Abstract
Background
Plasmodium vivax malaria is a major public health problem in French Guiana. Some cases of resistance to chloroquine, the first-line treatment used against P. vivax malaria, have been described in the Brazilian Amazon region. The aim of this study is to investigate a possible dispersion of chloroquine-resistant P. vivax isolates in French Guiana. The genotype, polymorphism and copy number variation, of the P. vivax multidrug resistance gene-1 (pvmdr1) have been previously associated with modification of the susceptibility to chloroquine.
Methods
The pvmdr1 gene polymorphism was evaluated by sequencing and copy number variation was assessed by real-time PCR, in P. vivax isolates obtained from 591 symptomatic patients from 1997 to 2013.
Results
The results reveal that 1.0% [95% CI 0.4–2.2] of French Guiana isolates carry the mutations Y976F and F1076L, and that the proportion of isolates with multiple copies of pvmdr1 has significantly decreased over time, from 71.3% (OR = 6.2 [95% CI 62.9–78.7], p < 0.0001) in 1997–2004 to 12.8% (OR = 0.03 [95% CI 9.4–16.9], p < 0.0001) in 2009–2013. A statistically significant relationship was found between Guf-A (harboring the single mutation T958M) and Sal-1 (wild type) alleles and pvmdr1 copy number.
Conclusions
Few P. vivax isolates harboring chloroquine-resistant mutations in the pvmdr1 gene are circulating in French Guiana. However, the decrease in the prevalence of isolates carrying multiple copies of pvmdr1 might indicate that the P. vivax population in French Guiana is evolving towards a decreased susceptibility to chloroquine.
Background
Plasmodium vivax remains the most geographically widespread of the five Plasmodium species infecting humans. As the second most common cause of malaria worldwide, P. vivax is the main cause of malaria in South America, where 390,000 cases were reported by the World Health Organization in 2015 [1]. Approximately 95% of these P. vivax malaria cases occur in nine countries of the Amazon Basin, namely Brazil, Bolivia, Colombia, Ecuador, French Guiana, Guyana, Peru, Suriname and Venezuela [1]. A total of 311 P. vivax cases were reported in French Guiana in 2014, representing 70% of the total number of malaria cases [2]. Since 1995, the treatments of uncomplicated P. falciparum malaria, mefloquine or halofantrine were used in monotherapy until 2002 when they were replaced by the association atovaquone–proguanil and in 2009 by the combination of artemether–lumefantrine [2–4]. Like the other countries across the continent, chloroquine is still recommended as the first-line treatment for P. vivax in French Guiana.
In 1989, the first cases of chloroquine-resistant P. vivax infection were reported in Papua New Guinea [5]. First cases of P. vivax resistance to chloroquine in South America were described in clinical studies of unsupervised chloroquine treatment in 1989 and 1992, in Colombia and Brazil, respectively [6, 7]. It was only in 1996 that the first confirmed clinical case of resistance was described in Brazil [8]. Chloroquine resistance has spread around the world over the last decade [9], and is now found in Southeast Asia [10–14] but also in Africa [15, 16], South America [8, 17–22] and the Middle East [23, 24].
Polymorphisms in pvmdr1 gene (P. vivax multidrug resistance-1 gene, PVX_080100), orthologous to pfmdr1 gene in Plasmodium falciparum (PF3D7_0523000) [25], has been associated with chloroquine resistance in many studies. Pvmdr1 Y976F and F1076L mutations are found in all malaria-endemic regions where chloroquine is used as the first-line treatment [13, 14, 26, 27]. Isolates bearing only F1076L mutations were identified but were not associated with a chloroquine resistance [28, 29]. This observation supports the argument that P. vivax chloroquine resistance requires the presence of both mutations. It has been suggested that F1076L is the prerequisite for the secondary acquisition of Y976F, which is responsible for the decrease in chloroquine susceptibility [29]. However, no other studies have observed this correlation between Y976F mutation and resistance phenotype [15, 18]. Several studies have pinpointed an increase in pvmdr1 gene copy number, which seems to be related to an increased susceptibility to chloroquine [13, 14, 30, 31]. Furthermore, a recent study suggested that chloroquine resistance and clinical severity in vivax malaria were associated with increased expression levels of pvmdr1 and pvcrt-o genes [32].
This study aims to estimate the possible emergence of chloroquine-resistant P. vivax in French Guiana. The work is divided into two parts. Firstly, pvmdr1 gene polymorphism and copy number were assayed in P. vivax isolates obtained from blood samples of patients collected since 1997 and the temporal evolution of pvmdr1 gene polymorphism and copy number were studied.
Methods
Sample collection
Between 1997 and 2013, samples were collected from 591 symptomatic patients presenting with malaria symptoms at health centres in French Guiana. Plasmodium vivax mono-infections were diagnosed by rapid diagnostic tests and/or microscopic examination of the blood. Blood samples were collected in EDTA-coated tubes and sent to the National Reference Centre (NRC) for Malaria at the Institut Pasteur de la Guyane for further analysis and biobanking.
DNA extraction, amplification and sequencing
Parasite DNA was extracted from whole blood samples using the QIAamp DNA Blood Mini Kit (Qiagen, Courtaboeuf, France), following manufacturer’s instructions. The pvmdr1 gene was amplified and sequencing using protocol (primers and amplification condition) previously described by Lekweiry et al. [33] except the polymerase, 0.025 U/μl of Ampli Taq Gold™, 2.5 mM MgCl2, 1× PCR Gold Buffer (Applied Biosystems). The amplification product was loaded on a 1.5% agarose gel and visualized after electrophoresis.
Sequencing of pvmdr1 PCR product was performed by using the nested primers [33] generating a product length of 547 bp (region between codon 931 and 1095). These sequence were compared with the reference sequence Sal-1 (Genbank accession number AY571984).
Pvmdr1 and pvaldolase cloning
As a reference sample with a known pvmdr1 copy number was not available, two reference plasmids containing one copy of the pvmdr1 and pvaldolase genes were generated, respectively, to use as positive controls for the real-time PCR (qPCR). They were created by PCR amplification of pvmdr1 and pvaldolase genes using A380 and A379 or A382 and A383 primers, respectively (Table 1). Each PCR product was cloned in pCR™4-TOPO® plasmid using the TOPO® TA Cloning® kit (Invitrogen), following manufacturer’s protocol.
Table 1.
Primers and probes for quantification of the pvmdr1 copy number by qPCR
| Primer and probe name | Sequence | Gene |
|---|---|---|
| A380 | 5′-GAG-AGG-ACG-TAA-ACG-TGC-TT-3′ | pvmdr1 |
| A379 | 5′-ACG-TTG-GTG-TCG-TAC-TGA-TTC-G-3′ | |
| A399 | 5′-FAM_TTT-GCC-GCA-ATT-GA_MGB/NFQ-3′ | |
| A382 | 5′-AGT-TTT-GTT-GGA-AGG-AGC-TTT-ATT-G-3′ | pvaldolase |
| A383 | 5′-TGG-TTT-TCA-CAG-CAC-AGT-CGT-AT-3′ | |
| A397 | 5′-FAM_CCC-AAC-ATG-GTG-ACC-G_MGB/NFQ-3′ |
MGB/NFQ minor groove binder/non-fluorescent quencher, pvmdr1 Plasmodium vivax multi-drug resistance 1
Real-time PCR to quantify the pvmdr1 gene copy number
The pvmdr1 gene copy number was measured by performing a qPCR in comparison to the reference gene pvaldolase, using the method previously described by Lekweiry et al. [33]. The reproducibility problems encountered were solved by designing new probes using Primer Express® software (Applied Biosystems). These primers and probes are listed in Table 1. The qPCR was carried out in a 25 μl reaction volume containing 1 μl of DNA, 300 nM of each primer, 200 nM of probe, 12.5 μl of TaqMan® Universal Master Mix II (Applied Biosystems) and water. Real-time PCR was performed under the following conditions: 95 °C for 10 min, then 40 cycles at 95 °C for 15 s and 65 °C for 1 min. Samples were set up in triplicate and experiments were repeated independently twice.
Results analysis was executed by StepOne™ software (Applied Biosystems). The signal from the pvmdr1 gene was normalized to the single copy pvaldolase reference gene, then copy number was determined using the mathematical model described by Pfaffl [34].
Statistical analysis
All statistical analyses were performed with R software (version 3.0.2). Percentages were calculated for each parameter studied, namely single nucleotide polymorphisms and increased pvmdr1 copy number, in comparison to the total sample size. Corresponding 95% confidence intervals (CI) were calculated using the exact (Clopper-Pearson interval) method [35]. In this study, the pvmdr1 copy number was analysed as a qualitative variable and two groups were considered: samples with one copy and samples with at least two copies of the pvmdr1 gene.
A Chi square test for trend allowed comparison of the temporal evolution of pvmdr1 allele frequencies and gene copy number. A logistic regression was used to determine the association between polymorphisms and the pvmdr1 gene copy numbers. A p value below 0.05 was considered significant.
Nucleotide sequence accession numbers
The Guf-A, Guf-B and Guf-C allele sequences of the pvmdr1 gene reported in this study were deposited in GenBank under accession numbers KU196660, KU196661 and KU196662, respectively.
Results
Demographic information
A total of 547 patients for 591 sample, 362 men and 185 women with the sex ratio of 1.96, were included in this study. The average age was 29.2 years (1 month to 76 years, including 88 children under 15 years) with parasitaemia between 0.001 and 4% with an average of 0.32% (Table 2). Three time periods (1997–2004, 2005–2008 and 2009–2013) were considered according to the years 2005 when the P. vivax became the dominant species diagnosed in French Guiana, and 2009 when the combination of artemether and lumefantrine was adopted for the treatment of P. falciparum and mixed infections (P. falciparum/P. vivax) in French Guiana (Table 2). No significant association on all parameters (age, sex ratio and parasitaemia) and the time period.
Table 2.
Demographic information
| Year | Number of patient | Age (mini–max) | Nb men | Nb women | Sex ratio | Parasitemia % (mini–max) |
|---|---|---|---|---|---|---|
| 1997–2004 | 136 | 26.7 (0.46–56) | 85 | 50 | 1.7 | 0.4 (0.01–2) |
| 2005–2008 | 120 | 22.9 (0.08–63) | 82 | 38 | 2.16 | 0.5 (0.01–4) |
| 2009–2013 | 291 | 31.2 (0.67–76) | 195 | 97 | 2.01 | 0.3 (0.001–2.3) |
| Total | 547 | 29.2 (0.08–76) | 362 | 185 | 1.96 | 0.32 (0.001–4) |
Age were indicated in year; the parasitemia were indicated on percentage of red blood cell infected by P. vivax
Polymorphism of the pvmdr1 gene
Among the 591 genotyped samples, four non-synonymous mutations (T958 M, Y976F, F1070L and F1076L) and one synonymous mutation (L1022L) were identified. The French Guiana strains were then divided into four alleles (Fig. 1). The Sal-1 wild-type allele was present in 11.2% (n = 66/591, CI 95% [8.7–14.0]) of the samples while 86.5% (n = 511/591, CI 95% [83.4–89.1]) of isolates carried the T958 M mutation and this predominant allele was called Guf-A. Only 1.4% (n = 8/591, CI 95% [0.6–2.7]) of isolates carried the T958M/F1070L mutations and this double mutant allele was referenced as Guf-B. Finally, 1.0% (n = 6/591, CI 95% [0.4–2.2]) of isolates carried the T958M/Y976F/F1076L, this triple mutant was named Guf-C (Fig. 1).
Fig. 1.

Pvmdr1 sequence polymorphism (codon 931–1095) of 591 isolates collected in French Guiana between 1997 and 2013. Only polymorphic codons are indicated. Open symbols denote the wild-type (Sal-1-type) nucleotide sequence, and black symbols indicate the presence of the mutant sequence shown at the top. The number of isolates observed for each allele is indicated in brackets
The temporal evolution of these alleles was then determined (1997–2004, n = 136; 2005–2008, n = 120 and 2009–2013, n = 335; Fig. 2). No significant association was found between the frequency of Guf-B and Guf-C alleles and the time period (p = 0.44 and p = 0.35, respectively, Chi square test for trend). A statistically significant increase in the frequency of the Sal-1 allele through time (p < 0.0001) has detected, along with a statistically significant decrease for the Guf-A allele (p < 0.002) (Fig. 2). Furthermore, relationship between epidemiological data and genotype was not observed.
Fig. 2.
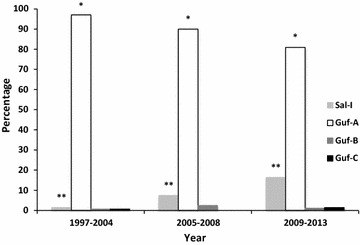
Temporal evolution of pvmdr1 allele frequencies. *p < 0.0002 and **p < 0.0001 denote significant differences in allele frequencies through time (Chi square test for trend)
Relationship between copy number and genotype of the pvmdr1 gene
The copy number of the pvmdr1 gene was determined for all samples and varied from 1 to 8 (mean 1.45, CI 95% [1.35–1.55]). The majority of the sample, 68.5% (n = 405/591, CI 95% [64.6–72.3]), had one copy while 31.5% (n = 186/591, CI 95% [27.7–35.4]) had two to eight copies. The frequency of isolates with multiple copies of the pvmdr1 gene was significantly higher in samples collected between 1997 and 2004 (97/136, 71.3%, CI 95% [62.9–78.7]) than in samples from 2005 to 2008 (45/120, 37.5%, CI 95% [28.8–46.8]) or even after 2008 (43/335, 12.8%, CI 95% [9.4–16.9]). This decrease over time was statistically significant (p < 0.0001, Chi square test for trend, see Fig. 3). Moreover, relationship between epidemiological data and copy number was not observed.
Fig. 3.
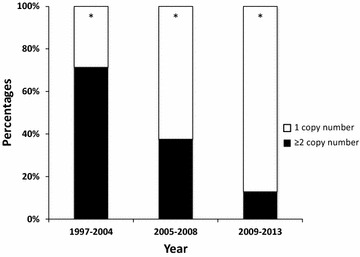
Temporal evolution of the proportion of isolates harbouring single or multiple pvmdr1 copy number. White represents the isolates with single copy number of pvmdr1 gene, and black indicates isolates with two or more pvmdr1 copy numbers. *p < 0.0001 for Chi square test for trend
The association between the copy number and genotype of the pvmdr1 gene was evaluated. The proportion of isolates harbouring multiple copies was not equally distributed among the different alleles. A statistically significant association was found between the Sal-1 allele and single copy pvmdr1 gene (p = 0.0006) as well as between the Guf-A allele and copy number greater than 1 (p = 0.0002). No statistically significant relationship was found between the Guf-B and Guf-C alleles and the pvmdr1 gene copy number (p = 0.93 and p = 0.58, respectively, Fig. 4).
Fig. 4.
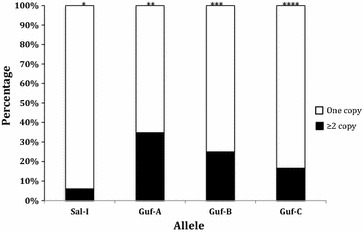
Evaluation of the relationship between pvmdr1 copy number and genotype. *OR = 0.12 [95% CI 0.04–0.034], p = 0.00006. **OR = 5.57 [95% CI 2.51–12.36], p = 0.00002. ***OR = 0.73 [95% CI 0.15–3.64], p = 0.70. ****OR = 0.44 [95% CI 0.05–3.76], p = 0.45
Discussion
Vivax malaria is a major public health problem in South America. In French Guiana, it is currently responsible for two-thirds of malaria cases. Resistance to chloroquine, the main treatment used against vivax malaria [2], has been reported in the Brazilian Amazon Region [4, 17, 18, 20, 21, 32, 36, 37]. In French Guiana, the development of gold-mining activities and the consequent human migration between French Guiana and neighbouring countries, Brazil and Suriname [4, 38, 39], have raised fears that chloroquine-resistant P. vivax isolates may spread. It is therefore important to follow the circulation of resistant isolates. There are previous studies suggesting that Y976F and F1076L mutations in the pvmdr1 gene are associated with in vitro resistance to chloroquine [13, 14]. Pvmdr1 Y976F mutation alone is not sufficient to cause the failure of chloroquine treatment, as observed in Madagascar [15], Brazil [21] and Honduras [40]. It can only affect treatment outcome when associated with the F1076L mutation. In French Guiana, these two mutations were carried by 1.0% of the isolates (i.e., Guf-C allele). Therefore, these parasites could potentially be resistant to chloroquine; however, an association with clinical drug response or in vitro susceptibility have not been investigated. This value is similar to the 1.8% prevalence reported in Brazil [41].
Sal-1 and Guf-A alleles were present in 11.2 and 86.5% of the samples in French Guiana, respectively, showing significant and inverse trends in the temporal evolution of their frequency; while the frequency of the Sal-1 allele significantly increased over time, the frequency of the Guf-A allele decreased. These temporal variations of allele frequencies could be explained by different factors, such as the increased circulation of isolates between French Guiana and Brazil or changes in drug policy for the treatment of P. falciparum. This is supported by a recent study showing that the T958M mutation allele is the majority among Brazilian isolates collected between 2010 and 2014 [42]. Many studies in Southeast Asia have shown that isolates with pvmdr1 gene amplification were characterized by increased susceptibility to chloroquine but decreased susceptibility to mefloquine [13, 31]. A significant decrease of the proportion of isolates with multiple copies of pvmdr1 over this 16-year study period has been found, decreasing from 71.3% between 1997 and 2004 to only 12.8% between 2009 and 2013. A recent study comparing P. vivax isolates from French Guiana (collected between 2001 and 2003) and Southeast Asia (collected in 2010) showed the number of isolates with multiple copies of pvmdr1 gene to be higher in French Guiana than in Cambodia [43]. Moreover, pvmdr1 gene amplifications were rare (fewer than 2%) in countries where mefloquine has never been used for malaria treatment, such as Madagascar and Sudan [43]. In P. falciparum, multiple copies of pfmdr1 were associated with mefloquine-resistant isolates [44]. This has been confirmed in French Guiana where isolates with amplified copy number of pfmdr1 gene were significantly correlated with resistance to mefloquine and halofantrine, both used in monotherapy against uncomplicated P. falciparum malaria until 2002 [45]. P. vivax isolates were therefore subjected to indirect selection pressure by mefloquine during the treatment of P. falciparum or mixed infections (P. falciparum and P. vivax). When the use of mefloquine and halofantrine ceased in French Guiana, P. falciparum isolates with one copy of pfmdr1 increased [45]. This loss of selective pressure would also explain the increased frequency of P. vivax isolates with a single copy of the pvmdr1 gene, a genotype associated to chloroquine resistance [13].
Recently two studies analyzing the whole genome sequences of isolates collected in America, Africa and Asia, have shown great diversity of P. vivax isolates according to their geographical origin in particular for malaria drug antifolate resistance genes involved in resistance to (pvdhfr and pvdhps) [46, 47]. Nevertheless, the role of pvmdr1 in conferring resistance to chloroquine is still elusive and controversial and was recently further challenged by global population genomic studies of P. vivax. Indeed, Schousboe et al. studied the prevalence of polymorphisms and the diversity in microsatellite markers flanking the pvmdr1 gene in P. vivax isolates from seven endemic countries worldwide (Pakistan, Afghanistan, Nepal, Sri Lanka, Ecuador, Sao Tomé and Sudan). Although they showed that Y976F and F1076L mutations in pvmdr1 gene have developed on multiple haplotype backgrounds by convergent evolution in these countries, they highlighted high levels of diversity around mutant alleles, suggesting these alleles were not subject to a selective sweep [48].
Conclusions
The present study indicates that P. vivax isolates with mutations in pvmdr1 previously described as associated with chloroquine resistance are present at low frequency in French Guiana. In addition, the number of copies of the gene decreases over time. A continuous surveillance of these genetic markers in the P. vivax population circulating in this region should be maintained to ensure public health.
Authors’ contributions
EF and EL carried out the molecular genetic studies; EF, EL and SB analysed the data; JC, EL, LM, SP and BV confirmed the diagnostic and updated the biobank collection; EL and LM supervised, carried out and coordinated field collections of patient isolates; LM, SB and EL conceived and coordinated the study; VC carried out the sequencing of PCR products; DM realized the sequencing and qPCR of sample collected between 2000 and 2003; EF, SB, SP, LM and EL drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors are grateful to Cayenne Hospital, Saint-Laurent du Maroni Hospital, the Health Centre coordination and all partners working in the diagnosis centres for their help collecting materials and data. The authors also thank the Institut Pasteur Clinical Research Department (PIRC) for the clinical research regulatory review.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data and material presented in this article are made available, unless otherwise stated.
Consent for publication
All authors approved the manuscript.
Ethics approval and consent to participate
All the samples analysed in the study came from blood collections required by standard medical care for any patient presenting fever on admission to hospital in French Guiana. According to French legislation (Article L.1211-2 and related, French Public Health Code), biobanking and the secondary use of remaining human clinical samples for scientific purposes are possible if the corresponding patient is informed and has not objected to such use. This requirement was fulfilled for the present study: each patient was informed via the hospital brochure entitled Information for Patients, and no immediate or delayed patient opposition was reported to the Malaria NRC by the clinicians. Moreover, in application of French legislation (Article L.1243-3 and related, French Public Health Code), samples received at the Malaria NRC had been registered for use in research in the NRC biobank, which was declared to and approved by the French Ministry for Research and a French Ethics Committee before its registration under declaration number DC-2010-1223; collection Nu2. French legislation does not require institutional review board approval.
Funding
This work benefited from funding from the Institut de Veille Sanitaire (French Ministry of Health) and the French Army (Grant LR607e). SP was financed by European grant managed by the European Commission (REGPOT-CT-2011-285837-STRONGER). EF was financed by the European Union (Lifelong Learning Programme).
Contributor Information
Emilie Faway, Email: emiliefaway@hotmail.com.
Lise Musset, Email: lmusset@pasteur-cayenne.fr.
Stéphane Pelleau, Email: spelleau@pasteur-cayenne.fr.
Béatrice Volney, Email: bvolney@pasteur-cayenne.fr.
Jessica Casteras, Email: casterasj@yahoo.fr.
Valérie Caro, Email: valerie.caro@pasteur.fr.
Didier Menard, Email: dmenard@pasteur-kh.org.
Sébastien Briolant, Email: sbriolant@wanadoo.fr.
Eric Legrand, Email: eric.legrand@pasteur.fr.
References
- 1.WHO . World Malaria Report 2015. Geneva: World Health Organization; 2016. pp. 1–255. [Google Scholar]
- 2.National Reference Centre for Malaria . National reference centre for malaria: 2014 annual activity report. Paris: National Reference Centre for Malaria; 2015. [Google Scholar]
- 3.Legrand E, Volney B, Meynard JB, Mercereau-Puijalon O, Esterre P. In vitro monitoring of Plasmodium falciparum drug resistance in French Guiana: a synopsis of continuous assessment from 1994 to 2005. Antimicrob Agents Chemother. 2008;52:288–298. doi: 10.1128/AAC.00263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musset L, Pelleau S, Girod R, Ardillon V, Carvalho L, Dusfour I, et al. Malaria on the Guiana Shield: a review of the situation in French Guiana. Mem Inst Oswaldo Cruz. 2014;109:525–533. doi: 10.1590/0074-0276140031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rieckmann KH, Davis DR, Hutton DC. Plasmodium vivax resistance to chloroquine? Lancet. 1989;2:1183–1184. doi: 10.1016/S0140-6736(89)91792-3. [DOI] [PubMed] [Google Scholar]
- 6.Arias AE, Corredor A. Low response of Colombian strains of Plasmodium vivax to classical antimalarial therapy. Trop Med Parasitol. 1989;40:21–23. [PubMed] [Google Scholar]
- 7.Garavelli PL, Corti E. Chloroquine resistance in Plasmodium vivax: the first case in Brazil. Trans R Soc Trop Med Hyg. 1992;86:128. doi: 10.1016/0035-9203(92)90535-K. [DOI] [PubMed] [Google Scholar]
- 8.Phillips EJ, Keystone JS, Kain KC. Failure of combined chloroquine and high-dose primaquine therapy for Plasmodium vivax malaria acquired in Guyana, South America. Clin Infect Dis. 1996;23:1171–1173. doi: 10.1093/clinids/23.5.1171. [DOI] [PubMed] [Google Scholar]
- 9.Baird JK. Resistance to therapies for infection by Plasmodium vivax. Clin Microbiol Rev. 2009;22:508–534. doi: 10.1128/CMR.00008-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dua VK, Kar PK, Sharma VP. Chloroquine resistant Plasmodium vivax malaria in India. Trop Med Int Health. 1996;1:816–819. doi: 10.1111/j.1365-3156.1996.tb00116.x. [DOI] [PubMed] [Google Scholar]
- 11.Guthmann JP, Pittet A, Lesage A, Imwong M, Lindegardh N, Min Lwin M, et al. Plasmodium vivax resistance to chloroquine in Dawei, southern Myanmar. Trop Med Int Health. 2008;13:91–98. doi: 10.1111/j.1365-3156.2007.01978.x. [DOI] [PubMed] [Google Scholar]
- 12.Phan GT, de Vries PJ, Tran BQ, Le HQ, Nguyen NV, Nguyen TV, et al. Artemisinin or chloroquine for blood stage Plasmodium vivax malaria in Vietnam. Trop Med Int Health. 2002;7:858–864. doi: 10.1046/j.1365-3156.2002.00948.x. [DOI] [PubMed] [Google Scholar]
- 13.Suwanarusk R, Chavchich M, Russell B, Jaidee A, Chalfein F, Barends M, et al. Amplification of pvmdr1 associated with multidrug-resistant Plasmodium vivax. J Infect Dis. 2008;198:1558–1564. doi: 10.1086/592451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suwanarusk R, Russell B, Chavchich M, Chalfein F, Kenangalem E, Kosaisavee V, et al. Chloroquine resistant Plasmodium vivax: in vitro characterisation and association with molecular polymorphisms. PLoS ONE. 2007;2:e1089. doi: 10.1371/journal.pone.0001089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barnadas C, Ratsimbasoa A, Tichit M, Bouchier C, Jahevitra M, Picot S, et al. Plasmodium vivax resistance to chloroquine in Madagascar: clinical efficacy and polymorphisms in pvmdr1 and pvcrt-o genes. Antimicrob Agents Chemother. 2008;52:4233–4240. doi: 10.1128/AAC.00578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ketema T, Bacha K, Birhanu T, Petros B. Chloroquine-resistant Plasmodium vivax malaria in Serbo town, Jimma zone, south-west Ethiopia. Malar J. 2009;8:177. doi: 10.1186/1475-2875-8-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chehuan YF, Costa MR, Costa JS, Alecrim MG, Nogueira F, Silveira H, et al. In vitro chloroquine resistance for Plasmodium vivax isolates from the Western Brazilian Amazon. Malar J. 2013;12:226. doi: 10.1186/1475-2875-12-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gama BE, Oliveira NK, Souza JM, Daniel-Ribeiro CT, Ferreira-da-Cruz Mde F. Characterisation of pvmdr1 and pvdhfr genes associated with chemoresistance in Brazilian Plasmodium vivax isolates. Mem Inst Oswaldo Cruz. 2009;104:1009–1011. doi: 10.1590/S0074-02762009000700012. [DOI] [PubMed] [Google Scholar]
- 19.Graf PC, Durand S, Alvarez Antonio C, Montalvan C, Galves Montoya M, Green MD, et al. Failure of supervised chloroquine and primaquine regimen for the treatment of Plasmodium vivax in the Peruvian Amazon. Malar Res Treat. 2012;2012:936067. doi: 10.1155/2012/936067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marques MM, Costa MR, Santana Filho FS, Vieira JL, Nascimento MT, Brasil LW, et al. Plasmodium vivax chloroquine resistance and anemia in the western Brazilian Amazon. Antimicrob Agents Chemother. 2014;58:342–347. doi: 10.1128/AAC.02279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orjuela-Sanchez P, de Santana Filho FS, Machado-Lima A, Chehuan YF, Costa MR, Alecrim M, et al. Analysis of single-nucleotide polymorphisms in the crt-o and mdr1 genes of Plasmodium vivax among chloroquine-resistant isolates from the Brazilian Amazon region. Antimicrob Agents Chemother. 2009;53:3561–3564. doi: 10.1128/AAC.00004-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soto J, Toledo J, Gutierrez P, Luzz M, Llinas N, Cedeno N, et al. Plasmodium vivax clinically resistant to chloroquine in Colombia. Am J Trop Med Hyg. 2001;65:90–93. doi: 10.4269/ajtmh.2001.65.90. [DOI] [PubMed] [Google Scholar]
- 23.Khatoon L, Baliraine FN, Bonizzoni M, Malik SA, Yan G. Prevalence of antimalarial drug resistance mutations in Plasmodium vivax and P. falciparum from a malaria-endemic area of Pakistan. Am J Trop Med Hyg. 2009;81:525–528. [PMC free article] [PubMed] [Google Scholar]
- 24.Kurcer MA, Simsek Z, Kurcer Z. The decreasing efficacy of chloroquine in the treatment of Plasmodium vivax malaria, in Sanliurfa, south-eastern Turkey. Ann Trop Med Parasitol. 2006;100:109–113. doi: 10.1179/136485906X86284. [DOI] [PubMed] [Google Scholar]
- 25.Foote SJ, Thompson JK, Cowman AF, Kemp DJ. Amplification of the multidrug resistance gene in some chloroquine-resistant isolates of P. falciparum. Cell. 1989;57:921–930. doi: 10.1016/0092-8674(89)90330-9. [DOI] [PubMed] [Google Scholar]
- 26.Lu F, Lim CS, Nam DH, Kim K, Lin K, Kim TS, et al. Genetic polymorphism in pvmdr1 and pvcrt-o genes in relation to in vitro drug susceptibility of Plasmodium vivax isolates from malaria-endemic countries. Acta Trop. 2011;117:69–75. doi: 10.1016/j.actatropica.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 27.Rungsihirunrat K, Muhamad P, Chaijaroenkul W, Kuesap J, Na-Bangchang K. Plasmodium vivax drug resistance genes; Pvmdr1 and Pvcrt-o polymorphisms in relation to chloroquine sensitivity from a malaria endemic area of Thailand. Korean J Parasitol. 2015;53:43–49. doi: 10.3347/kjp.2015.53.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim YK, Kim C, Park I, Kim HY, Choi JY, Kim JM. Therapeutic efficacy of chloroquine in Plasmodium vivax and the pvmdr1 polymorphisms in the Republic of Korea under mass chemoprophylaxis. Am J Trop Med Hyg. 2011;84:532–534. doi: 10.4269/ajtmh.2011.10-0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orjuela-Sanchez P, Karunaweera ND, da Silva-Nunes M, da Silva NS, Scopel KK, Goncalves RM, et al. Single-nucleotide polymorphism, linkage disequilibrium and geographic structure in the malaria parasite Plasmodium vivax: prospects for genome-wide association studies. BMC Genet. 2010;11:65. doi: 10.1186/1471-2156-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez-Becerra C, Pinazo MJ, Gonzalez A, Alonso PL, del Portillo HA, Gascon J. Increased expression levels of the pvcrt-o and pvmdr1 genes in a patient with severe Plasmodium vivax malaria. Malar J. 2009;8:55. doi: 10.1186/1475-2875-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imwong M, Pukrittayakamee S, Pongtavornpinyo W, Nakeesathit S, Nair S, Newton P, et al. Gene amplification of the multidrug resistance 1 gene of Plasmodium vivax isolates from Thailand, Laos, and Myanmar. Antimicrob Agents Chemother. 2008;52:2657–2659. doi: 10.1128/AAC.01459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melo GC, Monteiro WM, Siqueira AM, Silva SR, Magalhaes BM, Alencar AC, et al. Expression levels of pvcrt-o and pvmdr-1 are associated with chloroquine resistance and severe Plasmodium vivax malaria in patients of the Brazilian Amazon. PLoS ONE. 2014;9:e105922. doi: 10.1371/journal.pone.0105922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lekweiry KM, Boukhary AOMS, Gaillard T, Wurtz N, Bogreau H, Hafid JE, et al. Molecular surveillance of drug-resistant Plasmodium vivax using pvdhfr, pvdhps and pvmdr1 markers in Nouakchott, Mauritania. J Antimicrob Chemother. 2012;67:367–374. doi: 10.1093/jac/dkr464. [DOI] [PubMed] [Google Scholar]
- 34.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clopper C, Pearson S. The use of confidence or fiducial limits illustrated in the case of binomial. Biometrika. 1934;26:404–413. doi: 10.1093/biomet/26.4.404. [DOI] [Google Scholar]
- 36.de Santana Filho FS, Arcanjo AR, Chehuan YM, Costa MR, Martinez-Espinosa FE, Vieira JL, et al. Chloroquine-resistant Plasmodium vivax, Brazilian Amazon. Emerg Infect Dis. 2007;13:1125–1126. doi: 10.3201/eid1307.061386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gama BE, Lacerda MV, Daniel-Ribeiro CT, Ferreira-da-Cruz Mde F. Chemoresistance of Plasmodium falciparum and Plasmodium vivax parasites in Brazil: consequences on disease morbidity and control. Mem Inst Oswaldo Cruz. 2011;106(Suppl 1):159–166. doi: 10.1590/S0074-02762011000900020. [DOI] [PubMed] [Google Scholar]
- 38.Pommier de Santi V, Dia A, Adde A, Hyvert G, Galant J, Mazevet M, et al. Malaria in French Guiana linked to illegal gold mining. Emerg Infect Dis. 2016;22:344–346. doi: 10.3201/eid2202.151292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pommier de Santi V, Djossou F, Barthes N, Bogreau H, Hyvert G, Nguyen C, et al. Malaria hyperendemicity and risk for artemisinin resistance among illegal gold miners, French Guiana. Emerg Infect Dis. 2016;22:903–906. doi: 10.3201/eid2205.151957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jovel IT, Mejia RE, Banegas E, Piedade R, Alger J, Fontecha G, et al. Drug resistance associated genetic polymorphisms in Plasmodium falciparum and Plasmodium vivax collected in Honduras, Central America. Malar J. 2011;10:376. doi: 10.1186/1475-2875-10-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vargas-Rodriguez Rdel C, da Silva Bastos M, Menezes MJ, Orjuela-Sanchez P, Ferreira MU. Single-nucleotide polymorphism and copy number variation of the multidrug resistance-1 locus of Plasmodium vivax: local and global patterns. Am J Trop Med Hyg. 2012;87:813–821. doi: 10.4269/ajtmh.2012.12-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomes LR, Almeida-de-Oliveira NK, de Lavigne AR, de Lima SR, de Pina-Costa A, Brasil P, et al. Plasmodium vivax mdr1 genotypes in isolates from successfully cured patients living in endemic and non-endemic Brazilian areas. Malar J. 2016;15:96. doi: 10.1186/s12936-016-1141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khim N, Andrianaranjaka V, Popovici J, Kim S, Ratsimbasoa A, Benedet C, et al. Effects of mefloquine use on Plasmodium vivax multidrug resistance. Emerg Infect Dis. 2014;20:1637–1644. doi: 10.3201/eid2010.140411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Legrand E, Yrinesi J, Ekala MT, Peneau J, Volney B, Berger F, et al. Discordant temporal evolution of Pfcrt and Pfmdr1 genotypes and Plasmodium falciparum in vitro drug susceptibility to 4-aminoquinolines after drug policy change in French Guiana. Antimicrob Agents Chemother. 2012;56:1382–1389. doi: 10.1128/AAC.05280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hupalo DN, Luo Z, Melnikov A, Sutton PL, Rogov P, Escalante A, et al. Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax. Nat Genet. 2016;48:953–958. doi: 10.1038/ng.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pearson RD, Amato R, Auburn S, Miotto O, Almagro-Garcia J, Amaratunga C, et al. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat Genet. 2016;48:959–964. doi: 10.1038/ng.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schousboe ML, Ranjitkar S, Rajakaruna RS, Amerasinghe PH, Morales F, Pearce R, et al. Multiple origins of mutations in the mdr1 gene—a putative marker of chloroquine resistance in P. vivax. PLoS Negl Trop Dis. 2015;9:e0004196. doi: 10.1371/journal.pntd.0004196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and material presented in this article are made available, unless otherwise stated.


