Active JAK1 kinase domain was crystallized with MgADP and soaked in the presence of EDTA to allow displacement of the nucleotide by compounds of interest. This high-throughput method of structure determination may be widely applicable to other protein kinases and small-molecule ATP-competitive ligands.
Keywords: Janus kinase, kinases, crystal soaking, ligand exchange, structure-based drug design
Abstract
Crystals of phosphorylated JAK1 kinase domain were initially generated in complex with nucleotide (ADP) and magnesium. The tightly bound Mg2+-ADP at the ATP-binding site proved recalcitrant to ligand displacement. Addition of a molar excess of EDTA helped to dislodge the divalent metal ion, promoting the release of ADP and allowing facile exchange with ATP-competitive small-molecule ligands. Many kinases require the presence of a stabilizing ligand in the ATP site for crystallization. This procedure could be useful for developing co-crystallization systems with an exchangeable ligand to enable structure-based drug design of other protein kinases.
1. Introduction
The Janus kinase (JAK) family is comprised of four receptor-associated protein tyrosine kinases: JAK1, JAK2, JAK3 and TYK2 (Firmbach-Kraft et al., 1990 ▸; Harpur et al., 1992 ▸; Rane & Reddy, 1994 ▸; Velazquez et al., 1992 ▸; Wilks et al., 1989 ▸). This family of kinases is involved in interferon and cytokine signaling through interaction with signal transducers and activators of transcription (STATs; Schindler et al., 2007 ▸). The JAK–STAT pathway is important for driving biological responses to cytokines (Imada & Leonard, 2000 ▸). Upon cytokine-receptor stimulation, the JAK enzymes phosphorylate STAT proteins in the cytoplasm, inducing dimerization and translocation to the nucleus, where they mediate gene transcription (Leonard & O’Shea, 1998 ▸). Members of the JAK family are influential in a variety of inflammatory diseases and cancer (Wilks, 2008 ▸). Pan-JAK inhibitors have been successfully developed for the treatment of rheumatoid arthritis (Traynor, 2012 ▸) and myelofibrosis (Deisseroth et al., 2012 ▸). The design of inhibitors that are specific for various JAK isoforms has great potential for the development of safer and more efficacious drug therapies for many autoimmune disorders (Dymock et al., 2014 ▸).
The JAK kinases (120–130 kDa) are comprised of seven JAK homology domains (JH1–JH7; Harpur et al., 1992 ▸). The C-terminal kinase module (JH1) is the catalytically active domain that is responsible for the physiological activity of the protein. The JH2 domain is a catalytically inactive pseudokinase domain which has been shown to regulate the activity of the JH1 domain (Saharinen & Silvennoinen, 2002 ▸). The N-terminus contains two Src homology 2 (SH2) domains (JH3 and JH4), followed by the FERM domain (JH5–JH7). The JH1 domain contains the ATP-binding site, for which a variety of small-molecule inhibitors have been designed. All four family members have highly conserved kinase domains, especially at the ATP-binding site, making the design of specific inhibitors challenging. A robust crystallographic system for each member is essential in order to engineer isoform selectivity.
Crystal structures have been elucidated for the kinase domain of each JAK kinase family member (Lucet et al., 2006 ▸; Boggon et al., 2005 ▸; Chrencik et al., 2010 ▸; Williams et al., 2009 ▸), but apo JAK1 is unstable and therefore only a limited number of JAK1 structures are known. The first structures of the JAK1 kinase domain were determined by co-purifying the protein with stabilizing ligands, followed by crystallization and structure determination (Williams et al., 2009 ▸). More recently, structures have been solved with various ligands via co-crystallization methods without the need for ligand during the purification process (Kulagowski et al., 2012 ▸). Here, we report a high-throughput method for obtaining ligand-bound structures of the JAK1 kinase domain using a novel soaking system. Unlike JAK1, crystal structures of JAK2 and JAK3 have been determined by exchanging ligands in the crystal through soaking protocols (Lucet & Bamert, 2013 ▸). Purification of the JAK1 kinase domain in the presence of Mg2+-ATP results in an active, doubly phosphorylated form of the kinase. The ATP is hydrolyzed to ADP and this complex can readily be crystallized. Our early attempts to soak in ligands of interest failed as the ADP remained bound in the active site, despite the superior potency of the ligands in solution studies. The unique aspects of the crystalline environment somehow hamper the displacement of MgADP by synthetic ligands. This problem was eventually remedied by soaking ligands in the presence of EDTA to chelate the Mg2+ ion. The addition of the metal chelator is critical for freeing the active site of the nucleotide, allowing occupancy by the ligand of interest. This soaking method allows the solution of protein–ligand complex structures with a variety of ligands. The overall structure of the kinase domain in complex with PF-4950736 is shown in Fig. 1 ▸. These structures have provided a strong SBDD platform within Pfizer for the design of isoform-selective inhibitors of JAK1 and other JAK isoforms.
Figure 1.
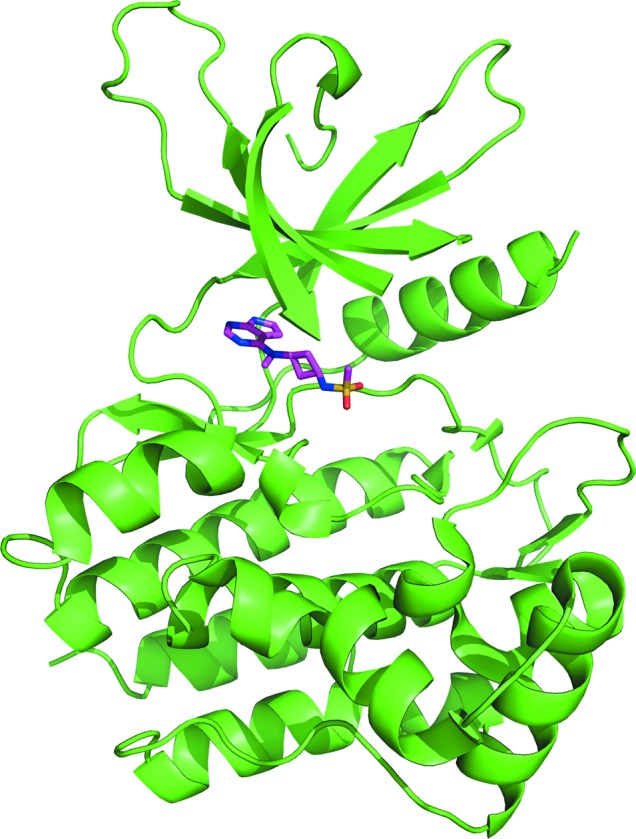
Overall structure of the JAK1 kinase domain in complex with PF-4950736.
2. Materials and methods
2.1. Protein expression and purification
The DNA encoding residues 841–1154 of the human JAK1 kinase domain was codon-optimized for insect-cell expression at GenScript and was cloned into a modified pFastBac vector (see Table 1 ▸). A recombinant baculovirus was prepared using the Bac-to-Bac method (Invitrogen).
Table 1. Expression of the JAK1 kinase domain.
| DNA source | Human synthetic codon-optimized |
| Cloning vector | pUC57 |
| Expression vector | Modified pFastBac |
| Expression host | Sf9 |
| Complete amino-acid sequence of the construct produced | MASHHHHHHDYDGATTENLYFQGSIMRDINKLEEQNPDIVSEKKPATEVDPTHFEKRFLKRIRDLGEGHFGKVELCRYDPEGDNTGEQVAVKSLKPESGGNHIADLKKEIEILRNLYHENIVKYKGICTEDGGNGIKLIMEFLPSGSLKEYLPKNKNKINLKQQLKYAVQICKGMDYLGSRQYVHRDLAARNVLVESEHQVKIGDFGLTKAIETDKEYYTVKDDRDSPVFWYAPECLMQSKFYIASDVWSFGVTLHELLTYCDSDSSPMALFLKMIGPTHGQMTVTRLVNTLKEGKRLPCPPNCPDEVYQLMRKCWEFQPSNRTSFQNLIEGFEALLK |
Spodoptera frugiperda (Sf9) cells were cultured in Sf-900 III SFM medium (Invitrogen) in Corning 3 l baffled flasks (VWR) at 65 rev min−1 on NBS #4430 shakers (two-inch throw) at 27°C. A 1 ml aliquot of baculovirus-infected insect cells [BIIC; Wasilko & Lee, 2006 ▸; preserved at 1 × 107 viable cells (vc) per millilitre in freezing medium: 90%(v/v) Sf-900 III SFM supplemented with 10 g l−1 bovine serum albumin (Sigma–Aldrich) and 10%(v/v) DMSO (Sigma–Aldrich)] was removed from preservation at −80°C, rapidly thawed and diluted into 99 ml Sf-900 III medium. 10 ml of the diluted BIIC baculovirus suspension was used to infect 1 l of log-phase Sf9 cells at a viable cell density of ∼1.5 × 106 vc ml−1, at >98% viability, in each Fernbach flask. Cell density, cell viability and cell diameter were determined daily using a Cedex cell analyzer (Roche Innovatis). The harvest time (72–96 h post-infection) was indicated by a coinciding decrease in percentage cell viability (80–90%) and increase in cell diameter (3–4 µm), at which time 5 mM sodium orthovanadate (FIVEphoton) was added 2 h prior to harvesting the cells. The cell paste was collected by refrigerated centrifugation at 3500g, flash-frozen in liquid nitrogen and stored at −80°C.
The cells expressing the JAK1 kinase domain were resuspended in 25 mM Tris pH 8.0, 300 mM NaCl, 20 mM imidazole, 1 mM ATP, 1 mM MgCl2, 1 mM sodium orthovanadate (NEB) with the addition of 1:100 Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific). The cells were lysed by microfluidization and clarified by centrifugation at 20 000g. The His-tagged JAK1 kinase domain was purified from the supernatant by Ni2+-affinity chromatography using a HisTrap FF column (GE Healthcare). The column was washed with 25 mM Tris pH 8.0, 300 mM NaCl, 10% glycerol, 20 mM imidazole, 1 mM ATP, 1 mM MgCl2 and eluted in the same solution with the addition of 300 mM imidazole. TEV protease was added to the eluate in a 1:100(w:w) ratio and the reaction was dialyzed against 25 mM Tris pH 8.0, 300 mM NaCl, 1 mM TCEP, 10% glycerol, 10 mM ATP, 10 mM MgCl2 overnight at 4°C to remove the His tag and imidazole. The cleaved protein was applied onto Ni ProBond Resin (Qiagen) and washed with dialysis buffer plus 20 mM imidazole. The flowthrough and wash fractions containing the JAK1 kinase domain were pooled and concentrated to 5 ml. Finally, the JAK1 kinase domain was purified by size-exclusion chromatography on a Superdex 75 16/60 prep-grade column (GE Healthcare) and eluted with 25 mM Tris pH 8.0, 250 mM NaCl, 0.25 mM TCEP, 10 mM ATP, 10 mM MgCl2. Fractions containing the JAK1 kinase domain were concentrated to 5 mg ml−1 in a centrifugal concentrator with a molecular-weight cutoff of 10 kDa (Millipore).
2.2. Crystallization
JAK1 kinase domain crystals were grown as described in Table 2 ▸. Microseeding was used to optimize crystal growth. Crystals grew in 2–3 d and were flash-cooled in liquid nitrogen after cryoprotection in reservoir solution supplemented with 30% glycerol. Complexes with inhibitors of interest were generated by soaking crystals in reservoir solution supplemented with 20 mM EDTA and 5 mM compound.
Table 2. Crystallization of the JAK1 kinase domain.
| Method | Hanging-drop vapor diffusion |
| Plate type | Hampton Research VDXm |
| Temperature (K) | 281 |
| Protein concentration (mg ml−1) | 5 |
| Buffer composition of protein solution | 25 mM Tris, 250 mM NaCl, 5 mM TCEP, 10 mM ATP, 10 mM MgCl2 pH 8.0 |
| Composition of reservoir solution | 30% PEG 1500, 8% MPD, 0.1 M Tris pH 9 |
| Volume and ratio of drop | 1 µl:1 µl |
| Volume of reservoir (µl) | 500 |
2.3. Data collection and processing
X-ray diffraction data were collected on the IMCA-CAT 17-ID beamline at the Advanced Photon Source (APS). The data sets were processed using autoPROC (Vonrhein et al., 2011 ▸). Data-collection statistics are given in Table 3 ▸.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| JAK1–ADP | JAK1–PF-4950736 | |
|---|---|---|
| Diffraction source | IMCA-CAT 17-ID, APS | IMCA-CAT 17-ID, APS |
| Wavelength (Å) | 1.0 | 1.0 |
| Temperature (K) | 100 | 100 |
| Detector | Dectris PILATUS 6M | Dectris PILATUS 6M |
| Crystal-to-detector distance (mm) | 400 | 400 |
| Rotation range per image (°) | 0.25 | 0.25 |
| Total rotation range (°) | 180 | 180 |
| Exposure time per image (s) | 0.25 | 0.25 |
| Space group | P21 | C2221 |
| a, b, c (Å) | 45.6, 146.6, 49.9 | 46.2, 88.3, 146.8 |
| α, β, γ (°) | 90.0, 116.7, 90.0 | 90.0, 90.0, 90.0 |
| Mosaicity (°) | 0.2 | 0.3 |
| Resolution range (Å) | 146–2.5 | 146–2.4 |
| Total No. of reflections | 70875 | 64014 |
| No. of unique reflections | 20949 | 11807 |
| Completeness (%) | 99.5 (99.5) | 97.3 (80.9) |
| Multiplicity | 3.4 (3.6) | 5.4 (3.3) |
| 〈I/σ(I)〉 | 10.2 (2.6) | 14.5 (2.3) |
| R r.i.m. | 0.06 (0.28) | 0.05 (0.29) |
| Overall B factor from Wilson plot (Å2) | 46.9 | 40.7 |
2.4. Structure solution and refinement
Data refinement and structure determination were performed with autoBUSTER (Blanc et al., 2004 ▸), Phaser (McCoy et al., 2007 ▸) and Coot (Emsley & Cowtan, 2004 ▸) using the JAK1 kinase domain crystal structure from PDB entry 3eyg (Williams et al., 2009 ▸) as the model. Crystallographic statistics are shown in Table 4 ▸. Refined coordinates and structure factors have been deposited in the Protein Data Bank as entries 5khw and 5khx for the complexes with ADP and PF-4950736, respectively.
Table 4. Structure solution and refinement.
| JAK1–ADP | JAK1–PF-4950736 | |
|---|---|---|
| Resolution range (Å) | 73.0–2.5 | 73.0–2.4 |
| Completeness (%) | 99.4 | 96.8 |
| σ Cutoff | 0.0 | 0.0 |
| No. of reflections, working set | 19797 | 11212 |
| No. of reflections, test set | 1072 | 561 |
| Final R cryst | 0.19 | 0.17 |
| Final R free | 0.23 | 0.24 |
| Cruickshank DPI | 0.68 | 0.43 |
| No. of non-H atoms | ||
| Protein | 4525 | 2271 |
| Ion | 2 | 0 |
| Ligand | 54 | 20 |
| Water | 84 | 79 |
| Total | 4665 | 2370 |
| R.m.s. deviations | ||
| Bonds (Å) | 0.01 | 0.01 |
| Angles (°) | 1.14 | 1.18 |
| Average B factors (Å2) | ||
| Protein | 49.9 | 50.4 |
| Ion | 31.4 | n/a |
| Ligand | 36.4 | 37.3 |
| Water | 42.0 | 45.0 |
| Ramachandran plot | ||
| Most favored (%) | 92.8 | 91.8 |
| Allowed (%) | 7.2 | 8.2 |
3. Results and discussion
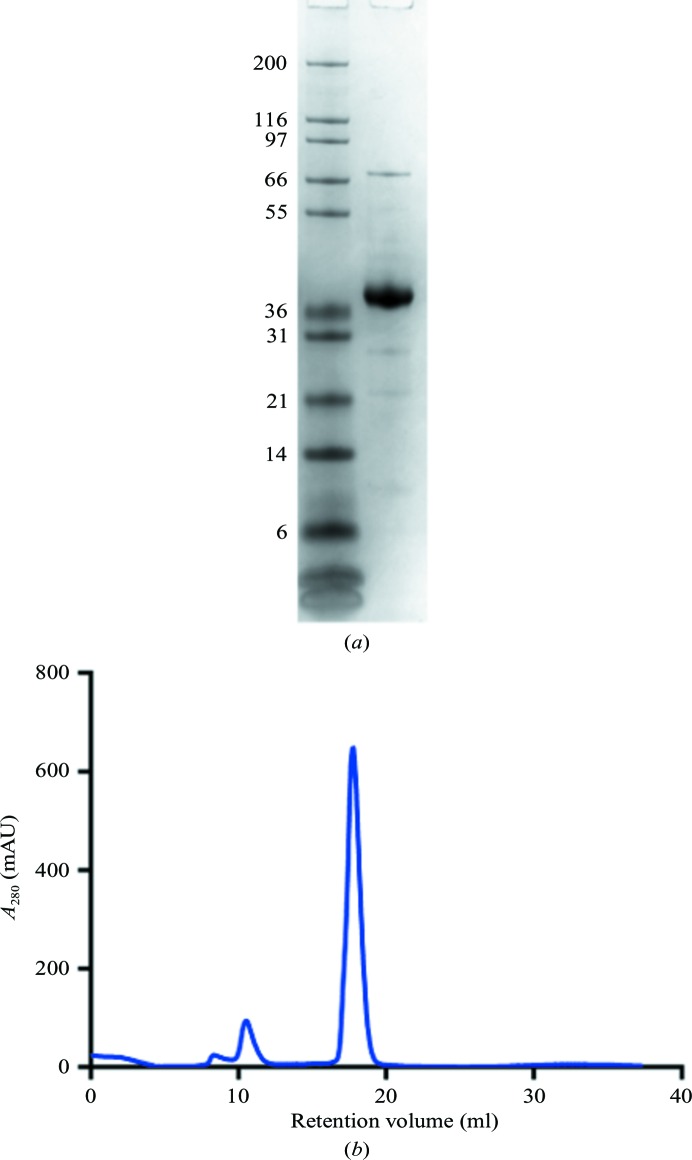
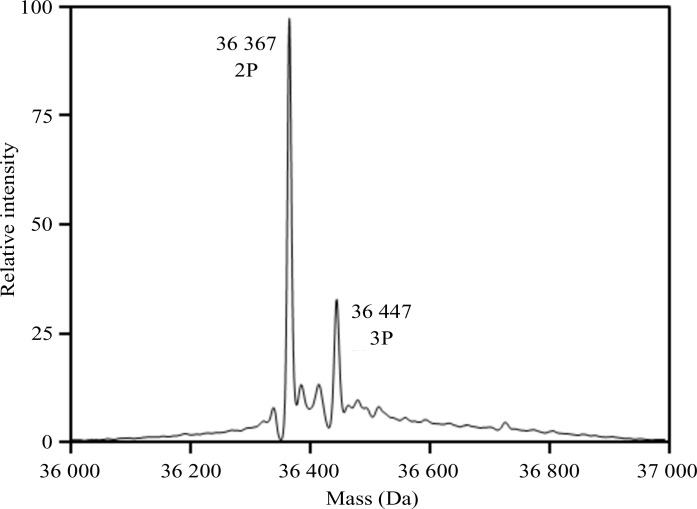
The JAK1 kinase domain was expressed in insect cells in the presence of the phosphatase inhibitor sodium orthovanadate, and purified by Ni2+-affinity chromatography followed by size-exclusion chromatography. The purification yielded about 4 mg of protein per litre of insect-cell expression medium. The protein purity was greater than 90% as judged by SDS–PAGE, and the protein eluted as a single, symmetrical peak from the size-exclusion column (Fig. 2 ▸). The addition of phosphatase inhibitor resulted in partial phosphorylation and activation of the kinase. The level of phosphorylation was increased by purifying the enzyme in the presence of MgATP, promoting autophosphorylation. The kinase domain autophosphorylates two Tyr residues in the activation loop (Tyr1034 and Tyr1035). Mass spectrometry reveals that the final purified JAK1 kinase domain had an additional 2–3 phosphate groups, with the bisphosphorylated protein as the primary species (Fig. 3 ▸). Peptide mapping confirmed that the majority of Tyr1034 and Tyr1035 are phosphorylated (data not shown). The doubly phosphorylated JAK1 kinase domain complex was readily crystallized by vapor-diffusion experiments. The crystals grew to a full size of about 80 µm in 2–3 d.
Figure 2.
Purity analysis of the JAK1 kinase domain. (a) 4–12% NuPAGE (Invitrogen) SDS denaturing gel. The left lane contains molecular-mass markers (labeled in kDa). (b) Size-exclusion chromatogram of the JAK1 kinase domain. The elution from SEC is later than expected owing to interaction of the protein with the stationary phase of the column.
Figure 3.
Intact mass analysis of the crystallized form of the JAK1 kinase domain. The theoretical mass for the bisphosphorylated (abbreviation 2P) form of the protein was 36 366 Da. The mass discrepancy between this and the major observed mass (36 367 Da) was 27 p.p.m.
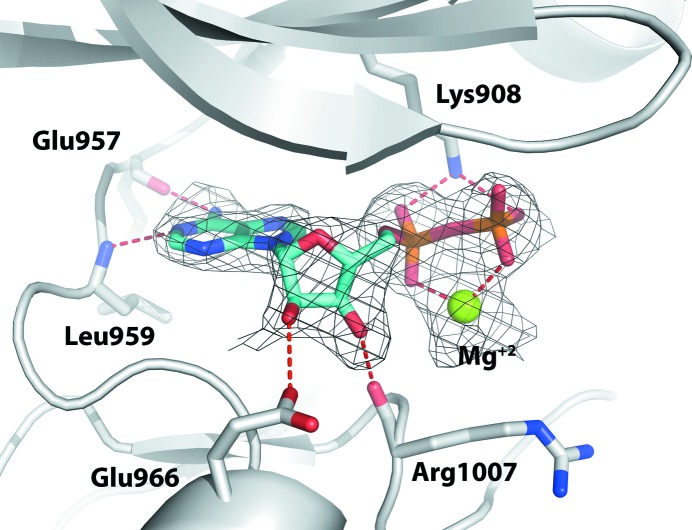
The structure of the JAK1 kinase domain purified with MgATP was solved at a resolution of 2.5 Å. The electron-density map (Fig. 4 ▸) clearly shows the presence of MgADP in the hinge region even though the crystallization experiments were set up with MgATP. This has been observed with other protein kinases and suggests hydrolysis of the triphosphate nucleotide by the ATPase activity of the kinase. The resulting product, ADP, is bound in the hinge region of the active site of the kinase domain, forming hydrogen-bond interactions with the backbones of Glu957 and Leu959. The ribose hydroxyl groups are anchored by the side chain of Glu966 and the carbonyl group of Arg1007. The catalytic Lys908 and magnesium ion place the phosphate groups of ADP in the active conformation.
Figure 4.
Crystal structure of the JAK1 kinase domain in complex with ADP and Mg2+ ion. ADP is shown in cyan and the (F o − F c) OMIT map is contoured at 2.5σ. Hydrogen bonds are depicted by red dashed lines.
Exchanging ADP for ligand using conventional soaking methods was not successful, as the ADP remained stubbornly bound to the active site despite the presence of high concentrations of potent ligands in the soaking solution. It is surprising that ADP should bind this tightly, as release of this ligand is essential for the normal kinase activity of the enzyme. It is likely that the crystalline environment promotes tighter binding than that in the solution state. We tried several methods to help displace ADP from the ATP site, including varying the concentration, the temperature and the length of the soaks, but all of these proved unsuccessful. We hypothesized that breaking up the MgADP complex using a strong metal chelator may reduce the affinity of ADP, promoting ligand displacement. Addition of a very high concentration of EDTA effectively removed the Mg2+ ion, which allowed facile displacement of ADP by the ligands of interest. We did not specifically evaluate the minimum concentration of EDTA required to dislodge MgADP from the ATP site of JAK1, but chose to always use a high concentration to ensure its displacement.
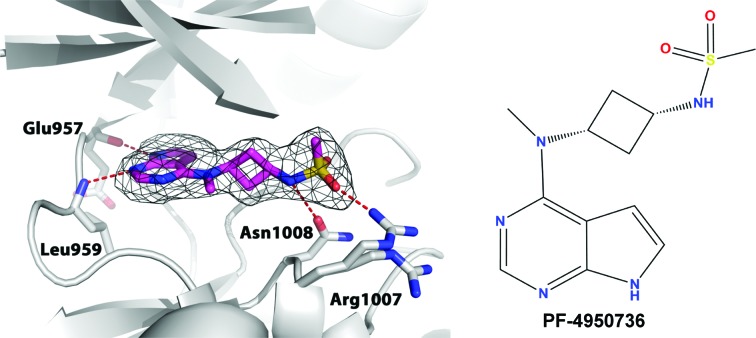
This soaking method was used to obtain a structure with PF-4950736 to 2.4 Å resolution (Fig. 5 ▸). This ligand has an IC50 of 14 nM. Upon addition of EDTA and the ligand, the space group of the crystals transformed from primitive monoclinic with two molecules per asymmetric unit to a related C-centered orthorhombic lattice with one monomer per asymmetric unit. Two of the unit-cell parameters remained more or less the same, while the third one nearly doubled in the C2221 lattice. The electron-density map unambiguously shows replacement of ADP by PF-4950736. In the active site, the P-loop is partially disordered owing to flexibility and the Arg1007 side chain shows two alternative conformations. The pyrrolopyrimidine core forms two direct hydrogen bonds to the hinge region between the N-terminal and C-terminal lobes of JAK1 (backbones of Glu957 and Leu959). The sulfonamide group is anchored by the side chains of Asn1008 and Arg1007. At the solvent-exposed surface in the N-terminal lobe of the kinase domain, continuous electron density connects the side chains of Cys944 and Lys953, indicating the formation of a lysinoalanine cross-link during X-ray irradiation. In the current model, we have included only Cys944 and Lys953 instead of lysinoalanine, as we observe a mixture of intact residues and the covalent side-chain adduct.
Figure 5.
Crystal structure of the JAK1 kinase domain in complex with PF-4950736. The compound is shown in magenta and the (F o − F c) OMIT map is contoured at 2.5σ. Hydrogen bonds are depicted by red dashed lines.
In conclusion, purification of the JAK1 kinase domain with MgATP yields a biochemically active protein that retains kinase and ATPase activities. Diffraction-quality crystals of this protein grow readily and contain MgADP bound at the active site. Soaking with ligand alone is insufficient to displace the ADP, whereas addition of the Mg2+-ion chelator EDTA renders the active site available for ligand binding. This has proven to be a useful approach for reproducible structure determination of JAK1 with a variety of ligands. The method described here could be a general method applicable to other protein kinases and ATPases, although this was not explicitly evaluated. Kinases often require the presence of a ligand in the active site to stabilize the enzyme for crystallization. Having a ligand that can be displaced from the crystal allows the introduction of ligands of interest for structure determination. Chelating the Mg2+ ion unlocks the nucleotide-bound state, allowing occupancy by a ligand of interest in the active site.
Supplementary Material
PDB reference: JAK1 kinase domain, complex with ADP, 5khw
PDB reference: complex with PF-4950736, 5khx
References
- Blanc, E., Roversi, P., Vonrhein, C., Flensburg, C., Lea, S. M. & Bricogne, G. (2004). Acta Cryst. D60, 2210–2221. [DOI] [PubMed]
- Boggon, T. J., Li, Y., Manley, P. W. & Eck, M. J. (2005). Blood, 106, 996–1002. [DOI] [PMC free article] [PubMed]
- Chrencik, J. E. et al. (2010). J. Mol. Biol. 400, 413–433. [DOI] [PubMed]
- Deisseroth, A. et al. (2012). Clin. Cancer Res. 18, 3212–3217. [DOI] [PubMed]
- Dymock, B. W., Yang, E. G., Chu-Farseeva, Y. & Yao, L. (2014). Future Med. Chem. 6, 1439–1471. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Firmbach-Kraft, I., Byers, M., Shows, T., Dalla-Favera, R. & Krolewski, J. J. (1990). Oncogene, 5, 1329–1336. [PubMed]
- Harpur, A. G., Andres, A. C., Ziemiecki, A., Aston, R. R. & Wilks, A. F. (1992). Oncogene, 7, 1347–1353. [PubMed]
- Imada, K. & Leonard, W. J. (2000). Mol. Immunol. 37, 1–11. [DOI] [PubMed]
- Kulagowski, J. J. et al. (2012). J. Med. Chem. 55, 5901–5921. [DOI] [PubMed]
- Leonard, W. J. & O’Shea, J. J. (1998). Annu. Rev. Immunol. 16, 293–322. [DOI] [PubMed]
- Lucet, I. S. & Bamert, R. (2013). Methods Mol. Biol. 967, 275–300. [DOI] [PubMed]
- Lucet, I. S., Fantino, E., Styles, M., Bamert, R., Patel, O., Broughton, S. E., Walter, M., Burns, C. J., Treutlein, H., Wilks, A. F. & Rossjohn, J. (2006). Blood, 107, 176–183. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Rane, S. G. & Reddy, E. P. (1994). Oncogene, 9, 2415–2423. [PubMed]
- Saharinen, P. & Silvennoinen, O. (2002). J. Biol. Chem. 277, 47954–47963. [DOI] [PubMed]
- Schindler, C., Levy, D. E. & Decker, T. (2007). J. Biol. Chem. 282, 20059–20063. [DOI] [PubMed]
- Traynor, K. (2012). Am. J. Health Syst. Pharm. 69, 2120.
- Velazquez, L., Fellous, M., Stark, G. R. & Pellegrini, S. (1992). Cell, 70, 313–322.
- Vonrhein, C., Flensburg, C., Keller, P., Sharff, A., Smart, O., Paciorek, W., Womack, T. & Bricogne, G. (2011). Acta Cryst. D67, 293–302. [DOI] [PMC free article] [PubMed]
- Wasilko, D. J. & Lee, S. E. (2006). Bioprocess J. 5, 29–32.
- Wilks, A. F. (2008). Semin. Cell Dev. Biol. 19, 319–328.
- Wilks, A. F., Kurban, R. R., Hovens, C. M. & Ralph, S. J. (1989). Gene, 85, 67–74. [DOI] [PubMed]
- Williams, N. K., Bamert, R. S., Patel, O., Wang, C., Walden, P. M., Wilks, A. F., Fantino, E., Rossjohn, J. & Lucet, I. S. (2009). J. Mol. Biol. 387, 219–232. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: JAK1 kinase domain, complex with ADP, 5khw
PDB reference: complex with PF-4950736, 5khx