Abstract
Toll-like receptor (TLR) family are receptors for extracellular or intracellular signaling, such as lipopolysaccharide (LPS), or 12-O-tetradecanoylphorbol-13-acetate. TLR induces the differentiation of human myeloid monocytic-leukemia cells (THP-1) to macrophages. However, the relationship between extracellular or intracellular signaling and the TLR protein level remain to be determined. Using RT-PCR and western blot analysis, the aim of the present study was to determine whether TLR4, a major TLR family member, could be moderately upregulated by high concentration of LPS and whether it promoted the maturation of THP1 cells. The results showed that, upregulated TLR4 at the protein level and mRNA level enriched the TLR4 modulation style. In addition, TLR4 expression was blocked by nuclear factor (NF)-κB inhibitor, and LPS stimulated NF-κB binding in the TLR4 gene promoter. Therefore, the increased expression of TLR4 in the responsiveness of LPS-treated THP1 cells occurred in response to the upregulation of their respective receptors, as well as a tight binding of NF-κB in the TLR4 gene promoter.
Keywords: THP1 cells, Toll-like receptor 4, nuclear factor-κB, lipopolysaccharide
Introduction
Dysregulation of the immune system leads to multiple pathophysiological processes due to loss of the capacity to detect and eliminate antigens by not distinguishing between self and non-self (1). Generally, the immune system can be divided into adaptive and innate immunity. It is known that the complex processes of the adaptive immune system enable B and T cells to detect non-self structures in B and T cells through antigen-specific receptors. The adaptive immune system exists only in vertebrates, whereas the innate immune system is present in all multicellular organisms, indicating that it is an evolutionarily conserved system (2). Sensing and excluding of invading pathogens by innate immune cells is dependent on the molecular structures that are shared by many pathogens but not expressed by the host. These structures are detected through multiple-encoded receptors or molecules, collectively named pattern recognition receptors (PRRs) (3).
An important class of PRRs is the Toll-like receptor (TLR) family that was identified recently in mammals and consists of multiple members. TLRs were found during investigations involving the search for proteins with sequence homology in the intracellular domain of the interleukin (IL)-1 receptor and Drosophila Toll molecules (4–6). Drosophila Toll molecules originally were characterized as a protein involved in the establishment of the dorsoventral polarity in developing fly embryos, and then in the resistance of adult flies to fungal infections. Since then, TLR genes have been identified in the mouse and human genomes (7). The effects of TLR in microbial product were first reported in 1998. Null mutations and missense mutations of TLR4 gene lead to loss of the capacity in response to lipopolysaccharide (LPS), which was the major component of gram-negative bacteria (8,9).
TLRs have been shown to recognize a broad range of microbial structures (10,11). Following activation, most TLRs induce a common intracellular signaling pathway that culminates in activation of the nuclear factor (NF)-κB, a key transcription factor involved in activating the expression of chemokines, cytokines, and cell-surface molecules such as adhesions, selectins, integrins, and co-stimulatory molecules (10–12). Initially, TLR4 was thought to be expressed solely in the immune system. However, kidney mesangial and tubular cells also express TLR4 (7,13).
Blood monocytes are an intermediate stage of development that further differentiates tissues into various types of macrophages populations. Tissue macrophages and other cells of the innate immune system are critical for these surveillance and defense activities (14). Monocytes are activated by microbial components such as LPS and 12-O-tetradecanoylphorbol-13-acetate (TPA) that are recognized by TLRs. After ligand binding, TLRs transduce extracellular signaling to the nucleus and induce cytokine production and ultimately clear the infection. Although currently 10 different human TLRs have been characterized (10–12,15), cytokine-mediated regulation of TLR expression remains poorly understood. TLR expression analysis in primary human leukocytes showed that professional phagocytes express the most varied TLR profile, with CD14+ mononuclear cells expressing the greatest amount of TLR-2, −4 and −8 (16,17). In monocytes, the constant expression of TLR4 is important in accepting bacterial infection signaling molecules such as LPS, which functions as the main ligand binding to TLR4 to induce inflammation. However, TLR4 signaling requires other binding partners, including myeloid differentiation factor 88 (MyD88) as an adaptor and NF-κB as a transcription factor. LPS binding occurs via the coordinated sequence of binding events between soluble and cell membrane proteins including LPS-binding protein, myeloid differentiation-2 (MD-2) protein and CD14 (15). CD14 is a key LPS co-receptor, pivotal in the initial binding of LPS and transfer of LPS to the MD2/TLR4 complex to initiate signal signaling cascades (16).
In the present study, constant stimulation by high concentration of LPS was used to determine whether LPS moderately upregulated TLR4 expression through a positive feedback pathway in a NF-κB-dependent manner. The results showed upregulated TLR4 at the protein and mRNA levels enriched through the TLR4 modulation style. However, cytokines such as tumor necrosis factor (TNF)-α and IL-2 were not detected. Thus, the positive feedback mechanism facilitates monocytes to differentiate into macrophages and rapidly enhances multiple cytokine expression (known as ‘a cytokine storm’) in order to react with the extracellular stress.
Materials and methods
Antibodies and reagents
CLI-095, also known as TAK-242, is a novel cyclohexene derivative that suppresses TLR4 signaling specifically. CLI-095 was purchased from InvivoGen (San Diego, CA, USA). Helenalin, an NF-κB inhibitor, was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA), and LPS (Escherichia coli) and β-actin were purchased from Sigma-Aldrich (St. Louis, MO, USA). The Alexa Fluor 488-labeled mouse anti-human antibodies against TLR4 and Alexa Fluor 488-labeled isotype controls (mouse IgG1) were obtained from BD Biosciences (Franklin Lakes, NJ, USA). Monoclonal rabbit NF-κB antibody (cat. no. 8242; dilution, 1:1,000), monoclonal rabbit NF-κB pho-Ser468 antibody (cat. no. 2642; dilution, 1:1,000) and tyrosine phosphorylation antibody were from Cell Signaling Technology, Inc. (Beverly, MA, USA). STAT3 inhibitor was STA-21 (sc-200757), 2 µmmol/l and NF-κB inhibitor was helenalin (sc-218579) (both from Santa Cruz Biotechnology, Inc.).
Cell culture
Human THP1 cells were treated in a time- and concentration-dependent manner. The cells were divided into the low LPS group, which was treated with 1 ng/ml LPS for 15 min; the high LPS group, treated with 20 ng/ml LPS for 2 h; and the mock group, treated with isovolumetric fetal bovine serum (FBS) for 15 min.
The cells were cultured with RPMI-1640 medium supplemented with 20% heat-inactivated FBS, 2 mmol/l L-glutamine, antibiotics (50 U/ml penicillin and 50 µg/ml streptomycin), and maintained at 37°C in a humidified 5% CO2 atmosphere. The differentiation of THP1 cells to macrophages involved a 6-day protocol and 3-day exposure to LPS (5 nmol/l), followed by 3 days of culture in the absence of LPS. Both concentrations of LPS were used to induce optimal macrophage differentiation.
Determination of TLR4 by FACS analysis
The levels of TLR4 were determined using FACScan analysis. Cells (1×106) were re-suspended in phosphate-buffered saline (PBS) containing 0.1% sodium azide and 5% FBS was incubated in ice for 30 min, followed by incubation with Alexa Fluor 488-conjugated anti-TLR4 antibody for 1 h. The cells were washed twice, fixed in 2% formaldehyde in PBS and analyzed using FACScan analysis. Negative controls were stained with isotype-matched Alexa Fluor 488-conjugated IgG and compensation was adjusted using the single-stained cell samples. Fluorescence intensities were determined using FlowJo software (Tree Star Inc., Ashland, OR, USA).
Semi-quantitative and RT-PCR
Reverse transcription was performed in a 20-µl reaction system with a total of 2 µg of RNA M-MLV reverse transcriptase (Toyobo, Osaka, Japan). Quantitative RT-PCR and RT-PCR were performed with an ABI PCR Thermal Cycler Dice detection system and SYBR-Green dye (Toyobo) according to the manufacturer's instructions. The primers used were: human TLR4 forward, TGGGCAACCTGCTCTACCTA and reverse, GCTGTAGCTCGTTGGCAGA; and GAPDH forward, ACAACTTTGGTATCGTGGAAGG and reverse, GCCATCACGCCACAGTTTC.
Chromatin immunoprecipitation (ChIP) and quantitative PCR
The method of treatment for THP1 cells is shown in Fig. 2. The cells were cross linked with 1% formaldehyde (Sigma-Aldrich) at 25°C for 15 min, rinsed twice with ice-cold PBS (pH 7.4), and collected in PBS prior to centrifugation at 2,000 × g for 5 min. Then cells were lysed in 1 ml of ChIP lysis buffer [50 mmol/l Tris-HCl (pH 8.1), 1% SDS and 10 mmol/l EDTA] and sonicated for 20 min with 30 sec on and off cycles at high settings (Bioruptor; Diagenode, Denville, NJ, USA) to produce chromatin fragments of 0.5 kb on average. Following centrifugation and quantification, 50 µl of the supernatants were used as inputs and the remainder was diluted 10-fold in IP buffer [10 mmol/l Tris-HCl (pH 8.1), 1% Triton X-100, 0.1% deoxycholate, 0.1% SDS, 90 mmol/l NaCl and 2 mmol/l EDTA] and an equivalent of 2×106 cells/time-point was subjected to IP overnight at 4°C, followed by 2 h pre-clearing with 20 µl of a 50% protein A/G-Sepharose beads (GE) slurry. These beads were prepared by two washings in IP buffer and a 3-h incubation with 25 µg shared salmon DNA and 200 µg BSA/ml of solution. The beads were then resuspended 1:1 in IP buffer and used for ChIPs. Anti-NF-κB (4 µl) and anti-RNA polymerase II (4 µl) were used in each ChIP experiment. Complexes were then recovered with 2-h incubation at 4°C with 20 µl of the protein A/G-Sepharose beads solution. The precipitates were washed, the beads removed and the DNA was precipitated by ethanol at −80°C for >2 h and then centrifuged at 9,000 × g for 10 min. The pellet was washed in 70% ethanol and recovered by TE buffer (pH 7.5).
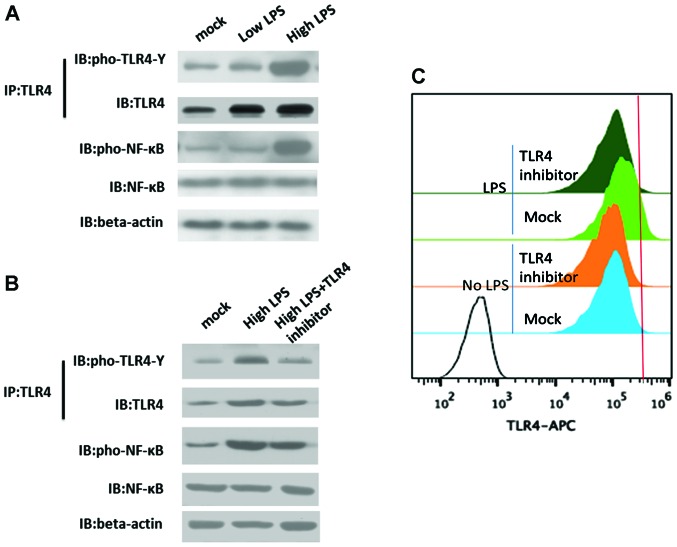
Figure 2.
LPS-induced activation of TLR4 is required to upregulate TLR4 expression. (A) LPS-induced TLR4 activation. (B) Inhibition of TLR4 activation prevents TLR4 feedback loop. (C) FACS showed TLR4 protein expression after treatment with TLR4 inhibitor in the absence or presence of LPS stimulation in the cell membrane. LPS stimulation condition was 4 h and at 20 ng/m LPS. TLR4 signaling inhibitor was CLI-095, 50 nmol/l. LPS, lipopolysaccharide; TLR4, Toll-like receptor 4.
Western blotting
Cells were collected and washed twice in PBS and the cell lysates were obtained by adding RIPA lysis buffer for 20 min, and centrifuging for 15 min at 9,000 × g. The supernatant was obtained and densitometric analysis was performed with Quantity One software (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The relative expression level of each protein was normalized by dividing the level of target protein by the level of GAPDH for each sample.
Statistical analysis
Statistical analysis was performed using the Statistical Package for Social Sciences (SPSS) for Windows (version 12.0; SPSS, Inc., Chicago, IL, USA). Student's t-tests were used to determine the statistical significance of the differences between the experimental groups. P<0.05 was considered to indicate a statistically significant difference. The graphs were created using Microcal Origin software (version 3.78; MicroCal Inc., Northampton, MA, USA).
Results
LPS moderately upregulates TLR4 expression in THP1 cells
Monocytes are activated by microbial components that are recognized by TLRs. After ligand binding, TLRs induce cytokine production and ultimately clear the infection. However, except for TLR activation, changes in the expression levels of TLRs following ligand binding were detected with difficulty. It was previously reported that the gene expression of TLR2 although not of TLR4 was induced by LPS in mouse macrophages (7).
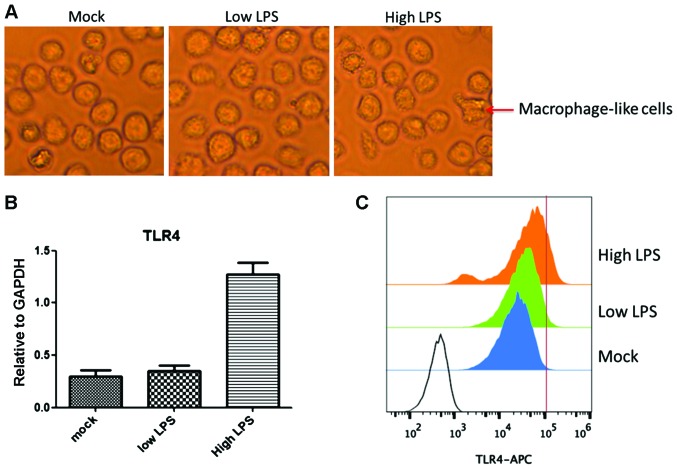
We found that TLR4 expression was upregulated by LPS treatment in a time- and concentration-dependent manner. Stimulation with 1 ng/ml LPS (low LPS) for 15 min had no obvious effect on TLR4 expression enhancement. By contrast, stimuli with 20 ng/ml LPS (high LPS) for 2 h increased TLR4 expression 4-fold compared to the level with low LPS (P<0.05) (Fig. 1B and C). Similarly, most THP1 cells in the high LPS group differentiated to macrophage-like cells, which did not occur in the low LPS group (Fig. 1A). A high concentration of LPS led to microbial infection and the rapid upregulation of TLR4 facilitates clearance of these extracellular invaders. These results suggested that the TLR4 transcriptional level was tightly regulated by a feedback mechanism following extracellular stimuli.
Figure 1.
LPS upregulated TLR4 expression, especially at a high concentration of LPS. (A) Morphology of THP1 cells under LPS stimulation. (B) RT-PCR tested TLR4 expression in the three treatment groups of THP1 cells, respectively. (C) TLR4 protein expression in the cell membrane using FACS. Three groups' treatment include: i) Mock group with PBS; ii) low LPS group with time of 15 min exposure and concentration of LPS at 1 ng/ml; and iii) high LPS group with time of 4 h exposure and concentration of LPS at 20 ng/ml. LPS, lipopolysaccharide; TLR4, Toll-like receptor 4.
LPS-induced TLR4 activation is required for upregulation of TLR4 expression
Activation of TLR4 by LPS only required 15 min, but the prolongation of stimulation had an obvious effect on upregulating TLR4 expression. The activation efficiency of TLR4 was dependent on the increased expression in the cell membrane, and continuous stimulation resulted in the monocytes obtaining the capacity to respond to external stimuli because of ever-increasing amounts of TLR4.
Compared to the mock and low LPS group, the tyrosine phosphorylation of TLR4 tyrosine in the high LPS group presented obvious enhancement. The downstream target of TLR4 signaling, NF-κB, in the high LPS group was also strongly activated (Fig. 2A). When treated with the TLR4 inhibitor, the expression of TLR4 decreased and the TLR4 activation was blocked (Fig. 2B and C). Suspended THP1 cell differentiation from monocyte to macrophage was also inhibited. The upregulation of TLR4 expression was increased by TLR4 activation, whereas the expression of TLR4 was decreased following stimulation by the TLR4 inhibitor. Thus, TLR4 activation was required to upregulate TLR4 expression.
NF-κB targets TLR4 in chromatin and promotes TLR4 expression
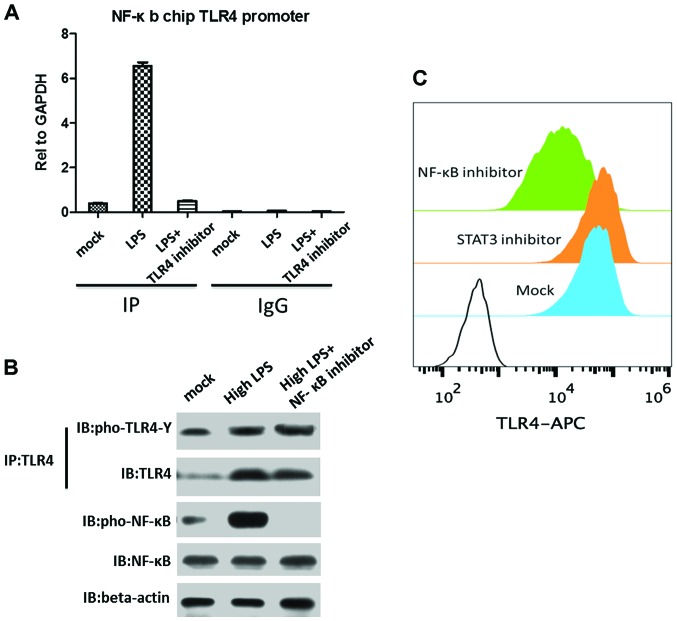
After TLR4 activation, MyD88 and other adaptors triggered a cascade of cell signals through the intracellular signal transduction domain such as NF-κB, signal transducer and activator of transcription (STAT)-3 and −1, or STAT1 and STAT3, thereby inducing the expression of inflammatory genes. To clarify TLR4 promoter, we used ChIP to study the regulation of TLR4 expression. LPS stimulation resulted in NF-κB targeting TLR4 promoter while STAT3 STAT1 and STAT1 STAT3 had no obvious binding to the TLR4 promoter (Fig. 3A), indicating that TLR4 activation was transduced by LPS signaling to NF-κB and not via STAT3 or STAT1 in THP1 cells. Furthermore, to identify the effect of NF-κB inhibitor on TLR4 expression by blocking the NF-κB transcription activity, we found that blocking NF-κB transcription activity resulted in markedly and rapidly decreased TLR4 expression. However, blocking NF-κB transcription activity had no effect on TLR4 tyrosine phosphorylation. NF-κB functioned as a transcription factor targeting chromatin, while TLR4 was stimulated by LPS and then phosphorylated.
Figure 3.
NF-κB targeted TLR4 in chromatin and promotes TLR4 expression. (A) ChIP assay for NF-κB binding to TLR4 gene promoter. (B) Blocking NF-κB activation blocked TLR4 expression. (C) TLR4 protein expression in the cell membrane by FACS. Three group treatment: i) Mock group, LPS stimulation without any inhibitor; ii) STAT3 inhibitor group, LPS stimulation with STAT3 inhibitor; and iii) NF-κB inhibitor group, LPS stimulation with NF-κB inhibitor. LPS stimulation condition was 4 h and at 20 ng/ml LPS. STAT3 inhibitor was STA-21 (sc-200757; Santa Cruz Biotechnology, Inc.), 2 µmmol/l. NF-κB inhibitor was helenalin (sc-218579; Santa Cruz Biotechnology, Inc.), 5 µmmol/l. NF-κB, nuclear factor-κB; TLR4, Toll-like receptor 4; ChIP, chromatin immunoprecipitation; LPS, lipopolysaccharide.
Discussion
In the present study, we investigated the gene expression of LPS signaling receptor TLR4 in THP1 cells. The gene expression of TLR4 was significantly increased in the presence of high LPS concentration. The results were consistent with the hypothesis that TLR4 is essential in LPS signaling. In non-stimulated THP1 cells, TLR4 mRNA was constitutively expressed and TLR4 may be essential to innate immunity in the first encounter with gram-negative bacteria. The higher concentration and longer constant LPS stimulation markedly promote TLR4 expression in the THP1 cell membrane during the process of differentiation to macrophages, indicating that a higher expression level of TLR4 has the ability to lead a ‘cytokine storm’.
Our findings were based on the measurements of mRNA and protein expression levels. However, the FACS results showed that the surface expression of TLR4 in THP1 cell membranes was less than the total TLR4 protein expression level. The surface expression of TLR4 may not necessarily correlate with that of the mRNA level, while the high LPS stimulation was able to supply sufficient signaling to reinforce TLR4 accumulation in the cell membrane.
TNF-α can upregulate TLR2 gene expression, and the serum TNF-α level increases in response to LPS. Thus, the same positive feedback mechanism may be present in the response of TLR4 expression to LPS, and as a consequence, TLR4 would probably contribute to accelerating immune responses in macrophages. If this occurs, the regulation of TLR4 expression may be one of the immune regulatory mechanisms commonly involved in host defense against many bacterial strains. THP1 cells were selected to regulate TLR4 expression with a positive-feedback loop in a high concentration of LPS for various reasons. First, according to the strength of external signals, the expression intensity of the downstream functional genes was induced by macrophages to prevent the invasion of external antigens and not waste energy. Second, the accumulation of TLR4 protein itself in THP1 cell membranes had the ability to transmit increased LPS signals. Third, the form of positive-feedback regulation of TLR4 signaling was efficient and rapid. To demonstrate this positive positive-feedback mechanism, the TLR4 inhibitor, CLI-095, was used to interrupt TLR4 signaling, and in turn, TLR4 protein in the THP1 cell membrane was almost depleted.
In macrophages, NF-κB activation seems to be essential for LPS-mediated TLR4 induction. LPS is a strong activator of NF-κB and all of the cytokines shown here that increase TLR4 expression are known as activators of NF-κB (2,3). Additionally, helenalin, an inhibitor of NF-κB transcriptional activity at high concentrations, completely inhibited LPS-mediated TLR4 induction at a concentration of 5 µmmol/l. Although helenalin is not a specific inhibitor of NF-κB activation, the dose response of TLR4 mRNA inhibition correlated well with that of NF-κB activation. Furthermore, STA-21, a specific inhibitor of STAT3 transcriptional activity was only slightly inhibited to LPS-mediated TLR4 induction. These results were noteworthy because NF-κB and STAT3 were in sharp contrast with regard to binding to TLR4 promoter in THP1 cells. This difference clearly suggested that the regulatory roles of NF-κB pathways in TLR4 mRNA exhibit conserved characteristics, while STAT3 may vary considerably in different cell types.
In conclusion, the data show that the expression of the LPS signaling receptor gene TLR4, was strictly regulated by LPS concentration and stimulation during THP1 cell differentiation into macrophages. TLR4 was constitutively expressed and remained constant after various stimulations, including LPS. However, a higher concentration of LPS significantly increased TLR4 expression. This suggests a positive-feedback mechanism. When gram-negative bacteria invade the host, macrophages first recognize LPS through the constitutively expressed TLR4, and then according to the strength of LPS signaling, which is accumulated by TLR4 expression. Recent studies have indicated that TLR4 mRNA expression in macrophages was decreased within a few hours of LPS treatment, whereas we did not observe any decrease in TLR4 mRNA until 4 h after the LPS treatment. This result may be due to the different concentration of LPS and cell types.
Limitations also existed in the present study. This study only revealed that NF-κB can inhibit TLR4 activity induced by LPS from the cellular and molecular level. However, how to transfer these findings to clinical applications remains unclear. In addition, the results of the present study should be confirmed in large-scale future studies.
Acknowledgements
The present study was supported by a grant from the Outstanding Subject Leader Training Plan of Health Bureau of Shanghai Pudong New Area (no. PWRd 2013-11).
References
- 1.Bellanti JA. Immunology; Proceedings of the Annual Meeting of the Medical Section of the American Life Insurance Association; Washington, D.C.. 1975; pp. 16–30. [PubMed] [Google Scholar]
- 2.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 3.Lien E, Sellati TJ, Yoshimura A, Flo TH, Rawadi G, Finberg RW, Carroll JD, Espevik T, Ingalls RR, Radolf JD, et al. Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J Biol Chem. 1999;274:33419–33425. doi: 10.1074/jbc.274.47.33419. [DOI] [PubMed] [Google Scholar]
- 4.Zeromski J, Mozer-Lisewska I, Kaczmarek M. Significance of toll-like receptors expression in tumor growth and spreading: a short review. Cancer Microenviron. 2008;1:37–42. doi: 10.1007/s12307-008-0005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.West AP, Brodsky IE, Rahner C, Woo DK, Erdjument-Bromage H, Tempst P, Walsh MC, Choi Y, Shadel GS, Ghosh S. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flo TH, Ryan L, Latz E, Takeuchi O, Monks BG, Lien E, Halaas Ø, Akira S, Skjåk-Braek G, Golenbock DT, et al. Involvement of toll-like receptor (TLR) 2 and TLR4 in cell activation by mannuronic acid polymers. J Biol Chem. 2002;277:35489–35495. doi: 10.1074/jbc.M201366200. [DOI] [PubMed] [Google Scholar]
- 7.Tsuboi N, Yoshikai Y, Matsuo S, Kikuchi T, Iwami K, Nagai Y, Takeuchi O, Akira S, Matsuguchi T. Roles of toll-like receptors in C-C chemokine production by renal tubular epithelial cells. J Immunol. 2002;169:2026–2033. doi: 10.4049/jimmunol.169.4.2026. [DOI] [PubMed] [Google Scholar]
- 8.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 9.Pfarr KM, Fischer K, Hoerauf A. Involvement of Toll-like receptor 4 in the embryogenesis of the rodent filaria Litomosoides sigmodontis. Med Microbiol Immunol. 2003;192:53–56. doi: 10.1007/s00430-002-0159-5. [DOI] [PubMed] [Google Scholar]
- 10.Lien E, Means TK, Heine H, Yoshimura A, Kusumoto S, Fukase K, Fenton MJ, Oikawa M, Qureshi N, Monks B, et al. Toll-like receptor 4 imparts ligand-specific recognition of bacterial lipopolysaccharide. J Clin Invest. 2000;105:497–504. doi: 10.1172/JCI8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300:1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 12.Zhang G, Ghosh S. Toll-like receptor-mediated NF-kappaB activation: a phylogenetically conserved paradigm in innate immunity. J Clin Invest. 2001;107:13–19. doi: 10.1172/JCI11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anders HJ, Banas B, Schlöndorff D. Signaling danger: toll-like receptors and their potential roles in kidney disease. J Am Soc Nephrol. 2004;15:854–867. doi: 10.1097/01.ASN.0000121781.89599.16. [DOI] [PubMed] [Google Scholar]
- 14.Territo MC, Cline MJ. Mononuclear phagocyte proliferation, maturation and function. Clin Haematol. 1975;4:685–703. [PubMed] [Google Scholar]
- 15.Suzuki Y, Ruiz-Ortega M, Lorenzo O, Ruperez M, Esteban V, Egido J. Inflammation and angiotensin II. Int J Biochem Cell Biol. 2003;35:881–900. doi: 10.1016/S1357-2725(02)00271-6. [DOI] [PubMed] [Google Scholar]
- 16.Wolf G, Wenzel U, Burns KD, Harris RC, Stahl RA, Thaiss F. Angiotensin II activates nuclear transcription factor-kappaB through AT1 and AT2 receptors. Kidney Int. 2002;61:1986–1995. doi: 10.1046/j.1523-1755.2002.00365.x. [DOI] [PubMed] [Google Scholar]
- 17.Leendertse M, Willems RJ, Giebelen IA, van den Pangaart PS, Wiersinga WJ, de Vos AF, Florquin S, Bonten MJ, van der Poll T. TLR2-dependent MyD88 signaling contributes to early host defense in murine Enterococcus faecium peritonitis. J Immunol. 2008;180:4865–4874. doi: 10.4049/jimmunol.180.7.4865. [DOI] [PubMed] [Google Scholar]