Abstract
Several studies point towards alteration in gut microbiota composition and function in coeliac disease, some of which can precede the onset of disease and/or persist when patients are on a gluten-free diet. Evidence also exists that the gut microbiota might promote or reduce coeliac-disease-associated immunopathology. However, additional studies are required in humans and in mice (using gnotobiotic technology) to determine cause–effect relationships and to identify agents for modulating the gut microbiota as a therapeutic or preventative approach for coeliac disease. In this Review, we summarize the current evidence for altered gut microbiota composition in coeliac disease and discuss how the interplay between host genetics, environmental factors and the intestinal microbiota might contribute to its pathogenesis. Moreover, we highlight the importance of utilizing animal models and long-term clinical studies to gain insight into the mechanisms through which host–microbial interactions can influence host responses to gluten.
Introduction
The gastrointestinal tract forms the body's largest interface with the external environment and is exposed to a vast amount of foreign material, including pathogenic and commensal bacteria, as well as food antigens. Oral tolerance is a key feature of the gut immune system, whereby a state of local and systemic unresponsiveness to food protein or systemic ignorance of commensal bacteria is maintained under homeostatic conditions. Intestinal homeostasis requires balanced interactions between the gut microbiota, dietary antigens and the host.1 Environmental factors that disrupt this relationship can contribute to a breakdown in intestinal homeostasis by directly influencing immune and barrier function, as well as the composition of the gut microbiota. This step in turn can lead to proinflammatory reactions to otherwise innocuous antigens and the development of chronic inflammation.2,3
Coeliac disease is a chronic immune-mediated enteropathy triggered by the ingestion of gluten, the water-insoluble protein fraction in wheat, rye and barley, in patients who are HLA-DQ2 or HLA-DQ8 positive. Patients with coeliac disease can experience a loss of oral tolerance to gluten any time throughout life.4 The increasing incidence of coeliac disease and the observation that only a small proportion of genetically susceptible individuals will develop active small intestinal inflammation suggest a role for additional environmental factors or host factors in disease pathogenesis.5–7 Genome-wide association studies (GWAS) have identified 39 non-HLA coeliac disease risk loci.8 Many of these loci harbour genes associated with immune function, including T-cell activation and development (that is, IL2 and IL21) and innate immunity (that is, TLR7 and TLR9).8 Another study published in 2012 identified an association between fucosyltransferase 2 (FUT2) and coeliac disease. The FUT2 gene encodes an enzyme that controls the expression of the A, B, and h blood group antigens in mucus and bodily secretions. In the intestinal mucosa, these antigens act as microbial anchors and carbon sources for the bacteria in the gastrointestinal tract,9 and defects in FUT2 are associated with altered intestinal microbiota.10 However, these non-HLA loci have been estimated to only contribute to 14% of the genetic variance of coeliac disease,8 suggesting a critical role for additional host and environmental factors in disease pathogenesis.
The awareness and understanding of coeliac disease has progressed in the past few decades; however, we are still far from fully understanding disease pathogenesis, the spectrum of disease manifestations and the factors involved in disease onset and progression. Coeliac disease is a unique autoimmune disease in that the trigger (gluten) and the major susceptibility genes have been identified. This knowledge makes it an ideal disease to study the environmental microbial factors that can act as disease modifiers, the interplay between microbiota and host genetics, and how these factors can be manipulated to prevent or improve treatment of this condition. The overall goal of this Review is to explore the current evidence for the intestinal microbiota as an environmental modifier in coeliac disease.
Gut microbiota in gut homeostasis
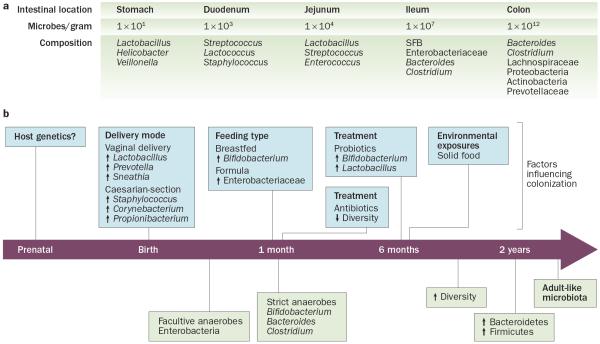
At birth, we are colonized with a complex community of microbes that reaches up to a density of 1 × 1012 bacterial cells per gramme of content in the adult colon (Figure 1).11 These microbes live in a symbiotic relationship with the host and are key determinants of health and disease by influencing nutrient absorption, barrier function and immune development. Even though the bacterial load in the colon is markedly higher than in the small bowel, evidence exists that the microbiota of the small bowel is in closer contact with the host because of a loose mucus layer, and that it has a critical role in shaping the immune system and inducing the production of antimicrobial peptides that in turn affect the colonic microbiota.11 A comprehensive study of the gut microbiota using culture-independent approaches determined that, unlike previously thought, the small intestine harbours facultative and strict anaerobes.12 Although less complex than the microbiota of the colon, Clostridium spp., Streptococcus spp. and coliforms are dominant groups in the small intestine. Moreover, this study indicated that the small intestinal microbial community rapidly responds metabolically to dietary changes.12
Figure 1.
Development of the gut microbiota. The composition and density of the microbiota varies along the length of the intestine as well as with age. a | Differences in microbial composition and density are observed along the length of the gastrointestinal tract, with much lower densities and greater variability in the proximal intestine. b | The gut microbiota fluctuates over the first 2–3 years of life, with high interindividual variability and low diversity, but becomes more stable over time. Immediately after birth, the neonatal intestine is colonized by facultative anaerobes and is dominated by Enterobacteriaceae. After the introduction of solid food, the gut microbiota continues to mature and the diversification of Bacteroides and Clostridium rapidly increases, whereas the proportion of Bifidobacterium stabilizes. By 2 years of age, the gut microbiota is dominated by members belonging to the Firmicutes and Bacteroidetes phyla and begins to resemble that of an adult-like microbiota. A number of factors, such as host genetics, birth delivery mode, diet, antibiotic or probiotic treatment and infections can influence the developing gut microbiota. Whether and how these host or environmentally triggered changes in gut microbiota also affect risk of inflammatory disease, including coeliac disease, remains controversial. Abbreviation: SFB, segmented filamentous bacteria.
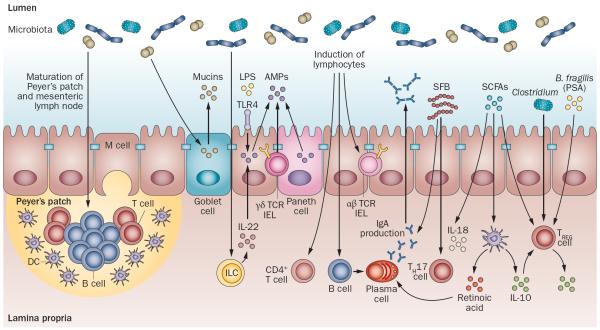
Studies using germ-free mice have demonstrated the importance of the microbiota on the development of host physiology and a functional immune system (Figure 2). In addition to gut morphological and functional differences,2,13,14 germ-free animals have immature organized lymphoid tissues,2,13,14 as well as reduced numbers of intestinal CD4+ T cells, small intestinal type 17 T helper (TH17) cells,15 colonic regulatory T (TREG) cells16 and T-cell receptor (TCR)αβ+ intraepithelial lymphocytes (IELs)17,18 compared with colonized mice. Differentiation of B cells to IgA-producing plasma cells is also dependent on the microbiota.19,20 Signals from the microbiota induce production of antimicrobial peptides (AMPs), such as RegIIIγ, from Paneth cells, γδTCR+ IELs and epithelial cells.14,21–23 The gut microbiota also stimulates the release of mucins from goblet cells.2 The microbiota might also be critical for the development of various innate lymphoid cell (ILC) subsets, and for the production of IL-22 from group 3 ILCs,24 which in turn stimulates AMP release from epithelial cells.25,26 Thus, microbiota–host interactions are key in the development of normal functional and immune responses to gut luminal antigens.
Figure 2.
Gut microbiota shapes host immunity. The gut microbiota induces maturation of the gastrointestinal lymphoid tissue (Peyer's patches, MLN). Signals from the microbiota induce production of AMPs, such as RegIIIγ, from Paneth cells, γδTCR+ IELs and epithelial cells. Microbial signals can also stimulate the development of ILC subsets, including IL-22-producing ILCs. Flagellin or LPS can stimulate AMP production from epithelial cells via IL-22 or TLR4, respectively. The gut microbiota also stimulates the release of mucins from goblet cells, and microbes influence the development of T-cell subsets, including CD4+ T cells, αβTCR+ IELs, and are critical for the induction of IgA-producing plasma cells. SFB are potent inducers of TH17 cells, whereas Clostridium, PSA derived from Bacteroides fragilis, and SCFAs stimulate TREG -cell differentiation. SCFAs can also promote IL-18 production from epithelial cells and promote IL-10 and retinoic acid production from DCs, which in turn promotes differentiation of TREG cells and IgA-producing plasma cells. Abbreviations: AMP, antimicrobial peptide; DC, dendritic cell; FasL, Fas ligand; IEL, intraepithelial lympocytes; ILC, innate lymphoid cell; LPS, lipopolysaccharide; MLN, mesenteric lymph node; NKG2D, NKG2-D type II integral membrane protein; PSA, polysaccharide A; SCFA, short-chain fatty acid; SFB, segmented filamentous bacteria; TCR, T-cell receptor; TH17 cell, type 17 T helper cell; TREG cell, regulatory T cell.
When studied individually, particular members of the gut microbiota can differentially modulate host responses (Figure 2). For example, flagellin, a bacterial structure that stimulates Toll-like receptor (TLR)5, can stimulate RegIIIγ production from epithelial cells via IL-22 release from dendritic cells.27 Flagella are associated with pathogenicity by promoting bacterial motility, cell adhesion and biofilm formation, constitute a virulence factor that can modulate host immune responses and are present in bacteria such as Escherichia coli and Salmonella.28,29 Furthermore, segmented filamentous bacteria (SFB) are potent inducers of TH17 cells in mice.30,31 A murine community of eight commensals, or altered Schaedler flora, induce balanced immune responses, which includes TREG cells as well as TH17 cells, but to a lesser extent than SFB.16 On the other hand, monocolonization of mice with Clostridium or Bacteroides fragilis induces colonic TREG-cell differentiation.16,32–34 Bacterial products, such as B. fragilis-derived polysaccharide A or short-chain fatty acids (SCFAs; for example, acetic acid, propionic acid and butyric acid), have also been shown to induce TREG cells.35–38 Products of bacterial metabolism (SCFAs) have also been shown to induce IL-18 production from epithelial cells and promote tolerogenic dendritic cells, which produce IL-10 and retinoic acid.39 Overall, these studies suggest that induction of immune responses by the gut microbiota is influenced not only by the presence or absence of live bacteria (germ-free versus colonized conditions), but also by the relative abundance of particular members of the microbiota and their by-products.
Thus, given the importance of host–microbial interactions on host immunity and physiology, disruptions in gut microbiota composition or function (dysbiosis) might have important implications for health and disease. Indeed, dysbiosis has been described in a number of chronic inflammatory diseases.3,40 However, the overall contribution of dysbiosis from disruption of homeostasis to disease development is not well understood.
The gut microbiota in coeliac disease
Approximately 30% of the general population carry the HLA-DQ2/8 coeliac disease susceptibility genes; however, only 2–5% of these individuals will go on to develop coeliac disease, suggesting that additional environmental factors contribute to disease development.41 The additional factors that influence coeliac disease development are unknown, but might include alterations in the intestinal microbiota. Indeed, some studies have demonstrated that patients with active coeliac disease have altered faecal and duodenal microbiota compositions compared with healthy individuals, which is partially restored after treatment with a gluten-free diet (Supplementary Table 1 online). Specifically, changes in the abundance of Firmicutes and Proteobacteria have been detected in children and adults with active coeliac disease.42,43 Other studies have reported decreases in the proportion of protective, anti-inflammatory bacteria such as Bifidobacterium, and increases in the proportion of Gram-negative bacteria such as Bacteroides and E. coli, in patients with active coeliac disease.44–46 Increases in the number of Staphylococcus44,46 and Clostridium,44,47 and decreases in Lactobacillus spp.46,48,49 have also been reported in children with coeliac disease. Altered diversity and altered metabolic function (SCFAs) of the microbiota have also been reported in patients with coeliac disease.46,50 A study demonstrated that the microbial composition of the gut in patients with coeliac disease was associated with the clinical manifestation of disease. The gut microbiota in patients experiencing gastrointestinal symptoms was dominated by Proteobacteria, whereas the microbiota of patients with dermatitis herpetiformis or individuals experiencing dyspepsia (controls) was dominated by Firmicutes.43 Increases in the number of Proteobacteria were also detected in patients with coeliac disease who were experiencing persistent symptoms, despite having normal histology and adhering to a gluten-free diet.51
The associations made between factors that influence microbial colonization, such as birth delivery mode, antibiotic use and breastfeeding and/or feeding practices (Figure 1b) and later disease development have shed more light on the potential role of microbes in coeliac disease. In Sweden, the annual incidence of coeliac disease in children was four times higher from 1985–1995 compared with previous years or children born in 1996–1997, which has been coined the “Swedish epidemic”. This increase in coeliac disease incidence coincided with changes in feeding practice guidelines, which included postponing gluten introduction from 4 months to 6 months, an increase in the average gluten consumption in children under 2 years of age, and with the identification of rod-shaped bacteria in small intestinal biopsy samples.52,53 These findings suggest that the interaction between feeding practices combined with subsequent changes induced in intestinal microbiota composition might have a role in coeliac disease development. Associations between delivery by Caesarean section and increased coeliac disease risk54,55 have been made, although are not consistent.56 Positive associations between early antibiotic use and later coeliac disease development have also been made.57 Breastfeeding might have a protective role, particularly if breastfeeding is maintained during gluten introduction.58 However, these findings are controversial, and several clinical trials found no association between early gluten introduction or duration of breastfeeding and subsequent coeliac disease risk.59–61
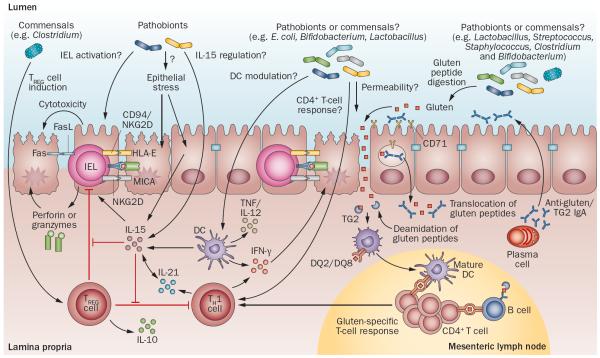
Together, these studies demonstrate that there are differences in microbial composition between patients with coeliac disease and healthy individuals as controls; however, the literature has not revealed a typical `coeliac microbiota signature' (Supplementary Table 1 online). This scenario is not unlike other chronic inflammatory gastrointestinal diseases, such as IBD or IBS, for which evidence supports an association between altered microbial composition and disease states.62–64 However, consensus across studies with respect to the specific changes involved is lacking and a disease-specific microbial signature has not yet been defined.65–67 Differences in the age of the study population (children versus adults), methodology (fluorescence in situ hybridization-PCR, denaturing gradient gel electrophoresis, 16s ribosomal RNA sequencing), sampling technique (biopsy versus faecal sample), length of gluten-free diet and the clinical presentation of disease could contribute to inconsistent findings in the literature. These differences make it difficult to compare across studies and determine whether the gut microbiota contributes to coeliac disease development or progression, or whether it is simply a consequence of the disease. Moreover, the exact mechanisms through which the gut microbiota might influence coeliac disease onset or progression is unknown, but could include activation of innate immune system, modulation of the epithelial barrier, or exacerbation of the gliadin-specific immune response (Figure 3).
Figure 3.
Potential microbial modulation of coeliac disease pathogenesis. Gluten peptides in the small intestinal lumen translocate the epithelial barrier, via transcellular or paracellular mechanisms and are deamidated by tissue TG2 in the lamina propria. Deamidated gliadin peptides are taken up by lamina propria DCs, inducing a proinflammatory gluten-specific CD4+ T-cell response, characterized by IFN-γ and IL-21 production, and anti-gliadin and anti-TG2 antibody production by B cells in genetically predisposed hosts. Activation of the innate immune response is also a key initial step in coeliac disease. Increased epithelial cell stress can upregulate stress molecules on epithelial cells (HLA-E, MICA/B) and induce IL-15 production from epithelial cells. IL-15 can induce IEL proliferation and activation and cytotoxic killing of epithelial cells, leading to tissue damage. IL-15 can also inhibit the regulatory effects of TREG cells and induce proinflammatory DCs. Microbes, both commensals or opportunistic pathogens (pathobionts), might contribute to the development of coeliac disease by influencing TREG-cell induction, epithelial cell stress, IEL activation or upregulation, IL-15 regulation, DC maturation and proinflammatory cytokine production, intestinal permeability modulation, gluten peptide digestion, and induction of CD4+ T-cell responses. Abbreviations: DC, dendritic cell; FasL, Fas ligand; IEL, intraepithelial lymphocyte; MICA, MHC class I polypeptide-related sequence A; NKG2D, NKG2-D type II integral membrane protein; TCR, T-cell receptor; TG2, tissue transglutaminase 2; TREG cell, regulatory T cell.
Genetic susceptibility and dysbiosis
In addition to environmental factors—such as birth delivery mode, diet and antibiotic use—host genetics can influence the composition of the gut microbiota (Figure 1). Although partly attributed to shared environmental influences, family members have more similar gut microbiotas than unrelated individuals.68–70 Human studies have shown gene–microbiota interactions at specific genetic loci, and associations between the abundance of bacterial taxa and genetic loci have been described.71–75 Similarly, in mice, the abundances of bacterial taxa have been linked to genotype76,77 and microbial differences have been detected between transgenic mice (for example, mice lacking pattern-recognition receptors) and wild-type controls.78,79 These microbial differences, however, might be due to a so-called maternal effect (mice from the same litter might have a more similar gut microbiota than mice from a different litter, even if they are genetically identical and reared in adjacent cages) or housing differences.78,80,81 Moreover, it was shown that diet dominates the host genome in shaping the gut microbiota in mice.82 These studies highlight the complexity and difficulty in delineating the influence of host genotype and/or environment interactions in shaping the gut microbiota, and emphasize the need for utilizing proper controls in experimental designs. Furthermore, human monozygotic and dizygotic twin pair studies have failed to detect a strong influence of genotype on microbial composition of the gut.69 Thus, the degree to which genotype can influence microbial composition or particular members of the gut microbiota is not fully understood, and this aspect remains a controversial topic. Moreover, how these gene–microbiota interactions might also be influenced by environmental factors and can contribute to disease susceptibility is unknown.
Nevertheless, a number of studies suggest that the interaction between genetics and the gut microbiota might play a part in coeliac disease susceptibility. In mice, the expression of different MHC genes were shown to influence the composition of the gut microbiota in otherwise genetically identical mice,83 suggesting that a coeliac-disease-associated genotype could influence gut microbial composition. In patients with coeliac disease, some changes in the gut microbiota are not restored after long-term treatment with a gluten-free diet, suggesting that these changes might be linked to the coeliac-disease-associated genotype (Supplementary Table 1). However, human data linking the high-risk genotypes, gut microbial composition and disease onset is lacking and several new studies have started to explore this aspect.
In a first study, infants at high genetic risk of coeliac disease were shown to have higher proportions of the Bacteroides–Prevotella group than infants at intermediate or low genetic risk of coeliac disease.84 Increased proportions of Gram-negative bacteria, E. coli, Streptococcus–Lactococcus, Eubacterium rectale–Clostridium coccoides, sulfate-reducing bacteria, C. lituseburense and C. histolyticum were also detected in high-risk infants compared with low-risk infants.84 However, the influence of environmental factors, such as milk-feeding type (breastfed versus formula fed) or birth delivery mode, in this study were not considered. A subsequent study also detected differences in the prevalence of Bacteroides spp. between infants at high-risk and low-risk of coeliac disease, some of which were independent from the influence of milk-feeding type.85 Similarly, in a cohort of 164 infants, those with increased genetic risk of developing coeliac disease had reduced numbers of Bifidobacterium spp. and B. longum and increased numbers of B. fragilis group and Staphylococcus spp. compared with infants with low risk of developing coeliac disease. However, differences in numbers of Bacteroides spp. and Bifidobacterium spp. were attenuated by breastfeeding whereas increased numbers of Staphylococcus spp. were found in high-risk breastfed and formula-fed infants. This finding suggests that the HLA-DQ genotype might have a more prominent role in Staphylococcus colonization.86 Another study found that the gut microbiota of infants at high risk of developing coeliac disease does not stabilize or resemble an adult microbiota by 2 years of age.87 Finally, a study of exclusively breastfed infants demonstrated that infants at high risk of coeliac disease (DQ2+) had increased proportions of Firmicutes (mainly due to the genera Clostridium), increased proportions of Proteobacteria (mainly due to Enterobacteriaceae and Raoultella) and decreased proportions of Actinobacteria (mainly due to Bifidobacterium and Corynebacterium).88 However, the precise role of these genotype-associated microbial profiles in the development of coeliac disease has not yet been determined and follow-up studies on these high-risk and low-risk cohorts are underway.
Other non-HLA genes associated with increased coeliac disease risk might also affect intestinal microbiota composition. For example, mutations leading to the absence of a functional FUT2 leads to a nonsecretor status, which has been associated with alterations in the composition of the gut microbiota in healthy individuals89 and in patients with Crohn's disease.73 The nonsecretor status was associated with decreases in faecal bifidobacteria,89 an increased risk of uropathogenic E. coli infections90 and is associated with coeliac disease.9
Evidence indicates that disease-associated microbiota profiles might be modulated by environmental factors. These existing studies are limited by a low number of patients, so future studies involving an increased number of participants and longer follow-up are needed to determine the interaction between the gut microbiota and disease-associated genotypes and whether this increased risk can be modified by environmental interventions. Animal models that express the human DQ2 or DQ8 genes can also be used to study gene–microbe interactions and the influence on gluten-induced immune responses (Table 1).
Table 1.
Animal models to study gluten-related disorders
| Model | Genetic background or association | Spontaneous or sensitization | Gluten-dependent enteropathy | AGA or anti-TG2 | Reference |
|---|---|---|---|---|---|
| Dog (Irish Setter) | No MHC class II association | Spontaneous | Partial villus atrophy | AGA | Batt et al. (1984)112; Hall & Batt (1992)113; Polvi et al. (1998)114 |
| Rhesus macaque | MHC class II association unknown | Spontaneous | Partial villus atrophy | AGA anti-TG2 | Sestak et al. (2011)115; Bethune et al.* (2008)116 |
| Horse | MHC class II association unknown | Spontaneous | Yes, partial villus atrophy | AGA anti-TG2 | van der Kolk et al. (2012)117 |
| Germ-free rats | Wistar-AVN, no MHC class II association | Gliadin-feeding following birth | Mild, number of IELs increased | No | Štepánková et al. (1996)96 |
| Mice | Balb/c | Gluten-feeding for 30 days | Partial villus atrophy, number of IELs increased | AGA | Papista et al. (2012)118 |
| Rag −/− | Transfer of T cells from sensitized C56BL/6 mice to Rag−/1 mice | Partial villus atrophy | AGA | Freitag et al. (2009)119 | |
| HLA-DQ8 transgenic, MHC class II dependent | Sensitization | Number of IELs increased | AGA | Black et al. (2002)120; Verdu et al. (2008)121 | |
| HLA-DQ2 transgenic, MHC class II dependent | Sensitization | No | AGA anti-TG2 | De Kauwe et al. (2009)122 | |
| NOD | Gluten-feeding | Mild, number of IELs increased | Not detected | Maurano et al. (2005)123 | |
| IL-15-DQ8 transgenic, MHC class II dependent | Gliadin-feeding for 10 days | Number of IELs increased | AGA anti-TG2 | DePaolo et al. (2011)104 |
Abbreviations: AGA, anti-gliadin antibody; IEL, intraepithelial lymphocyte; NOD, nonobese diabetic; TG2, tissue transglutaminase 2.
From association to causality
In vitro studies have demonstrated that microbes can influence immune responses to gluten. For example, the addition of Bifidobacterium, E. coli or Shigella to peripheral blood mononuclear cell and dendritic cell cultures can influence cytokine production after gliadin stimulation.91,92 Bifidobacterium strains have also been shown to prevent the generation of toxic gliadin peptides during in vitro digestion,93 and reduce gliadin-induced barrier and tight junction dysfunction in epithelial cell lines and intestinal loop models.94,95 Furthermore, E. coli and Shigella were shown to increase gliadin peptide translocation in intestinal loops.95 Although these in vitro studies demonstrate that microbes can influence gluten-induced immune responses and barrier function, the in vivo importance of these findings are unclear. Although associated with several limitations, animal models, including germ-free and gnotobiotic models (Table 1), will be of critical value to study whether the composition of the gut microbiota influences the loss of tolerance to gluten in genetically susceptible hosts and the mechanisms through which microbes can influence host responses to gluten.
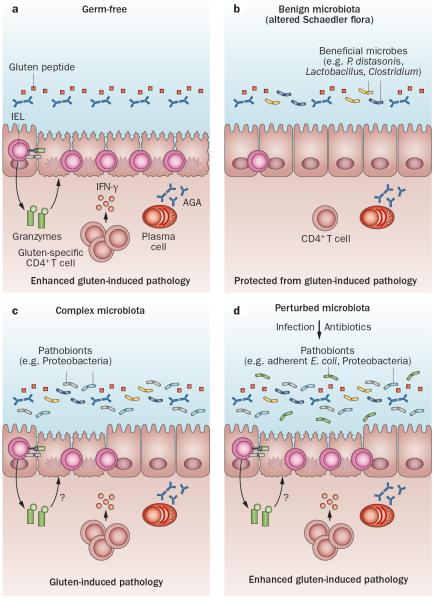
An early study in germ-free rats demonstrated that long-term gliadin, but not albumin, feeding from birth until day 63 induced moderate small intestinal damage, including crypt hyperplasia, villous atrophy and increased numbers of IELs, suggesting that gliadin, but not albumin, can induce mucosal damage in the absence of gut microbes.96 Transgenic nonobese diabetic-DQ8 mice, which express the human DQ8 gene,97 have been used to further study the influence of microbial colonization and composition on gluten-induced immune responses in genetically susceptible hosts. In agreement with the findings in germ-free rats, germ-free nonobese diabetic-DQ8 mice developed more severe gluten-induced pathology (characterized by increased numbers of IELs), reduced villus-to-crypt ratios, increased enterocyte cell death and increased IEL activation compared with colonized mice. In addition, germ-free mice developed a heightened proinflammatory, gluten-specific CD4+ T-cell response compared with colonized mice (Figure 4a). Altogether, these findings suggest that the gut microbiota might exert a beneficial effect by reducing the proinflammatory effects associated with gluten ingestion. In line with the concept that the gut microbiota can exert both a beneficial and harmful effect, the composition of the gut microbiota was found to influence gluten-induced pathology. Mice colonized with a benign microbiota, derived from the altered Schaedler flora that lacks any opportunistic pathogens, are protected from gluten-induced pathology, whereas mice colonized with a specific pathogen free (SPF) flora that includes Proteobacteria (Helicobacter and Escherichia) develop a moderate degree of gluten-induced pathology (Figure 4b,c). Supplementation of the benign microbiota with an adherent E. coli isolated from patients with coeliac disease led to gluten-induced pathology. Finally, perinatal antibiotic treatment, which led to increased numbers of Proteobacteria, including Escherichia, enhanced gluten-induced pathology98 (Figure 4d). These findings provide evidence that distinct changes in the gut microbiota can either ameliorate or worsen responses to gluten in genetically susceptible hosts and support the hypothesis that this microbiota might represent an environmental factor involved in coeliac disease development or progression. In addition, the data also provide evidence underlying the reported clinical association between antibiotic use and subsequent increased coeliac disease risk.57
Figure 4.
Modulation of host responses to gluten by the composition of the gut microbiota. a | In genetically susceptible hosts, the absence of bacteria can exacerbate gluten-induced immune responses, with increased IEL cytotoxicity (increased granzyme B production and NKG2D expression), increased proinflammatory gluten-specific CD4+ T-cell responses, and increased IgA AGA production. Together, this process leads to enhanced gluten-induced pathology characterized by reduced villous:crypt ratios and increased enterocyte cell death in the small intestine. b | In the presence of a limited, benign microbiota (altered Schaedler flora) that lacks any opportunistic pathogens, genetically susceptible hosts are protected from gluten-induced immune responses and pathology. c | In the presence of a complex microbiota that harbours opportunistic pathogens including Proteobacteria (Escherichia, Helicobacter), genetically susceptible hosts develop gluten-induced pathology, with increased IELs, proinflammatory gluten-specific T-cell responses and increased AGA production. However, the modulation of IEL cytotoxicity is unknown. d | Perturbation of a complex gut microbiota through antibiotic use or the presence of pathobionts can also exacerbate gluten-induced inflammation through unknown mechanisms. Abbreviations: AGA, anti-gliadin antibody; IEL, intraepithelial lymphocyte; NKG2D, NKG2-D type II integral membrane protein.
Another important question is whether dysbiosis could precede and promote development of coeliac disease by altering the intestinal environment and immune homeostasis. For instance, IL-15 was found to be a cytokine that promotes lymphokine killer activity in IELs from patients with active coeliac disease99–101 and patients with refractory sprue,102 promotes IEL survival,103 blocks inducible TREG-cell differentiation104 and blocks TREG cell and TGF-β mediated inhibition of effector T-cell function.105,106 Whether the gut microbiota contributes to upregulation of IL-15 in the intestinal mucosa remains to be determined, but this concept is supported by findings showing that TLR activation upregulates IL-15 expression.107,108 Furthermore, it remains to be determined whether IL-15 can alter the composition and/or function of the gut microbiota that in turn could enhance or reduce IL-15-mediated immunopathology.
Future research
In this Review, we have described the latest advances made in understanding the emerging role of the gut microbiota in the pathogenesis of coeliac disease. However, many issues remain unresolved. Although the studies in patients with coeliac disease clearly demonstrate an association between altered microbial composition and disease, a coeliac microbiota signature has not been identified and there is no consensus regarding bacterial species that are associated with active or inactive coeliac disease. These inconsistent findings make it difficult to elucidate the potential pathogenic role of gut microbes on the development or progression of coeliac disease in the clinical setting. Although proof-of-concept studies have demonstrated that the gut microbial composition can influence how a genetically susceptible host responds to gluten,98 mechanistic studies are lacking. In vitro evidence has provided some clues as to how certain microbes91,92 might influence gluten-induced immune responses. However, future studies utilizing a variety of genetically susceptible, gnotobiotically derived mouse models are needed to fully understand how and what specific microbes influence innate immunity, gluten-specific responses, barrier function and gluten metabolism, as well as whether these factors contribute to the development of coeliac disease. These types of studies are critical for determining causality, but their direct translational value might be limited due to the associated limitations with animal models of coeliac disease.109 Thus, clinical studies will be needed to provide translational value of basic studies. Long-term follow-up studies in high-risk versus low-risk infants within families could help to identify microbial signatures or species associated with disease onset, and will also help elucidate the role of genotype–environment–microbial interactions in coeliac disease. Finally, the identification of causal or coeliac-disease-promoting bacteria will provide opportunities for modulating the gut microbiota as a therapeutic or preventative approach. Indeed, early clinical trials (in both adults with active coeliac disease and children with newly diagnosed coeliac disease) with probiotics have reported some success in modulating varying aspects of gluten-induced inflammation and symptoms.110,111 Clinical studies have also shown associations between increased levels of Proteobacteria and persistent symptoms,51 suggesting modulation of the gut microbiota might improve ongoing symptoms, despite adhering to a gluten-free diet. However, a deeper understanding of the precise role of microbes in coeliac disease pathogenesis will aid in the development of appropriate and effective microbiota-modulating strategies.
Conclusions
Host–microbial interactions have a critical role in maintaining intestinal homeostasis by influencing host immunity and physiology. Disruption of these host–microbial interactions could contribute to the breakdown in intestinal homeostasis and to disease pathogenesis. However, the overall contribution of factors that disrupt homeostasis, such as dysbiosis, to disease development is poorly understood. Dysbiosis has been described in patients with coeliac disease, but studies have failed to find a distinct coeliac microbiota signature, making it difficult to determine the true pathogenic role of the gut microbiota in coeliac disease. However, the ongoing studies linking altered microbiota in high-risk infants to later coeliac disease development will provide insight into the influence of environmental factors on gene–microbe interactions and the potential role of gut microbes in coeliac disease onset or progression. Animal models have provided proof-of-concept studies demonstrating that host responses to gluten can be modified by the composition of the gut microbiota. The continued use of animal models will provide clues into the mechanisms by which the composition of the microbiota influences host responses to gluten. Ultimately, these studies could provide opportunities for modulating the gut microbiota as a therapeutic or preventative approach.
Supplementary Material
Key points.
-
▪
The intestinal microbiota coexists with its host in a continuum between homeostasis and pathogenicity; the upper gastrointestinal tract harbours a gut microbiota that is affected compositionally and metabolically by food components
-
▪
Coeliac disease is a chronic immune-mediated enteropathy caused by dietary gluten in genetically susceptible individuals
-
▪
The role of microbial factors in coeliac disease pathogenesis has been suggested
-
▪
Although clinical studies demonstrate that microbial changes are associated with coeliac disease, the individual microbes involved and underlying mechanisms remain elusive
-
▪
Emerging data in gnotobiotic models indicate that the intestinal microbiota has a complex modulatory role in host immune responses to gluten
-
▪
A deeper understanding of the precise role of microbes in coeliac disease pathogenesis will aid in the development of microbiota-modulating strategies, such as probiotics, to prevent or help treat the disease
Acknowledgements
The authors are supported by grants from the Canadian Institutes of Health Research to E.F.V. (MOP 123282); and the Digestive Diseases Research Core Centre (DK42086) at the University of Chicago and from the NIH (RO1DK67180) to B.J. E.F.V. holds a Canada Research Chair and H.J.G. a Canadian Celiac Association fellowship.
Footnotes
Competing interests The authors declare no competing interests.
Author contributions All authors contributed equally to all aspects of this manuscript.
Supplementary information is linked to the online version of the paper at www.nature.com/nrgastro.
References
- 1.Pabst O, Mowat A. Oral tolerance to food protein. Mucosal Immunol. 2012;5:232–239. doi: 10.1038/mi.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sommer F, Bäckhed F. The gut microbiota—masters of host development and physiology. Nat. Rev. Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 3.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catassi C, et al. Natural history of celiac disease autoimmunity in a USA cohort followed since 1974. Ann. Med. 2010;42:530–538. doi: 10.3109/07853890.2010.514285. [DOI] [PubMed] [Google Scholar]
- 5.Ludvigsson JF, et al. Increasing incidence of celiac disease in a North American population. Am. J. Gastroenterol. 2013;108:818–824. doi: 10.1038/ajg.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murray JA, et al. Trends in the identification and clinical features of celiac disease in a North American community, 1950–2001. Clin. Gastroenterol. Hepatol. 2003;1:19–27. doi: 10.1053/jcgh.2003.50004. [DOI] [PubMed] [Google Scholar]
- 7.White LE, et al. The rising incidence of celiac disease in Scotland. Pediatrics. 2013;132:e924–e931. doi: 10.1542/peds.2013-0932. [DOI] [PubMed] [Google Scholar]
- 8.Trynka G, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Gen. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parmar A, et al. Association study of FUT2 (rs601338) with celiac disease and inflammatory bowel disease in the Finnish population. Tissue Antigens. 2012;80:488–493. doi: 10.1111/tan.12016. [DOI] [PubMed] [Google Scholar]
- 10.Tong M, et al. Reprograming of gut microbiome energy metabolism by the FUT2 Crohn's disease risk polymorphism. ISME J. 2014;8:2193–2206. doi: 10.1038/ismej.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 2014;14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 12.Zoetendal EG, et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6:1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Aidy S, et al. Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol. 2012;5:567–579. doi: 10.1038/mi.2012.32. [DOI] [PubMed] [Google Scholar]
- 14.Natividad JM, et al. Differential induction of antimicrobial REGIII by the intestinal microbiota and Bifidobacterium breve NCC2950. Appl. Environ. Microbiol. 2013;79:7745–7754. doi: 10.1128/AEM.02470-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 17.Helgeland L, Vaage J, Rolstad B, Midtvedt T, Brandtzaeg P. Microbial colonization influences composition and T-cell receptor Vβ repertoire of intraepithelial lymphocytes in rat intestine. Immunology. 1996;89:494–501. doi: 10.1046/j.1365-2567.1996.d01-783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imaoka A, Matsumoto S, Setoyama H, Okada Y, Umesaki Y. Proliferative recruitment of intestinal intraepithelial lymphocytes after microbial colonization of germ-free mice. Eur. J. Immunol. 1996;26:945–948. doi: 10.1002/eji.1830260434. [DOI] [PubMed] [Google Scholar]
- 19.Uematsu S, et al. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat. Immunol. 2008;9:769–776. doi: 10.1038/ni.1622. [DOI] [PubMed] [Google Scholar]
- 20.Hapfelmeier S, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ismail AS, Behrendt CL, Hooper LV. Reciprocal interactions between commensal bacteria and γδ intraepithelial lymphocytes during mucosal injury. J. Immunol. 2009;182:3047–3054. doi: 10.4049/jimmunol.0802705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ismail AS, et al. γδ intraepithelial lymphocytes are essential mediators of host–microbial homeostasis at the intestinal mucosal surface. Proc. Natl Acad. Sci. USA. 2011;108:8743–8748. doi: 10.1073/pnas.1019574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaishnava S, et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanos SL, et al. RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22–producing NKp46+ cells. Nat. Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen VL, Kasper DL. Interactions between the intestinal microbiota and innate lymphoid cells. Gut Microbes. 2014;5:129–140. doi: 10.4161/gmic.27289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moro K, Koyasu S. Innate lymphoid cells, possible interaction with microbiota. Semin. Immunopathol. 2015;37:27–37. doi: 10.1007/s00281-014-0470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinnebrew MA, et al. Interleukin 23 production by intestinal CD103+ CD11b+ dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramos HC, Rumbo M, Sirard J-C. Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 2004;12:509–517. doi: 10.1016/j.tim.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Duan Q, Zhou M, Zhu L, Zhu G. Flagella and bacterial pathogenicity. J. Basic Microbiol. 2013;53:1–8. doi: 10.1002/jobm.201100335. [DOI] [PubMed] [Google Scholar]
- 30.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 31.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 33.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl Acad. Sci. USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hrncir T, Stepankova R, Kozakova H, Hudcovic T, Tlaskalova-Hogenova H. Gut microbiota and lipopolysaccharide content of the diet influence development of regulatory T cells: studies in germ-free mice. BMC Immunol. 2008;9:65. doi: 10.1186/1471-2172-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 38.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic TREG cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh N, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLean MH, Dieguez D, Miller LM, Young HA. Does the microbiota play a role in the pathogenesis of autoimmune diseases? Gut. 2014;64:332–341. doi: 10.1136/gutjnl-2014-308514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossi M, Schwartz KB. Editorial: Celiac disease and intestinal bacteria: not only gluten? J. Leukoc. Biol. 2010;87:749–751. doi: 10.1189/jlb.1209784. [DOI] [PubMed] [Google Scholar]
- 42.Sánchez E, Donat E, Ribes-Koninckx C, Fernández-Murga ML, Sanz Y. Duodenal-mucosal bacteria associated with celiac disease in children. Appl. Environ. Microbiol. 2013;79:5472–5479. doi: 10.1128/AEM.00869-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wacklin P, et al. The duodenal microbiota composition of adult celiac disease patients is associated with the clinical manifestation of the disease. Inflamm. Bowel Dis. 2013;19:934–941. doi: 10.1097/MIB.0b013e31828029a9. [DOI] [PubMed] [Google Scholar]
- 44.Collado MC, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Specific duodenal and faecal bacterial groups associated with paediatric coeliac disease. J. Clin. Pathol. 2009;62:264–269. doi: 10.1136/jcp.2008.061366. [DOI] [PubMed] [Google Scholar]
- 45.Collado M, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalances in faecal and duodenal Bifidobacterium species composition in active and non-active coeliac disease. BMC Microbiol. 2008;8:232. doi: 10.1186/1471-2180-8-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Cagno R, et al. Duodenal and faecal microbiota of celiac children: molecular, phenotype and metabolome characterization. BMC Microbiol. 2011;11:219. doi: 10.1186/1471-2180-11-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Palma G, et al. Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. 2010;10:63. doi: 10.1186/1471-2180-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanz Y, et al. Differences in faecal bacterial communities in coeliac and healthy children as detected by PCR and denaturing gradient gel electrophoresis. FEMS Immunol. Med. Microbiol. 2007;51:562–568. doi: 10.1111/j.1574-695X.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- 49.Nadal I, Donant E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J. Med. Microbiol. 2007;56:1669–1674. doi: 10.1099/jmm.0.47410-0. [DOI] [PubMed] [Google Scholar]
- 50.Schippa S, et al. A distinctive 'microbial signature'in celiac pediatric patients. BMC Microbiol. 2010;10:175. doi: 10.1186/1471-2180-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wacklin P, et al. Altered duodenal microbiota composition in celiac disease patients suffering from persistent symptoms on a long-term gluten-free diet. Am. J. Gastroenterol. 2014;109:1933–1941. doi: 10.1038/ajg.2014.355. [DOI] [PubMed] [Google Scholar]
- 52.Ivarsson A, et al. Epidemic of coeliac disease in Swedish children. Acta Paediatr. 2000;89:165–171. doi: 10.1080/080352500750028771. [DOI] [PubMed] [Google Scholar]
- 53.Ou G, et al. Proximal small intestinal microbiota and identification of rod-shaped bacteria associated with childhood celiac disease. Am. J. Gastroenterol. 2009;104:3058–3067. doi: 10.1038/ajg.2009.524. [DOI] [PubMed] [Google Scholar]
- 54.Decker E, et al. Cesarean delivery is associated with celiac disease but not inflammatory bowel disease in children. Pediatrics. 2010;125:e1433–e1440. doi: 10.1542/peds.2009-2260. [DOI] [PubMed] [Google Scholar]
- 55.Mårild K, Stephansson O, Montgomery S, Murray JA, Ludvigsson JF. Pregnancy outcome and risk of celiac disease in offspring: a nationwide case-control study. Gastroenterology. 2012;142:39–45. doi: 10.1053/j.gastro.2011.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emilsson L, Magnus MC, StØrdal K. Perinatal risk factors for development of celiac disease in children, based on the prospective Norwegian Mother and Child cohort study. Clin. Gastroenterol. Hepatol. 2015;13:921–927. doi: 10.1016/j.cgh.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Canova C, et al. Association of maternal education, early infections, and antibiotic use with celiac disease: a population-based birth cohort study in northeastern Italy. Am. J. Epidemiol. 2014;180:76–85. doi: 10.1093/aje/kwu101. [DOI] [PubMed] [Google Scholar]
- 58.Ivarsson A, et al. Prevalence of childhood celiac disease and changes in infant feeding. Pediatrics. 2013;131:e687–e694. doi: 10.1542/peds.2012-1015. [DOI] [PubMed] [Google Scholar]
- 59.Lionetti E, et al. Introduction of gluten, HLA status, and the risk of celiac disease in children. N. Engl. J. Med. 2014;371:1295–1303. doi: 10.1056/NEJMoa1400697. [DOI] [PubMed] [Google Scholar]
- 60.Vriezinga SL, et al. Randomized feeding intervention in infants at high risk for celiac disease. N. Engl. J. Med. 2014;371:1304–1315. doi: 10.1056/NEJMoa1404172. [DOI] [PubMed] [Google Scholar]
- 61.Jansen MA, et al. Infant feeding and anti-tissue transglutaminase antibody concentrations in the Generation R Study. Am. J. Clin. Nutr. 2014;100:1095–1101. doi: 10.3945/ajcn.114.090316. [DOI] [PubMed] [Google Scholar]
- 62.Collins SM. A role for the gut microbiota in IBS. Nat. Rev. Gastroenterol. Hepatol. 2014;11:497–505. doi: 10.1038/nrgastro.2014.40. [DOI] [PubMed] [Google Scholar]
- 63.Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel disease. Semin. Immunopathol. 2015;37:47–55. doi: 10.1007/s00281-014-0454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 65.Rajilić-Stojanović M, et al. Intestinal microbiota and diet in IBS: causes, consequences, or epiphenomena? Am. J. Gastroenterol. 2015;110:278–287. doi: 10.1038/ajg.2014.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simrén M, et al. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut. 2012;62:159–176. doi: 10.1136/gutjnl-2012-302167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walters WA, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014;588:4223–4233. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tims S, et al. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J. 2013;7:707–717. doi: 10.1038/ismej.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frank DN, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 2011;17:179–184. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khachatryan ZA, et al. Predominant role of host genetics in controlling the composition of gut microbiota. PLoS ONE. 2008;3:e3064. doi: 10.1371/journal.pone.0003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rausch P, et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc. Natl Acad. Sci. USA. 2011;108:19030–19035. doi: 10.1073/pnas.1106408108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rehman A, et al. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011;60:1354–1362. doi: 10.1136/gut.2010.216259. [DOI] [PubMed] [Google Scholar]
- 75.Goodrich JK, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Benson AK, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl Acad. Sci. USA. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McKnite AM, et al. Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS ONE. 2012;7:e39191. doi: 10.1371/journal.pone.0039191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 79.Natividad JMM, et al. Commensal and probiotic bacteria influence intestinal barrier function and susceptibility to colitis in Nod1−/−; Nod2−/− mice. Inflamm. Bowel Dis. 2012;18:1434–1446. doi: 10.1002/ibd.22848. [DOI] [PubMed] [Google Scholar]
- 80.Robertson SJ, et al. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes. 2013;4:222–231. doi: 10.4161/gmic.24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ubeda C, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med. 2012;209:1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carmody RN, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Toivanen P, Vaahtovuo J, Eerola E. Influence of major histocompatibility complex on bacterial composition of fecal flora. Infect. Immun. 2001;69:2372–2377. doi: 10.1128/IAI.69.4.2372-2377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Palma G. d., Nova E, Pozo T, Sanz Y. Interplay between human leukocyte antigen genes and the microbial colonization process of the newborn intestine. Curr. Issues Mol. Biol. 2010;12:1–10. [PubMed] [Google Scholar]
- 85.Sánchez E, et al. Influence of environmental and genetic factors linked to celiac disease risk on infant gut colonization by Bacteroides species. Appl. Environ. Microbiol. 2011;77:5316–5323. doi: 10.1128/AEM.00365-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.De Palma G, et al. Influence of milk-feeding type and genetic risk of developing coeliac disease on intestinal microbiota of infants: the PROFICEL study. PLoS ONE. 2012;7:e30791. doi: 10.1371/journal.pone.0030791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sellitto M, et al. Proof of concept of microbiome-metabolome analysis and delayed gluten exposure on celiac disease autoimmunity in genetically at-risk infants. PLoS ONE. 2012;7:e33387. doi: 10.1371/journal.pone.0033387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Olivares M, et al. The HLA-DQ2 genotype selects for early intestinal microbiota composition in infants at high risk of developing coeliac disease. Gut. 2014;64:406–417. doi: 10.1136/gutjnl-2014-306931. [DOI] [PubMed] [Google Scholar]
- 89.Wacklin P, et al. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS ONE. 2011;6:e20113. doi: 10.1371/journal.pone.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stapleton A, Nudelman E, Clausen H, Hakomori S-I, Stamm W. Binding of uropathogenic Escherichia coli R45 to glycolipids extracted from vaginal epithelial cells is dependent on histo-blood group secretor status. J. Clin. Invest. 1992;90:965–972. doi: 10.1172/JCI115973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.De Palma G, Cinova J, Stepankova R, Tuckova L, Sanz Y. Pivotal Advance: Bifidobacteria and Gram-negative bacteria differentially influence immune responses in the proinflammatory milieu of celiac disease. J. Leukoc. Biol. 2010;87:765–778. doi: 10.1189/jlb.0709471. [DOI] [PubMed] [Google Scholar]
- 92.De Palma G, et al. Modulation of phenotypic and functional maturation of dendritic cells by intestinal bacteria and gliadin: relevance for celiac disease. J. Leukoc. Biol. 2012;92:1043–1054. doi: 10.1189/jlb.1111581. [DOI] [PubMed] [Google Scholar]
- 93.Laparra JM, Sanz Y. Bifidobacteria inhibit the inflammatory response induced by gliadins in intestinal epithelial cells via modifications of toxic peptide generation during digestion. J. Cell. Biochem. 2010;109:801–807. doi: 10.1002/jcb.22459. [DOI] [PubMed] [Google Scholar]
- 94.Lindfors K, et al. Live probiotic Bifidobacterium lactis bacteria inhibit the toxic effects induced by wheat gliadin in epithelial cell culture. Clin. Exp. Immunol. 2008;152:552–558. doi: 10.1111/j.1365-2249.2008.03635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cinova J, et al. Role of intestinal bacteria in gliadin-induced changes in intestinal mucosa: study in germ-free rats. PLoS ONE. 2011;6:e16169. doi: 10.1371/journal.pone.0016169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Štepánková R, Tlaskalova-Hogenova H, Šinkora J, Jodl J, Fric P. Changes in jejunal mucosa after long-term feeding of germfree rats with gluten. Scand. J. Gastroenterol. 1996;31:551–557. doi: 10.3109/00365529609009127. [DOI] [PubMed] [Google Scholar]
- 97.Galipeau HJ, et al. Sensitization to gliadin induces moderate enteropathy and insulitis in nonobese diabetic-DQ8 mice. J. Immunol. 2011;187:4338–4346. doi: 10.4049/jimmunol.1100854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Galipeau H, et al. Gluten-induced responses in NOD/DQ8 mice are influenced by bacterial colonization [abstract Tu1749] Gastroenterology. 2014;146(Suppl. 1):S833. [Google Scholar]
- 99.Roberts AI, et al. Cutting edge: NKG2D receptors induced by IL-15 costimulate CD28-negative effector CTL in the tissue microenvironment. J. Immunol. 2001;167:5527–5530. doi: 10.4049/jimmunol.167.10.5527. [DOI] [PubMed] [Google Scholar]
- 100.Meresse B, et al. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity. 2004;21:357–366. doi: 10.1016/j.immuni.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 101.Tang F, et al. Cytosolic PLA2 is required for CTL-mediated immunopathology of celiac disease via NKG2D and IL-15. J. Exp. Med. 2009;206:707–719. doi: 10.1084/jem.20071887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mention JJ, et al. Interleukin 15: a key to disrupted intraepithelial lymphocyte homeostasis and lymphomagenesis in celiac disease. Gastroenterology. 2003;125:730–745. doi: 10.1016/s0016-5085(03)01047-3. [DOI] [PubMed] [Google Scholar]
- 103.Malamut G, et al. IL-15 triggers an antiapoptotic pathway in human intraepithelial lymphocytes that is a potential new target in celiac disease–associated inflammation and lymphomagenesis. J. Clin. Invest. 2010;120:2131–2143. doi: 10.1172/JCI41344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.DePaolo RW, et al. Co-adjuvant effects of retinoic acid and IL-15 induce inflammatory immunity to dietary antigens. Nature. 2011;471:220–224. doi: 10.1038/nature09849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Benahmed M, et al. Inhibition of TGF-β signaling by IL-15: a new role for IL-15 in the loss of immune homeostasis in celiac disease. Gastroenterology. 2007;132:994–1008. doi: 10.1053/j.gastro.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 106.Hmida NB, et al. Impaired control of effector T cells by regulatory T cells: a clue to loss of oral tolerance and autoimmunity in celiac disease. Am. J. Gastroenterol. 2011;107:604–611. doi: 10.1038/ajg.2011.397. [DOI] [PubMed] [Google Scholar]
- 107.Zhang X, Sun S, Hwang I, Tough DF, Sprent J. Potent and selective stimulation of memory-phenotype CD8+ T cells in vivo by IL-15. Immunity. 1998;8:591–599. doi: 10.1016/s1074-7613(00)80564-6. [DOI] [PubMed] [Google Scholar]
- 108.Mattei F, Schiavoni G, Belardelli F, Tough DF. IL-15 is expressed by dendritic cells in response to type I IFN, double-stranded RNA, or lipopolysaccharide and promotes dendritic cell activation. J. Immunol. 2001;167:1179–1187. doi: 10.4049/jimmunol.167.3.1179. [DOI] [PubMed] [Google Scholar]
- 109.Korneychuk N, Meresse B, Cerf-Bensussan N. Lessons from rodent models in celiac disease. Mucosal Immunol. 2015;8:18–28. doi: 10.1038/mi.2014.102. [DOI] [PubMed] [Google Scholar]
- 110.Olivares M, Castillejo G, Varea V, Sanz Y. Double-blind, randomised, placebo-controlled intervention trial to evaluate the effects of Bifidobacterium longum CECT 7347 in children with newly diagnosed coeliac disease. Br. J. Nutr. 2014;112:30–40. doi: 10.1017/S0007114514000609. [DOI] [PubMed] [Google Scholar]
- 111.Smecuol E, et al. Exploratory, randomized, double-blind, placebo-controlled study on the effects of Bifidobacterium infantis natren life start strain super strain in active celiac disease. J. Clin. Gastroenterol. 2013;47:139–147. doi: 10.1097/MCG.0b013e31827759ac. [DOI] [PubMed] [Google Scholar]
- 112.Batt R, Carter M, McLean L. Morphological and biochemical studies of a naturally occurring enteropathy in the Irish setter dog: a comparison with coeliac disease in man. Res. Vet. Sci. 1984;37:339–346. [PubMed] [Google Scholar]
- 113.Hall E, Batt R. Dietary modulation of gluten sensitivity in a naturally occurring enteropathy of Irish setter dogs. Gut. 1992;33:198–205. doi: 10.1136/gut.33.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Polvi A, et al. Genetic susceptibility to gluten sensitive enteropathy in Irish setter dogs is not linked to the major histocompatibility complex. Tissue Antigens. 1998;52:543–549. doi: 10.1111/j.1399-0039.1998.tb03085.x. [DOI] [PubMed] [Google Scholar]
- 115.Sestak K, et al. Recognition of epidermal transglutaminase by IgA and tissue transglutaminase 2 antibodies in a rare case of rhesus dermatitis. J. Vis. Exp. 2011;58:e3154. doi: 10.3791/3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bethune MT, et al. A non-human primate model for gluten sensitivity. PLoS ONE. 2008;3:e1614. doi: 10.1371/journal.pone.0001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.van der Kolk J, et al. Gluten-dependent antibodies in horses with inflammatory small bowel disease (ISBD) Vet. Q. 2012;32:3–11. doi: 10.1080/01652176.2012.675636. [DOI] [PubMed] [Google Scholar]
- 118.Papista C, et al. Gluten induces coeliac-like disease in sensitised mice involving IgA, CD71 and transglutaminase 2 interactions that are prevented by probiotics. Lab. Invest. 2012;92:625–635. doi: 10.1038/labinvest.2012.13. [DOI] [PubMed] [Google Scholar]
- 119.Freitag T, et al. Gliadin-primed CD4+CD45RBlowCD25-effector/memory T cells drive gluten-dependent small intestinal damage after adoptive transfer into lymphopenic mice. Gut. 2009;58:1597–1605. doi: 10.1136/gut.2009.186361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Black KE, Murray JA, David CS. HLA-DQ determines the response to exogenous wheat proteins: a model of gluten sensitivity in transgenic knockout mice. J. Immunol. 2002;169:5595–5600. doi: 10.4049/jimmunol.169.10.5595. [DOI] [PubMed] [Google Scholar]
- 121.Verdu EF, et al. Gliadin-dependent neuromuscular and epithelial secretory responses in gluten-sensitive HLA-DQ8 transgenic mice. Am J. Physiol. Gastrointest. Liver Physiol. 2008;294:G217–G225. doi: 10.1152/ajpgi.00225.2007. [DOI] [PubMed] [Google Scholar]
- 122.de Kauwe AL, et al. Resistance to celiac disease in humanized HLA-DR3-DQ2-transgenic mice expressing specific anti-gliadin CD4+ T cells. J. Immunol. 2009;182:7440–7450. doi: 10.4049/jimmunol.0900233. [DOI] [PubMed] [Google Scholar]
- 123.Maurano F, et al. Small intestinal enteropathy in non-obese diabetic mice fed a diet containing wheat. Diabetologia. 2005;48:931–937. doi: 10.1007/s00125-005-1718-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.