Abstract
Innate sensing of pathogens by pattern-recognition receptors (PRRs) plays essential roles in the innate discrimination between self and non-self components, leading to the generation of innate immune defense and inflammatory responses. The initiation, activation and resolution of innate inflammatory response are mediated by a complex network of interactions among the numerous cellular and molecular components of immune and non-immune system. While a controlled and beneficial innate inflammatory response is critical for the elimination of pathogens and maintenance of tissue homeostasis, dysregulated or sustained inflammation leads to pathological conditions such as chronic infection, inflammatory autoimmune diseases. In this review, we discuss some of the recent advances in our understanding of the cellular and molecular mechanisms for the establishment and regulation of innate immunity and inflammatory responses.
Keywords: dendritic cells, inflammation, innate lymphoid cells, innate signaling, pattern-recognition receptors (PRRs)
INTRODUCTION
Innate immune system constitutes the first critical line against microbial infection by discriminating self and non-self components. The innate immune system relies on the host pattern-recognition receptors (PRRs) expressed by innate immune cells such as macrophages and dendritic cells (DCs) to rapidly recognize and respond to signals derived from the invading pathogens or injured self-cells. PRRs such as Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) and nucleotide-binding domain and leucine-rich repeat containing molecules (NLRs) mediate the initial recognition of microbial components known as pathogen-associated molecular patterns (PAMPs).1, 2 This recognition triggers a series of signaling cascades that culminate in activation of transcriptional factors nuclear factor-κB (NF-κB), interferon regulatory factor (IRF) and activator protein-1 (AP-1), which induce numerous downstream genes encoding a broad range of inflammatory cytokines, chemokines, antimicrobial peptides, complement factors and interferons.3, 4
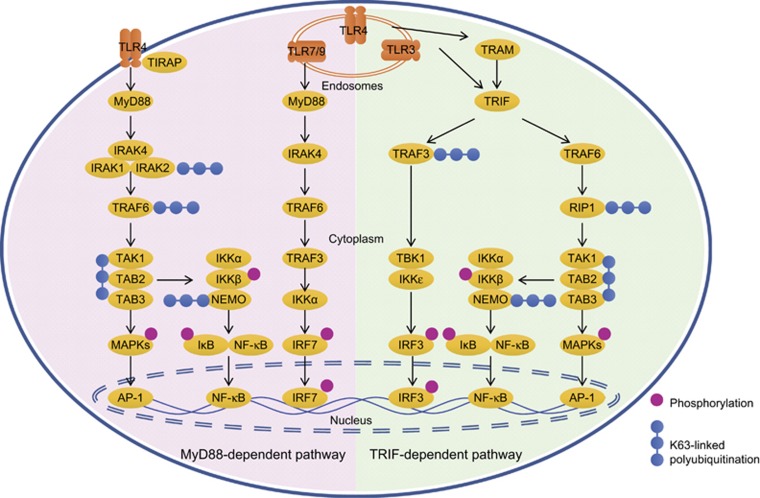
TLRs are type I transmembrane molecules which transduce their downstream signaling through the MyD88-dependent pathway or the MyD88-independent but TRIF-dependent pathway, subsequently leading to activation of mitogen-activated protein kinase (MAPK), NF-κB and IRF pathway, inducing the production of proinflammatory cytokines and IFNs (Figure 1).5, 6, 7, 8 Innate immune activation of phagocytes through TLRs also induces an Mst1–Mst2–Rac signaling axis to activate intracellular microbicidal killing.9, 10
Figure 1.
Schematic of signaling pathways of TLRs. Most TLRs with the exception of TLR3 initiate a MyD88-dependent pathway as shown in the left part. With the cooperation of Mal/TIRAP, TLR4 induces the MyD88–IRAK4 complex to recruit IRAK1 and IRAK2, which then interact with and induces K63-linked polyubiquitination of TRAF6 and TAK1/TAB2/TAB3 complexes. Activated TAK1 subsequently induce phosphorylation and activation of MAPKs and the IKK complex consisting of IKKα, IKKβ and K63-polyubiquitinated NEMO, finally promoting activation of AP-1 and NF-κB, and production of proinflammatory cytokines. TLR7 and TLR9 induces MyD88 activation to recruit signaling complex formed by IRAK4, TRAF6 and TRAF3, which induces phosphorylation and activation of IRF7 and production of type I IFN. Meanwhile, TLR3 and internalized TLR4 activate TRIF-dependent signaling as shown in the right part. TRAM is needed for the interaction between TLR4 and TRIF. TRIF recruits TRAF6 and RIP1, which induces downstream activation MAP kinases and NF-κB, similar to the MyD88-dependent pathway. TRIF also activates that TRAF3/TBK1/IKKɛ axis to promote IRF3-dependent expression of type I IFN.
RLRs, a family of cytoplasmic RNA helicases, are essential for innate recognition of viruses and are the key mechanism for the control of viral replication and dissemination. RIG-I11 and melanoma differentiation-associated protein 5 (MDA5)12 can recognize viral dsRNA and recruit the CARD containing adaptor protein MAVS (also known as IPS-1, CARDIF or VISA), leading to IRF activation and the production of type I IFN. In addition to RLRs, a group of cytosolic DNA sensors such as cyclic GMP-AMP synthase (cGAS),13, 14, 15 absent in melanoma 2 (AIM2),16, 17 DDX41,18, 19 Rad50,20, 21 LRRFIP1,22 DNA-dependent activator of IRFs (DAI),23 as well as various RNA sensors such as IFN-induced protein with tetratricopeptide repeats 1 (IFIT1)24 also play potent roles in inducing antiviral immune response, respectively via the adaptor protein stimulator of interferon genes (STING, also known as MITA, ERIS or MPYS) or the MAVS pathway. cGAS, which was previously thought to recognize cytoplasmic dsDNA over 40 bp in a sequence-independent manner, is recently shown to recognize unpaired guanosines flanking short (12–20 bp) dsDNA (Y-form DNA) found in human immunodeficiency virus type 1 to induce type I IFN production.25, 26 In neutrophils, transcription factor Sox2 could directly recognize microbial DNA through its high-mobility-group domain to activate innate immunity against microbial infection.27, 28, 29
NLRs consist of a large group of intracellular PRRs that includes NODs (nucleotide-binding oligomerization domains), NLRPs (LRR- and pyrin-domain (PYD)-containing protein), CIITA (Class II, major histocompatibility complex, transactivator), IPAF (ICE protease-activating factor) and NAIPs (neuronal apoptosis inhibitory protein), which vary in the effector domain they use to transduce downstream signals.30, 31 NOD1 and NOD2 respectively recognize meso-diamin-opimelic acid (Meso-DAP) and muramyl dipeptide of intracellular bacteria to trigger host defense against bacterial infection via the MAPK and NF-κB pathway. NOD1 and NOD2 are also shown to recruit ATG16L1 to the plasma membrane at the site of bacterial entry to induce autophagy, a process which is critical for the limitation of bacterium invasion.32, 33
Inflammasomes are large multi-molecular platforms that recruit the adaptor ASC to activate caspase-1, leading to the maturation and secretion of IL-18 and IL-1β and pyroptotic cell death, thus contributing to innate inflammatory responses to microbial or danger signals.34, 35 ASC particles could accumulate in the extracellular space to amplify activation of caspase-1 and maturation of IL-1β.36, 37 Recognition of dsDNA by AIM2 also recruits the inflammasome adaptor ASC to induce caspase-1-dependent inflammasome activation and IL-1β production. Activation of AIM2 inflammasome by Francisella tularensis subspecies novicida (F. novicida) is dependent on guanylate-binding proteins (GBPs)-mediated bacterium lysis for release of pathogenic DNA. GBPs are encoded by interferon-stimulated genes which is induced by F. novicida via the DNA sensor cGAS and its adaptor STING.38, 39 In addition, RNA viral infection also triggers inflammasome activation and IL-1β production. Recognition of RNA virus by RIG-I mediates ASC/caspase-1 inflammasome-dependent IL-1β procession in a manner independent of MAVS and NLRP3.40 RNA virus also initiates assembly of the receptor-interacting protein 1 (RIP1)–RIP3 complex to drive mitochondrial damage and activation of NLRP3 inflammasome via GTPase dynamin-related protein1 (DRP1).41
While efficient activation of PRR signaling is essential for the establishment of antimicrobial host defense and maintenance of tissue homeostasis, dysregulated or exaggerated innate immune response may cause pathological inflammation and even lead to pathogenesis of autoimmune diseases, inflammatory diseases, and cancer and so on. Thus, a delicate regulatory network is required to achieve the optimal signal output of innate immune responses, that is to efficiently eliminate invading pathogens while to avoid harmful immunological diseases. Comprehensive and multi-level mechanisms have evolved to tightly regulate the magnitude and duration of PRR signaling.42, 43 In this review, we summarize the molecular and cellular mechanisms underlying the activation and regulation of innate inflammatory responses.
Molecular regulation of innate immunity and inflammation
Gene-specific regulation of inflammatory responses
PRR-triggered inflammatory responses involve the activation and suppression of several thousands of genes with distinct functions.44 How to ensure the specific gene to be activated or silenced at the right time and space is a fundamental question in innate immune regulation. Epigenetic mechanisms such as DNA methylation, histone modifications and non-coding RNAs emerge to play essential roles in gene-specific transcriptional regulation of innate immunity via controlling chromatin status and gene expression.45, 46, 47 These chromatin modifiers perform coordinated actions to convert the extracellular stimuli into the complex gene expression patterns during innate inflammatory responses. At the steady state, the poised/inactive enhancers are occupied by lineage-determining transcription factors known as pioneers, such as PU.1 and marked with a combination of H3K4me1 and repressive H3K27me3. Upon TLR stimulation, the pioneer transcription factor PU.1 allows the binding of signal-dependent transcription factors such as NF-κB, IRFs, AP-1, and STAT and relaxes chromatin structure with acquisition of H3K27ac and removal of H3K27me3 marks.48, 49 Notably, various specific enzymes or mediators have been shown to regulate inflammatory gene expression via controlling chromatin status. In the antiviral immunity, DNA methyltransferase Dnmt3a upregulates histone deacetylase 9 (HDAC9) via epigenetic mechanisms to deacetylases the kinase Tank-binding kinase 1 (TBK1) for activation, contributing to enhanced IFN production.50 Viral infection also upregulates the expression of protein lysine methyltransferase Setdb2 to occupy and induce the repressive H3K9me3 of Cxcl1 promoters. The decreased CXCL1 inhibits infiltration of neutrophils and thus mediate sensitivity to bacterial superinfection after infection with influenza virus. Thus, chromatin modification provide molecular basis for the crosstalk between inflammatory responses against different pathogens.51 During the late phase of inflammatory response, induction of de novo 5-hydroxymethylation upstream of the Il6 locus by a methylcytosine dioxygenase TET2 recruits HDAC2 to inhibit Il6 transcription via histone deacetylation and is critical for termination of the high transcription of Il6.52 It will be intriguing to further investigate the molecular mechanisms and physiological relevance of epigenetic modification at different phases of innate inflammatory response.
In addition, a number of nuclear receptors, such as glucocorticoid receptor,53 peroxisome proliferator-activated receptor gamma,54 liver X receptor,55 small heterodimer partner (SHP),56 nuclear receptor subfamily 4, group A, members (NR4A1)57 and (NR4A2)58 inhibit TLR4-induced NF-κB downstream genes via inhibiting NF-κB activity and/or enhancing the NCoR/SMRT corepressor complex to limit inflammatory gene programs. NR4A1 enhances host resistance to lipopolysaccharide (LPS) sepsis in mice via inhibiting NF-κB binding on target gene promoter regions. Interestingly, LPS-activated p38α blocks the suppressive activity of NR4A1 by inducing its phosphorylation and therefore facilitates LPS-induced inflammatory response. It is also shown that NR4A1 limits the production of norepinephrine in macrophages through recruitment of the CoREST–histone deacetylase complex to the Th promoter, thus inhibiting the pathogenesis of experimental autoimmune encephalomyelitis, outlining a novel molecular link between sympathetic stress response and inflammation.59
Signal-specific regulation of inflammatory responses
Post-translational modifications (PTMs) constitute an essential layer of regulation of innate inflammatory signaling via affecting the function and activity of existing signaling molecules at post-translational level.60, 61 The conventional PTMs such as ubiquitination and phosphorylation and unconventional PTMs such as methylation, acetylation and sumoylation target nearly all critical components of PRR signaling, such as receptors, adaptors, enzymes and transcriptional factors to modulate the quality of PRR signals.62, 63, 64, 65
Taking PTM control of TLR-triggered tumor necrosis factor receptor-associated factor 6 (TRAF6)/NF-κB signaling pathway as an example. Removal of K63-linked polyubiquitination by A2066 and TRAF family member-associated NF-κB activator (TANK)67 is shown to modulate TRAF6 activity for inhibition of TLR signaling activation.68, 69, 70 Besides, phosphorylation of TRAF6 by germinal center kinase MST4 prevents TRAF6 oligomerization and autoubiquitination and consequently inhibits inflammatory responses.71 In addition, Rhbdd3, a member of rhomboid family of proteases, negatively regulates TLR-triggered activation of NF-κB and IL-6 production in DCs, contributing to balanced T-cell-mediated immunity and prevention of autoimmunity. Mechanistically, Rhbdd3 localizes in early endosomes in DCs and interacts with K27-linked ubiquitination of NF-κB essential modifier (NEMO) and subsequently recruits A20 to facilitate A20-mediated K63-linked deubiquitination of NEMO.72, 73 By contrast, iRhom2, a novel noncatalytic relative of rhomboid proteins, facilitates LPS and Listeria-induced TNF production by promoting the TNF convertase (TACE) maturation and trafficking,74, 75 and also enhances innate immunity to DNA viruses by mediating STING trafficking and stability.76 The differential regulation of innate immunity by Rhomboid family members awaits further investigation, and is likely to be related with their different subcellular localization that may provide specific biochemical and physical environment for specific signaling modifications.
Notably, the enzymes that mediate PTM control of PRR signaling, such as A20, are themselves being tightly controlled in a gene-specific manner, thus forming an intersecting network to modulate the quality of innate immune signaling. The transcriptional repressor downstream regulatory element antagonist modulator binds to the downstream regulatory elements (DREs) of A20 gene to repress A20 transcription, leading to enhancement of TLR-triggered NF-κB activation. By contrast, binding of the transcription factor USF1 to the DRE-associated E-box domain in the gene encoding A20 strengthens A20 expression in response to inflammatory stimuli.77 In addition, histone methyltransferase Ash1l binds to the promoter regions of A20 gene to enhance the H3K4 methylation level, thus inducing A20 gene transcription and expression, consequently suppressing TLR signaling and inflammatory autoimmune diseases.78 These studies further highlight the importance of crosstalk between PTM and chromatin modification in innate immune signaling regulation.
For the regulation of type I IFN-dependent antiviral immunity, protein PTM also plays an indispensable role via their effects in activity of signaling molecules such as RIG-I,79 NEMO and IRF3. The tumor suppressor PTEN-mediated negative phosphorylation at Ser97 of IRF3 controls the import of IRF3 into the nucleus, contributing to inhibition of IRF3 activation and type I IFN production.80, 81 E3 ligase TRIM29 inhibits IRF3 signaling via the transcription factor NF-κB by directing binding to NEMO and inducing its ubiquitination and proteolytic degradation.82 In addition, sumoylation contributes to repression of both inflammatory and antiviral responses, partially via targeting and suppressing activity of the Ifnb1 promoter.83 These data identify key negative regulators of innate immunity and might have important clinical implications for related inflammatory and infectious diseases.
Dysregulation of inflammasomes have been closely associated with diverse inflammatory diseases, therefore the negative regulation of inflammasomes are essential for prevention of excessive inflammation and maintenance of immune homeostasis.84, 85 PYD-only protein POP3 competes with ASC to bind AIM2-like receptors (ALRs), thus acting as an inhibitor of DNA virus-induced activation of ALR inflammasomes in monocytes and macrophages.86, 87 Neurotransmitter dopamine (DA)/dopamine D1 receptor (DRD1) signaling pathway negatively regulate NLRP3 inflammasome activation and NLRP3-dependent systemic inflammation via a second messenger cyclic adenosine monophosphate, which binds to NLRP3 and promotes its ubiquitination and degradation via the E3 ubiquitin ligase MARCH7.88 Moreover, protein kinase A directly phosphorylates NLRP3 at Ser295 and attenuates its ATPase function, accordingly, mutations in NLRP3-encoding residues adjacent to Ser295 are linked to the auto-inflammatory disease cryopyrin-associated periodic syndromes (CAPS).89 These studies reveal endogenous regulatory mechanisms of inflammasome regulation and suggest molecular basis and potential therapeutic targets of inflammatory and autoimmune diseases.
Interplay across different PRR signaling pathways
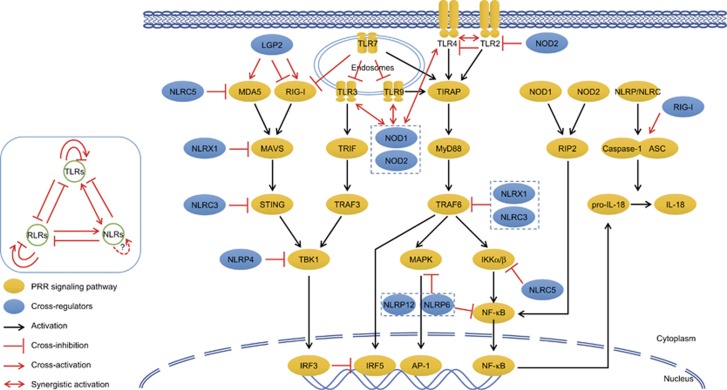
The TLRs, RLRs and NLRs trigger an intensively interacting network of downstream signaling to activate innate defense against invading pathogens (Figure 2). Distinct set of PRRs in different species, tissues, cells or cellular organelles display a partially overlapped and compensated recognition of PAMPs during inflammatory conditions. LPS from Gram-negative bacteria, is crucial for TLR4 activation, but also initiates innate immune signaling via detection by cytosol caspase-4/5/11 in mammals,90, 91 while in plants via sensing by transmembrane receptor kinase LORE. LORE confers recognition of LPS in plants and the subsequent induction of an immune response.92, 93 Whether additional proteins, e.g. LPS-binding protein is involved in LORE-mediated recognition of LPS remain to be elucidated.
Figure 2.
Interplay across TLR, RLR and NLR signaling pathways. TLR, RLR and NLR-triggered signaling pathways display complex interplay with each other. ① Crosstalk inside PRR family: LGP2 both positively and negatively regulate MDA5 and RIG-I signaling, so do TLR2 for TLR4. ② TLR-mediated cross-regulation: TLR7 agonists inhibit TLR3, TLR9 and RIG-I signaling; TLR3/4/9 synergize with NOD1/2 for amplified innate immune responses. ③ RLR-mediated cross-regulation: RIG-I signaling inhibits TLR signaling via competitive occupancy at Il12 promoter by IRF5 over IRF3, but promotes NLRP3 inflammasome activation via interacting with ASC. ④ NLR-mediated cross-regulation: NOD2 tolerates TLR2 signaling for prevention of colitis. NLRX1, NLRC3/5, NLRP4/6/12 suppress TLR and RLR signaling via targeting distinct molecules.
A mutual inhibition of RLR and TLR pathways has been shown. RLR-triggered IRF3 following viral stimulation inhibits TLR-induced IRF5 activation following bacterial stimulation via dominantly occupying Il12b promoter, thus explaining the molecular mechanisms of bacterial superinfection post-viral infection.94 Conversely, TLR7 small-molecule agonist inhibits nucleic acid-mediated TLR3, TLR7, TLR9 and RIG-I-dependent type I IFN signaling via inhibiting the formation of phosphorylated signal transducer and activator of transcription factor 1 (p-STAT1), p-STAT2 and IRF9 complex.95
Meanwhile, RIG-I plays a positive role in regulation of inflammasome and IL-1β secretion. Upon RNA viral stimulation, RIG-I could interact with adaptor ASC to trigger caspase-1-dependent inflammasome activation and IL-1β maturation.34 On the contrary, some NLR members play negative roles in regulating RLR-mediated type I IFN responses via targeting distinct signaling molecules: for NLR family CARD domain-containing protein 5 (NLRC5) via interaction with RIG-I and MDA5;96 for NLR family member X1 (NLRX1) via interaction with MAVS and disruption of RLR–MAVS interactions;97, 98 for NLRC3 via impeding the interaction between STING and TBK1 interaction;99 for NLRP4 via targeting TBK1 for degradation.100
NOD proteins and TLRs are both critical for host defense against bacterial infection. NOD1 and NOD2 agonists play synergistic effects with TLR2, TLR3, TLR4, and TLR9 agonists to promote maturation and activation of DCs and basophils.101, 102, 103 On the contrary, NOD2 deficiency increases TLR2-mediated activation of NF-κB and dysregulated TLR2 in NOD2-deficient mice causes the development of antigen-specific colitis.104, 105 Not only NOD2, many other NLRs play inhibitory roles in regulating the signaling of TLRs: for NLRX1 and NLRC3 via interfering with the TRAF6-NF-κB signaling;61, 106, 107 for NLRC5 via interacting with and blocking phosphorylation of IκB kinase α (IKKα) and IKKβ59 for NRLP6 and NLRP12 via targeting MAPK and NF-κB activation;108, 109 for NLRC4 via downregulating TLR5-mediated antibody immune responses against flagellin.110 These synergistic or antagonistic interactions across these three PRR families contribute to a cross-linked and finely tuned network of PRR signaling in response to the large repertoire of PAMPs and ensures the most effective and proper outcomes of innate immune responses.
Cellular regulation of innate immunity and inflammation
PRR-triggered inflammatory responses are mediated and regulated by a variety of immune and non-immune cells.111, 112 The endothelium system is critical player of innate defense via releasing inflammatory cytokines and chemokines in the first barrier to recruit monocytes, neutrophils, eosinophils and basophils from the circulation.113, 114, 115, 116 TLR stimulation induce phenotypic and functional maturation of DC into potent antigen-presenting cells which efficiently initiate and control adaptive immune responses, while in combination with IFN-γ could induce M1 macrophage polarization that is critical for phagocytosis and killing of invading pathogens. In this section, we focus on two important cell populations in innate immunity, DCs as the key bridge between innate and adaptive immunity, and innate lymphoid cells (ILCs) as the key mediator of effector responses during innate immunity.
Dendritic cells
DCs play important roles in the initiation and modulation of adaptive immune responses. DCs represent a complicated heterogeneous cell populations with distinct developmental origins, surface markers and effector/regulatory functions. Conventional DCs are further classified into subgroups according to the expression of CD8′ that is regulated by a complex network of cytokines and transcriptional factors.117, 118, 119, 120, 121 While CD8′− classical dendritic cells (cDCs) primarily promote antigen-specific CD4+ T-cell activation via the MHC II pathway, CD8′+ cDCs characteristically mediate cross-presentation of exogenous antigens on MHC I molecules to cytotoxic T lymphocytes (CTL). Ligation of TLRs by microbial products in DCs rapidly induces the expression of inflammatory cytokines, chemokines and chemokine receptors, co-stimulatory molecules and MHC molecules, allowing procession and presentation of antigens to T cells, and therefore play essential roles in determining the activation and differentiation of T-cell subsets.122, 123, 124, 125, 126, 127 The activation of DCs by TLR agonists is accompanied by a rapid increase in glycolysis DC which is dependent on signaling via the kinases TBK1, IKKɛ and Akt. TLR-driven glycolytic flux serves an essential role in supporting the de novo synthesis of fatty acids for activation and function of DCs.128, 129
Meanwhile, novel subsets of DC are also important for inducing immune tolerance toward harmless components via induction of immune tolerance under specific physiological or pathological conditions.130 The regulatory capacity of DCs at steady state is programmed by specific local microenvironment, such as stromal cells of the spleen, lung and liver or intestinal epithelial cells.131, 132, 133, 134, 135 Thymic DCs induce Treg-cell development via production of IL-2,136 and that CD11b(−) cDCs from the gut-draining lymph nodes were critical for induction of peripheral Treg cells and oral tolerance.137 The presence of exogenous immunosuppressive mediators, genetic manipulation, specific pathogenic stimuli also induce regulatory property of DCs through various mechanisms.138
The cross-presentation of exogenous antigens via MHC class I molecules to initiate CTL responses is essential for immunological defense against viruses, intracellular bacteria and tumors. While Rab11a activity recruits and keeps MHC-I within endosomal recycling compartment (ERC) under steady condition, MyD88-dependent TLR signals drive IKK2-mediated phosphorylation of phagosome-associated SNAP23, orchestrating ERC-phagosome fusion, promoting enrichment of phagosomes with ERC-derived MHC-I, and finally allowing cross-presentation during infection.139 Transcription factor TFEB,140 cell stress sensor IRE-1α,141, 142, 143 NF-κB-inducing kinase (NIK)144 and the lectin Siglec-G145 have been shown to regulate DC cross-presentation in initiating antigen-specific CTL responses via distinct molecular mechanisms. For example, Siglec-G inhibits cross-presentation by CD8α+ DC via impairing the formation of the MHC class I-exogenous antigen peptide complex, contributing to suppression of CTL responses to intracellular bacterial infection with Listeria monocytogenes or Mycobacterium bovis bacillus Calmette–Guérin and tumors. Siglec-G is associated with SHP-1 to inhibit the activation of NOX2, consequently leading to promotion of phagosomal acidification and less effective antigen cross-presentation. This study provides new mechanistic insight into DC-mediated regulation of innate and adaptive immune responses under inflammatory conditions.
Innate lymphoid cells
ILCs are most recently identified populations of innate immune cells that play an important role in lymphoid tissue development, metabolic homeostasis and innate immunity against microbial infection and are increasingly linked with diverse pathological conditions such as infection, chronic inflammation, metabolic disease and cancer.146, 147 Distinct ILC groups are defined on the basis of expression of surface markers, transcription factors and cytokine secretion profiles, and effector functions in homeostasis and inflammation.148 Whereas ILC1s and ILC3s are essential to host defense against infection by viruses, intracellular bacteria and parasites, ILC2s potently drive type 2 inflammation and mediate allergic inflammation, tissue repair and anti-helminth innate immunity.149, 150, 151, 152, 153 A novel population of IL-25-responsive inflammatory ILC2 (iILC2) could develop into IL-33-responsive natural ILC2 (nILC2)-like cells and contribute to immunity to both helminths and fungi.154, 155 Inflammatory cytokines such as IL-1 and IFN,156, 157 and crosstalk with stromal cells, epithelial cells and various immune cells such as DC,158 T cells159 and B cells160, 161 are important for regulation of ILC function and plasticity at the crossroad of homeostasis and inflammation. For example, ILC3 can enhance antibody production by splenic marginal zone B cells via integrating stromal and myeloid signals. Further investigations are required to uncover the detailed mechanism underlying the plasticity and flexibility in the functions of distinct ILCs under biological and pathological conditions.
The cellular and molecular events that underlie ILC fate specification and functional regulation attract much attention in the past few years. Key transcription factors and regulators that control the development of ILC subsets at different stages are being increasingly identified, such as TOX, TCF-1, NFIL3, Id2, Runx3, GATA-3 and so on.162, 163, 164 TOX-deficient mice have diminished numbers of LTi cells, NK cells, ILC1, ILC2 and ILC3 cells, indicating that TOX is required for in vivo differentiation of common lymphoid progenitors into ILC lineage-restricted cells.165, 166 Using novel reporter mice, researchers identified a novel subset of early ILC progenitors (EILPs) with distinctive expression of transcription factor TCF-1. EILPs exclusively and efficiently gives rise to NK cells and all known adult helper ILC lineages and therefore are perhaps the earliest ILC-committed progenitors identified so far.167, 168 The interaction of the TOX, TCF-1 and other transcription factors and epigenetic factors in the development of ILCs cells awaits further investigation.
Conclusions and perspectives
Increasing evidence reveal an essential role of PRRs in innate sensing of pathogens and initiation of innate inflammatory responses. A delicate regulatory network of PRR signaling both at molecular and cellular level contribute to an appropriate and effective host immune response under steady and inflammatory state. Though substantial progress have been made in depicting the initiation, activation and regulation of PRR-mediated innate immune response, some intriguing question still challenge further investigation. How does the immune system organize tissue-, cell- and gene-specific regulation of innate inflammatory responses? What is molecular basis for the combination, assembly and translocation of signaling molecule machinery? What is the developmental and functional characteristic of many other rare but important cell populations of innate immune systems, such as natural killer cells,169 invariant natural killer T cells, mast cells, plasmacytoid DCs and so on.
Excitingly, with the rapid development in immunological technological platforms and combination of immunology and many disciplines such as epigenetics, genetics, biochemistry and neurobiology, substantial progress are being achieved in evaluating how the immune system quickly and accurately respond to external and internal stimuli. Single-cell sequencing technologies have identified the complex heterogeneities and divergences of cell phenotype and function.170, 171, 172 Advances in genomics, transcriptomics, proteomics, metabolomics, interactomics, phenomics have illustrated the complex networks of immune cells and molecules in the settings of health and diseases.173 Development of new mouse strains such as the mouse strain lacking iNKT cells174 and new disease models such as the mice with myeloid-cell-specific deletion of A20 as a new model of rheumatoid arthritis175 have provided useful tools for immunological studies. Cell fate mapping and in vivo imaging technologies have enabled real-time, dynamic and in situ assessment of the activity and function of immune system.176,177,178 Future investigations will provide essential insights into the regulatory mechanisms of innate immunity and inflammation and outline potential clues for the development of effective therapeutic approaches of inflammatory diseases.
Acknowledgments
This work was supported by Grants from National Natural Science Foundation of China (31400777, 31622024, 81671546) and “Chen Guang” project supported by Shanghai Municipal Education Commission and Shanghai Education Development Foundation (13CG39).
The authors declare no conflict of interest.
References
- Kotas ME, Medzhitov R. Homeostasis, inflammation, and disease susceptibility. Cell 2015; 160: 816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010; 140: 805–820. [DOI] [PubMed] [Google Scholar]
- Hunter CA, Jones SA. IL-6 as a keystone cytokine in health and disease. Nat Immunol 2015; 16: 448–457. [DOI] [PubMed] [Google Scholar]
- Wack A, Terczyńska-Dyla E, Hartmann R. Guarding the frontiers: the biology of type III interferons. Nat Immunol 2015; 16: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Control of adaptive immunity by the innate immune system. Nat Immunol 2015; 16: 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 2011; 34: 637–650. [DOI] [PubMed] [Google Scholar]
- Ma Z, Zhang E, Yang D, Lu M. Contribution of Toll-like receptors to the control of hepatitis B virus infection by initiating antiviral innate responses and promoting specific adaptive immune responses. Cell Mol Immunol 2015; 12: 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Liu Y, Gao XX, Gao X, Cai H. TLR2 and TLR4 signaling pathways are required for recombinant Brucella abortus BCSP31-induced cytokine production, functional upregulation of mouse macrophages, and the Th1 immune response in vivo and in vitro. Cell Mol Immunol 2014; 11: 477–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Sun X, Wang P, Zhang S, Wang X, Wu H et al. Kinases Mst1 and Mst2 positively regulate phagocytic induction of reactive oxygen species and bactericidal activity. Nat Immunol 2015; 16: 1142–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart LM, Lacy-Hulbert A. De-Mst-ifying microbicidal killing. Nat Immunol 2015; 16: 1107–1118. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science 2006; 314: 997–1001. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006; 441: 101–105. [DOI] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 2013; 339: 826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013; 339: 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wu J, Chen ZJ, Chen C. Molecular basis for the specific recognition of the metazoan cyclic GMP-AMP by the innate immune adaptor protein STING. Proc Natl Acad Sci USA 2015; 112: 8947–8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 2009; 458: 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürckstümmer T, Baumann C, Blüml S, Dixit E, Dürnberger G, Jahn H et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol 2009; 10: 266–272. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol 2011; 12: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol 2012; 13: 1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S, Rottach A, Lotz-Havla AS, Laux V, Muschaweckh A, Gersting SW et al. Rad50-CARD9 interactions link cytosolic DNA sensing to IL-1β production. Nat Immunol 2014; 15: 538–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie AG. Rad50 and CARD9, missing links in cytosolic DNA-stimulated inflammation. Nat Immunol 2014; 15: 534–536. [DOI] [PubMed] [Google Scholar]
- Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y et al. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol 2010; 11: 487–494. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 2007; 448: 501–505. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Lassnig C, Eberle CA, Górna MW, Baumann CL, Burkard TR et al. IFIT1 is an antiviral protein that recognizes 5'-triphosphate RNA. Nat Immunol 2011; 12: 624–630. [DOI] [PubMed] [Google Scholar]
- Herzner AM, Hagmann CA, Goldeck M, Wolter S, Kübler K, Wittmann S et al. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat Immunol 2015; 16: 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JJ, Gack MU. Reading the fine print: sequence-specific activation of cGAS. Nat Immunol 2015; 16: 1009–1010. [DOI] [PubMed] [Google Scholar]
- Xia P, Wang S, Ye B, Du Y, Huang G, Zhu P et al. Sox2 functions as a sequence-specific DNA sensor in neutrophils to initiate innate immunity against microbial infection. Nat Immunol 2015; 16: 366–375. [DOI] [PubMed] [Google Scholar]
- Mankan AK1. Hornung V1. Sox2 as a servant of two masters. Nat Immunol 2015; 16: 335–336. [DOI] [PubMed] [Google Scholar]
- Yu Z, Chen T, Cao X. Neutrophil sensing of cytoplasmic, pathogenic DNA in a cGAS-STING-independent manner. Cell Mol Immunol 2016; 13: 411–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol 2003; 4: 702–707. [DOI] [PubMed] [Google Scholar]
- Rauch I, Tenthorey JL, Nichols RD, Al Moussawi K, Kang JJ, Kang C et al. NAIP proteins are required for cytosolic detection of specific bacterial ligands in vivo. J Exp Med 2016; 213: 657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhães JG et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol 2010; 11: 55–62. [DOI] [PubMed] [Google Scholar]
- Shibutani ST, Saitoh T, Nowag H, Münz C, Yoshimori T. Autophagy and autophagy-related proteins in the immune system. Nat Immunol 2015; 16: 1014–1024. [DOI] [PubMed] [Google Scholar]
- Henao-Mejia J, Elinav E, Strowig T, Flavell RA. Inflammasomes: far beyond inflammation. Nat Immunol 2012; 13: 321–324. [DOI] [PubMed] [Google Scholar]
- Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature 2012; 481: 278–286. [DOI] [PubMed] [Google Scholar]
- Franklin BS, Bossaller L, De Nardo D, Ratter JM, Stutz A, Engels G et al. The adaptor ASC has extracellular and 'prionoid' activities that propagate inflammation. Nat Immunol 2014; 15: 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroja-Mazo A, Martín-Sánchez F, Gomez AI, Martínez CM, Amores-Iniesta J, Compan V et al. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat Immunol 2014; 15: 738–748. [DOI] [PubMed] [Google Scholar]
- Man SM, Karki R, Malireddi RK, Neale G, Vogel P, Yamamoto M et al. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat Immunol 2015; 16: 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Rühl S et al. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol 2015; 16: 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for in terleukin 1 beta production. Nat Immunol 2010; 11: 63–69. [DOI] [PubMed] [Google Scholar]
- Wang X, Jiang W, Yan Y, Gong T, Han J, Tian Z et al. RNA viruses promote activation of the NLRP3 inflammasome through a RIP1-RIP3-DRP1 signaling pathway. Nat Immunol 2014; 15: 1126–1133. [DOI] [PubMed] [Google Scholar]
- Piccirillo CA, Bjur E, Topisirovic I, Sonenberg N, Larsson O. Translational control of immune responses: from transcripts to translatomes. Nat Immunol 2014; 15: 503–511. [DOI] [PubMed] [Google Scholar]
- Kafasla P, Skliris A, Kontoyiannis DL. Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nat Immunol 2014; 15: 492–502. [DOI] [PubMed] [Google Scholar]
- Schreiber J, Jenner RG, Murray HL, Gerber GK, Gifford DK, Young RA. Coordinated binding of NF-κB family members in the response of human cells to lipopolysaccharide. Proc Natl Acad Sci USA 2006; 103: 5899–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cao X. Long noncoding RNAs in innate immunity. Cell Mol Immunol 2016; 13: 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M, Galloway A, Vigorito E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat Immunol 2014; 15: 484–491. [DOI] [PubMed] [Google Scholar]
- Nishitsuji H, Ujino S, Yoshio S, Sugiyama M, Mizokami M, Kanto T et al. Long noncoding RNA #32 contributes to antiviral responses by controlling interferon-stimulated gene expression. Proc Natl Acad Sci USA 2016; 113: 10388–10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 2010; 32: 317–328. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang Q, Ding Y, Liu Y, Zhao D, Zhao K et al. Methyltransferase Dnmt3a upregulates HDAC9 to deacetylate the kinase TBK1 for activation of antiviral innate immunity. Nat Immunol 2016; 17: 806–815. [DOI] [PubMed] [Google Scholar]
- Schliehe C, Flynn EK, Vilagos B, Richson U, Swaminathan S, Bosnjak B et al. The methyltransferase Setdb2 mediates virus-induced susceptibility to bacterial superinfection. Nat Immunol 2015; 16: 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li X et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 2015; 525: 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrebi D, Bruscoli S, Cohen N, Foussat A, Migliorati G, Bouchet-Delbos L et al. Synthesis of glucocorticoid-induced leucine zipper (GILZ) by macrophages: an anti-inflammatory and immunosuppressive mechanism shared by glucocorticoids and IL-10. Blood 2003; 101: 729–738. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell 2005; 122: 707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG et al. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev 2009; 23: 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuk JM, Shin DM, Lee HM, Kim JJ, Kim SW, Jin HS et al. The orphan nuclear receptor SHP acts as a negative regulator in inflammatory signaling triggered by Toll-like receptors. Nat Immunol 2011; 12: 742–751. [DOI] [PubMed] [Google Scholar]
- Li L, Liu Y, Chen HZ, Li FW, Wu JF, Zhang HK et al. Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation. Nat Chem Biol 2015; 11: 339–346. [DOI] [PubMed] [Google Scholar]
- Saijo K, Winner B, Carson CT, Collier JG, Boyer L, Rosenfeld MG et al. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell 2009; 137: 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked I, Hanna RN, Shaked H, Chodaczek G, Nowyhed HN, Tweet G et al. Transcription factor Nr4a1 couples sympathetic and inflammatory cues in CNS-recruited macrophages to limit neuroinflammation. Nat Immunol 2015; 16: 1228–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowen KA, David M. Unconventional post-translational modifications in immunological signaling. Nat Immunol 2014; 15: 512–520. [DOI] [PubMed] [Google Scholar]
- Liu J, Qian C, Cao X. Post-translational modification control of innate immunity. Immunity 2016; 45: 15–30. [DOI] [PubMed] [Google Scholar]
- Harikumar KB, Yester JW, Surace MJ, Oyeniran C, Price MM, Huang WC et al. K63-linked polyubiquitination of transcription factor IRF1 is essential for IL-1-induced production of chemokines CXCL10 and CCL5. Nat Immunol 2014; 15: 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wang J. K63-linked polyubiquitination of IRF1: an essential step in the IL-1 signaling cascade. Cell Mol Immunol 2014; 11: 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawan M, Venkatesan N, Loh JT, Wong JF, Berger H, Neo WH et al. The methyltransferase Ezh2 controls cell adhesion and migration through direct methylation of the extranuclear regulatory protein talin. Nat Immunol 2015; 16: 505–516. [DOI] [PubMed] [Google Scholar]
- Chuprin A, Avin A, Goldfarb Y, Herzig Y, Levi B, Jacob A et al. The deacetylase Sirt1 is an essential regulator of Aire-mediated induction of central immunological tolerance. Nat Immunol 2015; 16: 737–745. [DOI] [PubMed] [Google Scholar]
- Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol 2004; 5: 1052–1060. [DOI] [PubMed] [Google Scholar]
- Kawagoe T, Takeuchi O, Takabatake Y, Kato H, Isaka Y, Tsujimura T et al. TANK is a negative regulator of Toll-like receptor signaling and is critical for the prevention of autoimmune nephritis. Nat Immunol 2009; 10: 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. Immune diseases caused by mutations in kinases and components of the ubiquitin system. Nat Immunol 2014; 15: 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton SM, Borg NA, Dixit VM. Ubiquitin in the activation and attenuation of innate antiviral immunity. J Exp Med 2016; 213: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, You M, Shi H, Hou Y. Ubiquitin-mediated NFκB degradation pathway. Cell Mol Immunol 2015; 12: 653–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao S, Zhang Z, Li C, Huang M, Shi Z, Wang Y et al. The kinase MST4 limits inflammatory responses through direct phosphorylation of the adaptor TRAF6. Nat Immunol 2015; 16: 246–257. [DOI] [PubMed] [Google Scholar]
- Liu J, Han C, Xie B, Wu Y, Liu S, Chen K et al. Rhbdd3 controls autoimmunity by suppressing the production of IL-6 by dendritic cells via K27-linked ubiquitination of the regulator NEMO. Nat Immunol 2014; 15: 612–622. [DOI] [PubMed] [Google Scholar]
- Liu J, Liu S, Xia M, Xu S, Wang C, Bao Y et al. Rhomboid domain-containing protein 3 is a negative regulator of TLR3-triggered natural killer cell activation. Proc Natl Acad Sci USA 2013; 110: 7814–7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrain C, Zettl M, Christova Y, Taylor N, Freeman M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science 2012; 335: 225–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain DR, Lang PA, Maretzky T, Hamada K, Ohishi K, Maney SK et al. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science 2012; 335: 229–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo WW, Li S, Li C, Lian H, Yang Q, Zhong B et al. iRhom2 is essential for innate immunity to DNA viruses by mediating trafficking and stability of the adaptor STING. Nat Immunol 2016; 17: 1057–1066. [DOI] [PubMed] [Google Scholar]
- Tiruppathi C, Soni D, Wang DM, Xue J, Singh V, Thippegowda PB et al. The transcription factor DREAM represses the deubiquitinase A20 and mediates inflammation. Nat Immunol 2014; 15: 239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Liu J, Wu X, Liu S, Li G, Han C et al. Histone methyltransferase Ash1l suppresses interleukin-6 production and inflammatory autoimmune diseases by inducing the ubiquitin-editing enzyme A20. Immunity 2013; 39: 470–481. [DOI] [PubMed] [Google Scholar]
- Wang W, Jiang M, Liu S, Zhang S, Liu W, Ma Y et al. RNF122 suppresses antiviral type I interferon production by targeting RIG-I CARDs to mediate RIG-I degradation. Proc Natl Acad Sci USA 2016; 113: 9581–9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhu M, Pan R, Fang T, Cao YY, Chen S et al. The tumor suppressor PTEN has a critical role in antiviral innate immunity. Nat Immunol 2016; 17: 241–249. [DOI] [PubMed] [Google Scholar]
- Champion BR, Fisher K, Seymour L. A PTENtial cause for the selectivity of oncolytic viruses? Nat Immunol 2016; 17: 225–226. [DOI] [PubMed] [Google Scholar]
- Xing J, Weng L, Yuan B, Wang Z, Jia L, Jin R et al. Identification of a role for TRIM29 in the control of innate immunity in the respiratory tract. Nat Immunol; e-pub ahead of print 3 October 2016; doi:10.1038/ni.3580. [DOI] [PMC free article] [PubMed]
- Decque A, Joffre O, Magalhaes JG, Cossec JC, Blecher-Gonen R, Lapaquette P et al. Sumoylation coordinates the repression of inflammatory and anti-viral gene-expression programs during innate sensing. Nat Immunol 2016; 17: 140–149. [DOI] [PubMed] [Google Scholar]
- Rathinam VA, Vanaja SK, Fitzgerald KA. Regulation of inflammasome signaling. Nat Immunol 2012; 13: 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo EK, Kim JK, Shin DM, Sasakawa C. Molecular mechanisms regulating NLRP3 inflammasome activation. Cell Mol Immunol 2016; 13: 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S, Ratsimandresy RA, de Almeida L, Cuda CM, Rellick SL, Misharin AV et al. The PYRIN domain-only protein POP3 inhibits ALR inflammasomes and regulates responses to infection with DNA viruses. Nat Immunol 2014; 15: 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy JK, Liu D, Eisenbarth SC. POP goes the inflammasome. Nat Immunol 2014; 15: 311–313. [DOI] [PubMed] [Google Scholar]
- Yan Y, Jiang W, Liu L, Wang X, Ding C, Tian Z et al. Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell 2015; 160: 62–73. [DOI] [PubMed] [Google Scholar]
- Mortimer L, Moreau F, MacDonald JA, Chadee K. NLRP3 inflammasome inhibition is disrupted in a group of auto-inflammatory disease CAPS mutations. Nat Immunol 2016; 17: 1176–1186. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 2014; 514: 187–192. [DOI] [PubMed] [Google Scholar]
- Liu W, Menoret A, Vella AT. Responses to LPS boost effector CD8 T-cell accumulation outside of signals 1 and 2. Cell Mol Immunol; e-pub ahead of print 20 July 2015; doi:10.1038/cmi.2015.69. [DOI] [PMC free article] [PubMed]
- Ranf S, Gisch N, Schäffer M, Illig T, Westphal L, Knirel YA et al. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat Immunol 2015; 16: 426–433. [DOI] [PubMed] [Google Scholar]
- Zipfel C. A new receptor for LPS. Nat Immunol 2015; 16: 340–341. [DOI] [PubMed] [Google Scholar]
- Negishi H, Yanai H, Nakajima A, Koshiba R, Atarashi K, Matsuda A et al. Cross-interference of RLR and TLR signaling pathways modulates antibacterial T cell responses. Nat Immunol 2012; 13: 659–666. [DOI] [PubMed] [Google Scholar]
- Forsbach A, Müller C, Montino C, Kritzler A, Nguyen T, Weeratna R et al. Negative regulation of the type I interferon signaling pathway by synthetic Toll-like receptor 7 ligands. J Interferon Cytokine Res 2012; 32: 254–268. [DOI] [PubMed] [Google Scholar]
- Cui J, Zhu L, Xia X, Wang HY, Legras X, Hong J et al. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell 2010; 141: 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature 2008; 451: 573–577. [DOI] [PubMed] [Google Scholar]
- Allen IC, Moore CB, Schneider M, Lei Y, Davis BK, Scull MA et al. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-κB signaling pathways. Immunity 2011; 34: 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Mo J, Swanson KV, Wen H, Petrucelli A, Gregory SM et al. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity 2014; 40: 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Li Y, Zhu L, Liu D, Songyang Z, Wang HY et al. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol 2012; 13: 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz JH, Girardin SE, Fitting C, Werts C, Mengin-Lecreulx D, Caroff M et al. Synergistic stimulation of human monocytes and dendritic cells by Toll-like receptor 4 and NOD1- and NOD2-activating agonists. Eur J Immunol 2005; 35: 2459–2470. [DOI] [PubMed] [Google Scholar]
- Tada H, Aiba S, Shibata K, Ohteki T, Takada H. Synergistic effect of Nod1 and Nod2 agonists with toll-like receptor agonists on human dendritic cells to generate interleukin-12 and T helper type 1 cells. Infect Immun 2005; 73: 7967–7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao D, Wong CK, Qiu HN, Dong J, Cai Z, Chu M et al. NOD2 and TLR2 ligands trigger the activation of basophils and eosinophils by interacting with dermal fibroblasts in atopic dermatitis-like skin inflammation. Cell Mol Immunol 2016; 13: 535–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Kitani A, Murray PJ, Strober W. NOD2 is a negative regulator of Toll-like receptor 2-mediated T helper type 1 responses. Nat Immunol 2004; 5: 800–808. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Kitani A, Murray PJ, Wakatsuki Y, Fuss IJ, Strober W. Nucleotide binding oligomerization domain 2 deficiency leads to dysregulated TLR2 signaling and induction of antigen-specific colitis. Immunity 2006; 25: 473–485. [DOI] [PubMed] [Google Scholar]
- Xia X, Cui J, Wang HY, Zhu L, Matsueda S, Wang Q et al. NLRX1 negatively regulates TLR-induced NF-κB signaling by targeting TRAF6 and IKK. Immunity 2011; 34: 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Zimmermann AG, Roberts RA, Zhang L, Swanson KV, Wen H et al. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-κB. Nat Immunol 2012; 13: 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand PK, Malireddi RK, Lukens JR, Vogel P, Bertin J, Lamkanfi M et al. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature 2012; 488: 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen IC, Wilson JE, Schneider M, Lich JD, Roberts RA, Arthur JC et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-κB signaling. Immunity 2012; 36: 742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Yang J, Zhang E, Zhong M, Xiao Y, Yu J et al. Activation of NLRC4 downregulates TLR5-mediated antibody immune responses against flagellin. Cell Mol Immunol 2016; 13: 514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera A, Siracusa MC, Yap GS, Gause WC. Innate cell communication kick-starts pathogen-specific immunity. Nat Immunol 2016; 17: 356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Lopez N, StLouis D, Lehr MA, Sanchez-Rodriguez EN, Arenas-Hernandez M. Immune cells in term and preterm labor. Cell Mol Immunol 2014; 11: 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitsett JA, Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat Immunol 2015; 16: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauseef WM, Borregaard N. Neutrophils at work. Nat Immunol 2014; 15: 602–611. [DOI] [PubMed] [Google Scholar]
- Lakschevitz FS, Visser MB, Sun C, Glogauer M. Neutrophil transcriptional profile changes during transit from bone marrow to sites of inflammation. Cell Mol Immunol 2015; 12: 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey S, Dempsey E, Long A. The role of chemokines in acute and chronic hepatitis C infection. Cell Mol Immunol 2014; 11: 25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Lugt B, Khan AA, Hackney JA, Agrawal S, Lesch J, Zhou M et al. Transcriptional programming of dendritic cells for enhanced MHC class II antigen presentation. Nat Immunol 2014; 15: 161–167. [DOI] [PubMed] [Google Scholar]
- Schlitzer A, Sivakamasundari V, Chen J, Sumatoh HR, Schreuder J, Lum J et al. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat Immunol 2015; 16: 718–728. [DOI] [PubMed] [Google Scholar]
- Grajales-Reyes GE, Iwata A, Albring J, Wu X, Tussiwand R, Kc W et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α(+) conventional DC clonogenic progenitor. Nat Immunol 2015; 16: 708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson TM, Keown AA, Cmero M, Yeo JH, Kumar A, Lew AM et al. Drosha controls dendritic cell development by cleaving messenger RNAs encoding inhibitors of myelopoiesis. Nat Immunol 2015; 16: 1134–1141. [DOI] [PubMed] [Google Scholar]
- van der Veen AG, Maillard PV. Reis e Sousa C. Drosha cuts the tethers of myelopoiesis. Nat Immunol 2015; 16: 1110–1112. [DOI] [PubMed] [Google Scholar]
- Natsuaki Y, Egawa G, Nakamizo S, Ono S, Hanakawa S, Okada T et al. Perivascular leukocyte clusters are essential for efficient activation of effector T cells in the skin. Nat Immunol 2014; 15: 1064–1069. [DOI] [PubMed] [Google Scholar]
- Mueller SN. Skin DCs cluster for efficient T cell activation. Nat Immunol 2014; 15: 1004–1005. [DOI] [PubMed] [Google Scholar]
- Ulvmar MH, Werth K, Braun A, Kelay P, Hub E, Eller K et al. The atypical chemokine receptor CCRL1 shapes functional CCL21 gradients in lymph nodes. Nat Immunol 2014; 15: 623–630. [DOI] [PubMed] [Google Scholar]
- Woodruff MC, Turley SJ. Chemokine 'grooming' by cLECs directs DC migration. Nat Immunol 2014; 15: 595–596. [DOI] [PubMed] [Google Scholar]
- Loschko J, Schreiber HA, Rieke GJ, Esterházy D, Meredith MM, Pedicord VA et al. Absence of MHC class II on cDCs results in microbial-dependent intestinal inflammation. J Exp Med 2016; 213: 517–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panek CA, Ramos MV, Mejias MP, Abrey-Recalde MJ, Fernandez-Brando RJ, Gori MS et al. Differential expression of the fractalkine chemokine receptor (CX3CR1) in human monocytes during differentiation. Cell Mol Immunol 2015; 12: 669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts B, Amiel E, Huang SC, Smith AM, Chang CH, Lam WY et al. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKɛ supports the anabolic demands of dendritic cell activation. Nat Immunol 2014; 15: 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LA. Glycolytic reprogramming by TLRs in dendritic cells. Nat Immunol 2014; 15: 314–315. [DOI] [PubMed] [Google Scholar]
- Liu J, Cao X. Regulatory dendritic cells in autoimmunity: a comprehensive review. J Autoimmun 2015; 63: 1–12. [DOI] [PubMed] [Google Scholar]
- Kopf M, Schneider C, Nobs SP. The development and function of lung-resident macrophages and dendritic cells. Nat Immunol 2015; 16: 36–44. [DOI] [PubMed] [Google Scholar]
- Watchmaker PB, Lahl K, Lee M, Baumjohann D, Morton J, Kim SJ et al. Comparative transcriptional and functional profiling defines conserved programs of intestinal DC differentiation in humans and mice. Nat Immunol 2014; 15: 98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring AJ, Ann D'Angelo J. Dissecting the dendritic cell controversy in chronic hepatitis B virus infection. Cell Mol Immunol 2015; 12: 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoletti A, Kennedy PT. The immune tolerant phase of chronic HBV infection: new perspectives on an old concept. Cell Mol Immunol 2015; 12: 258–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Cao X. Intratumoral dendritic cells in the anti-tumor immune response. Cell Mol Immunol 2015; 12: 387–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weist BM, Kurd N, Boussier J, Chan SW, Robey EA. Thymic regulatory T cell niche size is dictated by limiting IL-2 from antigen-bearing dendritic cells and feedback competition. Nat Immunol 2015; 16: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterházy D, Loschko J, London M, Jove V, Oliveira TY, Mucida D. Classical dendritic cells are required for dietary antigen-mediated induction of peripheral T(reg) cells and tolerance. Nat Immunol 2016; 17: 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanets-Korbut O, Kovalevska LM, Seya T, Sidorenko SP, Horvat B. Measles virus hemagglutinin triggers intracellular signaling in CD150-expressing dendritic cells and inhibits immune response. Cell Mol Immunol; e-pub ahead of print 15 June 2015; doi:10.1038/cmi.2015.55. [DOI] [PMC free article] [PubMed]
- Nair-Gupta P, Baccarini A, Tung N, Seyffer F, Florey O, Huang Y et al. TLR signals induce phagosomal MHC-I delivery from the endosomal recycling compartment to allow cross-presentation. Cell 2014; 158: 506–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samie M, Cresswell P. The transcription factor TFEB acts as a molecular switch that regulates exogenous antigen-presentation pathways. Nat Immunol 2015; 16: 729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio F, Tavernier SJ, Hoffmann E, Saeys Y, Martens L, Vetters J et al. The unfolded-protein-response sensor IRE-1α regulates the function of CD8α+ dendritic cells. Nat Immunol 2014; 15: 248–257. [DOI] [PubMed] [Google Scholar]
- Subramanian M, Tabas I. A new RIDDle in DC-mediated cross-presentation. Nat Immunol 2014; 15: 213–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S, Pulendran B, Lambrecht BN. Emerging functions of the unfolded protein response in immunity. Nat Immunol 2014; 15: 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katakam AK, Brightbill H, Franci C, Kung C, Nunez V, Jones C 3rd et al. Dendritic cells require NIK for CD40-dependent cross-priming of CD8+ T cells. Proc Natl Acad Sci USA 2015; 112: 14664–14669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Guo Z, Liu Y, Li X, Zhang Q, Xu X et al. The lectin Siglec-G inhibits dendritic cell cross-presentation by impairing MHC class I-peptide complex formation. Nat Immunol 2016; 17: 1167–1175. [DOI] [PubMed] [Google Scholar]
- Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat Immunol 2011; 12: 21–27. [DOI] [PubMed] [Google Scholar]
- Klose CS, Artis D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol 2016; 17: 765–774. [DOI] [PubMed] [Google Scholar]
- Artis D, Spits H. The biology of innate lymphoid cells. Nature 2015; 517: 293–301. [DOI] [PubMed] [Google Scholar]
- Hernández PP, Mahlakõiv T, Yang I, Schwierzeck V, Nguyen N, Guendel F et al. Interferon-λ and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat Immunol 2015; 16: 698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjösberg JM, Trifari S, Crellin NK, Peters CP, van Drunen CM, Piet B et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol 2011; 12: 1055–1062. [DOI] [PubMed] [Google Scholar]
- Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol 2011; 12: 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm C, Harrison OJ, Schmitt V, Pelletier M, Spencer SP, Urban JF Jr et al. Critical role of fatty acid metabolism in ILC2-mediated barrier protection during malnutrition and helminth infection. J Exp Med 2016; 213: 1409–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antignano F, Braam M, Hughes MR, Chenery AL, Burrows K, Gold MJ et al. G9a regulates group 2 innate lymphoid cell development by repressing the group 3 innate lymphoid cell program. J Exp Med 2016; 213: 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Guo L, Qiu J, Chen X, Hu-Li J, Siebenlist U et al. IL-25-responsive, lineage-negative KLRG1(hi) cells are multipotential 'inflammatory' type 2 innate lymphoid cells. Nat Immunol 2015; 16: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyasu S. Inflammatory ILC2 cells: disguising themselves as progenitors? Nat Immunol 2015; 16: 133–134. [DOI] [PubMed] [Google Scholar]
- Ohne Y, Silver JS, Thompson-Snipes L, Collet MA, Blanck JP, Cantarel BL et al. IL-1 is a critical regulator of group 2 innate lymphoid cell function and plasticity. Nat Immunol 2016; 17: 646–655. [DOI] [PubMed] [Google Scholar]
- Duerr CU, McCarthy CD, Mindt BC, Rubio M, Meli AP, Pothlichet J et al. Type I interferon restricts type 2 immunopathology through the regulation of group 2 innate lymphoid cells. Nat Immunol 2016; 17: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim TY, Hwang YY, Scanlon ST, Zaghouani H, Garbi N, Fallon PG et al. Group 2 innate lymphoid cells license dendritic cells to potentiate memory TH2 cell responses. Nat Immunol 2016; 17: 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature 2013; 498: 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri G, Miyajima M, Bascones S, Mortha A, Puga I, Cassis L et al. Innate lymphoid cells integrate stromal and immunological signals to enhance antibody production by splenic marginal zone B cells. Nat Immunol 2014; 15: 354–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD. ILCs in the zone. Nat Immunol 2014; 15: 313–314. [DOI] [PubMed] [Google Scholar]
- Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol 2015; 16: 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach A. Profiling the diversity of innate lymphoid cells. Nat Immunol 2015; 16: 222–224. [DOI] [PubMed] [Google Scholar]
- Ebihara T, Song C, Ryu SH, Plougastel-Douglas B, Yang L, Levanon D et al. Runx3 specifies lineage commitment of innate lymphoid cells. Nat Immunol 2015; 16: 1124–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehus CR, Aliahmad P, de la Torre B, Iliev ID, Spurka L, Funari VA et al. The development of innate lymphoid cells requires TOX-dependent generation of a common innate lymphoid cell progenitor. Nat Immunol 2015; 16: 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H. TOX sets the stage for innate lymphoid cells. Nat Immunol 2015; 16: 594–595. [DOI] [PubMed] [Google Scholar]
- Yang Q, Li F, Harly C, Xing S, Ye L, Xia X et al. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat Immunol 2015; 16: 1044–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J ILC. development: TCF-1 reporting. in. Nat Immunol 2015; 16: 1011–1012. [DOI] [PubMed] [Google Scholar]
- Sun C, Sun H, Zhang C, Tian Z. NK cell receptor imbalance and NK cell dysfunction in HBV infection and hepatocellular carcinoma. Cell Mol Immunol 2015; 12: 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay PK, Gierahn TM, Roederer M, Love JC. Single-cell technologies for monitoring immune systems. Nat Immunol 2014; 15: 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke P, Reyes A, Pinto S, Rattay K, Nguyen M, Küchler R et al. Single-cell transcriptome analysis reveals coordinated ectopic gene-expression patterns in medullary thymic epithelial cells. Nat Immunol 2015; 16: 933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton CM, Fucile CF, Darce J, Chida A, Ichikawa T, Gregoretti I et al. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat Immunol 2015; 16: 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd BA, Peters LA, Schadt EE, Dudley JT. Unifying immunology with informatics and multiscale biology. Nat Immunol 2014; 15: 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S, Zhao M, Budelsky A, de Mingo Pulido A, Day J, Fu Z et al. A new mouse strain for the analysis of invariant NKT cell function. Nat Immunol 2015; 16: 799–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Walle L, Van Opdenbosch N, Jacques P, Fossoul A, Verheugen E, Vogel P et al. Negative regulation of the NLRP3 inflammasome by A20 protects against arthritis. Nature 2014; 512: 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff S, Brewitz A, Gerner MY, Klauschen F, Komander K, Hemmi H et al. Robust anti-viral immunity requires multiple distinct T cell-dendritic cell interactions. Cell 2015; 162: 1322–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher B, Schlitzer A, Chen J, Mair F, Sumatoh HR, Teng KW et al. High-dimensional analysis of the murine myeloid cell system. Nat Immunol 2014; 15: 1181–1189. [DOI] [PubMed] [Google Scholar]
- Hirata E, Girotti MR, Viros A, Hooper S, Spencer-Dene B, Matsuda M et al. Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin β1/FAK signaling. Cancer Cell 2015; 27: 574–588. [DOI] [PMC free article] [PubMed] [Google Scholar]