Abstract
Hepatocellular carcinoma (HCC) remains one of the most common types of cancer worldwide and prognosis remains poor. Previous studies have suggested that long non-coding RNAs (lncRNAs) may be key regulators of tumor development and progression in HCC. It has been determined that 61–72% of transcribed regions contain lncRNAs in the antisense orientation (aslncRNAs). However, the function of aslncRNAs in HCC remains to be elucidated. The present study investigated the function of the aslncRNA zinc finger E-box binding homeobox 2 antisense RNA 1 (ZEB2-AS1) in 40 HCC tissues and 5 different human HCC cell lines using reverse transcription-quantitative polymerase chain reaction. Additionally, the expression levels of ZEB2-AS1 were downregulated by transfection of small interfering RNAs (siRNAs) to determine whether ZEB2-AS1 is capable of affecting cell proliferation, invasion and metastasis by regulating ZEB2, vimentin, fibronectin, E-cadherin and N-cadherin expression levels. The results of the present study demonstrated that the expression levels of ZEB2-AS1 were greater in HCC tissues when compared with the adjacent normal tissues. Furthermore, ZEB2-AS1 expression was significantly associated with the size of the primary tumor, intrahepatic metastasis and tumor-node-metastasis stage. The Kaplan-Meier survival curves suggested that patients with high ZEB2-AS1 expression levels experienced the lowest overall and recurrence-free survival rates compared with those that had low expression levels. In addition, the current study demonstrated that the downregulation of ZEB2-AS1 was associated with decreased tumor growth and metastasis in HCC by the regulation of the expression levels of epithelial mesenchymal transition-induced markers. In conclusion, lncRNA ZEB2-AS1 may be used as a valuable biomarker in patients with HCC.
Keywords: antisense orientation long non-coding RNA, zinc finger E-box binding homeobox 2 antisense RNA 1, tumor growth, metastasis, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) has been deemed the fifth most common type of malignant tumor. HCC has the third highest cancer-related mortality rate and a previous study have estimated that 500,000–1,000,000 novel cases of HCC are diagnosed annually (1). Invasion, metastasis and postoperative recurrence are predominant causes of mortality in patients with HCC and are the primary factors that affect clinical treatment and prognosis. Therefore, due to aggressive metastasis and invasion, HCC is frequently diagnosed at an advanced stage in patients and has a high lethality (2). A previous study demonstrated that aberrant activation of epithelial mesenchymal transition (EMT) is important for the process of tumor cell invasion and metastasis. EMT triggers multiple biochemical changes in normal hepatic epithelial cells, which ultimately enable them to assume a mesenchymal cell phenotype. This leads to enhanced migratory capacity, invasiveness, elevated resistance to apoptosis and increased production of ECM components (3). EMT is involved in multiple molecular mechanisms and signal transduction pathways. Their main function being the downregulation of the cell adhesion molecule E-cadherin and the upregulation of the mesenchymal molecule vimentin. In addition, a number of transcriptional factors have been identified as crucial for the induction of EMT, including Snail homologues, basic helix-loop-helix and zinc-finger E-box-binding homeobox (ZEB) (4). The ZEB family (consisting of ZEB1 and ZEB2) has a consistent inverse correlation with E-cadherin and vimentin in numerous types of cancer (5).
Long non-coding RNAs (lncRNAs) are a type of RNA molecule that are over 200 nucleotides in length and do not have protein-coding potential (6). They have been identified to affect multiple biological regulatory processes, including development, differentiation and carcinogenesis (7,8). A previous study suggested that the dysregulation of lncRNAs may be associated with the development and metastasis of various types of cancer (9). Antisense long non-coding RNAs (aslncRNAs) are a type of lncRNAs that are oriented in antisense direction with respect to a protein coding locus. They are transcribed frequently in human cells and 61–72% of all transcribed regions possess aslncRNAs (10). Additionally, aslncRNAs have been associated with tumor recurrence, metastasis and prognosis in various types of cancer.
A recent study by Li et al (11) identified the ZEB 1 antisense RNA 1 (ZEB1-AS1) gene as a non-coding oncogene involved in the tumorigenesis of HCC (11). It was determined that higher expression levels of lncRNA ZEB1-AS1 were observed in HCC tissues, compared with the adjacent normal tissues. Li et al (11) also described the effects of the ZEB1-AS1 lncRNA on cell proliferation, migration and invasion, and cell cycle regulation. In addition, it was determined that ZEB1-AS1 may control ZEB1 expression levels and promote tumor growth and metastasis in patients with HCC. Sequence analysis was performed and revealed that there was also a non-coding antisense transcript in the promoter sequence of ZEB2 (12). The function of ZEB2 antisense RNA 1 (ZEB2-AS1) remains to be elucidated. Therefore, the present study investigated whether lncRNA ZEB2-AS1 contributes to HCC development and how it may alter cell growth and metastasis by regulating ZEB2, vimentin, fibronectin, E-cadherin and N-cadherin expression levels. Additionally, the clinical characteristics of patients that may influence the expression levels of ZEB2-AS1 and provide data for the prognosis and targeted therapy of HCC were determined.
Materials and methods
Patient and tumor samples
HCC and adjacent normal tissues were collected following curative surgical resection from 40 patients with HCC in the Zhongnan Hospital of Wuhan University (Wuhan, China) between May 2010 and May 2012. The present study was approved by the ethics committee of the Zhongnan Hospital of Wuhan University and written informed consent was obtained from all patients. Curative resection was defined as the removal of all recognizable tumor tissue with a clear microscopic margin. Patients were not treated using any preoperative therapy, such as transcatheter arterial chemoembolization, radiofrequency ablation or percutaneous ethanol injection. The collected tissue samples were immediately frozen in liquid nitrogen and stored at −80°C until they were used for RNA extraction. Tumor tissues were divided into low-expression (n=20) and high expression (n=20) groups based on the mean expression level of all tumor tissues (n=40). Using the patient medical documents, a 48-month follow-up survival survey was conducted.
Cell lines and culture
The Hep3B, HepG2, Huh-7 and SMMC-7721 HCC cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). The HCCLM9 cell line was obtained from the Liver Cancer Institute, Fudan University (Shanghai, China) (13). All cells were cultured in RPMI-1640 medium, which was supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 1% penicillin and streptomycin in a humidified atmosphere of 5% CO2 at 37°C.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from HCC and normal adjacent tissues, and human HCC cell lines using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RNA was converted into cDNA (124 µg/20 µl) using a Takara RNA PCR kit (Takara Biotechnology Co., Ltd., Dalian, China). qPCR was performed using a iQ SYBR-Green Supermix (Bio-Rad Laboratories, Inc., Hercules, CA, USA) with 124 ng/2 µl cDNA. All reactions were performed in triplicate on iCycler IQ multi-color Detection System (Bio-Rad Laboratories, Inc.). The thermocycling conditions were as follows: 95°C for 5 min; 40 cycles of denaturation at 95°C for 30 sec; annealing at 60°C for 30 sec and extension at 72°C for 20 sec. The following primers were used: ZEB2-AS1 forward (F) 5′-ATGAAGAAGCCGCGAAGTGT-3′ and reverse (R) 5′-CACACCCTAATACACATGCCCT-3′; and β-actin F 5′-AGTTGCGTTACACCCTTTCTTGAC-3′ and R 5′-GCTCGCTCCAACCGACTGC-3′. The comparative cycle quantification (Cq) method (14) was used to quantify the relative expression levels of mRNA. The relative amount was calculated using the equation 2−ΔΔCq where ΔCq = (Cq of interest mRNA - Cq of β-actin). All the qPCR reactions were performed in duplicate.
ZEB2-AS1 small interfering RNAs (siRNAs) and transfection
The siRNA-ZEB2-AS1 sequence used was 5′-CAAAGGACACCTTTGGTTACCTGAA-3′. Synthetic sequence-scrambled siRNA (Santa Cruz Biotechnology Inc., Dallas, TX, USA) was used as a negative control (siRNA-NC). The siRNAs were transfected into Huh-7 cells during the logarithmic growth phase using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Cells were incubated with the siRNA transfection complex for 24 h. Next, cells were harvested in order to assess mRNA and protein expression level changes using RT-qPCR and western blot analysis at 24 h. For the conventional PCR, the relative mRNA expression level was represented as the grayscale ratio of the mRNA of interest to β-actin.
Western blot analysis
Proteins from HCC human cell lines were extracted using a radioimmunoprecipitation lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China). The proteins were quantified by the bicinchoninic acid method and samples (7530 µg/ml) were then separated on 10% sodium dodecyl sulfate-polyacrylamide gel and transferred onto polyvinylidene fluoride membranes. The membranes were blocked with 5% skimmed milk powder in Tris-buffered saline and Tween 20 (TBST; 10 mM Tris-HCl, pH 7.5, 150 mM NaCl and 0.1% Tween-20) at room temperature for 1 h. The following primary antibodies were added and membranes were incubated overnight at 4°C: Mouse monoclonal anti-ZEB2 antibody (1:200; Santa Cruz Biotechnology, Inc.; cat. no. sc-271984), mouse monoclonal anti-vimentin (1:200; Santa Cruz Biotechnology, Inc.; cat. no. sc-6260), mouse monoclonal E-cadherin (1:500; Abcam, Cambridge, UK; cat. no. ab1416), rabbit polyclonal anti-N-cadherin (1:1,000; Abcam; cat. no. ab18203) and mouse monoclonal anti-fibronectin (1:1,000; Abcam; cat. no. ab6328). After washing with TBST, goat anti-mouse (cat. no. SA00001-1) and goat anti-rabbit (SA00001-2) horseradish peroxidase-conjugated secondary antibody (1:2,000; Proteintech Group, Inc., Chicago, IL, USA) were added and the membrane was incubated at room temperature for 1 h. The proteins were visualized using autoradiography with an enhanced chemiluminescence reagent (Wuhan Boster Biological Technology, Ltd., Wuhan, China). The relative protein expression levels were determined by using the grayscale ratio of the protein of interest to glyceraldehyde 3-phosphate dehydrogenase and the results were analyzed using GraphPad Prism version 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA).
In vitro cell proliferation assay
Huh-7 cells were seeded at a density of 8.0×103 cells/well in a 96-well plate and incubated for 18 h. Water-soluble tetrazolium salts-8 (10 µl) from the Cell Counting Kit-8 (Wuhan Boster Biological Technology, Ltd., Wuhan, China) was then added to each well. The plates were incubated at 37°C in 5% CO2 for 12, 24, 48, 72 and 96 h, and the absorbance of each sample was determined at a wavelength of 450 nm using a microplate reader.
Bromodeoxyuridine (BrdU) labeling assay
The Huh-7 cells (1×106/cm2) were seeded on a slide and incubated in RPMI-1640 with 10% FBS with or without siRNA-ZEB1 transfection for 24 h. BrdU (10 µM; Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) was added to the culture for 1 h. Cells were then fixed with 4% paraformaldehyde and exposed to ice-cold methanol for 2 min. Cells were incubated in 2 M HCl for 1 h at room temperature and then incubated in 0.1 M sodium borate for 2 min. Finally, immunocytochemistry for BrdU labeling of proliferative Huh-7 cells was performed. Nuclei were stained with 1 µg DAPI solution for 5 min. Images were captured using an epifluorescence microscope (Zeiss AG, Oberkochen, Germany) with a charge-coupled device camera and AxioVision software SE64 4.9.1 (Zeiss AG).
In vitro invasion assay
In order to perform the invasion assay, Transwell chambers precoated with Matrigel (BD Bioscience, San Jose, CA, USA) were used. Huh-7 cells were cultured in serum-free medium (GE Healthcare Life Sciences, Logan, UT, USA), where the upper chambers of a Transwell plate (Corning, Corning, NY, USA) were separated from the lower chambers with a permeable 8 µm polycarbonate membrane. Medium with 10% FBS was added into the lower chambers as a chemoattractant. Cells were incubated for 24 h, cells remaining on the upper membrane were carefully removed, those that had invaded through the membrane were fixed in 75% ethanol and stained with crystal violet. The number of stained cells was counted under a light microscope from five different visual fields of each filter. Three independent experiments were performed.
In vitro migration assay
The Huh-7 cells were seeded at a density of 1.0×106 cells/well in 6-well plates. Confluent cell monolayers were disrupted by standardized wound scratching using a sterile 10 µl pipette tip and incubated in serum-free medium for 60 h. Migration of cells into the disrupted area and the recovery of the monolayer was assessed and photographed every 12 h for 60 h using a phase contrast microscope.
Statistical analysis
Data are presented as the mean ± standard deviation and analyzed using GraphPad Prism 5.0 software (GraphPad Software, Inc.). Continuous variables were compared using a Kruskal-Wallis test and categorical data were compared using χ2 or Fisher exact tests. The Kaplan-Meier method was used for survival analysis and differences in survival were determined using the log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression level of ZEB2-AS1 in HCC tissues
In order to determine whether there was a difference in the expression level of lncRNA ZEB2-AS1 in HCC tissues and the adjacent normal tissues the ZEB2-AS1 expression levels in 40 paired tissue samples were detected using RT-qPCR. The expression of lncRNA ZEB2-AS1 was significantly upregulated in HCC tissues when compared with adjacent normal tissues (P<0.001; Fig. 1A). Additionally, the correlation between lncRNA ZEB2-AS1 expression level and clinicopathological variables of patients with HCC patients was assessed in order to evaluate its clinical significance. As presented in Table I, ZEB2-AS1 expression levels in HCC were significantly associated with a primary tumor size of ≥5 cm (P=0.027), intrahepatic metastasis (P=0.025) and TNM stage (P=0.011). However, ZEB2-AS1 expression was not correlated with other clinicopathological variables, including age (P=0.342), liver cirrhosis (P=0.113) or differentiation (P=0.784). Additionally, patients with HCC and high ZEB2-AS1 expression were revealed to have significantly shorter overall survival (P=0.038; Fig. 1B) and significantly higher recurrence rates (P=0.037; Fig. 1C) compared with those with low expression of ZEB2-AS1. These findings demonstrate that the upregulation of lncRNA ZEB-AS1 may be associated with tumor metastasis and poor prognosis in patients with HCC.
Figure 1.
Expression levels of ZEB2-AS1 in HCC tissues and cell lines. (A) ZEB2-AS1 expression was higher in HCC tissues compared with the paired adjacent normal tissues (n=40). Expression was normalized to glyceraldehyde 3-phosphate dehydrogenase expression. Kaplan-Meier analysis of (B) overall survival and (C) recurrence-free survival based on ZEB2-AS1 expression levels in 40 patients with HCC. The median level of ZEB2-AS1 was used as the cut off. Patients with HCC were divided into high ZEB2-AS1 expression group and low ZEB2-AS1 expression group. (D) ZEB2-AS1 expression levels in different human HCC cell lines. Huh-7 cells had the highest ZEB2-AS1 expression level. *P<0.05. HCC, hepatocellular carcinoma; ZEB2-AS1, Zeb2 antisense RNA 1.
Table I.
Correlation between ZEB2-AS1 expression and clinicopathological variables of patients with hepatocellular carcinoma.
| Relative ZEB2-AS1 expression | ||||
|---|---|---|---|---|
| Variable | Total (n=40) | High (n=20) | Low (n=20) | P-value |
| Gender | ||||
| Male | 29 | 16 | 13 | 0.479 |
| Female | 11 | 4 | 7 | |
| Age (years) | ||||
| <60 | 19 | 11 | 8 | 0.342 |
| ≥60 | 21 | 9 | 12 | |
| Size of primary tumor (cm) | ||||
| <5 | 19 | 6 | 13 | 0.027 |
| ≥5 | 21 | 14 | 7 | |
| PVTT | ||||
| Present | 6 | 4 | 2 | 0.658 |
| Absent | 34 | 16 | 18 | |
| Intrahepatic metastasis | ||||
| Present | 17 | 12 | 5 | 0.025 |
| Absent | 23 | 8 | 15 | |
| Serum AFP (ng/ml) | ||||
| <200 | 15 | 5 | 10 | 0.103 |
| ≥200 | 25 | 15 | 10 | |
| Liver cirrhosis | ||||
| Present | 19 | 12 | 7 | 0.113 |
| Absent | 21 | 8 | 13 | |
| Differentiation | ||||
| Well | 7 | 3 | 4 | 0.784 |
| Moderate | 28 | 15 | 13 | |
| Poor | 5 | 2 | 3 | |
| TNM stage | ||||
| I–II | 22 | 7 | 15 | 0.011 |
| III–IV | 18 | 13 | 5 | |
ZEB2-AS1, Zeb2 antisense RNA 1; PVTT, portal vein tumor thrombosis; AFP, α-fetoprotein; TNM, tumor-node-metastasis.
Expression levels of ZEB2-AS1 in various human HCC cell lines
RT-qPCR was performed to determine ZEB2-AS1 expression in 5 human HCC cell lines, SMMC-7721, Hep3B, HepG2, HCCLM9 and Huh-7. The highest expression level of ZEB2-AS1 was detected in Huh-7 cells when compared with the remaining cell lines (P<0.05; Fig. 1D). Therefore, the Huh-7 HCC cell line was selected for the remaining experiments in the present study.
Effects of ZEB2-AS1 siRNAs on the expression levels of EMT-induced markers
The present study aimed to determine the association between ZEB2-AS1 expression levels and EMT-induced markers, and determine the function of ZEB2-AS1 in human HCC cells. Therefore, siRNAs were used to knock down ZEB2-AS1 in the Huh-7 HCC cell line. Huh-7 cells were divided into three treatment groups: i) siRNA-ZEB2-AS1-transfected group; ii) siRNA-NC group; and iii) blank control group. The knockdown effect of siRNA-ZEB2-AS1 was confirmed by conventional PCR. This revealed that the expression levels of ZEB1-AS1 were significantly reduced in the siRNA-ZEB1-transfected group compared with the siRNA-NC and the blank control groups (P<0.05; Fig. 2A), indicating the interference ability of siRNA on ZEB2-AS1 was effective and specific. Subsequently, the result of the western blot analysis demonstrated that the successful knock down of ZEB2-AS1 may lead to a significant downregulation of ZEB2 (P<0.05; Fig. 2B), vimentin (P<0.05; Fig. 2B), fibronectin (P<0.05; Fig. 2B) and N-cadherin (P<0.05; Fig. 2B), and an upregulation of E-cadherin (P<0.05; Fig 2B), which suggested that the expression of EMT-induced markers may be regulated by ZEB2-AS1.
Figure 2.
Effect of ZEB2-AS1 siRNAs on the expression levels of EMT-induced markers. (A) ZEB2-AS1 expression was significantly downregulated in the siRNA-ZEB2-AS1 transfected group compared with the siRNA-NC and blank control groups. (B) Expression of EMT markers was significantly deregulated in Huh-7 cells transfected with siRNA-ZEB2-AS1. *P<0.05. siRNA, small interfering RNA; ZEB2-AS1, zinc finger E-box binding homeobox 2 antisense RNA 1; NC, negative control; NS, not significant; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
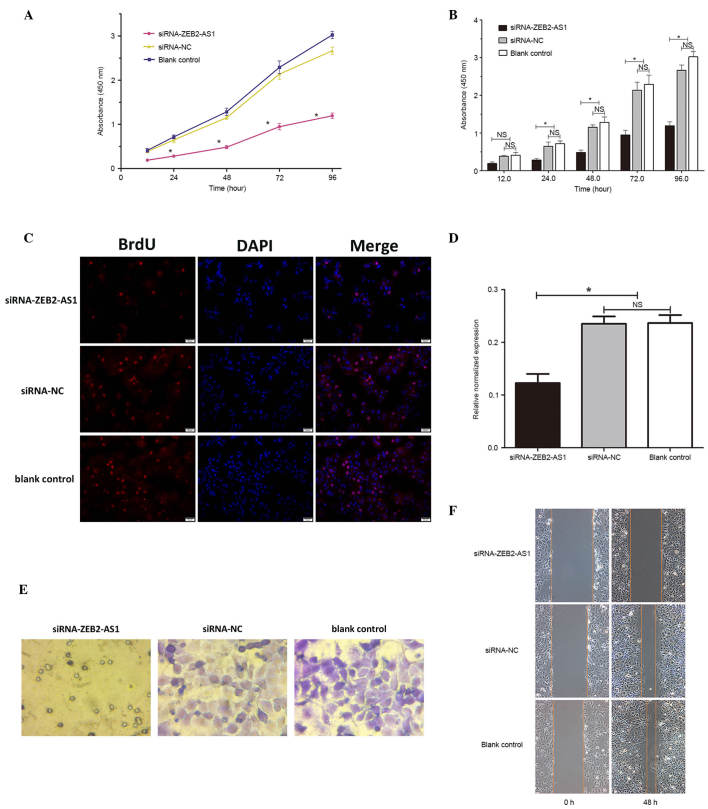
Effects of ZEB2-AS1 siRNAs on cell proliferation and cell cycle in vitro
CCK-8 assay was used to assess the effect of ZEB2-AS1 suppression on cell vitality over a period of 96 h. It was determined that cell viability was significantly reduced in the siRNA-ZEB2-AS1-transfected group compared with the negative and blank control groups (P<0.05; Fig. 3A and B). Therefore, it is possible that cell viability was suppressed by the knockdown of ZEB2-AS1. The BrdU labeling assay revealed that DNA synthesis was also significantly inhibited in the siRNA-ZEB2-AS1-transfected group compared with the remaining control groups (P<0.05; Fig. 3C and D). These findings reveal that Huh-7 cell proliferation was inhibited by the silencing of ZEB2-AS1.
Figure 3.
Effects of siRNA-ZEB2-AS1 on cell proliferation, cell cycle, invasion and migration in vitro. (A and B) Proliferation of Huh-7 cells was assayed subsequent to ZEB2-AS1 downregulation using Cell Counting kit-8 according to the manufacturer's protocol. (C and D) DNA synthesis was reduced in the siRNA-ZEB2-AS1 transfected group vs. the siRNA-NC group and blank control groups. Cell (E) invasion (magnification, ×250) and (F) migration assay magnification, 100) of Huh-7 cells following ZEB2-AS1 downregulation for 24 h. *P<0.05. siRNA, small interfering RNA; ZEB2-AS1, Zeb2 antisense RNA 1; NC, negative control; NS, not significant.
Effects of ZEB2-AS1 siRNAs on invasion and migration of Huh-7 cells in vitro
Cell invasion and migration assays were performed to determine how ZEB2-AS1 affected these characteristics in Huh-7 cells. The in vitro invasiveness of Huh-7 cells was detected using a Transwell invasion chamber assay. Crystal violet staining revealed that the numbers of Huh-7 cells per high power field (three fields analyzed) in siRNA-ZEB2-AS1, siRNA-NC and blank control groups were 7.33±2.49, 70.33±3.86 and 66.33±3.68, respectively, this suggests that the number of Huh-7 cells that had passed through the polycarbonate membrane of the Transwell invasion chamber was significantly reduced in the siRNA-ZEB2-AS1-transfected group compared with the negative and blank control groups (P<0.05; Fig. 3E). No significant difference was identified between the invasiveness of the Huh-7 cells siRNA-NC group and the blank control group (P>0.05; Fig. 3E). The wound-healing assays were performed to investigate the migration ability of Huh-7 cells in the three treatment groups. It was determined, from three high power fields that the movement of cells in siRNA-ZEB2-AS1, siRNA-NC and blank control groups were 1.93±0.26, 3.53±0.33 and 3.23±0.21 cm, respectively which demonstrates that the migration ability of cells the siRNA-ZEB2-AS1-transfected group was significantly reduced compared with the two remaining control groups, 48 h after wounding (Fig. 3F). These findings demonstrated that the downregulation of ZEB2-AS1 reduced the migration and invasion abilities of Huh-7 cells.
Discussion
Previous studies have demonstrated that lncRNAs have important functions in the regulation of gene expression and their dysregulation may participate in the proliferation and metastasis of cancer cells (15–17). Notably, aslncRNAs, which are oriented in antisense direction with respect to a protein coding loci, usually act as regulators of the gene located on the opposite strand (18). Previous studies have determined that aslncRNAs are involved in the complex network of signaling pathways in cancer cells and may function as enhancers of tumor-associated gene expression (19,20).
ZEB has been identified as a transcriptional factor and has been demonstrated to be important for the process of EMT, which is closely associated with carcinogenesis. Previous studies have revealed that abnormal expression of ZEB in various types of cancer has been associated with aggressive disease, low differentiation, the development of metastases and poor prognosis in patients (21,22). Li et al (11) reported that the expression levels of ZEB1-AS1 were upregulated in HCC tissues when compared with paired adjacent normal tissues. ZEB1-AS1 expression was observed to increase with HCC progression. Patients with HCC that had high ZEB1-AS1 expression were identified to have shorter overall survival and higher recurrence rates compared with those that had low expression levels of ZEB-AS1. ZEB1-AS1 promoted the proliferation and metastasis of HCC in vitro and in vivo; therefore, acting as an oncogene in HCC. Using the Pubmed gene database, it was determined that there was a non-coding antisense transcript located from the promoters of ZEB2 (ZEB2-AS1) (23). The present study determined that the expression of ZEB2-AS1 in HCC tissues was significantly higher compared with adjacent normal tissues and this was significantly associated with the size of the primary tumor (P=0.027), intrahepatic metastasis (P=0.025) and TNM stage (P=0.011). Additionally, according to the 48-month follow-up survival survey the present study demonstrated that patients with HCC and high ZEB2-AS1 expression levels had shorter overall survival and higher recurrence rates compared with those with low expression levels of ZEB2-AS1 (P<0.05). Therefore, ZEB2-AS1 expression levels may serve as a prognostic factor for HCC pathogenesis. Furthermore, the successful downregulation of ZEB2-AS1 reduced HCC cell viability significantly and DNA synthesis was also inhibited. Simultaneously, the invasion and migration ability of HCC cells was suppressed by the knockdown of the ZEB2-AS1 gene. Therefore, the current study indicates that ZEB2-AS1 may serve as an oncogene and may be important for HCC development and progression.
In conclusion, the present study demonstrated that the downregulation of aslncRNA ZEB2-AS1 was associated with the reduction of tumor growth and metastasis in HCC tissues. The importance of ZEB2-AS1 in tumor progression and survival prediction in HCC patients was elucidated, indicating that ZEB2-AS1 may be a valuable prognostic biomarker and a novel therapeutic target for HCC.
Acknowledgements
The present study was supported by the Science and Technology Project of Wuhan (grant no. 2013060501010153).
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Lau WY, Lai EC. Hepatocellular carcinoma: Current management and recent advances. Hepatobiliary Pancreat Dis Int. 2008;7:237–257. [PubMed] [Google Scholar]
- 3.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwatsuki M, Mimori K, Yokobori T, Ishi H, Beppu T, Nakamori S, Baba H, Mori M. Epithelial-mesenchymal transition in cancer development and its clinical significance. Cancer Sci. 2010;101:293–299. doi: 10.1111/j.1349-7006.2009.01419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo F, Kerrigan BC Parker, Yang D, Hu L, Shmulevich I, Sood AK, Xue F, Zhang W. Post-transcriptional regulatory network of epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions. J Hematol Oncol. 2014;7:19. doi: 10.1186/1756-8722-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Huang JL, Zheng L, Hu YW, Wang Q. Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis. 2014;35:507–514. doi: 10.1093/carcin/bgt405. [DOI] [PubMed] [Google Scholar]
- 8.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 9.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35:1575–1584. doi: 10.1038/onc.2015.223. [DOI] [PubMed] [Google Scholar]
- 12.Zhuang J, Lu Q, Shen B, Huang X, Shen L, Zheng X, Huang R, Yan J, Guo H. TGFβ1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Sci Rep. 2015;5:11924. doi: 10.1038/srep11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Tang ZY, Ye SL, Liu YK, Chen J, Xue Q, Chen J, Gao DM, Bao WH. Establishment of cell clones with different metastatic potential from the metastatic hepatocellular carcinoma cell line MHCC97. World J Gastroenterol. 2001;7:630–636. doi: 10.3748/wjg.v7.i5.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao T, Xu J, Liu L, Bai J, Xu C, Xiao Y, Li X, Zhang L. Identification of cancer-related lncRNAs through integrating genome, regulome and transcriptome features. Mol Biosyst. 2015;11:126–136. doi: 10.1039/C4MB00478G. [DOI] [PubMed] [Google Scholar]
- 18.Nolasco S, Bellido J, Gonçalves J, Tavares A, Zabala JC, Soares H. The expression of tubulin cofactor A (TBCA) is regulated by a noncoding antisense Tbca RNA during testis maturation. PloS One. 2012;7:e42536. doi: 10.1371/journal.pone.0042536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lepoivre C, Belhocine M, Bergon A, Griffon A, Yammine M, Vanhille L, Zacarias-Cabeza J, Garibal MA, Koch F, Maqbool MA, et al. Divergent transcription is associated with promoters of transcriptional regulators. BMC Genomics. 2013;14:914. doi: 10.1186/1471-2164-14-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurihara M, Shiraishi A, Satake H, Kimura AP. A conserved noncoding sequence can function as a spermatocyte-specific enhancer and a bidirectional promoter for a ubiquitously expressed gene and a testis-specific long noncoding RNA. J Mol Biol. 2014;426:3069–3093. doi: 10.1016/j.jmb.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 21.Sánchez-Tilló E, de Barrios O, Siles L, Amendola PG, Darling DS, Cuatrecasas M, Castells A, Postigo A. ZEB1 Promotes invasiveness of colorectal carcinoma cells through the opposing regulation of uPA and PAI-1. Clin Cancer Res. 2013;19:1071–1082. doi: 10.1158/1078-0432.CCR-12-2675. [DOI] [PubMed] [Google Scholar]
- 22.Graham TR, Zhau HE, Odero-Marah VA, Osunkoya AO, Kimbro KS, Tighiouart M, Liu T, Simons JW, O'Regan RM. Insulin-like growth factor-I-dependent up-regulation of ZEB1 drives epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2008;68:2479–2488. doi: 10.1158/0008-5472.CAN-07-2559. [DOI] [PubMed] [Google Scholar]
- 23.Beltran M, Puig I, Peña C, García JM, Alvarez AB, Peña R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]