Abstract
Reduced brain-derived neurotrophic factor (BDNF) may underlie age-related synaptic loss, in turn contributing to cerebral atrophy, cognitive decline, and increased risk for psychiatric disorders. However, the specific contribution of BDNF to the age-related expression changes in synaptic markers and their temporal trajectories remain uncharacterized. Using microarray data from orbitofrontal cortex of control subjects (n=209; 16–96 years), we identified genes whose expression positively correlates with BDNF (r>0.575; n=200 genes) and analyzed them for enriched biological pathways. qPCR was performed to measure the expression level of transcript variants of BDNF, NTRK2, and selected BDNF-coexpressed genes in younger and older subjects. We confirmed age-related downregulation of BDNF and show 78 of the top 200 BDNF-coexpressed genes are associated with synaptic function. Both excitatory and inhibitory synaptic genes show decreased expression with age and are positively correlated with BDNF and NTRK2 expression and negatively correlated with dominant-negative truncated NTRK2 level. Results were validated at the RNA level in an independent cohort and at the protein level for selected findings. We next tested the causal link between the correlative human findings using mice with conditional blockade of BDNF/NTRK2 signaling. Blockade of NTRK2 activity in adult mice recapitulate the age-like pattern in the expression of markers for inhibitory presynaptic but notably not for excitatory synaptic genes. Together, these findings suggest that age-dependent decrease in BDNF signaling may cause synaptic alterations through an initial and preferential effect on GABA presynaptic genes. These results have implications for neuropsychiatric disorders characterized by accelerated aging molecular profiles, such as major depression.
INTRODUCTION
Normal brain aging is associated with progressive cellular and structural changes, cognitive decline, and increased vulnerability to neurobiological diseases. With dramatic growth of the older population, the need to understand the mechanism and consequence of aging on the brain has become critically important. Imaging studies report smaller prefrontal cortex (PFC) volume in elderly subjects without obvious neurological disease (Tisserand et al, 2002). Consistent with functional impairment and brain atrophy, a decrease in neuronal body size, dendritic length, and loss of synapses without loss of neurons have been observed in postmortem studies (Morrison and Baxter, 2012; Rajkowska, 2000; Stockmeier et al, 2004).
The effect of aging on the incidence of psychiatric disease remains controversial (Blazer, 1994; Glaesmer et al, 2011; Kessler et al, 2010). For instance, while subthreshold depression and depressive symptoms are more common in later life (Beekman et al, 1995), they are often misattributed to aging per se, leading to underdiagnosis of depression. Animal studies show that the effects of stress on cognitive, neuroendocrine, biochemical, and anatomical changes are augmented in older subjects (Juster et al, 2010; Lupien et al, 2009). Recent advances in understanding the molecular and cellular bases of altered mood regulation in adult depressed subjects, and the observation of a considerable overlap between normal aging and depression-related brain changes (McKinney et al, 2012; McKinney and Sibille, 2013), suggest that aspects of mood-regulatory mechanisms may be selectively vulnerable to early homeostatic changes during normal aging. Indeed, older age is a potent risk factor for the functional decline of the PFC, which is important for cognitive function, working memory, and emotion regulation across species, ranging from rodents to humans.
BDNF, a small secreted protein, and its receptor, NTRK2 (also known as TRKB), have important roles in neuronal development, differentiation, maintenance, and plasticity throughout life. In the brain, two isoforms of NTRK2 are abundantly expressed: full-length (NTRK2-FL) and truncated form (NTRK2-T1). In contrast to NTRK2-FL, which binds to BDNF and activates downstream kinase cascades, NTRK2-T1 lacks intracellular catalytic domain and therefore acts as an endogenous inhibitor of NTRK2-FL by competing for the available BDNF pool (Eide et al, 1996; Gupta et al, 2013). Low BDNF and/or NTRK2 expression has been reported in multiple brain disorders and during normal brain aging, which are often accompanied by mild brain atrophy, reduced neuronal function, and synaptic loss (Guilloux et al, 2012; Hattiangady et al, 2005; Howells et al, 2000; Romanczyk et al, 2002; Thompson Ray et al, 2011; Wong et al, 2013). Brain-specific deletion of BDNF or NTRK2 in mouse induces neuronal shrinkage, dendritic retraction in the cortex, and cognitive and learning deficit (Gorski et al, 2003a; Gorski et al, 2003b; Xu et al, 2000). These data imply that normal aging-associated BDNF and NTRK2 changes in the PFC might compromise synaptic integrity and function, although this link has not been tested in humans.
The biological role of BDNF has been thoroughly studied; however, investigating BDNF-driven changes in vivo and/or in human subjects is challenging owing to its low endogenous level and complex regulation by multiple promoters. Most studies were conducted with in vitro models, sometimes with the aid of overexpression or exogenous application of BDNF. Moreover, whereas changes in BDNF levels seem an obvious candidate biological event for age-related structural and functional changes, numerous other age-related complex processes simultaneously occur, including among others, increased inflammation, reduced blood flow, and accumulated free radical damage to macromolecules, all potentially contributing to the molecular, structural, and functional alterations of the brain.
In the present study, we investigated age-related BDNF change in the human PFC and specifically focused on its putative contribution to GABA and glutamate synaptic-related transcriptome changes occurring during normal aging. We then used an inducible genetic mouse model to directly test the putative causal role of BDNF/NTRK2 signaling in aging-associated gene expression alterations. We predicted that reduced BDNF would directly contribute to low synaptic function through altered expression of markers for inhibitory and excitatory neurons and synaptic function-related genes.
MATERIALS AND METHODS
Human Postmortem Subjects
Postmortem brains were collected from the Allegheny County Coroner's Office (Pittsburgh, PA) after consent from the next of kin. A committee of experienced clinical scientists examined clinical records, toxicology, and standardized psychological autopsy data for all cases. Individuals were also screened for the absence of neurodegenerative disorders by neuropathological examination. All procedures were approved by the University of Pittsburgh's Committee for the Oversight of Research Involving the Dead and Institutional Review Board for Biomedical Research. After careful examination of clinical and technical parameters, 209 control subjects without DSM-IV diagnosis were selected. Because of the large cohort size, a subcohort was generated for qPCR validation (n=40/group) and immunoblot (n=10/group) based on groups with clear age separation between younger (⩽42 years) and older (⩾60 years) age.
To validate our findings with independent samples, fresh-frozen postmortem brain tissues were obtained from the Douglas–Bell Canada brain bank (Douglas Mental Health University Institute, Montreal, Canada). Psychological autopsy data and medical and demographic information were thoroughly examined to generate a cohort of 13 younger and 13 older subjects. The effects of age on gene expression are mostly linear (Erraji-Benchekroun et al, 2005) so the chosen age thresholds were arbitrary but they provided two balanced groups that did not differ in other measures (Supplementary Table S1).
Animals and Drug Treatment
NTRK2F616A heterozygote mice, which harbor a point mutation in the ATP-binding pocket of NTRK2 that is selectively blocked by ATP competitive kinase inhibitor 1NMPP1, resulting in a blockade of NTRK2-mediated signaling (Chen et al, 2005), were obtained from Jackson laboratories (Bar Harbor, ME) and intercrossed to generate homozygote mice. Male homozygote mice (9–10 weeks old) were fed with 25 μM 1NMPP1 or vehicle (0.0003% DMSO) via drinking water for 3 weeks and killed. Medial PFCs were collected and stored at −80 °C until RNA isolation.
RNA Extraction
Human RNA was isolated from all six cortical layers of orbitofrontal cortex (Brodmann area 47) using TRIzol (Invitrogen Life Technologies, Carlsbad, CA) and further purified with RNeasy spin columns (QIAGEN, Valencia, CA). To isolate total RNA from mouse cortices, RNeasy Micro Plus Kit (QIAGEN) was directly used.
Gene Arrays and Expression Analysis
RNA samples were processed for microarray analysis using Affymetrix GeneChip Human Gene 1.1ST, according to the manufacturer's protocol (Affymetrix, Santa Clara, CA). Gene expression data were extracted using Expression Console build 1.2.1.20 and normalized with quantile normalization method to eliminate batch effects. To identify the effect of age on gene expression, each expression value was fitted to a regression model using potential confounding covariates. The residuals of each gene were further adjusted by a power function regression model for age effect only.
BDNF Coexpression Network Analysis
Pearson's correlation values were calculated between BDNF and each transcript examined by array. The top 200 genes positively correlated with BDNF (r>0.575) were analyzed and visualized using Cytoscape (version 3.1.1) (Shannon et al, 2003) with ClueGO plugin (version 2.1.7) (Bindea et al, 2009) and further examined with DAVID Functional Annotation Clustering Tool (Huang da et al, 2009a, b). The group p-values that are reported in the article are adjusted with Bonferroni step down method.
Real-Time Quantitative PCR (qPCR)
cDNA was synthesized with total RNA using qScript cDNA supermix (Quanta BioSciences, Gaithersburg, MD). PCR products were amplified in triplicate on a Mastercycler real-time PCR machine (Eppendorf, Hamburg, Germany) using universal PCR conditions. Results were calculated as the geometric mean of threshold cycles normalized to three validated internal controls (actin, glyceraldehyde-3-phosphate dehydrogenase, and cyclophilin G). Based on the BDNF coexpression network analysis, candidate genes for qPCR verification were selected: (i) BDNF transcript variants (exon1, exon2, exon4, exon6, protein-coding sequence); (ii) two NTRK2 isoforms (NTRK2-FL, NTRK2-T1); (iii) five excitatory synapse-related genes: solute carrier family 17; member 7 (SLC17A7, also known as vGLUT1); glutamate receptor, AMPA1 (GRIA1); glutamate receptors, NMDA 2A and NMDA 2B (GRIN2A, GRIN2B); discs large homolog 4 (DLG4, also known as PSD95); (iv) eight inhibitory neuronal genes (presynaptic markers): solute carrier family 32, member 1 (SLC32A1, also known as vGAT); and seven interneuron markers (SST, NPY, CORT, PVALB, CCK, GAD1, and GAD2); and (v) three postsynaptic GABA receptors (GABRA4, GABRA5, GABRB3).
Protein Isolation and Western Blotting
About 15 mm3 of orbitofrontal cortices were collected in RIPA lysis buffer (50 mM Tris-Cl, 150 mM NaCl, 1% Triton-X100, 0.5% sodium deoxycholate, 0.1% SDS, phosphatase inhibitor cocktails (04906837001; Sigma-Aldrich, Oakville, ON, Canada), protease inhibitors (539131; EMD Millipore, Etobicoke, ON, Canada)). In all, 20–50 μg of total protein samples were separated on 4–15% gradient gel (4561083; Bio-Rad Laboratories, Mississauga, ON, Canada) and blotted to PVDF membrane. After blocking with 5% non-fat milk, membranes were incubated overnight at 4 °C with the following primary antibodies: anti-NTRK2 (sc-8316; 1:2,000, Santa Cruz Biotechnology, Santa Cruz, CA), anti-β-actin (A2228; 1:10,000, Sigma-Aldrich), anti-GRIN2A (4205; 1:1,000, Cell Signaling Technology, Danvers, MA), anti-GRIN2B (06-600; 1:1,000, EMD Millipore), anti-DLG4 (2507; 1:2,000, Cell Signaling Technology), anti-SLC32A1 (131011, 1:1,000, Synaptic systems, Goettingen, Germany), and anti-GAD1 (MAB5406, 1:5,000, EMD Millipore). After rinse, membranes were incubated with peroxidase-conjugated secondary antibodies (Vector Laboratories, Burlington, ON, Canada) and developed with chemiluminescence substrate (34096; Fisher Scientific). Immuno-reactive bands were scanned using ChemiDoc XRS imaging system (Bio-Rad Laboratories). The intensity of each band was normalized to the intensity of corresponding β-actin band.
Statistical Analysis
Analyses were performed using SPSS (SPSS, Chicago, IL). Correlations between two genes or between gene and age were calculated using Pearson's correlation coefficient. Gene expression differences between two age groups were determined by analysis of covariance (ANCOVA) as described elsewhere (Tripp et al, 2011). To determine relevant covariates, each nominal factor was tested as the main factor in ANOVA, scale covariates were tested by Pearson's correlation. Results of covariate factor analyses were corrected by Bonferroni–Holm method. The final ANCOVA models included only significant co-factors and were then applied to our genes of interest as the dependent variable and subject group as the main effect. For the rodent drug studies, we performed and replicated our assays using two separate cohorts. Unpaired t-tests were performed for the separate cohorts, and p-values were combined using Stouffer's z-trend method (Whitlock, 2005) and reported in the article.
RESULTS
BDNF and BDNF-Associated Transcriptome Changes in the Aging Human Brain
Large-scale gene expression data were obtained by microarray analysis in the orbitofrontal cortex of 209 postmortem samples from subjects without psychiatric or neurological illness, ranging from 16 to 96 years of age. BDNF expression gradually decreased with age (Pearson's correlation value to age (r)=−0.36, p<0.0000001; Figure 1a). To identify BDNF-related transcriptome changes, we calculated Pearson's correlation values between BDNF and each gene probeset. Overall, genes with higher correlation to BDNF showed larger age-related changes (Figure 1b).
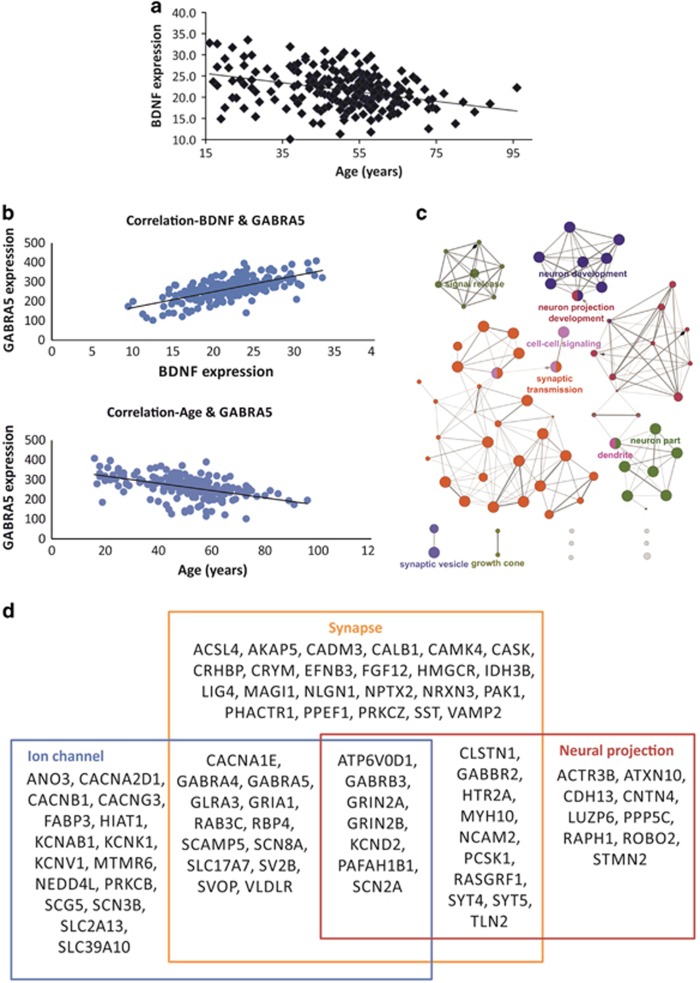
Figure 1.
Age-dependent changes in BDNF expression and identification of BDNF-coexpressed genes. (a) Age-related reduction of BDNF in human frontal cortex revealed by array (r=−0.36, p<0.0000001). (b) Example of opposite correlations of BDNF and age for genes of interest: GABRA5 expression is positively correlated with BDNF and negatively with age. (c) Sets of GO terms enriched in top 200 genes correlated with BDNF in human frontal cortex as identified by Cytoscape with ClueGO. (d) Synapse-related genes enriched in BDNF coexpression network as identified by DAVID functional analysis.
To investigate biological pathways potentially affected by BDNF, genes positively correlated to BDNF were analyzed with Cytoscape with ClueGO (Figure 1c). As similar results were obtained with the top 100 and 200 genes, we focused the report on the top 200 genes (r>0.575, full gene list is available in Supplementary Table S2). Among 75 GO terms with p<0.01, 9 functional groups were identified: synaptic transmission (66 genes, p=2.4E-22), neuron projection development (47 genes, p=3.2E-15), neuron part (49 genes, p=1.3E-17), neuron development (46 genes, p=5.7E-16), signal release (20 genes, p=2.9E-07), dendrite (24 genes, p=1.6E-10), cell–cell signaling (54 genes, p=3.8E-18), synaptic vesicle (12 genes, p=3.7E-08), and growth cone (9 genes, p=1.9E-05) (full annotations are presented in Supplementary Figure S1). DAVID Functional Annotation Clustering Tool confirmed that the most enriched gene cluster in the BDNF coexpression network included synapse-related genes (enrichment score=6.93). The second and third clusters identified by DAVID analysis were also associated with synapse (Figure 1d and Supplementary Table S3). Genes in top three clusters were related to voltage-gated channels (CACNA2D1, KCND2, SCN3B, KCNAB1, SCN2A, CACNB1, CACNA1E, CACNG3, SCN8A, KCNK1, KCNV1), neurotransmitter receptors (GABRB3, GRIN2B, GABRA4, GRIA1, GLRA3, GABRA5, GRIN2A, GABBR2, HTR2A), synaptic vesicle-related molecules (SLC17A7, SVOP, RAB3C, GRIN2B, SYT4, SYT5, SV2B, ATP6V0D1), and synaptic cell adhesion molecules (NLGN1, NRXN3) (Figure 1d).
As most investigated genes showed age effects, an important control analysis is to rule out a general effect of age underlying the correlations between BDNF and the expression of other genes. Accordingly, we performed the same analysis using data in which the effect of age was analytically subtracted (ie, using age-residual values). Results indicated very similar patterns of correlation values at the level of individual genes and for the functional clustering analysis: 145 of the top 200 were overlapping with the previous analysis and synapse-related genes were enriched in BDNF coexpressed genes (Supplementary Figure S2). This demonstrated a role for BDNF beyond, or in addition to, the effect of age in the observed gene expression trajectories.
Excitatory and Inhibitory Synaptic Genes Demonstrate Age-Related Changes that Parallel Changes in BDNF
DAVID functional annotation tool revealed that GABA as well as glutamate receptor activity-related genes were significantly enriched in the top 200 BDNF coexpression network: GABA receptor activity-related genes (GO:16917): GABBR2, GABRA4, GABRA5, GABRB3 (fold enrichment=13.7, p=0.003), and glutamate receptor activity-related genes (GO:0008066): GABBR2, GRIA1, GRIN2A, GRIN2B (fold enrichment=10.6, p=0.006). To investigate whether additional markers of excitatory and inhibitory synapses undergo aging-related changes, we performed the same analysis on the full expression data set (ie, not limited to BDNF-coexpressed genes). Overall, age-related downregulation was observed across excitatory and inhibitory synaptic markers (percentage of genes with significant negative correlation to age: 68.4% of GABA synaptic genes, 63.0% of glutamate synaptic genes, Table 1), suggesting a general suppression of neurotransmission. Although the degrees of correlation to BDNF expression were slightly reduced using age-residual values, 79 out of the 84 synaptic genes showed the same directionality and significance of correlation using residual or uncorrected expression values (94% matching; Table 1).
Table 1. Correlation Between Age-Related BDNF Expression and Expression Level or Age-Adjusted Residual Expression Values of Synapse-Related Genes.
|
Correlation and
p-values |
|||||||
|---|---|---|---|---|---|---|---|
| Gene symbol | mRNA accession | Genes to age | p-Value | Genes to BDNF | p-Value | Residuals to BDNF | p-Value |
| Excitatory synpse related | |||||||
| DLG1 | NM_001098424 | 0.12 | 0.043 | 0.11 | 0.056 | 0.16 | 0.012 |
| DLG2 | NM_001364 | −0.40 | <1.E−07 | 0.52 | <1.E−07 | 0.39 | <1.E−07 |
| DLG3 | NM_021120 | −0.41 | <1.E−07 | 0.64 | <1.E−07 | 0.54 | <1.E−07 |
| DLG4 | NM_001365 | −0.47 | <1.E−07 | 0.48 | <1.E−07 | 0.31 | 3.E−06 |
| GLS | NM_014905 | −0.09 | 0.087 | 0.50 | <1.E−07 | 0.44 | <1.E−07 |
| GLUL | NM_002065 | 0.18 | 0.005 | −0.46 | <1.E−07 | −0.35 | <1.E−07 |
| GLUL | NM_002065 | 0.23 | 3.E−04 | −0.46 | <1.E−07 | −0.33 | 5.E−07 |
| GRIA1 | NM_000827 | −0.33 | 1.E−06 | 0.63 | <1.E−07 | 0.52 | <1.E−07 |
| GRIA2 | NM_001083619 | −0.29 | 1.E−05 | 0.51 | <1.E−07 | 0.42 | <1.E−07 |
| GRIA3 | NM_007325 | −0.09 | 0.087 | 0.48 | <1.E−07 | 0.41 | <1.E−07 |
| GRIA4 | NM_000829 | −0.41 | <1.E−07 | 0.54 | <1.E−07 | 0.42 | <1.E−07 |
| GRID1 | NM_017551 | 0.06 | 0.213 | −0.19 | 0.003 | −0.14 | 0.019 |
| GRID2 | NM_001510 | −0.34 | <1.E−07 | 0.48 | <1.E−07 | 0.37 | <1.E−07 |
| GRIK1 | NM_175611 | −0.60 | <1.E−07 | 0.52 | <1.E-07 | 0.32 | 1.E−06 |
| GRIK2 | NM_175768 | 0.11 | 0.050 | −0.09 | 0.109 | −0.03 | 0.321 |
| GRIK3 | NM_000831 | −0.50 | <1.E−07 | 0.55 | <1.E−07 | 0.40 | <1.E−07 |
| GRIK4 | NM_014619 | −0.18 | 0.004 | 0.12 | 0.042 | 0.07 | 0.162 |
| GRIK5 | NM_002088 | −0.32 | 2.E−06 | 0.23 | 0.001 | 0.14 | 0.026 |
| GRIN1 | NM_007327 | −0.29 | 9.E−06 | 0.39 | <1.E−07 | 0.29 | 1.E−05 |
| GRIN2A | NM_001134407 | −0.51 | <1.E−07 | 0.61 | <1.E−07 | 0.47 | <1.E−07 |
| GRIN2B | NM_000834 | −0.52 | <1.E−07 | 0.62 | <1.E−07 | 0.49 | <1.E−07 |
| GRIN2C | NM_000835 | 0.10 | 0.082 | -0.38 | <1.E−07 | −0.33 | 5.E−07 |
| GRIN2D | NM_000836 | −0.12 | 0.041 | −0.06 | 0.197 | −0.08 | 0.127 |
| GRIN3A | NM_133445 | −0.57 | <1.E−07 | 0.57 | <1.E−07 | 0.44 | <1.E−07 |
| GRIN3B | NM_138690 | 0.00 | 0.497 | −0.27 | 5.E−05 | −0.27 | 5.E−05 |
| GRINA | NM_000837 | −0.16 | 0.011 | 0.29 | 1.E−05 | 0.21 | 0.001 |
| GRIP1 | NM_021150 | 0.05 | 0.241 | 0.21 | 0.001 | 0.20 | 0.002 |
| GRIP2 | NM_001080423 | −0.13 | 0.027 | −0.12 | 0.037 | −0.19 | 0.003 |
| GRM1 | NM_001114329 | −0.22 | 0.001 | 0.55 | <1.E−07 | 0.46 | <1.E−07 |
| GRM2 | NM_000839 | −0.43 | <1.E−07 | 0.41 | <1.E−07 | 0.29 | 1.E−05 |
| GRM3 | NM_000840 | −0.57 | <1.E−07 | 0.32 | 2.E−06 | 0.15 | 0.015 |
| GRM4 | NM_000841 | −0.06 | 0.185 | −0.06 | 0.185 | −0.09 | 0.108 |
| GRM5 | NM_001143831 | −0.27 | 5.E−05 | 0.44 | <1.E−07 | 0.34 | <1.E−07 |
| GRM6 | NM_000843 | 0.06 | 0.179 | −0.12 | 0.037 | −0.11 | 0.062 |
| GRM7 | NM_181874 | −0.32 | 2.E−06 | 0.28 | 2.E−05 | 0.19 | 0.003 |
| GRM8 | NM_001127323 | −0.18 | 0.006 | 0.43 | <1.E−07 | 0.40 | <1.E−07 |
| HOMER1 | NM_004272 | −0.67 | <1.E−07 | 0.55 | <1.E−07 | 0.44 | <1.E−07 |
| HOMER2 | NM_199330 | −0.19 | 0.003 | 0.07 | 0.167 | 0.01 | 0.429 |
| HOMER3 | NM_004838 | 0.04 | 0.266 | −0.40 | <1.E−07 | −0.39 | <1.E−07 |
| SLC17A1 | NM_005074 | 0.15 | 0.013 | −0.33 | 5.E−07 | −0.23 | 4.E−04 |
| SLC17A2 | NM_005835 | −0.01 | 0.450 | −0.07 | 0.172 | −0.07 | 0.153 |
| SLC17A6 | NM_020346 | −0.19 | 0.004 | 0.45 | <1.E−07 | 0.34 | <1.E−07 |
| SLC17A7 | NM_020309 | −0.40 | <1.E−07 | 0.60 | <1.E−07 | 0.50 | <1.E−07 |
| SLC17A8 | NM_139319 | −0.08 | 0.114 | 0.01 | 0.450 | −0.02 | 0.368 |
| SLC1A1 | NM_004170 | −0.26 | 8.E−05 | 0.34 | <1.E−07 | 0.25 | 1.E−04 |
| SLC1A4 | NM_003038 | 0.08 | 0.129 | −0.21 | 0.001 | −0.17 | 0.008 |
| Inhibitory synapse related | |||||||
| APITD1-CORT | NM_198544 | −0.45 | <1.E−07 | 0.55 | <1.E−07 | 0.40 | <1.E−07 |
| CALB1 | NM_004929 | −0.68 | <1.E−07 | 0.59 | <1.E−07 | 0.47 | <1.E−07 |
| CALB2 | NM_001740 | −0.40 | <1.E−07 | 0.39 | <1.E−07 | 0.25 | 1.E−04 |
| CCK | NM_000729 | −0.18 | 0.005 | 0.42 | <1.E−07 | 0.35 | <1.E−07 |
| GABARAP | NM_007278 | −0.05 | 0.216 | 0.28 | 2.E−05 | 0.23 | 5.E−04 |
| GABARAPL1 | NM_031412 | −0.20 | 0.002 | 0.40 | <1.E−07 | 0.32 | 2.E−06 |
| GABARAPL2 | NM_007285 | −0.01 | 0.418 | 0.26 | 6.E−05 | 0.21 | 0.001 |
| GABBR1 | NM_001470 | −0.31 | 2.E−06 | 0.36 | <1.E−07 | 0.21 | 0.001 |
| GABBR1 | NM_001470 | −0.30 | 4.E−06 | 0.36 | <1.E−07 | 0.23 | 4.E−04 |
| GABBR1 | NM_001470 | −0.29 | 9.E−06 | 0.34 | 5.E−07 | 0.23 | 4.E−04 |
| GABBR2 | NM_005458 | −0.50 | <1.E−07 | 0.59 | <1.E−07 | 0.48 | <1.E−07 |
| GABRA1 | NM_000806 | −0.21 | 0.001 | 0.54 | <1.E−07 | 0.46 | <1.E−07 |
| GABRA2 | NM_000807 | −0.18 | 0.004 | 0.11 | 0.054 | 0.08 | 0.123 |
| GABRA3 | NM_000808 | −0.26 | 8.E−05 | 0.57 | <1.E−07 | 0.47 | <1.E−07 |
| GABRA4 | NM_000809 | −0.35 | <1.E−07 | 0.63 | <1.E−07 | 0.53 | <1.E−07 |
| GABRA5 | NM_000810 | −0.50 | <1.E−07 | 0.67 | <1.E−07 | 0.58 | <1.E−07 |
| GABRA6 | NM_000811 | 0.07 | 0.157 | −0.12 | 0.044 | −0.09 | 0.088 |
| GABRB1 | NM_000812 | 0.06 | 0.192 | 0.14 | 0.019 | 0.23 | 4.E−04 |
| GABRB2 | NM_021911 | −0.22 | 0.001 | 0.53 | <1.E−07 | 0.44 | <1.E−07 |
| GABRB3 | NM_021912 | −0.39 | <1.E−07 | 0.63 | <1.E−07 | 0.51 | <1.E−07 |
| GABRD | NM_000815 | −0.22 | 0.001 | 0.48 | <1.E−07 | 0.35 | <1.E−07 |
| GABRE | NM_004961 | 0.11 | 0.053 | −0.19 | 0.004 | −0.09 | 0.100 |
| GABRG1 | NM_173536 | 0.05 | 0.259 | −0.37 | <1.E−07 | −0.30 | 6.E−06 |
| GABRG2 | NM_198904 | 0.05 | 0.250 | 0.44 | <1.E−07 | 0.43 | <1.E−07 |
| GABRG3 | NM_033223 | −0.24 | 3.E−04 | 0.33 | 5.E−07 | 0.26 | 7.E−05 |
| GABRP | NM_014211 | −0.04 | 0.262 | -0.06 | 0.195 | -0.08 | 0.139 |
| GABRQ | NM_018558 | −0.15 | 0.015 | 0.42 | <1.E−07 | 0.38 | <1.E−07 |
| GABRR1 | NM_002042 | 0.04 | 0.285 | −0.23 | 5.E−04 | -0.22 | 0.001 |
| GABRR2 | NM_002043 | 0.10 | 0.075 | −0.23 | 5.E−04 | −0.17 | 0.007 |
| GABRR3 | NM_001105580 | 0.10 | 0.072 | −0.09 | 0.098 | −0.09 | 0.108 |
| GAD1 | NM_000817 | −0.20 | 0.002 | 0.50 | <1.E−07 | 0.39 | <1.E−07 |
| GAD2 | NM_000818 | −0.27 | 3.E−05 | 0.51 | <1.E−07 | 0.39 | <1.E−07 |
| GPHN | NM_020806 | −0.15 | 0.016 | 0.16 | 0.010 | 0.13 | 0.031 |
| NPY | NM_000905 | −0.23 | 4.E−04 | 0.36 | <1.E−07 | 0.21 | 0.001 |
| PVALB | NM_002854 | −0.09 | 0.088 | 0.30 | 5.E−06 | 0.19 | 0.003 |
| SLC32A1 | NM_080552 | −0.30 | 6.E−06 | 0.44 | <1.E−07 | 0.28 | 2.E−05 |
| SST | NM_001048 | −0.50 | <1.E−07 | 0.59 | <1.E−07 | 0.45 | <1.E−07 |
| VIP | NM_003381 | −0.46 | <1.E−07 | 0.42 | <1.E−07 | 0.29 | 1.E−05 |
Note that correlations to BDNF are maintained after aging effect on gene expression is analytically removed.
Validation and Extension of Age-Dependent Changes in BDNF-, GABA-, and Glutamate Signaling-Related Genes
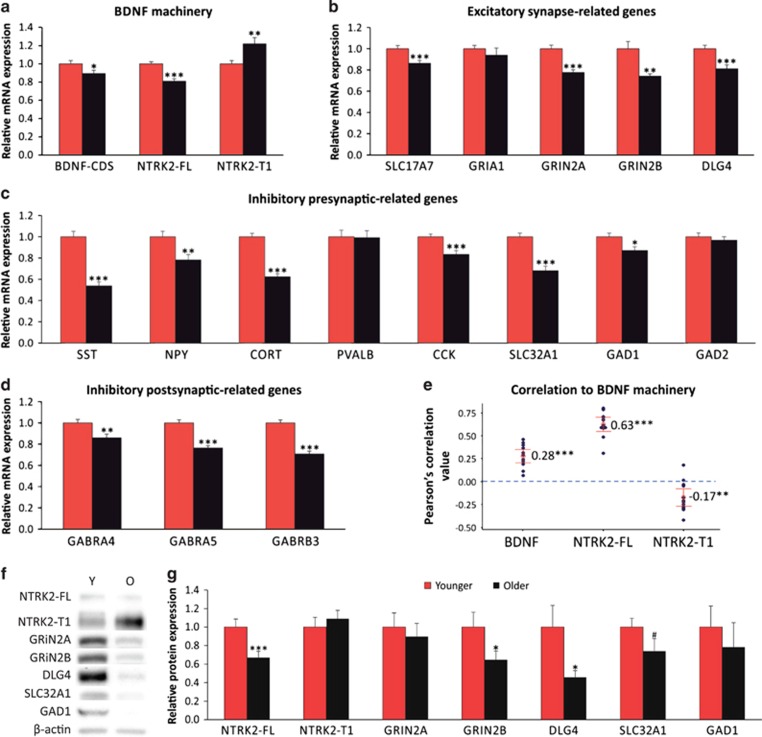
To extend and validate the microarray results, we performed qPCR with primers for BDNF machinery and synaptic gene transcripts in selected subcohorts of 40 younger and 40 older subjects. In addition to 16 glutamatergic and GABAergic synapse-related genes, we included primers targeting the protein-coding sequence and four different 5′ exons of BDNF as well as two isoforms of NTRK2 as the array data did not differentiate splice variants. Total BDNF level, which was measured by coding sequence-targeting primers, was significantly reduced (−10.7±3.6% compared with young, F=4.44, p=0.038) with age. Similar age-associated decreases were observed in BDNF exon2 (−49.6±13.0%, F=5.83, p=0.018) and exon4 (+) transcripts (31.0±3.2%, F=27.10, p=1.5E-06) but not in exon1 and exon6 (Supplementary Figure S3). Whereas the expression of full-length NTRK2 decreased (−19.1±2.7%, F=27.44, p=1.4E-06), the expression of the truncated form increased in the older brain (+21.9±6.8%, F=8.92, p=0.004). Most examined synaptic markers displayed lower expression in older subjects (Figures 2a–d): SLC17A7 (−13.8±2.4%, F=12.58, p=0.001), GRIN2A (−22.4±3.8%, F=19.07, p=3.8E-05), GRIN2B (−25.9±5.2%, F=9.38, p=0.003), DLG4 (−18.9±2.6%, F=19.04, p=3.9E-05), SST (−46.2±3.8%, F=49.04, p=8.2E-10), NPY (−21.7±5.0%, F=9.49, p=0.003), CORT (−37.5±2.7%, F=74.02, p=6.4E-13), CCK (−16.6±3.6%, F=14.23, p=3.2E-04), SLC32A1 (−31.9±4.3%, F=35.33, p=7.9E-08), GAD1 (−12.9±3.6%, F=5.68, p=0.020), GABRA4 (−14.1±3.6%, F=8.53, p=0.005), GABRA5 (−23.7±2.1%, F=43.36, p=4.8E-09), and GABRB3 (−29.3±2.8%, F=52.73, p=2.7E-10). Overall, the qPCR data replicated the array findings with high positive correlation (r=0.74, p=4.8E-04; Supplementary Figure S4).
Figure 2.
Validation of age-related gene expression changes in two different age groups. (a) Relative mRNA expression level (old/young) of BDNF signaling (BDNF, NTRK2-FL, NTRK2-T1) and (b–d) selected sets of synapse-related genes in aged subjects compared with the younger subjects (Younger (Red): ⩽42 years, Older (Black): ⩾60 years, n=40/group). Statistical significance of NTRK2-FL, NTRK2-T1, GRIN2A, DLG4, SLC32A1, SST, NPY, CCK, GAD1, and GABRB3 is corrected for multiple comparisons. (e) Pearson's correlation between BDNF signaling- and synaptic-associated genes in aging cohort (one-sample t-test to 0). (f) Representative immunoblots of NTRK2 and selected synaptic proteins in younger (Y) and older (O) subjects. (g) Relative protein level in each age group (n=10/group, #p<0.1 *p<0.05 **p<0.01 ***p<0.001).
Notably, the expression of synaptic genes showed the highest correlation to NTRK2-FL expression among BDNF signaling-associated genes (average r=0.63, p<0.0000001; Figure 2e), including NTRK2 isoforms and total and BDNF transcript variants (data of BDNF transcript variants were not shown), suggesting that low NTRK2-FL expression in the older brain may drive age-related changes of synaptic genes.
To further validate our findings, we attempted to replicate selected results in an independent cohort of younger and older adult subjects from a different brain bank (n=13/group). qPCR assessment of expression of 23 genes or exons show significant age-associated changes for BDNF machinery and synaptic genes. Results across the two cohorts were highly correlated (r=0.86, p<0.0000001; Supplementary Figure S5).
Finally, we assessed changes at the protein level in a subgroup of the original cohort (n=10/group). Significant age-associated downregulation of NTRK2-FL, GRIN2B, and DLG4 was also observed by quantitative western blotting analysis: NTRK2-FL (−32.9±6.9%, p=0.005), GRIN2B (−35.2±9.3%, p=0.042), and DLG4 (−54.1±7.1%, p=0.025). Although significant group differences were not found in other proteins, most likely because of the small group size and higher standard deviation in western blotting analysis, the direction of changes was the same with that of mRNA and the protein levels were overall highly correlated with mRNA levels (n=10/group; 7 genes; r=0.53, p=0.000001; Figures 2f and g).
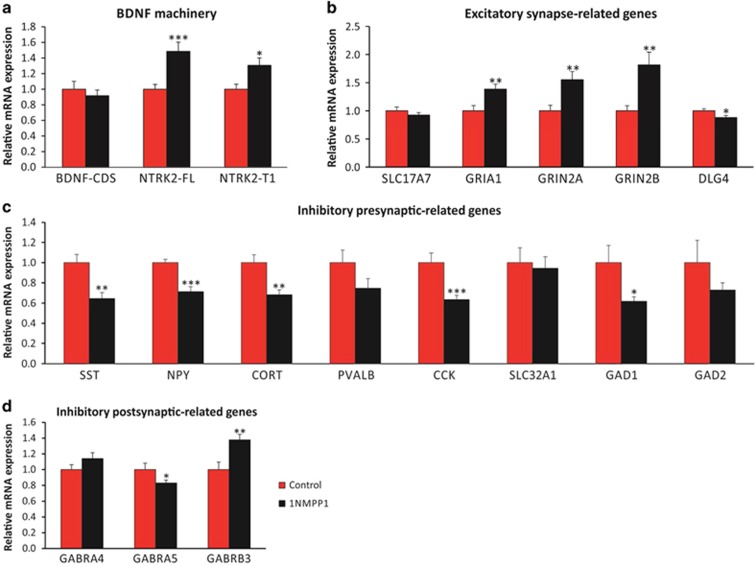
Temporal Blockade of BDNF Signaling is Sufficient to Induce Age-Like Changes in Inhibitory Synapse-Related Genes
To investigate whether low BDNF signaling is sufficient to induce the pattern of gene changes observed during aging, we utilized a pharmacogenetic approach using NTRK2F616A transgenic mice that harbor a point mutation in the ATP-binding domain of NTRK2. The modified NTRK2 is fully functional and does not cause detectable phenotype in the absence of 1NMPP1, a cell permeable kinase inhibitor (Chen et al, 2005). This model allows precise control of BDNF-NTRK2 signaling in adult animals, hence bypassing potential developmental compensations. Three weeks of 1NMPP1 treatment did not affect BDNF expression but resulted in increased expression of NTRK2-FL (+48.5±11.9%, p=0.001) and NTRK2-T1 (+30.6±9.5%, p=0.014) in the medial frontal cortex of NTRK2F616A homozygote mice (Figure 3a), as putative adaptations to reduced NTRK2 signaling. Downstream from reduced BDNF-NTRK2 signaling, five of the tested inhibitory presynaptic markers showed significant decreased expression changes that matched the human age-related changes: SST (−35.6±6.0%, p=0.001), NPY (−28.8±4.8%, p=2.4E-05), CORT (−31.7±4.7%, p=0.003), CCK (−36.6±4.4%, p=0.001), and GAD1 (−38.2±4.5%, p=0.016), (Figure 3c). In contrast, markers of excitatory synapse and inhibitory postsynaptic genes showed mixed patterns: GRIA1 (+38.6±8.7%, p=0.008), GRIN2A (+55.3±14.4%, p=0.006), GRIN2B (+81.6±22.7%, p=0.001), DLG4 (−12.0±3.4%, p=0.016), GABRA5 (−17.0±3.8%, p=0.041), and GABRB3 (+37.8±7.1%, p=0.003) (Figures 3b and d).
Figure 3.
Gene expression changes by temporal blockade of NTRK2. (a) Relative mRNA expression level (1NMPP1/vehicle) of BDNF signaling (BDNF, NTRK2-FL, NTRK2-T1) and (b–d) selected sets of synapse-related genes in 1NMPP1-treated adult mice compared with the control (n=12/each group; *p<0.05 **p<0.01 ***p<0.001).
These results indicate that the temporal inhibition of NTRK2 activity is sufficient to induce an aging-associated pattern in the expression of markers for inhibitory presynaptic but not excitatory synaptic genes in the frontal cortex of adult mice.
DISCUSSION
Reduced BDNF function has been suggested as a contributor to the molecular and cellular underpinnings of age-related brain deficits, but its specific contribution to age-related gene expression changes for excitatory and inhibitory synaptic markers remains uncharacterized. In the present study, we observed a progressive reduction of BDNF expression with age in the control human brain (Figure 1). The analysis of the BDNF gene coexpression network shows that the expression of multiple synapse-related genes is closely associated with BDNF expression (Figure 1) and also downregulated with aging (Table 1). Our isoform-specific qPCR investigation revealed that all BDNF signaling genes, total BDNF, NTRK2-FL, and NTRK2-T1, changed in the direction of lower BDNF function in the older human brain, which was also supported by decreased protein level of NTRK2-FL (Figure 2). Finally, using transgenic mice, we demonstrated that transient blockade of BDNF signaling, through NTRK2 inhibition in the adult brain, is sufficient to cause an aging-like gene expression profile, although restricted to GABA-related presynaptic markers (Figure 3). Thus we speculate that other synaptic-related changes observed during aging, namely, affecting glutamatergic markers, may occur as a long-term consequence of BDNF effects on GABA markers. These results significantly extend previous correlative findings on age-associated decrement of BDNF and NTRK2 in elderly humans and animal models (Hattiangady et al, 2005; O'Callaghan et al, 2009; Romanczyk et al, 2002; Silhol et al, 2007) in two ways: first, by delineating in the human brain the extent of the putative impact of reduced BDNF signaling on the age-related transcriptome, and second, by demonstrating in rodents a causal link between reduced neurotrophic support and the expression of genes that code inhibitory synaptic-associated proteins and that are affected during aging. Notably, the preferential impact of reduced BDNF signaling on GABA-related markers (Figure 3) provides insights into the dynamics of gene changes with aging (ie, GABA changes occurring first) and has implications for mechanisms of brain disorders.
It is widely accepted that synaptic impairments, rather than neuronal loss, underlie brain aging. Human studies have reported breakdown in network connectivity as well as impaired white matter integrity with aging (Bishop et al, 2010). Non-human primate studies have provided more direct evidence that cognitive impairment might result from the loss of cortico-cortical synapses in PFC: age-associated synaptic loss is most prominent for axospinous synapses in layer 3 where cortico-cortical synapses predominate and the extent of synaptic loss correlates with the degree of cognitive impairments (Dumitriu et al, 2010). In agreement with this report, we observed in older brain decrements of expression for markers of presynaptic boutons, such as SLC17A7 (vGLUT1), which is enriched in cortico-cortical axon terminals. In vitro studies have shown that BDNF has a key role in synaptogenesis and synaptic maturation not only for excitatory synapses but also for inhibitory synapses (Kohara et al, 2007). The present transcriptome and coexpression network analysis confirmed the close link between reduced BDNF and the downregulated expression of many synapse-related genes during aging. These correlations remained significant even after gene expression was normalized by aging effect (ie, using age-residual expression values), implying that age-dependent changes in BDNF expression mediate, rather than moderate, specific age-related transcriptome changes in the human brain, potentially contributing to the observed structural, connectivity, and functional changes during aging.
Notably, we show that GABA receptors (GABBR2, GABRA4, GABRA5, and GABRB3), glutamate receptors (GRIA1, GRIN2A, GRIN2B), and multiple genes associated with both excitatory and inhibitory functions are closely correlated with BDNF expression and downregulated with age. BDNF regulates many cellular functions related to differentiation, maintenance, and plasticity and is thus considered a valid biological candidate to orchestrate such complex changes in the human brain. Our postmortem human study provides supporting evidence for low BDNF leading to decreased synaptic markers during brain aging; however, these findings do not demonstrate causation. In previous studies, high BDNF dependency was observed for interneuron markers in constitutive (Guilloux et al, 2012; Tripp et al, 2012) and induced (Glorioso et al, 2006) BDNF knockout mice. In addition, constitutive BDNF heterozygous mice showed greater age-related excitatory (Carretón et al, 2012) and inhibitory synaptic marker changes (Saylor et al, 2006). Yet, constitutive or chronic reduction of BDNF can lead to biological compensations and the observed gene expression changes could themselves be the result of those compensatory mechanisms. Further, age-related gene expression changes in humans seem to be more highly affected by NTRK2 changes than by BDNF as supported by the closest association of age-related synaptic genes to NTRK2-FL among BDNF signaling genes and transcripts (Figure 2). Utilization of NTRK2F616A mice allowed us to address causality while circumventing those limitations. Contrary to our expectation that both excitatory and inhibitory synaptic genes would be decreased by a temporal NTRK2 blockade, we observed aging-like changes mainly in the markers of GABA neuron inhibitory presynaptic functions: SST, NPY, CORT, CCK, and GAD1.
Several reasons might account for the observed differences between the effects of aging and of temporal blockade of BDNF signaling. First, species differences might account for the discrepancy; indeed, age-related alterations in gene expression patterns differ between mice and humans (Loerch et al, 2008). Second, interneurons might be more dependent on BDNF than pyramidal cells and consequently are more likely to exhibit a response to low BDNF signaling. This is consistent with prior findings from constitutive KO lines (Glorioso et al, 2006; Guilloux et al, 2012; Tripp et al, 2012). GABAergic cells do not produce BDNF and rely on BDNF supply from other cell population, such as excitatory neurons (Cellerino et al, 1996; Gorba and Wahle, 1999; Jin et al, 2003; Kohara et al, 2007; Marty et al, 2000; Rocamora et al, 1996). Third and related, 1NMPP1-treated NTRK2F616A mice are known to exhibit a NTRK2-null phenotype (Chen et al, 2005), so contrary to an aging-related gradual decrease of BDNF function, the BDNF/NTRK2 activity change may be too dramatic and/or too short for evoking the gradual and long-term compensatory changes that occur during aging. We believe that this temporal de-coupling allowed us here to differentiate the effects of BDNF on markers of distinct cell populations within an intact complex brain. Integrating the findings from human postmortem study and the animal data, we propose a model of healthy brain aging in which age-repressed BDNF signaling may result in lower GABA function, which with time may lead to glutamate signaling downregulation to preserve excitatory–inhibitory balance (Figure 4).
Figure 4.
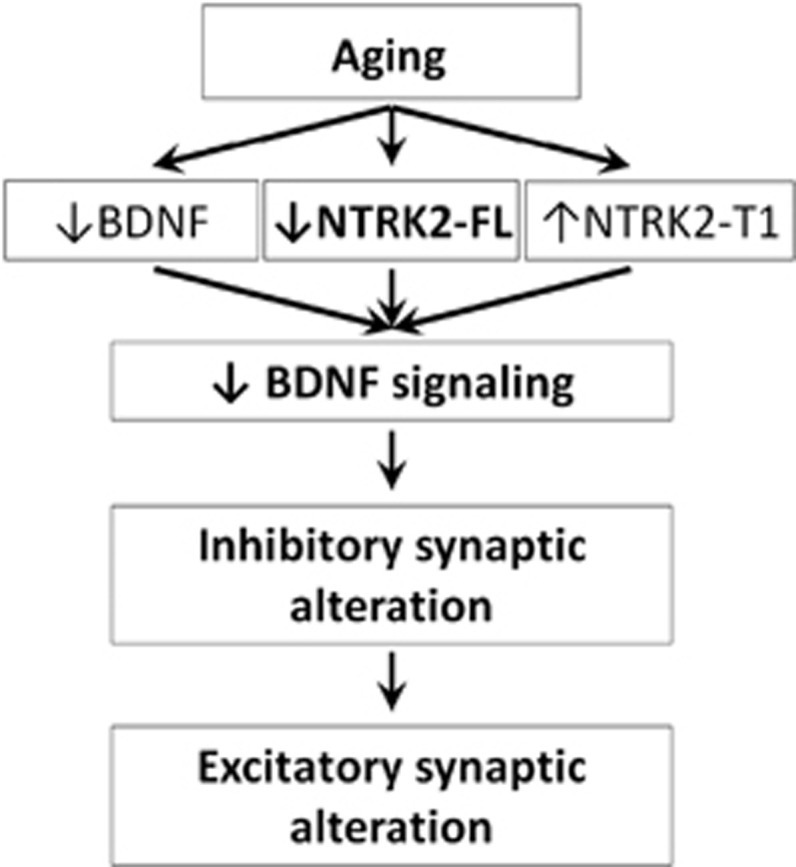
Proposed sequence of BDNF-induced brain changes during normal aging. Results from this study show that brain aging suppresses BDNF and NTRK2-FL expression and promotes NTRK2-T1 expression, all together in the direction of repressing BDNF signaling. Studies in rodents with temporal blockade in NTRK2 suggest that low BDNF function may initially cause reduced GABA synaptic activity which with prolonged time (ie, aging) may lead to a compensatory downregulation of excitatory synaptic genes.
One can assume that this mechanism of low BDNF function—driving synaptic alterations without breaking the excitatory–inhibitory balance—is crucial for healthy aging. On the other hand, deregulation in this homeostatic mechanism may lead to pathological changes and brain disorders. Interestingly, MDD has been associated with reduced central BDNF function (Guilloux et al, 2012; Tripp et al, 2012) and with an accelerated pattern of age-related transcriptome changes (Douillard-Guilloux et al, 2013). Moreover, these MDD- and BDNF-related changes have been preferentially associated with altered/reduced presynaptic GABA function rather than excitatory glutamatergic-related functions (Ding et al, 2015; Guilloux et al, 2012). This suggests that MDD may result from a failure for excitation to compensate for BDNF and GABA-related gene changes. Similar expression and BDNF correlation studies performed in the postmortem brains of MDD and control subjects could begin testing this hypothesis. This would be consistent with an age by disease interaction model, where the normal trajectory of age-dependent changes provides substrates for pathological changes. This model suggests that whenever those changes occur out of their expected trajectories and biological context (eg, too early) or do not lead to appropriate homeostatic adaptations, they may lead to biological imbalance and pathophysiological entities (McKinney et al, 2012; Sibille, 2013). Here the mechanism underpinning reduced BDNF during aging was not investigated; however, hypermethylation in BDNF gene was observed in older subjects of the same cohort (McKinney et al, 2015). It would be interesting to investigate how those (and other) age-related regulatory mechanisms are affected in MDD.
In summary, the findings from human postmortem study suggest that age-related reduction in BDNF/NTRK2 signaling is closely associated with alterations in both excitatory and inhibitory synapses in the PFC. The selective downregulation of GABA presynaptic markers by temporal NTRK2 blockade in mouse model suggests that the early age-related changes may involve alterations in GABA system, whereas excitatory synaptic changes may follow to maintain the excitation/inhibition balance.
Limitations
First, no gene or cellular functional analysis was performed in this study. Although animal and imaging studies imply that synaptic disturbance is the underpinning mechanism of age-related cognitive decline, we cannot directly determine whether synaptic gene expression changes are responsible for functional deficit. We cannot address this issue with our animal model either because chronic 1NMPP1 treatment induces degeneration of motor neuron terminals (Chen, 2007), which may compromise behavioral test results. Second, gene expression profiles were analyzed in tissue homogenate containing all six layers of gray matter. Considering that age-associated cognitive impairment has been suggested to correlate with layer three pyramidal neurons, the molecular changes might be diluted and information such as cell specific-gene alteration pattern would be lost. Third, additional BDNF-related changes may occur through non-NTRK2 signaling, which were not investigated here.
FUNDING AND DISCLOSURE
This work was supported by National Institute of Mental Health (NIMH) MH093723 (ES). DAL currently receives investigator-initiated research support from Pfizer and in 2012–2014 served as a consultant in the areas of target identification, validation and new compound development for Autifony, Bristol-Myers Squibb, Concert Pharmaceuticals, and Sunovion. The other authors declare no conflict of interest.
Acknowledgments
We thank Chien-Wei Lin for statistical assistance.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Beekman ATF, Deeg DJH, vanTilburg T, Smit JH, Hooijer C, vanTilburg W (1995). Major and minor depression in later life: a study of prevalence and risk factors. J Affect Disord 36: 65–75. [DOI] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A et al (2009). ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25: 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Lu T, Yankner BA (2010). Neural mechanisms of ageing and cognitive decline. Nature 464: 529–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazer DG (1994). Is depression more frequent in late-life - an honest look at the evidence. Am J Geriat Psychiat 2: 193–199. [DOI] [PubMed] [Google Scholar]
- Carretón O, Giralt A, Torres-Peraza JF, Brito V, Lucas JJ, Ginés S et al (2012). Age-dependent decline of motor neocortex but not hippocampal performance in heterozygous BDNF mice correlates with a decrease of cortical PSD-95 but an increase of hippocampal TrkB levels. Exp Neurol 237: 335–345. [DOI] [PubMed] [Google Scholar]
- Cellerino A, Maffei L, Domenici L (1996). The distribution of brain-derived neurotrophic factor and its receptor trkB in parvalbumin-containing neurons of the rat visual cortex. Eur J Neurosci 8: 1190–1197. [DOI] [PubMed] [Google Scholar]
- Chen XNeurotrophin Function During Maintenance and Regeneration of the Adult Nervous System. The Johns Hopkins University: Baltimore, MD, 2007. [Google Scholar]
- Chen X, Ye H, Kuruvilla R, Ramanan N, Scangos KW, Zhang C et al (2005). A chemical-genetic approach to studying neurotrophin signaling. Neuron 46: 13–21. [DOI] [PubMed] [Google Scholar]
- Ding Y, Chang LC, Wang X, Guilloux JP, Parrish J, Oh H et al (2015). Molecular and genetic characterization of depression: overlap with other psychiatric disorders and aging. Mol Neuropsychiatry 1: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillard-Guilloux G, Guilloux JP, Lewis DA, Sibille E (2013). Anticipated brain molecular aging in major depression. Am J Geriatr Psychiatry 21: 450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitriu D, Hao J, Hara Y, Kaufmann J, Janssen WG, Lou W et al (2010). Selective changes in thin spine density and morphology in monkey prefrontal cortex correlate with aging-related cognitive impairment. J Neurosci 30: 7507–7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide FF, Vining ER, Eide BL, Zang KL, Wang XY, Reichardt LF (1996). Naturally occurring truncated trkB receptors have dominant inhibitory effects on brain-derived neurotrophic factor signaling. J Neurosci 16: 3123–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erraji-Benchekroun L, Underwood MD, Arango V, Galfalvy H, Pavlidis P, Smyrniotopoulos P et al (2005). Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biol Psychiatry 57: 549–558. [DOI] [PubMed] [Google Scholar]
- Glaesmer H, Riedel-Heller S, Braehler E, Spangenberg L, Luppa M (2011). Age- and gender-specific prevalence and risk factors for depressive symptoms in the elderly: a population-based study. Int Psychogeriatr 23: 1294–1300. [DOI] [PubMed] [Google Scholar]
- Glorioso C, Sabatini M, Unger T, Hashimoto T, Monteggia LM, Lewis DA et al (2006). Specificity and timing of neocortical transcriptome changes in response to BDNF gene ablation during embryogenesis or adulthood. Mol Psychiatry 11: 633–648. [DOI] [PubMed] [Google Scholar]
- Gorba T, Wahle P (1999). Expression of TrkB and TrkC but not BDNF mRNA in neurochemically identified interneurons in rat visual cortex in vivo and in organotypic cultures. Eur J Neurosci 11: 1179–1190. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Balogh SA, Wehner JM, Jones KR (2003. a). Learning deficits in forebrain-restricted brain-derived neurotrophic factor mutant mice. Neuroscience 121: 341–354. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Zeiler SR, Tamowski S, Jones KR (2003. b). Brain-derived neurotrophic factor is required for the maintenance of cortical dendrites. J Neurosci 23: 6856–6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilloux JP, Douillard-Guilloux G, Kota R, Wang X, Gardier AM, Martinowich K et al (2012). Molecular evidence for BDNF- and GABA-related dysfunctions in the amygdala of female subjects with major depression. Mol Psychiatry 17: 1130–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta VK, You Y, Gupta VB, Klistorner A, Graham SL (2013). TrkB receptor signalling: implications in neurodegenerative, psychiatric and proliferative disorders. Int J Mol Sci 14: 10122–10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattiangady B, Rao MS, Shetty GA, Shetty AK (2005). Brain-derived neurotrophic factor, phosphorylated cyclic AMP response element binding protein and neuropeptide Y decline as early as middle age in the dentate gyrus and CA1 and CA3 subfields of the hippocampus. Exp Neurol 195: 353–371. [DOI] [PubMed] [Google Scholar]
- Howells DW, Porritt MJ, Wong JY, Batchelor PE, Kalnins R, Hughes AJ et al (2000). Reduced BDNF mRNA expression in the Parkinson's disease substantia nigra. Exp Neurol 166: 127–135. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009. a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009. b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Jin X, Hu H, Mathers PH, Agmon A (2003). Brain-derived neurotrophic factor mediates activity-dependent dendritic growth in nonpyramidal neocortical interneurons in developing organotypic cultures. J Neurosci 23: 5662–5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juster RP, McEwen BS, Lupien SJ (2010). Allostatic load biomarkers of chronic stress and impact on health and cognition. Neurosci Biobehav R 35: 2–16. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Birnbaum HG, Shahly V, Bromet E, Hwang I, McLaughlin KA et al (2010). Age differences in the prevalence and co-morbidity of DSM-IV major depressive episodes: results from the Who World Mental Health Survey Initiative. Depress Anxiety 27: 351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara K, Yasuda H, Huang Y, Adachi N, Sohya K, Tsumoto T (2007). A local reduction in cortical GABAergic synapses after a loss of endogenous brain-derived neurotrophic factor, as revealed by single-cell gene knock-out method. J Neurosci 27: 7234–7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerch PM, Lu T, Dakin KA, Vann JM, Isaacs A, Geula C et al (2008). Evolution of the aging brain transcriptome and synaptic regulation. PLoS One 3: e3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien SJ, McEwen BS, Gunnar MR, Heim C (2009). Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat Rev Neurosci 10: 434–445. [DOI] [PubMed] [Google Scholar]
- Marty S, Wehrle R, Sotelo C (2000). Neuronal activity and brain-derived neurotrophic factor regulate the density of inhibitory synapses in organotypic slice cultures of postnatal hippocampus. J Neurosci 20: 8087–8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney BC, Lin CW, Oh H, Tseng GC, Lewis DA, Sibille E (2015). Hypermethylation of BDNF and SST genes in the orbital frontal cortex of older individuals: a putative mechanism for declining gene expression with age. Neuropsychopharmacology 40: 2604–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney BC, Oh H, Sibille E (2012). Age-by-disease biological interactions: implications for late-life depression. Front Genet 3: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney BC, Sibille E (2013). The age-by-disease interaction hypothesis of late-life depression. Am J Geriatr Psychiatry 21: 418–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison JH, Baxter MG (2012). The ageing cortical synapse: hallmarks and implications for cognitive decline. Nat Rev Neurosci 13: 240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan RM, Griffin EW, Kelly AM (2009). Long-term treadmill exposure protects against age-related neurodegenerative change in the rat hippocampus. Hippocampus 19: 1019–1029. [DOI] [PubMed] [Google Scholar]
- Rajkowska G (2000). Postmortem studies in mood disorders indicate altered numbers of neurons and glial cells. Biol Psychiatry 48: 766–777. [DOI] [PubMed] [Google Scholar]
- Rocamora N, Welker E, Pascual M, Soriano E (1996). Upregulation of BDNF mRNA expression in the barrel cortex of adult mice after sensory stimulation. J Neurosci 16: 4411–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanczyk TB, Weickert CS, Webster MJ, Herman MM, Akil M, Kleinman JE (2002). Alterations in trkB mRNA in the human prefrontal cortex throughout the lifespan. Eur J Neurosci 15: 269–280. [DOI] [PubMed] [Google Scholar]
- Saylor AJ, Meredith GE, Vercillo MS, Zahm DS, McGinty JF (2006). BDNF heterozygous mice demonstrate age-related changes in striatal and nigral gene expression. Exp Neurol 199: 362–372. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibille E (2013). Molecular aging of the brain, neuroplasticity, and vulnerability to depression and other brain-related disorders. Dialogues Clin Neurosci 15: 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhol M, Arancibia S, Maurice T, Tapia-Arancibia L (2007). Spatial memory training modifies the expression of brain-derived neurotrophic factor tyrosine kinase receptors in young and aged rats. Neuroscience 146: 962–973. [DOI] [PubMed] [Google Scholar]
- Stockmeier CA, Mahajan GJ, Konick LC, Overholser JC, Jurjus GJ, Meltzer HY et al (2004). Cellular changes in the postmortem hippocampus in major depression. Biol Psychiatry 56: 640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson Ray M, Weickert CS, Wyatt E, Webster MJ (2011). Decreased BDNF, trkB-TK+ and GAD67 mRNA expression in the hippocampus of individuals with schizophrenia and mood disorders. J Psychiatry Neurosci 36: 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisserand DJ, Pruessner JC, Arigita EJS, van Boxtel MPJ, Evans AC, Jolles J et al (2002). Regional frontal cortical volumes decrease differentially in aging: An MRI study to compare volumetric approaches and voxel-based morphometry. Neuroimage 17: 657–669. [PubMed] [Google Scholar]
- Tripp A, Kota RS, Lewis DA, Sibille E (2011). Reduced somatostatin in subgenual anterior cingulate cortex in major depression. Neurobiol Dis 42: 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp A, Oh H, Guilloux JP, Martinowich K, Lewis DA, Sibille E (2012). Brain-derived neurotrophic factor signaling and subgenual anterior cingulate cortex dysfunction in major depressive disorder. Am J Psychiatry 169: 1194–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC (2005). Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J Evol Biol 18: 1368–1373. [DOI] [PubMed] [Google Scholar]
- Wong J, Rothmond DA, Webster MJ, Weickert CS (2013). Increases in two truncated TrkB isoforms in the prefrontal cortex of people with schizophrenia. Schizophr Bull 39: 130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Zang K, Ruff NL, Zhang YA, McConnell SK, Stryker MP et al (2000). Cortical degeneration in the absence of neurotrophin signaling: dendritic retraction and neuronal loss after removal of the receptor TrkB. Neuron 26: 233–245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.