Abstract
Background
The outermost layer of mycobacterial cell wall is rich in lipids and glycolipids, surface molecules which differ among species. Mycobacterium smegmatis, an attractive model for the study of both pathogenic and non-pathogenic mycobacteria, presents glycopeptidolipids (GPLs). All the genes necessary for the biosynthesis of such molecules are clustered in a single region of 65 kb and among them, the msmeg_0412 gene has not been characterized yet. Here we report the isolation and subsequent analysis of a MSMEG_0412 null mutant strain.
Results
The inactivation of the msmeg_0412 gene had a drastic impact on bacterial surface properties which resulted in the lack of sliding motility, altered biofilm formation and enhanced drug susceptibility. The GPLs analysis showed that the observed mutant phenotype was due to GPLs deficiencies on the mycobacterial cell wall. In addition, we report that the expression of the gene is enhanced in the presence of lipidic substrates and that the encoded protein has a membrane localization.
Conclusion
msmeg_0412 plays a crucial role for GPLs production and translocation on M. smegmatis surface. Its deletion alters the surface properties and the antibiotic permeability of the mycobacterial cell barrier.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-016-0888-z) contains supplementary material, which is available to authorized users.
Keywords: Mycobacterium smegmatis, msmeg_0412, GPLs, Lipolytic, Cell wall, Biofilm
Background
Mycobacterial cell wall is a unique well organized barrier which confers low permeability to different chemical agents and common antibiotics [1]. The envelope is waxy, hydrophobic, and thicker compared to that of other bacteria and is organized in two compartments, the inner layer, covalently linked to the plasma membrane and formed by peptidoglycan, arabinogalactan and mycolic acid, and, the outer layer, which is essentially formed by glycolipids and proteins [2, 3]. Glycopeptidolipids (GPLs) are the major surface exposed molecules found in various mycobacterial species including non-pathogenic (e.g M. smegmatis) and non–tuberculous pathogenic mycobacteria (e.g. M. avium, M. intracellulare, M. abscessus) [4, 5]. GPLs have a common fatty acyl-tetrapeptide core which is glycosylated with 6-deoxy-l-talose and variable O-methyl-l-rhamnose residues [6–8]. This is known as non-serovar-specific GPL (nsGPLs) and represents the main product of M. smegmatis GPLs. In M. avium GPLs present a more complicated structure in which an additional rhamnose and oligosaccharide residues are added to 6-deoxy-l-talose, resulting in serovar-specific GPLs [7, 9, 10]. These molecules, which may represent more than 70 % of total surface lipids [11], are required for cell aggregation, sliding motility and biofilm formation [12–15]. Moreover several studies reported that slight modifications of the GPLs structure are able to affect the mycobacterial ability to stimulate the host immune response, suggesting a role as antigenic molecules [16–19].
The fast growing M. smegmatis is frequently used as a model organism to study the molecular and physiological mechanisms used by slow-growing pathogenic mycobacteria. The M. smegmatis GPLs biosynthetic locus has been previously characterized either experimentally or by in silico prediction [20–23]. Most genes of the M. smegmatis GPL locus are conserved in two clinically significant mycobacterial species such as M. abscessus and M. avium subsp. paratuberculosis [5, 24].
Among them, the gene msmeg_0412 is still uncharacterized and its product shares 66 % and 67 % identity with the predicted protein MAB_0402, and MAP2244 from M. abscessus and M. avium subsp. paratuberculosis, respectively. Interestingly, M. tuberculosis, a mycobacterium which does not produce GPLs on the cell wall, presents Rv1184c which shares 45 % identity with MSMEG_0412. Rv1184c has been recently characterized as the Acyltransferase Chp2, essential for the final biosynthetic steps of polyacyltrehalose (PAT), a pentaacylated, trehalose-based glycolipids only found on the surface of pathogenic mycobacteria [25, 26].
In this study we show that the inactivation of msmeg_0412 gene alters the lipid profile surface of M. smegmatis and enhances the bacterial susceptibility to antibiotics. In addition, we report that msmeg_0412 gene expression is enhanced in the presence of lipidic substrates and that the encoded protein localizes on the cell wall. Taken together, our results indicate that MSMEG_0412 is an important player in the complex GPLs biosynthetic process in M. smegmatis and allow speculating that MSMEG_0412 is translocated to the cell wall where it contributes to GPLs synthesis.
Results
Deletion of the msmeg_0412 gene alters M. smegmatis surface properties, enhances suscepitibility to antibiotics and affects GPLs synthesis
The msmeg_0412 gene (Fig. 1a) belongs to a gene cluster devoted to the biosynthesis and transport of GPLs to mycobacterial cell wall surface. Most of the genes inside the cluster have been previously characterized [5, 15]. In particular, mps1 and mps2 genes encode the mycobacterial peptide synthetase required for the formation of tripeptide-amino-alcohol moiety which is then linked to the 3-hydroxy/methoxy C26-C34 acyl chain, the lipid moiety of GPLs. The latter is synthesized by the combined action of Pks, Fad23 and PapA3 gene products. Finally, the action of various glycosyltransferases (Gtf), methyltransferases (Mtf) and acetyltransferases (Atf) leads to the building of the glycosyl side of GPLs. Once synthesized, GPLs are transferred to GPL addressing protein (Gap), a small integral membrane protein, required for the translocation of GPLs on the cell surface. As shown in Fig. 1a, the msmeg_0412 gene is close to the genes involved in the synthesis of lipid portion of GPLs. In order to understand the role of the msmeg_0412 gene product, we constructed a null mutant strain by using the pGOAL/pNIL procedure as reported in methods. A PCR confirmation of gene deletion in GM1 strain is shown in Fig. 1b. The deletion mutant, GM1, was analyzed for some phenotypes . Compared to its isogenic wild type strain, GM1 displayed altered biofilm formation (Fig. 2a), lack of sliding motility (Fig. 2b), and rough colony phenotype (Fig. 2c).
Fig. 1.
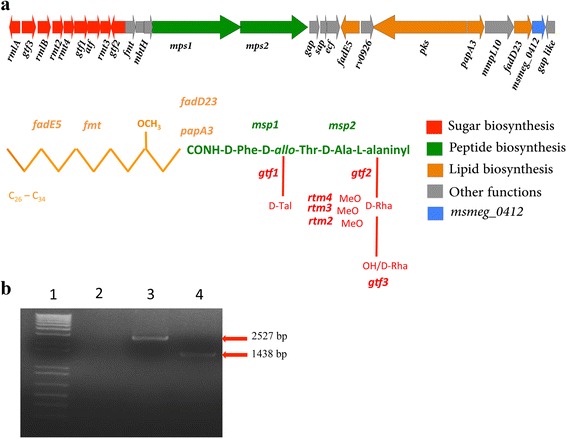
a) Schematic representation of the GPLs structure from M. smegmatis and its biosynthetic locus on the chromosome. The genes involved in the synthesis of lipid moiety (orange), peptide (green) and glycosylation (red) of GPLs are indicated. b) Agarose gel electrophoresis showing PCR-based validation of the msmeg_0412 gene deletion in the GM1 strain. Lanes: 1) DNA Ladder marker (1Kb); 2) PCR negative control; 3) wt Chromosomal DNA used as template (2527 bp-amplicon expected using 0412-F1 and 0412-R2 as primers); 4) GM1 Chromosomal DNA used as template (1438 bp-amplicon expected using 0412-F1 and 0412-R2 as primers)
Fig. 2.
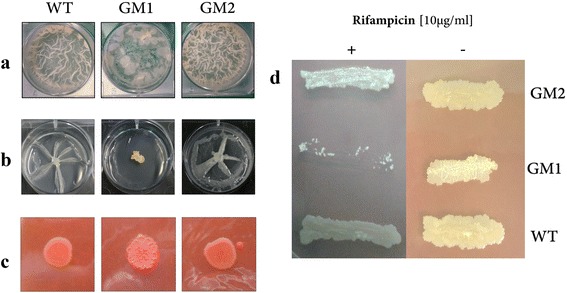
Phenotypic analysis of M. smegmatis strains. Wild-type strain was compared to msmeg_0412 null mutant (GM1) and complemented (GM2) strains for the following phenotypes: Biofilm production (a), Sliding motility (b) Colony morphology (c). The same strains were streaked on LB agar plates supplemented with or without Rifampicin (10 μg/ml) to evaluate the Antimicrobial susceptibility towards this antibiotic (d)
To confirm that these phenotypes were due to msmeg_0412 gene inactivation, we performed a complementation analysis. A wild type allele of the msmeg_0412 gene was PCR-amplified, cloned into pMV10-25 vector [27] without the gfp gene (methods), and transformed into strain GM1, yielding strain GM2. As shown in Fig. 2 all phenotypes of strain GM1 were rescued in strain GM2, that appeared identical to the wild type strain.
The drastic impact of msmeg_0412 deletion on M. smegmatis surface properties was also evidenced by a slower growth rate in liquid media with a generation time of the mutant of 4.5 h compared to 3 h of the wt strain (see Additional file 1: Figure S1) and by the enhanced susceptibility to antimicrobials. As shown in Fig. 2d, when streaked on LB agar medium containing Rifampicin at a sub-MIC concentration (10 μg/ml) [28], the GM1 strain was unable to grow compared to the wild type and complemented strains. The same result was obtained using Erythromycin as antibiotic (13 μg/ml) [29]. Since both antibiotics have cytosolic targets, it is possible to hypothesize that the mutation induces changes in the cell wall permeability that lead to an increase in antibiotics penetration.
To better characterize the effects of msmeg_0412 deletion we extracted and analyzed the GPLs content from the wild type, mutant and complemented strains. TLC analysis of the three organic extracts (Fig. 3a) indicated that GPLs were absent only in GM1 while they were equally abundant in the remaining strains. Conversely, the occurrence of the lipid fraction was detected in both wt, GM1 and GM2 strains (see Additional file 2: Figure S2). An even more accurate picture was provided by ESI-MS analysis (Fig. 3b). In this case, wt and GM2 strains provided identical MS profiles, which could be referred to the occurrence of 6-deoxy-l-talose- and O-methyl-l-rhamnose-containing GPLs and their methylated congeners [23]. Conversely, no traces of any sugar-containing compound at the same m/z range was found in GM1. The lack of GPLs in the msmeg_0412 deletion mutant apparently suggests that msmeg_0412 gene product is involved in the synthesis and assembly of GPLs on the M. smegmatis cell wall.
Fig. 3.
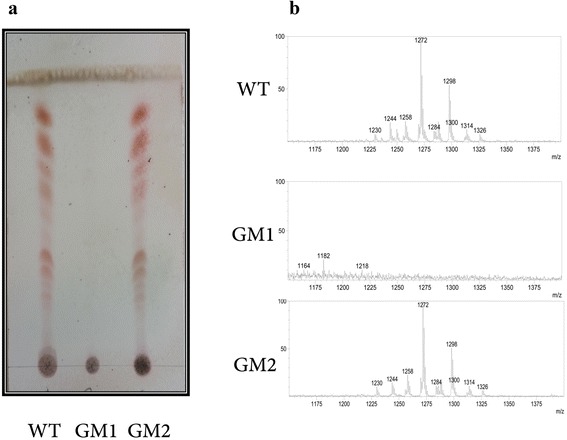
Chemical analysis of GPLs. GPLs content from the wild-type, GM1 and GM2 strains as indicated by TLC analysis (a) and ESI-MS profile of the organic extracts (b)
The msmeg_0412 gene expression is induced by lipid substrates
In order to evaluate the role played by MSMEG_0412 protein during GPLs production a bioinformatics approach was initially followed. MSMEG_0412 has the canonical penta-peptide sequence motif GXSXG/S and the conserved Ser, Asp and His catalytic triad (indicated in bold in Fig. 4) typical of proteins with lipase/esterase activity and identified as hydrolases involved in lipid metabolism [30, 31].
Fig. 4.

Amino acid sequence of MSMEG_0412. In bold is reported the conserved pentapeptide sequence motif GXSXG/S. Aminoacid residues potentially involved in the catalytic triad are indicated by an asterisk (*). Underlined is the putative N-terminal signal peptide
To experimentally validate the bioinformatic prediction we monitored the effects of two widely used lipid substrates, tributyrin and tween 80, on the transcriptional profile of the msmeg_0412 gene. To this purpose M. smegmatis wild type strain was grown in minimal medium containing glucose, tributyrin or tween 80 as the only carbon source and total RNA was extracted and reverse transcribed as reported in methods. As shown in Fig. 5, the transcriptional level of the gene was induced approximately 50-fold in the presence of tween 80 and 2-fold in the presence of tributyrin.
Fig. 5.
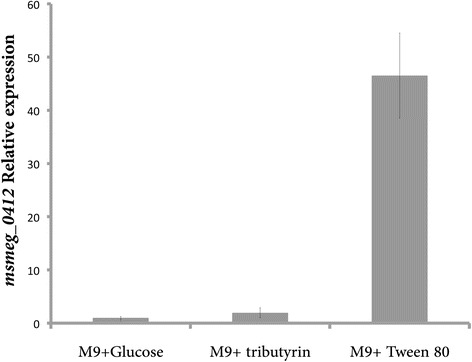
Real-Time PCR analysis of msmeg_0412 gene under different growth conditions. Cells of M. smegmatis mc2155 wild type strain were grown in aerated conditions in minimal medium containing glucose, tributyrin or tween 80 to early exponential growth phase. The ∆∆Ct method was used to calculate the relative amount of specific RNA present in each sample, and the transcriptional induction determined by comparison to values from cells growing in minimal medium containing glucose
Based on this result we decided to evaluate the effect of tween 80 on the growth of the mutant strain. To this purpose wt, GM1 and GM2 strains were grown in minimal medium containing 0.2 % (w/v) glucose or 1%t ween 80 as the only carbon source. The result (see Additional file 3: Figure S3) clearly indicated that the mutant was unable to grow in presence of tween 80. Although this result, together with the bioinformatic analysis, allows speculating a lipolytic function for MSMEG_0412, a detailed biochemical characterization is needed to decipher its function.
The MSMEG_0412 protein is localized on the cell wall
In silico analysis suggested that MSMEG_0412 contains a N-terminal signal sequence (underlined in Fig. 4) with a putative cleavage site between residues A29 and D30, most likely involved in targeting the MSMEG_0412 to the cell membrane. To validate this prediction we constructed two gene fusions between gfp, the gene encoding for the Green Fluorescent Protein, and the msmeg_0412 gene with and without the sequence coding for the putative signal peptide (Methods). The gene fusions were independently used to transform M. smegmatis wild type cells, yielding strains GM7 (carrying the entire msmeg_0412 gene) and GM8 (carrying the msmeg_0412 gene without the signal peptide). A fluorescence microscopy analysis showed that the fluorescence signal was localized around the cells of strain GM7 and inside the cells of strain GM8 (Fig. 6). This result indicates that MSMEG_0412 is a membrane protein and that the removal of the signal peptide induces protein aggregation and its retention within the cytosol.
Fig. 6.
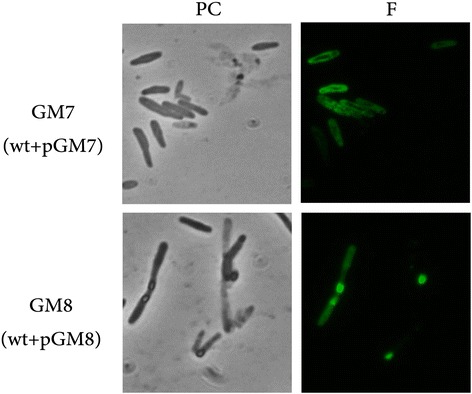
Fluorescence analysis. M. smegmatis strains carrying gene fusions between gfp and msmeg_0412 gene with (GM7) and without (GM8) the region coding for the putative signal peptide were analysed by phase contrast (PC) and Fluorescent (F) microscopy. Exposure time was 200 ms in both cases
Discussion
Mycobacteria comprise bacilli with a specialized cell wall divided in two compartments: the inner- and the outer- layer. The latter is rich in lipids and glycolipids which differ among species. M. smegmatis shares a richness in glycopeptidolipids (GPLs) with pathogenic non tubercle mycobacteria, such as M. abscessus and M. avium.
Msmeg_0412, a gene located in the GPLs biosynthetic locus, is highly conserved among mycobacteria, and its product shares 45 % identity with Rv1184c from the human pathogen M. tuberculosis. The gene encodes a protein which primary structure has features of proteins annotated as putative lipases or esterases.
Lipase/esterase-like enzymes belong to a family of proteins involved in lipid metabolism, which play a crucial role during mycobacterial infection and persistence inside the host [30, 32]. In fact, it has been reported that such enzymes contribute to the destruction of the host tissues thereby supplying nutrients to the pathogens [33, 34]. Therefore, a better understanding of these proteins will surely help in developing new strategies against mycobacterial infections.
Earlier reports have clearly indicated that the loss of GPLs on M. smegmatis surface induce an increased cell wall hydrophobicity [14, 15, 23] which results in the alteration of surface properties such as a change of colony morphotype (from smooth to rough), an altered biofilm formation at liquid/air interface, and the inability to slide on soft agar plates. Here we report the construction and analysis of a M. smegmatis strain carrying a null mutation in msmeg_0412 gene.
Our results show that MSMEG_0412 is required for the correct processing of the GPLs on mycobacterial surface and that its deletion induced a surface perturbation. Furthermore, in line with the bioinformatic predictions, the msmeg_0412 gene expression is enhanced in the presence of lipid substrates, and the protein encoded localizes on the cell wall. Taken together our results allow to speculate that once synthesized, MSMEG_0412 is translocated to the cell wall, where plays a crucial role for the correct GPLs processing and translocation on M. smegmatis surface. A further biochemical approach is needed to better clarify other aspects of the function and the localization of this important protein.
Conclusions
The analysis of msmeg_0412 mutant highlighted that this gene is important for Mycobacterium smegmatis surface organization and resulted in different important phenotypes, as lack of sliding motility, altered biofilm formation and enhanced drug susceptibility. The analysis of Glycopeptidolipids (GPLs) component of bacterial surface showed that the msmeg_0412 mutant was strongly impaired for GPLs presence in the mycobacterial cell wall. In addition, based on in-silico predictions, we report that the expression of the gene is enhanced by the presence of lipidic substrates and that the encoded protein has a membrane localization.
Finally, generation time of the mutant was shorter than the one of the wild type strain.
Mutations affecting the organization of the cell wall have often pleiotropic effects. This is the first experimental evidence for an involvement of msmeg_0412 gene in the synthesis of the GPLs.
Taken together, the results presented in this manuscript offer the possibility to consider MSMEG_0412 as a new target to impair the mycobacterial growth and to increase drug susceptibility.
Methods
Bacterial strains, media and growth conditions
M. smegmatis mc2155 [35] was the parental strain of all the recombinant strains described below. DH5α (supE44 ΔlacU169 [ϕ80ΔlacZM15] hsdR17 recA1) [36] was used for all cloning experiments.
The recombinant strain GM1, carrying a msmeg_0412 gene deletion was engineered using the p2NIL/pGOAL19-based flexible cassette method as previously reported [37].
M. smegmatis mc2155 and derivatives were grown in 7H9 medium (Difco) containing 2 % glycerol and 0.05 % tween 80 (Applichem) or in M9 containing 1 mM Mg2SO4 and 0.2 % glucose. Where indicated, glucose was replaced by 1 % glycerol-tributyrate (tributyrin) or 1 % tween 80.
E. coli strains were grown in LB medium. When required, antibiotics were added to the medium at the following final concentrations: ampicillin 100 μg/ml, hygromycin at 200 μg/ml for E. coli and 50 μg/ml for M. smegmatis mc2155, respectively.
For the antimicrobial susceptibility test, Erythromycin and Rifampicin were added to LB agar medium at the following final concentrations: 13 μg/ml and 10 μg/ml respectively.
For the Microscopy analysis, samples were observed using an Olympus BX51 fluorescence microscope using a FITC filter. The Images were captured using an Olympus DP70 digital camera and processed. Standard acquisition times were 200 ms.
Bioinformatic approach
A multiple sequence alignment was carried out using PRALINE (http://www.ibi.vu.nl/programs/pralinewww) as program. MSMEG_0412 aminoacid sequence was used as query against the earlier identified M. tuberculosis proteins [31] showing the conserved pentapetide sequence motif (GXGXG) and the conserved Ser, Asp and His catalytic triad characteristic of lipase/esterase activities.
The signal peptide server, (http://www.cbs.dtu.dk/services/SignalP), was used to predict the presence and location of the signal peptide cleavage site inside MSMEG_0412.
Sliding motility assay
5 μl of a liquid culture in stationary phase was dispensed on plate containing 7H9 medium plus 0.3 % agar with no added carbon source. The plate was then incubated at 37 °C for 1 week [15, 23].
Congo red assays
5 μl of a liquid culture in stationary phase was dispensed on plate containing LB medium supplemented with 2 % agar and 100 μg/ml Congo Red (Sigma) and incubated at 37 °C for 3 days [15, 23].
Biofilm assay
5 μl of a liquid culture in stationary phase was inoculated into PVC well plates containing 5 ml Sauton liquid medium and incubated in standing condition at 37 °C for 1 week [15, 23].
DNA manipulation
Plasmid and chromosomal DNA preparation, restriction digestion, ligation, bacterial transformation and agarose gel electrophoresis were performed as described [36].
Plasmid pGM5 carries a gene deletion cassette (Δ0412 cassette) containing 689-bp segment upstream of msmeg_0412 gene + first 4 codon (5’ fragment) followed by last 15 codons + 685-bp segment downstream of the gene (3’ fragment). The 5’ fragment was PCR amplified using the primer pairs 0412-1 F and 0412-1R listed in Table 1 and the genomic DNA of wild type mc2155 strain as template and the obtained PCR product was cloned in pGEMT-easy vector, giving the plasmid pGM2. The 3’ fragment was PCR amplified using the primer pairs 0412-2 F and 0412-2R listed in Table 1 and the genomic DNA of wild type mc2155 strain as template and the obtained PCR product was digested with KpnI and cloned in pGM2 vector at the unique KpnI restriction site, giving plasmid pGM3. The deletion cassette was then excised from pGM3 vector using the restriction enzymes BamHI and SalI and transferred into the p2NIL plasmid previously digested by the same restriction enzymes. The resulting plasmid, named pGM4 and the plasmid pGOAL19 were digested with PacI, and the PacI selectable marker cassette of pGOAL19, carrying the sacB, LacZ and Hyg genes, was ligated to the linearized pGM4 to create the suicide vector pGM5.
Table 1.
Synthetic oligonucleotides
| Name | Sequence (5′ - 3′)a | Position of Annealingb |
|---|---|---|
| 0412-S | CATGATCCCCGAGGAGCAC | +857/+875 |
| 0412-A | TTGCCGTTCAAGTACCTCGG | +889/+90 |
| 0412-Fw | AAAGCTAGCATGAGGAGACGACTTCTCGCGTTC | +1/+24 |
| 0412-spFw | AAAGCTAGCGATGACACCAAACCGGTCG | +85/+103 |
| 0412-Rv | AAAGCTAGCTCAGGGCTTGGGCGCCGGTTG | +1132/+1152 |
| 0412-gfpRv | AAAGCTAGCGGGCTTGGGCGCCGGTTG | +1132/+1149 |
| 0412-1F | CTAGAAGCTTGCACCGCACTGGTGAGATA | -690/-672 |
| 0412-1R | CTAGGGTACCCGCTGTAGTCCTCTGCT | -5/+12 |
| 0412-2F | CTAGGGTACCGGACTGCAGTACGTCGTGAACA | +1108/+1129 |
| 0412-2R | CTAGGGTACCACCATGTGGATCCCTCTCCT | +1818/+1837 |
aUnderlined is an unpaired tail carrying a restriction site
bPosition of annealing refers to msmeg_0412 gene sequence, with the first base of the translational initiation codon as +1
Plasmid pGM6, used for complementation analysis, was constructed as follow: the wild type msmeg_0412 gene from M. smegmatis wild type strain was PCR amplified with Pfu Turbo high Fidelity DNA polymerase (Stratagene) using chromosomal DNA as template and the oligo 0412-Fw and 0412-Rv (Table 1) as primers. The last ones contained the engineered NheI and BamHI restriction sites respectively.
The PCR product of expected size was purified with a PCR Purification Kit (Qiagen), digested with NheI and BamHI restriction enzymes and cloned under the hsp60 promoter into the shuttle vector pMV10-25 [27], previously digested with the same enzyme pairs which allowed the excision of the gfp encoding gene. The resulting plasmid was controlled by sequencing and then used to transform GM1 cells, giving the strain GM2.
Plasmid pGM7 and pGM8 containing the entire version or the truncated version of the msmeg_0412 gene fused to the gfp reporter gene, respectively, were constructed as follow: the msmeg_0412 gene except the stop codon was PCR amplified with Pfu Turbo high Fidelity DNA polymerase (Stratagene) using chromosomal DNA as template and priming the reaction with two different couples of primers: 0412-Fw and 0412-gfpRv or 0412-spFw and 0412-gfpRv (Table 1). The first couple of primers has allowed to amplify the entire copy of the gene, the other combination of primers has allowed to amplify the gene that misses of 84 bp, encoding a putative N-terminal signal peptide. All primers contained a 5’ engineered NheI restriction site. The PCR products of expected sizes were purified with the PCR purification Kit (Qiagen) digested with the NheI restriction enzyme and cloned in the plasmid pMV10-25 [27], under the control of hsp60 promoter and in frame with GFP coding gene using the NheI restriction site located at 5’ end of gfp.
The resulting plasmids were controlled by sequencing and then used to transform M. smegmatis wild type cell, giving the strains GM7 and GM8 respectively.
Isolation of Δmsmeg_0412 mutant strain
The msmeg_0412 null mutant strain, named GM1, was obtained by using the allelic replacement system reported previously [37].
The pGM5 suicide vector was used to transform cells of M. smegmatis mc2155 strain and transformants were selected on LB agar plates supplemented with hygromicin (200 mg/ml), kanamycin (25 mg/ml) and X-Gal (50 mg/ml). Blue colonies were restreaked onto LB agar plates; a loopful of cells was then resuspended in LB broth, serially diluted and plated on LB agar containing 2 % sucrose (Suc) and X-gal. White KanS, HygS and SucR colonies were isolated and identified by PCR analysis. One KanS, HygS and SucR colony with a deleted msmeg_0412 gene was identified as GM1 strain.
RNA extraction
Total RNA was extracted by using the RNeasy Mini Kit (QIAGEN) according to manufacturer’s protocol. RNAs were resuspended in 100 ml of RNAse-free water and treated with DNAse I (Fermentas, 1U/ml) for 30 minutes at 37 °C. The quality of RNA samples were estimated using the RNA nanochip on the Thermoscientific Nanodrop 1000. The concentration of RNA was determined by measuring the absorbance at 260 nm.
Real Time PCR analysis
cDNA was synthesized by using PrimeScript Enzyme mix (Takara) and random hexamers as primers. Primers 0412-S and 0412-A (Table 1), and those for the normalizing gene 16S, 16S-fw (5′-GGCGAACGGGTGAGTAACA-3′) and 16S-Rv (5′-GCCCTGCACTTTGGGATAAG-3′), were designed with ABI PRISM Primer Express software (PE Applied Biosystems). Real-Time PCR was performed by using SYBR green RT-PCR (Takara). Reactions were performed as previously described [38]. Values reported here were the average of at least three independent experiments and were expressed as the mean ± SEM (Standard Error of the Mean). The fluorescence signal due to SYBR Green intercalation was monitored to quantify the double-stranded DNA product formed in each PCR cycle. Statistical significance was determined by Student’s unpaired t-test and the significance levels were reported in the text.
Extraction and analysis of mycobacterial lipids
Total lipids were extracted from whole cells as previously reported [16]. In brief, cells from wt, mutant and complemented strains were grown in LB medium until stationary phase (OD600: 2.5), harvested 10 minutes at 7000 rpm and resuspended in PBS, pH 7.4. Lipids were extracted with CHCl3:CH3OH (1:2 v/v) for 24 hours, separately dried under vacuum and partitioned between water and chloroform (1:1 v/v).
The organic phases were further washed with distilled water and evaporated to dryness. For analytical thin-layer chromatography (TLC), the crude residues (1.0 mg) were dissolved in CHCl3:CH3OH (0.75 mL, 9:1 v/v), spotted on silica gel 60-precoated plates (0.25-mm thickness, Merck) and eluted with CHCl3:CH3OH (9:1 v/v). Sugar-containing compounds were visualized by spraying TLC plates with a chromosulfuric acid staining solution and charring them at 200 °C for 2 min. The GPLs were detected as brownish-red spots. Conversely, the lipid content in the three strains was detected by extracting the crudes with hexane (0.3 mL). The resulting solutions were spotted on TLC plates which were eluted in toluene:acetone (99:1 v/v). In this case, lipids were visualized by spraying the plates with a cerium molybdate staining solution and charring them at 200 °C for 2 min. The lipids were detected as dark blue spots.
ESI-MS analysis of the organic extracts was performed using a Shimadzu LC-MS-2010EV system with ESI interface, Q-array-octapole-quadrupole mass analyser and a Shimadzu LC-MS solution Workstation software for data processing. The optimized MS parameters were selected as follows: CDL (curved desolvation line) temperature 250 °C; heat block temperature 250 °C; probe temperature 250 °C; detector gain 1.6 kV; probe voltage +4.5 kV; CDL voltage -15 V. Nitrogen served as nebulizing (flow rate: 1.5 L/min) and drying gas (flow rate: 15 L/min). Each sample (0.5 mg) was dissolved in 1 mL of acetonitrile (ACN)/0.1 % trifluoroacetic acid (TFA); 2 μL aliquots of the resulting solutions were injected using ACN/0.1 % TFA as solvent (flow rate: 0.2 mL/min). MS spectra were acquired in positive ion mode.
Acknowledgements
We would like to acknowledge Prof. Ezio Ricca for critical reading of the manuscript.
Funding
This work was financially supported by the Department of Biology of the University of Naples “Federico II”.
Availability of data and material
The data supporting the conclusion of this work are included within the article.
Authors’ contributions
AC conceived the study; AZ, AM, and AC performed mutant isolation, strains construction and phenotypic analysis, Real-Time PCR experiments and Fluorescence microscopy analysis; DD and AL carried out lipid extraction and analysis; AZ, MV and AC analyzed data and wrote the manuscript. All authors read and approved the final manuscript.
Competing interest
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This work did not involve human subjects, human material, or human data and therefore does not require ethical approval.
Abbreviations
- ESI-MS
Electrospray Ionization Mass Spectometer
- GPLs
Glycopeptidolipids
- TLC
Thin Layer chromatography
Additional files
Growth curve of M. smegmatis strains in LB medium. M. smegmatis wild type and GM1 strains were grown in LB medium containg 0,05 % tween 80 and OD600nm determined every 3 hours. For each strains the data reported in graph are the mean of three independent experiments. (JPG 601 kb)
Chemical analysis of total lipids. Total lipids were extracted from whole wt, GM1 and GM2 cells and analyzed by TLC. (JPG 483 kb)
Growth curve of M. smegmatis strains in Minimal medium. M. smegmatis wild type, GM1, GM2 strains were grown in minimal medium containing 0.2 % (w/v) glucose or 1%t ween 80 as the only carbon source. and OD600nm determined every 3 hours. For each strains the data reported in graph are the mean of three independent experiments. (JPG 710 kb)
Contributor Information
Anna Zanfardino, Email: anna.zanfardino@unina.it.
Adriana Migliardi, Email: ad.migliardi@studenti.unina.it.
Daniele D’Alonzo, Email: dandalonzo@unina.it.
Angela Lombardi, Email: angelina.lombardi@unina.it.
Mario Varcamonti, Email: mario.varcamonti@unina.it.
Angela Cordone, Email: angelina.cordone@unina.it.
References
- 1.Hett EC, Rubin J. Bacterial growth and cell division. Microbiol Mol Biol Rev. 2008;72:126–156. doi: 10.1128/MMBR.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan PJ. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis (Edinburgh) 2003;83:91–97. doi: 10.1016/S1472-9792(02)00089-6. [DOI] [PubMed] [Google Scholar]
- 3.Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 4.Howard ST, Byrd TF. The rapidly growing mycobacteria: Saprophytes and parasites. Microbes Infect. 2000;2:1845–1853. doi: 10.1016/S1286-4579(00)01338-1. [DOI] [PubMed] [Google Scholar]
- 5.Ripoll F, Deshayes C, Pasek S, Laval F, Beretti JL, Biet F, Risler JL, et al. Genomics of glycopeptidolipid biosynthesis in Mycobacterium abscessus and M. chelonae. BMC Genomics. 2007;8:114. doi: 10.1186/1471-2164-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daffè M, Laneelle MA, Puzo G. Structural elucidation by field desorption and electron-impact mass spectrometry of the C-mycosides isolated from Mycobacterium smegmatis. Biochim Biophys Acta. 1983;751:439–443. doi: 10.1016/0005-2760(83)90304-1. [DOI] [PubMed] [Google Scholar]
- 7.Aspinall GO, Chatterjee D, Brennan PJ. The variable surface glycolipids of mycobacteria: structures, synthesis of epitopes, and biological properties. Adv Carbohydr Chem Biochem. 1995;51:169–242. doi: 10.1016/S0065-2318(08)60194-8. [DOI] [PubMed] [Google Scholar]
- 8.Miyamoto Y, Mukai T, Maeda Y, Kai M, Naka T, Yano I, Makino M. Characterization of the Fucosylation Pathway in the Biosynthesis of Glycopeptidolipids from Mycobacterium avium Complex. J Bacteriol. 2007;189:5515–22. doi: 10.1128/JB.00344-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vergne I, Daffe M. Interaction of mycobacterial glycolipids with host cells. Front Biosci. 1998;3:865–876. doi: 10.2741/A330. [DOI] [PubMed] [Google Scholar]
- 10.Chatterjee D, Khoo KH. The surface glycopeptidolipids of mycobacteria: structures and biological properties. Cell Mol Life Sci. 2001;58:2018–2042. doi: 10.1007/PL00000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Billman-Jacobe H. Glycopeptidolipid synthesis in Mycobacteria. Curr Sci. 2004;86:11–114. [Google Scholar]
- 12.Recht J, Martinez A, Torello S, Kolter R. Genetic analysis of sliding motility in Mycobacterium smegmatis. J Bacteriol. 2000;182:4348–4351. doi: 10.1128/JB.182.15.4348-4351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Recht J, Kolter R. Glycopeptidolipid acetylation affects sliding motility and biofilm formation in Mycobacetrium smegmatis. J Bacteriol. 2001;183:5718–5724. doi: 10.1128/JB.183.19.5718-5724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Etienne G, Villeneuve C, Billman-Jacobe H, Astarie-Dequeker C, Dupont MA, Daffè M. The impact of the absence of glycopeptidolipids on the ultrastructure, cell surface and cell wall properties, and phagocytosis of Mycobacterium smegmatis. Microbiology. 2002;148:3089–100. [DOI] [PubMed]
- 15.Deshayes C, Laval F, Montrozier H, Daffè M, Etienne G, Reyrat JM. A Glycosyltransferase Involved in Biosynthesis of Triglycosylated Glycopeptidolipids in Mycobacterium smegmatis: Impact on Surface Properties. J Bacteriol. 2005;187:7283–7291. doi: 10.1128/JB.187.21.7283-7291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villeneuve C, Etienne G, Abadie V, Montrozier H, Bordier C, Laval F, Daffè M, et al. Surface-exposed glycopeptidolipids of Mycobacterium smegmatis specifically inhibit the phagocytosis of mycobacteria by human macrophages. Identification of a novel family of glycopeptidolipids. JBC. 2003;278:51291–51300. doi: 10.1074/jbc.M306554200. [DOI] [PubMed] [Google Scholar]
- 17.Sweet L, Schorey JS. Glycopeptidolipids from Mycobacterium avium promote macrophage activation in a TLR2- and MyD88-dependent manner. J Leukoc Biol. 2006;80:415–423. doi: 10.1189/jlb.1205702. [DOI] [PubMed] [Google Scholar]
- 18.Sweet L, Zhang W, Torres-Fewell H, Serianni A, Boggess W, Schorey J. Mycobacterium avium glycopeptidolipids require specific acetylation and methylation patterns for signaling through Toll-like Receptor 2. JBC. 2008;238:33221–33231. doi: 10.1074/jbc.M805539200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nessar R, Reyrat J, Davidson LB, Byrd TF. Deletion of the mmpL4b gene in the Mycobacterium abscessus glycopeptidolipid biosynthetic pathway results in loss of surface colonization capability, but enhanced ability to replicate in human macrophages and stimulate their innate immune response. Microbiology. 2011;157:1187–1195. doi: 10.1099/mic.0.046557-0. [DOI] [PubMed] [Google Scholar]
- 20.Patterson JH, McConville MJ, Haites RE, Coppel RL, Billman-Jacobe H. Identification of a methyltransferase from Mycobacterium smegmatis involved in glycopeptidolipid synthesis. JBC. 2000;275:24900–24906. doi: 10.1074/jbc.M000147200. [DOI] [PubMed] [Google Scholar]
- 21.Jeevarajah D, Patterson JH, McConville MJ, Billman-Jacobe H. Modification of glycopeptidolipids by an O-methyltransferase of Mycobacterium smegmatis. Microbiology. 2002;148:3079–3087. doi: 10.1099/00221287-148-10-3079. [DOI] [PubMed] [Google Scholar]
- 22.Trivedi OA, Arora P, Sridharan V, Tickoo R, Mohanty D, Gokhale RS. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature. 2004;428:441–445. doi: 10.1038/nature02384. [DOI] [PubMed] [Google Scholar]
- 23.Sonden B, Kocincova D, Deshayes C, Euphrasie D, Rhayat L, Laval F, Frehel C, et al. Gap, a mycobacterial specific integral membrane protein, is required for glycolipid transport to the cell surface. Mol Microbiol. 2005;54:426–440. doi: 10.1111/j.1365-2958.2005.04847.x. [DOI] [PubMed] [Google Scholar]
- 24.Biet F, Bay S, Thibault VC, Euphrasie D, Grayon M, Ganneau C, Lanotte P, et al. Lipopentapeptide induces a strong host humoral response and distinguishes Mycobacterium avium subsp. paratuberculosis from M. avium subsp. avium. Vaccine. 2008;26:257–268. doi: 10.1016/j.vaccine.2007.10.059. [DOI] [PubMed] [Google Scholar]
- 25.Touchette MH, Holsclaw CM, Previti ML, Solomon VC, Leary JA, Bertozzi CR, Seeliger JC. The rv1184c Locus Encodes Chp2, an Acyltransferase in Mycobacterium tuberculosis Polyacyltrehalose Lipid Biosynthesis. J Bacteriol. 2015;197:201–210. doi: 10.1128/JB.02015-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belardinelli JM, Larrouy-Maumus G, Jones V, de Sorio de Carvalho LP, McNeil MR, Jackson M. Biosynthesis and Translocation of Unsulfated Acyltrehaloses in Mycobacterium tuberculosis. JBC. 2014;289:27952–27965. doi: 10.1074/jbc.M114.581199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delogu G, Pusceddu C, Bua A, Fadda G, Brennan MJ, Zanetti S. Rv1818c-encoded PE_PGRS protein of Mycobacterium tuberculosis is surface exposed and influences bacterial cell structure. Mol Microbiol. 2004;52:725–733. doi: 10.1111/j.1365-2958.2004.04007.x. [DOI] [PubMed] [Google Scholar]
- 28.Larrouy-Maumus G, Kovierová HS, Dhouib R, Angala SK, Zuberogoitia S, Pham H, et al. A Small Multidrug Resistance-like Transporter Involved in the Arabinosylation of Arabinogalactan and Lipoarabinomannan in Mycobacteria. JBC. 2012;47:39933–39941. doi: 10.1074/jbc.M112.400986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu T, Zhao X, Li X, Hansen G, Blondeau J, Drlica K. Effect of chloramphenicol, erythromycin, moxifloxacin, penicillin and tetracycline concentration on the recovery of resistant mutants of Mycobacterium smegmatis and Staphylococcus aureus. J Antimicrobial Chemother. 2003;52:61–64. doi: 10.1093/jac/dkg268. [DOI] [PubMed] [Google Scholar]
- 30.Singh G, Singh G, Jadeja D, Kaur J. Lipid hydrolizing enzymes in virulence: Mycobacterium tuberculosis as a model system. Crit Rev Microbiol. 2010;36:259–269. doi: 10.3109/1040841X.2010.482923. [DOI] [PubMed] [Google Scholar]
- 31.Sultana R, Tanneeru K, Guruprasad L. The PE-PPE domain in mycobacterium reveals a Serine α/β Hydrolase fold and function: an in-silico analysis. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen G, Singh K, Chandra D, Serveau-Avesque C. maurin D, Canaan S, Singla R, Behera D, Laal S. LipC (Rv0220) is an immunogenic cell surface esterase of Mycobacterium tuberculosis. Infect Immun. 2012;80:243–253. doi: 10.1128/IAI.05541-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deb C, Daniel J, Sirakova TD, Abomoelak B, Dubey VS, Kolattukudy PE. A novel lipase belonging to the Hormone-sensitive Lipase Family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. JBC. 2006;281:3866–3875. doi: 10.1074/jbc.M505556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cotes K, Bakala N'goma CJ, Dhouib R, Douchet I, Maurin D, Carrière F, Canaan S. Lipolytic enzymes in Mycobacterium tuberculosis. Appl Microbiol Biotechnol. 2008;78:741–749. doi: 10.1007/s00253-008-1397-2. [DOI] [PubMed] [Google Scholar]
- 35.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fitsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Press; 1989.
- 37.Parish T, Soker NG. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology. 2000;146:1969–1975. doi: 10.1099/00221287-146-8-1969. [DOI] [PubMed] [Google Scholar]
- 38.Cordone A, Mauriello EMF, Pickard DJ, Dougan DJ, De Felice M, Ricca E. The lrp Gene and Its Role in Type I Fimbration in Citrobacter rodentium. J Bacteriol. 2005;187:7009–7017. doi: 10.1128/JB.187.20.7009-7017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


