Windows: an update. Journal of Applied Crystallography. 2012;45(4):849–854. doi: 10.1107/s0021889812029111. [DOI] [Google Scholar]
- 18.Houghton P., Fang R., Techatanawat I., Steventon G., Hylands P. J., Lee C. C. The sulphorhodamine (SRB) assay and other approaches to testing plant extracts and derived compounds for activities related to reputed anticancer activity. Methods. 2007;42(4):377–387. doi: 10.1016/j.ymeth.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Veltzé S., Egdal R. K., Johansson F. B., Bond A. D., Mckenzie C. J. Coordinative flexibility in an acyclic bis(sulfonamide) ligand. Dalton Transactions. 2009;(47):10495–10504. doi: 10.1039/b912617a. [DOI] [PubMed] [Google Scholar]
- 20.Perera T., Marzilli P. A., Fronczek F. R., Marzilli L. G. NH NMR shifts of new structurally characterized fac-[Re(CO)3(polyamine)]n+ complexes probed via outer-Sphere hydrogen-bonding interactions to anions, including the paramagnetic [ReIVBr6]2− anion. Inorganic Chemistry. 2010;49:5560–5572. doi: 10.1021/ic100386c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottlieb H. E., Kotlyar V., Nudelman A. NMR chemical shifts of common laboratory solvents as trace impurities. The Journal of Organic Chemistry. 1997;62(21):7512–7515. doi: 10.1021/jo971176v. [DOI] [PubMed] [Google Scholar]
- 22.Fulmer G. R., Miller A. J. M., Sherden N. H., et al. NMR chemical shifts of trace impurities: common laboratory solvents, organics, and gases in deuterated solvents relevant to the organometallic chemist. Organometallics. 2010;29(9):2176–2179. doi: 10.1021/om100106e. [DOI] [Google Scholar]
- 23.Banerjee S. R., Levadala M. K., Lazarova N., et al. Bifunctional single amino acid chelates for labeling of biomolecules with the Tc(CO)3 + and Re(CO)3 + cores. Crystal and molecular structures of [ReBr(CO)3(H2NCH2C5H4N)], [Re(CO)3(C5H4NCH2)2NH]Br, [Re(CO)3(C5H4NCH2)2NCH2CO2H]Br, [Re(CO)3X(Y)NCH2CO2CH2CH3]Br (X = Y = 2-pyridylmethyl; X = 2-pyridylmethyl, Y = 2-(1-methylimidazolyl)methyl; X = Y = 2-(1-methylimidazolyl)methyl), [ReBr(CO)3(C5H4NCH2)NH(CH2C4H3S)], and [Re(CO)3(C5H4NCH2)N(CH2C4H3S)(CH2CO2)] Inorganic Chemistry. 2002;41(24):6417–6425. doi: 10.1021/ic020476e. [DOI] [PubMed] [Google Scholar]
- 24.Guo X.-Q., Castellano F. N., Li L., Szmacinski H., Lakowicz J. R., Sipior J. A long-lived, highly luminescent Re(I) metal-ligand complex as a biomolecular probe. Analytical Biochemistry. 1997;254(2):179–186. doi: 10.1006/abio.1997.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullice L. A., Laye R. H., Harding L. P., Buurma N. J., Pope S. J. A. Rhenium complexes of chromophore-appended dipicolylamine ligands: syntheses, spectroscopic properties, DNA binding and X-ray crystal structure. New Journal of Chemistry. 2008;32(12):2140–2149. doi: 10.1039/b800999f. [DOI] [Google Scholar]
- 26.Lambert J. B. Introduction to Organic Spectroscopy. 1987. [Google Scholar]
- 27.Tanaka Y., Tanaka Y. Infrared absorption spectra of organic sulfur compounds. II. Studies on S-N stretching bands of methanesulfonamide derivatives. Chemical and Pharmaceutical Bulletin. 1965;13(7):858–861. doi: 10.1248/cpb.13.858. [DOI] [PubMed] [Google Scholar]
- 28.Van Outersterp J. W. M., Hartl F., Stufkens D. J. Variable temperature IR spectroelectrochemical investigation of the stability of the metal-metal-bonded radical anions [(CO)5MnRe(CO)3(L)]. bul.-(L= 2, 2′-Bipyridine (BPY), 2, 2′-Bipyrimidine (BPYM), 2, 3-Bis (2-pyridyl) pyrazine (DPP)) and [(CO)5MnRe(CO)3(L)Re(Br)(CO)3]. bul.-(L= BPYM, DPP) controlled by the lowest. pi.(. alpha.-Diimine) orbital energ. Organometallics. 1995;14:3303–3310. [Google Scholar]