Abstract
Numerous studies have demonstrated that interferon-γ (IFN-γ) is an important inflammatory cytokine, which may activate the immunomodulatory abilities of mesenchymal stem cells (MSCs), and may influence certain other functions of these cells. MicroRNAs are small non-coding RNAs that regulate the majority of the biological functions of cells and are important in a variety of biological processes. However, few studies have been performed to investigate whether IFN-γ affects the microRNA profile of MSCs. The aim of the present study was to analyze the microRNA profile of MSCs derived from the umbilical cord (UC-MSCs) cultured in the presence or absence of IFN-γ (IFN-UC-MSCs). An array that detects 754 microRNAs was used to determine the expression profiles. Statistical analysis of the array data revealed that 8 microRNAs were significantly differentially expressed in UC-MSCs and IFN-UC-MSCs. Reverse transcription-quantitative polymerase chain reaction validated the differential expression of the 8 identified microRNAs. The target genes of the 8 microRNAs were predicted through two online databases, TargetScan and miRanda, and the predicted results were screened by bioinformatics analysis. The majority of the target genes were involved in the regulation of transcription, signal transduction, proliferation, differentiation and migration. These results may provide insight into the mechanism underlying the regulation of the biological functions of MSCs by IFN-γ, in particular the immunomodulatory activity.
Keywords: umbilical cord-derived mesenchymal stem cells, interferon-γ, microRNA array
Introduction
Mesenchymal stem cells (MSCs) are multipotent stem cells with the capacity for self-renewal and multi-lineage differentiation. They are obtained from various tissues, including the human umbilical cord. The isolation and culture of human umbilical cord-derived MSCs (UC-MSCs) has been previously demonstrated by Lu et al (1).
MSCs are promising pluripotent cells for use in regenerative medicine due to their biological characteristics, in particular their immunomodulatory ability (2,3). Although the underlying immunomodulatory mechanism of MSCs remains to be fully elucidated, it is commonly thought that immunomodulation of MSCs is controlled by inflammation (4). Interferon-γ (IFN-γ) is a critical inflammatory cytokine, which may activate the immunomodulation of MSCs, and is involved in inflammation and autoimmune disease (5). Krampera et al (6) demonstrated that following MSC co-culture with IFN-γ-secreting CD4+ T cells, CD8+ T cells or natural killer cells, MSCs inhibited the proliferation of these cells. Following MSC co-culture with non-IFN-γ-secreting B cells and T helper 2 cells, there was minimal inhibition of proliferation. IFN-γ was demonstrated to induce the secretion of prostaglandin E2, indoleamine 2,3-dioxygenase (IDO1), hepatocyte growth factor and transforming growth factor-β1 (TGF-β) in MSCs (5,6). Notably, IDO1 exhibits important immunomodulatory properties (7). The IDO1 level in MSCs increased significantly in mixed lymphocyte reaction cultures compared with normal culture conditions, resulting in consumption of tryptophan and the accumulation of kynurenine in the medium, thus inhibiting the proliferation of lymphocytes (8). In addition, IFN-γ may have other effects on MSCs. The tryptophan depletion mediated by IDO1 inhibited the proliferation of MSCs in serum-free medium containing IFN-γ. However, in the presence of serum containing an IDO1 inhibitor, 1-methyl-tryptophan, proliferation of MSCs was not markedly inhibited by IFN-γ. The mechanism underlying the inhibition of MSC proliferation by IFN-γ remains to be fully elucidated (9). Furthermore, IFN-γ may suppress the differentiation of MSCs. For example, IFN-γ affected adipogenesis and osteogenesis of MSCs by inhibiting adipsin, adipoQ and secreted phosphoprotein 1, integrin-binding sialoprotein expression levels (10). These inhibitory effects were verified by in vivo experiments; however, the underlying mechanisms remain unclear. IFN-γ may also influence the apoptosis, phenotype and chemotactic factors of MSCs (11,12).
MicroRNAs (miRNAs) are small, non-coding RNAs, typically ~22 nucleotides in length. They regulate gene expression by degrading mRNA or inhibiting mRNA translation (13). miRNAs are involved in the regulation of diverse cellular processes, including apoptosis, proliferation, differentiation and metabolism (14,15). Previous studies have confirmed that miRNAs contribute to the regulation of the functions of MSCs, including chondrogenesis, adipogenic differentiation, osteogenic differentiation, proliferation and senescence (16,17). For example, miR-449a may recognize and inhibit the expression of lymphoid enhancer-binding factor-1, resulting in regulation of the chondrogenesis of human MSCs (18). In addition, numerous other miRNAs are involved in the regulation of MSCs, including miR-193, miR-138, miR-486-5p and miR-22 (19). Although numerous studies have demonstrated that miRNAs are key regulators of the majority of the biological processes of MSCs, the miRNAs involved in the regulation of MSCs by IFN-γ have rarely been systematically studied (20).
In the present study, UC-MSCs were incubated with IFN-γ (IFN-UC-MSCs). The miRNA expression profiles of UC-MSCs and IFN-UC-MSCs were subsequently analyzed using miRNA arrays. The results of the present study demonstrated that there were significant differences between the miRNA profiles in UC-MSCs and IFN-UC-MSCs, and that various miRNAs may be involved in the regulation of MSCs by IFN-γ.
Materials and methods
Ethics statement
This study was performed in accordance with the ethical standards of the institutional review board on human experimentation, the Institutional Review Board of the Institute of Hematology & Blood Diseases Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College (Tianjin, China), and was approved by this review board. Human umbilical cords (n=11) were obtained from healthy donors with written informed consent.
Generation of UC-MSCs
UC-MSCs were isolated and expanded from human umbilical cords as described previously (1). UC-MSCs were cultured in Dulbecco's modified Eagle medium (DMEM) with nutrient mixture F-12 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal calf serum (FCS; HyClone; GE Healthcare Life Sciences, Logan, UT, USA), 100 U/ml penicillin (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany), 100 µg/ml streptomycin (Sigma-Aldrich; Merck Millipore), 2 mM glutamine and 10 ng/ml epidermal growth factor (PeproTech, Inc., Rocky Hill, NJ, USA). This mixture medium is referred to as culture medium. Cells were cultured at 37°C in a humidified atmosphere containing 5% carbon dioxide in a Forma™ Steri-Cycle™ CO2 Incubator (Thermo Fisher Scientific, Inc.). UC-MSCs were stained with rhodamine phalloidin and DAPI and observed under a confocal microscope.
Immunophenotyping of UC-MSCs
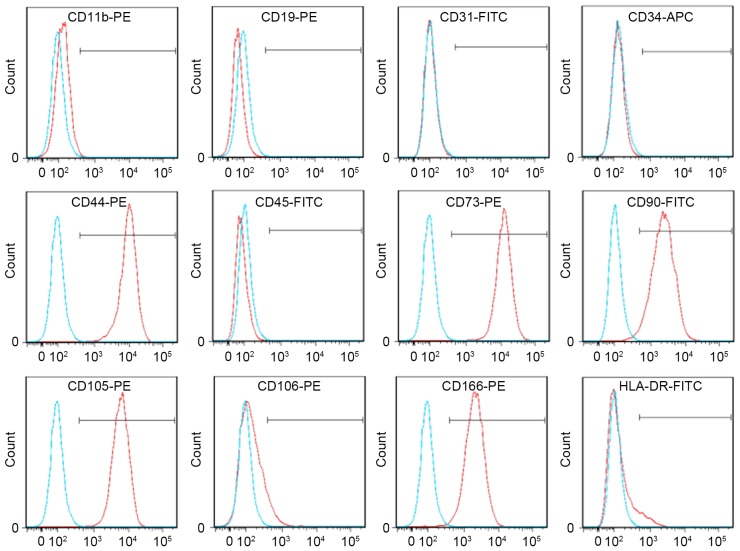
UC-MSCs were harvested and placed into tubes at a density of 1×105 cells/tube. Cells were washed twice in phosphate-buffered saline (PBS) by centrifugation at 180 × g for 5 min at room temperature, and labeled with the following antibodies (10 µl/tube) purchased from BD Pharmingen (San Diego, CA, USA) for 1 h at 4°C: Mouse anti-cluster of differentiation (CD) 11b-phycoerythrin (PE; catalog no. 555388), mouse anti-CD19-PE (catalog no. 555413), mouse anti-CD31-fluorescein isothiocyanate (FITC; catalog no. 555445), mouse anti-CD34-allophycocyanin (APC; catalog no. 555824), mouse anti-CD44-PE (catalog no. 555479), mouse anti-CD45-FITC (catalog no. 555482), mouse anti-CD73-PE (catalog no. 550257), mouse anti-CD90-FITC (catalog no. 555595), mouse anti-CD105-PE (catalog no. 560839), mouse anti-CD106-PE (catalog no. 555647), mouse anti-CD166-PE (catalog no. 559263) and human leukocyte antigen D related (HLA-DR)-FITC (catalog no. 555811). FITC-(catalog no. 555748), APC-(catalog no. 555751) or PE-conjugated (catalog no. 555749) isotype control antibodies served as controls. Following incubation, UC-MSCs were washed with PBS, resuspended in 2% paraformaldehyde and analyzed on a BD™ LSR II flow cytometer (BD Biosciences) using BD FACSDiva™ software version 6.1.3 (BD Biosciences).
Osteogenic differentiation
UC-MSCs were cultured in 24-well cell culture plates (Corning Incorporated, Corning, NY, USA) until they reached 60% confluence. Osteogenic medium was then used to culture UC-MSCs for three weeks. Osteogenic medium comprised low-glucose (LG)-DMEM (Gibco; Thermo Fisher Scientific, Inc.), 10% FCS, 0.1 µM dexamethasone (Sigma-Aldrich; Merck Millipore), 0.2 µM ascorbic acid-2-phosphate (AsA; Sigma-Aldrich; Merck Millipore) and 10 mM glycerol-2-phosphate (Sigma-Aldrich; Merck Millipore).
Adipogenic differentiation
UC-MSCs were cultured in 24-well cell culture plates until they reached 60% confluence. Adipogenic medium was then used to culture UC-MSCs for three weeks. Adipogenic medium comprised LG-DMEM (Gibco; Thermo Fisher Scientific, Inc.), 10% FCS, 1 µM dexamethasone, 0.5 µM 3-isobutyl-1-methylxanthine (Sigma-Aldrich; Merck Millipore), 10 µg/ml insulin (Sigma-Aldrich; Merck Millipore) and 100 µM indomethacin (Sigma-Aldrich; Merck Millipore).
Chondrogenic differentiation
UC-MSCs were transferred into 15-ml centrifuge tubes (Corning Incorporated) at 5×105 cells/tube and centrifuged at 110 × g for 3 min at room temperature. The pelleted micromass of UC-MSCs formed at the bottom of each tube was treated with chondrogenic medium for three weeks. Chondrogenic medium comprised high-glucose (HG)-DMEM (Gibco; Thermo Fisher Scientific, Inc.), 0.1 µM dexamethasone, 50 µg/ml AsA, 100 µg/ml sodium pyruvate (Lonza Group, Basel, Switzerland), 40 µg/ml proline (Sigma-Aldrich; Merck Millipore), 10 ng/ml TGF-β1 (PeproTech, Inc.) and 1% insulin-transferrin-selenium-A (Gibco; Thermo Fisher Scientific, Inc.).
Cellular staining
UC-MSCs were stained with Von Kossa reagents to evaluate mineralized matrix formation following osteogenic differentiation, and were stained with oil-red O to evaluate lipid droplets following adipogenic differentiation. Osteogenic and adipogenic differentiation were observed under an Olympus IX71-22-000–2 (Olympus Corporation, Tokyo, Japan) inverted microscope. The cellular pellets obtained from the chondrogenic differentiation assay were fixed in 2% paraformaldehyde for 24 h, embedded in optimal cutting temperature compound (Thermo Fisher Scientific, Inc.) and frozen at −80°C. Samples were sectioned into 6 µm-thick slices on a cryostat and stained with Alcian blue 8GX (Sigma-Aldrich; Merck Millipore) to evaluate sulfated proteoglycans. The analysis of chondrogenic differentiation was performed using a Nikon Eclipse 80i microscope (Nikon Corporation, Tokyo, Japan).
IFN-γ treatment
UC-MSCs obtained from one donor were seeded in two T75 culture flasks (Corning Incorporated) in culture medium at a density of 2×106 cells per flask. A total of 24 h later, the culture medium was replaced, with one flask receiving fresh medium and the other ‘IFN-γ-medium’, in which 30 ng/ml recombinant IFN-γ (PeproTech, Inc.) was added to the culture medium. Cells were cultured for 24 h (5).
RNA extraction
Total RNA was extracted with mirVana™ miRNA Isolation kit (Ambion; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The quality and quantity of the total RNA was assessed by NanoDrop™ 2000 spectrophotometry (Thermo Fisher Scientific, Inc.).
miRNA array
TaqMan Array Human MicroRNA A+B Cards Set version 3.0 was purchased from Applied Biosystems (Thermo Fisher Scientific, Inc.). This two-card set includes Card A, which focuses on more highly characterized miRNAs and Card B, containing more recently discovered miRNAs. This makes a total of 754 unique assays specific to human miRNAs, and each card contains four control assays (three carefully selected candidate endogenous control assays and one negative control assay). The miRNA array was performed by Invitrogen (Thermo Fisher Scientific, Inc.; cnservice.invitrogen.com). Two assays were performed in parallel, using UC-MSCs obtained from two separate donors.
Validation of miRNA expression levels by reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
UC-MSCs obtained from five donors were cultured and treated with IFN-γ. RNA was extracted and assessed as described above. Total RNA (500 ng) was used to generate the single-stranded cDNA of a specific regulated miRNA using the TaqMan™ MicroRNA RT kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) and the miRNA-specific stem-loop RT primer according to the manufacturer's instructions. The primers used were as follows: hsa-miR-7-5p stem-loop RT primer, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAACAA-3′; hsa-miR-34a-3p stem-loop RT primer, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGGGCAG-3′; hsa-miR-145-3p stem-loop RT primer, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGAACAG-3′; hsa-miR-301a-3p stem-loop RT primer, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCTTTGA-3′; hsa-miR-323a-3p stem-loop RT primer, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGAGGTC-3′; hsa-miR-331-5p stem-loop RT primer, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGATCCC-3′; hsa-miR-382-5p stem-loop RT primer, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCGAATCC-3′; hsa-miR-424-3p stem-loop RT primer, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACATAGCAG-3′. qPCR was performed with a 7900HT Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the SYBR™ Select Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.). The primers used were as follows: hsa-miR-7-5p forward, 5′-GGCGGTGGAAGACTAGTGATT-3′ (64 bp); hsa-miR-34a-3p forward, 5′-GGCGGCAATCAGCAAGTATAC-3′ (63 bp); hsa-miR-145-3p forward, 5′-GCGGGGATTCCTGGAAATAC-3′ (62 bp); hsa-miR-301a-3p forward, 5′-TTCGGCAGTGCAATAGTATTGTC-3′ (63 bp); hsa-miR-323a-3p forward, 5′-GCGGCACATTACACGGTCG-3′ (61 bp); hsa-miR-331-5p forward, 5′-CGGCTAGGTATGGTCCCAG-3′ (61 bp); hsa-miR-382-5p forward, 5′-TCGGGAAGTTGTTCGTGGTG-3′ (62 bp); hsa-miR-424-3p forward, 5′-ATTGGCAAAACGTGAGGCGC-3′ (62 bp); the common reverse primer, 5′-CAGTGCAGGGTCCGAGGTAT-3′. The RT-qPCR data were normalized using the ΔΔCq method (21) and the endogenous reference gene, U6. The U6 primers were as follows: Forward, 5′-CTCGCTTCGGCAGCACA-3′; reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
Statistical and bioinformatics analysis
Fold-change was used to identify the miRNAs that were significantly differentially expressed. miRNA information was obtained by referencing the 18th human miRBase (www.mirbase.org/).
The target genes of significantly regulated miRNAs were predicted using two online databases, miRanda (www.microrna.org/microrna/home.do) and TartgetScan (www.targetscan.org/vert_71/), to avoid the overpredicting limitation of individual programs and reduce the potential for false positives. The intersection of genes was selected as the preliminary screening target genes.
The significant, accurate and targeting functions of these genes were analyzed through gene ontology (GO) analysis (geneontology.org/). All functions of the preliminary screening target genes were obtained, and then the Fisher's exact test was used to calculate the P-value and the chi-square test was used to calculate the Pk-value. The false discovery rate (FDR) was calculated to correct the P-value, as the smaller the FDR, the smaller the error in judging the P-value. The FDR was defined as FDR=1-Nk/T, where Nk refers to the number of P-values less than Pk-values and T refers to the number of Pk-values less than P-values. P<0.001 was considered to indicate a statistically significant difference. In addition, enrichment analysis was performed. When the P-values of the functions were identical, the lower enrichment degree represented that the function was described in more detail in the GO system. Through GO analysis the significant functions of target genes were determined. The pathways of the preliminary screening target genes were analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) system. The P-value and FDR of these target gene pathways was calculated with the Fisher's exact test and multiple comparison test. P<0.05 was considered to indicate a statistically significant difference. Through pathway analysis the significant pathways regulated by target genes were determined. The analysis was performed by Genminix Informatics Ltd., Co. (Shanghai, China).
Results
Identification of UC-MSCs
Fibroblast-like cells adhered to the plastic bottom of flasks were observed under an inverted light microscope (Fig. 1A). Rhodamine phalloidin, an actin filament fluorescent dye, was used to stain cells to identify the shape of fibroblast-like cells under a confocal fluorescent microscope (Fig. 1B).
Figure 1.
Morphology of UC-MSCs. (A) Fibroblast-like cells adherent to the plastic bottom of flasks were observed under an inverted light microscope. Scale bar=500 µm. (B) Actin filaments and nuclei of cells were stained with rhodamine phalloidin and DAPI, respectively and cells were observed under a confocal fluorescent microscope. Scale bar=40 µm. Cells adherence to plastic conformed to the minimal criteria of MSCs. UC-MSCs, umbilical cord-mesenchymal stem cells.
Flow cytometry of UC-MSCs revealed them to be positive for CD44, CD73, CD90, CD105 and CD166, and negative for CD11b, CD19, CD31, CD34, CD45, CD106 and HLA-DR (Fig. 2). These findings are in accordance with the minimal criteria for the definition of MSCs (22). In addition, the minimal criteria state that MSCs must differentiate into osteoblasts, adipocytes and chondroblasts in vitro. Therefore, differentiation experiments were performed.
Figure 2.
Phenotype of UC-MSCs. The phenotype of UC-MSCs was analyzed by flow cytometry. UC-MSCs were positive for CD44, CD73, CD90, CD105 and CD166, and negative for CD11b, CD19, CD31, CD34, CD45, CD106 and HLA-DR. The blue line in each histogram represents staining with the appropriate isotype control antibody. UC-MSCs, umbilical cord-mesenchymal stem cells; CD, cluster of differentiation; PE, phycoerythrin; FITC, fluorescein isothiocyanate; HLA-DR, human leukocyte antigen-D related.
Following treatment with osteogenic medium for three weeks, the shape of UC-MSCs altered from fibroblast-like to polygon. The deposition of mineralized matrix was detected with Von Kossa reagents (Fig. 3A). Lipid droplets gradually appeared during treatment with adipogenic medium for three weeks, and were detected by oil red O staining (Fig. 3B). During chondrogenic differentiation, UC-MSCs gradually formed a pellet suspended in medium, and sulfated proteoglycans were detected by Alcian blue 8GX staining following three weeks (Fig. 3C).
Figure 3.
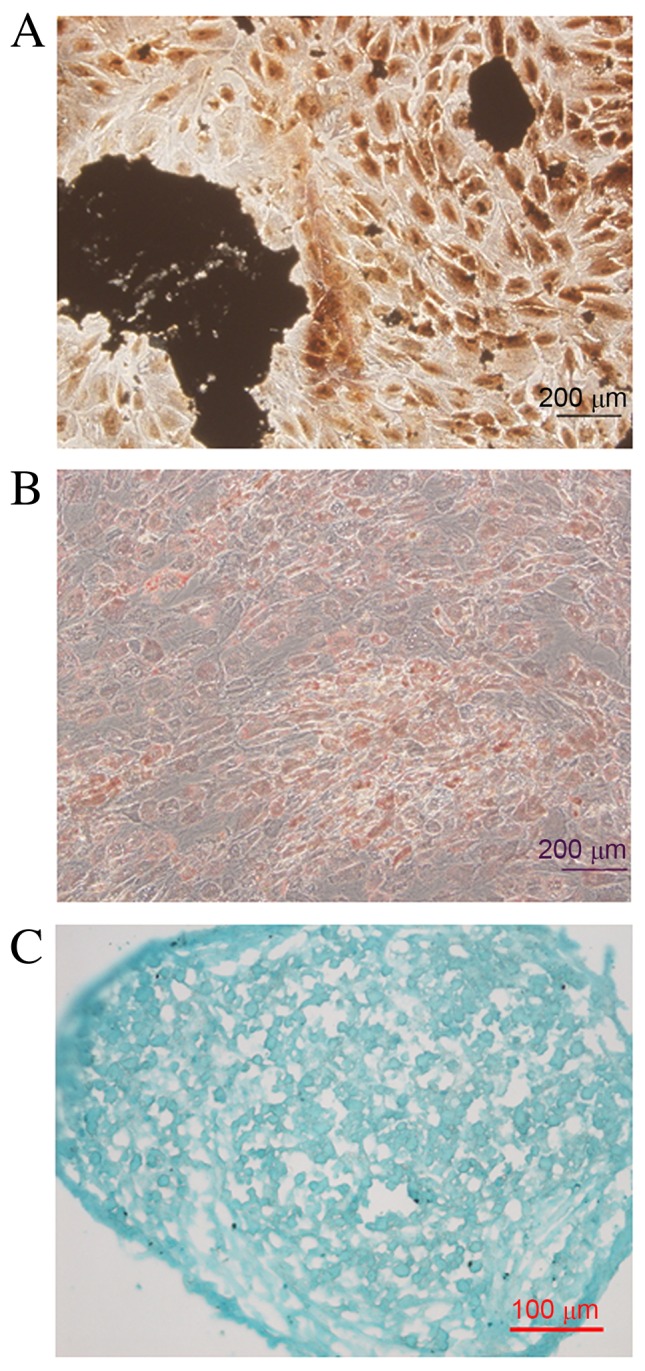
Differentiation of UC-MSCs. (A) Following conditioned culture, the deposition of mineralized matrix was detected with Von Kossa reagents in UC-MSCs. Scale bar=200 µm. (B) Oil red O staining revealed lipid droplets in UC-MSCs following treatment with adipogenic medium. Scale bar=200 µm. (C) Alcian blue 8GX staining revealed sulfated proteoglycans in UC-MSCs following treatment with chondrogenic medium. Scale bar=100 µm. UC-MSCs, umbilical cord-mesenchymal stem cells.
Culture with IFN-γ significantly alters the miRNA profile of UC-MSCs
The miRNA array data revealed significant changes in miRNA expression levels in UC-MSCs following treatment with IFN-γ. A total of 256 miRNAs were detected in UC-MSCs or IFN-UC-MSCs; of these, 5 miRNAs were detected only in UC-MSCs and 6 miRNAs were detected only in IFN-UC-MSCs. As the expression levels of these 11 miRNAs were not significantly different between the two groups (Cq>35), they were not analyzed further. The remaining 245 miRNAs were expressed in UC-MSCs and IFN-UC-MSCs. The expression levels of these 245 miRNAs were analyzed; 110 miRNAs were upregulated and 135 miRNAs were downregulated in UC-MSCs following treatment with IFN-γ (Table I).
Table I.
miRNAs expressed in UC-MSCs and IFN-UC-MSCs.
| miRNA category | Number | miRNAs |
|---|---|---|
| Upregulated miRNAs | 110 | hsa-let-7g; hsa-miR-10b; hsa-miR-15b; hsa-miR-16; hsa-miR-17; hsa-miR-18a; hsa-miR-19a; hsa-miR-19b; hsa-miR-20a; hsa-miR-20b; hsa-miR-25; hsa-miR-28-3p; hsa-miR-29a; hsa-miR-29b; hsa-miR-92a; mmu-miR-93; hsa-miR-106a; hsa-miR-128a; hsa-miR-130b; mmu-miR-134; mmu-miR-137; hsa-miR-148b; hsa-miR-192; hsa-miR-199a; hsa-miR-214; hsa-miR-218; hsa-miR-223; hsa-miR-299-5p; hsa-miR-301; hsa-miR-323-3p; hsa-miR-330; hsa-miR-331-5p; hsa-miR-337-5p; hsa-miR-339-5p; hsa-miR-345; hsa-miR-361; hsa-miR-362-3p; hsa-miR-369-5p; hsa-miR-370; hsa-miR-376a; mmu-miR-379; hsa-miR-381; hsa-miR-382; hsa-miR-410; hsa-miR-411; hsa-miR-422a; hsa-miR-424; hsa-miR-425-5p; hsa-miR-450b-5p; hsa-miR-454; hsa-miR-455-3p; hsa-miR-485-3p; hsa-miR-487b; hsa-miR-493; hsa-miR-494; mmu-miR-495; hsa-miR-500; hsa-miR-501; hsa-miR-502-3p; hsa-miR-503; hsa-miR-532-3p; hsa-miR-532; hsa-miR-539; hsa-miR-590-5p; hsa-miR-598; hsa-miR-618; hsa-miR-652; hsa-miR-654-3p; hsa-miR-655; hsa-miR-671-3p; hsa-miR-744; hsa-miR-758; hsa-miR-885-5p; hsa-miR-886-3p; hsa-miR-886-5p; hsa-miR-376c; hsa-miR-432; hsa-miR-656; hsa-miR-550; hsa-miR-629; hsa-miR-454#; hsa-miR-770-5p; hsa-miR-505#; hsa-miR-136#; hsa-miR-130b#; hsa-miR-93#; hsa-miR-99a#; hsa-miR-149#; hsa-miR-29b-1#; hsa-miR-99b#; hsa-miR-378; hsa-miR-10a#; hsa-miR-214#; hsa-miR-22#; hsa-miR-424#; hsa-miR-409-3p; hsa-miR-543; hsa-miR-125b-1#; hsa-miR-15a#; hsa-miR-18a#; hsa-miR-19b-1#; hsa-miR-20a#; hsa-miR-21#; hsa-miR-27a#; hsa-miR-29a#; hsa-miR-1227; hsa-miR-1254; hsa-miR-1275; hsa-miR-320B; hsa-miR-1300 |
| Downregulated miRNAs | 135 | hsa-let-7a; hsa-let-7c; hsa-let-7d; hsa-let-7e; hsa-let-7f; hsa-miR-10a; hsa-miR-21; hsa-miR-24; hsa-miR-26a; hsa-miR-26b; hsa-miR-27a; hsa-miR-27b; hsa-miR-28; hsa-miR-29c; hsa-miR-30b; hsa-miR-30c; hsa-miR-31; hsa-miR-34a; hsa-miR-98; hsa-miR-99a; hsa-miR-99b; hsa-miR-100; hsa-miR-101; hsa-miR-103; hsa-miR-106b; hsa-miR-125a-5p; hsa-miR-125b; hsa-miR-126; hsa-miR-127; hsa-miR-130a; hsa-miR-132; hsa-miR-138; hsa-miR-140-3p; mmu-miR-140; hsa-miR-143; hsa-miR-145; hsa-miR-146a; hsa-miR-146b; hsa-miR-148a; hsa-miR-149; hsa-miR-152; hsa-miR-181a; hsa-miR-185; hsa-miR-186; hsa-miR-191; hsa-miR-193a-3p; hsa-miR-193a-5p; hsa-miR-193b; hsa-miR-194; hsa-miR-195; hsa-miR-196b; hsa-miR-197; hsa-miR-199a-3p; hsa-miR-204; hsa-miR-210; hsa-miR-221; hsa-miR-222; hsa-miR-224; hsa-miR-296; hsa-miR-302b; hsa-miR-320; hsa-miR-324-3p; hsa-miR-324-5p; hsa-miR-328; hsa-miR-331; hsa-miR-335; hsa-miR-339-3p; hsa-miR-340; hsa-miR-155; hsa-let-7b; hsa-miR-342-3p; hsa-miR-362; hsa-miR-365; hsa-miR-374; mmu-miR-374-5p; hsa-miR-423-5p; hsa-miR-452; hsa-miR-455; hsa-miR-483-5p; hsa-miR-484; hsa-miR-489; mmu-miR-491; hsa-miR-542-3p; hsa-miR-574-3p; hsa-miR-582-5p; hsa-miR-625; hsa-miR-628-5p; hsa-miR-660; hsa-miR-708; hsa-miR-212; dme-miR-7; hsa-miR-30a-3p; hsa-miR-30a-5p; hsa-miR-30d; hsa-miR-30e-3p; hsa-miR-126#; hsa-miR-206; hsa-miR-213; rno-miR-7#; hsa-miR-589; hsa-miR-766; hsa-miR-769-5p; hsa-miR-222#; hsa-miR-34b; hsa-miR-31#; hsa-miR-33a#; hsa-miR-100#; hsa-miR-145#; hsa-miR-27b#; hsa-miR-335#; hsa-miR-942; hsa-miR-875-5p; hsa-miR-151-3p; hsa-miR-425#; hsa-miR-30d#; hsa-miR-10b#; hsa-miR-34a#; hsa-miR-181a-2#; hsa-miR-550; hsa-miR-625#; hsa-miR-24-2#; hsa-miR-26a-1#; hsa-miR-151-5P; hsa-miR-590-3P; hsa-miR-1233; hsa-miR-1271; hsa-miR-1285; hsa-miR-1183; hsa-miR-1180; hsa-miR-1290; hsa-miR-1274A; hsa-miR-1274B; hsa-miR-720; hsa-miR-1260; hsa-miR-664 |
miRNA, microRNA; UC-MSCs, umbilical cord-mesenchymal stem cells; IFN-UC-MSCs, interferon-γ-treated umbilical cord-mesenchymal stem cells.
A total of 21 miRNAs exhibited >2-fold upregulation and 41 miRNAs exhibited >2-fold downregulation. miRNAs whose fold-changes were <2-fold in one of the two experiments or that altered in opposite directions in the two experiments were excluded from further analysis. A total of 10 miRNAs were significantly regulated by >2-fold and were altered in the same direction in the two experiments. As 2 miRNAs (miRNA-886-5p and miRNA-1300) had been removed from the 18th human miRBase, further analysis was performed on 8 significantly regulated miRNAs. RT-qPCR was performed to validate the results of the miRNA array; 5 of the miRNAs were upregulated and 3 were downregulated in IFN-UC-MSCs compared with UC-MSCs. (Fig. 4; Table II).
Figure 4.
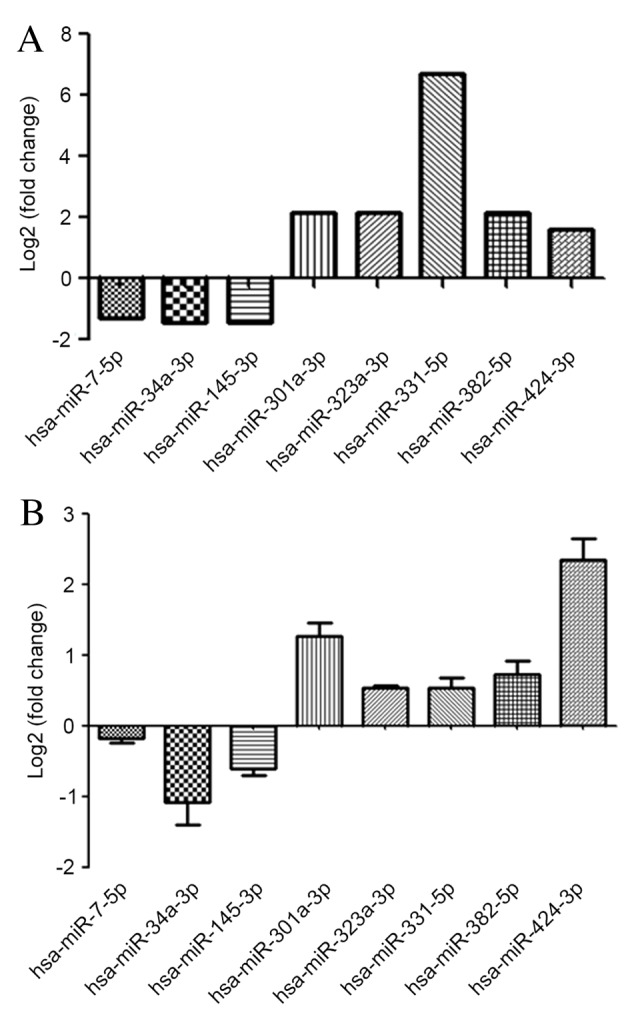
Significantly regulated miRNAs. (A) The 8 miRNAs differentially expressed in interferon-γ-treated UC-MSCs compared with untreated UC-MSCs, as identified by miRNA array. Of these, 3 miRNAs were downregulated and 5 upregulated. (B) The miRNA array data was validated by reverse transcription-quantitative polymerase chain reaction analysis of the expression levels of the 8 miRNAs. miRNA, microRNA; UC-MSCs, umbilical cord-mesenchymal stem cells.
Table II.
Significantly regulated miRNAs.
| miRNA name | miRNA array | RT-qPCR | |||
|---|---|---|---|---|---|
| Human 18th miRBase ID | Assay ID | Fold-change | Direction | Fold-change | Direction |
| hsa-miR-7-5p | dme-miR-7 | 2.488 | Down | 1.12 | Down |
| hsa-miR-34a-3p | hsa-miR-34a# | 2.688 | Down | 1.91 | Down |
| hsa-miR-145-3p | hsa-miR-145# | 2.71 | Down | 1.51 | Down |
| hsa-miR-301a-3p | hsa-miR-301 | 4.414 | Up | 2.494 | Up |
| hsa-miR-331-5p | hsa-miR-323-3p | 4.44 | Up | 1.457 | Up |
| hsa-miR-382-5p | hsa-miR-331-5p | 105.664 | Up | 1.489 | Up |
| hsa-miR-323a-3p | hsa-miR-382 | 4.456 | Up | 1.727 | Up |
| hsa-miR-424-3p | hsa-miR-424# | 3.024 | Up | 5.5 | Up |
miRNA, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Target genes of significantly regulated miRNAs and the ‘microRNA-Gene-Network’
Target gene prediction for the 8 differentially expressed miRNAs was performed using two online databases, TargetScan and miRanda. A total of 2,494 predicted genes were obtained from TargetScan, and 2,271 predicted genes were acquired from miRanda. To increase the specificity, only genes predicted by the two databases were examined, leaving 816 predicted target genes (Fig. 5).
Figure 5.

Venn diagram demonstrating the number of target genes predicted by the online databases TargetScan (total, 2,494) and miRanda (total, 2,271), and the overlapping predicted genes (861).
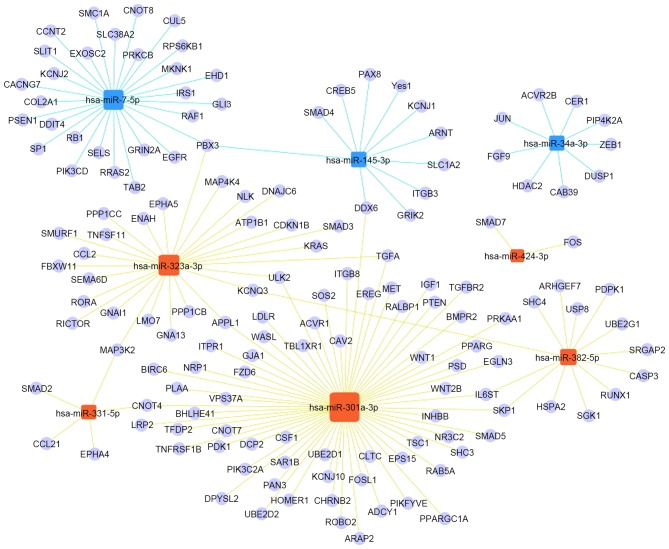
GO and pathway analyses were applied to identify the key genes in the significant functions and pathways from the predicted target genes. A total of 149 significant overlapping genes were obtained following these two analyses (Table III). The miRNA-Gene-Network was used to evaluate the status of miRNAs and genes (Fig. 6). The functions of the identified genes are listed in Table IV.
Table III.
Predicted target genes of significantly regulated miRNAs.
| miRNA | Target genes |
|---|---|
| hsa-miR-145-3p | smad4; arnt; pax8; kcmj1; yes1; itgb3; creb5; pbx3; ddx6; grik2; slc1a2 |
| hsa-miR-34a-3p | fgf9; jun; hdac2; acvr2b; cab39; cer1; dusp1; pip4k2a; zeb1 |
| hsa-miR-7-5p | egfr; rb1; gli3; pik3cd; raf1; prkcb; rps6kb1; sp1; irs1; ddit4; ehd1; psen1; mknk1; col2a1; tab2; cacng7; rras2; kcnj2; grin2a; slit1; cul5; ccnt2; pbx3; sels; exosc2; cnot8; smc1a; slc38a2 |
| hsa-miR-301a-3p | fzd6; igf1; pparg; appl1; pten; tgfa; wnt1; tgfbr2; sos2; egln3; ralbp1; wnt2b; met; inhbb; bmpr2; smad5; skp1; acvr1; nr3c2; ereg; shc3; prkaa1; tsc1; ulk2; vps37a; rab5a; cav2; eps15; psd; arap2; ldlr; cltc; tbl1xr1; fosl1; wasl; ppargc1a; itgb8; pikfyve; gja1; adcy1; itpr1; chrnb2; nrp1; robo2; dpysl2; kcnj10; ube2d2; ube2d1; birc6; sar1b; plaa; pdk1; bhlhe41; pan3; ddx6; cnot7; cnot4; dcp2; lrp2; pik3c2a; tfdp2; homer1; tnfrsf1b; csf1; il6st |
| hsa-miR-323a-3p | appl1; tgfa; smad3; kras; cdkn1b; smurf1; atp1b1; rictor; ulk2; dnajc6; fbxw11; nlk; lmo7; ppp1cc; ppp1cb; map4k4; map3k2; gna13; enah; gnai1; kcnq3; epha5; sema6d; pbx3; roar; ccl2; tnfsf11 |
| hsa-miR-331-5p | smad2; map3k2; epha4; cnot4; ccl21 |
| hsa-miR-382-5p | runx1; casp3; skp1; sgk1; pdpk1; shc4; usp8; hspa2; arhgef7; kcnq3; srgap2; ube2g1; il6st |
| hsa-miR-424-3p | fos; smad7 |
miRNA, microRNA.
Figure 6.
miRNA-Gene-Network. The regulatory association between the significantly regulated miRNAs and the corresponding gene functions. Blue squares represent downregulated miRNAs, and orange squares represent upregulated miRNAs, in interferon-γ-treated UC-MSCs compared with untreated UC-MSCs. The size of the miRNA square reflects the number of regulated genes. miRNA, microRNA; UC-MSCs, umbilical cord-mesenchymal stem cells.
Table IV.
The functions of the predicted target genes.
| Gene | P-value |
|---|---|
| Proliferation | |
| cav2 | 1.80E-10 |
| cdkn1b | 1.80E-10 |
| cer1 | 1.80E-10 |
| cnot8 | 1.80E-10 |
| ereg | 1.80E-10 |
| gja1 | 1.80E-10 |
| gli3 | 1.80E-10 |
| igf1 | 1.80E-10 |
| jun | 1.80E-10 |
| pparg | 1.80E-10 |
| smad2 | 1.80E-10 |
| smad3 | 1.80E-10 |
| smad4 | 1.80E-10 |
| tsc1 | 1.80E-10 |
| zeb1 | 1.80E-10 |
| birc6 | 1.40E-06 |
| csf1 | 1.40E-06 |
| egfr | 1.40E-06 |
| fgf9 | 1.40E-06 |
| hdac2 | 1.40E-06 |
| il6st | 1.40E-06 |
| irs1 | 1.40E-06 |
| tgfbr2 | 1.40E-06 |
| wnt1 | 1.40E-06 |
| appl1 | 7.95E-06 |
| bhlhe41 | 7.95E-06 |
| cul5 | 7.95E-06 |
| eps15 | 7.95E-06 |
| lrp2 | 7.95E-06 |
| met | 7.95E-06 |
| pten | 7.95E-06 |
| raf1 | 7.95E-06 |
| tgfa | 7.95E-06 |
| usp8 | 7.95E-06 |
| bmpr2 | 8.76E-05 |
| egln3 | 8.76E-05 |
| nr3c2 | 8.76E-05 |
| sgk1 | 8.76E-05 |
| Differentiation | |
| arnt | 7.21E-14 |
| bhlhe41 | 7.21E-14 |
| csf1 | 7.21E-14 |
| dpysl2 | 7.21E-14 |
| ereg | 7.21E-14 |
| gna13 | 7.21E-14 |
| inhbb | 7.21E-14 |
| nrp1 | 7.21E-14 |
| pax8 | 7.21E-14 |
| pik3cd | 7.21E-14 |
| robo2 | 7.21E-14 |
| sema6d | 7.21E-14 |
| slit1 | 7.21E-14 |
| smurf1 | 7.21E-14 |
| tnfsf11 | 7.21E-14 |
| zeb1 | 7.21E-14 |
| Apoptosis | |
| acvr1 | 9.54E-08 |
| cdkn1b | 9.54E-08 |
| col2a1 | 9.54E-08 |
| gli3 | 9.54E-08 |
| hdac2 | 9.54E-08 |
| prkaa1 | 9.54E-08 |
| pten | 9.54E-08 |
| rps6kb1 | 9.54E-08 |
| smad3 | 9.54E-08 |
| wnt1 | 9.54E-08 |
| appl1 | 0.00026423 |
| arhgef7 | 0.00026423 |
| birc6 | 0.00026423 |
| ddit4 | 0.00026423 |
| egln3 | 0.00026423 |
| gja1 | 0.00026423 |
| prkcb | 0.00026423 |
| psen1 | 0.00026423 |
| raf1 | 0.00026423 |
| sos2 | 0.00026423 |
| tgfbr2 | 0.00026423 |
| tnfrsf1b | 0.00026423 |
| ppargc1a | 0.000494088 |
| tgfa | 0.000494088 |
| Migration | |
| igf1 | 1.73E-05 |
| il6st | 1.73E-05 |
| nrp1 | 1.73E-05 |
| rps6kb1 | 1.73E-05 |
| gja1 | 0.000551751 |
| met | 0.000551751 |
| psen1 | 0.000551751 |
| TGF-β/BMP signaling pathway | |
| acvr1 | 4.69E-09 |
| ccl2 | 4.69E-09 |
| fos | 4.69E-09 |
| jun | 4.69E-09 |
| nlk | 4.69E-09 |
| smad2 | 4.69E-09 |
| smad3 | 4.69E-09 |
| smad4 | 4.69E-09 |
| smad7 | 4.69E-09 |
| smurf1 | 4.69E-09 |
| tgfbr2 | 4.69E-09 |
| ube2d1 | 4.69E-09 |
| acvr2b | 1.25E-06 |
| bmpr2 | 1.25E-06 |
| cer1 | 1.25E-06 |
| smad5 | 1.25E-06 |
| zeb1 | 1.68E-06 |
| wnt1 | 0.000231202 |
| FGF signaling pathway | |
| cdkn1b | 0.000165506 |
| fgf9 | 0.000165506 |
| irs1 | 0.000165506 |
| rictor | 0.000165506 |
| EGFR signaling pathway | |
| egfr | 2.75E-06 |
| raf1 | 2.75E-06 |
| ereg | 2.75E-06 |
| shc3 | 2.75E-06 |
| eps15 | 2.75E-06 |
| cltc | 2.75E-06 |
| kras | 2.75E-06 |
| arhgef7 | 2.75E-06 |
Results were obtained by Gene Ontology Analysis. P-values indicate the significance of the functions of target genes. P<0.001 was considered to indicate a statistically significant difference. TGF-β, transforming growth factor β; BMP, bone morphogenetic protein; FGF, fibroblast growth factor; EGFR, epidermal growth factor receptor.
Discussion
MSCs affect innate and adaptive immunity; these immunomodulatory effects are induced by certain inflammatory cytokines, including IFN-γ (4). In addition, IFN-γ affects numerous other biological characteristics of MSCs (10–12). However, the mechanisms underlying the effect of IFN-γ on MSCs remain to be fully elucidated. As intracellular miRNAs are essential regulators of proliferation, survival, migration, differentiation and paracrine functions of MSCs (19), it was hypothesized that miRNAs are involved in the effects of IFN-γ on MSCs.
Through miRNA arrays, it was demonstrated that the miRNA expression profile of UC-MSCs was significantly altered following stimulation with IFN-γ. Expression of 256 microRNAs were detected in UC-MSCs or IFN-UC-MSCs of the 754 analyzed. Of these, 5 miRNAs were detected only in UC-MSCs and 6 miRNAs were detected only in IFN-UC-MSCs; these were excluded from further analysis due to their weak expression (Cq>35). Following statistical analysis, 8 significantly differentially expressed miRNAs were identified from the 245 miRNAs detected in UC-MSCs and IFN-UC-MSCs. RT-qPCR analysis of these 8 miRNAs demonstrated results consistent with the miRNA array data. Bioinformatics analysis of the 8 miRNAs identified 149 significant target genes. In addition to regulation of transcription, the functions of these genes were associated with cell growth, proliferation, apoptosis, differentiation and migration.
Hsa-miR-34a-3p, hsa-miR-7-5p and hsa-miR-145-3p were downregulated in IFN-UC-MSCs compared with UC-MSCs. Guennewiq et al (23) demonstrated that hsa-miR-34a-3p may influence cellular apoptosis via targeting SP4 transcription factor, which is involved in TGF-β signaling. Epidermal growth factor receptor (EGFR) is an important target gene of hsa-miR-7-5p. Vasanthan et al (24) revealed that knockdown of hsa-miR-7-5p resulted in increased EGFR expression. Numerous studies have reported that EGFR regulates cellular proliferation. For example, Li et al (25) suggested the EGFR/mitogen-activated protein kinase signaling has a principle function in regulating the proliferation of renal and nephric stem cells. Hsa-miR-301a-3p was one of 5 miRNAs, (hsa-miR-301a-3p, hsa-miR-323a-3p, hsa-miR-331-5p, hsa-miR-382-5p and hsa-miR-424-3p) upregulated in IFN-UC-MSCs compared with UC-MSCs. Lu et al (26) demonstrated that the miR-301 and miR-302 families target serine and arginine rich splicing factor 2 and methyl-CpG binding domain protein 2a, and are involved in human pluripotent stem cell self-renewal and reprogramming. In our previous study, miR-301a was upregulated by Toll-like receptor signaling in UC-MSCs. Attenuation or enhancement of the expression of miR-301a altered the quantity of inflammation-associated cytokines secreted by UC-MSCs accordingly (27). Previous studies have indicated that hsa-miR-323a-3p is a potential therapeutic target for rheumatoid arthritis, and it has been associated with Wnt/cadherin signaling (28,29). Seok et al (30) revealed that hsa-miR-382-5p may be involved in vascular endothelial cell proliferation, migration and tube formation in hypoxic conditions via the regulation of phosphatase and tensin homolog. The results of GO analysis in the present study suggested that numerous other predicted target genes may contribute to cellular proliferation, apoptosis, differentiation and migration. Further studies are required to determine whether the predicted genes are authentic targets of the regulated miRNAs.
Through gene function analysis, various important signaling pathways were identified. Hsa-miR-34a-3p, hsa-miR-145-3p, hsa-miR-301a-3p, hsa-miR-323a-3p, hsa-miR-331-5p and hsa-miR-424-3p were predicted to regulate the TGF-β/bone morphogenetic protein (BMP) signaling pathway. The TGF-β/BMP signaling pathway has been reported to be associated with osteogenic differentiation of MSCs (31). Hsa-miR-34a-3p, hsa-miR-7-5p and hsa-miR-323a-3p were predicted to regulate the fibroblast growth factor signaling pathway, which has been reported to be important for differentiation, growth and proliferation of MSCs (32,33). Hsa-miR-7-5p, hsa-miR-301a-3p and hsa-miR-323a-3p were predicted to affect the EGFR signaling pathway, which may regulate the proliferation, differentiation and paracrine capability of MSCs (34,35). miRNAs upregulated and downregulated in IFN-UC-MSCs compared with UC-MSCs are predicted to target genes involved in the same signaling pathway. This suggests that the mechanism underlying the involvement of miRNAs in the effect of IFN-γ on MSCs are complex and require further investigation.
In conclusion, the present study systematically analyzed the effect of IFN-γ on the miRNA profile of UC-MSCs, to the best of our knowledge for the first time. The results of the present study suggest that miRNAs may be crucial in mediating the effects of IFN-γ on the biological functions of MSCs. The identification of these miRNAs may indicate a novel direction for research of the molecular mechanism underlying the effect of IFN-γ on MSCs, which may provide insight into the influence of inflammatory environment on MSCs and their immunomodulatory function.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant nos. 81330015 and 31470951) and the National Science and Technology Major Project (grant no. 2014ZX09508002-003).
References
- 1.Lu LL, Liu YJ, Yang SG, Zhao QJ, Wang X, Gong W, Han ZB, Xu ZS, Lu YX, Liu D, et al. Isolation and characterization of human umbilical cord mesenchymal stem cells with hematopoiesis-supportive function and other potentials. Haematologica. 2006;91:1017–1026. [PubMed] [Google Scholar]
- 2.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 3.Salem HK, Thiemermann C. Mesenchymal stromal cells: Current understanding and clinical status. Stem Cells. 2010;28:585–596. doi: 10.1002/stem.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Chen X, Cao W, Shi Y. Plasticity of mesenchymal stem cells in immunomodulation: Pathological and therapeutic implications. Nat Immunol. 2014;15:1009–1016. doi: 10.1038/ni.3002. [DOI] [PubMed] [Google Scholar]
- 5.Chen K, Wang D, Du WT, Han ZB, Ren H, Chi Y, Yang SG, Zhu D, Bayard F, Han ZC. Human umbilical cord mesenchymal stem cells hUC-MSCs exert immunosuppressive activities through a PGE2-dependent mechanism. Clin Immunol. 2010;135:448–458. doi: 10.1016/j.clim.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 6.Krampera M, Cosmi L, Angeli R, Pasini A, Liotta F, Andreini A, Santarlasci V, Mazzinghi B, Pizzolo G, Vinante F, et al. Role for interferon-gamma in the immunomodulatory activity of human bone marrow mesenchymal stem cells. Stem Cells. 2006;24:386–398. doi: 10.1634/stemcells.2005-0008. [DOI] [PubMed] [Google Scholar]
- 7.Ryan JM, Barry F, Murphy JM, Mahon BP. Interferon-gamma does not break, but promotes the immunosuppressive capacity of adult human mesenchymal stem cells. Clin Exp Immunol. 2007;149:353–363. doi: 10.1111/j.1365-2249.2007.03422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DelaRosa O, Lombardo E, Beraza A, Mancheño-Corvo P, Ramirez C, Menta R, Rico L, Camarillo E, García L, Abad JL, et al. Requirement of IFN-gamma-mediated indoleamine 2,3-dioxygenase expression in the modulation of lymphocyte proliferation by human adipose-derived stem cells. Tissue Eng Part A. 2009;15:2795–2806. doi: 10.1089/ten.tea.2008.0630. [DOI] [PubMed] [Google Scholar]
- 9.Chan WK, Lau AS, Li JC, Law HK, Lau YL, Chan GC. MHC expression kinetics and immunogenicity of mesenchymal stromal cells after short-term IFN-gamma challenge. Exp Hematol. 2008;36:1545–1555. doi: 10.1016/j.exphem.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Croitoru-Lamoury J, Lamoury FM, Caristo M, Suzuki K, Walker D, Takikawa O, Taylor R, Brew BJ. Interferon-γ regulates the proliferation and differentiation of mesenchymal stem cells via activation of indoleamine 2,3 dioxygenase (IDO) PLoS One. 2011;6:e14698. doi: 10.1371/journal.pone.0014698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Wang L, Kikuiri T, Akiyama K, Chen C, Xu X, Yang R, Chen W, Wang S, Shi S. Mesenchymal stem cell-based tissue regeneration is governed by recipient T lymphocytes via IFN-γ and TNF-α. Nat Med. 2011;17:1594–1601. doi: 10.1038/nm.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laschober GT, Brunauer R, Jamnig A, Singh S, Hafen U, Fehrer C, Kloss F, Gassner R, Lepperdinger G. Age-specific changes of mesenchymal stem cells are paralleled by upregulation of CD106 expression as a response to an inflammatory environment. Rejuvenation Res. 2011;14:119–131. doi: 10.1089/rej.2010.1077. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Shenoy A, Blelloch RH. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat Rev Mol Cell Biol. 2014;15:565–576. doi: 10.1038/nrm3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han ZB, Yang Z, Chi Y, Zhang L, Wang Y, Ji Y, Wang J, Zhao H, Han ZC. MicroRNA-124 suppresses breast cancer cell growth and motility by targeting CD151. Cell Physiol Biochem. 2013;31:823–832. doi: 10.1159/000350100. [DOI] [PubMed] [Google Scholar]
- 16.Wen Z, Zheng S, Zhou C, Yuan W, Wang J, Wang T. Bone marrow mesenchymal stem cells for post-myocardial infarction cardiac repair: MicroRNAs as novel regulators. J Cell Mol Med. 2012;16:657–671. doi: 10.1111/j.1582-4934.2011.01471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin KK, Kim YS, Kim JY, Bae YC, Jung JS. miR-137 controls proliferation and differentiation of human adipose tissue stromal cells. Cell Physiol Biochem. 2014;33:758–768. doi: 10.1159/000358650. [DOI] [PubMed] [Google Scholar]
- 18.Paik S, Jung HS, Lee S, Yoon DS, Park MS, Lee JW. miR-449a regulates the chondrogenesis of human mesenchymal stem cells through direct targeting of lymphoid enhancer-binding factor-1. Stem Cells Dev. 2012;21:3298–3308. doi: 10.1089/scd.2011.0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark EA, Kalomoiris S, Nolta JA, Fierro FA. Concise review: MicroRNA function in multipotent mesenchymal stromal cells. Stem Cells. 2014;32:1074–1082. doi: 10.1002/stem.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomé M, López-Romero P, Albo C, Sepúlveda JC, Fernández-Gutiérrez B, Dopazo A, Bernad A, González MA. miR-335 orchestrates cell proliferation, migration and differentiation in human mesenchymal stem cells. Cell Death Differ. 2011;18:985–995. doi: 10.1038/cdd.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop Dj, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 23.Guennewig B, Roos M, Dogar AM, Gebert LF, Zagalak JA, Vongrad V, Metzner KJ, Hall J. Synthetic pre-microRNAs reveal dual-strand activity of miR-34a on TNF-α. RNA. 2014;20:61–75. doi: 10.1261/rna.038968.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasanthan P, Govindasamy V, Gnanasegaran N, Kunasekaran W, Musa S, Abu Kasim NH. Differential expression of basal microRNAs' patterns in human dental pulp stem cells. J Cell Mol Med. 2015;19:566–580. doi: 10.1111/jcmm.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Liu S, Cai Y. EGFR/MAPK signaling regulates the proliferation of Drosophila renal and nephric stem cells. J Genet Genomics. 2015;42:9–20. doi: 10.1016/j.jgg.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 26.Lu Y, Loh YH, Li H, Cesana M, Ficarro SB, Parikh JR, Salomonis N, Toh CX, Andreadis ST, Luckey CJ, et al. Alternative splicing of MBD2 supports self-renewal in human pluripotent stem cells. Cell Stem Cell. 2014;15:92–101. doi: 10.1016/j.stem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma F, Chen D, Chi Y, Chen F, Li X, Han Z. The expression and role of miR-301a in human umbilical cord-derived mesenchymal stromal cells. Cytotherapy. 2013;15:1511–1516. doi: 10.1016/j.jcyt.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 28.Pandis I, Ospelt C, Karagianni N, Denis MC, Reczko M, Camps C, Hatzigeorgiou AG, Ragoussis J, Gay S, Kollias G. Identification of microRNA-221/222 and microRNA-323-3p association with rheumatoid arthritis via predictions using the human tumour necrosis factor transgenic mouse model. Ann Rheum Dis. 2012;71:1716–1723. doi: 10.1136/annrheumdis-2011-200803. [DOI] [PubMed] [Google Scholar]
- 29.Xu T, Huang C, Chen Z, Li J. MicroRNA-323-3p: A new biomarker and potential therapeutic target for rheumatoid arthritis. Rheumatol Int. 2014;34:721–722. doi: 10.1007/s00296-013-2767-3. [DOI] [PubMed] [Google Scholar]
- 30.Seok JK, Lee SH, Kim MJ, Lee YM. MicroRNA-382 induced by HIF-1α is an angiogenic miR targeting the tumor suppressor phosphatase and tensin homolog. Nucleic Acids Res. 2014;42:8062–8072. doi: 10.1093/nar/gku515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang R, Lee P, Lui VC, Chen Y, Liu X, Lok CN, To M, Yeung KW, Wong KK. Silver nanoparticles promote osteogenesis of mesenchymal stem cells and improve bone fracture healing in osteogenesis mechanism mouse model. Nanomedicine. 2015;11:1949–1959. doi: 10.1016/j.nano.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 32.Ng F, Boucher S, Koh S, Sastry KS, Chase L, Lakshmipathy U, Choong C, Yang Z, Vemuri MC, Rao MS, Tanavde V. PDGF, TGF-beta, and FGF signaling is important for differentiation and growth of mesenchymal stem cells (MSCs): Transcriptional profiling can identify markers and signaling pathways important in differentiation of MSCs into adipogenic, chondrogenic, and osteogenic lineages. Blood. 2008;112:295–307. doi: 10.1182/blood-2007-07-103697. [DOI] [PubMed] [Google Scholar]
- 33.Mojsilović S, Krstić A, Ilić V, Okić-Đorđević I, Kocić J, Trivanović D, Santibañez JF, Jovčić G, Bugarski D. IL-17 and FGF signaling involved in mouse mesenchymal stem cell proliferation. Cell Tissue Res. 2011;346:305–316. doi: 10.1007/s00441-011-1284-5. [DOI] [PubMed] [Google Scholar]
- 34.Rodrigues M, Blair H, Stockdale L, Griffith L, Wells A. Surface tethered epidermal growth factor protects proliferating and differentiating multipotential stromal cells from FasL-induced apoptosis. Stem Cells. 2013;31:104–116. doi: 10.1002/stem.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamama K, Barbeau DJ. Early growth response genes signaling supports strong paracrine capability of mesenchymal stem cells. Stem Cells Int. 2012;2012:428403. doi: 10.1155/2012/428403. [DOI] [PMC free article] [PubMed] [Google Scholar]