Abstract
Our conception of the human genome, long focused on the 2% that codes for proteins, has profoundly changed since its first draft assembly in 2001. Since then, an unanticipatedly expansive functionality and convolution has been attributed to the majority of the genome that is transcribed in a cell-type/context-specific manner into transcripts with no apparent protein coding ability. While the majority of these transcripts, currently annotated as long non-coding RNAs (lncRNAs), are functionally uncharacterized, their prominent role in embryonic development and tissue homeostasis, especially in the context of the heart, is emerging. In this review, we summarize and discuss the latest advances in understanding the relevance of lncRNAs in (re)building the heart.
Keywords: long non-coding RNAs, embryonic development, homeostasis, cardiac development, cardiac regeneration, cell-fate commitment
“His very existence was improbable, inexplicable, and altogether bewildering. He was an insoluble problem. It was inconceivable how he had existed, how he had succeeded in getting so far, how he had managed to remain – why he did not instantly disappear.”
—Joseph Conrad, Heart of Darkness.
Over the last 20 years, the advent of novel sequencing technologies and initiatives such as ENCODE have revealed that the majority of the mammalian genome, despite not being translated into proteins, is pervasively transcribed into myriad RNAs of different sizes and characteristics, collectively referred to as non-coding RNAs (ncRNAs). To date, thousands of ncRNAs have been described in humans, yet the precise functional roles of a great majority remain unclear and controversial. Non-coding RNAs are traditionally classified based on their size into two major classes, small non-coding RNAs and long non-coding (lnc) RNAs. Small non-coding RNAs were first described in 1993 (Lee et al., 1993), and have been studied extensively since then, along with their molecular mechanisms of action and function. Although much is left to be learnt from them (e.g., piRNAs) they are outside the scope of this review, but readers can consult the excellent reviews (Bartel, 2004, 2009; Carthew and Sontheimer, 2009; Malone and Hannon, 2009; Mendell and Olson, 2012).
Long non-coding RNAs are defined as a large and diverse group of non-protein coding transcripts longer than 200 nucleotides (Rinn and Chang, 2012). Currently, they represent one of the most prominent but least understood genes in vertebrates. Overall, more than ~100,000 genomic loci are predicted to produce lncRNAs in the human genome (Zhao et al., 2016) and account for the largest class of genes, giving an idea of the potential relevance of this class of molecules. The consistent observation of cell type/tissue specific expression, together with their increased presence in evolutionarily more complex organisms, lncRNAs have been proposed to play key roles in affecting diverse cellular functions from modulating chromatin states to protein synthesis. Ultimately, this imparts a new layer of sophistication to the control of fundamental biological processes. In line with this, their roles in health and disease, including embryonic development, homeostasis, cancer, neurodegenerative, and metabolic disorders are rapidly becoming clear.
As evident from the exponentially increasing body of literature, lncRNAs are particularly interesting in the context of cardiac biology. The hierarchy and ontogeny of cardiac specification is becoming increasingly complex with the identification of primary, secondary [and potentially tertiary (Bressan et al., 2013)] heart fields that together give rise to the multitude of highly specialized cell types that constitute the mammalian heart [for review, see (Buckingham et al., 2005; Brade et al., 2013)]. Among multiple modes of gene regulation that fine tune cardiac development, recent studies reveal a prominent role for lncRNAs (Grote et al., 2013; Klattenhoff et al., 2013; Ounzain et al., 2014, 2015a,b; Kurian et al., 2015). Additionally, dysregulated lncRNAs have been implicated in cardiovascular diseases (Yap et al., 2010; Han et al., 2014; Michalik et al., 2014; Ounzain et al., 2014, 2015a,b; Wang et al., 2014a,b; Yan et al., 2015), including coronary artery and other heart dysfunctions, which remain the leading cause of mortality in the developed world, ahead of all cancer types combined. In the United States alone, 80 million people suffer from cardiovascular disorders, and cardiovascular diseases are projected to be the leading cause of death in the world by 2020, highlighting the importance of a better understanding of their etiology. Cardiac injuries, such as myocardial infarction (MI), usually caused by an occlusion of a coronary artery, can lead to immediate death due to the loss of oxygenation in the ventricular muscle. Such an occlusion quickly results in ischemia/reperfusion injury and necrosis of the tissue. Patients who survive a cardiac episode face progressive deterioration due to compensatory responses over several years, resulting in heart failure and eventually death. Although the molecular and cellular mechanisms involved in these pathologic responses are now better understood, many puzzles remain to be solved and the treatment of cardiac disorders remains poor. Investigating the regulatory functions of lncRNAs in cardiac homeostasis and disease might open new avenues in the treatment of cardiac disorders. In this review, we will summarize the latest progress in understanding the molecular mechanisms of lncRNA function in cardiac biology and their potential as novel therapeutic targets in the future.
Evolution of the lncRNA paradigm—from junk to rosetta stone
The study of lncRNAs originates from the interest in the non-protein coding (hereafter termed non-coding) part of the genome, its evolution and its potential function in the 1950s. Scientists quickly discovered the C-value paradox, that is, the lack of correlation between organismal size or developmental complexity and the DNA content of a cell. Lower animals—from an anthropocentric perspective—such as salamanders and fish, can have a genome many times larger than higher order organisms, such as humans. The paradox seemed solved when it was found that most of the genome does not encode for protein-coding genes (Ohno, 1972). The non-coding portion of the genome was prematurely termed “junk DNA” due to the high presence of transposons, pseudogenes, repeats, and other elements of dubious or unknown function. Interestingly, this part of the genome accounts for 50–70% of the mammalian genome (de Koning et al., 2011). However, even then geneticists were still puzzled by the C-paradox. Although a part of the non-coding genome accounted for structural regions in the DNA, the majority remained unexplained. So the question remained: why would organisms maintain such a large genome—at high energy cost—without having any major function? Indeed, many of the early pioneers suspected something more was at stake and expressed their belief that the non-coding genome might not be entirely useless.
The first evidence for the potential function of the non-coding part of the genome was that the transcriptional activity in cells was too high to be entirely attributable to coding genes and ribosomal RNAs. Heterogeneous nuclear RNAs were soon discovered as RNAs originating from repetitive and heterochromatic regions with no protein-coding ability (Yunis and Tsai, 1978). Some years later, more non-coding RNAs involved in post-transcriptional regulation were found, including snRNAs and snoRNAs. However, it was not until the advent of microarrays and next-generation sequencing methodologies in the last decades that our picture of non-coding RNAs became more complete. The scale of pervasive transcription is much larger than previously appreciated, with estimates ranging from 70 to 90% of the human genome being transcribed at one point or another during development, homeostasis or disease (Bertone et al., 2004; Mercer and Mattick, 2013). Some of these transcripts are present at very low copy numbers and controversy still exists on their actual relevance. Indeed, a number of putative lncRNA transcripts might be the effect of pervasive transcription and transcriptional “noise.” However, detailed RNA-sequencing analysis and chromatin signatures (histone modifications, TF binding, DNase I hypersensitivity assays) indicate strong control of regulatory elements in these loci, suggesting that a significant percentage of lncRNA transcripts indeed constitute novel non-coding genes with currently unknown function (Guttman et al., 2009; Cabili et al., 2011; Iyer et al., 2015). It would seem now that ncRNAs play more important roles than anticipated and might constitute a “Rosetta Stone,” a new layer of gene regulation that might offer insight into the complexity of higher organisms.
Classes of lncRNAs and their genomic architecture
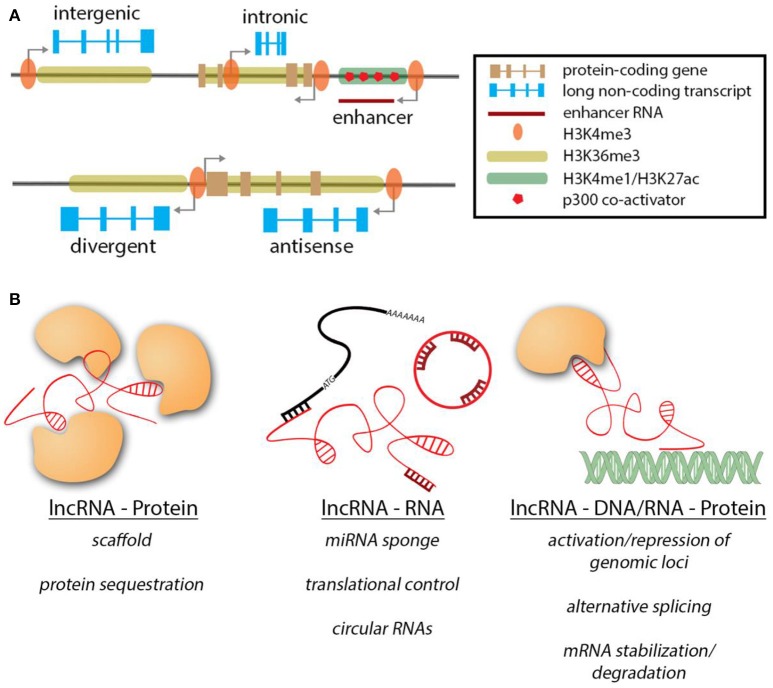
Being defined solely by size and a lack of coding potential, lncRNAs constitute a large and heterogeneous group of RNA molecules. It is therefore not surprising that their genomic contexts show a similar variability (Figure 1A; Cabili et al., 2011). Strikingly, a large fraction of lncRNA loci and transcripts show strong similarities to protein-coding transcripts with regards to histone marks occupying their promoters and gene body, intron-exon structure and poly-adenylation. Based on their genomic architecture, lncRNAs are categorized as long intergenic non-coding RNAs (lincRNAs), antisense lncRNAs, intronic lncRNAs, circular RNAs, divergent lncRNAs, and enhancer RNAs. A major class of lncRNAs, long intergenic non-coding RNAs (lincRNAs), is located in barren gene deserts or between two protein-coding genes. This particular class operates as stand-alone genes, often having their own regulatory landscape. Another class of lncRNAs, intronic lncRNAs, is located within introns of protein-coding genes similar to what has been reported for many microRNAs. Antisense lncRNAs are transcribed from the opposite strand of a protein-coding RNA. They can also exist outside the gene body of their associated coding gene, resulting in various degrees of transcriptional overlap, ranging from short stretches to a complete overlap. A different form of antisense transcripts is divergent lncRNAs. These genes are controlled by the same promoter region as the coding neighbor, but are transcribed in the opposite direction to the coding gene, without any transcriptional overlap. The function of this class of lncRNAs is very poorly understood. In contrast, enhancer RNA (eRNA) and circular RNAs (ciRNAs) are uniquely distinct in their genomic architecture. eRNAs are transcribed by active enhancers and are often single exonic and not poly-adenylated. Furthermore, during active transcription the gene body is not marked by H3K3me3, but H3K4 monomethylation and H3K27 acetylation in combination with abundant binding of the p300 co-activator (De Santa et al., 2010; Kim et al., 2010). The function of eRNA transcripts currently remains controversial. Recently, a novel class of lncRNAs arising from circularization of exons has been described (Zhang et al., 2013). Interestingly these circular RNAs (ciRNAs) can emerge from protein-coding genes by covalently linking separate exons.
Figure 1.
Genomic contexts of lncRNAs & modes of action. (A) LncRNA genes (blue) can exist in various genomic contexts. The histone marks are similar to protein-coding genes. NcRNAs transcribed from enhancer regions are unique in their features. (B) Overview of the different interactions of lncRNAs and their target molecules.
Molecular mechanisms of lncRNA-mediated gene regulation
Similar to the diverse genomic contexts in which lncRNA genes can be found, lncRNAs can exert their regulatory function through a wide variety of mechanisms (Figure 1B). In contrast to the well-studied microRNAs, lncRNAs are not limited to interactions with other transcripts, but can exert their function along with proteins, RNAs and DNA, thereby expanding the dimensions by which they can operate. When interacting with proteins, lncRNAs can act as guides for targeting a protein/protein complex to specific sites/subcellular locations (Rinn et al., 2007; Yap et al., 2010; Grote et al., 2013), as scaffolds to bring different subunits of protein complexes together and maintain them in close proximity (Wang et al., 2004; Tsai et al., 2010; Yang et al., 2011) or act as decoys to sequester proteins away from their site of action (Kino et al., 2010; Tripathi et al., 2010; Hung et al., 2011). In the context of the heart, lncRNA Myheart (Mhrt) directly binds to Brg1 in order to inhibit its helicase function (Han et al., 2014). Similarly, lncRNAs carrying repeats of corresponding complimentary sequences to miRNAs can sequester them acting as miRNA sponges [e.g., CARL (Wang et al., 2014b), CHRF (Wang et al., 2014a), HULC (Cui et al., 2015), linc-MD1(Legnini et al., 2014), H19(Giovarelli et al., 2014; Tao et al., 2016)]. A very recently discovered example of lncRNA-RNA interaction is mediated by circular RNAs that can act as miRNA sponges [e.g., ciRS-7(Xu et al., 2015; Geng et al., 2016; Yu et al., 2016)]. It remains to be determined if ciRNAs can impart their function via other mechanisms.
In addition, lncRNAs can regulate protein coding transcripts by controlling their translation [Uchl1-as (Carrieri et al., 2012), lincRNA-p21(Yoon et al., 2012)] or by regulating transcript stability. For example, the lncRNA TINCR binds Staufen 1 (Stau1) in order to stabilize mRNAs containing the TINCR binding motif (Kretz et al., 2013). Besides interacting with other RNA molecules, lncRNAs can also bind to DNA, often in complex with proteins, resulting in multi-layered regulatory complexity. Many cases of activation or repression of genomic loci by lncRNAs have been described to date [HOTAIR (Gupta et al., 2010), Braveheart (Klattenhoff et al., 2013), FENDRR (Grote et al., 2013)]. In this scenario, lncRNAs recruit epigenetic modifiers to target specific genomic loci, leading to either activation or inactivation. LncRNAs have also been shown to regulate gene activity co-transcriptionally, e.g., by aiding alternative splicing of mRNA isoforms (Gonzalez et al., 2015) or binding to newly synthesized mRNA, resulting in their stabilization or degradation (Matsui et al., 2008; Gong and Maquat, 2011; Wang et al., 2016a). eRNAs have been demonstrated to act in cis to regulate expression of target (protein-coding) genes (Ounzain et al., 2014). Another example for lncRNAs acting in cis is during genomic imprinting, an important process during development, where bi-allelic expression is changed to monoallelic and viceversa. Several lncRNAs have been demonstrated to mediate imprinting of neighboring genomic loci [Xist & Tsix (Brown et al., 1992; Clemson et al., 1996; Lee et al., 1999; Bell et al., 2014; Chu et al., 2015; McHugh et al., 2015), KCNQ1OT1 (Thakur et al., 2004), Airn (Sleutels et al., 2002)].
Recently, a handful of reports indicated that some lncRNAs work by more conventional means than previously expected. Several groups have independently demonstrated that a fraction of genes currently annotated as lncRNAs can be translated to code for short peptides (Chng et al., 2013; Pauli et al., 2014, 2015). This also explains why some lncRNAs appear preferentially located in the cytoplasm and in association with ribosomes in a manner similar to typical mRNAs. In an elegant study, Pauli et al. identified 28 candidate signaling proteins expressed during zebrafish embryogenesis, including toddler [(Pauli et al., 2015); also identified as ELABELA (Chng et al., 2013)], a previously annotated lncRNA which in reality codes for a short, conserved, secreted peptide. Impaired expression of Toddler reduces the motility of mesendodermal cells during zebrafish gastrulation and leads to grave developmental defects, including cardiovascular defects. Toddler promotes cell movement locally, suggesting that it is neither an attractant nor a repellent, but acts as a motogen. Toddler promotes the internalization of G-protein-coupled APJ/Apelin receptors and activation of APJ/Apelin signaling rescues toddler mutants. Intriguingly, toddler and similar short peptides might constitute a previously unrecognized family of signaling molecules crucial for vertebrate development. Recently, the Olson lab reported another short peptide (DWORF) translated from a previously annotated long non-coding RNA (Nelson et al., 2016). DWORF enhances muscle contractility by regulating Ca2+ levels in skeletal muscle cells. Based on these studies, the genes annotated as lncRNAs should be studied carefully to discriminate between a transcript specific function, a function by the act of transcription or potential peptides/novel proteins encoded by them. Taken together, lncRNAs regulate cellular events in impressively wide mechanistic avenues.
Role of lncRNAs in modulating cardiac development
In recent years, lncRNAs have been extensively studied in both development and disease. The majority of novel lncRNAs have been associated to carcinogenesis and cancer progression. However, several studies have also demonstrated a crucial role for lncRNAs in development and disease, their contribution being particularly prominent in cardiac development and cardiac disorders (see Table 1). These findings probably reflect more the active efforts by the cancer and cardiac community to tackle these prominent diseases, rather than a biological overabundance of lncRNAs in cancer or cardiac-related cells. While thousands of lncRNAs have been reported to be expressed exclusively in the heart during development or disease, their function remains to be explored (Ounzain et al., 2014, 2015b).
Table 1.
Summary of lncRNAs involved in cardiac development and disease.
| Name | Species | Genomic context | Function and mechanism | Location (human hg38, mouse mm10) | References |
|---|---|---|---|---|---|
| ALIEN | Human | Intergenic | Cardiovascular commitment | chr20:22,560,553–22,578,642 | Kurian et al., 2015 |
| ANRIL | Human | Antisense | Metabolism, coronary artery disease, myocardial infarction | chr9:21994790–22121097 | Yap et al., 2010; Vausort et al., 2014 |
| Bvht | Mouse | Intergenic | Cardiac mesoderm commitment Decoy | chr18:61639653–61647503 | Klattenhoff et al., 2013 |
| Carl | Mouse | Intergenic | Mitochondria, cardiomyocyte apoptosis; miRNA sponge | chr2:18799244–18801291 | Wang et al., 2014b |
| CARMEN | Mouse/Human | Intergenic | Cardiac specification & homeostasis; enhancer-associated, cis- & trans-regulation | chr5:149,406,845–149,432,836 (human) chr18:61,645,878–61,665,538 (mouse) | Ounzain et al., 2015a |
| CHAST | Mouse/Human | Antisense | Pro-hypertrophic; cis-regulation | chr17:64,783,199–64,783,552 (human) chr11:103,363,213–103,364,651 (mouse) | Viereck et al., 2016 |
| Chrf | Mouse | Intergenic | Cardiac hypertrophy; miRNA sponge | chr18:72164057–72165898 | Wang et al., 2014a |
| Fendrr | Mouse/Human | Intergenic | Cardiac development; cis- & trans-regulation, Guide | chr16:86,474,529–86,508,860 (human) chr8:121059119–121083032 (mouse) | Grote et al., 2013 |
| H19 | Mouse/Human | Antisense | Cardiac fibroblast proliferation, negative regulator of hypertrophy | chr11:1995176–1997835 (human) chr7:142575532–142578146 (mouse) | Tao et al., 2016 |
| HIF1A-AS2 | Human | Antisense | Myocardial infarction | chr14:61747039–61749089 | Vausort et al., 2014 |
| KCNQ1OT1 | Human | Antisense | Cardiovascular development, arrhythmia, myocardial infarction; Guide | chr11:2,608,328–2,699,994 | Thakur et al., 2004; Korostowski et al., 2012; Vausort et al., 2014 |
| LIPCAR | Human | Unknown | Heart failure | chrM:7586–15887 | Kumarswamy et al., 2014 |
| Malat1 | Mouse/Human | Intergenic | Endothelial cell identity, myocardial infarction; Decoy | chr11:65,497,762–65,506,469 (human) chr19:5,795,690–5,802,671 (mouse) | Michalik et al., 2014; Vausort et al., 2014 |
| MEG3 | Mouse/Human | Intergenic | Modulation of TGF-β pathway; Guide | chr14:100,826,134–100,861,008 (human) chr12:109542023–109568650 (mouse) | Mondal et al., 2015 |
| Mhrt | Mouse/Human | Intergenic | Cardiac hypertrophy; Decoy | chr14:23,415,450–23,417,595 (human) chr14:54,968,787–54,974,349 (mouse) | Han et al., 2014 |
| MIAT | Human | Intergenic | Myocardial infarction; miRNA sponge | chr22:26657482–26676477 (human) chr5:112220925–112228948 (mouse) | Yan et al., 2015 |
| Mm67/77/85/130/132 | Mouse | Intergenic | Cardiac development & remodeling; Enhancer-associated lncRNA Cis-regulation | Several loci (see original publication) | Ounzain et al., 2014 |
| Novlncs | Mouse | Intergenic | Cardiac remodeling; Enhancer-associated | Several loci (see original publication) | Ounzain et al., 2015b |
| NRF | Mouse | Intergenic | Regulated necrosis of cardiomyocytes; miRNA sponge | chr3:45,438,398–45,440,956 | Wang et al., 2016b |
| PANCR | Human | Intergenic | Cardiac differentiation | chr4:110,595,513–110,615,458 | Gore-Panter et al., 2016 |
| PUNISHER | Human | Antisense | Endothelial cell identity | chr12:57,726,240–57,728,356 | Kurian et al., 2015 |
| PVT1 | Mouse | Antisense | Cardiomyocyte cell size, possibly regulating hypertrophy | chr15:62,037,986–62,260,212 | Yu et al., 2015 |
| RNCR3 | Mouse/Human | Intergenic | Atheroprotective; miRNA sponge | chr13:90,060,247–90,119,719 (human) chr14:64,588,115–64,593,961 (mouse) | Shan et al., 2016 |
| ROR | Human | Intergenic | Pro-Hypertrophic; miRNA sponge | chr18:57,054,559–57,072,119 | Jiang et al., 2016 |
| SENCR | Human | Antisense | Smooth muscle contractility | chr11:128,691,672–128,696,023 | Bell et al., 2014 |
| Smad7-lncRNA | Mouse | Antisense | Enhancer-associated lncRNA, Cis-regulation | chr18:75522879–75528680 | Ounzain et al., 2014 |
| SMILR | Human | Intergenic | Vascular smooth muscle cell proliferation | chr8:122,414,332–122,428,551 | Ballantyne et al., 2016 |
| TERMINATOR | Human | Intergenic | Pluripotency, cardiovascular development | chr1:200,411,800–200,475,513 | Kurian et al., 2015 |
| uc.167 | Mouse | Antisense | Involved in cardiac differentiation | chr5:88,889,445–89,465,982 | Song et al., 2016 |
| UCA1 | Human | Biomarker for acute myocardial infarction, anti-apoptotic (rat model) | chr19:15,828,947–15,836,321 | Liu et al., 2015; Yan et al., 2016 |
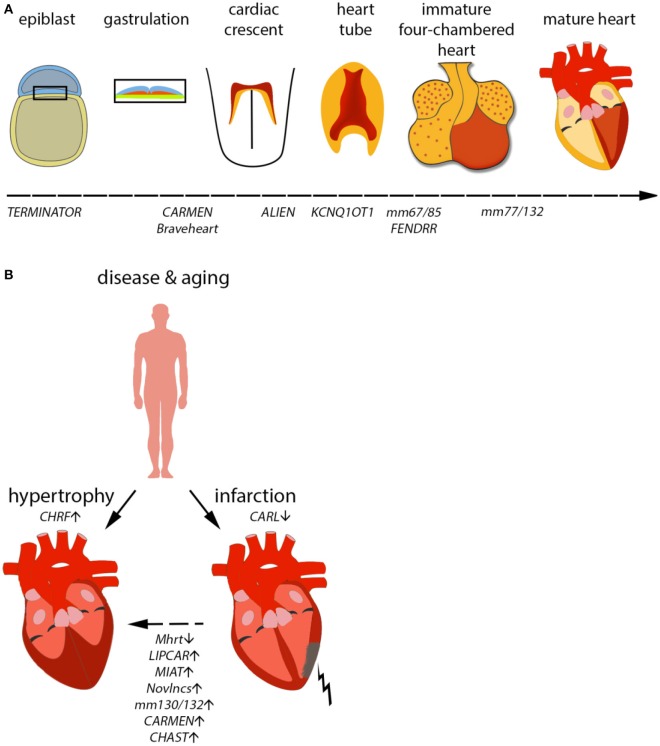
The heart is the first organ to form and function in an embryo, and survival of the embryo and all subsequent events strongly depends on its uninterrupted function. Heart development is an extraordinarily complex process involving the migration and integration of multiple cell lineages into a three-dimensional organ and its adequate connection with the vascular system (Figure 2A). Abnormalities in heart development lead to congenital heart disease, the most common human birth defect, which results in a series of functional defects and a host of potentially fatal disorders, including arrhythmias, cardiomyopathies, heart failure, and sudden death (Olson and Schneider, 2003; Bruneau, 2008).
Figure 2.
Mammalian cardiac development in vivo/in vitro and consequences of aging on the heart. (A) Development of the mammalian heart is a complex series of events, marked by distinct morphological changes. Expression of selected lncRNAs is indicated along the developmental timeline. (B) During aging, the heart becomes hypertrophic, i.e., the heart muscle increases by an increase in cell size, not number. Furthermore, the risk of myocardial infarction (MI) increases with age. MI can eventually result in hypertrophy in order to compensate for the loss in heart muscle. Selected lncRNAs identified to play a role in infarction or hypertrophy are depicted.
Due to its importance in early survival, it is not surprising that the formation of the cardiovascular system and its function are precisely controlled by integrated networks of transcription factors that link signaling systems with the protein-coding genes required for cardiac myogenesis, morphogenesis, and contractility (Pipes et al., 2006). Surprisingly, until recently we were unaware that these pathways are intimately intertwined with dozens of critical non-coding genes, including microRNAs and long non-coding RNAs (Pipes et al., 2006; Aguirre et al., 2014; Kurian et al., 2015; see Table 1). These non-coding RNAs act as fine switches to modulate and choreograph multiple aspects of cardiac development, function, disease, and injury recovery. The increasing number of non-coding elements identified to date illustrates the importance of ncRNAs in modulating and creating complex phenotypes at the cellular and organ level, especially in the context of the heart.
The first lncRNA identified as a critical regulator of cardiac development was Braveheart (Klattenhoff et al., 2013). Braveheart controls the cardiac gene network in multipotent cardiac progenitors derived from mouse ESCs and functions upstream of mesoderm posterior basic helix-loop-helix transcription factor 1 (MESP1), a master regulator of cardiac development. Braveheart acts as an epigenetic regulator of cardiac commitment by interacting with SUZ12, a member of the polycomb repressive complex 2 (PRC2). As of now, it remains to be seen if Braveheart is required for normal heart development in vivo. Unfortunately, Braveheart seems to be a mouse-specific lncRNA and no homologs have been found in other species. Its expression is well-documented in mouse ESCs and heart samples using RNA sequencing. However, the orthologous human and rat genomic regions are not actively transcribed. The lack of a Braveheart homolog in humans begs the question of how well its knowledge will translate into the human cardiac setting. Given its critical importance in cardiac lineage commitment, it is however possible that an undiscovered functional Braveheart homolog gets transcribed from a different genomic locus.
Very few lncRNAs have been functionally interrogated in vivo in mammalian models. Fendrr, a novel lncRNA expressed in the heart, has been explored using mouse genetics. Two independent studies demonstrated that loss of Fendrr is embryonic lethal in mice (Grote et al., 2013; Sauvageau et al., 2013). Mutants display numerous developmental defects, including disrupted cardiac morphogenesis, consistent with the finding that Fendrr is expressed in the mouse lateral plate mesoderm and developing hearts. However, there is a difference in the phenotype severity associated with the two mutant mouse lines developed; while one mutant line dies at embryonic day E13.75(Grote et al., 2013), the other one dies postnatally (Sauvageau et al., 2013). One possible explanation for this discrepancy is the different targeting strategies used to remove the Fendrr gene. Mechanistically, Fendrr has been shown to interact with the PRC2 complex to modulate the epigenetic regulation of gene expression, and might be involved in the control of the activating H3K4me3 mark on a subset of promoters, thus modifying the expression level of those genes (Grote et al., 2013). However, the mechanisms of Fendrr-dependent molecular events remain to be fully understood.
Embryonic stem cell (ESC) based differentiation models provide a powerful tool for discovery and functional identification of novel long non-coding RNAs in humans. Current differentiation protocols allow for the precise in vitro recapitulation of the early developmental steps leading to cardiac commitment and cardiac or endothelial cell generation (Kurian et al., 2013; Bhattacharya et al., 2014; Burridge et al., 2014; Zhang et al., 2015). Using human ESCs and zebrafish as a model of vertebrate development, we recently were able to identify 76 novel human lncRNAs partially conserved across vertebrates, and characterized three of them in cardiovascular development in vertebrates (Kurian et al., 2015). These three lncRNAs, named TERMINATOR, ALIEN [also known as linc00261 and DEANR1(Jiang et al., 2015)] and PUNISHER, are essential at different stages of mesodermal and cardiovascular development, albeit their specific mechanisms of action still remain poorly understood. TERMINATOR is an abundant transcript in pluripotent stem cells and is essential for early embryonic survival, pluripotency, and early mesendodermal commitment both in zebrafish and mouse embryos. In contrast, ALIEN is more specifically expressed in mesendodermal tissues and its loss-of-function leads to disrupted vasculature and heart formation. Interestingly, ALIEN is also an endodermal lncRNA participating in pancreatic differentiation through its interaction with FOXA2 and SMAD2/3 (Jiang et al., 2015).
Regulation of cardiac function and disease by lncRNAs in the adult
Besides their involvement in cell fate decisions and developmental programs, lncRNAs are also important regulators of homeostasis in the adult cardiovascular system, and their dysregulation appears associated with a variety of disease states.
Chromatin remodeling and epigenetic state regulation by lncRNAs in the diseased heart
A prominent feature of the aging or failing heart is hypertrophy, a process involving a pathological increase in cardiomyocyte size in order to cope with inefficient cardiac function (Figure 2B). Two lncRNAs have been demonstrated to influence cardiac hypertrophy in murine models. The first of them was named Myheart (Mhrt, for myosin heavy chain-associated RNA transcript) and constitutes a cardioprotective lncRNA cluster transcribed antisense from the Myh7 locus in mouse (Han et al., 2014). Mhrt is specific to cardiac tissue and highly expressed in the heart. During pathological stress, such as pressure overload, Mhrt expression is silenced by a chromatin remodeling complex (Brg1-Hdac-Parp). This repression is an essential step in the progression of dilated cardiomyopathy. Restoring Mhrt expression to pre-stress levels is enough to protect from disease progression and heart failure. Interestingly, it seems that Mhrt antagonizes Brg1 (also known as Smarca4) directly by binding to its helicase domain and preventing Brg1 recognition of its DNA targets. A human version of MHRT exists and is repressed in cardiomyopathy cases, suggesting the same mechanism is present in humans. Another lncRNA involved in the same pathological process of heart remodeling and hyperthrophy is lncRNA CHRF (cardiac hypertrophy-related factor; Wang et al., 2014a). However, CHRF regulates hypertrophy by very different mechanisms compared to Mhrt. During hypertension, angiotensin-II production stimulates paracrine and hormonal release of pro-hypertrophic factors, such as endothelin-1 and TGF-β1. Under these conditions, microarray profiling experiments were able to identify microRNA-489 as a significantly downregulated transcript. Overexpression of this microRNA was enough to attenuate the hypertrophic phenotype in cardiomyocytes. Under cardiomyopathy conditions, lncRNA CHRF is expressed and acts as a sponge for microRNA-489, preventing it from exerting its function, which is the downregulation of Myd88 (a known inducer if cardiac hypertrophy). In another very promising recent study, the pro-hypertrophic lncRNA CHAST was identified (Viereck et al., 2016). CHAST was found to be functionally conserved and in vivo silencing of this lncRNA resulted in an improved recovery after cardiac injury, demonstrating a potential treatment of pathological heart remodeling processes.
Cardiac metabolism, mitochondrial function, and lncRNAs
Metabolism is an essential aspect of cellular physiology, even more so in cardiomyocytes and the heart, an organ with an extraordinary bio-energetic demand. Metabolic remodeling is a hallmark of heart failure and cardiomyopathy and is characterized by a reversion to a fetal-like glycolytic metabolism and a capacity to efficiently employ fatty acid oxidation—the preferred energy source for the adult heart. Very few lncRNAs related to cardiac metabolism have been identified to date. CARL (cardiac apoptosis-related lncRNA) is a lncRNA identified during anoxic treatment of cardiomyocytes (Wang et al., 2014b). During anoxia, PHB2, an important regulator of mitochondrial fission and fusion dynamics, is strongly downregulated by the increased expression of miR-539, leading to mitochondrial fission and cardiomyocyte apoptosis. Interestingly, this dramatic upregulation of miR-539 is the direct consequence of lncRNA CARL silencing. CARL acts as a sponge for miR-539, and is highly expressed in the heart as well as other tissues, indicating this mechanism of apoptosis might be relevant outside the cardiac setting. Forced overexpression of CARL is sufficient to counter the effects of anoxia and rescue a significant proportion of cardiomyocytes from apoptosis. The trigger for CARL downregulation remains unknown.
Recently, a lncRNA profiling study carried out in plasma from patients suffering from heart failure identified a number of circulating lncRNAs associated with LV (left ventricular) remodeling (Kumarswamy et al., 2014). The authors were able to detect hundreds of lncRNAs, and in an interesting turn of events, found out that the most abundant majority of plasma lncRNAs originate from the mitochondrial genome (77%). One lncRNA was significantly upregulated in heart failure and predicted cardiac remodeling with high probability. This lncRNA, named LIPCAR (for long intergenic non-coding RNA predicting cardiac remodeling), increases significantly during late-stage post-myocardial infarction remodeling and during chronic heart failure, thus constituting a prognostic marker for heart failure. The mechanisms of action of LIPCAR and its function at the cellular and tissue level remain to be described.
Control of hypertension and endothelial cell dysfunction by lncRNAs
ANRIL (antisense non-coding RNA in the INK4 locus) plays a critical role in hypertension and atherosclerosis (Yap et al., 2010). ANRIL was discovered accidentally by several independent genome-wide association studies (GWAS) indicating the presence of a coronary artery disease (CAD) risk locus in chromosome 9p21. No known protein-coding elements were present in the haplotype block, however a transcript of unknown function did originate and was very active in atherosclerotic patients. Little is known about the mechanisms by which ANRIL promotes CAD, except that it directly regulates the expression of ADIPOR1, VAMP3, and an orphan protein, C11ORF10. Another example of a vascular-associated lncRNA involved in disease is MALAT1 (metastasis-associated lung adenocarcinoma transcript 1; Liu et al., 2014; Michalik et al., 2014). MALAT1 is significantly upregulated during hyperglycemia and appears associated with diabetes mellitus and microvascular endothelial cell dysfunction. MALAT1 knockdown restores vessel function and alleviates inflammation in diabetic rats (Liu et al., 2014). Upon further inspection, it was found that MALAT1 accumulates in the nucleus of endothelial cells, and it is necessary for cell survival, migration, and angiogenesis in a similar fashion to lncRNA PUNISHER, as described before.
Translational strategies
Although much remains to be learned about lncRNA biology and their mechanisms of gene regulation, lncRNAs are quickly emerging as interesting therapeutic targets in a very similar fashion to their other non-coding relatives, microRNAs. MicroRNAs have been the focus of intense translational and clinical research in the last decade due to their therapeutic promise, and a few successful examples exist and/or are close to commercialization (Kole et al., 2012; Sharma et al., 2014; Lundin et al., 2015). Most approaches are based on specific silencing of microRNA expression using short antisense oligonucleotides (ASOs) with patented derivatives resistant to cellular degradation. ASOs work on similar principles as those of microRNA-mRNA binding and degradation (Walder and Walder, 1988; Dagle et al., 1990; Braasch et al., 2002; Kurreck et al., 2002; Grünweller et al., 2003). ASOs are exciting in the context of lncRNA therapeutics because the same validated approaches as those tested for microRNAs are applicable. As a matter of fact, many of the reports described in this review have made use of ASOs to knockdown lncRNAs successfully for functional studies in mice or rats, bringing lncRNA therapeutics one step closer to the clinic. Overexpression of cardioprotective or otherwise beneficial lncRNAs is a much more complicated matter, given the length of these molecules. Efficient delivery would be complicated by the inability of the long modified transcript to cross the membrane barrier, not to mention synthesis costs and potential toxicity. As of now, alternatives would include the use of gene delivery vectors, such as engineered adeno-associated virus (AAVs), to provide exogenous expression of the desired gene (Asokan et al., 2012; Kotterman and Schaffer, 2014; Samulski and Muzyczka, 2014). Numerous clinical trials using gene therapies are underway (Ginn et al., 2013) and the European Commission already approved its first gene therapy [targeting lipoprotein lipase deficiency (LPLD)] in 2012. Additionally, the first gene therapy for children [for treatment of severe combined immunodeficiency (ADA-SCID)] was very recently approved in by the European commission. Thus therapeutic strategies aimed at lncRNA overexpression might be closer than expected.
Conclusions and future prospects
Non-coding RNAs have emerged as critical regulators of gene expression and function. Understanding their molecular mechanisms of action is providing important novel insights into (cardiac) development, homeostasis, and disease. Judging from the number of predicted lncRNA genes, it is conceivable that future studies will clarify their role and expand our knowledge on the regulatory networks underlying cardiac development and disease. Eventually, these advances will translate into novel therapeutic approaches targeting the leading cause of death, and thus providing a substantial benefit to human healthcare.
Author contributions
SF and AA contributed equally to the review article. They compiled the literature, conceptualized ideas, and prepared the manuscript under the supervision of LK. JH gave critical comments to the review during the preparatory phase.
Conflict of interest statement
AA is a consultant for Jaan Biotherapeutics. The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to those authors whose work could not be cited due to space constraints. The Kurian lab is supported by the NRW Stem Cell Network Independent Group Leader Grant, CECAD, German Heart Association (DZHK) and University of Cologne (Grant No: 3681000801 & 2681101801).
References
- Aguirre A., Montserrat N., Zacchigna S., Nivet E., Hishida T., Krause M. N., et al. (2014). In vivo activation of a conserved microRNA program induces mammalian heart regeneration. Cell Stem Cell 15, 589–604. 10.1016/j.stem.2014.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asokan A., Schaffer D. V., Samulski R. J. (2012). The AAV vector toolkit: poised at the clinical crossroads. Mol. Ther. 20, 699–708. 10.1038/mt.2011.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballantyne M. D., Pinel K., Dakin R., Vesey A. T., Diver L., Mackenzie R., et al. (2016). Smooth muscle enriched long noncoding RNA (SMILR) regulates cell proliferation. Circulation 133, 2050–2065. 10.1161/CIRCULATIONAHA.115.021019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell R. D., Long X., Lin M., Bergmann J. H., Nanda V., Cowan S. L., et al. (2014). Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler. Thromb. Vasc. Biol. 34, 1249–1259. 10.1161/ATVBAHA.114.303240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P., Stolc V., Royce T. E., Rozowsky J. S., Urban A. E., Zhu X., et al. (2004). Global identification of human transcribed sequences with genome tiling arrays. Science 306, 2242–2246. 10.1126/science.1103388 [DOI] [PubMed] [Google Scholar]
- Bhattacharya S., Burridge P. W., Kropp E. M., Chuppa S. L., Kwok W. M., Wu J. C., et al. (2014). High efficiency differentiation of human pluripotent stem cells to cardiomyocytes and characterization by flow cytometry. J. Vis. Exp. 91:52010. 10.3791/52010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch D. A., Liu Y., Corey D. R. (2002). Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucleic Acids Res. 30, 5160–5167. 10.1093/nar/gkf651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brade T., Pane L. S., Moretti A., Chien K. R., Laugwitz K. L. (2013). Embryonic heart progenitors and cardiogenesis. Cold Spring Harb. Perspect. Med. 3:a013847. 10.1101/cshperspect.a013847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan M., Liu G., Mikawa T. (2013). Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field. Science 340, 744–748. 10.1126/science.1232877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. J., Hendrich B. D., Rupert J. L., Lafrenière R. G., Xing Y., Lawrence J., et al. (1992). The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71, 527–542. 10.1016/0092-8674(92)90520-M [DOI] [PubMed] [Google Scholar]
- Bruneau B. G. (2008). The developmental genetics of congenital heart disease. Nature 451, 943–948. 10.1038/nature06801 [DOI] [PubMed] [Google Scholar]
- Buckingham M., Meilhac S., Zaffran S. (2005). Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 6, 826–835. 10.1038/nrg1710 [DOI] [PubMed] [Google Scholar]
- Burridge P. W., Matsa E., Shukla P., Lin Z. C., Churko J. M., Ebert A. D., Lan F., et al. (2014). Chemically defined generation of human cardiomyocytes. Nat. Methods 11, 855–860. 10.1038/nmeth.2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili M. N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., et al. (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927. 10.1101/gad.17446611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri C., Cimatti L., Biagioli M., Beugnet A., Zucchelli S., Fedele S., et al. (2012). Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457. 10.1038/nature11508 [DOI] [PubMed] [Google Scholar]
- Carthew R. W., Sontheimer E. J. (2009). Origins and Mechanisms of miRNAs and siRNAs. Cell 136, 642–655. 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng S. C., Ho L., Tian J., Reversade B. (2013). ELABELA: a hormone essential for heart development signals via the apelin receptor. Dev. Cell 27, 672–680. 10.1016/j.devcel.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Chu C., Zhang Q. C., da Rocha S. T., Flynn R. A., Bharadwaj M., Calabrese J. M., et al. (2015). Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416. 10.1016/j.cell.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C. M., McNeil J. A., Willard H. F., Lawrence J. B. (1996). XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 132, 259–275. 10.1083/jcb.132.3.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M., Xiao Z., Wang Y., Zheng M., Song T., Cai X., et al. (2015). Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 75, 846–857. 10.1158/0008-5472.CAN-14-1192 [DOI] [PubMed] [Google Scholar]
- Dagle J. M., Walder J. A., Weeks D. L. (1990). Targeted degradation of mRNA in Xenopus oocytes and embryos directed by modified oligonucleotides: studies of An2 and cyclin in embryogenesis. Nucleic Acids Res. 18, 4751–4757. 10.1093/nar/18.16.4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning A. P., Gu W., Castoe T. A., Batzer M. A., Pollock D. D. (2011). Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 7:e1002384. 10.1371/journal.pgen.1002384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F., Barozzi I., Mietton F., Ghisletti S., Polletti S., Tusi B. K., et al. (2010). A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 8:e1000384. 10.1371/journal.pbio.1000384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H. H., Li R., Su Y. M., Xiao J., Pan M., Cai X. X., et al. (2016). The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS ONE 11:e0151753. 10.1371/journal.pone.0151753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginn S. L., Alexander I. E., Edelstein M. L., Abedi M. R., Wixon J. (2013). Gene therapy clinical trials worldwide to 2012 - an update. J. Gene Med. 15, 65–77. 10.1002/jgm.2698 [DOI] [PubMed] [Google Scholar]
- Giovarelli M., Bucci G., Ramos A., Bordo D., Wilusz C. J., Chen C. Y., et al. (2014). H19 long noncoding RNA controls the mRNA decay promoting function of KSRP. Proc. Natl. Acad. Sci. U.S.A. 111, E5023–E5028. 10.1073/pnas.1415098111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C., Maquat L. E. (2011). lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470, 284–288. 10.1038/nature09701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez I., Munita R., Agirre E., Dittmer T. A., Gysling K., Misteli T., et al. (2015). A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat. Struct. Mol. Biol. 22, 370–376. 10.1038/nsmb.3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore-Panter S. R., Hsu J., Barnard J., Moravec C. S., Van Wagoner D. R., Chung M. K., et al. (2016). PANCR, the PITX2 adjacent noncoding RNA, is expressed in human left atria and regulates PITX2c expression. Circ. Arrhythm Electrophysiol. 9:e003197. 10.1161/CIRCEP.115.003197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote P., Wittler L., Hendrix D., Koch F., Währisch S., Beisaw A., et al. (2013). The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 24, 206–214. 10.1016/j.devcel.2012.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünweller A., Wyszko E., Bieber B., Jahnel R., Erdmann V. A., Kurreck J. (2003). Comparison of different antisense strategies in mammalian cells using locked nucleic acids, 2′-O-methyl RNA, phosphorothioates and small interfering RNA. Nucleic Acids Res. 31, 3185–3193. 10.1093/nar/gkg409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. A., Shah N., Wang K. C., Kim J., Horlings H. M., Wong D. J., et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076. 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Amit I., Garber M., French C., Lin M. F., Feldser D., et al. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227. 10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P., Li W., Lin C. H., Yang J., Shang C., Nurnberg S. T., et al. (2014). A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514, 102–106. 10.1038/nature13596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T., Wang Y., Lin M. F., Koegel A. K., Kotake Y., Grant G. D., et al. (2011). Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 43, 621–629. 10.1038/ng.848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer M. K., Niknafs Y. S., Malik R., Singhal U., Sahu A., Hosono Y., et al. (2015). The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208. 10.1038/ng.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Zhou X., Huang J. (2016). Long non-coding RNA-ROR mediates the reprogramming in cardiac hypertrophy. PLoS ONE 11:e0152767. 10.1371/journal.pone.0152767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Liu Y., Liu R., Zhang K., Zhang Y. (2015). The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep. 11, 137–148. 10.1016/j.celrep.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. K., Hemberg M., Gray J. M., Costa A. M., Bear D. M., Wu J., et al. (2010). Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187. 10.1038/nature09033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T., Hurt D. E., Ichijo T., Nader N., Chrousos G. P. (2010). Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 3:ra8. 10.1126/scisignal.2000568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C. A., Scheuermann J. C., Surface L. E., Bradley R. K., Fields P. A., Steinhauser M. L., et al. (2013). Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152, 570–583. 10.1016/j.cell.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole R., Krainer A. R., Altman S. (2012). RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 11, 125–140. 10.1038/nrd3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostowski L., Sedlak N., Engel N. (2012). The Kcnq1ot1 long non-coding RNA affects chromatin conformation and expression of Kcnq1, but does not regulate its imprinting in the developing heart. PLoS Genet. 8:e1002956. 10.1371/journal.pgen.1002956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotterman M. A., Schaffer D. V. (2014). Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 15, 445–451. 10.1038/nrg3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M., Siprashvili Z., Chu C., Webster D. E., Zehnder A., Qu K., et al. (2013). Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493, 231–235. 10.1038/nature11661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarswamy R., Bauters C., Volkmann I., Maury F., Fetisch J., Holzmann A., et al. (2014). Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ. Res. 114, 1569–1575. 10.1161/CIRCRESAHA.114.303915 [DOI] [PubMed] [Google Scholar]
- Kurian L., Aguirre A., Sancho-Martinez I., Benner C., Hishida T., Nguyen T. B., et al. (2015). Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation 131, 1278–1290. 10.1161/CIRCULATIONAHA.114.013303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian L., Sancho-Martinez I., Nivet E., Aguirre A., Moon K., Pendaries C., et al. (2013). Conversion of human fibroblasts to angioblast-like progenitor cells. Nat. Methods 10, 77–83. 10.1038/nmeth.2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurreck J., Wyszko E., Gillen C., Erdmann V. A. (2002). Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 30, 1911–1918. 10.1093/nar/30.9.1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. T., Davidow L. S., Warshawsky D. (1999). Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 21, 400–404. 10.1038/7734 [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The, C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- Legnini I., Morlando M., Mangiavacchi A., Fatica A., Bozzoni I. (2014). A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol. Cell 53, 506–514. 10.1016/j.molcel.2013.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Y., Yao J., Li X. M., Song Y. C., Wang X. Q., Li Y. J., et al. (2014). Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 5:e1506. 10.1038/cddis.2014.466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhou D., Li G., Ming X., Tu Y. F., Tian J., et al. (2015). Long non coding RNA-UCA1 contributes to cardiomyocyte apoptosis by suppression of p27 expression. Cell. Physiol. Biochem. 35, 1986–1998. 10.1159/000374006 [DOI] [PubMed] [Google Scholar]
- Lundin K. E., Gissberg O., Smith C. I. (2015). Oligonucleotide therapies: the past and the present. Hum. Gene Ther. 26, 475–485. 10.1089/hum.2015.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C. D., Hannon G. J. (2009). Small RNAs as guardians of the genome. Cell 136, 656–668. 10.1016/j.cell.2009.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K., Nishizawa M., Ozaki T., Kimura T., Hashimoto I., Yamada M., et al. (2008). Natural antisense transcript stabilizes inducible nitric oxide synthase messenger RNA in rat hepatocytes. Hepatology 47, 686–697. 10.1002/hep.22036 [DOI] [PubMed] [Google Scholar]
- McHugh C. A., Chen C. K., Chow A., Surka C. F., Tran C., McDonel P., et al. (2015). The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236. 10.1038/nature14443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J. T., Olson E. N. (2012). MicroRNAs in stress signaling and human disease. Cell 148, 1172–1187. 10.1016/j.cell.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T. R., Mattick J. S. (2013). Understanding the regulatory and transcriptional complexity of the genome through structure. Genome Res. 23, 1081–1088. 10.1101/gr.156612.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalik K. M., You X., Manavski Y., Doddaballapur A., Zörnig M., Braun T., et al. (2014). Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 114, 1389–1397. 10.1161/CIRCRESAHA.114.303265 [DOI] [PubMed] [Google Scholar]
- Mondal T., Subhash S., Vaid R., Enroth S., Uday S., Reinius B., et al. (2015). MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 6:7743. 10.1038/ncomms8743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B. R., Makarewich C. A., Anderson D. M., Winders B. R., Troupes C. D., Wu F., et al. (2016). A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271–275. 10.1126/science.aad4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. (1972). So much “junk” DNA in our genome. Brookhaven Symp. Biol. 23, 366–370. [PubMed] [Google Scholar]
- Olson E. N., Schneider M. D. (2003). Sizing up the heart: development redux in disease. Genes Dev. 17, 1937–1956. 10.1101/gad.1110103 [DOI] [PubMed] [Google Scholar]
- Ounzain S., Micheletti R., Arnan C., Plaisance I., Cecchi D., Schroen B., et al. (2015a). CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell. Cardiol. 89, 98–112. 10.1016/j.yjmcc.2015.09.016 [DOI] [PubMed] [Google Scholar]
- Ounzain S., Micheletti R., Beckmann T., Schroen B., Alexanian M., Pezzuto I., Crippa S., et al. (2015b). Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur. Heart J. 36, 353–368a. 10.1093/eurheartj/ehu180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ounzain S., Pezzuto I., Micheletti R., Burdet F., Sheta R., Nemir M., et al. (2014). Functional importance of cardiac enhancer-associated noncoding RNAs in heart development and disease. J. Mol. Cell. Cardiol. 76, 55–70. 10.1016/j.yjmcc.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A., Norris M. L., Valen E., Chew G. L., Gagnon J. A., Zimmerman S., et al. (2014). Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science 343:1248636. 10.1126/science.1248636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A., Valen E., Schier A. F. (2015). Identifying (non-)coding RNAs and small peptides: challenges and opportunities. Bioessays 37, 103–112. 10.1002/bies.201400103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipes G. C., Creemers E. E., Olson E. N. (2006). The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 20, 1545–1556. 10.1101/gad.1428006 [DOI] [PubMed] [Google Scholar]
- Rinn J. L., Chang H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J. L., Kertesz M., Wang J. K., Squazzo S. L., Xu X., Brugmann S. A., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samulski R. J., Muzyczka N. (2014). AAV-mediated gene therapy for research and therapeutic purposes. Annu. Rev. Virol. 1, 427–451. 10.1146/annurev-virology-031413-085355 [DOI] [PubMed] [Google Scholar]
- Sauvageau M., Goff L. A., Lodato S., Bonev B., Groff A. F., Gerhardinger C., et al. (2013). Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife 2:e01749. 10.7554/eLife.01749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan K., Jiang Q., Wang X. Q., Wang Y. N., Yang H., Yao M. D., et al. (2016). Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction. Cell Death Dis. 7:e2248. 10.1038/cddis.2016.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V. K., Sharma R. K., Singh S. K. (2014). Antisense oligonucleotides: modifications and clinical trials. MedChemComm 5, 1454–1471. 10.1039/C4MD00184B [DOI] [Google Scholar]
- Sleutels F., Zwart R., Barlow D. P. (2002). The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415, 810–813. 10.1038/415810a [DOI] [PubMed] [Google Scholar]
- Song G., Shen Y., Ruan Z., Li X., Chen Y., Yuan W., et al. (2016). LncRNA-uc.167 influences cell proliferation, apoptosis and differentiation of P19 cells by regulating Mef2c. Gene 590, 97–108. 10.1016/j.gene.2016.06.006 [DOI] [PubMed] [Google Scholar]
- Tao H., Cao W., Yang J. J., Shi K. H., Zhou X., Liu L. P., et al. (2016). Long noncoding RNA H19 controls DUSP5/ERK1/2 axis in cardiac fibroblast proliferation and fibrosis. Cardiovasc. Pathol. 25, 381–389. 10.1016/j.carpath.2016.05.005 [DOI] [PubMed] [Google Scholar]
- Thakur N., Tiwari V. K., Thomassin H., Pandey R. R., Kanduri M., Göndör A., et al. (2004). An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Mol. Cell. Biol. 24, 7855–7862. 10.1128/MCB.24.18.7855-7862.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V., Ellis J. D., Shen Z., Song D. Y., Pan Q., Watt A. T., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938. 10.1016/j.molcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M. C., Manor O., Wan Y., Mosammaparast N., Wang J. K., Lan F., et al. (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693. 10.1126/science.1192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vausort M., Wagner D. R., Devaux Y. (2014). Long noncoding RNAs in patients with acute myocardial infarction. Circ. Res. 115, 668–677. 10.1161/CIRCRESAHA.115.303836 [DOI] [PubMed] [Google Scholar]
- Viereck J., Kumarswamy R., Foinquinos A., Xiao K., Avramopoulos P., Kunz M., et al. (2016). Long noncoding RNA Chast promotes cardiac remodeling. Sci. Transl. Med. 8:326ra322. 10.1126/scitranslmed.aaf1475 [DOI] [PubMed] [Google Scholar]
- Walder R. Y., Walder J. A. (1988). Role of RNase H in hybrid-arrested translation by antisense oligonucleotides. Proc. Natl. Acad. Sci. U.S.A. 85, 5011–5015. 10.1073/pnas.85.14.5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. Q., Wang Y., Xiong Y., Chen X. C., Ma M. L., Cai R., et al. (2016a). Sirt1 AS lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci. Rep. 6:21865. 10.1038/srep21865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., et al. (2004). Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878. 10.1038/nature02985 [DOI] [PubMed] [Google Scholar]
- Wang K., Liu F., Liu C. Y., An T., Zhang J., Zhou L. Y., et al. (2016b). The long noncoding RNA NRF regulates programmed necrosis and myocardial injury during ischemia and reperfusion by targeting miR-873. Cell Death Differ. 23, 1394–1405. 10.1038/cdd.2016.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Liu F., Zhou L. Y., Long B., Yuan S. M., Wang Y., et al. (2014a). The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ. Res. 114, 1377–1388. 10.1161/CIRCRESAHA.114.302476 [DOI] [PubMed] [Google Scholar]
- Wang K., Long B., Zhou L. Y., Liu F., Zhou Q. Y., Liu C. Y., et al. (2014b). CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat. Commun. 5, 3596. 10.1038/ncomms4596 [DOI] [PubMed] [Google Scholar]
- Xu H., Guo S., Li W., Yu P. (2015). The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep. 5:12453. 10.1038/srep12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B., Yao J., Liu J. Y., Li X. M., Wang X. Q., Li Y. J., et al. (2015). lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ. Res. 116, 1143–1156. 10.1161/CIRCRESAHA.116.305510 [DOI] [PubMed] [Google Scholar]
- Yan Y., Zhang B., Liu N., Qi C., Xiao Y., Tian X., et al. (2016). Circulating long noncoding RNA UCA1 as a novel biomarker of acute myocardial infarction. Biomed Res. Int. 2016:8079372. 10.1155/2016/8079372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Lin C., Liu W., Zhang J., Ohgi K. A., Grinstein J. D., et al. (2011). ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147, 773–788. 10.1016/j.cell.2011.08.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K. L., Li S., Muñoz-Cabello A. M., Raguz S., Zeng L., Mujtaba S., et al. (2010). Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 38, 662–674. 10.1016/j.molcel.2010.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J. H., Abdelmohsen K., Srikantan S., Yang X., Martindale J. L., De S., et al. (2012). LincRNA-p21 suppresses target mRNA translation. Mol. Cell 47, 648–655. 10.1016/j.molcel.2012.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Gong X., Sun L., Zhou Q., Lu B., Zhu L. (2016). The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting mir-7 expression. PLoS ONE 11:e0158347. 10.1371/journal.pone.0158347 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yu Y.-H., Hu Z.-Y., Li M.-H., Li B., Wang Z.-M., Chen S.-L. (2015). Cardiac hypertrophy is positively regulated by long non-coding RNA PVT1. Int. J. Clin. Exp. Pathol. 8, 2582–2589. [PMC free article] [PubMed] [Google Scholar]
- Yunis J. J., Tsai M. Y. (1978). Mapping of polysomal messenger RNA and heterogeneous nuclear RNA to the lightly staining G-bands of human chromosomes. Cytogenet. Cell Genet. 22, 364–367. 10.1159/000130973 [DOI] [PubMed] [Google Scholar]
- Zhang M., Schulte J. S., Heinick A., Piccini I., Rao J., Quaranta R., Zeuschner D., et al. (2015). Universal cardiac induction of human pluripotent stem cells in two and three-dimensional formats: implications for in vitro maturation. Stem Cells 33, 1456–1469. 10.1002/stem.1964 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang X. O., Chen T., Xiang J. F., Yin Q. F., Xing Y. H., et al. (2013). Circular intronic long noncoding RNAs. Mol. Cell 51, 792–806. 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Li H., Fang S., Kang Y., Wu W., Hao Y., et al. (2016). NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 44, D203–D208. 10.1093/nar/gkv1252 [DOI] [PMC free article] [PubMed] [Google Scholar]