Abstract
Background
Over the past several years, thousands of microRNAs (miRNAs) have been identified in the genomes of various insects through cloning and sequencing or even by computational prediction. However, the number of miRNAs identified in anopheline species is low and little is known about their role. The mosquito Anopheles funestus is one of the dominant malaria vectors in Africa, which infects and kills millions of people every year. Therefore, small RNA molecules isolated from the four life stages (eggs, larvae, pupae and unfed adult females) of An. funestus were sequenced using next generation sequencing technology.
Results
High throughput sequencing of four replicates in combination with computational analysis identified 107 mature miRNA sequences expressed in the An. funestus mosquito. These include 20 novel miRNAs without sequence identity in any organism and eight miRNAs not previously reported in the Anopheles genus but are known in non-anopheles mosquitoes. Finally, the changes in the expression of miRNAs during the mosquito development were determined and the analysis showed that many miRNAs have stage-specific expression, and are co-transcribed and co-regulated during development.
Conclusions
This study presents the first direct experimental evidence of miRNAs in An. funestus and the first profiling study of miRNA associated with the maturation in this mosquito. Overall, the results indicate that miRNAs play important roles during the growth and development. Silencing such molecules in a specific life stage could decrease the vector population and therefore interrupt malaria transmission.
Keywords: MicroRNA, Non-coding RNA, Anopheles funestus, Vector-borne diseases, Malaria
Background
MicroRNAs (miRNAs) comprise a large family of endogenous, evolutionarily conserved, non-coding RNA that post-transcriptionally regulate gene expression in plants and animals [1, 2]. Many studies have indicated that miRNAs play a pivotal role in most critical biological events [3–17]. Therefore, identifying and characterization of miRNAs to understand their physiological and pathological roles have become popular research topics.
After the discovery of the first miRNA in the nematode, Caenorhabditis elegans, thousands of miRNAs have been identified [18–21]. The major approaches to identifying miRNAs include genetic screening, direct cloning, bioinformatic analysis, and deep sequencing [22, 23]. The majority of known miRNAs have been identified through traditional direct cloning, which is both time-consuming and labour-intensive. Bioinformatic analysis is limited because the majority of computational programs require genomic sequence as a template. Next generation sequencing (NGS) has provided an innovative tool to look into the genome with an unprecedented depth of coverage. It allows for a comprehensive coverage of miRNAs in many species [24–33]. As a consequence, this new approach has opened the door to functional genomic analyses of non-model species. It is widely used for profiling miRNAs in populations in various developmental stages, in either normal or diseased states [34–50].
Although there are over 30 species of Anopheles which transmit malaria in the world [51], miRNAs have so far only been experimentally identified in three malaria vectors, the African vector (Anopheles gambiae) and in two Asian vectors (Anopheles stephensi and Anopheles anthropophagus) [7, 52, 53]. Among the African vectors, Anopheles funestus is one of the most proficient malaria vectors, mainly because of its remarkable ability to populate a wide range of ecological settings across Africa [54–58]. Therefore, experimental identification of miRNAs controlling key genes required for mosquitoes to complete their life-cycle will not only help to better understand the vector biology, but it may uncover novel approaches to control this mosquito. In this context, the main objective of this study was to identify miRNAs expressed in the four main life stages (eggs, larvae, pupae and unfed adult females) of the mosquito An. funestus using high throughput sequencing technology and bioinformatics approaches.
Methods
Mosquito strain and rearing condition
The experimental work was performed using a colony of An. funestus (FUMOZ) that originates from southern Mozambique [59]. The mosquitoes were reared in the insectary of the Vector Control Reference Laborator at the National Institute for Communicable Diseases (NICD), Johannesburg, South Africa since 2000. The insectary is kept at 25 °C, 80% relative humidity with a 12-h day/night lighting regime and 45-min dusk/dawn cycles.
RNA extraction and small RNA sequencing
Total RNA was isolated separately from the four different life cycle stages of An. funestus (eggs, larvae, pupae and unfed adult females) using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s protocol. Four different biological replicates for each life stage for RNA extraction were prepared. In order to obtain a large and broad miRNA transcriptome data set, RNA was extracted from 5 egg patches, 100 fourth instar larvae, 100 pupae, and 100 unfed adult females for each single batch. The quality and quantity of resulting RNA was assessed using a spectrophotometer (Nanodrop Technologies), and quality assessment determined by using Agilent 2100 Bioanalyzer RNA Nano 6000 kit. No rRNA depletion was performed. The RNA extracts from the four life stages were sent to Macrogen Inc (South Korea) for small RNA sequencing. Sequencing libraries were generated from 1 μg each of the total RNA samples by ligation of adapter RNAs at 5′ and 3′ ends, followed by reverse transcription and PCR using Illumina TruSeq Small RNA Sample Preparation protocol (Illumina Inc., USA). The libraries were size-selected for sequencing of RNA fragments of 18–30 nucleotides. Sequencing was performed on an Illumina HiSeq 2000 platform to obtain single-end reads of 50 bases. Each batch sequenced independently.
Sequencing data processing and analysis
Read quality check and filtering
Following sequencing, the quality of the four sequenced libraries were checked using FastQC [60]. Later, all sequencing reads with low quality tags and shorter than 18 nucleotides were removed using FASTX-Toolkit [61]. For all following analysis, all reads from each stage were combined into a single input.
Mapping the reads to the reference genome
All reads were mapped to the An. funestus strain FUMOZ genome [GenBank: KB668221]. The sequence reads were mapped to the genome using miRDeep2 mapper module [62].
Small non-coding RNAs detection
All reads were aligned to the RNA families database (Rfam) version 11 [63] using the BLASTn algorithm allowing for a 2 nucleotide mismatch and e-value lower than 0.01 in order to annotate the small non-coding RNAs present in the libraries.
Identification of miRNA sequences
For identification of miRNA sequences present in the four life stage datasets, the miRDeep2 package was employed [62]. The package scripts detect known or novel miRNAs from deep sequencing data. It looks for the pattern that the miRNA processing machinery leaves in the sequencing data. The most important pattern that miRDeep2 considers are clusters of reads that align along the reference genome that is compatible with the mature miRNA sequence, the loop sequence, and the star sequence structure of the miRNA precursor molecule. If such a pattern is found, miRDeep2 cuts out the potential miRNA precursor sequence from the reference genome and utilizes an RNA folding algorithm (randfold) from the Vienna package [64] to assess if the sequence can be folded into a hairpin structure. Furthermore, the prediction software searches for potential cleavage sites of Drosha and Dicer. The phylogenetic conservation and filtering of other known small non-coding RNA species can be optionally used to improve the predictions. All identified miRNAs are named according to their most similar miRNAs in the miRNA database (miRBase) release 21 (June 2014) [18–21] match.
Differential expression of the miRNAs
Differentially expressed miRNAs between two sequential life stages (egg-larva, larva-pupa, pupa-unfed adult female) were determined by log2 fold change >2 for the normalized reads +1. This analysis was computed using gtools package for the R programming language.
Results
Sequence quality of the four libraries
Small RNAs were sequenced in quadruplicate using Illumina sequencing from eggs, larvae, pupae and unfed adult females batches to identify An. funestus miRNAs expressed during development. More than 150 million raw reads were produced from each life stage (Table 1). The length distribution of the reads was between 18 and 30 nucleotides (data not shown). After filtering the impurities and reads of length smaller than 18 nucleotides, 75.92, 71.52, 80.85 and 75.39%, high-quality reads had Phred quality values (PQV) of 20 obtained from the eggs, larvae, pupae, and unfed adult females libraries, respectively (Table 1). The PQV has previously been reported to be an indicator of base call accuracy and therefore sequence quality [65].
Table 1.
Summary of small RNA sequencing data analysis for the four life stage libraries of An. funestus
| Stage | Eggs | Larvae | Pupae | Unfed adult females |
|---|---|---|---|---|
| Total raw reads (%) | 151,229,067 | 165,299,940 | 165,782,104 | 154,104,200 |
| High quality reads (%) | 114,821,916 (75, 92) | 118,232,206 (71, 52) | 134,044,428 (80, 85) | 116,182,404 (75, 39) |
| Mapped reads to the genome (%) | 71,894,278 (62, 61) | 75,552,909 (63, 90) | 98,996,560 (73, 85) | 69,037,524 (59, 42) |
Mapping reads from the four libraries
Mapping reads over the unmasked genome represents an unbiased option, allowing the detection of known and still undiscovered miRNAs. The total number of the reads mapped to An. funestus genome constitutes only 62.61, 63.90, 73.85 and 59.42% of the total high-quality reads from eggs, larvae, pupae, and unfed adult females libraries, respectively (Table 1).
Annotation of small non-coding RNAs in the four libraries
In order to annotate other small non-coding RNAs in the four libraries, all clean reads were aligned against Rfam version 11. As shown in Fig. 1, the most abundant class of small non-coding RNAs in the eggs library were miRNAs. However, rRNAs were most abundant in the larvae and pupae, and tRNAs in the unfed adult females library.
Fig. 1.
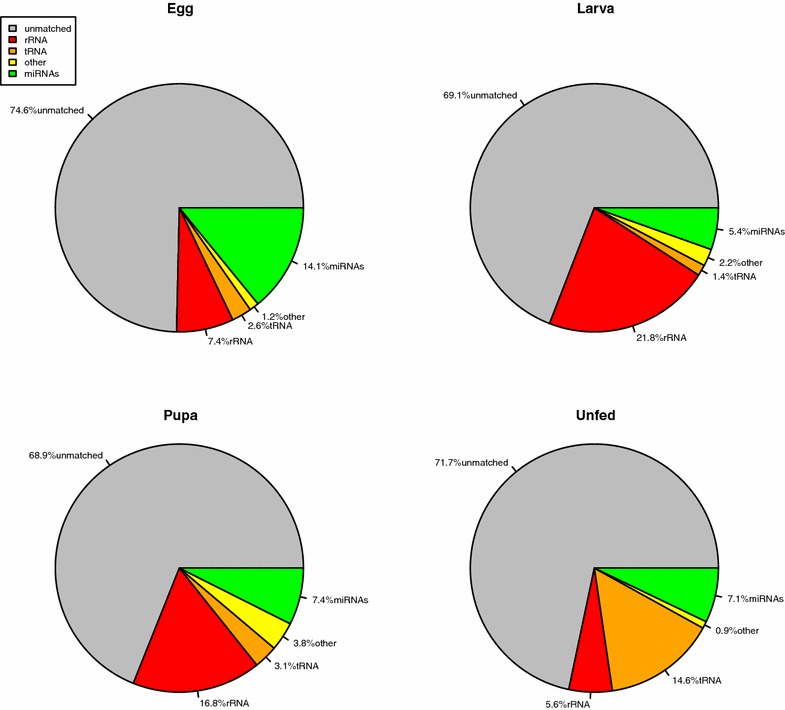
Small non-coding RNAs annotated from the four life stage libraries of An. funestus. From aligning the high quality reads to the RNA families database (Rfam) version 11. The total reads can be divided into five categories; miRNAs, tRNAs, rRNAs, others and unmatched. The others and unmatched referred to the other class of small non-coding RNAs and the reads not aligned to RNA families database , respectively
Identification of miRNAs in the four life stages libraries of An. funestus
Of the 65 previously known An. gambiae mature miRNAs in miRBase release 21 (June 2014), 64 mature miRNAs were detected in the sequenced short reads of An. funestus (Table 2). The two miR-276 precursors in An. gambiae generate two different mature miRNAs whereas in An. funestus they produce the same mature miRNA. These include all the 46 miRNA sequences computationally predicted in the An. funestus strain FUMOZ genome hosted by the invertebrate vectors of human pathogens database (VectorBase) [66]. The full precursor structure (mature, loop and star sequence) or parts of it were detected. Only star sequences were found for miR-12, miR-306, miR-965, miR-989, miR-1174 and miR-1889 and the precursor locations for these miRNAs were not determined. Reads that do not match any known An. gambiae miRNA sequences from miRBase and mapped to the An. funestus genome were screened for precursor or hairpin structures. The miRDeep2 algorithm was employed to investigate if non-annotated sequences mapping to the An. funestus genome demonstrated folding properties of precursors or hairpins. A total of 43 mature miRNA candidates expressed from 42 precursor candidates were identified (Table 3). Among these miRNAs, 15 miRNAs have been described in the Anopheles genus (An. gambiae and/or An. anthropophagus) but not registered in miRBase database, eight have not been previously reported in the Anopheles genus but are known in other mosquitoes such as Aedes aegypti and/or Culex quinquefasciatus, and the remaining 20 miRNAs have not been described before in any species. For all the mature miRNA candidates, the full or parts of the precursor were detected in the four life stage libraries except for miR-2943 that was detected only in the eggs library. MiRNAs frequently exhibit sequence differences when compared to a reference mature sequence, generating multiple variations that are known as isoforms. New isoforms were identified for miR-927 (miR-927-3p) and miR-980 (miR-980-5p). The new isoforms of miR-980 were expressed only in the eggs library. Our analysis uncovered a third stem-loop precursor for miR-2.
Table 2.
Known miRNAs identified in the four life stages libraries of An. funestus
| miRNA name | Sequences | Length | Location | Egg | Larva | Pupa | Unfed adult females |
|---|---|---|---|---|---|---|---|
| afu-bantam | UGAGAUCACUUUGAAAGCUGAU | 22 | KB669192.1_supercont1.62:255022..255087 | ||||
| afu-let-7 | UGAGGUAGUUGGUUGUAUAGU | 21 | KB669047.1_supercont1.49:698595..698654 | ||||
| afu-miR-1 | UGGAAUGUAAAGAAGUAUGGAG | 22 | KB668556.1_supercont1.130:34984..35060 | ||||
| afu-miR-10 | ACCCUGUAGAUCCGAAUUUGUU | 22 | KB668870.1_supercont1.33:182453..182515 | ||||
| afu-miR-100 | AACCCGUAGAUCCGAACUUGUG | 22 | KB669047.1_supercont1.49:703012..703075 | ||||
| afu-miR-1000 | AUAUUGUCCUGUCACAGCAGUA | 22 | KB669169.1_supercont1.6:205456..205544 | ||||
| afu-miR-11 | CAAGAACUUGGUACUGUGACCUGUG | 25 | KB669169.1_supercont1.6:410485..410557 | ||||
| afu-miR-1175 | AAGUGGAGUAGUGGUCUCAUCGCU | 24 | KB669358.1_supercont1.77:114656..114718 | ||||
| afu-miR-124 | UAAGGCACGCGGUGAAUGCCA | 21 | KB668556.1_supercont1.130:267597..267658 | ||||
| afu-miR-125 | UCCCUGAGACCCUAACUUGUGAC | 23 | KB669047.1_supercont1.49:697981..698042 | ||||
| afu-miR-133 | UUGGUCCCCUUCAACCAGCUGU | 22 | KB668872.1_supercont1.331:18310..18396 | ||||
| afu-miR-137 | UAUUGCUUGAGAAUACACGUAG | 22 | KB669203.1_supercont1.63:461201..461262 | ||||
| afu-miR-13b | UAUCACAGCCAUUUUGACGA | 20 | KB668788.1_supercont1.256:85269..85330 | ||||
| afu-miR-14 | UCAGUCUUUUUCUCUCUCCUAU | 22 | KB668222.1_supercont1.10:1664927..1664991 | ||||
| afu-miR-184 | UGGACGGAGAACUGAUAAGGGC | 22 | KB668984.1_supercont1.432:34990..35049 | ||||
| afu-miR-1890 | UGAAAUCUUUGAUUAGGUCUGG | 22 | KB668768.1_supercont1.238:330029..330096 | ||||
| afu-miR-1891 | UGAGGAGUUAAUUUGCGUGUUU | 22 | KB668738.1_supercont1.210:313240..313298 | ||||
| afu-miR-190 | AGAUAUGUUUGAUAUUCUUGGUUG | 24 | KB668683.1_supercont1.161:159767..159832 | ||||
| afu-miR-2-1/2 | UAUCACAGCCAGCUUUGAUGAGC | 23 | KB668788.1_supercont1.256:84051..84114 | ||||
| afu-miR-210 | CUUGUGCGUGUGACAACGGCUAU | 23 | KB668844.1_supercont1.306:43398..43456 | ||||
| afu-miR-219 | AGAGUUGUGACUGGACAUCCGUG | 23 | KB668767.1_supercont1.237:131113..131179 | ||||
| afu-miR-263a | AAUGGCACUGGAAGAAUUCACGGG | 24 | KB668222.1_supercont1.10:692930..692993 | ||||
| afu-miR-263b | CUUGGCACUGGGAGAAUUCACAG | 23 | KB668812.1_supercont1.278:190692..190757 | ||||
| afu-miR-275 | UCAGGUACCUGAAGUAGCGCGCG | 23 | KB668400.1_supercont1.116:398864..398930 | ||||
| afu-miR-276 | UAGGAACUUCAUACCGUGCUCU | 22 | KB668722.1_supercont1.197:198964..199032 | ||||
| afu-miR-277 | UAAAUGCACUAUCUGGUACGACA | 23 | KB669169.1_supercont1.6:1092927..1092992 | ||||
| afu-miR-278 | ACGGACGAUAGUCUUCAACGACC | 23 | KB668223.1_supercont1.100:30655..30714 | ||||
| afu-miR-279 | UGACUAGAUCCACACUCAUUAA | 22 | KB668289.1_supercont1.106:353827..353890 | ||||
| afu-miR-281 | AAGAGAGCUAUCCGUCGACAGU | 22 | KB669503.1_supercont1.90:359505..359565 | ||||
| afu-miR-282 | UAGCCUCUUCUAGGCUUUGUCU | 22 | KB669503.1_supercont1.90:578663..578726 | ||||
| afu-miR-283 | AAAUAUCAGCUGGUAAUUCUAGG | 23 | KB668836.1_supercont1.3:514580..514646 | ||||
| afu-miR-286 | UGACUAGACCGAACACUCGCGUCCU | 25 | KB668445.1_supercont1.120:344766..344839 | ||||
| afu-miR-305 | AUUGUACUUCAUCAGGUGCUCUGG | 24 | KB668400.1_supercont1.116:390085..390147 | ||||
| afu-miR-307 | UCACAACCUCCUUGAGUGAGCGA | 23 | KB668223.1_supercont1.100:429991..430050 | ||||
| afu-miR-308 | CGCAGUAUAUUCUUGUGAACUUG | 23 | KB668933.1_supercont1.387:20015..20073 | ||||
| afu-miR-309 | UCACUGGGCAAAGUUUGUCGCA | 22 | KB668445.1_supercont1.120:344139..344203 | ||||
| afu-miR-315 | UUUUGAUUGUUGCUCAGAAAGCC | 23 | KB668747.1_supercont1.219:307337..307401 | ||||
| afu-miR-317 | UGAACACAUCUGGUGGUAUCUCAGU | 25 | KB669169.1_supercont1.6:1109058..1109126 | ||||
| afu-miR-34 | UGGCAGUGUGGUUAGCUGGUUGU | 23 | KB669169.1_supercont1.6:1091079..1091145 | ||||
| afu-miR-375 | UUUGUUCGUUUGGCUCGAGUUA | 22 | KB669281.1_supercont1.70:314839..314901 | ||||
| afu-miR-7 | UGGAAGACUAGUGAUUUUGUUGUU | 24 | KB668798.1_supercont1.265:312209..312271 | ||||
| afu-miR-79 | AUAAAGCUAGAUUACCAAAGCAU | 23 | KB668930.1_supercont1.384:9585..9649 | ||||
| afu-miR-8 | UAAUACUGUCAGGUAAAGAUGUC | 23 | KB668666.1_supercont1.146:27512..27573 | ||||
| afu-miR-87 | CCAGCCUGAAAUUUGCUAAACCU | 23 | KB669058.1_supercont1.5:469321..469384 | ||||
| afu-miR-927-5p | UUUAGAAUUCCUACGCUUUACC | 22 | KB668660.1_supercont1.140:64296..64359 | ||||
| afu-miR-929 | AAAUUGACUCUAGUAGGGAGU | 21 | KB668836.1_supercont1.3:1318319..1318378 | ||||
| afu-miR-92a | UAUUGCACUUGUCCCGGCCUAU | 22 | KB668836.1_supercont1.3:1691007..1691065 | ||||
| afu-miR-92b | AAUUGCACUUGUCCCGGCCUGC | 22 | KB668836.1_supercont1.3:1704691..1704754 | ||||
| afu-miR-957 | UGAAACCGUCCAAAACUGAGGC | 22 | KB669514.1_supercont1.91:212823..212889 | ||||
| afu-miR-970 | UCAUAAGACACACGCGGCUAU | 21 | KB668797.1_supercont1.264:230410..230478 | ||||
| afu-miR-981 | UUCGUUGUCGACGAAACCUGCA | 22 | KB668389.1_supercont1.115:58097..58172 | ||||
| afu-miR-988 | CCCCUUGUUGCAAACCUCACGC | 22 | KB669391.1_supercont1.8:63805..63867 | ||||
| afu-miR-993 | GAAGCUCGUUUCUAUAGAGGUAUC | 24 | KB668870.1_supercont1.33:220731..220822 | ||||
| afu-miR-996 | UGACUAGAUUACAUGCUCGUCU | 22 | KB668289.1_supercont1.106:354280..354344 | ||||
| afu-miR-9a | UCUUUGGUUAUCUAGCUGUAUGA | 23 | KB668816.1_supercont1.281:204094..204152 | ||||
| afu-mir-9b | ACUUUGGUGAUUUAGCUGUAUGU | 23 | KB668930.1_supercont1.384:9150..9215 | ||||
| afu-miR-9c | UCUUUGGUAUUCUAGCUGUAGA | 22 | KB668930.1_supercont1.384:12370..12438 | ||||
| afu-miR-iab-4 | ACGUAUACUGAAUGUAUCCUGA | 22 | KB668694.1_supercont1.171:357100..357157 | ||||
| afu-miR-12 | UGAGUAUUACAUCAGGUACUGGU | 23 | Unknown | ||||
| afu-miR-306 | UCAGGUACUGGAUGACUCUCAG | 22 | Unknown | ||||
| afu-mir-965 | UAAGCGUAUAGCUUUUCCCAUU | 22 | Unknown | ||||
| afu-miR-989 | UGUGAUGUGACGUAGUGGUAC | 21 | Unknown | ||||
| afu-miR-1174 | UCAGAUCUACUUCAUACCCAUG | 22 | Unknown | ||||
| afu-miR-1889 | ACACAUUACAGAUUGGGAUUA | 21 | Unknown |
The three columns for each life stage correspond to detection of the full precursor structure namely mature, loop and star sequence, respectively. indicates that miRNA full precursor structure was detected in the library where indicates the the miRNA was not detected in the library
Table 3.
Novel miRNAs identified in the four life stages libraries of An. funestus
| miRNA name | Sequences | Length | Location | Egg | Larva | Pupa | Unfed adult females | Description | |
|---|---|---|---|---|---|---|---|---|---|
| afu-miR-2-3 | UAUCACAGCCAGCUUUGAAGAGC | 23 | KB668788.1_supercont1.256:85435..85508 | aae-miR-2a | Identified in An. anthropophagus and An. gambiae [53, 100] | ||||
| afu-miR-252 | CUAAGUACUAGUGCCGCAGGAG | 22 | KB669170.1_supercont1.60:199766..199821 | dme-miR-252-5p | Identified in An. anthropophagus and An. gambiae [53, 100] | ||||
| afu-miR-932 | UCAAUUCCGUAGUGCAUUGCAGU | 23 | KB668896.1_supercont1.353:66093..66159 | tca-mir-932 | Identified in An. anthropophagus and An. gambiae [53, 100] | ||||
| afu-miR-2796 | GUAGGCCGGCGGAAACUACUUGC | 23 | KB669425.1_supercont1.83:106051..106111 | bmo-miR-2796-3p | Identified in An. anthropophagus and An. gambiae [53, 100] | ||||
| afu-miR-971 | UUGGUGUUAUAUCUUACAGUGAG | 23 | KB669081.1_supercont1.52:713511..713586 | ame-miR-971 | Identified in An. gambiae [53] | ||||
| afu-miR-998 | UAGCACCAUGAGAUUCAGCUC | 21 | KB669169.1_supercont1.6:410177..410245 | aae-miR-998 | Identified in An. gambiae [53] | ||||
| afu-mir-2942 | UAUUCGAGACCUUCACGAGUUAA | 23 | KB668333.1_supercont1.11:1154487..1154559 | aae-miR-2942 | Identified in An. gambiae [53] | ||||
| afu-miR-31 | UAGCUAUUCAACUUCUUGUUUAU | 23 | KB668245.1_supercont1.102:383384..383442 | cqu-miR-31-3p | Identified in An. gambiae [100] | ||||
| afu-mir-33 | GUGCAUUGUAGUUGCAUUGCA | 21 | KB668389.1_supercont1.115:38232..38297 | aae-miR-33 | Identified in An. gambiae [100] | ||||
| afu-miR-285 | UAGCACCAUUCGAAAUCAGUAC | 22 | KB668666.1_supercont1.146:348761..348825 | aae-miR-285 | Identified in An. gambiae [100] | ||||
| afu-miR-980-5p | CGGUCGUUCACCAGGUCAUCUAGC | 24 | KB668730.1_supercont1.203:371780..371842 | aae-miR-980-5p | Identified in An. gambiae [100] | ||||
| afu-miR-999 | UGUUAACUGUAAGACUGUGUCU | 22 | KB668862.1_supercont1.322:158491..158552 | aae-miR-999 | Identified in An. gambiae [100] | ||||
| afu-miR-2940 | GUCGACAGAGAGAUAGAUCACU | 22 | KB668668.1_supercont1.148:483999..484065 | aae-miR-2940 | Identified in An. gambiae [100] | ||||
| afu-miR-2944a | GAAGGAACUUCUGCUGUGAUCU | 22 | KB668445.1_supercont1.120:344303..344359 | aae-miR-2944a-5p | Identified in An. gambiae [100] | ||||
| afu-miR-2944b | GAAGGAACUCCCGGUGUGAUAUA | 23 | KB668456.1_supercont1.121:76523..76595 | aae-miR-2944b-5p | Identified in An. gambiae [100] | ||||
| afu-miR-71 | UCUCACUACCUUGUCUUUCAUG | 22 | KB668788.1_supercont1.256:83429..83494 | aae-miR-71-3p | Novel in Anopheles spp. | ||||
| afu-miR-193 | UACUGGCCUACUAAGUCCCAAC | 22 | KB668718.1_supercont1.193:152692..152757 | aae-miR-193 | Novel in Anopheles spp. | ||||
| afu-miR-316 | UGUCUUUUCCUGCUUACUGCCG | 22 | KB668714.1_supercont1.19:760291..760355 | aae-miR-316 | Novel in Anopheles spp. | ||||
| afu-miR-927-3p | UAAAGCGUAGGAAUUCUAAAAC | 22 | KB668660.1_supercont1.140:64294..64357 | dme-miR-927-5p | Novel in Anopheles spp. | ||||
| afu-miR-980-3p | UAGCUGCCUAGUGAAGGGC | 19 | KB668730.1_supercont1.203:371784..371839 | aae-miR-980-3p | Novel in Anopheles spp. | ||||
| afu-miR-2765 | UGGUAACUCCACCACCGUUGGC | 22 | KB668981.1_supercont1.43:346454..346513 | aae-miR-2765 | Novel in Anopheles spp. | ||||
| afu-miR-2941 | UAGUACGGAUAAGUAACACACU | 22 | KB668992.1_supercont1.44:163365..163440 | aae-miR-2941 | Novel in Anopheles spp. | ||||
| afu-miR-2943 | UUAAGUAGGCACUUGCAGGCAA | 22 | KB668790.1_supercont1.258:358408..358469 | aae-miR-2943 | Novel in Anopheles spp. | ||||
| afu-miR-a | UGAGGUAAACCGCGCUUAAAGA | 22 | KB668367.1_supercont1.113:186839..186900 | - | Novel | ||||
| afu-miR-b | AACGGACGGGUACCUUCGCACC | 22 | KB668478.1_supercont1.123:544003..544072 | - | Novel | ||||
| afu-miR-c | UCAACAUACAUUCUCGUUCUGU | 22 | KB668522.1_supercont1.127:174494..174560 | - | Novel | ||||
| afu-miR-d | UAAUACAUUUGCUUACGGCAGAGU | 24 | KB668522.1_supercont1.127:49592..49655 | - | Novel | ||||
| afu-miR-e | UCUGCCGUAGGAAUGUAUUUACC | 23 | KB668522.1_supercont1.127:51191..51253 | - | Novel | ||||
| afu-miR-f | UGCUGUAGGAAGUGAUUUACCU | 22 | KB668522.1_supercont1.127:51545..51607 | - | Novel | ||||
| afu-miR-g | AGUCAUCGUCGUCACUCGAUCGA | 23 | KB668728.1_supercont1.201:241585..241642 | - | Novel | ||||
| afu-miR-h | CCAUCUGACGGACACCACCGGA | 22 | KB668739.1_supercont1.211:143895..143961 | - | Novel | ||||
| afu-miR-i | AUGGCAGUCAACGUAUACCCAUU | 23 | KB668781.1_supercont1.25:515317..515377 | - | Novel | ||||
| afu-miR-j | UUCCGAGUGGACAAGUGGAACCU | 23 | KB668806.1_supercont1.272:68360..68416 | - | Novel | ||||
| afu-miR-k | UACCUGAGUUCGUUUAAACUGA | 22 | KB668821.1_supercont1.286:244740..244793 | - | Novel | ||||
| afu-miR-l | UCUAUCAUUUGAGUACCAUGA | 21 | KB668837.1_supercont1.30:192923..192989 | - | Novel | ||||
| afu-miR-m | CUAACCUGUAAGGAGCUUUGGCGGC | 25 | KB668906.1_supercont1.362:184842..184933 | - | Novel | ||||
| afu-miR-n | AUGGCACAAGCAACAUCAGCUG | 22 | KB668909.1_supercont1.365:72126..72172 | - | Novel | ||||
| afu-miR-o | UCUUGAGCGUAUUUGGCACUGCU | 23 | KB668992.1_supercont1.44:163580..163641 | - | Novel | ||||
| afu-miR-p | UAGCUCAACAAACAUCUGGAGGA | 23 | KB669059.1_supercont1.50:521609..521669 | - | Novel | ||||
| afu-miR-q | UACAACCGGACGGUACACUGCAUAG | 25 | KB669214.1_supercont1.64:671164..671237 | - | Novel | ||||
| afu-miR-r | AACGAAGUUGAUCCGCUGAAGCU | 23 | KB669480.1_supercont1.88:541508..541571 | - | Novel | ||||
| afu-miR-s | ACUUGAAACGUAGUCCGGGAACCC | 24 | KB669502.1_supercont1.9:563831..563898 | - | Novel | ||||
| afu-miR-t | AUUAGAAUGUGGAAUCUGUUUUU | 23 | KB668820.1_supercont1.285:93655..93723 | - | Novel |
The three columns for each life stage correspond to detection of the full precursor structure namely mature, loop and star sequence, respectively. indicates that miRNA full precursor structure was detected in the library where indicates the the miRNA was not detected in the library
Expression profiling of miRNAs during An. funestus development
To obtain insight into possible stage dependent roles of miRNAs in An. funestus, the expression patterns of miRNAs in different life stages were examined based on the number of reads obtained. The heatmaps (Fig. 2) summarize the expression of the miRNAs identified in the four life stage libraries. The majority of miRNAs were sequenced between 1–66 times. The expression profiles of miRNAs varied from highly specific to ubiquitous during the four stages. Among the known miRNAs, miR-263a was the most frequently expressed miRNA in the eggs, the larvae, and the pupae libraries (>1 million reads), whereas miR-8 was the dominant miRNA in the unfed adult females library (>300 thousand reads). Nevertheless, miR-87 had the lowest expression level in the egg and the pupa, similarly miR-189 in the larva, and miR-309 and miR-929 in the unfed adult females library. The highly expressed miRNA amongst the new miRNAs in the eggs library was miR-2944a (>1.5 million reads), miR-2765 (>20 thousand reads) in the larvae and pupae libraries and miR-999 (>30 thousand reads) in the unfed adult females library. To analyse the changes in the expression of miRNAs during the mosquito life stages, all read counts within a dataset were normalized, then log2 (fold-changes) were calculated between each two stages. The results of the comparisons (egg-larva, larva-pupa and pupa-unfed adult female) of the expression level of all the miRNAs in the four libraries are shown in Figs. 3 and 4. Between the egg to the larva, 29 miRNAs were up-regulated and 12 other miRNAs were down-regulated. Six miRNAs were up-regulated and another 13 down-regulated between the larva and pupa stage. However, seven miRNAs were up-regulated and four down-regulated between the pupa and the unfed adult female.
Fig. 2.
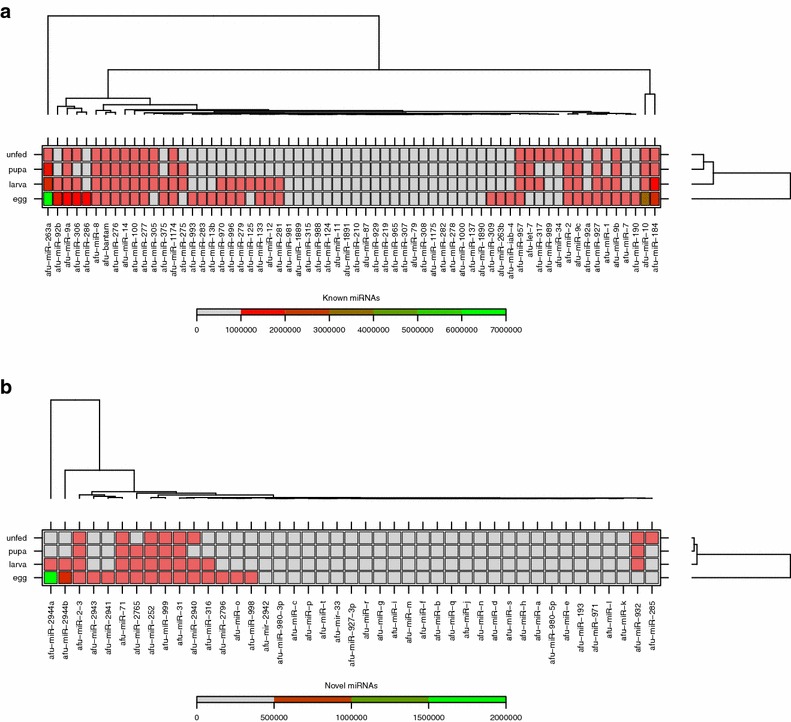
Heatmaps clustering of miRNAs expressed in the four life stage libraries of An. funestus. The clustering was performed on all known (a) and novel (b) miRNAs based on a raw read copy number (sequencing frequency) and the four life stage samples. Each row represents a stage and each column represents one miRNA. The miRNA clustering is shown on top. The colour scale (shown on the left) illustrates the number of the reads of a miRNA across the life stages
Fig. 3.
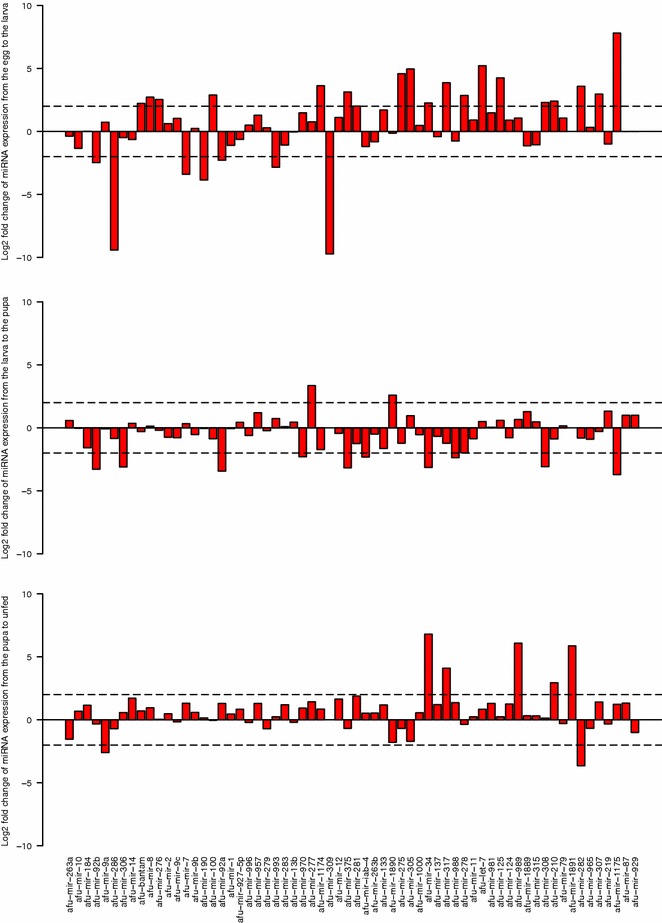
Fold-change of the known miRNAs during the development of An. funestus. The fold-changes were calculated for each miRNA (x-axis) using normalized reads +1 (y-axis, bars)
Fig. 4.
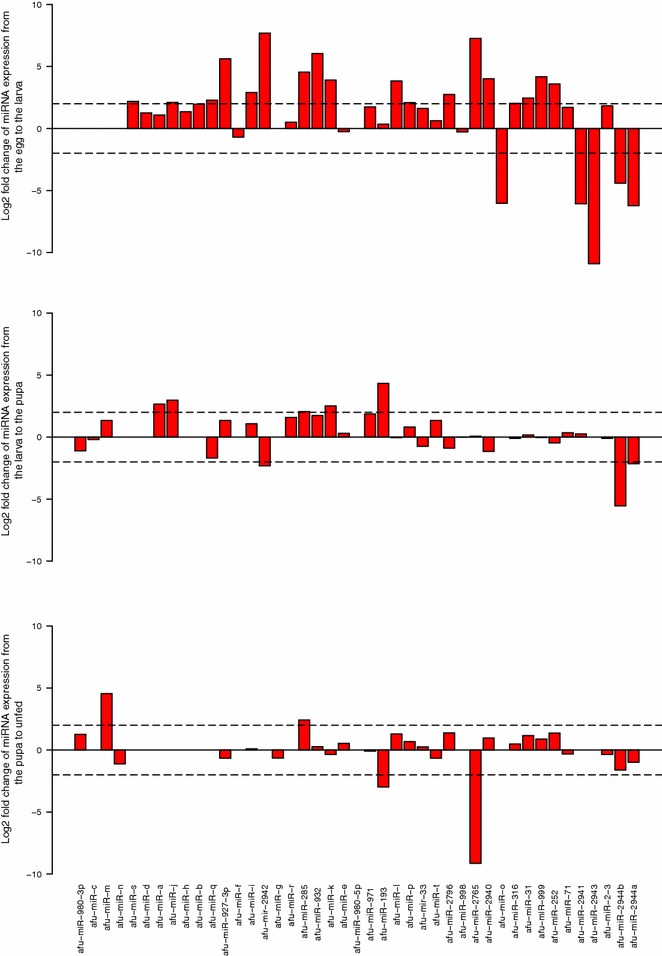
Fold-change of the novel miRNAs during the development of An. funestus. The fold-changes were calculated for each miRNA (x-axis) using normalized reads +1 (y-axis, bars)
Discussion
Although thousands of small RNAs have been identified [18–21], the challenge remains to fully identify all small RNAs, especially very low abundance miRNAs and to determine their individual functions. The majority of known miRNAs have been identified through the traditional cloning method [67]. The Illumina sequencing approach is one of high throughput technologies by which miRNAs can be detected in any organism without prior sequence or secondary structure information sequence or secondary structure information. Here, 107 miRNA sequences expressed in the four life stages of the African malaria vector An. funestus were identify and characterize using the deep sequencing approach. This method is more powerful than other conventional technologies previously used in mosquitoes [7, 11, 52], as it is able to identify new miRNAs that are beyond the capabilities of older traditional methods.
In this study, approximately 100 million high-quality reads from each life stage by deep sequencing were obtained. The size distribution of sequenced reads showed peaks between 20–30 nucleotides compare to the average miRNA length of 22 nucleotides in animals [1]. Using similar deep sequencing techniques, other studies on insects show small RNA sequence size distributions with a peak at 22 nucleotides [11, 34, 68, 69]. All the high-quality reads were mapped on to the An. funestus genome with low percentage (average of 65%) identity. Such a mapping bias has been reported in several studies [21, 70–72]. This approach has the weakness that it might favour alignment ambiguities due to the limited alignment specificity given by the small length of mature miRNAs (18–25 nucleotides) detected by the short reads, and to the size and high complexity of an unmasked reference genome [70].
As expected, most of the known miRNAs identified in An. funestus were highly conserved across diverse animal species, suggesting that the ancient regulatory pathways mediated by evolutionary conserved miRNAs are present in mosquitoes. The full stem-loop precursor structure was detected in all four life stage libraries for miR-8, miR-9c, miR-100, miR-275, miR-277, miR-317, miR-1000 and miR-1890. In the majority of cases, mature miRNAs are more abundant than loop and star sequences. Additionally, miR-309 and miR-988 were not detected in the pupae and unfed adult females libraries, miR-iab-4 was not found in eggs and pupae libraries and miR-308 was not identified from the larvae library. These results indicate that the expression of some miRNAs are probably stage specific. It can be speculated that a miRNA may be involved in regulation of function and dysfunction, differentiation, growth and development of a specific stage [73].
The identification of novel miRNAs is an eminent and challenging problem for the understanding of post-transcriptional gene regulation. The characteristic hairpin structure of miRNA precursors can be used to predict novel miRNAs. With this feature and using miRDeep2, novel miRNAs were predicted by exploring the secondary structure, the Dicer cleavage site, and the minimum free energy of the unannotated small RNA tags that could be mapped to the An. funestus genome. Based on the analysis, 43 mature sequences were identified as novel miRNA candidates because they were not captured in any RNA database. The full precursor structure for some of these novel miRNAs was identified. Detection of the miRNA star is a strong clue, albeit not infallible, for the formation of precursor hairpin structures. This adds weight to the authenticity of the predicted candidates [74, 75]. However, the evolution and function of the star miRNAs remains unclear. Two studies proposed that these star miRNAs might differ from their sense partners by acting on different mRNA targets [76, 77].
The miRBase databases release 21 (June 2014) was searched for homologs to determine whether these novel miRNAs are conserved among other animal species. Some candidates are conserved in other insect species but not in the Anopheles genus, suggesting that these are insect-specific miRNAs. Among the newly identified Anopheles miRNAs, four mosquito specific miRNAs (miR-2940, miR-2941, miR-2942 and miR-2943) were detected [68]. This is the first description of miR-2941 and miR-2943 in the anopheline species using the high-throughput sequencing technology.
The NGS technology was sensitive enough to detect a new variant for miR-927 and miR-980, miRNAs found only in insects. These results are congruent with the existence of multiple variants for both miRNAs in other insects such as Drosophila [18]. A third stem-loop precursor for miR-2 was also detected which produces different mature miR-2. The miR-2 family is widespread in invertebrates, and it is the largest family of miRNAs in the model species Drosophila melanogaster (8 family members), Capitella teleta (7 family members), Apis mellifera (6 family members), Bombyx mori (5 members) [20].
Counting redundant miRNA reads revealed that expression varies significantly among different miRNAs. In the three pre-adult stages, insect-specific miRNA (miR-263a) was found to be the most abundant miRNA. However, miR-8 was the common miRNA in the unfed adult females library. Furthermore, the four libraries shared five out of the top ten most frequently occurring miRNAs: miR-263a, miR-10, miR-184, bantam and miR-8. In Drosophila, miRNA miR-263a confers robustness during development by protecting nascent sense organs from apoptosis [9]. Moreover, a recent study on Nilaparvata lugens reported that miR-263a was found as high abundant miRNAs in the last instar female nymph females [78]. Both miR-8 and miR-184 were reported in the embryos of Drosophila [79, 80], mosquitoes [68, 81], silk worm [82], Schistosoma japonicum [83], zebrafish [84], Asian seabass[85], mouse [86–88] and humans [15, 89]. This suggest a conservative developmental function for these two miRNAs across different animals. High number of reads for miR-2941, miR-2943 miR-2944a, and miR-2944b in the eggs library were also observed, which indicate an embryonic roles for these insect specific miRNAs. High expression for miR-2941 and miR-2943 in the embryonic stage was reported previously [53, 90].
The expression of miRNAs varies across different developmental stages [82, 91]. In this study, bantam, miR-8, miR-31 and miR-278 were among the up-regulated miRNAs between the egg and larva stage. In D. melanogaster, overexpression of bantam induces tissue overgrowth. Related to growth, and also in D. melanogaster, miR-8 and miR-278 have been implicated in insulin receptor signaling , thus contributing to regulation of the energy balance [6]. Embryos depleted for miR-31 can complete development, but were affected by severe segmentation defects [92]. In the gregarious phase of locust, canonical miRNAs were expressed at levels between 1.5- and 2-fold higher than in the solitary phase. The most prominent differences were found in miR-276, miR-125, and miR-315 [34]. Interestingly, the same change in these miRNAs between the egg and the larva stage except for miR-315 were observed. As is known, the genes that encode for miRNA are distributed across the chromosome either individually, or in clusters in which two or more miRNA genes are located within a short distance on the same segment of a chromosome [93, 94]. Therefore, it is assumed that miRNA genes located in a gene cluster are first transcribed as a single primary transcript that is subsequently processed to generate the individual miRNAs [7]. In the larvae library, very high relative expression levels of the let-7-complex locus (miR-125, let-7 and miR-100), the cluster of the two mosquito-specifc miRNAs; miR-1174 and miR-1175, miR-278 and miR-307, miR-305 and miR-275, miR-210 and miR-927 were observed. Down-regulation of the miR-92 cluster (miR-92a and miR-92b) and miR-309 cluster (miR-309 and miR-286) were also observed. Similarly in Drosophila, miR-309 and miR-286 (which was processed from a single 1.5 kb primary transcript) displayed a dynamic pattern of expression in the early embryo [95].
Between the larva and the pupa, miR-193, miR-277 and miR-1890 were significantly up-regulated. miR-34, miR-308, miR-375, miR-1175 and and miR-2942 were down-regulated in the pupa after increasing in the larva stage. Again, down-regulation of the miR-92a cluster was noticed. The expression of the members of this cluster has been related to the embryonic development in Ae. aegypti [68] and B. mori [69].
Among the up-regulated miRNA between the pupa and the unfed adult female is miR-1891 the mosquito-specific miRNA which displayed changes in its expression levels in the unfed adult females library, suggesting a significant role for this miRNA during development beside its function in the host response to infection [14]. In the unfed adult females library, up-regulation of miR-34 was noticed compared to significant down-regulation in the pupae library, and a major change in miR-989 expression for the first time. The expression of these miRNAs has been studied in different fly species, and results have shown that miR-989 expression is restricted to females, and predominantly to the ovary in An. anthropophagus, An. stephensi, Ae. aegypti and Drosophila [52, 53, 68, 96], although it was later detected in the midgut of An. gambiae [7]. Both miRNAs (miR-34 and miR-989) with two other miRNAs displayed changes in the expression levels during Plasmodium parasite invasion [7]. For unknown reasons, some miRNAs such as miR-87 and the arthropod-specific miR-929 showed low read counts in the four life stage libraries, suggesting that miRNAs might be involved in other process but not development. Further studies are needed to reveal the function of these miRNAs.
The predicted novel miRNAs exhibited much lower expression levels, consistent with the evidence that non-conserved miRNAs are often expressed at a lower level than conserved miRNAs [97–99].
It is important to note that the expression profile analysis in this study are based only on the sequence read counts and required further experimental validation. However, this result is a key step towards improving our understanding of the complexity and regulation mode of miRNAs in mosquitoes. Changes in the expression profiles were noted in all stages indicating a role for these small RNAs in the mosquito maturation. Knockdown or blocking the biogenesis pathway of one of these miRNAs may limit the mosquito’s development at a crucial stage thereby leading to novel approaches to combat this mosquito in the early development stages.
Conclusion
In this study, 107 mature miRNA sequences in the four developing stages (eggs, larvae, pupae, and unfed adult females) of An. funestus were identified, one of the most prevalent malaria vector on the African continent using the high throughput sequencing and described their expression patterns during development.
Authors’ contributions
MA conducted the experiments and data analyses, interpreted results, and drafted the first version of the manuscript. BLS and HA reared the mosquitoes and carried out the RNA isolation. DM performed the statistical analysis. LLK provided technical support, supervised the experiments and contributed to the editing of the manuscript. AC conceived the project, coordinated the study, provided funding for the study and helped with revision of the manuscript. All authors read and commented on the final manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and material
The sequencing reads are available in ftp://ftp.sanbi.ac.za/anopheles funestus_miRNA_sequencing_reads. All miRNA sequences identified in this study are provided in the manuscript.
Funding
This work was supported by the South African Research Chairs Initiative of the Department of Science and Technology and National Research Foundation of South Africa.
Contributor Information
Mushal Allam, Email: mushalallam@gmail.com.
Belinda L. Spillings, Email: belinda@whitesci.co.za
Hiba Abdalla, Email: hibabakri@gmail.com.
Darlington Mapiye, Email: darlingtonmapiye@gmail.com.
Lizette L. Koekemoer, Email: lizettek@nicd.ac.za
Alan Christoffels, Email: alan@sanbi.ac.za.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 3.Macdonald PM, Struhl G. A molecular gradient in early Drosophila embryos and its role in specifying the body pattern. Nature. 1986;324:537–545. doi: 10.1038/324537a0. [DOI] [PubMed] [Google Scholar]
- 4.Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 5.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/S0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 6.Teleman AA, Maitra S, Cohen SM. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev. 2006;20:417–422. doi: 10.1101/gad.374406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winter F, Edaye S, Hüttenhofer A, Brunel C. Anopheles gambiae miRNAs as actors of defence reaction against Plasmodium invasion. Nucleic Acids Res. 2007;35:6953–6962. doi: 10.1093/nar/gkm686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Zhou X, Ge X, Jiang J, Li M, Jia S, et al. Insect-specific microRNA involved in the development of the silkworm Bombyx mori. PLoS ONE. 2009;4:e4677. doi: 10.1371/journal.pone.0004677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilgers V, Bushati N, Cohen SM. Drosophila microRNAs 263a/b confer robustness during development by protecting nascent sense organs from apoptosis. PLoS Biol. 2010;8:e1000396. doi: 10.1371/journal.pbio.1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hussain M, Asgari S. Functional analysis of a cellular microRNA in insect host-ascovirus interaction. J Virol. 2010;84:612–620. doi: 10.1128/JVI.01794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skalsky RL, Vanlandingham DL, Scholle F, Higgs S, Cullen BR. Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus. BMC Genom. 2010;11:119. doi: 10.1186/1471-2164-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeiner GM, Norman KL, Thomson JM, Hammond SM, Boothroyd JC. Toxoplasma gondii infection specifically increases the levels of key host microRNAs. PLoS ONE. 2010;5:e8742. doi: 10.1371/journal.pone.0008742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dkhil M, Abdel-Baki AA, Delić D, Wunderlich F, Sies H, Al-Quraishy S. Eimeria papillata: upregulation of specific miRNA-species in the mouse jejunum. Exp Parasitol. 2011;127:581–586. doi: 10.1016/j.exppara.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Hussain M, Frentiu FD, Moreira LA, O’Neill SL, Asgari S. Wolbachia uses host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc Natl Acad Sci USA. 2011;108:9250–9255. doi: 10.1073/pnas.1105469108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vallejo DM, Caparros E, Dominguez M. Targeting notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells. EMBO J. 2011;30:756–769. doi: 10.1038/emboj.2010.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marco A, Hooks K, Griffiths-Jones S. Evolution and function of the extended miR-2 microRNA family. RNA Biol. 2012;9:242–248. doi: 10.4161/rna.19160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi IK, Hyun S. Conserved microRNA miR-8 in fat body regulates innate immune homeostasis in Drosophila. Dev Comp Immunol. 2012;37:50–54. doi: 10.1016/j.dci.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 20.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang WC, Lin FM, Chang WC, Lin KY, Huang HD, Lin NS. miRExpress: analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinform. 2009;10:328. doi: 10.1186/1471-2105-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 24.Creighton CJ, Benham AL, Zhu H, Khan MF, Reid JG, Nagaraja AK, et al. Discovery of novel microRNAs in female reproductive tract using next generation sequencing. PLoS ONE. 2010;5:e9637. doi: 10.1371/journal.pone.0009637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung CH, Hansen MA, Makunin IV, Korbie DJ, Mattick JS. Identification of novel non-coding RNAs using profiles of short sequence reads from next generation sequencing data. BMC Genom. 2010;11:77. doi: 10.1186/1471-2164-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hackl M, Jakobi T, Blom J, Doppmeier D, Brinkrolf K, Szczepanowski R, et al. Next-generation sequencing of the Chinese hamster ovary microRNA transcriptome: identification, annotation and profiling of microRNAs as targets for cellular engineering. J Biotechnol. 2011;153:62–75. doi: 10.1016/j.jbiotec.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Havecker ER. Detection of small RNAs and microRNAs using deep sequencing technology. Methods Mol Biol. 2011;732:55–68. doi: 10.1007/978-1-61779-083-6_5. [DOI] [PubMed] [Google Scholar]
- 28.Keller A, Backes C, Leidinger P, Kefer N, Boisguerin V, Barbacioru C, et al. Next-generation sequencing identifies novel microRNAs in peripheral blood of lung cancer patients. Mol Biosyst. 2011;7:3187–3199. doi: 10.1039/c1mb05353a. [DOI] [PubMed] [Google Scholar]
- 29.Kong BW. Identification of virus encoding microRNAs using 454 FLX sequencing platform. Methods Mol Biol. 2011;733:81–91. doi: 10.1007/978-1-61779-089-8_6. [DOI] [PubMed] [Google Scholar]
- 30.Persson H, Kvist A, Rego N, Staaf J, Vallon-Christersson J, Luts L, et al. Identification of new microRNAs in paired normal and tumor breast tissue suggests a dual role for the ERBB2/Her2 gene. Cancer Res. 2011;71:78–86. doi: 10.1158/0008-5472.CAN-10-1869. [DOI] [PubMed] [Google Scholar]
- 31.Gébelin V, Argout X, Engchuan W, Pitollat B, Duan C, Montoro P, et al. Identification of novel microRNAs in Hevea brasiliensis and computational prediction of their targets. BMC Plant Biol. 2012;12:18. doi: 10.1186/1471-2229-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gunaratne PH, Coarfa C, Soibam B, Tandon A. miRNA data analysis: next-gen sequencing. Methods Mol Biol. 2012;822:273–288. doi: 10.1007/978-1-61779-427-8_19. [DOI] [PubMed] [Google Scholar]
- 33.Pérez-Quintero ÁL, Quintero A, Urrego O, Vanegas P, López C. Bioinformatic identification of cassava miRNAs differentially expressed in response to infection by Xanthomonas axonopodis pv. manihotis. BMC Plant Biol. 2012;12:29. doi: 10.1186/1471-2229-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei Y, Chen S, Yang P, Ma Z, Kang L. Characterization and comparative profiling of the small RNA transcriptomes in two phases of locust. Genome Biol. 2009;10:R6. doi: 10.1186/gb-2009-10-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buchold GM, Coarfa C, Kim J, Milosavljevic A, Gunaratne PH, Matzuk MM. Analysis of microRNA expression in the prepubertal testis. PLoS ONE. 2010;5:e15317. doi: 10.1371/journal.pone.0015317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikopoulos K, Gilissen C, Hoischen A, van Nouhuys CE, Boonstra FN, Blokland EAW, et al. Next-generation sequencing of a 40 Mb linkage interval reveals TSPAN12 mutations in patients with familial exudative vitreoretinopathy. Am J Hum Genet. 2010;86:240–247. doi: 10.1016/j.ajhg.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borges F, Pereira PA, Slotkin RK, Martienssen RA, Becker JD. MicroRNA activity in the Arabidopsis male germline. J Exp Bot. 2011;62:1611–1620. doi: 10.1093/jxb/erq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krawetz SA, Kruger A, Lalancette C, Tagett R, Anton E, Draghici S, et al. A survey of small RNAs in human sperm. Hum Reprod. 2011;26:3401–3412. doi: 10.1093/humrep/der329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohorianu I, Schwach F, Jing R, Lopez-Gomollon S, Moxon S, Szittya G, et al. Profiling of short RNAs during fleshy fruit development reveals stage-specific sRNAome expression patterns. Plant J. 2011;67:232–246. doi: 10.1111/j.1365-313X.2011.04586.x. [DOI] [PubMed] [Google Scholar]
- 40.Wu Q, Lu Z, Li H, Lu J, Guo L, Ge Q. Next-generation sequencing of microRNAs for breast cancer detection. J Biomed Biotechnol. 2011;2011:597145. doi: 10.1155/2011/597145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boeri M, Pastorino U, Sozzi G. Role of microRNAs in lung cancer: microRNA signatures in cancer prognosis. Cancer J. 2012;18:268–274. doi: 10.1097/PPO.0b013e318258b743. [DOI] [PubMed] [Google Scholar]
- 42.Cutting AD, Bannister SC, Doran TJ, Sinclair AH, Tizard MVL, Smith CA. The potential role of microRNAs in regulating gonadal sex differentiation in the chicken embryo. Chromosome Res. 2012;20:201–213. doi: 10.1007/s10577-011-9263-y. [DOI] [PubMed] [Google Scholar]
- 43.Gilabert-Estelles J, Braza-Boils A, Ramon LA, Zorio E, Medina P, Espana F, et al. Role of microRNAs in gynecological pathology. Curr Med Chem. 2012;19:2406–2413. doi: 10.2174/092986712800269362. [DOI] [PubMed] [Google Scholar]
- 44.Kang M, Zhao Q, Zhu D, Yu J. Characterization of microRNAs expression during maize seed development. BMC Genom. 2012;13:360. doi: 10.1186/1471-2164-13-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leidner RS, Ravi L, Leahy P, Chen Y, Bednarchik B, Streppel M, et al. The microRNAs, MiR-31 and MiR-375, as candidate markers in Barrett’s esophageal carcinogenesis. Genes Chromosomes Cancer. 2012;51:473–479. doi: 10.1002/gcc.21934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu F, Peng W, Li Z, Li W, Li L, Pan J, et al. Next-generation small RNA sequencing for microRNAs profiling in Apis mellifera: comparison between nurses and foragers. Insect Mol Biol. 2012;21:297–303. doi: 10.1111/j.1365-2583.2012.01135.x. [DOI] [PubMed] [Google Scholar]
- 47.Nikaki A, Piperi C, Papavassiliou AG. Role of microRNAs in gliomagenesis: targeting miRNAs in glioblastoma multiforme therapy. Expert Opin Investig Drugs. 2012;21:1475–1488. doi: 10.1517/13543784.2012.710199. [DOI] [PubMed] [Google Scholar]
- 48.Shamimuzzaman M, Vodkin L. Identification of soybean seed developmental stage-specific and tissue-specific miRNA targets by degradome sequencing. BMC Genom. 2012;13:310. doi: 10.1186/1471-2164-13-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei C, Salichos L, Wittgrove CM, Rokas A, Patton JG. Transcriptome-wide analysis of small RNA expression in early zebrafish development. RNA. 2012;18:915–929. doi: 10.1261/rna.029090.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao MJ, Chen G, Zhao PP, Lu MH, Jian J, Liu MF, et al. Transcriptome analysis of microRNAs in developing cerebral cortex of rat. BMC Genom. 2012;13:232. doi: 10.1186/1471-2164-13-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kiszewski A, Mellinger A, Spielman A, Malaney P, Sachs SE, Sachs J. A global index representing the stability of malaria transmission. Am J Trop Med Hyg. 2004;70:486–498. [PubMed] [Google Scholar]
- 52.Mead EA, Tu Z. Cloning, characterization, and expression of microRNAs from the Asian malaria mosquito Anopheles stephensi. BMC Genom. 2008;9:244. doi: 10.1186/1471-2164-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu W, Huang H, Xing C, Li C, Tan F, Liang S. Identification and characterization of the expression profile of microRNAs in Anopheles anthropophagus. Parasit Vectors. 2014;7:159. doi: 10.1186/1756-3305-7-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiff CJ, Minjas JN, Hall T, Hunt RH, Lyimo S, Davis JR. Malaria infection potential of anopheline mosquitoes sampled by light trapping indoors in coastal Tanzanian villages. Med Vet Entomol. 1995;9:256–262. doi: 10.1111/j.1365-2915.1995.tb00131.x. [DOI] [PubMed] [Google Scholar]
- 55.Hargreaves K, Koekemoer LL, Brooke BD, Hunt RH, Mthembu J, Coetzee M. Anopheles funestus resistant to pyrethroid insecticides in South Africa. Med Vet Entomol. 2000;14:181–189. doi: 10.1046/j.1365-2915.2000.00234.x. [DOI] [PubMed] [Google Scholar]
- 56.Kamau L, Koekemoer LL, Hunt RH, Coetzee M. Anopheles parensis: the main member of the Anopheles funestus species group found resting inside human dwellings in Mwea area of central Kenya toward the end of the rainy season. J Am Mosq Control Assoc. 2003;19:130–133. [PubMed] [Google Scholar]
- 57.Coetzee M, Fontenille D. Advances in the study of Anopheles funestus, a major vector of malaria in Africa. Insect Biochem Mol Biol. 2004;34:599–605. doi: 10.1016/j.ibmb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 58.Ayala D, Caro-Riaño H, Dujardin JP, Rahola N, Simard F, Fontenille D. Chromosomal and environmental determinants of morphometric variation in natural populations of the malaria vector Anopheles funestus in Cameroon. Infect Genet Evol. 2011;11:940–7. doi: 10.1016/j.meegid.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hunt RH, Brooke BD, Pillay C, Koekemoer LL, Coetzee M. Laboratory selection for and characteristics of pyrethroid resistance in the malaria vector Anopheles funestus. Med Vet Entomol. 2005;19:271–275. doi: 10.1111/j.1365-2915.2005.00574.x. [DOI] [PubMed] [Google Scholar]
- 60.FastQC. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 61.FASTX-Toolkit. http://hannonlab.cshl.edu/fastx_toolkit/.
- 62.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res. 2011;39:D141–D145. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer S, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatshefte f Chemie. 1994;125:167–188. doi: 10.1007/BF00818163. [DOI] [Google Scholar]
- 65.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred I. accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 66.The invertebrate vectors of human pathogens database. https://www.vectorbase.org. [DOI] [PMC free article] [PubMed]
- 67.Tian G, Yin X, Luo H, Xu X, Bolund L, Zhang X, et al. Sequencing bias: comparison of different protocols of microRNA library construction. BMC Biotechnol. 2010;10:64. doi: 10.1186/1472-6750-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li S, Mead EA, Liang S, Tu Z. Direct sequencing and expression analysis of a large number of miRNAs in Aedes aegypti and a multi-species survey of novel mosquito miRNAs. BMC Genomics. 2009;10:581. doi: 10.1186/1471-2164-10-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu S, Li D, Li Q, Zhao P, Xiang Z, Xia Q. MicroRNAs of Bombyx mori identified by Solexa sequencing. BMC Genom. 2010;11:148. doi: 10.1186/1471-2164-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cordero F, Beccuti M, Arigoni M, Donatelli S, Calogero RA. Optimizing a massive parallel sequencing workflow for quantitative miRNA expression analysis. PLoS ONE. 2012;7:e31630. doi: 10.1371/journal.pone.0031630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oshlack A, Wakefield MJ. Transcript length bias in RNA-seq data confounds systems biology. Biol Direct. 2009;4:14. doi: 10.1186/1745-6150-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pelaez P, Trejo MS, Iniguez LP, Estrada-Navarrete G, Covarrubias AA, Reyes JL, et al. Identification and characterization of microRNAs in Phaseolus vulgaris by high-throughput sequencing. BMC Genom. 2012;13:83. doi: 10.1186/1471-2164-13-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang Y, Liu D, Zhang L, Ingvarsson S, Chen H. Quantitative analysis of miRNA expression in seven human foetal and adult organs. PLoS ONE. 2011;6:e28730. doi: 10.1371/journal.pone.0028730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of miRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu JK. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 2008;8:25. doi: 10.1186/1471-2229-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang B, Pan X, Stellwag EJ. Identification of soybean microRNAs and their targets. Planta. 2008;229:161–182. doi: 10.1007/s00425-008-0818-x. [DOI] [PubMed] [Google Scholar]
- 77.Chi X, Yang Q, Chen X, Wang J, Pan L, Chen M, et al. Identification and characterization of microRNAs from peanut (Arachis hypogaea L.) by high-throughput sequencing. PLoS ONE. 2011;6:e27530. doi: 10.1371/journal.pone.0027530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen Q, Lu L, Hua H, Zhou F, Lu L, Lin Y. Characterization and comparative analysis of small RNAs in three small RNA libraries of the brown planthopper (Nilaparvata lugens) PLoS ONE. 2012;7:e32860. doi: 10.1371/journal.pone.0032860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li P, Peng J, Hu J, Xu Z, Xie W, Yuan L. Localized expression pattern of miR-184 in Drosophila. Mol Biol Rep. 2011;38:355–358. doi: 10.1007/s11033-010-0115-1. [DOI] [PubMed] [Google Scholar]
- 80.Iovino N, Pane A, Gaul U. miR-184 has multiple roles in Drosophila female germline development. Dev Cell. 2009;17:123–133. doi: 10.1016/j.devcel.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 81.Zheng Pm Wu, Jb Jy, Gu, Zj Tu, Xg Chen. Isolation, identification and analysis of the expression profile of miRNAs in Aedes albopictus. Nan Fang Yi Ke Da Xue Xue Bao. 2010;30:677–680. [PubMed] [Google Scholar]
- 82.Liu S, Zhang L, Li Q, Zhao P, Duan J, Cheng D, et al. MicroRNA expression profiling during the life cycle of the silkworm (Bombyx mori) BMC Genom. 2009;10:455. doi: 10.1186/1471-2164-10-455. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 83.Huang J, Hao P, Chen H, Hu W, Yan Q, Liu F, et al. Genome-wide identification of Schistosoma japonicum microRNAs using a deep-sequencing approach. PLoS ONE. 2009;4:e8206. doi: 10.1371/journal.pone.0008206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Flynt AS, Thatcher EJ, Burkewitz K, Li N, Liu Y, Patton JG. miR-8 microRNAs regulate the response to osmotic stress in zebrafish embryos. J Cell Biol. 2009;185:115–127. doi: 10.1083/jcb.200807026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xia JH, He XP, Bai ZY, Yue GH. Identification and characterization of 63 MicroRNAs in the Asian seabass Lates calcarifer. PLoS ONE. 2011;6:e17537. doi: 10.1371/journal.pone.0017537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wenguang Z, Jianghong W, Jinquan L, Yashizawa M. A subset of skin-expressed microRNAs with possible roles in goat and sheep hair growth based on expression profiling of mammalian microRNAs. OMICS. 2007;11:385–396. doi: 10.1089/omi.2006.0031. [DOI] [PubMed] [Google Scholar]
- 87.Juhila J, Sipilä T, Icay K, Nicorici D, Ellonen P, Kallio A, et al. MicroRNA expression profiling reveals miRNA families regulating specific biological pathways in mouse frontal cortex and hippocampus. PLoS ONE. 2011;6:e21495. doi: 10.1371/journal.pone.0021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu J, Bao J, Wang L, Hu Y, Xu C. MicroRNA-184 downregulates nuclear receptor corepressor 2 in mouse spermatogenesis. BMC Dev Biol. 2011;11:64. doi: 10.1186/1471-213X-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, et al. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell. 2009;139:1096–1108. doi: 10.1016/j.cell.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 90.Gu J, Hu W, Wu J, Zheng P, Chen M, James AA, et al. miRNA genes of an invasive vector mosquito Aedes albopictus. PLoS ONE. 2013;8:e67638. doi: 10.1371/journal.pone.0067638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu H, Wang X, Du Z, Li N. Identification of microRNAs from different tissues of chicken embryo and adult chicken. FEBS Lett. 2006;580:3610–3616. doi: 10.1016/j.febslet.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 92.Leaman D, Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, et al. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 2005;121:1097–1108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 93.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, et al. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang X, Azhar G, Wei JY. The expression of microRNA and microRNA clusters in the aging heart. PLoS ONE. 2012;7:e34688. doi: 10.1371/journal.pone.0034688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Biemar F, Zinzen R, Ronshaugen M, Sementchenko V, Manak JR, Levine MS. Spatial regulation of microRNA gene expression in the Drosophila embryo. Proc Natl Acad Sci USA. 2005;102:15907–15911. doi: 10.1073/pnas.0507817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kugler JM, Verma P, Chen YW, Weng R, Cohen SM. miR-989 is required for border cell migration in the Drosophila ovary. PLoS ONE. 2013;8:e67075. doi: 10.1371/journal.pone.0067075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ling KH, Brautigan PJ, Hahn CN, Daish T, Rayner JR, Cheah PS, et al. Deep sequencing analysis of the developing mouse brain reveals a novel microRNA. BMC Genom. 2011;12:176. doi: 10.1186/1471-2164-12-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Inukai S, de Lencastre A, Turner M, Slack F. Novel MicroRNAs differentially expressed during aging in the mouse brain. PLoS ONE. 2012;7:e40028. doi: 10.1371/journal.pone.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang JZ, Ai XY, Guo WW, Peng SA, Deng XX, Hu CG. Identification of miRNAs and their target genes using deep sequencing and degradome analysis in trifoliate orange [Poncirus trifoliate (L.) Raf] Mol Biotechnol. 2012;51:44–57. doi: 10.1007/s12033-011-9439-x. [DOI] [PubMed] [Google Scholar]
- 100.Castellano L, Rizzi E, Krell J, Di Cristina M, Galizi R, Mori A, et al. The germline of the malaria mosquito produces abundant miRNAs, endo-siRNAs, piRNAs and 29-nt small RNAs. BMC Genom. 2015;16:100. doi: 10.1186/s12864-015-1257-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


