Abstract
This chapter describes the utility of ion mobility-mass spectrometry (IM-MS) for the detection and characterization of glycoproteins and associated glycoconjugates. IM-MS provides separations in two dimensions, one on the basis of molecular surface area, or structure, and the other on molecular mass which creates the ability to differentiate biomolecular classes and isobaric species. When applied to the characterization of glycoproteins, IM-MS separates peptides from the associated glycans in the same digest without purification, and can also be used to separate different isomeric glycans which is a significant challenge in current glycomic studies. The chapter will detail the methodologies to use IM-MS for the study of glycans and glycoproteins for an audience ranging from new and potential practitioners to those already utilizing the technique.
Keywords: Ion mobility, ion mobility-mass spectrometry, IM-MS, structural separations, MALDI, IM-MS/MS, glycomics, glycoproteomics
1. Introduction
Glycomics has progressed into a critical area of study due to the implications of carbohydrate participation in many biological functions, and variations in glycosylation being associated with many disease states (1–3). Protein glycosylation is one of the more intricate forms of post-translational modification (PTM) and is estimated to be present on over 50% of eukaryotic proteins (4). Glycoproteins have vital functions inside various organisms, and their associated glycans assist in the structure, function, and stability of proteins. Glycoproteins are involved in many important biological functions (e.g. embryonic development and the recognition of hormones, toxins, and other signals on the cell surface) and processes (e.g. coordination of immune function, cell division, and protein regulation and interactions) (5). With all of these important tasks of glycosylation, detrimental effects may occur from variation or defects in glycosylation patterns. Several disease states such as Alzheimer’s disease, HIV, cancer, and diabetes have characteristic defects in glycosylation patterns or unique glycoproteins associated with the disease (3). Further information about the functions of glycans and glycoconjugates can be explored in several excellent texts (6, 7) along with other chapters in this book. Overall, the function of carbohydrates are derived from their composition and structure necessitating rapid and efficient structural determination from complex mixtures, including glycoconjugates such as glycoproteins and glycolipids.
Collectively these challenges motivate the development of higher-throughput, more accurate, and minimal sample manipulation strategies for carbohydrate structure elucidation. Recently, 2D ion mobility-mass spectrometry (IM-MS) has been applied to the field of biological analysis (8–10). Ion mobility separates ions based on their apparent surface area or ion-neutral collision cross section (11). When merged with MS, IM can separate gas-phase ions in one dimension based on their structure, and a second dimension related to their mass to charge (m/z). The advantages provided by IM-MS would likely be of great utility in the field of glycoproteomics.
This chapter focuses on using IM-MS technologies for the study of carbohydrates and glycoproteins in the pursuit of combining omics (e.g. simultaneous glycomics, proteomics, lipidomics, etc.). Identification and conformational characterization of glycoproteins is pursued through studies of carbohydrate standards and separation of glycoprotein digests provided by the structural dimension of IM-MS. In this introductory chapter, IM-MS structural characterization will be summarized along with the theoretical background and instrumentation. The following sections describe an overview of IM-MS instrumentation, the theory of IM separations, different types of IM separations, and data interpretation in IM-MS conformation space. Previous studies of carbohydrates and glycoproteins using IM-MS, and methods for the characterization of glycoproteins using IM-MS will also be discussed.
1.1. Ion mobility applications to the life sciences
Ion mobility (IM), which has existed for over a century, is a well-developed separation technique that has been used extensively in the rapid detection of drugs and warfare agents due to its ease of use, low cost, speed, and sensitivity (12, 13). The coupling of IM to MS was first performed in the early 1960s (14, 15), but the utility of IM-MS for biomolecular separations was not fully realized until combined with soft ionization techniques, such as electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI) (16, 17) which were not developed until the late 1980s. The first pioneering studies which used IM-MS to determine peptide and protein structures were performed in the late 1990s (18–20). Following these early studies, research over the past decade has extended IM-MS techniques to the study of complex biological samples, such as whole cell lysates (21), plasma (22–25), homogenized tissue (21, 26, 27), non-covalent complexes (28–30), or directly from thin tissue sections (31, 32). However, IM–MS was essentially limited to a few laboratories where custom instrumentation was constructed. The recent introduction of commercially available IM–MS instruments, in several forms, has further stimulated the use of IM–MS for life sciences research. The following sections describe an overview of IM-MS separations (Section 1.2), IM-MS instrumentation (Section 1.3), the theory of IM separations (Section 1.4), data interpretation in 2D IM-MS conformation space (Section 1.4). Materials and methods for characterizing carbohydrates and glycoproteins using IM-MS are then detailed (Sections 2 and 3).
1.2. Overview of ion mobility separations
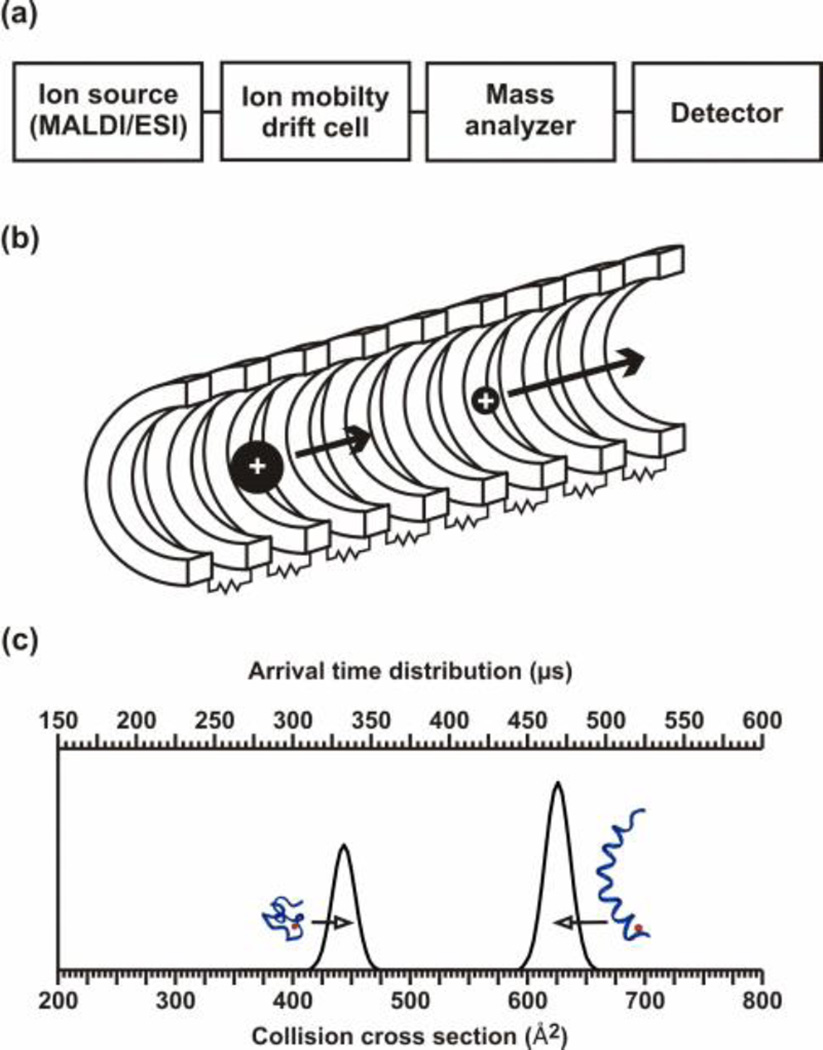
Most ion mobility-mass spectrometers have the same general layout. They are similar to mass spectrometers with the IM region inserted between the source and mass analyzer, hence IM is a post-ionization separation technique [Fig. 1(a)]. From this general layout, instruments can vary due to the type of IM used, the choice of mass analyzer or ionization source (i.e. ESI, nESI, MALDI), the insertion of a quadrupole for mass selection before IM or MS analysis, etc. There are two main methods for differentiating ions using ion mobility, either separating the ions using space or time. The main focus of this chapter will be time-dispersion, but we will also highlight several carbohydrate studies utilizing space-dispersion.
Figure 1.
(a) A block diagram of the primary components of biological IM-MS instrumentation. (b) A conceptual depiction of an IM drift cell. A stack of ring electrodes are connected via resistors in series to form a voltage divider, which is typically designed to generate a relatively uniform electrostatic field along the axis of ion propagation. Ions of larger apparent surface area experience more collisions with the neutral drift gas and therefore elute slower than ions of smaller apparent surface area. (c) A hypothetical IM separation for peptide ions exhibiting two distinct structural sub-populations corresponding to globular (left) and to helical (right) conformations. The arrival time distribution data (top axis), or what is measured, can be transformed to a collision cross-section profile (bottom axis) via equation [4] and described in Section 1.4 and 3.1. Adapted with kind permission from Springer Science+Business Media: Anal. Bioanal. Chem., Biomolecular structural separations by ion mobility-mass spectrometry, 391, 2008, 906, L.S. Fenn and J.A. McLean, Fig. 1.
The types of IM that use a time-dispersion of the ions are drift tube or traveling wave ion mobility spectrometers (DTIM and TWIM, respectively). DTIM and TWIM separate ions on the basis of molecular surface area due to interactions with a neutral buffer gas present in the IM drift cell. These interactions are not like high energy ion-neutral gas-phase collisions used in collision induced dissociation (CID) but are low energy gas-phase elastic collisions akin to the collisions of billiard balls. Ions are injected into the IM drift tube and migrate under the influence of a weak electrostatic field gradient [Fig. 1(b)] where they interact with the neutral drift gas. This field is electrostatic for drift tube and electrodynamic for traveling wave separations, respectively. Smaller ions have a higher mobility than larger ions which result in shorter drift times versus longer drift times, respectively. While the ions traverse the drift cell, their migration is impeded by collisions with the neutral drift gas, typically helium or nitrogen, to a degree that is proportional to apparent surface area or collision cross section. The actual experimental parameter obtained from IM separations is the ion arrival time distribution (tATD), or the time between ion injection and ion detection. It can be converted to collision cross section or apparent surface area as illustrated in Fig. 1(c) for DTIM. The difference between the separations for DTIM and TWIM is attributed to their instrumental design which will be examined in the next sections.
1.3. IM-MS Instrumentation
There are two main methods of separating ions with IM, through space or time-dispersion. The most common techniques used for separation though space is differential mobility and field asymmetric ion mobility spectrometry (FAIMS) whereas the most common methods for separating ions through time-dispersion is with drift tube or traveling wave IM (DTIM or TWIM, respectively). The methods presented here for the characterization of glycoproteins concentrates on the use of TWIM and DTIM.
1.3.1. Drift tube ion mobility
The first IM instruments utilized a drift tube (33). DTIM-MS has the basic design described previously and depicted in Fig. 1(b) in that it has a series of stacked ring electrodes that create an electrostatic field to create a forward force that is impeded by collisions with a buffer gas. IM resolution typically ranges from 30–50 (r=t/Δt at FWHM), whereas longer, cryogenically cooled, or higher pressure drift tubes have been reported with resolutions exceeding 100 (34–36). The drift time can be understood based on the kinetic theory of gases and used to calculate the ions absolute collision cross section without the need for standards (37–40). The calculation of collision cross sections is detailed in Section 1.4 and 2.5. For a derivation of ion-neutral collision cross section theory, the reader is directed to several excellent texts and reviews (11, 41, 42)
1.3.2. Traveling wave ion mobility
Traveling wave IM (TWIM) is a recently developed technique in comparison to DTIM. The recent commercial availability of TWIM instrumentation (Waters, Corp.) has made IM-MS accessible to the glycobiology community, not just those labs capable of building the instrumentation. Similar to drift tube instruments, TWIM separates ions by time dispersion through collisions with a background buffer gas, but in contrast, it uses electrodynamic fields rather than electrostatic fields (43, 44). This is accomplished by transmitting voltage pulses sequentially across a stack of ring electrodes (similar to Fig. 1(b)), which creates the travelling wave (45). Conceptually, TWIM separations are performed based on the susceptibility of different ions to the influence of the specific wave characteristics and have been described as the ability of ions to "surf" on waves (44). Since traveling wave separations utilize dynamic electric fields, presently TWIM measurements can only provide estimated collision cross sections based on internal standards from DTIM absolute collision cross sections (46, 47).
The first commercial platform (Synapt HDMS, now referred to as G1) is comprised of an interchangeable ESI and MALDI source, a mass resolving quadrupole, a trapping region for injecting pulses of ions into the TWIM, the TWIM drift cell, an ion transfer region, and an orthogonal TOFMS (r=m/Δm at FWHM of >17,500). Adjustable wave parameters include: travelling wave pulse height, wave velocity, and ramping either of these variables. CID can be performed in the regions before and after the TWIM drift cell (See Note 1). Generally resolution in the TWIM of the G1 is <15, but this is sufficient for the separation of many molecular classes of interest. For example TWIM has been used to separate biomolecular signals from complex samples (48) and to study the structure of peptides following CID in the trapping region (49). Recently, the Synapt G2 HDMS was released which has improved TWIM resolution (≥ 40) and improved mass resolution (≥ 50,000). It also can be easily interfaced with many different ionization sources and combined with other separation techniques prior to ionization [high-performance liquid chromatography (HPLC), ultra-performance liquid chromatography (UPLC), etc.]
1.4. Ion mobility theory: Converting drift time to collision cross section
This section details the methodology currently used to determine ion-neutral collision cross sections from data acquired with uniform electrostatic field DTIM. For estimating collision cross sections from TWIM data, see procedures described elsewhere (46, 47). Directions for implementing these measurements and equations experimentally will be detailed in Section 2.
In order to calculate the collision cross section of an ion, the ion has to traverse the drift cell under the influence of a weak electrostatic field (E, ca. 20–30 V cm−1 Torr−1) which provides “low-field” conditions (i.e. constant IM proportionality constant, K). The separation of the ions is measured as ion drift velocity which is determined by:
| [1] |
In order to standardize the value of K for comparison across different instruments, the pressure (p, Torr) of the neutral drift gas and the temperature (T, Kelvin) of separation must be considered which creates a standard or reduced mobility (K0):
| [2] |
This reduced mobility value is normalized to standard temperature and pressure (i.e. 0 °C and 760 Torr) and can be related to the ion-neutral collision cross section through the kinetic theory of gases:
| [3] |
Where ze is the charge of the ion, mi and mn are the mass of the ion and neutral, respectively, kb is Boltzmann’s constant, N0 is the number density of the drift gas at STP (2.69 × 1019 cm−3), and Ω the ion-neutral collision cross section. This assumes that the collisions are completely elastic. For an IM drift cell of fixed length (L), the drift time (td) of the packet across the cell can be used to solve for the ion-neutral collision cross section by substituting for K0 in Eqn. [3] and rearranging:
| [4] |
which is the form used to solve for collision cross sections from IM data (See Section 3.1). Further, since the collision cross sections follow the hard sphere model, molecular dynamics simulations can be performed to interpret structures consistent with the empirical data (50–53).
1.5. Data interpretation for glycomics in conformation space
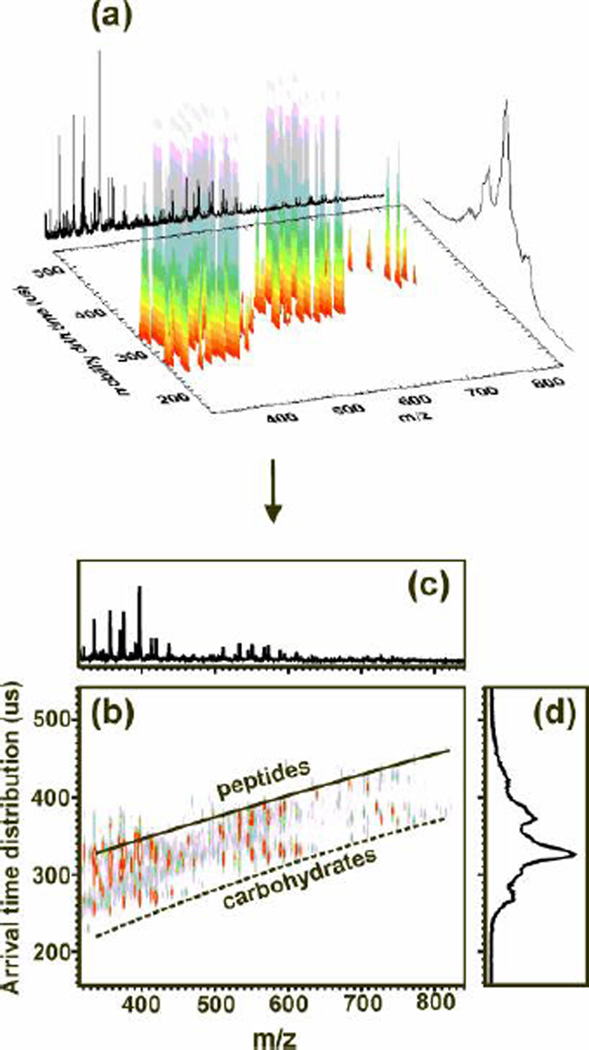
Typical data for an IM-MS experiment is acquired in three dimensions; m/z, IM arrival time distribution, and relative abundance of the signal. However, to analyze the data and centroid the peaks, the data is presented in a 2D plot with intensity being represented by false coloring. This conversion from 3D figure to 2D plot is presented in Fig. 2(a and b) for the separation of glycans and peptides from a glycoprotein digest. We refer to the 2D IM-MS plot as conformation space because it represents biomolecular structure, or conformation, as a function of m/z (See Note 2). An integrated mass spectrum over all arrival time distributions is shown in Fig. 2(c), which is what would be observed in the absence of IM. An integrated IM arrival time distribution is illustrated by the curve of Fig. 2(d) which would be obtained by placing the detector directly after the IM drift cell. By plotting the data in 2D conformation space two distinct correlations are observed, one for peptides and one for carbohydrates, respectively. Note that either extracted mass spectra or arrival time distributions can be derived from conformation space data.
Figure 2.
(a) A 3D representation of IM-MS data obtained for human glycoprotein (HGP) digested with Pronase. (b) A 2D IM-MS conformation space plot for the analysis of the HGP digest. This data illustrates the variation of gas-phase packing efficiencies for different types of biomolecules. Even though the glycans may still have amino acids attached, a clear differentiation between the peptides and glycans can be noted. (c) An integrated mass spectrum over all arrival time distributions. (d) The integrated arrival time distribution over the full mass range which would be obtained if a detector was placed after the IM drift cell.
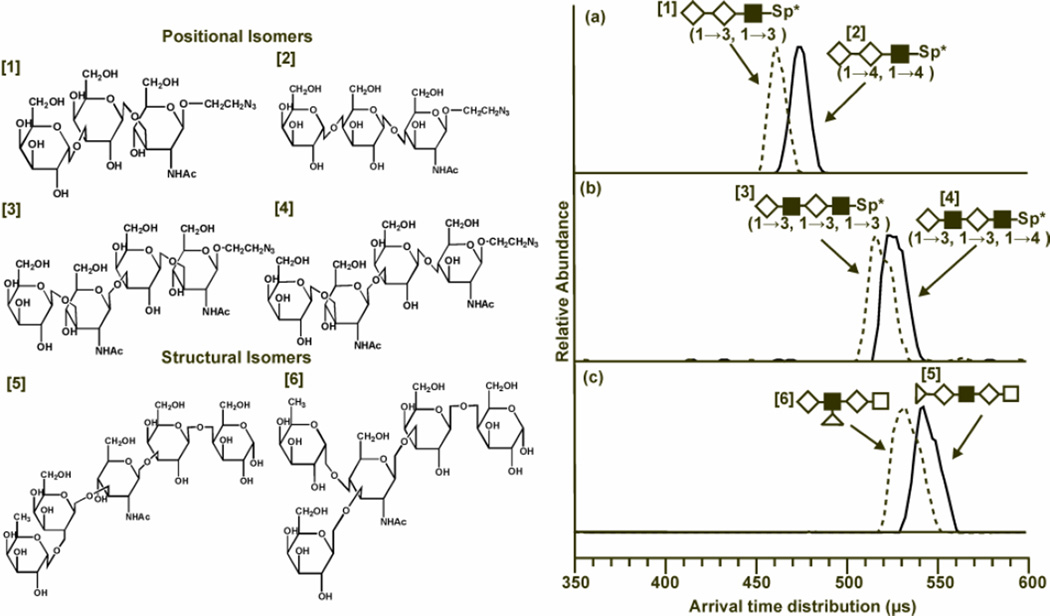
One of the main challenges in glycomics is the high probability of carbohydrates and glycans with different structures having the same mass, therefore being isobaric. When using MS alone, these isobaric molecules cannot be differentiated by the intact mass. However, with the addition of the structural separations of IM, some isobaric carbohydrates can be differentiated. This has been demonstrated with DTIM (54) and TWIM (55) and is seen in Fig. (3). In this figure, three pairs of isobaric structural and positional isomers were separated using the additional dimension of IM with MS.
Figure 3.
Structures of the isobaric sets of positional and structural isomers (left) and the associated drift time profiles (right). (Left) Note the difference in structures between glycans 1 and 2 are two 1→3 glycosidic linkages being replaced with two 1→4 linkages. Glycans 3 and 4 have one linkage variation, and glycans 5 and 6 (LNFP1 and LNFP2) vary in the location of fucose from galactose to N-acetylglucoseamine. (Right) Drift time profiles at an electrostatic field strength of 20.6 volts cm−1 Torr within the ion mobility drift cell. Structures of the oligosaccharides are replaced with shape representations. Drift times are related to the ion structure in that larger, more elongated ions experience more collisions with the neutral buffer gas present in the drift cell causing a longer drift time than more compact structures. (a) In the comparison between glycans 1 (dotted line) and 2 (solid line), the 1→3 linkages of glycan 1 cause it to have a shorter drift time which indicates a more compact structure that glycan 2, which is more elongated. (b) Glycans 3 (dotted line) and 4 (solid line) have differing drift times due to the change in one glycosidic linkage. The 1→3 linkages allow glycan 3 to adopt a more compact conformation when compared to its positional isomer, which has one 1→4 linkage. (c) Drift time profiles for glycans 5 (solid line) and 6 (dotted line) are compared. LNFP2 has a shorter drift time than LNFP1 at both voltages. This is attributed to increased branching in LNFP2 that allows the glycan to adopt a more compact structure. Individual monosaccharide representations are as follows: ◊ - galactose; ■-N-acetylglucoseamine; Δ -fucose; with linkage information for the positional isomers provided in the parenthesis below each representation (54). Fenn, L. S., and McLean, J. A. (2011) Structural resolution of carbohydrate positional and structural isomers based on gas-phase ion mobility-mass spectrometry. Phys Chem Chem Phys 13, 2196–2205- Reproduced by permission of the PCCP Owner Societies.
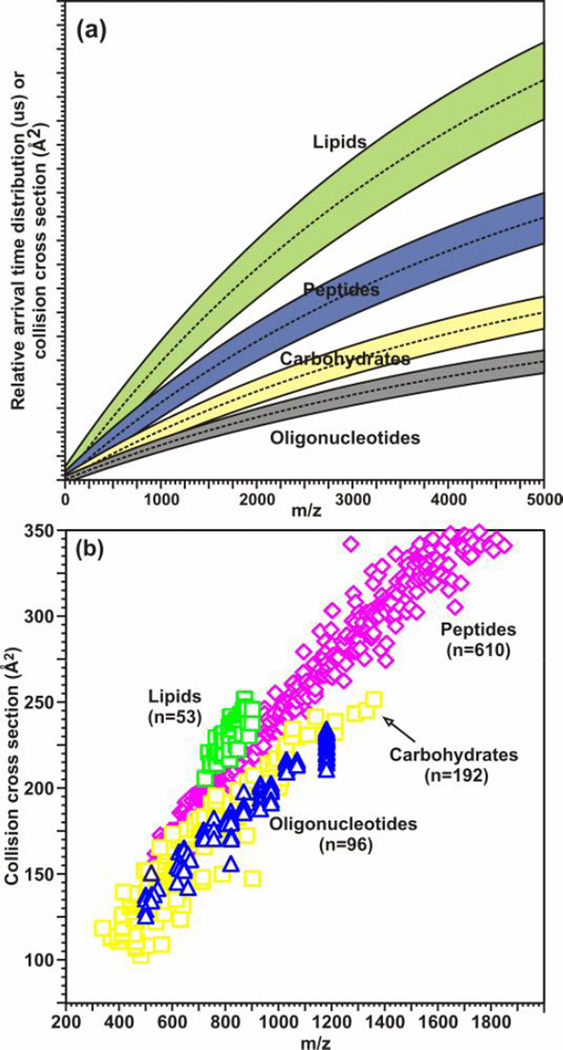
In parallel with the separation of different carbohydrates, the structural separations provided by IM can also be used to differentiate isobaric species belonging to different biomolecular classes [Fig. (4)]. Although biomolecules are generally composed of a limited combination of elements (e.g. C, O, H, N, S, and P), different biomolecular classes preferentially adopt structures at a given m/z correspondent to the prevailing intermolecular folding forces for that class. A representative plot delineating regions of conformation space for which different biomolecular classes (e.g. nucleotides, carbohydrates, peptides, lipids) are predicted to occur is presented in Fig. 4(a). These separations are a result of the different gas-phase packing efficiencies of the different classes (nucleotides > carbohydrates > peptides > lipids) (56). This plot is reinforced through the calculation of collision cross sections for standards of each biomolecular class [Fig. 4(b)]. The separation of different biomolecular classes can be utilized in glycoproteomics through the ability to identify peptides and glycans present in a complex sample simultaneously which will be further discussed in Section 2.
Figure 4.
(a) A hypothetical plot illustrating the differences in IM-MS conformation space for different molecular classes based on different gas-phase packing efficiencies. (b) A plot of collision cross section as a function of m/z for different biologically-relevant molecular classes, including: oligonucleotides (n = 96), carbohydrates (n = 192), peptides (n = 610), and lipids (53). All species correspond to singly-charged ions generated by using MALDI, where error ±1σ is generally within the data point. Values for peptides species are from Ref. (40). (a) is adapted with kind permission from Springer Science+Business Media: Anal. Bioanal. Chem., Biomolecular structural separations by ion mobility-mass spectrometry, 391, 2008, 906, L.S. Fenn and J.A. McLean, Fig. 2(a). (b) is adapted with kind permission from Springer Science+Business Media: Anal. Bioanal. Chem., Characterizing ion mobility-mass spectrometry conformation space for the analysis of complex biological samples, 394, 2009, 235, L.S. Fenn, M. Kliman, A. Mahsutt, S.R. Zhao, and J.A. McLean, Fig. 1(a).
1.6. IM-MS for the characterization of glycans and glycoproteins
Although the use of MS to characterize glycoproteins has been performed extensively for many years, the use of IM-MS for the characterization of carbohydrates and glycoproteins has only recently become increasingly prominent. This again is attributed to the limitation of IM-MS instrumentation to those labs which could construct it. Most early IM-MS studies concentrated on peptides and proteins. However there were a few number of experiments evaluating the use of IM-MS for glycomic studies. The first carbohydrate analyses conducted in the late 1990s aimed at examining short linear polysaccharides and cyclodextrins using DTIM-MS and comparing their collision cross sections to those obtained from molecular dynamic simulations (57). These studies also investigated at the interaction of carbohydrates with Na+ along with the resulting effect of metal coordination on the overall carbohydrate structure. Additional studies examined ways to enhance the ionization and improve sensitivity for oligosaccharides in ESI-IM-MS instruments by utilizing an ion trap interface and different injection energies (58, 59). These were followed by studies to examine variations in conformation of hexose complexes with zinc ligands (60) through collision cross section determinations and theoretical computational interpretation.
More recent studies have centered on structurally differentiating and determining stereochemical information about monomeric or small di- and trisaccharide structures using DTIM (54, 55, 61–64), TWIM (55, 65, 66), and FAIMS (67). N-linked and O-linked glycans removed from glycoproteins have also been characterized by IM-MS from purified samples (68, 69) after separation and extensive purification from serum (22) or urine (48). In addition, sulfated glycans were resolved by IM-MS through the differentiation of isomers (70), interpretation of collision cross sections with molecular modeling (71), and interactions with defensin inspired peptides (72). N-glycan structure and glycosylation sites for IgG have been determined using IM-MS/MS (68), and intact glycosylated IgG antibodies have also been analyzed using IM-MS to differentiate IgG1 and two different isoforms of IgG2 (73). Most of these studies removed the glycans from the glycoprotein and purified prior to the carbohydrates prior to analysis which is similar to other contemporary MS methodologies for the characterization of glycoproteins which analyze the glycans, peptides, or glycopeptides separately. In this work, we focus on the rapid characterization of biomolecules in complex samples without time-consuming purification steps before analysis.
1.6.1 Simultaneous glycoproteomics using IM-MS
The use of IM combined with MS allows for the simultaneous detection of different biomolecular classes (i.e. lipids, peptides, carbohydrates, and oligonucleotides) with little or no purification needed (Fig. 5). In these methods, we concentrate on the characterization of carbohydrate standards using DTIM-MS for the determination of the collision cross sectional area and region of 2D IM-MS space occupied by carbohydrates when compared to other biomolecules such as lipids, peptides, and oligonucleotides. We then describe the simultaneous separation and characterization of peptides and glycans from glycoprotein digests (Fig. 6,7) and glycans from lipids in a human milk sample without the need for extensive purification (Fig. 8).
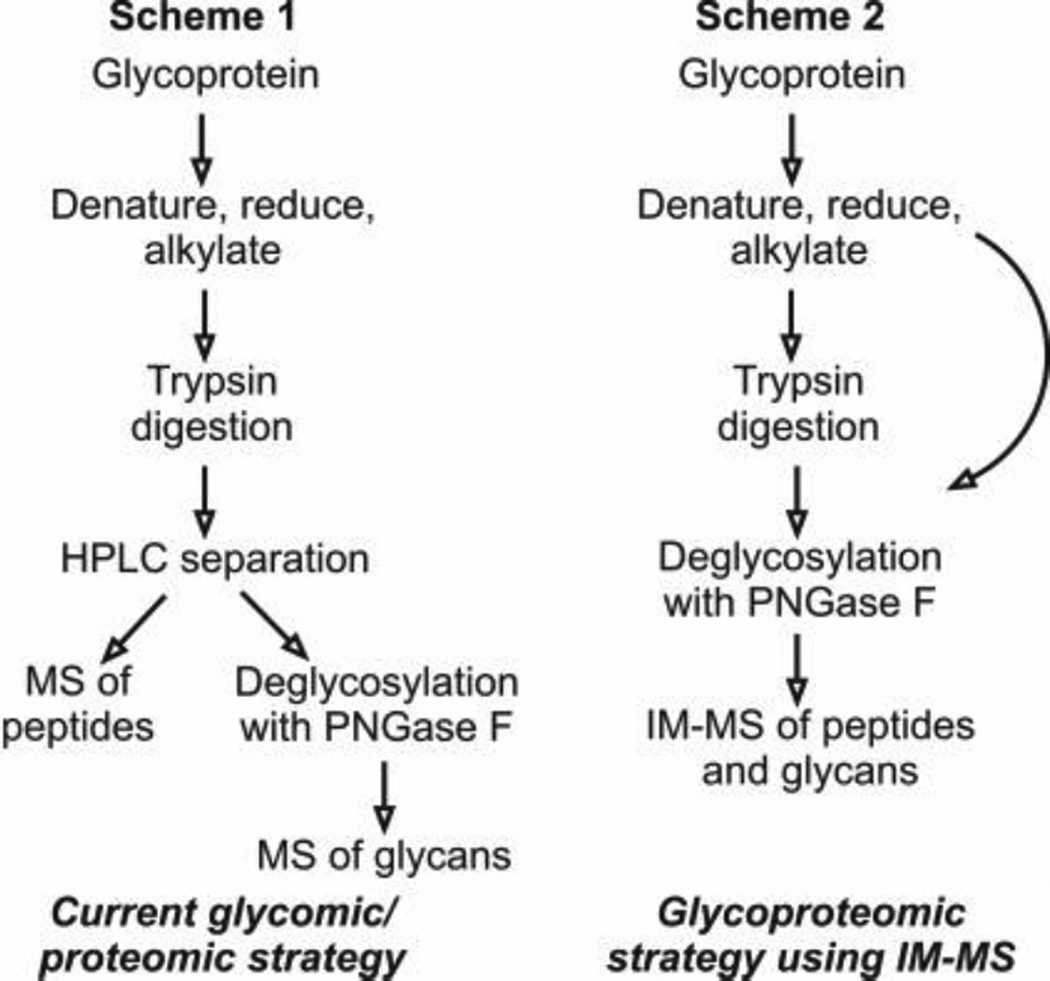
Figure 5.
Schemes 1 and 2 present a comparison of current glycomic and proteomic protocols versus the glycoproteomic strategy using IM-MS (88). Fenn, L. S., and McLean, J. A. (2009) Simultaneous glycoproteomics on the basis of structure using ion mobility-mass spectrometry. Molecular BioSystems 5, 1298–302- Reproduced by permission of The Royal Society of Chemistry (RSC).
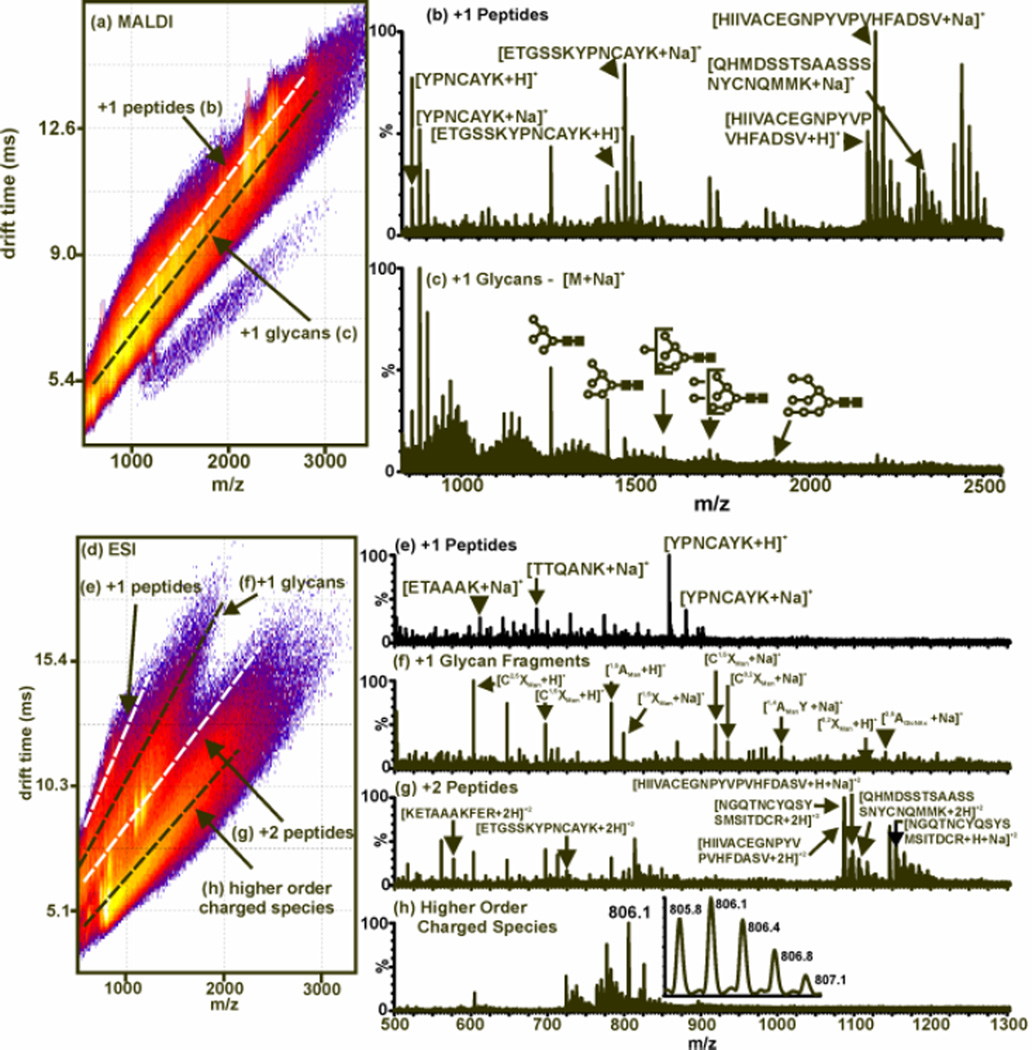
Figure 6.
MALDI- and ESI-IM-MS plot and extracted mass spectra from RNAse B digested and deglycosylated with trypsin and PNGase F, respectively. (a) A 2D MALDI-IM-MS plot of conformation space. Structural separations are observed for peptides [labeled (b)] and glycans [labeled (c)]. Since MALDI is used, all identified peaks correspond to singly-charged species as sodium-coordinated glycans and protonated peptides. (b) An extracted mass spectrum corresponding to peptides (along white dashed-line in (a)). (c) An extracted mass spectrum corresponding to glycans (along black dashed-line in (a)). Open circles and filled boxes correspond to mannose and N-acetylglucoseamine, respectively. Unidentified peaks seen at lower masses are due to in-source fragmentation of the glycans present. (d) A 2D ESI-IM-MS plot of conformation space. Structural separations are observed for singly-charged peptides [labeled (e)], singly-charged glycans [labeled (f)], doubly-charged peptides [labeled (g)], and higher order charged species [labeled (h)]. (e) An extracted mass spectrum corresponding to singly-charged peptides (along top white dashed-line in (d)). (f) An extracted mass spectrum corresponding to singly-charged glycans with identification of fragments by Domon and Costello nomenclature (along top black dashed-line in (d)). (g) An extracted mass spectrum corresponding to doubly-charged peptides (along bottom white dashed-line in (d)). (h) An extracted mass spectrum corresponding to higher order charged species (along bottom black dashed-line in (d)). The inset illustrates the isotopic pattern for a triply-charged analyte (88). Fenn, L. S., and McLean, J. A. (2009) Simultaneous glycoproteomics on the basis of structure using ion mobility-mass spectrometry. Molecular BioSystems 5, 1298–302 - Reproduced by permission of The Royal Society of Chemistry (RSC).
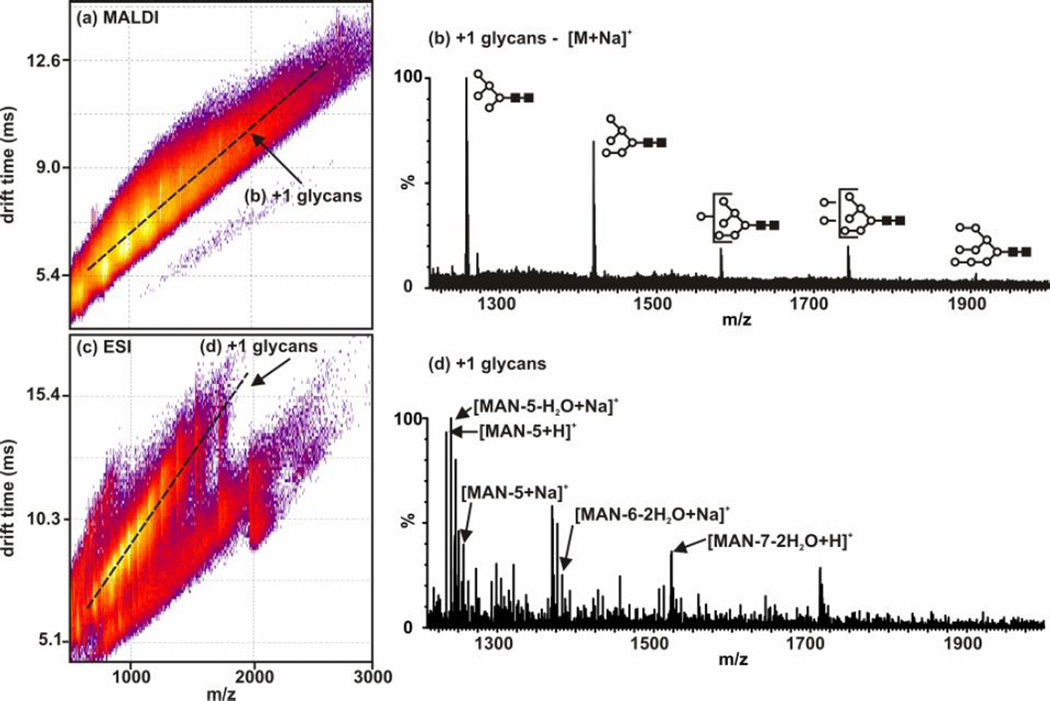
Figure 7.
Plots and extracted mass spectra from intact RNAse B that has been deglycosylated with PNGase F and analysed using MALDI-IM-MS (a,b) and ESI-IM-MS (c,d). Note that the protein was not proteolytically digested and remained intact (Mr ~ 13,700 Da). (a) A 2D MALDI-IM-MS plot of conformation space. Structural separations are observed for singly-charged glycans [labeled (b)] which are then compared to those identified in Figure 6. (b) An extracted mass spectrum corresponding to singly-charged glycans (along dashed-line in (a)). (c) A 2D ESI-IM-MS plot of conformation space. Structural separations are noted for singly-charged glycans [labeled (d)] which are then compared to those identified in Figure 6. (d) An extracted mass spectrum corresponding to to singly-charged glycans (along dashed-line in (c)). (88) - Reproduced by permission of The Royal Society of Chemistry (RSC).
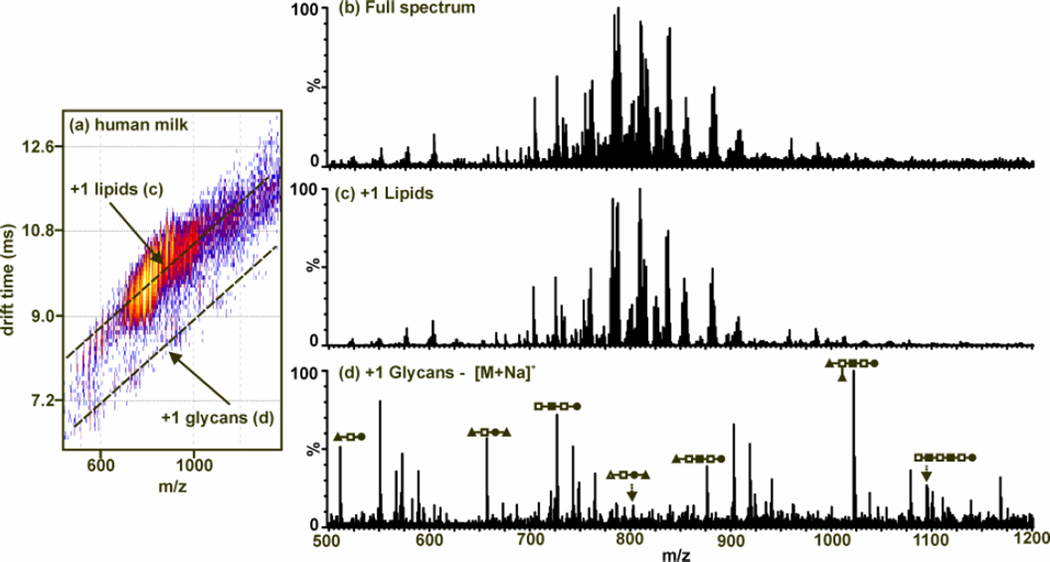
Figure 8.
MALDI-IM-MS plot and extracted mass spectra from human milk with no prior purification. (a) A 2D IM-MS plot of conformation space. Structural separations are observed for lipids [labeled (c)] and glycans [labeled (d)]. Since MALDI is used, all identified peaks correspond to singly-charged species. (b) An integrated mass spectrum for all of conformation space. This is what would be seen if using MS alone to characterize the human milk sample. (c) An extracted mass spectrum corresponding to lipids [along top dashed-line in (a)]. (d) An extracted mass spectrum corresponding to glycans [along bottom dashed-line in (a)]. Carbohydrate structure representations are as follows: ●-glucose, Δ-sialic acid, ■-N-acetylglucosamine, □-galactose, and ▲-fucose. Adapted from (89).
In Fig. 6, the simultaneous characterization of glycans and peptides in a digest of RNAse B using ESI and MALDI is presented. The protein is first digested with trypsin and subsequently with PNGase F to produce the peptides and glycans. In Fig. 7, the confidence in the identifications can be increased through performing only the deglycosylation of RNAse B to analyze glycans only in the mass range of interest. In both of these figures, the peptides and glycans are ionized using both ESI and MALDI. There are serveral differences between ESI and MALDI that would guide an investigator to select one source versus the other. Firstly, ESI generally produces ions of multiple charge states (e.g. [M+nH]n+) while MALDI generally produces singly-changed species (e.g. [M+H]+). For fragmentation-based MS/MS studies, multiply charged species are advantageous, however by partitioning the signal into multiply-charged channels can result in complicated spectra. Secondly, MALDI is more generally tolerant of salts than is ESI. Thirdly, MALDI is inherently an off-line ionization source, while ESI is more easily coupled with additional separation steps such as liquid chromatography. A more detailed description of the advantages and limitations of MALDI and ESI for glycoproteomics is presented in Ref. (88).
1.6.2 IM-MS/MS measurements for confident glycan identification
Most current MS methodologies for carbohydrate characterization require the use of collision-induced dissociation (CID) or tandem MS (MS/MS or MSn) to confirm the carbohydrate sequence and branching patterns (74–77). Similar to traditional MS/MS, structural information can be obtained through the use of IM-MS/MS. The collision cell for fragmentation can be inserted before, or after, the IM drift cell determined by the desired information. An advantage to fragmenting after IM separation is that all fragment ions will have the same drift time as the parent. Commercial IM-MS instruments currently available have the ability to isolate a certain mass through the use of a resolving quadrupole, to fragment ions before or after the IM separation region, and to fragment ions up to MS5 (45). These abilities are very useful when characterizing complex samples like those encountered in glycomics research. In addition to parent ion selection by mass, the parent ion can be selected by mobility or structure using time dispersion in the drift cell (8).
2. Materials
2.1 Collision cross section measurements for MALDI-IM-MS
Sample for which collision cross section is desired. This can be a pure compound or within a mixture but must be abundant enough to obtain sufficient signal for five measurements at different voltage settings in the IM dimension. For carbohydrates in Fig. 3 and 4, standards were used at a concentration of 1 mg/mL in DDI water prepared for MALDI analysis. Standards can be purchased from several companies including those that specialize in carbohydrates such as Dextra (Reading, UK) and V-Labs, Inc. (Covington, LA). Many of the carbohydrates used for the presented collision cross section database were obtained from the Consortium for Functional Glycomics.
-
Drift tube IM standards/calibrants. Mass standards are species, usually peptides and proteins, that span the mass range of interest. For DTIM structural standards, C60 and C70 (fullerenes) are typically used due to their existence in one structural form. Additionally, fullerenes are sometimes used as mass standards since they are structurally separated from biomolecules in 2D IM-MS space and provide numerous gas-phase reaction products resulting in peaks spanning a large mass range in increments of 24 Da. These standards can also be used to evaluate DTIM resolution and instrument performance.
Generally, the standard peptide bradykinin (RPPGFSPFR) is used to validate gas pressure in DTIM by comparison of the collision cross section measurement with the accepted value of 242 ± 2 Å2 (20). Bradykinin can be mixed with matrix of choice or a standard solution of 1 mg/mL in H2O can be combined 1:1 v/v with 20mg/mL α-cyano-4-hydroxycinnamic acid in 50% methanol.
Traveling wave IM standards/calibrants. As discussed in Section 1.3, TWIM provides relative collision cross sections therefore requiring internal standards with corresponding DTIM obtained absolute collision cross section values. Published absolute collision cross sections can be obtained from several databases, including: (i) peptide collision cross sections determined by ESI (78, 79), (ii) intact protein collision cross sections determined by ESI (80) (iii) peptide collision cross sections determined by MALDI (40), and (iv) biologically relevant carbohydrate, lipid, and oligonucleotide collision cross sections determined by MALDI (56). For these comparative measurements, it is necessary to have standards in the same biomolecular class as the samples being measured (81).
2.2 Simultaneous glycomics and proteomics using IM-MS
Purified glycoprotein containing N-linked glycans or glycan-containing sample of interest (i.e. human milk)
Drift tube IM standards/calibrants. See Section 2.1 for details.
Peptide-N4-(acetyl-b-glucosaminyl)-asparagine amidase F (PNGase F), usually from Chryseobacterium meningosepticum, for the removal of N-linked glycans can be obtained through Prozyme Glyko, Calbiochem, or other vendors.
3. Methods
3.1 Performing collision cross section measurements using MALDI-DTIM-MS
In order to take measurements, the samples should be prepared for MALDI analysis. In these experiments, the 1 mg/mL carbohydrate standards were combined 1:1 by volume (200:1 molar ratio) with saturated 2,5-dihydroxybenzoic acid (DHB) in 50% ethanol:DDI water.
Following insertion of the sample target into the instrument, mass and IM standard/ calibrants are measured. In particular, in using MALDI-IM-MS methods, the laser pulse serves as the start signal (t0) for measuring the IM arrival time distribution (tATD). These time distinctions are necessary for the calculations in Step 4.
Following structural separation in the IM drift cell filled with an inert gas (usually 1–10 Torr of He or N2, see Note 3) ions are directed through a skimming and differential pumping region where the pressure is reduced from 1–10 Torr to ~10−8 Torr for mass analysis in the orthogonal TOFMS. The stop time for tATD corresponds to the ion injection time for the TOFMS measurement.
-
To perform the collision cross section calculations as described in Section 1.3 (e.g. Eqn. [4]), the arrival time distribution must be corrected for time spent in regions outside of the drift cell (i.e. time spent traversing from the MALDI plate into the drift cell, in skimming and differential pumping regions, and ion optic regions prior to insertion into the TOFMS). This will result in the drift time (td) of the ions within the IM drift cell used in the calculation of collision cross section:
[5] To determine the value of tdtc, IM separations are performed by varying the voltage across the drift cell while maintaining all other experimental parameters constant. The tATD measured at each drift voltage and are then plotted versus the inverse of drift voltage (1/V). Provided the range of voltages used maintains ion separations under low field conditions, this plot will result in a linear correlation. If non-linearity is observed, a calculation of the low field limit should be performed, because curvature in this plot indicates that mobility is not constant over the voltage range used. A linear regression of this data results in a y-intercept corresponding to tdtc (See Note 4). Preferably at least five voltages should be used to define this line although for high precision measurements as many voltages as is practical should be used.
After the td has been determined, it can now be used to calculate the collision cross section, Ω, of the ion of interest through the Eqn. [5] (See Notes 5,6) (11).
After calculation of the collision cross section, the value can be further related to the structure using molecular dynamic simulations. Detailed information about these computational methods can be found in other resources (50–53).
For calculating relative collision cross sections using TWIM-MS, the two main procedures used can be found in the literature (46, 47).
3.2 Performing simultaneous glycoproteomics using IM-MS (See Note 7)
The N-linked glycan-containing glycoprotein is prepared by making a 1 mg/mL solution in DDI water or 50 mM ammonium acetate at pH ~7.5 (See Note 8). An aliquot (~1nmol) is pipetted into a microcentrifuge tube.
Thermal denaturation is performed by heating the sample at 90°C for 15 minutes (See Note 9) (82). To quench denaturation, the sample is placed in a −20°C freezer for ~15 minutes.
To reduce disulfide bonds in the protein, dithiothreitol (DTT) is added to make the final concentration 5 mM and reacts at 60°C for 30 minutes (83) (See Note 10).
To alkylate free cysteines, iodoacetamide is added to a final concentration of 50 mM and reacts for 1 hour at 37°C in the dark (84) (See Note 10).
For proteolytic digestion of the glycoprotein, trypsin is added to the sample (approximately 20 : 1 wt of substrate per wt of trypsin) and allowed to digest at 37°C for 24 hours. This step is omitted for control digests where only glycans are in the mass range of interest. The sample is placed in a −20°C freezer for at least 15 minutes to stop the enzyme activity (can be stored overnight).
For removal of the N-linked glycans of interest, 1 µU (microunit) of PNGase F is added to the sample and incubated at 37°C for at least 12 hours (See Note 11). The samples is then placed in a −20°C freezer until analysis.
To prepare for MALDI analysis, the glycoprotein digests are combined 1:1 by volume (200:1 molar ratio) with saturated 2,5-dihydroxybenzoic acid (DHB) in 50% ethanol:DDI water and then spotted on a MALDI plate.
In preparation for ESI analysis, the glycoprotein digest was dissolved in 50 : 50 (v/v) water : methanol to a final concentration of 25 mM (See Note 12).
-
The samples are then analyzed using a Synapt HDMS G1 or G2 (Waters Corp.) equipped with TWIM drift cell and operated with MassLynx software. For both ionization sources, the ion guide T-wave is operated at 300 m/s and linearly ramped in amplitude from 5–20 V over each experiment. The transfer guide T-wave is operated at 248 m/s and with a constant 3 V amplitude. Ion injection voltages in the Trap and Transfer were set at 6 and 4 V, respectively.
For MALDI and ESI, it is advantageous to optimize all source settings for the sample of interest paying particular attention to the optimization of glycan signal. Cone voltage in ESI along with laser energy in MALDI should be tuned to suppress carbohydrate in source-fragmentation.
For data analysis, MassLynx software is also used along with Driftscope for the visualization of 2D data. In Driftscope, the regions associated with different biomolecular classes, in particular carbohydrates and peptides, can be selected and extracted in order to identify the peaks associated with each class (See Figures 6,7). In order to increase confidence in identifications, IM-MS/MS can be used (See Section 1.6.2).
3.3 Performing simultaneous glycolipidomics using IM-MS
Dilute sample with DDI water. For the human milk example presented, the optimal dilution was 1:10 milk to DDI water by volume.
Mix diluted sample with matrix as described in Section 3.2, Step 6. The type of matrix, concentration, or matrix to analyte ratio can be varied to optimize for carbohydrate signal in the complex mixture if desired.
Analyze using MALDI-IM-MS for best results using same settings as above (optimized for glycan signal). An example of the plot obtained is presented in Figure 8.
Acknowledgments
Financial support for this work was provided by the National Institutes of Health (1R01GM092218-01 and RC2DA028981), the U.S. Defense Threat Reduction Agency (HDTRA-09-1-0013), Vanderbilt University College of Arts and Sciences, Vanderbilt Institute of Chemical Biology, and Vanderbilt Institute for Integrative Biosystems Research and Engineering. We also thank Richard M. Caprioli (Vanderbilt University, Department of Biochemistry) and the Vanderbilt University Mass Spectrometry Research Center for use of the Synapt HDMS. The carbohydrate compounds, LacNAc, Lec, Lac, B tetra type 1, 2’F-B type 2, LNT, LeALex, Di-Lec, Di-LeA, LNnT, Galα3-type1, B2-tri, Pk, P1 tri, Tri-LacNAc, H-type2-LN-LN, P1 penta, P1 antigen, GNLNLN, and 3’SLN-Lec, were provided by the Carbohydrate Synthesis/Protein Expression Core of The Consortium for Functional Glycomics funded by the National Institute of General Medical Sciences grant GM62116.
Footnotes
The Synapt G1 HDMS has activation/dissociation regions to perform up to MS5, but usually MS3 is the practical maximum.
IM-MS 2D data is presented in one of two ways with either m/z on the abscissa and arrival time distribution on the ordinate axes (in which all the data in this work is presented) or the reverse.
Our DTIM drift cells use He due to its low polarizability and low mass relative to other inert gases. However, most TWIM drift cells utilize N2, and other drift gases or drift gas additives can be used to promote interactions between the ion and drift gas.
The plot of arrival time distribution versus 1/V has a y-intercept that correlates to tdtc or drift time correction because it represents the limit of td→0 at infinite drift cell voltage.
When high accuracy collision cross section measurements are needed, the drift time correction should be evaluated for each species. This is due to correcting for the additional ion-neutral collisions in the differential pumping regions at the exit and/or entrance of the IM drift cell.
When calculating collision cross section, much care should be taken in the dimensionality of the units used. This is due to the equation being derived from classical electrodynamics, and the units for E should be expressed in cgs Gaussian units, i.e. statvolts cm−1, where 1 statvolt equals 299.79 V. Note that statvolts cm−1 is equivalent to statcoulombs cm−2 and that elementary charge, e, is 4.80 × 10−10 statcoulombs.
Simultaneous glycoproteomics methodologies are focused on the characterization of N-linked glycans. However, these experiments can be adapted for the characterization of O-linked glycans and glycoproteins treated with other enzymes such as Pronase.
There has been some debate against using ammonium-containing buffers (85). However, for these studies, ammonium acetate was used and acceptable glycan signal was obtained.
Other methods of protein denaturation can be used [i.e. chemical (86, 87)] but have not been tested.
Reduction and alkylation are performed in typical proteomic digestions and directions are given here if the procedure is desired. However, for the presented examples (Fig. 6,7), reduction and alkylation were not performed.
One unit is defined as the amount of enzyme that will completely catalyze the release of N-linked oligosaccharides from 1 µmol denatured ribonuclease in 1 min at 37°C, pH 7.5.
ESI analysis is performed based on high-throughput methodology (minimum time per sample) and does not utilize derivatization strategies known to enhance ionization and decrease fragmentation of glycans. These can be done if time allows.
References
- 1.Bahl OP. Glycoconjugates: Composition, Structure, and Function. New York, NY: Marcel Dekker; 1992. [Google Scholar]
- 2.Gorelik E, Galili U, Raz A. On the Role of Cell Surface Carbohydrates and their Binding Proteins (lectins) in Tumor Metastasis. Cancer and Metastasis Reviews. 2001;20:245–277. doi: 10.1023/a:1015535427597. [DOI] [PubMed] [Google Scholar]
- 3.Montreuil J, Vliegenthart JFG, Schachter H. Glycoproteins I. New York, NY: Elsevier Science; 1995. [Google Scholar]
- 4.Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochimica et Biophysica Acta (BBA) - General Subjects. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 5.Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the Immune System. Science. 2001;291:2370–2376. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- 6.Taylor ME, Drickamer K. Introduction to Glycobiology. Oxford, U.K.: Oxford University Press; 2006. [Google Scholar]
- 7.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. Second. Woodbury, NY: Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- 8.McLean JA, Ruotolo BT, Gillig KJ, Russell DH. Ion mobility-mass spectrometry: a new paradigm for proteomics. Int J Mass Spectrom. 2005;240:301–315. [Google Scholar]
- 9.Clemmer DE, Jarrold MF. Ion mobility measurements and their applications to clusters and biomolecules. J Mass Spectrom. 1997;32:577–592. [Google Scholar]
- 10.Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH., Jr Ion mobility-mass spectrometry. J Mass Spectrom. 2008;43:1–22. doi: 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- 11.Mason EA, McDaniel EW. Transport Properties of Ions in Gases. Indianapolis, IN: John Wiley and Sons; 1988. [Google Scholar]
- 12.Baumbach J. Process analysis using ion mobility spectrometry. Anal Bioanal Chem. 2006;384:1059–1070. doi: 10.1007/s00216-005-3397-8. [DOI] [PubMed] [Google Scholar]
- 13.Eiceman GA, Karpas Z. Ion Mobility Spectrometry. (Second) 2005 [Google Scholar]
- 14.McAfee KB, Jr, Edelson D. Identification and mobility of ions in a Townsend discharge by time-resolved mass spectrometry. Proc Phys Soc, London. 1963;81:382–384. [Google Scholar]
- 15.Barnes WS, Martin DW, McDaniel EW. Mass spectrographic identification of the ion observed in hydrogen mobility experiments. Phys Rev Lett. 1961;6:110–111. [Google Scholar]
- 16.Gieniec J, Mack LL, Nakamae K, Gupta C, Kumar V, Malcolm D. Electrospray mass spectroscopy of macromolecules: Application of an ion-drift spectrometer. Biomed Spectrom. 1984;11:259–268. [Google Scholar]
- 17.von Helden G, Wyttenbach T, Bowers MT. Inclusion of a MALDI ion source in the ion chromatography technique: conformational information on polymer and biomolecular ions. Int J Mass Spectrom Ion Processes. 1995;146–147:349–364. [Google Scholar]
- 18.Shelimov KB, Clemmer DE, Hudgins RR, Jarrold MF. Protein structure in vacuo: Gas-phase confirmations of BPTI and cytochrome c. J Am Chem Soc. 1997;119:2240–2248. [Google Scholar]
- 19.von Helden G, Wyttenbach T, Bowers MT. Conformation of Macromolecules in the Gas-Phase - Use of Matrix-Assisted Laser-Desorption Methods in Ion Chromatography. Science. 1995;267:1483–1485. doi: 10.1126/science.267.5203.1483. [DOI] [PubMed] [Google Scholar]
- 20.Wyttenbach T, vonHelden G, Bowers MT. Gas-phase conformation of biological molecules: Bradykinin. J Am Chem Soc. 1996;118:8355–8364. [Google Scholar]
- 21.Myung S, Lee YJ, Moon MH, Taraszka J, Sowell R, Koeniger S, Hilderbrand AE, Valentine SJ, Cherbas L, Cherbas P, Kaufmann TC, Miller DF, Mechref Y, Novotny MV, Ewing MA, Sporleder CR, Clemmer DE. Development of high-sensitivity ion trap ion mobility spectrometry time-of-flight techniques: A high-throughput nano-LC-IMS-TOF separation of peptides arising from a Drosophila protein extract. Anal Chem. 2003;75:5137–5145. doi: 10.1021/ac030107f. [DOI] [PubMed] [Google Scholar]
- 22.Isailovic D, Kurulugama RT, Plasencia MD, Stokes ST, Kyselova Z, Goldman R, Mechref Y, Novotny MV, Clemmer DE. Profiling of Human Serum Glycans Associated with Liver Cancer and Cirrhosis by IMS-MS. J Prot Res. 2008;7:1109–1117. doi: 10.1021/pr700702r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Plasencia M, Ragg S, Valentine SJ, Clemmer DE. Development of high throughput dispersive LC-ion mobility-TOFMS techniques for analysing the human plasma proteome. Brief Funct Genomic Proteomic. 2004;3:177–186. doi: 10.1093/bfgp/3.2.177. [DOI] [PubMed] [Google Scholar]
- 24.Liu XY, Valentine SJ, Plasencia MD, Trimpin S, Naylor S, Clemmer DE. Mapping the human plasma proteome by SCX-LC-IMS-MS. J Am Soc Mass Spectrom. 2007;18:1249–1264. doi: 10.1016/j.jasms.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valentine SJ, Plasencia MD, Liu XY, Krishnan M, Naylor S, Udseth HR, Smith RD, Clemmer DE. Toward plasma proteome profiling with ion mobility-mass spectrometry. J Prot Res. 2006;5:2977–2984. doi: 10.1021/pr060232i. [DOI] [PubMed] [Google Scholar]
- 26.Liu XY, Miller BR, Rebec GV, Clemmer DE. Protein expression in the striatum and cortex regions of the brain for a mouse model of Huntington's disease. J Prot Res. 2007;6:3134–3142. doi: 10.1021/pr070092s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taraszka JA, Kurulugama R, Sowell RA, Valentine SJ, Koeniger SL, Arnold RJ, Miller DF, Kaufman TC, Clemmer DE. Mapping the proteome of Drosophila melanogaster: Analysis of embryos and adult heads by LC-IMS-MS methods. J Prot Res. 2005;4:1223–1237. doi: 10.1021/pr050038g. [DOI] [PubMed] [Google Scholar]
- 28.Benesch JLP, Ruotolo BT, Simmons DA, Robinson CV. Protein complexes in the gas phase: Technology for structural genomics and proteomics. Chem Rev. 2007;107:3544–3567. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
- 29.Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, Robinson CV. Evidence for macromolecular protein rings in the absence of bulk water. Science. 2005;310:1658–1661. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- 30.Ruotolo BT, Hyung SJ, Robinson PM, Giles K, Bateman RH, Robinson CV. Ion mobility-mass spectrometry reveals long-lived, unfolded intermediates in the dissociation of protein complexes. Angew Chem, Int Ed Engl. 2007;46:8001–8004. doi: 10.1002/anie.200702161. [DOI] [PubMed] [Google Scholar]
- 31.Jackson SN, Ugarov M, Egan T, Post JD, Langlais D, Schultz JA, Woods AS. MALDI-ion mobility-TOFMS imaging of lipids in rat brain tissue. J Mass Spectrom. 2007;42:1093–1098. doi: 10.1002/jms.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLean JA, Ridenour WB, Caprioli RM. Profiling and imaging of tissues by imaging ion mobility-mass spectrometry. J Mass Spectrom. 2007;42:1099–1105. doi: 10.1002/jms.1254. [DOI] [PubMed] [Google Scholar]
- 33.Mason EA, McDaniel EW. Transport Properties of Ions in Gases. New York, NY: John Wiley & Sons; 1988. pp. 31–102. [Google Scholar]
- 34.Dugourd P, Hudgins RR, Clemmer DE, Jarrold MF. High-resolution ion mobility measurements. Rev Sci Instrum. 1997;68:1122–1129. [Google Scholar]
- 35.Merenbloom SI, Glaskin RS, Henson ZB, Clemmer DE. High-Resolution Ion Cyclotron Mobility Spectrometry. Anal Chem. 2009;81:1482–1487. doi: 10.1021/ac801880a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wyttenbach T, Kemper PR, Bowers MT. Design of a new electrospray ion mobility mass spectrometer. Int J Mass Spectrom. 2001;212:13–23. [Google Scholar]
- 37.Dwivedi P, Wu P, Klopsch SJ, Puzon GJ, Xun L, Hill HH., Jr Metabolic profiling by ion mobility mass spectrometry (IMMS) Metabolomics. 2008;4:63–80. [Google Scholar]
- 38.Furche F, Ahlrichs R, Weis P, Jacob C, Gilb S, Bierweiler T, Kappes MM. The structures of small gold cluster anions as determined by a combination of ion mobility measurements and density functional calculations. J Chem Phys. 2002;117:6982–6990. [Google Scholar]
- 39.Ruotolo BT, Verbeck, Thomson LM, Woods AS, Gillig KJ, Russell DH. Distinguishing between Phosphorylated and Nonphosphorylated Peptides with Ion Mobility-Mass Spectrometry. J Prot Res. 2002;1:303–306. doi: 10.1021/pr025516r. [DOI] [PubMed] [Google Scholar]
- 40.Tao L, McLean JR, McLean JA, Russell DH. A Collision Cross-Section Database of Singly-Charged Peptide Ions. J Am Soc Mass Spectrom. 2007;18:1232–1238. doi: 10.1016/j.jasms.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 41.Mason EA. In: Plasma Chromatography. Carr TW, editor. New York, NY: Plenum Press; 1984. pp. 43–93. [Google Scholar]
- 42.Revercomb HE, Mason EA. Theory of plasma chromatography/gaseous electrophoresis. Review. Anal Chem. 1975;47:970–983. [Google Scholar]
- 43.Fenn LS, McLean JA. Biomolecular structural separations by ion mobility-mass spectrometry. Anal Bioanal Chem. 2008;391:905–909. doi: 10.1007/s00216-008-1951-x. [DOI] [PubMed] [Google Scholar]
- 44.Giles K, Pringle SD, Worthington KR, Little D, Wildgoose JL, Bateman RH. Applications of a travelling wave-based radio-frequency-only stacked ring ion guide. Rapid Commun Mass Spectrom. 2004;18:2401–2414. doi: 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- 45.Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole/travelling wave IMS/oa-ToF instrument. Int J Mass Spectrom. 2007;261:1–12. [Google Scholar]
- 46.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung S-J, Robinson CV. Ion mobility-mass spectrometry analysis of large protein complexes. Nat Protocols. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 47.Williams JP, Scrivens JH. Coupling desorption electrospray ionisation and neutral desorption/extractive electrospray ionisation with a travelling-wave based ion mobility mass spectrometer for the analysis of drugs. Rapid Commun Mass Spectrom. 2008;22:187–196. doi: 10.1002/rcm.3346. [DOI] [PubMed] [Google Scholar]
- 48.Vakhrushev SY, Langridge J, Campuzano I, Hughes C, Peter-Katalinic J. Ion Mobility Mass Spectrometry Analysis of Human Glycourinome. Anal Chem. 2008;80:2506–2513. doi: 10.1021/ac7023443. [DOI] [PubMed] [Google Scholar]
- 49.Riba-Garcia I, Giles K, Bateman RH, Gaskell SJ. Evidence for Structural Variants of a- and b-Type Peptide Fragment Ions Using Combined Ion Mobility/Mass Spectrometry. J Am Soc Mass Spectrom. 2008;19:609–613. doi: 10.1016/j.jasms.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 50.Gidden J, Bowers MT. Gas-phase conformations of deprotonated trinucleotides (dGTT(−), dTGT(−), and dTTG(−)): The question of zwitterion formation. J Am Soc Mass Spectrom. 2003;14:161–170. doi: 10.1016/S1044-0305(02)00866-8. [DOI] [PubMed] [Google Scholar]
- 51.Gidden J, Bowers MT. Gas-phase conformations of deprotonated and protonated mononucleotides determined by ion mobility and theoretical modeling. J Phys Chem B. 2003;107:12829–12837. [Google Scholar]
- 52.Shvartsburg AA, Jarrold MF. An exact hard-spheres scattering model for the mobilities of polyatomic ions. Chem Phys Lett. 1996;261:86–91. [Google Scholar]
- 53.Wyttenbach T, Witt M, Bowers MT. On the Stability of Amino Acid Zwitterions in the Gas Phase: The Influence of Derivatization, Proton Affinity, and Alkali Ion Addition. J Am Chem Soc. 2000;122:3458–3464. [Google Scholar]
- 54.Fenn LS, McLean JA. Structural resolution of carbohydrate positional and structural isomers based on gas-phase ion mobility-mass spectrometry. Phys Chem Chem Phys. 2011;13:2196–2205. doi: 10.1039/c0cp01414a. [DOI] [PubMed] [Google Scholar]
- 55.Williams JP, Grabenauer M, Holland RJ, Carpenter CJ, Wormald MR, Giles K, Harvey DJ, Bateman RH, Scrivens JH, Bowers MT. Characterization of simple isomeric oligosaccharides and the rapid separation of glycan mixtures by ion mobility mass spectrometry. Int J Mass Spectrom. 2010 [Google Scholar]
- 56.Fenn LS, Kliman M, Mahsut A, Zhao SR, McLean JA. Characterizing ion mobility-mass spectrometry conformation space for the analysis of complex biological samples. Anal Bioanal Chem. 2009;394:235–244. doi: 10.1007/s00216-009-2666-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee S, Wyttenbach T, Bowers MT. Gas phase structures of sodiated oligosaccharides by ion mobility/ion chromatography methods. Int J Mass Spectrom Ion Processes. 1997;167–168:605–614. [Google Scholar]
- 58.Hoaglund CS, Valentine SJ, Clemmer DE. An Ion Trap Interface for ESI Ion Mobility Experiments. Anal Chem. 1997;69:4156–4161. [Google Scholar]
- 59.Liu Y, Clemmer DE. Characterizing Oligosaccharides Using Injected-Ion Mobility/Mass Spectrometry. Anal Chem. 1997;69:2504–2509. doi: 10.1021/ac9701344. [DOI] [PubMed] [Google Scholar]
- 60.Leavell MD, Gaucher SP, Leary JA, Taraszka JA, Clemmer DE. Conformational studies of Zn-ligand-hexose diastereomers using ion mobility measurements and density functional theory calculations. J Am Soc Mass Spectrom. 2002;13:284–293. doi: 10.1016/S1044-0305(01)00362-2. [DOI] [PubMed] [Google Scholar]
- 61.Clowers BH, Dwivedi P, Steiner WE, Hill HH, Bendiak B. Separation of Sodiated Isobaric Disaccharides and Trisaccharides Using Electrospray Ionization-Atmospheric Pressure Ion Mobility-Time of Flight Mass Spectrometry. J Am Soc Mass Spectrom. 2005;16:660–669. doi: 10.1016/j.jasms.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 62.Clowers BH, Hill HH., Jr Mass Analysis of Mobility-Selected Ion Populations Using Dual Gate, Ion Mobility, Quadrupole Ion Trap Mass Spectrometry. Anal Chem. 2005;77:5877–5885. doi: 10.1021/ac050700s. [DOI] [PubMed] [Google Scholar]
- 63.Dwivedi P, Bendiak B, Clowers BH, Hill HH., Jr Rapid Resolution of Carbohydrate Isomers by Electrospray Ionization Ambient Pressure Ion Mobility Spectrometry-Time-of-Flight Mass Spectrometry (ESI-APIMS-TOFMS) J Am Soc Mass Spectrom. 2007;18:1163–1175. doi: 10.1016/j.jasms.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 64.Zhu M, Bendiak B, Clowers B, Hill H. Ion mobility-mass spectrometry analysis of isomeric carbohydrate precursor ions. Anal Bioanal Chem. 2009;394:1853–1867. doi: 10.1007/s00216-009-2865-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamagaki T, Sato A. Peak width-mass correlation in CID MS/MS of isomeric oligosaccharides using traveling-wave ion mobility mass spectrometry. J Mass Spectrom. 2009;44:1509–1517. doi: 10.1002/jms.1641. [DOI] [PubMed] [Google Scholar]
- 66.Yamagaki T, Sato A. Isomeric Oligosaccharides Analyses Using Negative-ion Electrospray Ionization Ion Mobility Spectrometry Combined with Collision-induced Dissociation MS/MS. Anal Sci. 2009;25:985–988. doi: 10.2116/analsci.25.985. [DOI] [PubMed] [Google Scholar]
- 67.Gabryelski W, Froese KL. Rapid and sensitive differentiation of anomers, linkage, and position isomers of disaccharides using High-Field Asymmetric Waveform Ion Mobility Spectrometry (FAIMS) J Am Soc Mass Spectrom. 2003;14:265–277. doi: 10.1016/S1044-0305(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 68.Olivova P, Chen W, Chakraborty AB, Gebler JC. Determination of N-glycosylation sites and site heterogeneity in a monoclonal antibody by electrospray quadrupole ion-mobility time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2008;22:29–40. doi: 10.1002/rcm.3330. [DOI] [PubMed] [Google Scholar]
- 69.Plasencia MD, Isailovic D, Merenbloom SI, Mechref Y, Clemmer DE. Resolving and Assigning N-Linked Glycan Structural Isomers from Ovalbumin by IMS-MS. J Am Soc Mass Spectrom. 2008;19:1706–1715. doi: 10.1016/j.jasms.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schenauer MR, Meissen JK, Seo Y, Ames JB, Leary JA. Heparan Sulfate Separation, Sequencing, and Isomeric Differentiation: Ion Mobility Spectrometry Reveals Specific Iduronic and Glucuronic Acid-Containing Hexasaccharides. Anal Chem. 2009;81:10179–10185. doi: 10.1021/ac902186h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin L, Barran PE, Deakin JA, Lyon M, Uhrin D. Conformation of glycosaminoglycans by ion mobility mass spectrometry and molecular modelling. Phys Chem Chem Phys. 2005;7:3464–3471. doi: 10.1039/b508644b. [DOI] [PubMed] [Google Scholar]
- 72.McCullough BJ, Kalapothakis JM, Taylor K, Clarke DJ, Eastwood H, Campopiano D, MacMillan D, Dorin J, Barran PE. Binding a heparin derived disaccharide to defensin inspired peptides: insights to antimicrobial inhibition from gas-phase measurements. Phys Chem Chem Phys. 12:3589–3596. doi: 10.1039/b923784d. [DOI] [PubMed] [Google Scholar]
- 73.Bagal D, Valliere-Douglass JF, Balland A, Schnier PD. Resolving Disulfide Structural Isoforms of IgG2 Monoclonal Antibodies by Ion Mobility Mass Spectrometry. Anal Chem. 2010;82:6751–6755. doi: 10.1021/ac1013139. [DOI] [PubMed] [Google Scholar]
- 74.Harvey DJ. Matrix-assisted laser desorption/ionization mass spectrometry of carbohydrates and glycoconjugates. Int J Mass Spectrom. 2003;226:1–35. [Google Scholar]
- 75.Harvey DJ. Matrix-assisted laser desorption/ionization mass spectrometry of carbohydrates. Mass Spectrom Rev. 1999;18:349–450. doi: 10.1002/(SICI)1098-2787(1999)18:6<349::AID-MAS1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 76.Harvey DJ. Identification of protein-bound carbohydrates by mass spectrometry. Proteomics. 2001;1:311–328. doi: 10.1002/1615-9861(200102)1:2<311::AID-PROT311>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 77.An HJ, Lebrilla CB. Structure elucidation of native N- and O-linked glycans by tandem mass spectrometry (tutorial) Mass Spectrom Rev. 2010 doi: 10.1002/mas.20283. [DOI] [PubMed] [Google Scholar]
- 78.Henderson SC, Valentine SJ, Counterman AE, Clemmer DE. ESI/Ion Trap/Ion Mobility/Time-of-Flight Mass Spectrometry for Rapid and Sensitive Analysis of Biomolecular Mixtures. Anal Chem. 1998;71:291–301. doi: 10.1021/ac9809175. [DOI] [PubMed] [Google Scholar]
- 79.Hoaglund CS, Valentine SJ, Sporleder CR, Reilly JP, Clemmer DE. Three-Dimensional Ion Mobility/TOFMS Analysis of Electrosprayed Biomolecules. Anal Chem. 1998;70:2236–2242. doi: 10.1021/ac980059c. [DOI] [PubMed] [Google Scholar]
- 80.Clemmer Cross Section Database. [Accessed August 30th, 2010]; http://www.indiana.edu/~clemmer/Research/cross%20section%20database/cs%20database.htm. [Google Scholar]
- 81.Ridenour WB, Kliman M, McLean JA, Caprioli RM. Structural Characterization of Phospholipids and Peptides Directly from Tissue Sections by MALDI Traveling-Wave Ion Mobility-Mass Spectrometry. Anal Chem. 2010;82:1881–1889. doi: 10.1021/ac9026115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Park Z-Y, Russell DH. Thermal Denaturation: A Useful Technique in Peptide Mass Mapping. Anal Chem. 2000;72:2667–2670. doi: 10.1021/ac991444k. [DOI] [PubMed] [Google Scholar]
- 83.Cleland WW. Dithiothreitol, a New Protective Reagent for SH Groups*. Biochemistry. 1964;3:480–482. doi: 10.1021/bi00892a002. [DOI] [PubMed] [Google Scholar]
- 84.Smythe CV. The reaction of iodoacetate and of iodoacetamide with various sulfhydryl groups, with urease, and with yeast preparations. 1936;114:601–612. [Google Scholar]
- 85.Kuster B, Harvey DJ. Ammonium containing buffers should be avoided during enzymatic release of glycans from glycoproteins when followed by reducing terminal derivatization. Glycobiology. 1997;7:vii–ix. doi: 10.1093/glycob/7.2.161-a. [DOI] [PubMed] [Google Scholar]
- 86.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 87.Shirley BA. Urea and Guanidine Hydrochloride Denaturation Curves. Methods in Molecular Biology. 1995;40:177–190. doi: 10.1385/0-89603-301-5:177. [DOI] [PubMed] [Google Scholar]
- 88.Fenn LS, McLean JA. Simultaneous glycoproteomics on the basis of structure using ion mobility-mass spectrometry. Molecular BioSystems. 2009;5:1298–1302. doi: 10.1039/b909745g. [DOI] [PubMed] [Google Scholar]
- 89.Fenn LS, McLean JA. Simultaneous glycomics, proteomics, and lipidomics using ion mobility-mass spectrometry. Anal Chem. 2010 submitted. [Google Scholar]