Abstract
Protocadherin genes (PCDHs) have been suggested to act as tumor suppressor genes in various tumor types. Previous studies have demonstrated the upregulation of certain PCDH-γ subfamily genes in nodal and duodenal follicular lymphoma (FL) using gene expression analyses. However, the mechanisms and associated molecular function of PCDH-γ subfamily gene upregulation in FL remain to be elucidated. The present study examined the expression of PCDHGA3, an upregulated PCDH-γ gene subfamily member, in B-cell lymphoma 2 (BCL2)-positive and -negative FL, and evaluated its association with tumor cell proliferation in an FL-derived cell line. Immunohistochemical analysis demonstrated that the majority of FL grade 1–2 samples (19/20; 95%) and over half of grade 3A FL samples (5/9; 56%) were PCDHGA3-positive, whereas only 1/17 reactive lymphoid hyperplasia samples was positive. Notably, this positivity was widely observed in samples of BCL2-negative FL (13/15; 87%) and FL with diffuse area (10/10; 100%). The FL-derived cell line FL18 exhibited strong PCDHGA3 expression, similar to the patient samples, and its proliferation was suppressed by PCDHGA3 gene knockdown. Genes expressed concomitantly with PCDHGA3 were selected from gene expression data, and TNFRSF6B, a member of the tumor necrosis factor receptor superfamily, was among the top five most strongly correlated genes. Coexpression of TNFRSF6B and PCDHGA3 was observed immunohistochemically in FL18 cells, suggesting potential cooperation in tumor cell maintenance. In conclusion, the results of the present study indicated that PCDHGA3 was expressed in FL irrespective of BCL2 status and grading and was associated with cell proliferation. Further studies involving molecular genetic analyses are required to elucidate the mechanisms underlying the activity of PCDHGA3 in FL.
Keywords: protocadherin γ A3, follicular lymphoma, tumor cell growth
Introduction
Follicular lymphoma (FL) is the second most common form of non-Hodgkin's B-cell lymphoma (NHL) and the most common subtype of indolent NHL. The majority of FL cases (~85%) harbor the t(14;18)(q32;q21) translocation (1), which juxtaposes the B-cell lymphoma 2 (BCL2) gene on chromosome 18 to the immunoglobulin heavy chain gene (IGH) locus, resulting in constitutive expression of BCL2. Therefore, BCL2 is an important diagnostic marker of FL. However, 10–20% of cases are BCL2-negative (2), and this loss of BCL2 expression is frequent in high-grade cases (grades 3A and B) (1). Of these BCL2-negative cases, FL is difficult to diagnose even when other immunohistochemical markers are investigated.
It was previously reported that upregulation of certain protocadherin-γ (PCDH-γ) subfamily genes in nodal and duodenal FL using gene expression analysis (3). PCDH-γ subfamily genes, which are predominantly expressed in the nervous system, constitute the largest subgroup (~80 members) of the cadherin superfamily of cell-adhesion molecules (4,5). In various tumor types, PCDH genes are downregulated or silenced via promoter hypermethylation-mediated gene inactivation, including PCDH8 (6) in breast carcinoma, PCDH9 in glioblastoma (7), PCDH17 in esophageal carcinoma (8) and PCDH10 in Burkitt and diffuse large B-cell lymphoma (9). However, the mechanisms and associated molecular function of PCDH-γ subfamily gene upregulation in FL remain to be elucidated. In the present study, PCDHGA3 was investigated as a potentially useful diagnostic marker, even in BCL2-negative FL cases. Furthermore, a functional analysis of PCDHGA3 was performed in an FL-derived cell line.
Materials and methods
Patients and cell lines
A total of 71 formalin-fixed paraffin-embedded tissue (FFPET) samples were selected, including 17 nodal reactive lymphoid hyperplasia (RLH) samples and 54 FL samples, obtained from the routine and consultation files of the Department of Pathology, Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences (Okayama, Japan). Of the 54 FL cases, 20 were grade 1 or 2, 9 were grade 3A, 10 were FL with diffuse area and 15 were BCL2-negative.
Patient samples were diagnosed on the basis of morphological and immunophenotypical observations according to the current World Health Organization classification (1), and all cases were reviewed by three skilled hematopathologists (Dr Katsuyoshi Takata, Dr Yasuharu Sato and Professor Tadashi Yoshino). The study protocol was approved by the Institutional Review Board of Okayama University Graduate School of Medicine, Dentistry, and Pharmaceutical Sciences (Okayama, Japan). All study procedures were conducted in accordance with the guidelines of the Declaration of Helsinki.
The human FL-derived cell lines, FL18 and FL318 were provided by Dr Ohno of Kyoto University (Kyoto, Japan). The cells were cultured at 37°C in 5% CO2 in RPMI-1640 (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) supplemented with 10% fetal bovine serum (Biological Industries, Cromwell, CT, USA) and 10,000 U/ml streptomycin and 10,000 µg/ml penicillin (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The cells were passaged every 3 days and used at 60–70% confluence; 10–20 passages were used for subsequent experiments.
Microarray data analyses
Gene expression data of 18 nodal FL and 8 RLH samples from a previous study (3) (GSE48047) were re-analyzed. Expressed genes correlating with PCDHGA3 expression were identified using GeneSpring software version 11.0.2 (Agilent Technologies, Inc., Santa Clara, CA, USA), as previously described (3).
Hematoxylin and eosin staining and immunohistochemistry
Patient samples were fixed with 10% formaldehyde for 24 h at room temperature. The tissues were then embedded in paraffin and cut into 3 µm sections. The slides were soaked in xylene, dipped into a Coplin jar containing Mayer's hematoxylin and agitated for 30 sec. The slides were then rinsed in H2O for 1 min, then counterstained with 1% eosin Y solution for 10–30 sec with agitation. The sections were then dehydrated with two changes of 95% alcohol and two changes of 100% alcohol for 30 sec each, then 1–2 drops of mounting medium was added, and covered with a coverslip.
Formalin-fixed paraffin-embedded tissue sections (3 µm thick) were immunohistochemically stained using a BOND-MAX autostainer (Leica Biosystems, Melbourne, Australia), with immune complexes visualized by the polymer method, according to the manufacturer's protocol and as previously described (10). The following primary antibodies were used: Rabbit anti-human PCDHGA3 (1:50; cat. no. RB33029; Abgent, Inc., San Diego, CA, USA) and mouse anti-human CD21 (1:20; cat. no. IR608; Dako Cytomation, Glostrup, Denmark). For the indirect double immunofluorescence study, cytospin slides of FL18 cells were subjected to staining with the primary antibodies against PCDHGA3 and mouse anti- tumor necrosis factor receptor superfamily member 6B (TNFRSF6B; 1:100; cat. no. ab57956; Abcam, Cambridge, UK), as described previously (10).
Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from cells using the miRNeasy Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's protocols. RNA concentration was measured using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE, USA) and reverse-transcribed into cDNA using SuperScript III First-Strand Synthesis System kit (Invitrogen; Thermo Fisher Scientific, Inc.) and a Bio-Rad T100 Thermal Cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA), according to the manufacturer's protocol. PCR was performed using GeneAmp Fast (2X) PCR Master Mix (Thermo Fisher Scientific, Inc.), using the following PCDHGA3-specific primer sequences: Forward, 5′-CTCACAAGCTCAGTCCCAGA-3′ and reverse, 5′-CATAAGTGATGCGGGCGTTG-3′. β-actin (ACTB) was also amplified as a control, using the following primers: Forward, 5′-CATGTACGTTGCTATCCAGGC-3′ and reverse, 5′-CTCCTTAATGTCACGCACGAT-3′ (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany). Reactions were performed using a Bio-Rad T100 Thermal Cycler and thermal cycling conditions consisting of: 94°C for 2 min, 94°C for 30 sec, 55°C annealing temperature for 30 sec, and 72°C for 1 min for 30 cycles, followed by 72°C for 10 min. Following amplification, PCR products were separated on 1.5% agarose gels and visualized by ethidium bromide fluorescence using a Gel Print 2000i VGA, USA (BioImage; Thermo Fisher Scientific, Inc.) The resulting bands were compared to β-actin bands produced using the same cDNA isolated from each cell line.
PCDHGA3 gene knockdown assay
Three PCDHGA3 small interfering (si)RNAs (s31922, s31923 and s31924) and one negative control siRNA (Silencer Select Negative Control #1) were obtained from Thermo Fisher Scientific, Inc. FL18 cells (5×105) were transfected with 200 nM PCDHGA3 or negative control siRNA using a Neon Transfection System (Invitrogen; Thermo Fisher Scientific, Inc.) under the conditions of 1400 V, 20 ms and 2 pulses. At 24 h post-transfection, the medium was replaced with fresh medium. The cells were grown for a further 48 h and harvested for subsequent analysis.
Total RNA was extracted from cells using a miRNeasy Mini kit (Qiagen, Inc.) according to the manufacturer's protocol. RNA purity and concentration were determined spectrophotometrically. RNA was reverse-transcribed into cDNA using SuperScript III First-Strand Synthesis System kit (Invitrogen; Thermo Fisher Scientific, Inc.) and a Bio-Rad T100 Thermal Cycler (Bio-Rad Laboratories, Inc.), according to the manufacturer's protocol. Quantitative PCR (qPCR) was performed using a StepOnePlus Real-Time PCR System and TaqMan Gene Expression assays (Thermo Fisher Scientific, Inc.). Specific probes for PCDHGA3 (Hs00259230_s1), GAPDH (Hs02758991_g1) and ACTB (Hs99999903_m1) were obtained from Thermo Fisher Scientific, Inc. The relative degree of change for each gene was calculated using the 2−ΔΔCq method (11), with GAPDH as the endogenous control.
Cell proliferation assay
At 24 h post-siRNA transfection, cells were seeded in a 24-well culture plate at a density of 1×105 cells/ml. At 48 and 72 h post-transfection, cell numbers were manually counted via trypan blue staining.
Western blotting
Cultured FL18 cells (1×106) were harvested, washed in cold PBS, and lysed using Ambion KDalert lysis buffer (Thermo Fisher Scientific, Inc.). Equal amounts of protein (15 µg per lane) from each sample were separated by 8% SDS-PAGE (Bolt Bis-Tris Plus Gel, Thermo Fisher Scientific, Inc.), the membranes were blocked with PBS-T (PBS with 0.1% Tween-20) containing 5% skimmed milk power at room temperature for 1 h, then transferred onto a polyvinylidene difluoride membrane (Invitrogen; Thermo Fisher Scientific, Inc.). Membranes were incubated at 4°C overnight with a rabbit anti-human PCDHGA3 antibody (1:500; cat. no. AP13611C; Abgent, Inc., San Diego, CA, USA); a mouse anti-human β-actin antibody (1:50,000; cat. no. sc47778; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) served as a loading control, as previously described (12). The membrane was washed in PBS-T, and incubated with horseradish peroxidase-conjugated secondary antibody at room temperature for 1 h. The following secondary antibodies were used: Donkey anti-rabbit (1:50,000; cat. no. NA9340V; GE Healthcare Life Sciences, Chalfont, UK) and sheep anti-mouse (1:50,000; cat. no. NA9310V, GE Healthcare Life Sciences). The membrane was washed with PBS-T, and immune complexes were visualized using an enhanced chemiluminescence Prime Detection System (GE Healthcare Life Sciences).
Statistical analysis
The Chi-squared test, Fisher's exact test and one-way analysis of variance were conducted using SPSS software, version 14.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of PCDH-γ subfamily genes is upregulated in nodal FL compared with RLH
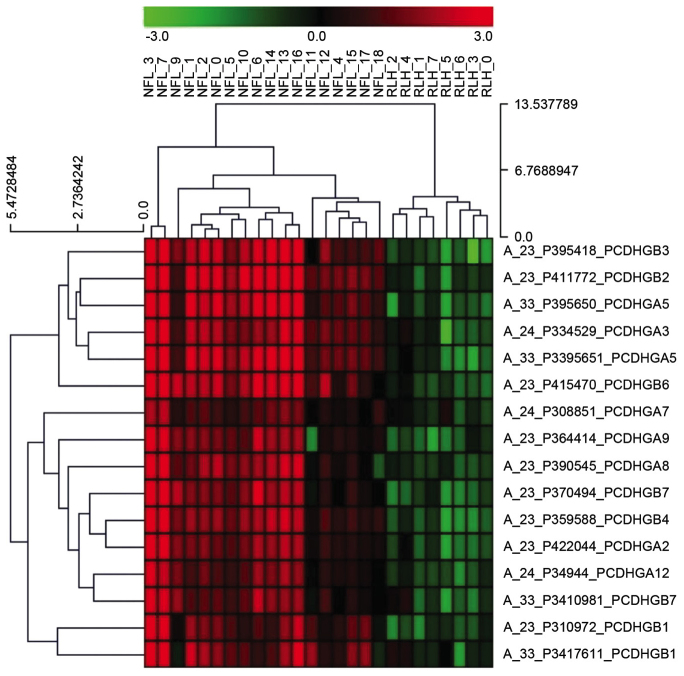
Of the differentially expressed genes in terms of gene ontology between nodal FL and RLH, the top five upregulated genes were PCDH-γ subfamily members B4, B3, B2, A5 and A3. A heatmap of the PCDH-γ subfamily genes is presented in Fig. 1. Therefore, PCDHGA3, as a representative gene, was selected for further analysis.
Figure 1.
Heatmap of PCDHG gene subfamilies in FL and RLH. The top five upregulated genes were PCDH-γ subfamily members B4, B3, B2, A5 and A3. Red indicates gene upregulation and green indicates gene downregulation. PCDHG, protocadherin γ; FL, follicular lymphoma; RLH, reactive lymphoid hyperplasia.
PCDHGA3 is strongly expressed in FL, including high-grade and BCL2-negative cases
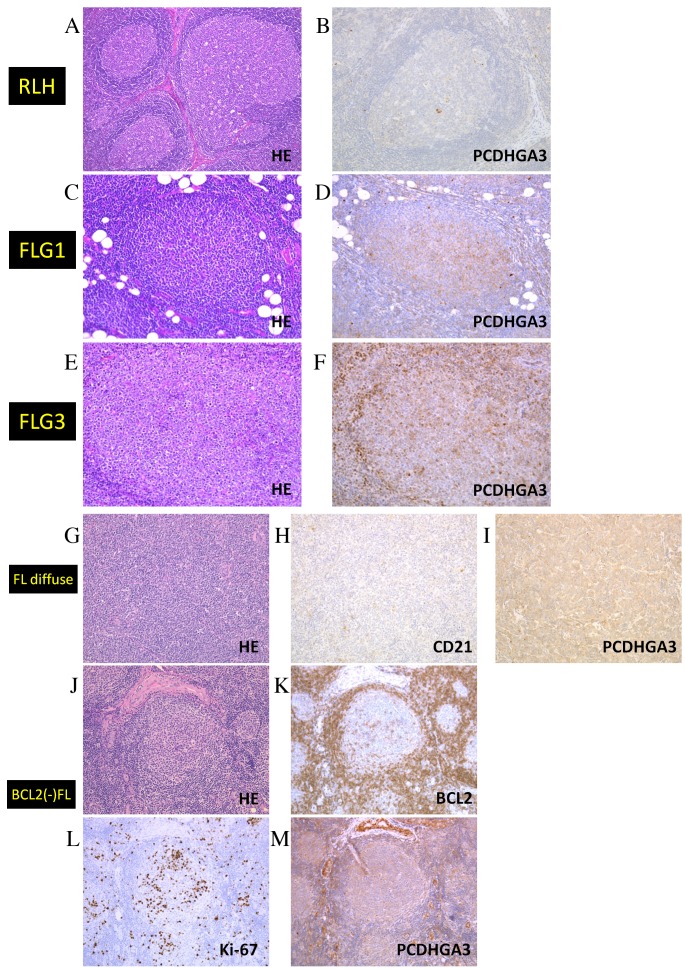
Results of the immunohistochemical analysis of RLH and FL samples are presented in Fig. 2 and Table I. Only 1/17 RLH samples was weakly PCDHGA3-positive (Fig. 2A and B), whereas 19/20 (95%) FL grade 1–2 samples were positive (Fig. 2C and D). In addition, approximately half of the FL grade 3A samples were positive (Fig. 2E and F). Notably, all 10 samples of FL with diffuse area were PCDHGA3-positive (Fig. 2G-I) and 13 of 15 (87%) BCL2-negative FL samples were PCDHGA3-positive (Fig. 2J-M). Ki-67 staining (a marker of proliferation) was performed to diagnose BCL2-negative neoplastic follicles. These results revealed that FL cells express PCDHGA3 irrespective of BCL2 status or grade.
Figure 2.
Immunohistochemical analysis of PCDHGA3 in FL samples. (A) HE staining of RLH. (B) PCDHGA3 was negative in RLH. (C) HE staining of an FL grade 1 sample, which was (D) positive for PCDHGA3. (E) HE staining of an FL grade 3 sample, which was (F) positive for PCDHGA3. (G) HE staining of an FL with diffuse area sample, which was (H) negative for CD21-expressing follicular dendritic cells and (I) positive for PCDHGA3. (J) HE staining of a BCL2-negative FL sample, which was (K) negative for BCL2, (L) positive for Ki-67 and (M) positive for PCDHGA3. All these images were representative of their groups. Original magnification A and G-M, ×100. Original magnification B-F, ×200. PCDHG, protocadherin γ; FL, follicular lymphoma; HE, hematoxylin and eosin; RLH, reactive lymphoid hyperplasia; CD, cluster of differentiation; BCL2, B-cell lymphoma 2.
Table I.
PCDHGA3 expression in RLH and FL samples.
| Histological subtype | No. studied | Positive (%) | P-value (vs. RLH) |
|---|---|---|---|
| RLH | 17 | 1 (5.9) | – |
| FL (Grades 1 and 2) | 20 | 19 (95) | <0.001 |
| FL (Grade 3) | 9 | 5 (55.6) | 0.028 |
| FL (Diffuse area) | 10 | 10 (100) | <0.001 |
| FL (BCL2 negative) | 15 | 13 (86.7) | <0.001 |
RLH, reactive lymphoid hyperplasia; FL, follicular lymphoma; BCL2, B-cell lymphoma 2.
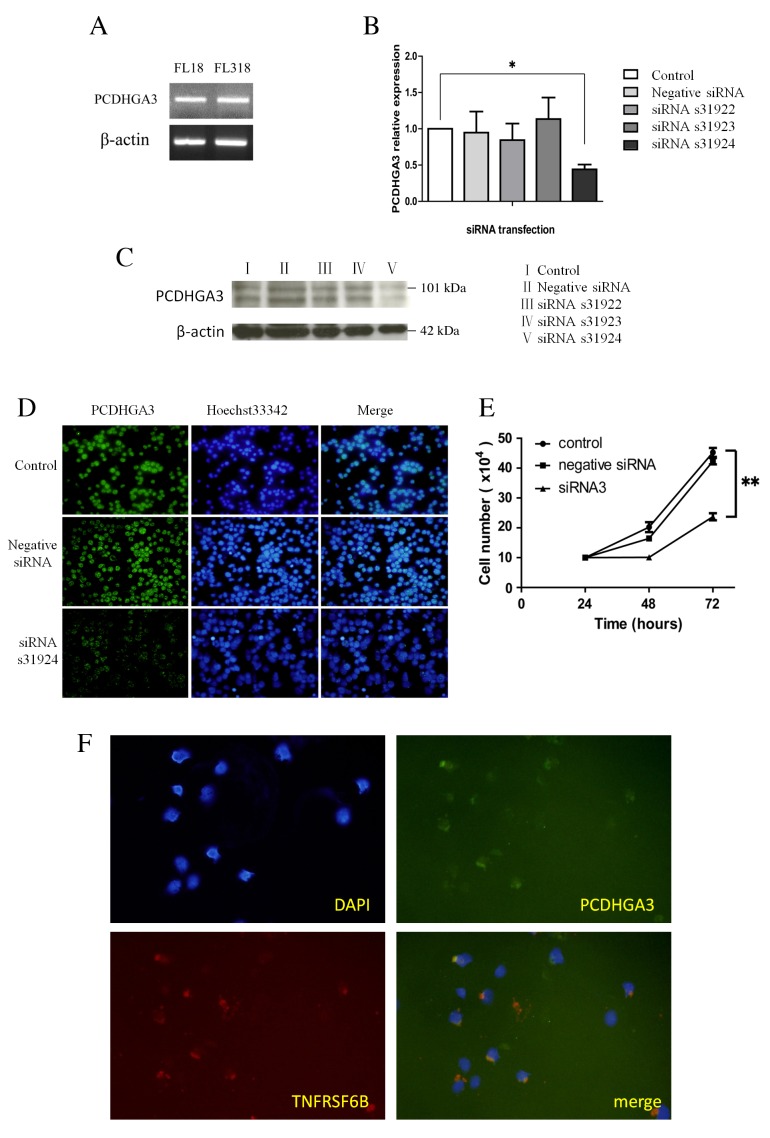
PCDHGA3 silencing reduces cell proliferation in the FL18 cell line
As a large proportion of nodal FL cases expressed PCDHGA3, PCDHGA3 expression status was examined in the FL cell lines, FL18 and FL318. The two cell lines expressed PCDHGA3 mRNA, as determined by RT-qPCR (Fig. 3A). PCDHGA3 gene knockdown with PCDHGA3 siRNAs was performed in the FL18 cell line. The silencing effects of three different PCDHGA3-specific siRNAs in FL18 cells were evaluated using RT-qPCR and western blotting. The s31924 siRNA was selected for further analysis as it yielded the greatest PCDHGA3 mRNA inhibition ratio (55.9%) relative to untreated FL18 cells (P=0.005 Fig. 3B and C). Following PCDHGA3 knockdown, proliferation curves were generated for untreated FL18 cells, and FL18 cells transfected with negative siRNA or PCDHGA3 s31924 siRNA. Proliferation was significantly suppressed in FL18 cells transfected with PCDHGA3 siRNA at 72 h compared with untreated FL18 cells. (P<0.001; Fig. 3D).
Figure 3.
PCDHGA3 gene knockdown. (A) PCDHGA3 mRNA was detected in the FL-derived cell lines, FL18 and FL318. Knockdown of the PCDHGA3 gene was confirmed by (B) reverse transcription-quantitative polymerase chain reaction, (C) western blotting and (D) immunofluorescence. The siRNA, s31924, was selected for further analysis because it yielded the greatest PCDHGA3 mRNA inhibition ratio (55.9%) relative to untreated FL18 cells, original magnification ×100. (E) Cell proliferation was significantly suppressed in PCDHGA3 siRNA-treated FL18 cells at 72 h. (F) Coexpression of PCDHGA3 and TNFRSF6B was observed in the FL18 cell line by double immunofluorescence staining, original magnification ×200. *P<0.01; **P<0.001. PCDHG, protocadherin γ; FL, follicular lymphoma; siRNA, small interfering RNA; TNFRSF6B, tumor necrosis factor receptor superfamily member 6B; DAPI, 4,6-diamidino-2-phenylindole.
Coexpression of TNFRSF6B and PCDHGA3 in the FL18 cell line
Genes expressed concomitantly with PCDHGA3 were selected from the gene expression data of nodal FL and RLH (Table II). TNFRSF6B was among the top five correlated genes. Furthermore, the protein encoded by this gene may be involved in regulating the suppression of Fas ligand-mediated cell death (13). Coexpression of TNFRSF6B and PCDHGA3 was observed in the FL18 cell line by double immunostaining (Fig. 3E).
Table II.
Top 10 genes correlated with PCDHGA3.
| Gene name | Gene symbol | Median value |
|---|---|---|
| Opiate receptor-like 1 | OPRL1 | 0.6731 |
| Tumor necrosis factor receptor superfamily, member 6b, decoy | TNFRSF6B | 0.6721 |
| ADAMTS-like 5 | ADAMTSL5 | 0.6635 |
| 1-Aminocyclopropane-1-carboxylate synthetase homolog | ACCSL | 0.6401 |
| Carbohydrate sulfotransferase 4 | CHST4 | 0.6375 |
| Transmembrane channel-like 6 | TMC6 | 0.6331 |
| Solute carrier family 16, member 12 | SLC16A12 | 0.6299 |
| MAM domain containing 4 | MAMDC4 | 0.6281 |
| PRKC, apoptosis, WT1, regulator | PAWR | 0.6271 |
| Cullin-associated and neddylation-dissociated 1 | CAND1 | 0.6288 |
Expressed genes correlating with PCDHGA3 expression were identified using GeneSpring software. Median value represents the correlation index between the gene and PCDHGA3. PCDHG, protocadherin γ.
Discussion
PCDHs, which are downregulated via epigenetic mechanisms in various tumor types, have been suggested as candidate tumor suppressor genes (6–9,14). It has been previously demonstrated that expression of PCDH-γ subfamily genes is upregulated in low-grade FL (3). In the present study, PCDHGA3 expression was investigated in various grades of FL using immunohistochemistry, and it was examined whether this gene product may contribute to cell proliferation in an FL-derived cell line. As an antibody against PCDHGA3 suitable for use in immunohistochemical studies was available, this gene and protein was selected for further analysis.
For the pathological diagnosis of FL, detection of aberrant BCL2 expression in neoplastic follicles is important and useful, as 80–90% of cases of low-grade FL harbor the IGH-BCL2 translocation. Although only 10–20% cases of low-grade FL lack BCL2 expression, cases of high-grade FL less commonly harbor this translocation and therefore less frequently express BCL2 (15). As it may be difficult to diagnose BCL2-negative FL, other diagnostic markers have been investigated. For example, Masir et al (16) reported that a rabbit monoclonal BCL2 antibody (E17) facilitated the diagnosis of BCL2-negative FLs, whereas Zamo et al (17) reported the lack of TUBB3 expression in BCL2-negative FLs. The present study hypothesized that PCDHGA3 is a useful diagnostic marker for all cases of FL, as PCDHGA3 expression was observed irrespective of BCL2 status. The positivity of PCDHGA3 in FL Grade 3 (55.6%) was lower than the rest of the listed FL group (86.7–100%) although it was still high enough to use as a diagnostic tool. This may be associated with the high-grade feature of the tumor cells, and should be investigated further in future studies.
Similarly high levels of PCDHGA3 expression were detected in FL18 cells and samples from FL cases, and FL18 cell proliferation was suppressed following PCDHGA3 gene knockdown, suggesting that PCDHGA3 is associated with FL tumor cell growth.
To investigate the mechanism underlying the effect of PCDHGA3 on tumor cell proliferation, gene expression profiles were reanalyzed to identify genes that were differentially expressed concomitantly with PCDHGA3. TNFRSF6B was among the top five coexpressed genes. TNFRSF6B overexpression has been reported in various types of malignant tumors (18–20) and its protein product inhibits Fas ligand-mediated cell death. With regards to lymphoma, a study by Bedewy et al (21) revealed that TNFRSF6B expression was associated with shorter event-free survival in an aggressive subtype of B-cell lymphoma. In the present study, coexpression of TNFRSF6B and PCDHGA3 proteins was observed in the FL18 cell line. The association between these genes remains to be elucidated, although the proteins may interact with each other. Further investigation is required to clarify this.
In conclusion, the results of the present study revealed that PCDHGA3 was expressed in FL, irrespective of the BCL2 status and tumor grade, and was demonstrated to be associated with cell proliferation. Further studies involving molecular genetic analysis are required to clarify the mechanisms underlying the effect of PCDHGA3 in FL.
Acknowledgements
Dr Xueyan Zhang was supported by a grant from the China Scholarship Council.
References
- 1.NL SS Harris, Jaffe ES, Ott G, Nathwani BN, et al. World Health Organization Classification of Tumours, WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th. International Agency for Research on Cancer (IARC); Lyon: 2008. pp. 220–226. [Google Scholar]
- 2.Lai R, Arber DA, Chang KL, Wilson CS, Weiss LM. Frequency of bcl-2 expression in non-Hodgkin's lymphoma: A study of 778 cases with comparison of marginal zone lymphoma and monocytoid B-cell hyperplasia. Mod Pathol. 1998;11:864–869. [PubMed] [Google Scholar]
- 3.Takata K, Tanino M, Ennishi D, Tari A, Sato Y, Okada H, Maeda Y, Goto N, Araki H, Harada M, et al. Duodenal follicular lymphoma: Comprehensive gene expression analysis with insights into pathogenesis. Cancer Sci. 2014;1015:608–615. doi: 10.1111/cas.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morishita H, Yagi T. Protocadherin family: Diversity, structure, and function. Curr Opin Cell Biol. 2007;19:584–592. doi: 10.1016/j.ceb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Chen WV, Alvarez FJ, Lefebvre JL, Friedman B, Nwakeze C, Geiman E, Smith C, Thu CA, Tapia JC, Tasic B, et al. Functional significance of isoform diversification in the protocadherin gamma gene cluster. Neuron. 2012;75:402–409. doi: 10.1016/j.neuron.2012.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu J, Koujak S, Nagase S, Li CM, Su T, Wang X, Keniry M, Memeo L, Rojtman A, Mansukhani M, et al. PCDH8, the human homolog of PAPC, is a candidate tumor suppressor of breast cancer. Oncogene. 2008;27:4657–4665. doi: 10.1038/onc.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Tayrac M, Etcheverry A, Aubry M, Saïkali S, Hamlat A, Quillien V, Le Treut A, Galibert MD, Mosser J. Integrative genome-wide analysis reveals a robust genomic glioblastoma signature associated with copy number driving changes in gene expression. Genes Chromosomes and Cancer. 2009;48:55–68. doi: 10.1002/gcc.20618. [DOI] [PubMed] [Google Scholar]
- 8.Haruki S, Imoto I, Kozaki K, Matsui T, Kawachi H, Komatsu S, Muramatsu T, Shimada Y, Kawano T, Inazawa J. Frequent silencing of protocadherin 17, a candidate tumour suppressor for esophageal squamous-cell carcinoma. Carcinogenesis. 2010;31:1027–1036. doi: 10.1093/carcin/bgq053. [DOI] [PubMed] [Google Scholar]
- 9.Ying J, Gao Z, Li H, Srivastava G, Murray PG, Goh HK, Lim CY, Wang Y, Marafioti T, Mason DY, et al. Frequent epigenetic silencing of protocadherin 10 by methylation in multiple haematologic malignancies. Br J Haematol. 2007;136:829–832. doi: 10.1111/j.1365-2141.2007.06512.x. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi E, Takata K, Sato Y, Tashiro Y, Tachiyama Y, Sawada-Kitamura S, Hiramatsu Y, Sugiguchi S, Nose S, Hirokawa M, et al. Distinct morphologic, phenotypic, and clinical-course characteristics of indolent peripheral T-cell lymphoma. Hum Pathol. 2013;44:1927–1936. doi: 10.1016/j.humpath.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Takata K, Sato Y, Nakamura N, Tokunaka M, Miki Y, Kikuti Y Yukie, Igarashi K, Ito E, Harigae H, Kato S, et al. Duodenal follicular lymphoma lacks AID but expresses BACH2 and has memory B-cell characteristics. Mod Pathol. 2013;26:22–31. doi: 10.1038/modpathol.2012.127. [DOI] [PubMed] [Google Scholar]
- 13.Curtin JF, Cotter TG. Live and let die: Regulatory mechanisms in Fas-mediated apoptosis. Cell Signal. 2003;15:983–992. doi: 10.1016/S0898-6568(03)00093-7. [DOI] [PubMed] [Google Scholar]
- 14.Zheng L, Sharma R, Gaskin F, Fu SM, Ju ST. A novel role of IL-2 in organ-specific autoimmune inflammation beyond regulatory T cell checkpoint: Both IL-2 knockout and Fas mutation prolong lifespan of Scurfy mice but by different mechanisms. J Immunol. 2007;179:8035–8041. doi: 10.4049/jimmunol.179.12.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leich E, Salaverria I, Bea S, Zettl A, Wright G, Moreno V, Gascoyne RD, Chan WC, Braziel RM, Rimsza LM, et al. Follicular lymphomas with and without translocation t(14;18) differ in gene expression profiles and genetic alterations. Blood. 2009;114:826–834. doi: 10.1182/blood-2009-01-198580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masir N, Campbell LJ, Goff LK, Jones M, Marafioti T, Cordell J, Clear AJ, Lister TA, Mason DY, Lee AM. BCL2 protein expression in follicular lymphomas with t(14;18) chromosomal translocations. Br J Haematol. 2009;144:716–725. doi: 10.1111/j.1365-2141.2008.07528.x. [DOI] [PubMed] [Google Scholar]
- 17.Zamo A, Erdini F, Malerba G, Chilosi M. Lack of expression of TUBB3 characterizes both BCL2-positive and BCL2-negative follicular lymphoma. Mod Pathol. 2014;27:808–813. doi: 10.1038/modpathol.2013.182. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y, Guo E, Yu J, Xie Q, Chen J. High DcR3 expression predicts stage pN2 in gastric cancer. Hepatogastroenterology. 2007;54:2172–2176. [PubMed] [Google Scholar]
- 19.Connor JP, Felder M. Ascites from epithelial ovarian cancer contain high levels of functional decoy receptor 3 (DcR3) and is associated with platinum resistance. Gynecol Oncol. 2008;111:330–335. doi: 10.1016/j.ygyno.2008.07.089. [DOI] [PubMed] [Google Scholar]
- 20.Weissinger D, Tagscherer KE, Macher-Göppinger S, Haferkamp A, Wagener N, Roth W. The soluble Decoy Receptor 3 is regulated by a PI3K-dependent mechanism and promotes migration and invasion in renal cell carcinoma. Mol Cancer. 2013;12:120. doi: 10.1186/1476-4598-12-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bedewy AM, Elgammal MM, Bedewy MM, El-Maghraby SM. Assessing DcR3 expression in relation to survivin and other prognostic factors in B cell non-Hodgkin's lymphoma. Ann Hematol. 2013;92:1359–1367. doi: 10.1007/s00277-013-1775-4. [DOI] [PubMed] [Google Scholar]