Abstract
Scientific models are abstractions that aim to explain natural phenomena. A successful model shows how a complex phenomenon arises from relatively simple principles while preserving major physical or biological rules and predicting novel experiments. A model should not be a facsimile of reality; it is an aid for understanding it. Contrary to this basic premise, with the 21st century has come a surge in computational efforts to model biological processes in great detail. Here we discuss the oxymoronic, realistic modeling of single neurons. This rapidly advancing field is driven by the discovery that some neurons don't merely sum their inputs and fire if the sum exceeds some threshold. Thus researchers have asked what are the computational abilities of single neurons and attempted to give answers using realistic models. We briefly review the state of the art of compartmental modeling highlighting recent progress and intrinsic flaws. We then attempt to address two fundamental questions. Practically, can we realistically model single neurons? Philosophically, should we realistically model single neurons? We use layer 5 neocortical pyramidal neurons as a test case to examine these issues. We subject three publically available models of layer 5 pyramidal neurons to three simple computational challenges. Based on their performance and a partial survey of published models, we conclude that current compartmental models are ad hoc, unrealistic models functioning poorly once they are stretched beyond the specific problems for which they were designed. We then attempt to plot possible paths for generating realistic single neuron models.
Keywords: dendrites, compartmental model, cable theory, ion channel, channel kinetics
“I come to bury Caesar, not to praise him.” [Mark Antony, in Shakespeare's Julius Caesar (1599)]
the brain computes, ergo, solving the puzzle of the brain requires a computer. This is one credo of modern computational neuroscience. A computer, however, is a golem that requires a human-written list of instructions, i.e., a program, to compute. Thus, day in and day out, computational neuroscientists write computer code. We vainly call this inglorious activity modeling. More formally perhaps, the ultimate goal of computational neuroscience, as expressed by Sejnowski et al. (1988) is “…to explain how electrical and chemical signals are used in the brain to represent and process information.” Modeling can be abstract, describing a neuron as a simple summation device (Abeles 1991; Gerstein and Mandelbrot 1964; Izhikevich 2007; Knight 1972). The allure of this methodology is the ability to use simple tractable rules to study neuronal computation. By contrast, other models, named, or perhaps misnamed, “realistic,” generate complex outputs, rich with biological detail. The basic assumption behind these realistic models is that expressing the biological details of the single neuron in silico creates a virtual double of the neuron of which its computational properties will emerge like Athena jumping out of Zeus's head. Recently, these biologically detailed models have been applied to simulate the activity of large neuronal populations (Markram 2006, 2012; Markram et al. 2015).
Between integrate and fire models and biologically detailed ones, there is a spectrum of neuronal modeling techniques (Herz et al. 2006). Here we remain agnostic regarding most modeling dogmas and take a critical look at the current state of biologically detailed or “realistic” neuronal modeling. We ask: have computers and biology ripened to the extent that we can build full, biologically detailed neural models or not? This question has been gathering importance since studies of voltage-gated channels, dendritic synaptic integration, axonal properties, and synaptic plasticity have shown that single neurons perform computations more complex than integrate-and-fire. This observation leads to a simple yet unanswered question: if single neurons do not just integrate and fire, what computation do they do (Koch 1997; Koch and Segev 2000; Silver 2010)?
We note, that at the heart of realistic neuronal modeling dwells the assumption that computations performed by a real neuron are more complex than those described by a point neuron. It is also assumed that this complex computation affects the computation of the network. The former assumption is supported by many studies demonstrating the complex nature of postsynaptic integration (Koch 1999; London and Hausser 2005; Major et al. 2013; Migliore and Shepherd 2002; Poirazi et al. 2003b; Polsky et al. 2004), the latter one less so, although some network simulations using complex neurons predict complex network activity (Kording and Konig 2000, 2001a, 2001b; Markram et al. 2015). Nevertheless, accepting these assumptions forces us to acknowledge that single neuron computation is complex.
One established strategy for approaching this question is to use abstract or partial models of an individual cellular function. This approach attempts to distill, one by one, a set of cellular computational properties. This approach has advanced rapidly in great part due to exposure to larger and larger data sets and more powerful computing resources. Indeed, progress is amazing. In less than two decades, we have gone from endless hours running crude simulations on 486 processors to automatically optimizing models using supercomputers, from modeling networks of just a few hundred virtual neurons to networks of billions of units, and from modeling single biochemical reactions to simulating entire cytoplasmic networks. Researchers now have the luxury of performing very large and complex simulations (Ananthanarayanan and Modha 2007; Eliasmith et al. 2012; Eliasmith and Trujillo 2014; Izhikevich and Edelman 2008; Markram 2006, 2012; Markram et al. 2015). Likewise, the availability of computing power and wealth of biological data themselves suggest biologically detailed modeling. And mightn't we hope that by better describing the underlying biology, we will also better approximate the functionality of these systems, in particular, their computations? We must take heed and remember that, as the adage runs, with great power comes great responsibility. Accordingly, some scientists have asked, given such mastodonic resources, how may they best be used for understanding brain function (Eliasmith and Trujillo 2014). Here we ask a more basic question, not how the gargantuan computer power and data sets delivered into our hands can best be deployed, but rather can we use these resources to understand the brain, or are such resources by themselves insufficient and we are unprepared to wield this power? While it is impossible to cover the entire field of computational neuroscience in one review, we can gain insight into this general question by focusing on recent attempts at “realistic” compartmental models for dendritic and axonal integration in individual, neocortical, pyramidal neurons in layer 5.
Complex Single-Neuron Computation
Complex single-neuron computation is best exhibited in two areas, and it is in them that we will focus our review of computational studies: dendritic synaptic integration (Branco et al. 2010; Branco and Hausser 2010; Cash and Yuste 1998, 1999; Gasparini et al. 2004; Gulledge et al. 2005; Hausser et al. 2000; Larkum et al. 2009; Losonczy and Magee 2006; Makara and Magee 2013; Mel 1993; Nettleton and Spain 2000; Oviedo and Reyes 2012; Polsky et al. 2004; Remy et al. 2009; Stuart et al. 1999; Yuste 2011) and axonal excitation (Bender and Trussell 2012; Clark et al. 2005; Foust et al. 2010; Hu et al. 2009; Khaliq and Raman 2006; Martina et al. 2000; Meeks and Mennerick 2007; Palmer et al. 2010; Palmer and Stuart 2006; Popovic et al. 2011; Stuart et al. 1997a; Stuart and Palmer 2006).
The properties and functions of dendrites have been extensively studied over the past two decades (for reviews, see Johnston 1999; Johnston et al. 2003; Kastellakis et al. 2015; London and Hausser 2005; Magee and Johnston 2005; Major et al. 2013; Migliore and Shepherd 2002; Ramaswamy and Markram 2015; Stuart et al. 1999), mainly due to the success of patch-clamp recording from visually identifiable dendrites in brain slices (Stuart et al. 1993) and of new imaging techniques (Antic 2003; Antic et al. 1999; Denk et al. 1994; Lasser-Ross et al. 1991; Tsien 1989). Conflicting with the early assumption that dendrites were passive, it has been shown that action potentials initiated at the axon either actively back-propagate into the dendritic tree (Bischofberger and Jonas 1997; Chen et al. 1997, 2002; Hausser et al. 1995; Spruston et al. 1995; Stuart et al. 1997a; Stuart and Sakmann 1994), or else fail to propagate effectively into the dendritic tree, for example, in cerebellar Purkinje cells, where the amplitude of the action potential decreased to just a few millivolts at the proximal dendrite (Llinas and Sugimori 1980; Stuart and Hausser 1994; Vetter et al. 2001). Furthermore, dendrites generate complex regenerative calcium and sodium spikes (Amitai et al. 1993; Antic 2003; Ariav et al. 2003; Bischofberger and Jonas 1997; Chen et al. 1997; Golding and Spruston 1998; Johnston et al. 1996, 2003; Kamondi et al. 1998; Korogod et al. 1996; Llinas and Sugimori 1980; Magee et al. 1998; Martina et al. 2000; Migliore and Shepherd 2002; Pouille et al. 2000; Schiller et al. 1997; Schwindt and Crill 1998; Stuart et al. 1997b; Zhu 2000), modulate synaptic potentials (Magee 1999; Magee and Johnston 1995), contain electrically- and chemically-defined compartments (Bekkers 2000a; Gasparini and Magee 2006; Hoffman et al. 1997; Korngreen and Sakmann 2000; Larkum et al. 1999b, 2001; Losonczy and Magee 2006; Magee 1999; Schiller et al. 1997, 2000), and influence the induction and expression of synaptic plasticity (Golding et al. 2002; Kim et al. 2012; Lavzin et al. 2012; Losonczy and Magee 2006; Losonczy et al. 2008; Magee and Johnston 1997; Makara and Magee 2013; Markram et al. 1997; Nevian et al. 2007; Polsky et al. 2004; Smith et al. 2013). Moreover, recent in vivo studies demonstrated complex dendritic computations (Grienberger et al. 2014, 2015; Palmer et al. 2014; Smith et al. 2013). These results revitalized the discussion of the computational properties of the single neuron (Hausser and Mel 2003; Koch 1999; London and Hausser 2005; Major et al. 2013; Migliore and Shepherd 2002; Oviedo and Reyes 2012; Poirazi et al. 2003b; Polsky et al. 2004; Silver 2010).
The neuron's computational capabilities depend on various parameters, such as its morphology (dendritic/axonal arborization, length, diameter, membrane area) and the distribution of ion channels throughout its membrane (Hausser et al. 2000; Kim and Connors 1993; Mainen and Sejnowski 1996; Roberts et al. 2009; Schaefer et al. 2003b; Serodio and Rudy 1998; Talley et al. 1999; van Elburg and van Ooyen 2010; Vetter et al. 2001; Weaver and Wearne 2008). The neuron's ability to couple inputs originating in different compartments also depends on the specific features of each compartment, and its threshold for attenuating postsynaptic potentials as defined by its cable properties, together with the different active properties of the dendrites (London and Hausser 2005; Rall et al. 1995). Pyramidal neurons in layer 5 couple and decouple proximal and distal areas through morphology of proximal oblique dendrites (Schaefer et al. 2003b). Coupling lowers the threshold for initiating dendritic calcium spikes and leads to axonal burst firing (Helmchen et al. 1999; Larkum et al. 1999a, 1999b; Williams and Stuart 1999).
Computational properties, however, are only rarely defined in a simple manner by physical properties. The efficacy of back-propagating action potentials, for example, is lower when dendrites have complex structure, but this can be compensated with an increase in the density of voltage-gated channels (Vetter et al. 2001). The relation between action potential propagation and dendritic complexity has the implication that distal synapses and axonal output are less coupled in neurons with highly branching geometries. Yet, once again, this can be compensated (Baer and Rinzel 1991; Jaslove 1992) by changes in spine density during development (Gould et al. 1990; Harris et al. 1992) and by synaptic plasticity (Engert and Bonhoeffer 1999; Maletic-Savatic et al. 1999). The computational significance of dendritic spines is underscored by the fact that dopaminergic neurons and various interneurons, which have minimal dendritic branching, tend to be aspiny, whereas Purkinje neurons have both highly branching dendrites and high spine density (Vetter et al. 2001).
The biophysics of dendrites too can nonlinearly modulate integration of synaptic potentials on one hand (Cash and Yuste 1998, 1999; Yuste 2011) and on another support local electrogenesis of dendritic spikes, leading to a nonlinear integration of synaptic potentials (Gasparini et al. 2004; Gulledge et al. 2005; Hausser et al. 2000; Losonczy and Magee 2006; Makara and Magee 2013; Mel 1993; Nettleton and Spain 2000; Polsky et al. 2004) and allowing the neuron to respond over a longer timescale. The nonlinear integration of synaptic inputs occurs either in response to functional or anatomical synapse clusters (Druckmann et al. 2014; Losonczy and Magee 2006; Makino and Malinow 2011; McBride et al. 2008; Poirazi et al. 2003b; Polsky et al. 2004; Yadav et al. 2012). In the case of functional clustering, synchronous stimulation of nearby synapses within the same branch results in a supralinear summation, while stimulation of the same number of synapses located between branches sum in a linear way (Branco et al. 2010; Branco and Hausser 2010; Cash and Yuste 1998; Hausser and Mel 2003; Larkum et al. 2009; Losonczy and Magee 2006; Mel 1993; Oviedo and Reyes 2012; Poirazi et al. 2003a; Polsky et al. 2004), suggesting that a single dendritic branch acts as a computational compartment of the neuron.
Furthermore, supralinear synaptic integration at distal dendritic branches can lower the driving force for activating voltage-gated ion channels (Hausser and Mel 2003; Magee 2000; Mel 1993; Spruston 2008; Williams and Stuart 2003). Since the driving force allows the dendrites to regenerate their spikes and propagate them forward to the axonal action potential initiation zone, its modulation serves as an additional mechanism for nonlinear dendritic computation. In layer 5 pyramidal neurons, synaptic conductance is highly compartmentalized, and the dendritic tree performs axo-somatic and dendritic integration in high-conductance conditions (Williams 2004, 2005). It has been suggested that the axon initial segment plays a significant role in neuronal processing through a mechanism of activity-dependent plasticity, where the neuron can modulate its excitation by changing the location of subcellular sections along the axon initial segment. This mechanism shifts the input-output function of the neuron according to the level of excitation (Burrone and Murthy 2003; Grubb and Burrone; Marder and Prinz 2002; Turrigiano and Nelson 2000). Combining axonal plasticity together with dendritic synaptic plasticity can maximize the neuron computational process and stabilize adaptation. A recent study has shown that not only does the axon initial segment influence layer 5 pyramidal neuron excitability, but also the distal axon plays a role in facilitating high-frequency burst firing (Kole 2011).
New morphological data too suggest that the traditional neuron computation model is oversimplified. Axons can emerge from dendrites in neuroendocrine cells (Herde et al. 2013), dopaminergeic neurons (Hausser et al. 1995), interneurons (Martina et al. 2000), hippocampal and cortical pyramidal neurons (Lorincz and Nusser 2010; Peters et al. 1968; Sloper and Powell 1979; Thome et al. 2014), and Thome et al. (2014) showed that this anatomical difference in CA1 pyramidal neurons causes different functional responses in neurons with axon-carrying dendrites. Dendritic spikes are more common in axon-carrying dendrites rather than in nonaxon-carrying dendrites. This enhances supralinear integration of synaptic inputs and tightly couples synchronous timing of synaptic inputs to action potential output (Ariav et al. 2003; Losonczy and Magee 2006; Losonczy et al. 2008; Thome et al. 2014). So neuronal morphology affects neuronal computation and broadens our view of how the outputs of single neurons are related to their inputs.
In the Beginning There Was the Hodgkin and Huxley Model
Neuronal modeling, as well as many other subdisciplines within computational neuroscience, grew directly out of Hodgkin and Huxley's seminal investigation of the action potential in the squid giant axon (Hodgkin and Huxley 1952d). One could even make a case that this study culminated with the first compartmental model. Using the newly invented voltage-clamp technique, Hodgkin and Huxley recorded ionic currents from the squid giant axon and, using clever ionic substitution and novel voltage-clamp experiments, isolated two voltage-gated conductances (Hodgkin and Huxley 1952a, 1952b, 1952c; Hodgkin et al. 1952). They then performed two stages of data reduction. First, they used standard chemical kinetics to analyze the voltage-dependence of the sodium and potassium conductances, thereby obtaining the rate constants for opening, closing, and inactivation of the conductances. Second, they reduced these rate constants to a small set of algebraic and differential equations describing the time and voltage dependence of the channels. This model explained and predicted the basis of the action potential in the squid giant axon (Hodgkin and Huxley 1952d). The entire model was derived from experimental results and contains almost no parameters of unknown value (the values for the maximal conductances and number of gates being manually adjusted by Hodgkin and Huxley). Any aspiring neuroscientist in general, and computational neuroscientists in particular, should find the time to read Hodgkin and Huxley's 1952 papers in the Journal of Physiology.
As a first step in our discussion, we reproduce Hodgkin and Huxley's model. We do so, however odd, to highlight several aspects of the model that are relevant to our discussion. Figure 1 presents the parallel conductance model of neural excitation. This graphical representation can be formally expressed using the following equations (we deliberately present the equations in the form given by Hodgkin and Huxley to calculate the propagating action potential, since they are the most relevant to the modern form of compartmental modeling):
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
Fig. 1.
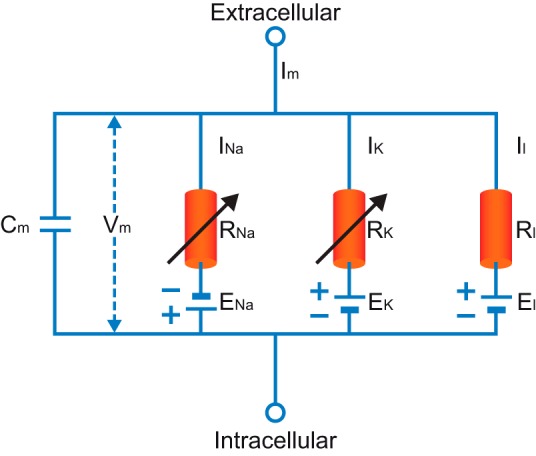
The parallel conductance circuit model of the squid giant axon. Im, membrane current; Vm, membrane potential; INa, sodium current; RNa, membrane resistance to sodium; IK, potassium current; RK, membrane resistance to potassium; Il, leak current; Rl, nonspecific membrane resistance.
where a is diameter of the axon; Ri is cytoplasmic resistance; Cm is membrane capacitance; ḡk is maximal potassium conductance density; n is potassium activation gate; V is membrane potential, Ek is potassium Nernst potential; ḡNa is maximal sodium conductance density; Ena is sodium Nernst potential; m is sodium activation gate; h is sodium inactivation gate; ḡl is maximal leak conductance density; El is leak Nernst potential; αn is forward rate constant for potassium activation; βn is backward rate constant for potassium activation; αm is forward rate constant for sodium activation; βm is backward rate constant for sodium activation; αh is forward rate constant for sodium inactivation; and βh is backward rate constant for sodium inactivation.
Two aspects of the model must be highlighted. First, it is remarkable that these 7 equations contain 26 parameters. Three are passive parameters Ri, Cm, and gl (today we use the membrane resistance Rm, which relates to 1/gl); one is a structural parameter (a), and three reversal potentials. The other 19 parameters define the kinetic properties of the voltage-gated sodium and potassium conductances. All of the parameters were derived from experiments on the squid giant axon. We call attention to the surprisingly high number of parameters at this early stage of the discussion to give us a point of reference for comparison below with today's compartmental models. Second, we must note, the Hodgkin and Huxley's model is a purely electrical one. It makes no reference to biochemistry, changes to cytoplasmic ionic concentrations, or other intracellular processes. Even today, compartmental models remain electro-centric, a legacy partly responsible for the lamentable lack of common language between computational neuroscience and chemo-centric systems biology (De Schutter 2008).
Then Came Compartmental Modeling
The Hodgkin and Huxley model describes the physiology of one cylindrical structure. Wilfred Rall blazed a new path in modeling quantifying the laws of electrical current flow in neurons with arbitrary structure (Rall et al. 1995). In a series of classic papers during the 1950s, he provided deep mechanistic insight into the cable properties of neurons with complex dendritic trees (Rall 1957, 1959, 1960). Then in 1964 he described a numerical approach for solving cable equations and demonstrated that juxtaposed synapses summate sublinearly (Rall 1964). This made him the first to use a computer for simulating neuronal physiology and among the first who applied computers to problems in biology. The numerical technique developed by Rall in his 1964 paper is the foundation of modern compartmental modeling. Of the multitude of contributions made by Rall, this work, in our opinion, truly distinguishes him as a founding father of computational neuroscience.
The numerical method presented by Rall takes its kernel from the analysis of compartmentalized chemical reactions. Reactants enter a first compartment (or subvolume of space), their products are funneled to the next compartment, and so forth. The reaction in each compartment is described with a discrete set of differential equations, while the flow of material from compartment to compartment, and reaction to reaction, can be expressed as mass flux. Within each compartment it is assumed that the activity is homogenous. This makes it possible to formalize it as an ordinary first order differential equation. Spatial changes are expressed only as differences between compartments.
Rall adopted this formalism to describe neuronal electrical activity in a distributed neuronal morphology (Rall 1964). The neuronal tree is segmented into cylindrical compartments. The length of each cylindrical segment is set so that the axial changes in potential are small enough to comply, within acceptable error, with the assumption of compartment homogeneity. Segmenting in this way often results in a series of hundreds, and even thousands, of compartments describing a single neuronal morphology. Since each compartment is defined by its own set of differential equations, the number of equations scales linearly with the number of compartments. The dynamics of membrane currents in each segment are described, assuming the parallel conductance model with ordinary differential equations. Segments are connected by resistors through which axial current flows. We call this numerical modeling scheme compartmental modeling, commemorating its chemical origins; it is one of the most widely applied techniques in computational neuroscience today.
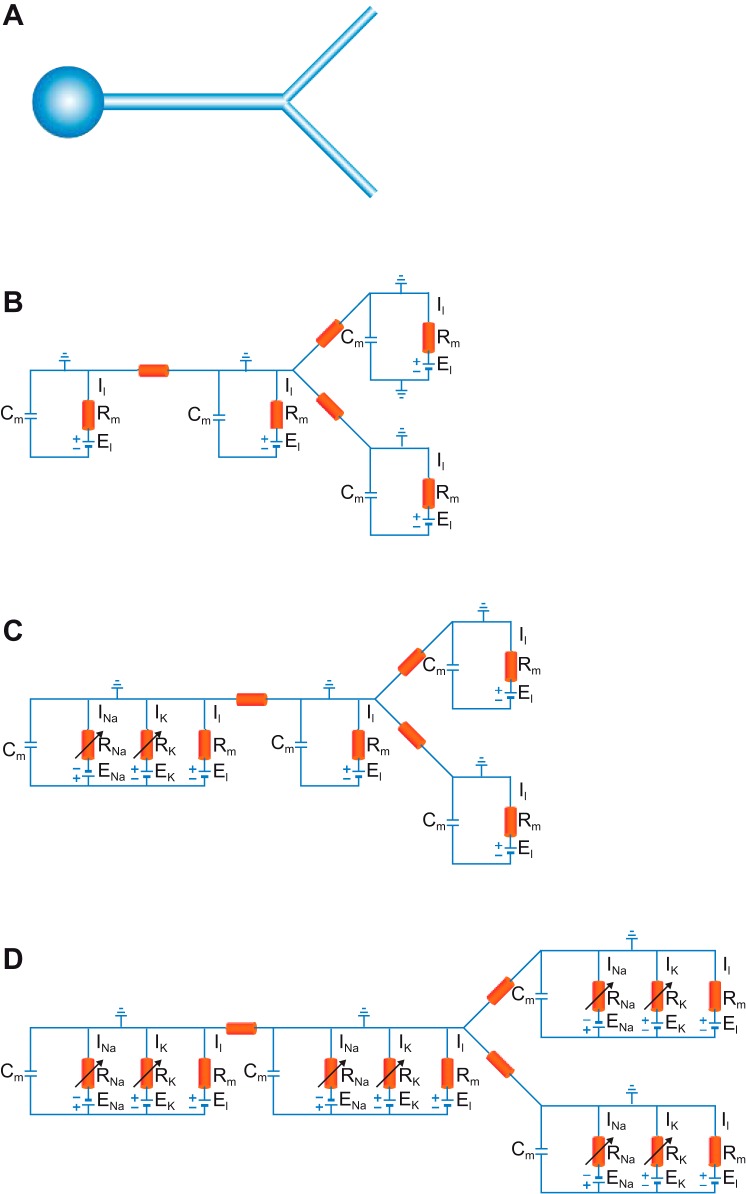
We noted above that the Hodgkin-Huxley model, which is by definition a single-compartment model, has 26 parameters. We now estimate the number of parameters in a simple compartmental model of a small neuron with one somatically connected dendrite bifurcating into two branches (Fig. 2A). Let the structure be represented by four compartments. The first functional model we impose on the structure is the passive one (Fig. 2B). Each compartment is represented as an equivalent membrane circuit of a capacitor connected in parallel to a resistor and the passive driving force of the membrane, and so it is endowed with three parameters. The entire neuron, then, is represented with four ordinary differential equations. With 4 compartments and 3 connections among them, the total number of parameters is 15. It is standard to normalize the passive parameters to the diameter of the compartment (Koch 1999) and to assume that these normalized parameters have the same values throughout the dendritic tree. This reduces the number of parameters which require tuning to just four (Cm, Rm, Ri, El). The actual number of parameters, however, scales with the number of compartments.
Fig. 2.
A simple compartmental model. A: a schematic drawing of a neuron with a soma and one bifurcating dendrite. B: an equivalent circuit of the same neuron assuming only passive membrane parameters. C: an equivalent circuit of the same neuron assuming passive dendrites and an active soma. D: an equivalent circuit of the same neuron assuming that both soma and dendrites are excitable. Rm, membrane resistance.
Consider now a second model (Fig. 2C) for this structure. It has the same morphological parameters but an excitable soma represented with the Hodgkin-Huxley parallel conductance model. Since the Hodgkin-Huxley model has 26 parameters, this small compartmental neuronal model with the Hodgkin-Huxley model embedded inside it has 38 parameters. As in the above example, we can make several assumptions to reduce the set of parameters. We assume that the passive properties are homogenous throughout the somato-dendritic tree. We also assume that the kinetics of the Hodgkin-Huxley model do not require any tuning, leaving us only with the maximal conductances of the sodium and potassium channels to tune. These assumptions allow us to define a set of just six parameters (GNa,soma, GK,soma, Cm, Rm, Ri, El) to make this model of a neuron with active soma and passive dendrites function reasonably well. Now, to model the dendrites as active, we embed the Hodgkin-Huxley model into each compartment of the structure (Fig. 2D). The total number of parameters thus rises to 107! With similar assumptions to those used above, we can reduce the number of those parameters requiring human tuning to ∼11. Compared with the Hodgkin-Huxley model (26 parameters), it may appear that we have achieved parameter reduction. This is not so. Similar reduction of the Hodgkin-Huxley model leaves us with just two parameters (GNa,soma, GK,soma). Thus we experience parameter inflation in both the full and reduced model.
Parameter inflation, we contend, is the Achilles' heel of compartmental modeling. While even in the simple structure we considered and even with the simplifying assumptions we made, the number of parameters rapidly inflated. Today, most compartmental models are constructed for complex cellular morphologies. They contain large numbers of voltage-gated channels, and their ion channels are not distributed along the neuronal membrane homogeneously. As a result, they contain many parameters of unknown value. And due to the limitations of modern recording techniques, we do not have experimental access to thin dendritic and axonal branches, so we are ignorant of the properties and distributions of many ion channels in the systems we are modeling. Therefore, much of the time spent making a compartmental model is dedicated to guessing parameter values, testing the model, guessing again, testing again, a cycle of hand-tuning that can loop for a depressingly long time. It should be emphasized that most of the reductions and assumptions that are commonly being made in constructing a compartmental model are due to a lack of available experimental measurements of specific parameters, rather than ignorance of such data on the modelers' part. Anyone who has hand-tuned a compartmental model knows that after many tuning cycles one loses the mental capacity to improve the model: every change made, it comes to seem, is bad. This outcome, almost inevitable, is a direct consequence of the rapid parameter inflation. Malaise accompanies many scientific and engineering problems whose character is defined by their large numbers of parameters. It is a malaise which emerges from an evil curse, the curse of dimensionality.
Multiparameter Models and the Curse of Dimensionality
“What casts the pall over our victory celebration? It is the curse of dimensionality, a malediction that has plagued the scientist from earliest days.” (Bellman 1961)
Richard Bellman coined the phrase “curse of dimensionality” to describe how adding dimensions to a mathematical space, practically building a multidimensional hypercube, exponentially increasing the volume of that space (Bellman 1957). In our discussion, the curse is that, as the number of model parameters increases, the time required for efficiently searching parameter space increases exponentially. Furthermore, when dimensionality increases, the volume of space containing a given number of data points increases so rapidly, the samples become sparse; as a result, the number of samples needed to reach statistical significance also grows exponentially. Moreover, while it is often important to find correlated parameters while analyzing parameter space, in a high-dimensional space all parameters pairings appear sparse, and this drastically hampers standard parameter organization schemes (Bellman 1957).
To put this in perspective, consider a simple model with 10 independent parameters of unknown value each with just 10 possible values. The number of parameter combinations to sift through is 1010. If each combination can be tested in just 1 s, a single-core processor still requires over 300 years to test each point in the space. And unfortunately, due to sparseness, local gradient decent methods are not efficient. Note that in nature one rarely finds completely independent parameters due to large parameter dependence. This reduces solution space considerably. Yet many compartmental models have parameter spaces vastly larger, with close to 20 parameters of unknown value each changing over a wide physiological range. Even conservative estimates of the possible permutations reach astronomical numbers. Moreover, it is highly likely that reducing computing time by randomly sampling such a vast space will lead to under-sampling and ill-representation of the space.
To demonstrate this problem let us consider how much of the parameter space need be sampled to study the neighborhood around one specific set of parameter vector x̄. This may tell us in which direction we may find more optimal parameters. We assume that points in the space are uniformly distributed in an n-dimensional cube of unit length; that is, all parameters range from 0 to 1 or were normalized to this range. Now we sample some portion of the total volume of the parameter space in a neighborhood around x̄. This neighborhood is a hypercube around x̄. It has volume V = ln, and thus side length l = V1/n. Now, given a proportion of the parameter space which we are willing to sample, we may calculate the proportion of each parameter's range which must be sampled around x̄. Let us sample just 1% of the parameter space in model with only five parameters of unknown value. Since the volume of the parameter space is 1, the volume of the neighborhood around x̄ which we must sample is V = 0.01, and the side of the hypercube is l = 0.011/5 ≈ 0.4. In other words, almost 40% of the possible values for each parameter must be sampled to understand the neighborhood around x̄. To sample just 0.1% of space in a neighborhood around x̄, we still need to sample 25% of that range for the same model. The proportion of each parameter's range that must be sampled increases exponentially with the number of parameters of unknown value (Fig. 3). This is a direct consequence of the curse of dimensionality. As the number of parameters increases, we are driven to sample practically the entire parameter space. Thus, when we construct a compartmental model with several unknown parameters and start to fiddle with them, we are fighting unwittingly a mighty curse, a curse which Hodgkin and Huxley elegantly averted by avoiding parameters of unknown value through carefully reducing their data, and a curse which casts its pall over all current attempts to realistically model neuronal physiology.
Fig. 3.
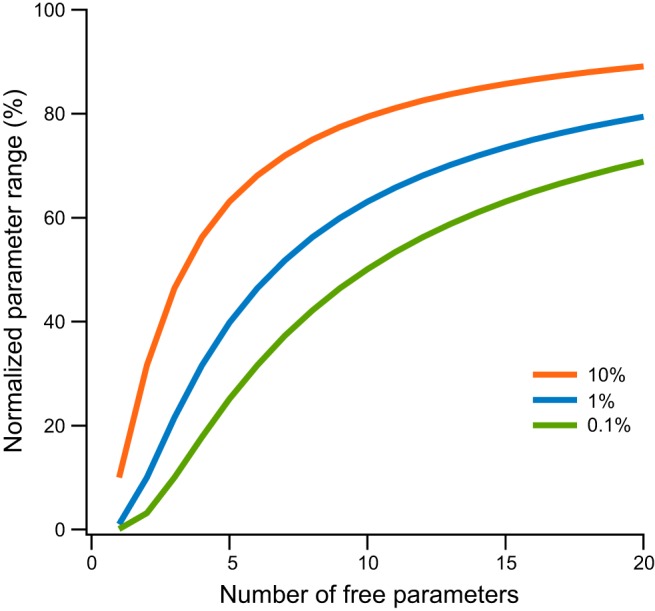
Sampling parameter neighborhood in a parameter hypercube. We assume that parameter values in the model are uniformly distributed in an n-dimensional cube of unit length. We sample some portion of the total volume of the parameter space in a neighborhood around x̄. This neighborhood is a hypercube around x̄. It has volume V = ln, and thus side length l = V1/n. The range of variation for each parameter in the model is plotted as a function of the number of model parameters. The lines represent the range of parameter variation one needs to sample to properly sample the volume around a specific solution x̄. The lines were calculated for sampling of 10%, 1%, and 0.1% of the total volume around x̄.
Methods in Compartmental Modeling
To demonstrate the complexity and the challenges which lay before the modeling community, we consider as a test case the quest to model the neocortical pyramidal neuron. First, although we will review the current state of compartmental modeling software and techniques. In Software, Constructing a compartmental model, and Tuning the model sections below, we give a fairly technical introduction to the techniques currently used for compartmental modeling. We then resume discussion of whether compartmental models can realistically model single neurons when we introduce the layer 5 pyramidal neuron in The Layer 5 Pyramidal Neuron as a Model System section below.
Software.
During the 1970s and early 1980s, researchers creating compartmental models wrote their own code using generic programming languages. By the end of the 1980s, several software packages dedicated for compartmental modeling had emerged. A brief literature search summons the names SPICE (Bunow et al. 1985; Segev et al. 1985), NEURON (Hines 1989), GENESIS (Bower and Beeman 1995), SABER (Carnevale et al. 1990), AXONTREE (Manor et al. 1991), and NODUS (De Schutter 1989).
Two programming languages among this early batch are actively maintained today, NEURON (Carnevale and Hines 2006; Hines and Carnevale 1997, 2000, 2001) and GENESIS (Bower and Beeman 1995). Both are environments for modeling individual neurons as well as networks of neurons. They contain tools for building and using models that are numerically sound. In our laboratory we have used NEURON for over a decade (Almog and Korngreen 2014; Brody and Korngreen 2013; Gurkiewicz and Korngreen 2007; Gurkiewicz et al. 2011; Keren et al. 2005, 2009; Lavian and Korngreen 2016; Pashut et al. 2011, 2014; Schaefer et al. 2003a, 2007). While our comments are based on our intimate acquaintance with NEURON, we have heard other computational neuroscientists make uncannily similar comments about GENESIS.
NEURON, probably for historical reasons, is constructed from two programming languages. Ion channel mechanisms (Hodgkin-Huxley-like kinetics, Markov chain kinetics, calcium-dependent kinetics, synaptic properties, and others) are programmed using templates in MOD (Carnevale and Hines 2006; Hines and Carnevale 2000). These templates are then parsed and translated into functions in C, compiled, and bound into a dynamic library. All other operations in NEURON are performed using scripts written in HOC and executed in an interpreter (Carnevale and Hines 2006). Since NEURON's advent several decades ago, computer programming has greatly advanced such that today some parts of its architecture lack standard features. The most prominent drawback in our opinion is the lack of formal debugger and profiler. Other problems are the tendency to rely on global variables, outdated graphic library, and the HOC language itself, which was not developed for this purpose and is simply too primitive [in an attempt to solve some of NEURON's intrinsic problems, the command line interpreter, once in HOC, is being switched to Python (Hines et al. 2009)]. Nevertheless, while many of NEURON's intrinsic problems require attention, it is among the best tools for compartmental modeling. Most importantly, it has a large and vibrant user community, mostly readable documentation (Carnevale and Hines 2006), and an extensive online database ModelDB (Hines et al. 2004; McDougal et al. 2015; Migliore et al. 2003; Morse 2008). In recent years many investigators have voluntarily contributed models to ModelDB. It is now possible to find code relevant to almost any project in ModelDB, thus cutting time for model development.
While some compartmental modeling software packages atrophied, others have sprung up in their place. In recent years, simulation packages have increased in number and variety. We will describe only a few. More elaborate lists can be found online.1 PSICS simulates neuronal activity, taking into account stochastic properties of ion channels (Cannon et al. 2010). This is an important aspect of neuronal physiology that is difficult to implement in other software packages. Successfully using PSICS to model the physiology of a CA1 neuron suggests that stochastic gating of dendritic ion channels may account for a major fraction of the probabilistic somatic and dendritic spikes (Cannon et al. 2010). CONICAL, a completely different package, is a C++ class library focusing on compartmental modeling. Contrary to the interpreter and GUI style of other packages, this allows direct programming in C++, and thus allows fast execution of the compiled program. As part of the OpenWorm project (Szigeti et al. 2014), an international collaboration aiming at understanding the behavior of the worm C. elegans using fundamental physiological processes, the Geppetto simulator was developed as the core modular simulation engine of this project to create computational models of the worm (Szigeti et al. 2014). Another ambitious simulation environment is MOOSE (http://moose.ncbs.res.in) whose ancestry can be traced to GENESIS (Ray and Bhalla 2008). MOOSE spans different scales, enabling simulations of chemical signaling models, reaction-diffusion models, single-neuron models, network models, and multiscale models (Ray and Bhalla 2008). It is compatible with GENESIS, Python and NeuroML. There are also more generic tools that have extensions allowing compartmental modeling. One of them is PyDSTool (Clewley 2012), a differential equation solver containing various toolboxes, including one allowing compartmental modeling.
Many of the recent software packages use Python as their main scripting language. Moreover, many of these simulation environments are accessible using an XML-based description language called NeuroML that provides a common data format for defining and exchanging descriptions of neuronal cell and network models that is developed as part of the Open Source Brain international collaboration (Cannon et al. 2014; Crook et al. 2007; Gleeson et al. 2007, 2010; Vella et al. 2014). NeuroML, acting as an über-language, allows the scientist to define a model using several levels of description starting at the channel level (ChannelML), through the cellular morphology (MorphML) and all the way to the network (NetworkML). The resulting XML templates, which can be converted to a variety of simulator specific scripts, are then executed using the appropriate tool (NEURON, GENESIS, MOOSE, PSICS, etc.). In contrast, the ongoing Global Neuroinformatic Project aims at providing a framework for simulator independent code generation in the hope of standardizing the diverse field of neuronal modeling. This heralds the transition from venerable, yet now somewhat outdated software, to more modern programming standards. In particular, it facilitates for debugging, which is sourly missed in older simulation packages. Modern debugging is of the utmost importance since it will allow assessing not only the logical correctness of the code, but also model comparisons to experiments, final model validation, parameter sensitivity analysis, and many other currently unavailable tests. In conclusion, there are many simulation environments available for compartmental modeling. A user, who is flexible enough to know or learn several platforms, can pick among them that which is best for his or her project (Gewaltig and Cannon 2014).
Constructing a compartmental model.
Constructing a compartmental model consists of three basic stages: defining the morphology and passive membrane parameters, building a kinetic model for each compartment, and inserting these kinetic models into the morphology. In the early days of compartmental modeling, all of these stages involved hand-tuning. Today hand-tuning has been reduced with the availability of online resources and raw computing power.
In the first stage, we decide on a cellular morphology, then adjust it to the requirements of the model and the limits of computing power. In one of his famous papers, Rall painstakingly reduced the morphology of an α-motoneuron to an equivalent cylinder (Rall 1959). This allowed him to analytically solve the model's equations and demonstrate the dendritic load on somatic synaptic integration. More recently, reducing the number of compartments in the model has been useful for other reasons. For a time, reduction was necessary to deal with the computing power limits of desktop computers. As the power of desktop computers has grown, this need has diminished such that today even models with several hundred compartments can be executed rapidly. Reduction is also a sensible strategy in light of aspiration to use efficient models of single neurons as building blocks for models of neural networks. Reducing the number of compartments can greatly increase the speed of network computation (Bahl et al. 2012; Bush and Sejnowski 1993; Destexhe 2001; Jackson and Cauller 1997). Due to a lack of experimentally reconstructed morphologies, abstract morphologies were used. Today this difficulty has been obviated to a great extent thanks to online databases such as NeuroMorpho (Ascoli et al. 2007; Halavi et al. 2008) which freely provide simulator-ready neuronal morphologies. That said, morphological reduction is still investigated as a tool for enhancing computational efficacy (Destexhe 2001; Wybo et al. 2015). It is important to note that morphology is not a constant thing but rather highly variable even between neurons of the same type (Cuntz et al. 2007, 2010, 2011; Hay et al. 2011; Torben-Nielsen and De Schutter 2014).
So, while reduction remains an important strategy, it is no longer forced upon our models of single neurons by the limits of our computers. The extent to which we may reduce neuronal morphology depends upon our research questions and the level of abstraction that is reasonable and in conformance with our assumptions. Since it is realistic neuronal modeling discussed here, we henceforth discuss models which do not employ reduction.
In the second stage of model construction, we assign passive parameters to the structure. This may be done by assigning average values given in physiology textbooks, by searching the literature for experimental measures of the parameters in the salient kind of neuron, or by estimating passive parameters experimentally. Using average values for the passive parameters can be beneficial when attempting to explore or demonstrate a basic subthreshold mechanism. In this way, we rapidly generate models and quickly gain a sense of their qualitative characteristics, which can be very helpful in explaining experimental results or suggesting working hypotheses. Searching the literature, either for specific values for the neuron simulated or for population values (Tripathy et al. 2015) can provide a good estimate of passive parameters and, of no less importance, their range of change. This physiological range of the parameters can be used to set the search space for the optimization algorithm discussed later in the text.
Simple experimental protocols (e.g., whole-cell patch-clamp recording combined with intracellular dye-staining and computer-aided morphological reconstruction) can also be used to estimate the values of the passive parameters. (Clements and Redman 1989; Major et al. 1994; Roth and Häusser 2001; Stuart and Spruston 1998). Typically, these measurements are made from the soma. We then posit that the passive parameters are similar throughout the dendritic tree (save for changes due to spine density). Note, though, that even this straightforward assumption should be verified per the investigated system. In layer 5 pyramidal neurons it has been shown that passive parameters change along the apical dendrite (Stuart and Spruston 1998). Once we have set the passive parameters, we can test the model's response to subthreshold stimulation. This can reveal problems in the code and verify that, at least for the subthreshold zone, the model behaves similarly to the neuron modeled.
In the third stage, we write and debug the voltage- and ligand-gated ion channels that will be inserted into the model. Most compartmental models use the Hodgkin-Huxley formalism to describe ion channel kinetics (Hodgkin and Huxley 1952d). It must be remembered, though, that this kinetic scheme is phenomenological and assumes that each activation and inactivation gate acts independently of other gates. The mechanisms underlying membrane excitation have revealed themselves over the years in much greater detail than Hodgkin and Huxley could see, and several differences from their model must be emphasized. Whereas the Hodgkin-Huxley model has no connectivity between the activating and inactivating gates of the voltage-dependent sodium channel, more recent studies have shown that the voltage-dependence of the inactivation gate is due to its coupling to the activation process, i.e., it can close only after the activation gate opens (Armstrong and Bezanilla 1977; Bezanilla and Armstrong 1977). Moreover, additional recording and analysis of single channels have shown that Markov chain models better describe ion channel kinetics (Horn and Lange 1983; Lampert and Korngreen 2014; Magleby 1992; McManus and Magleby 1989; McManus et al. 1989; Qin et al. 2000a, 2000b; Sakmann and Neher 1995; Venkataramanan and Sigworth 2002). Several studies have confronted cases where the Hodgkin-Huxley formalism proved insufficient requiring the use of Markov chain kinetics (Khaliq et al. 2006; Schmidt-Hieber and Bischofberger 2010). Functional Markov models can be extracted by curve fitting whole traces (Gurkiewicz and Korngreen 2006, 2007; Gurkiewicz et al. 2011; Lampert and Korngreen 2014; Menon et al. 2009; Milescu et al. 2005) not only from single channel records. Furthermore, an algorithm for automatically optimizing the Markov model by addition and subtraction of states has been proposed (Menon et al. 2009). It is, however, important to point out that Markov models are not a magic bullet. Replacing the description of ion channel kinetics from Hodgkin-Huxley type models to Markov models does not solve any of the problems we discuss in this review. Thus, even though Markov models are superior, physiological modeling of neuronal activity mostly uses the simpler Hodgkin-Huxley formalism. This is probably due to the lack of good tutorials for creating Markov models and their significantly longer execution time. In our hands Markov models executed up to four times slower than the Hodgkin-Huxley models for the same channel (unpublished observations). Interestingly, Hodgkin himself noted that the phenomenological gating scheme was adopted because he and Huxley could not provide a solution stemming from basic principles (Hodgkin 1976). Indeed no ab initio solution is possible even today because of the complexity of the problem we face when we try solving the ion channel structure-function problem.
Unfortunately, the vast genetic information available to us about the sequences of most voltage-gated channel subunits provides only partial kinetic information for compartmental modeling. Expressing these subunits in various expression systems enables generating functional channels from which gating kinetics can be extracted using the voltage-clamp technique. However, using these kinetic models for modeling neuronal physiology is problematic. For example, in our favorite model system, the apical dendrite of layer 5 pyramidal neurons, studies reveal the expression of the voltage-gated potassium channel subunits Kvα1.2, Kvα1.4, Kvα2.1, Kvβ1.1, Kvβ2.1, Kvα4.2 and Kvα4.3 (Rhodes et al. 1995, 1996; Serodio and Rudy 1998; Sheng et al. 1994; Trimmer and Rhodes 2004; Tsaur et al. 1997). However, voltage-clamp experiments have detected only three voltage-gated channels in these neurons (Bekkers 2000a, 2000b; Bekkers and Delaney 2001; Korngreen and Sakmann 2000; Schaefer et al. 2007). This variety of channel subunits leads to the conclusion that native potassium channels are heteromers, containing probably both α- and β-subunits. It has been shown that subunit composition greatly affects the properties of potassium channels (Rettig et al. 1994; Ruppersberg et al. 1990; Stuhmer et al. 1989). Similar disparity between the number of expressed subunits and the final number of channels has been observed in other neuronal types and may be the rule rather than the exception. And this is just the tip of the iceberg. We note in passing the modulation of channels by signaling pathways and auxiliary subunits. Thus the molecular identity of the channel cannot be used to predict its kinetic properties. Therefore, kinetics of voltage-gated channels extracted from artificial expression systems should be used with utmost caution in compartmental models.
The most reasonable alternative to using kinetic models extracted from expression systems is to use kinetic analysis of voltage-clamp experiments performed directly on the cell under investigation. It is important to caution that in vitro brain slice preparations too have their problems, many times void of neuromodulators with known and unknown effects on channel kinetics. In many cases, code for the appropriate channel based on such experiments is available on ModelDB. Nevertheless, such code must be checked against the original study on which it was based. Not all models we downloaded from the web were faithful descriptions of the experimental data. Another alternative is to generate a kinetic model by extracting the kinetic scheme from published papers. Still another alternative, one which can only be commended, is to perform the experiments yourself, or at least have a trusted and proximate collaborator do the experiments and hand you the data. Simply extract membrane patches, employ the voltage-clamp protocols, and use the Hodgkin-Huxley formalism or Markov chain modeling to generate a kinetic model. Nothing replaces working with the experimental data. Once we have the kinetic models, we need only to insert them into the passive model according to published conductance gradients.
Tuning the model.
At this stage of model construction we have made assumptions about the morphology, electrotonic structure, channel kinetics, and channel distribution. We assume that these have been conscious assumptions based on prior experience in modeling. It is vital to dredge these assumptions into consciousness as they determine the strength of the conclusions we can draw from the model. These assumptions are expressed in many parameters of unknown values. Unfortunately, we cannot yet fill in these values experimentally: our current technology cannot fully access fine dendritic and axonal processes. To overcome this knowledge gap, there are several strategies. Different strategies are appropriate for different kinds of research questions.
One straightforward strategy is general simplification. For example, we may assume that the dendrites are passive and the soma is excitable (like in the example presented in Fig. 2C). When investigating subthreshold activity, this is a fair and powerful assumption [although in some cases the involvement of voltage-gated ion channels in subthreshold activity cannot be neglected (Devor and Yarom 2002; Jacobson et al. 2005)]. We recently applied a similar assumption with success to qualitatively investigate dendritic inhibitory synaptic transmission (Lavian and Korngreen 2016). Such simplifying assumptions can reduce the compartmental model to a form that can help accept or reject a working hypothesis for a given physiological question.
Another strategy has to be adopted when, even after making such additional assumptions, there remain important parameters of unknown values. No values having been determined experimentally, one may try to hand-tune the model until it appears to function properly. This trial-and-error process requires that we keep in mind an image of the desired behavior or functionality. Simply modify the parameters, execute the model, check the results against the ideal, and repeat until the model fits the image. Since this is done by mentally scoring the performance of the model, the process is no better than your meat-brain and estimates, i.e., probably highly inaccurate, nearly impossible to reproduce, heuristic, almost arbitrary, erratic, and subject to sleep disturbances. Furthermore, due to the curse of dimensionality, parameter space cannot be searched exhaustively, and probably not even very well. The process is solipsistic: it's not at all clear that one can bootstrap beyond what is in essence assumed at the beginning. That said, when the model aims to address a conceptual problem or when it is very simple, hand-tuning may suffice. Otherwise, when the model is complex, or aims at realistic behavior, the curse of dimensionality damns hand-tuning to sisyphean looping and failure.
Computer-tuning the model.
This third strategy, or set of strategies, for tuning the model merits special attention. We are living in the information age of the 21st century. Perhaps there is salvation in silicon; perhaps computer algorithms can successfully search the parameter space and find the best models. The computer must perform the same heuristic process we would perform when hand-tuning the model. The hope is that the computer does it more rapidly, more thoroughly, and more objectively. The strategy for automatic model optimization is based on multiple quantitative comparisons of possible models to experimental, or surrogate, data. This approach has three basic components: the optimization algorithm, the comparison function (a.k.a., cost or score function), and the data set to which the model is compared. Each component affects the efficiency of the search through parameter space (Bahl et al. 2012; Druckmann et al. 2008, 2011; Keren et al. 2005; LeMasson and Maex 2001). Let us consider each in turn, surveying currently available techniques.
the optimization algorithm.
The simplest automatic approach to model optimization is brute force: the computer checks all possible parameter permutations. This is extremely cumbersome, even with large supercomputers. Thus this method can only be used sparingly and with models having small numbers of parameters where rastering through the whole parameter space is possible. This brute-force tuning algorithm has been used to great effect in studying models of the somatogastric ganglion demonstrating that identical network or neuron activities can be obtained from disparate modeling parameters (Goldman et al. 2001; Golowasch et al. 2002; Prinz et al. 2004). It has been suggested to use grid sampling of parameter space to tune compartmental models (Prinz et al. 2003). Brute force sampling, however, is probably not applicable to more complex models due to the almost infinite number of calculations needed to fully sample parameter space. This probably applies also to a variant of the brute-force approach in which a subset of the possible vectors is randomly selected (Marder and Taylor 2011; Taylor et al. 2006, 2009).
The alternative to grid sampling or random sampling is to use an algorithm to sample the parameter space in a smarter manner. The general idea behind such an algorithm is to use a set of heuristic rules and an iterative process to search within parameter space in the vicinity of a given solution to find a better solution. There are many such search algorithms. All of them draw their core from natural phenomena that search vast parameter spaces.
Simulated annealing takes its core from crystal formation and has proven highly effective for solving discrete problems, such as the traveling salesman problem (Kirkpatrick et al. 1983). A simple simulated annealing algorithm starts with a random parameter vector representing the system in the hot liquid state. A “family” of successor solutions is generated by randomly perturbing one or several parameters in the original parameter vector. Each new family member is tested and compared using a score function (see below). Family members that scored the best generate the next families of solutions. As this iterative process continues, the algorithms adjust a “temperature” parameter. This reduces the size of parameter perturbations according to a predetermined annealing schedule. As the iterations continue, random perturbations slowly decrease, converging on a selected solution. Simulated annealing performs well for compartmental models with a small numbers of parameters, but less effectively for models with many compartments (Vanier and Bower 1999). A combination of simulated annealing with the deterministic simplex optimization algorithm (Cardoso et al. 1996; Press 1992) was effective in determining the relative contribution of morphology and active conductances in models of neurons of the goldfish hindbrain (Weaver and Wearne 2008). It is worth noting that both random sampling and simulated annealing are forms of Gibbs sampling which theoretically is guaranteed to recover the true distribution, given sufficient sampling (Casella and George 1992).
Particle swarm optimization draws on the behavior of schools of fish and flocks of birds (Kennedy et al. 2001). The optimization method assumes that each individual in the swarm, i.e., each candidate solution, has two basic behaviors, conforming to the direction of the swarm and moving independently toward a potential food source. The optimization process begins with a swarm of random parameter vectors traveling at random “velocities.” Individuals are scored according to a cost function, and, using a linear combination of the best (lowest) scoring individuals, other individuals adjust their positions (i.e., their parameters). Although particle swarm optimization does not guarantee finding a global minimum, it does find local minima, and sometimes gets stuck in them, neglecting proximate better food sources (i.e., parameter sets) just over hills in the cost function. An important advantage of particle swarm optimization is that its convergence to a manifold of solutions allows for analysis of solution variability.
Genetic algorithms contain operators from both genetics and evolutionary biology (Mitchell 1996). Like simulated annealing and particle swarm optimization, genetic algorithms are initiated with a large population of random parameter vectors, each of which, in our context, represents the parameters of a compartmental model. Each vector is used to generate a simulation and is scored according to how well that simulation resembles the target data set. A first operator is then applied to this primordial soup of solutions descends from the concept in evolutionary biology of “survival of the fittest.” Parameter vectors, which generate simulations that are most similar to the target data, are considered to have metaphoric good sets of genes and allowed to transfer their “genetic material” to the next generation, while vectors generating simulations that do not measure up against this survival condition are taken to have bad sets of genes and not allowed to reproduce. The individuals with good scores are then paired and “mated” to produce a new population. During the mating process, the individuals, i.e., the parameter vectors, interchange parts of their genetic code, i.e., their values for particular parameters, using a crossover operator. This operator takes the form of randomly selecting one point along one parent parameter vector and exchanging the values from that point onwards with those of the other parent parameter vector (two-point crossover operators are also in use). At the same time, another operator is also active, a mutation operator, which, with a given probability, replaces or perturbs the value of one parameter in one individual. This next generation, then, is used to generate simulations, scored according to how well it resembles the target data set, and mated. This process is repeated for many generations. Many variants of the genetic-evolutionary algorithm have been presented in the general optimization literature. Of the stochastic search algorithms presented here, genetic algorithm has emerged as the most popular algorithm for optimizing compartmental neuronal models (Almog and Korngreen 2014; Bahl et al. 2012; Druckmann et al. 2007, 2008, 2011; Gerken et al. 2005; Gurkiewicz and Korngreen 2007; Gurkiewicz et al. 2011; Hay et al. 2011; Keren et al. 2005, 2009; Tabak and Moore 1998; Tabak et al. 2000; Vanier and Bower 1999).
These three stochastic optimization algorithms, as well as others we have not discussed here, attempt to reduce computational load while fully sampling parameter space. In other words, they aim to overcome, at least partially, the curse of dimensionality, by searching parameter space in a systematic yet heuristic manner. Indeed, it has been rigorously proven that, given infinite computing time, all these algorithms locate the best possible solution in parameter space. Unfortunately, since infinite computing time is hard to come by, they practically never reach optimal solutions (Keren et al. 2005, 2009). Furthermore, they are not immune to the curse of dimensionality (Chen et al. 2015) and cannot be considered magic bullets for compartmental model optimization and generation of realistic models. Moreover, they demand so much iteration, they require computing power orders of magnitude beyond that available in a single desktop computer. So, to actually implement these algorithms, parallel computing must be used. Fortunately, all of the above-mentioned search algorithms could be parallelized easily using simple, embarrassingly parallel methods. There are many flavors of parallel computing machines. The most widely distributed are Linux clusters that can be constructed and configured easily, even with old computers (Hoffman and Hargrove 1999). Parallel clusters of computers are now available in almost every university, drastically reducing computing time. Many companies, such as Amazon and Google, provide time on their immense clusters at reasonable rates. Commercially available graphic processing units (GPUs), which contain many parallel mathematical processing cores, can also be used to considerably speed up computation time, possibly by several orders of magnitude (John et al. 2009; Liu et al. 2009; Manavski and Valle 2008; Nageswaran et al. 2009; Pinto et al. 2009; Rossant et al.; Stone et al. 2007; Won-Ki et al. 2010). We have shown that parallelizing genetic algorithm optimization of ion channel kinetics (Ben-Shalom et al. 2012) or the execution of the compartmental model (Ben-Shalom et al. 2013) on a Fermi graphic computing engine from NVIDIA, using the CUDA extension of the C programming language, increased the algorithm's speed over a hundredfold. However, the use of GPUs is still on a small scale compared with Linux clusters, probably due to the nontrivial programming required to utilize these cards.
The cost function.
As we noted above, hand-tuning may be conceived as an optimization process in which the scientist scores the simulations produced by different parameters mentally with criteria ranging from explicit to subjective. To translate this scoring process to the computer, a cost function is defined and used to quantitatively compare the output of the simulations with a target data set. As suggested by their name, these functions measure the distance between the simulation and the data and are used to score the aptness of a specific simulation relative to many other simulations. The cost function used for optimizing compartmental models usually measure this distance using root mean squares:
| (8) |
In Eq. 8, N is the total number of points in the target data set, and data and simulation are, respectively, vectors of the experimental or surrogate data and of the simulation output matched appropriately. We have left Eq. 8 in a general and informal form because target data sets can differ radically between different optimization studies of compartmental models, and the methods by which different cost functions find the difference between these vectors too can differ in crucial ways.
In its simplest variant, the cost function subtracts the experimental traces from the simulated membrane potential traces (Keren et al. 2005, 2009). This point-by-point subtraction is highly sensitive to the precise timing of action potentials (LeMasson and Maex 2001), so that, if the simulated action potential is slightly only too early or too late, its score will be closer to a poor simulation than to an ideal simulation. This sharp and narrow sensitivity to timing makes this direct geometrical comparison between simulation and data function poorly in genetic algorithm optimization processes (Keren et al. 2005). Several solutions have been suggested. One may represent the phase plane of the membrane potential, and this solves the problem of sharpness (LeMasson and Maex 2001). It is also possible to add information about action potential timing and to construct combinations of several cost functions to help a genetic algorithm better search the parameter space (Keren et al. 2005) or to align action potentials before calculating the cost function (Weaver and Wearne 2006). Another approach is to extract average features from a group of experimentally recorded action potentials, such as amplitude, half-width, threshold, and amplitude after-hyperpolarization, and then to use these as the salient points for comparison (Druckmann et al. 2007, 2008; Hay et al. 2011). This has the advantage of using values representing a neuronal population rather than a single neuron. It also takes experimental variance into account a priori (Hay et al. 2011). Another powerful strategy is to combine several cost functions and a multiobjective algorithm (Druckmann et al. 2007, 2008). Extracting features from the target data, however, greatly reduces the information available from the target data. This can result in simulations which have, for example, the desired amplitude after-hyperpolarization, if this is a feature in the cost function, but not the correct shape of the after-hyperpolarization (Druckmann et al. 2007, 2008).
The cost function can be made to play another important role in the optimization algorithm, namely, as a measure of its progress (Bahl et al. 2012; Ben-Shalom et al. 2013; Druckmann et al. 2007, 2008; Keren et al. 2005). This can result in convergence that is either too fast or too slow. Extremely fast convergence, with the score rapidly dropping within a few generations and then leveling off (Keren et al. 2005), is less likely the result of an extremely efficient search than it is an indication that the parameter space was inadequately sampled. Conversely, extremely slow convergence can indicate that the algorithm became stuck in a local minimum, but not low enough minimum, to terminate. In several studies, the score function displayed an initial moderate decay followed by a slow one (Achard and De Schutter 2006; Gurkiewicz and Korngreen 2007; Keren et al. 2005; Vanier and Bower 1999). This problem we have shown elsewhere can be solved by greatly increasing the size of the population in a genetic algorithm (Ben-Shalom et al. 2012).
In any case, the multidimensional surface defined by the cost function can be very complex (Achard and De Schutter 2006; Bhalla and Bower 1993; Goldman et al. 2001; Golowasch et al. 2002; Taylor et al. 2006, 2009), including, for example, distinct regions with similar scores for similarly behaving simulations. Although it is nice in some sense that very different sets of parameters may yield reasonable behavior, such results can mislead us altogether, if we interpret parameter values as biological realities. So we must carefully analyze our cost functions and their sensitivity to perturbation of parameter values (Achard and De Schutter 2006; Bhalla and Bower 1993). In many cases, setting up the cost function properly is far from trivial and requires in-depth understanding of the goal of the optimization.
the target data set.
The third foundation of automatic model optimization is the target data set used by the algorithm. For obvious reasons, the selected target data set profoundly affects the output of the cost function. For all automatic compartmental model optimization to date, the target data sets have comprised membrane potential recordings or features extracted from such recordings (Bahl et al. 2012; Druckmann et al. 2007, 2008, 2011; Gerken et al. 2005; Hay et al. 2011; Keren et al. 2009; Vanier and Bower 1999; Weaver and Wearne 2008). It is worth noting that simulated data sets have occasionally been used as surrogate data to test the performance of optimization algorithms or specific features of the optimization process (Druckmann et al. 2008; Keren et al. 2005, 2009). Moreover, several studies have been dedicated to determining features of the data sets optimal for optimization algorithm searches of parameter space (Druckmann et al. 2011; Gurkiewicz and Korngreen 2007; Huys et al. 2006; Huys and Paninski 2009; Keren et al. 2005). Our laboratory has previously shown that to optimize a compartmental model containing dendritic and axonal conductance gradients a data set including recordings from the axon, soma, and dendrites is required (Keren et al. 2005). These studies suggest a simple rule of thumb: the larger and richer in features the target data set, the better.
Existing methods for fitting a curve over a large domain are highly sensitive to the size and diversity of the data set (Motulsky and Christopoulos 2004). To attempt such global curve fitting requires, in particular, that our data set contain traces sensitive to changes in all the parameters undergoing optimization. For otherwise, no matter how clever the optimization algorithm, although a parameter changes, and so too the model's simulations, the changes do not register compared with the target data, their simulations' scores do not differ, and the parameter is left free and unconstrained: it falls through a crack in the target data set and escapes optimization. In control theory when a parameter cannot be estimated using a given data set, it is said to be unobservable. Varying the value of such a parameter over a large interval, demonstrating that model behavior does not change significantly, can serve as a test for the parameter's unobservability.
The Layer 5 Pyramidal Neuron as a Model System
There are many approaches to optimizing compartmental models. This makes it difficult to compare between models and assessing their success in modeling neurons realistically. To avoid comparing apples and oranges, we designed several numerical challenges, focused on models of layer 5 neocortical pyramidal neurons, and applied our challenges to publically available models. The challenges required that the model reproduce an experimentally observed behavior of layer 5 pyramidal neurons. The models were not explicitly tuned to generate these behaviors at their time of creation.
Layer 5 neocortical pyramidal neurons are one of the primary output cells from the cortex; the main axonal stem projects to intrahemispheric, interhemispheric, and subcortical targets, including the spinal cord, pons, medulla, tectum, thalamus and the striatum (Feldman 1984). Efforts to record from these neurons have achieved noteworthy successes. Regenerative calcium spikes have been recorded from the apical dendrite of layer 5 pyramidal neurons (Amitai et al. 1993; Larkum et al. 1999a, 1999b, 2001; Schiller et al. 1997; Zhu 2000). And today a number of important phenomena have been revealed. When back-propagating action potentials coincide with distal synaptic inputs, dendritic calcium spikes are generated, and this in turn generates bursts of action potentials at the soma, a process nicknamed “BAC firing” (Larkum et al. 1999b). Under some conditions, calcium spikes can be isolated in the dendrites, whereas under others, the spikes can spread to the soma (Larkum et al. 1999a; Schiller et al. 1997). Action potential bursts can be triggered by the proximal apical dendrite as well as by the distal apical dendrite (Larkum et al. 2001; Williams and Stuart 1999). Depolarization of the proximal apical dendrite facilitates the forward propagation of dendritic sodium and calcium spikes (Larkum et al. 2001). Thin tuft and basal dendrites preferentially initiate local N-methyl-d-aspartate (NMDA) and weak sodium spikes (Larkum et al. 2009; Nevian et al. 2007; Polsky et al. 2004; Schiller et al. 2000), whereas the main trunk of the apical dendrite is dominated by calcium spikes (Larkum et al. 1999b; Schiller et al. 1997). NMDA spikes have been shown to confer increased computational abilities to basal and tuft dendrites, allowing clustered synaptic inputs to define a local region of nonlinear synaptic integration (Larkum et al. 2009; Nevian et al. 2007; Polsky et al. 2004; Schiller et al. 2000). Thus the dendritic tree of layer 5 pyramidal neurons may be viewed as a multilayer, constantly reconfiguring computing device (Poirazi et al. 2003a, 2003b; Poirazi and Mel 2001). Over the years, many studies have attempted to simulate the complex physiology of layer 5 pyramidal neurons. Rather than review them all, we will draw a short time line of the past 20 years, taking three models as test cases.
The first model to which we direct our attention is the Mainen and Sejnowski (1996) model. It was among the first efforts to model (Rapp et al. 1996) layer 5 pyramidal neurons following the discovery of the back-propagating action potential (Stuart and Hausser 1994; Stuart and Sakmann 1994). The Mainen and Sejnowski (1996) model is a fully developed, hand-tuned compartmental model containing a real morphology of a cortical neuron and several voltage-gated channels. The morphology has about 400 compartments, each with several voltage-gated channels. This brings the number of differential equations defining the compartmental model to ∼3,000 and the total number of parameters to over 10,000. Using homogenizing assumptions, the model was hand-tuned using about 15 parameters (Mainen and Sejnowski 1996). It faithfully reproduced the back-propagation of the action potential by introducing a low density of voltage-gated sodium channels in the apical dendrite (Mainen and Sejnowski 1996). While the model does contain a voltage-gated calcium channel in the apical dendrite, it does not generate dendritic calcium spikes. Instead, back-propagating action potentials can generate calcium electrogenesis in the distal apical dendrite and lead to a burst of action potentials. An important feature of this model, one that many later models adopted, is the stylized axon initial segment replacing the natural one. This model of the spike initiation zone is highly robust, functioning well in a variety of compartmental models. The weakness of this model is the high density of voltage-gated sodium channels in the axon initial segment; this introduces an unrealistically large local shunt.
The Mainen and Sejnowski (1996) model was modified a few years later by Schaefer et al. (2003b). Like the Mainen and Sejnowski (1996) model, the Schaefer et al. (2003b) model (henceforth: the Schaefer model) uses a fully reconstructed morphology and inflates to over 10,000 parameters which are then reduced using standard assumptions to about 17 parameters of unknown value. It adds an ionic mechanism for dendritic calcium spikes in the distal apical dendrite (Schaefer et al. 2003b). Together with other models (Acker and Antic 2009; Grewe et al. 2010; Hay et al. 2013; Vetter et al. 2001), the Schaefer model has been instrumental in examining the relationship between morphology, excitation, and coupling of somatic and dendritic action potential initiation sites. By adding a hot-spot of T-type voltage-gated calcium channels in the distal apical dendrite, this model acquires a local low-threshold site for the initiation of dendritic spikes. It was also able to reproduce BAC-firing and to illustrate how variability in BAC-firing is linked to morphology and the number of oblique dendrites (Schaefer et al. 2003b). Similar to the Mainen and Sejnowski (1996) model, the Schaefer model was hand-tuned to phenomenologically reproduce a specific neuronal physiology. There are other hand-tuned models describing dendritic excitability of layer 5 pyramidal neurons (Larkum et al. 2009; Rapp et al. 1996). Instead of discussing all of them, we will proceed to discuss computer-tuned models.
The second contender in our computational challenge is the Hay et al. (2011) (henceforth: the Hay model), a computer-optimized model designed to capture a wide range of dendritic and somatic properties. The Hay model was tuned using a multiobjective genetic algorithm (Deb et al. 2002), and the target data set comprised features extracted from somatic and dendritic membrane potential recordings (Hay et al. 2011). Similar to the Mainen and Sejnowski (1996) model, it uses a full reconstructed morphology, and, similarly, it inflates to over 10,000 parameters, which were then reduced using standard assumptions to 22 parameters of unknown value. In their elegant modeling study, Hay et al. (2011) smartly extracted features of somatic and dendritic spikes from several data sets and, in the fitting process, took the variability of each feature into account. They were thus able to generate a family of possible models for layer 5 pyramidal neurons falling within the same range of physiological activity. In this model, the conductance densities of the high- and low-voltage activated calcium channels were elevated in the distal apical dendrite. In contrast with the Mainen and Sejnowski (1996) model and the Schaefer model, the Hay model did not model an initiation zone in detail; rather, it discarded the axon and loaded the soma with high density of potassium and sodium conductances. The Hay model, then, departed from physiology, and generates action potentials at the soma. It faithfully reproduced BAC-firing and the somatic response to current injection (Hay et al. 2011).
The third contender enjoys home-field advantage; it is our own Almog and Korngreen (2014) model, henceforth, the Almog model. We used experimental target data containing membrane potential traces simultaneously recorded from the soma and apical dendrite of layer 5 pyramidal neurons and a simple genetic algorithm. Ours differs from the Hay model in several important respects. First, the target data set of the Almog model comprised actual membrane potentials recorded from the dendrite, not extracted features. Furthermore, to ameliorate the effects of the curse of dimensionality, we adopted a divide-and-conquer method and segmented parameter space (Keren et al. 2009; Roth and Bahl 2009). We recorded experimental traces in the presence of different cocktails of ion channel blockers so that each trace would address only four to seven of the model's parameters of unknown value. We then used the genetic algorithm to search each of these four- to seven-dimension spaces for parameter values that optimally fit the salient trace (Almog and Korngreen 2014; Keren et al. 2009). The model was optimized only for the apical dendrite and the soma. Thus it does not contain an axon initial segment. To investigate parameter variability, we performed the same optimization on data from several neurons and averaged the resulting parameters. This “divide-and-conquer” approach presents a simple solution to the curse of dimensionality by segmenting parameter space. Furthermore, a careful analysis of time-scales, for example, can be used to reduce the parameter set to be tuned for a particular data set. This can reduce the parameter space tremendously. Without doing this, search will probably be unsuccessful. Following our parameter-peeling optimization process, we attached a hand-tuned artificial axon initial segment derived from the Mainen and Sejnowski (1996) model. Contrary to the Schaefer and the Hay models, this model was optimized without a distal hot spot of calcium channels. In the optimized Almog model, the density of voltage-gated calcium channels decreased linearly from the soma, and leveled off at the distal apical dendrite. The density of the small-conductance calcium-activated channels decreased along the apical dendrite, whereas the density of the large-conductance calcium-gated potassium channel was homogenous throughout the apical dendrite. The model predicted an ionic mechanism for the generation of a dendritic spike, the interaction of this spike with the back-propagating action potential, the mechanism responsible for the ability of the proximal apical dendrite to control the coupling between the axon and the dendrite, and the generation of NMDA spikes in the distal apical tuft. It is important to note, and we will return to this point, that all the models here mentioned have far from realistic mechanisms for action potential generation at the axon initial segment.
What happens when models of layer 5 pyramidal neurons step out of their comfort zone?
To demonstrate the problems in the construction and optimization of realistic compartmental models, we drafted the three compartmental models of layer 5 pyramidal neurons into a computational reality show. Pass three computational challenges and claim victory.
challenge i: the proximal apical dendrite can support action potential back-propagation and generate an isolated dendrite spike.
It has been shown that action potentials are triggered at the axon initial segment by injecting current via a whole-cell electrode into the apical dendrite of a layer 5 pyramidal neuron at a proximal location (∼300 μm from the soma) (Stuart et al. 1997a). It has also been shown that an isolated dendritic calcium spike is triggered by similarly injecting current at a distal location (∼600 μm from the soma) (Schiller et al. 1997), and, according to our predictions, once voltage-gated sodium channels are blocked at the axon initial segment and action potentials thus muted, a dendritic calcium spike should be triggered, even when current is injected at proximal locations (Almog and Korngreen 2014). Can the three contestant models reproduce these behaviors?
In the Schaefer model, injecting suprathreshold current into the apical dendrite 300 μm from the soma triggered three somatic action potentials that back-propagated into the apical dendrite (Fig. 4A), but, when the axonal voltage-gated sodium conductance was set to zero, current injection at the same dendritic location generated only a small, nonlinear change to the membrane potential (Fig. 4B). In the Hay model, a similar simulated experiment also generated several somatic action potentials. Back-propagation, however, did not resemble experimental data (Fig. 4C). Nevertheless, after muting action potentials at the axon initial segment, proximal dendritic current injection generated a realistic isolated dendritic calcium spike (Fig. 4D). In the Almog model, the same stimulus (again, of amplitude suited to the model's assumptions) yielded realistic back-propagation (Fig. 4E) and a realistic local calcium spike (Fig. 4F).
Fig. 4.
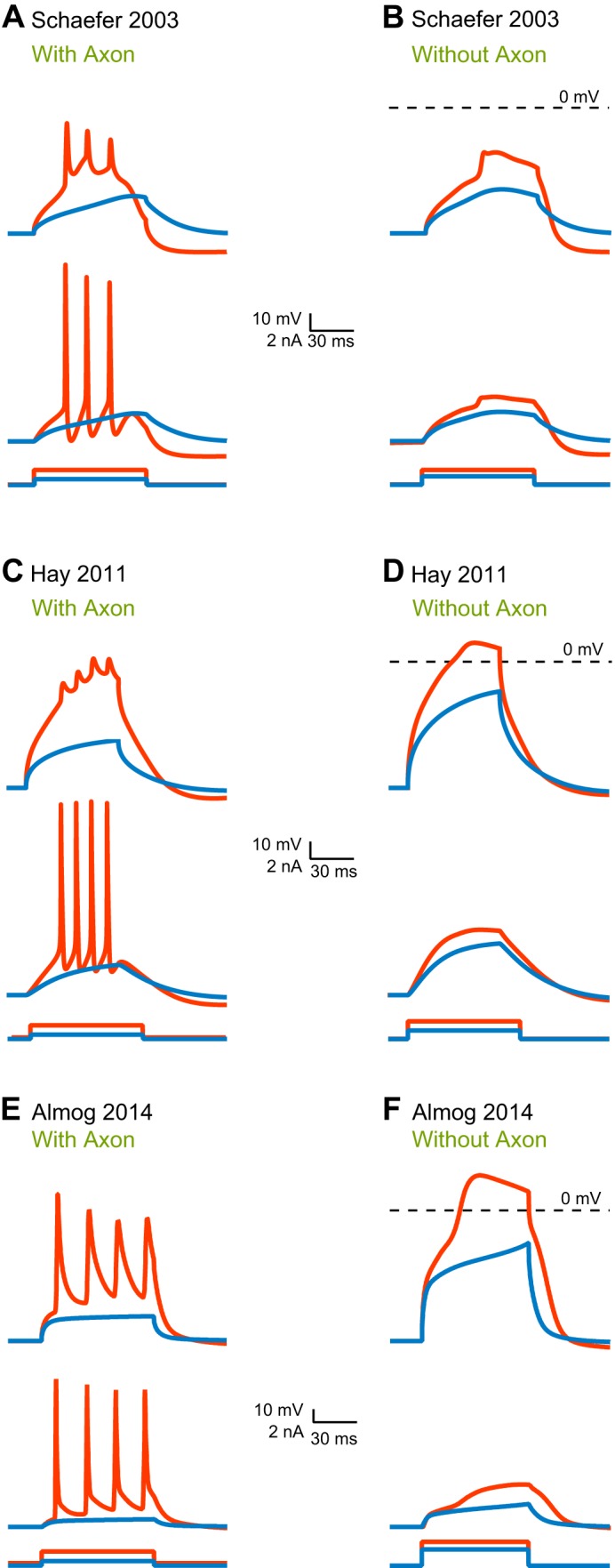
Initiation of a proximal dendritic calcium spike by different models. A, C, and E: three models of layer 5 pyramidal neuron's somatic and dendritic voltage responses to simulated current injection by dendritic pipette 300 μm from soma. Current injection was 50 ms; 0.8 nA and 1.5 nA in A, 0.5 nA and 1 nA in C, 1 nA and 1.5 nA in E. Stimulation evoked forward propagation of action potentials toward the soma in all models. In all panels, the dendritic response is shown at the top and the somatic at the bottom. Color codes correspond to the level of current injected in each case. B, D, and F: the three model responses to simulated current injection by dendritic pipette at 300 μm from soma after eliminating the axon from the models. Curent injection was 50 ms; 1 nA and 1.5 nA in B, 1 nA and 1.5 nA in D, 2.5 nA and 3 nA in F. A local dendritic spike was evoked in both Hay model (D) and Almog model (F), whereas in Schaefer model only a small response was observed (B).
To test the models' performance experimentally, we reproduced the simulated experiment experimentally (all methods are identical to Almog and Korngreen 2014). There can be no doubt: it is absolutely necessary to validate a model's predictions experimentally. This should be self-evident. Models of the physical world should predict practical experiments that can be used to accept or reject the working hypothesis used to generate the model and test the strength of its assumptions. Thus, when a current step into the apical dendrite via a whole-cell electrode positioned 300 μm from the soma, action potentials were generated at the axon initial segment, back-propagated, and recorded by the dendritic electrode (Fig. 5B). When the axon initial segment was exposed to tetrodotoxin (1 μM) via a second patch pipette, thus blocking voltage-gated sodium channels, axonal action potentials were not generated. Injecting current via the dendritic electrode generated an isolated dendritic spike in the proximal dendrite (Fig. 5C). Based on these results, the winners of the first challenge are the Almog and Hay models.
Fig. 5.
Initiation of a local dendritic regenerative potential at the proximal dendrite. A: reconstruction of a layer 5 pyramidal neuron stained with biocytin illustrating electrode placement. B: dendritic voltage responses to 50-ms current injection (0.8 and 1.1 nA) through the dendritic pipette at 300 μm, as illustrated in A. C: dendritic voltage responses to 50-ms current injection (1.6 and 1.8 nA) through the dendritic pipette at 300 μm when an application puffing pipette with an application solution (artificial cerebrospinal fluid and 10 μM tetrodotoxin) was placed on the axon initial segment, as illustrated in A. A local dendritic spike was evoked by a 1.8-nA current. Color codes correspond to the level of current injected in each case.
challenge ii: even when dendritic calcium channels are partially blocked, it is still possible to generate dendritic calcium spikes.
It has been shown that even when dendritic calcium channels are partially blocked, dendritic calcium spikes are still generated, if the amount of current injected via the dendritic pipette is increased (Perez-Garci et al. 2013). Furthermore, the same study showed the spike timing of somatic action potentials is preserved following dendritic spike regeneration (see Fig. 2 in Perez-Garci et al. 2013). In this way, dendritic calcium spikes are similar to action potentials generated at the axon initial segment that can still be generated even when low doses of tetrodotoxin are applied if the amount of current injected is increased.
When injected distally (∼600 μm from the soma) with an excitatory postsynaptic potential-like waveform, the Schaefer model neuron regenerated dendritic spikes and somatic firing after current was increased, although the simulated voltage responses compared poorly with experimental results (Perez-Garci et al. 2013) (Fig. 6, A and B; A, C, and E show responses when simulated high-voltage-activated channels are disabled; B, D, and F show when medium-voltage-activated channels are disabled; and in all panels top traces show dendritic response and bottom trances somatic response). In contrast to the hand-tuned Schaefer model, the Hay model nicely regenerated dendritic spikes (Fig. 6, C and D, top traces). The timing of somatic action potentials, however, was not similar to experimental data (Fig. 6, C and D, bottom traces). Finally, the Almog model regenerated dendritic spikes (Fig. 6, E and F, top traces) and maintained the timing of somatic spikes as in the physiological experiments (Fig. 6, E and F, bottom traces). Based on these simulations, the Hay model passes the challenge, and the Almog model passes with flying colors.
Fig. 6.
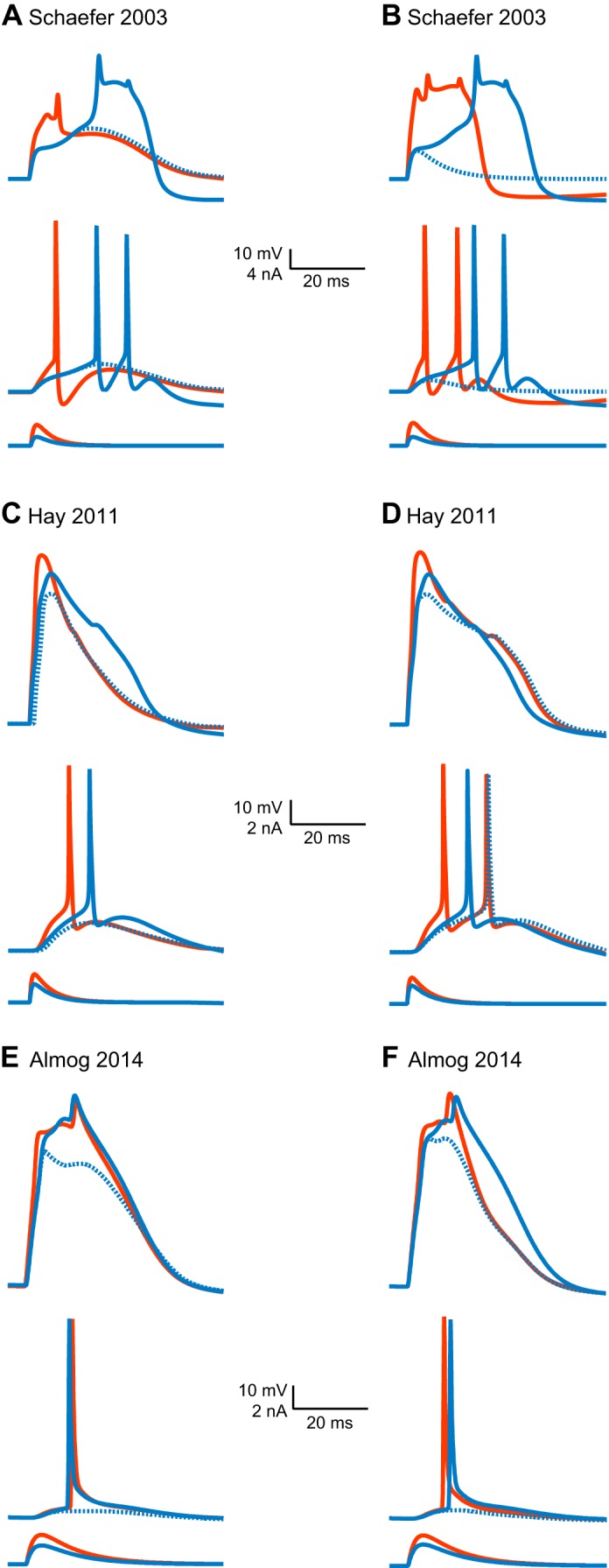
Regeneration of dendritic spike following partial block of voltage-gated calcium channels. A, C, and E: three layer 5 pyramidal neuron models' voltage response to distal injection (600 μm from the soma) of current with excitatory postsynaptic potential-like waveform (A: 1.8 nA, rising τ = 0.8 ms, declining τ = 4 ms; C: 1.5 nA, rising τ = 0.5 ms, declining τ = 5 ms; E: 1 nA, rising τ = 2 ms, declining τ = 10 ms). In each, current injection evoked a dendritic calcium spike, which forward propagated toward the soma and generated action potentials. In all panels, the dendritic response is shown at the top and the somatic at the bottom. Color codes correspond to the level of current injected in each case. Changing the permeability of the high-voltage-activated calcium channel (PHVA) calcium channel conductivity to zero in all models reduced the amplitude and the shape of the calcium spike. However, larger injections of current (A: 3 nA, rising τ = 0.8 ms, declining τ = 4 ms; C: 2 nA, rising τ = 0.5 ms, declining τ = 5 ms; E: 1.35 nA, rising τ = 2 ms, declining τ = 10 ms) regenerated the dendritic calcium spike. B, D, and F: just as described above for A, C, and E, except it was the permeability of the medium-voltage activated calcium channel (PMVA) that was changed to zero. It reduced the amplitude and the shape of the calcium spike only in the Hay and Almog models; in Schaefer model a subthreshold response was generated. Larger current injection (B: 4.7 nA, rising τ = 0.8 ms, declining τ = 4 ms; D: 2 nA, rising τ = 0.5 ms, declining τ = 5 ms; F: 1.2 nA, rising τ = 2 ms, declining τ = 10 ms) regenerated the dendritic calcium spike.
challenge iii: a relative refractory period follows the dendritic calcium spike.
It has been shown that, like the action potential, the dendritic calcium spike in layer 5 pyramidal neurons has a relative refractory period of ∼250 ms (see Fig. 2 in Larkum and Zhu 2002). We challenged the three models to reproduce this finding. The Schaefer model neuron had a refractory period of ∼100 ms (Fig. 7A), the Hay model neuron a refractory period of ∼150 ms (Fig. 7B), and the Almog model neuron a refractory period of ∼400 ms (Fig. 7C). None of the models passed this challenge. It should be noted, given experimental variability, the level of reproducibility one demands from the model, and the fact we compare the model to results from a single paper, that the models might be considered as passing this challenge. However, since no model reproduced both spike timing and shape, we will sustain our verdict of fail.
Fig. 7.
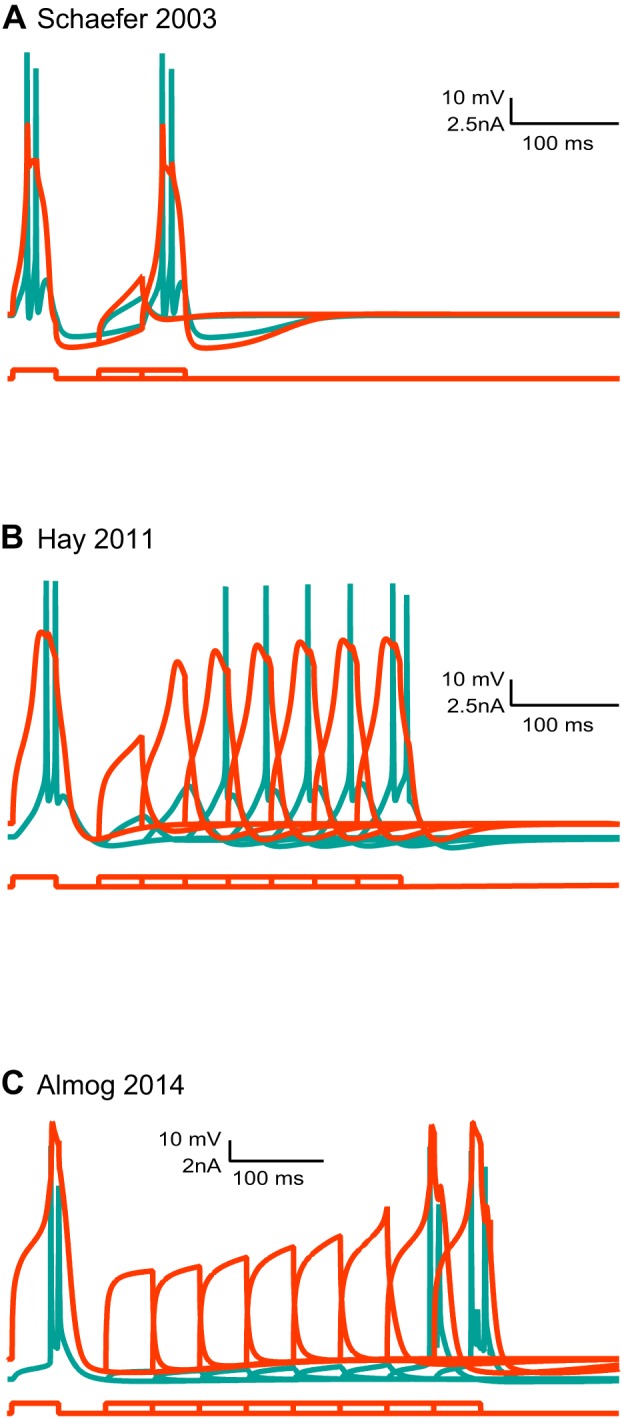
The refractory period of dendritic calcium spikes in different models. A paired depolarizing current of 50 ms with an increasing time interval (Δt = 50 ms) was injected to the distal apical dendrite (600 μm) of 3 different layer 5 pyramidal models (A: 1 nA; B: 0.6 nA; C: 1 nA). The somatic recording is depicted in cyan, and the dendritic one in red. A: Schaefer model reproduced a short refractory period of dendritic calcium spikes. B: Hay model reproduced a short refractory period of dendritic calcium spikes. C: Almog model reproduced a long refractory period of dendritic calcium spikes.
These three challenges, as well as other challenges not reported here, demonstrate that the two computer-optimized models performed substantially better than the hand-tuned model. None of the models, though, passed all of the challenges. While the three models we challenged had full neuronal morphologies, we also tested a recent, computer-optimized, model of layer 5 pyramidal neurons with a reduced morphology (Bahl et al. 2012). This model, performed similarly to the Almog and Hay models, passing two out of three challenges.
Modeling of the axon initial segment.
All three of the models featured above have poorly functioning axons. This is remarkable for its stark contrast to the relative realistic reconstruction of soma and dendrites. While early conceptual modeling of the axon initial segment demonstrated that the generation of action potentials and their invasion into the dendritic tree depends on the density of sodium channels in the axon initial segment, as well as the axon initial segment's shape and passive properties and the somato-dendritic compartment's weak excitability (Dodge and Cooley 1973; Moore et al. 1983), the Mainen and Sejnowski (1996) model axon has an unrealistically high conductance density of 30,000 pS/μm2 in the axon initial segment and, as a result, has an unrealistic shunt in the axon initial segment that increases filtration induced by the membrane of the axon initial segment and leads to poor somatic invasion of the axonal action potential. And this is the unrealistic axon used, as we mentioned above, in the Schaefer and Almog models. By contrast, but even more unrealistic, in the original Hay model, there was no axon.
Extending out from the soma, unmyelinated, roughly 50 μm, the axon initial segment is as biophysically complex as the apical dendrite of layer 5 pyramidal neurons. (When acolytes err in judging the axon initial segment far more complex than the apical dendrite, they underestimate the apical dendrite but do not overestimate the axon initial segment.) In the early days of intracellular recording, it was suggested that the action potential is generated at the axon initial segment, and that it then propagates forward through the axon and backward through into the soma (Araki and Otani 1955; Coombs et al. 1957a, 1957b; Fuortes et al. 1957). These suggestions were verified almost half a century later with simultaneous recordings from layer 5 pyramidal neurons' somata and axon initial segments (Colbert and Johnston 1996; Hausser et al. 1995; Stuart et al. 1997a; Stuart and Hausser 1994; Stuart and Sakmann 1994). Indeed, today it seems that, in layer 5 pyramidal neurons, it is at the distal end of the axon initial segment that the action potential is generated (Hu et al. 2009; Palmer and Stuart 2006; Popovic et al. 2011), although under some conditions it is generated at the first node of Ranvier. Distal initiation of the action potential has also been observed in CA3 pyramidal neurons (Meeks and Mennerick 2007), Purkinje neurons (Foust et al. 2010; Palmer et al. 2010), and in neocortical inhibitory interneurons (Li et al. 2014). That said, several studies have clearly shown complex crosstalk between the initiation zone and the soma (Foust et al. 2010; Khaliq and Raman 2006; Palmer and Stuart 2006).
The complexity of the axon initial segment derives from the composition and distribution of its voltage-gated ion channels, especially the voltage-gated sodium ion channels. [While some studies have found other voltage-gated channels to be important, we pass over them here to keep from straying too far from realistic modeling (for reviews see Bender and Trussell 2012; Kole and Stuart 2012).] Early patch-clamp recordings suggested that the voltage-gated sodium channel density in the axon initial segment is similar to that of the soma (Colbert and Johnston 1996; Colbert and Pan 2002), but recently it was shown that the patch-clamp technique is probably inappropriate for determining the density of ion channels in the axon initial segment because of how these channels are anchored in the cytoskeletal with Ankyrin-G, and that this suggests that, as models of the action potential predicted, the sodium channel density is high in the axon initial segment (Kole et al. 2008). Indeed, converging findings from labeling studies (Lorincz and Nusser 2008; Royeck et al. 2008) and sodium imaging (Baranauskas et al. 2013; Fleidervish et al. 2010; Palmer and Stuart 2006) suggest that the sodium channel density in the axon initial segment may be substantially larger than the soma, roughly ∼4,000 pS/μm2. Furthermore, using electrophysiology and immunohistochemistry, it was elegantly demonstrated that the axon initial segment has two different voltage-gated sodium channels, Nav1.2 and Nav1.6, that these channels have graded distributions along the membrane there (Hu et al. 2009), Nav1.6 has a higher density in the distal part of the initial segment, and that this anatomical complexity explains much of the axon initial segment's physiological complexity.
Several recent models have aimed at understanding the biophysics of the axon initial segment. Hu and collaborators, in the same study where they showed the graded densities of Nav1.2 and Nav1.6 (Hu et al. 2009), constructed a compartmental model of the initial segment using the Mainen and Sejnowski (1996) model as a backbone. The two sodium conductances were described using Hodgkin-Huxley formalism, inserted into the initial segment, and the model was hand-tuned. The model suggested that Nav1.6 is responsible for action potential initiation in the distal initial segment, while somatic Nav1.2 promotes action potential back-propagation. Unfortunately, the model lacked any description of the dendritic calcium spike initiation zone. Other modeling studies of layer 5 pyramidal neurons have been conducted in the same vein. Colbert and Pan (2002) observed functional differences between Nav1.6 and Nav1.2 in the axon initial segment, in particular, the prominent shift in the channels' activation curves, and interpreted them using a compartmental model; they manually adjusted the axon initial segment of a previously hand-tuned model of a CA1 hippocampal pyramidal to account for the observed changes in channel kinetics (Colbert and Pan 2002). In two important studies using sodium ion imaging in the axon initial segment (Baranauskas et al. 2013; Fleidervish et al. 2010), Fleidervish and Baranauskas (2010) and their collaborators used a compartmental model with a reduced, artificial morphology and a small set of voltage-gated ion channels to aid interpretation of the imaging data. The study by Kole and collaborators (Kole et al. 2008) applied a hand-tuned modification of the Mainen and Sejnowski (1996) model in which several of the ion channels were replaced to better tune the model to the dynamics of the axon initial segment, while a more recent study from the same laboratory demonstrated that Kv7 channels regulate the initiation and propagation of the action potential (Battefeld et al. 2014) in part by using a hand-tuned modification of a hippocampal granule cell initiation zone model (Schmidt-Hieber and Bischofberger 2010) containing a Markov model for the voltage-gated sodium channel. All these models (as well as others not mentioned here) were hand-tuned to perform a specific task and accordingly must be regarded as limited, perhaps suffering from wildly unrealistic parameter values and, in a sense, ad hoc. Several reduced models of action potential firing are available. See Brette (2015) for a recent intelligent comparison between several of these models.
But hand-tuning these models and tuning them for the sake of a specific task are not the only problems. Whereas the apical dendrite models of Hay and Almog succeeded because they were tuned using experimental data, no model of the axon has used such a data set. And yet such data are necessary to automatically constrain a model of the axon initial segment (Huys et al. 2006; Keren et al. 2005). So, although researchers have made great strides in understanding axonal excitability in cortical neurons over the past decade, we have made little progress in realistically modeling the axon initial segment and action potential generation in layer 5 pyramidal neurons.
Remarkably, the Hay model, although it lacks an axon, is able to generate bursts of high-frequency action potentials (Hay et al. 2011), a capability rather limited in the Almog and Schaefer models (Almog and Korngreen 2014; Schaefer et al. 2003b). It is able to do so because it includes a slowly inactivating, or persistent, voltage-gated sodium channel. Persistent sodium currents have been observed in layer 5 pyramidal neurons from the early days of whole-cell recordings (Crill 1996; Schwindt and Crill 1980; Stafstrom et al. 1982, 1984a, 1984b) and, together with resurgent voltage-gated sodium currents, greatly increase the contribution of sodium conductance to the action potential (Lewis and Raman 2014; Raman and Bean 2001, 1997; Raman et al. 1997). Layer 5 pyramidal neurons' persistent sodium currents are localized primarily in the axon initial segment (Astman et al. 2006; Stuart and Sakmann 1995). They affect action potential threshold (Kole et al. 2008; Kole and Stuart 2008) and contribute to the prominent spike observed after depolarization in these neurons (Astman et al. 2006; Brumberg et al. 2000; Fleidervish et al. 1996; Fleidervish and Gutnick 1996). Detailed kinetic analysis of Nav1.2 and Nav1.6 reveals that each of these channels displays both fast and slow kinetics (Rush et al. 2005). For these reasons, models of the axon initial segment must take into account these persistent sodium currents. The Hay model, as well as other models, treats the persistent sodium current and the transient sodium current as two distinct kinetic mechanisms; in fact, practically speaking, since it was programmed using NEURON, the two conductances were expressed using two separate MOD files. To avoid splitting kinetic features of the same channels into two separate MOD files, it may be possible to use Markov chain models, as we have discussed above.
In summary, it appears as though models of the axon initial segment are currently inferior to those of dendrites. However, this may be due to slower scientific progress in this area. High-quality recording of the membrane potential in the axon have been available for only ∼20 years. This, and the slow adaptation of automatic optimization methods, may be a primary reason why we have yet to see a computer-optimized model for the axon initial segment.
Can We Realistically Model Single Neurons?
Our opinion is that we currently can model single neurons effectively, but not realistically. But what is it to model effectively? What is it to model realistically? These are philosophical questions (Rosenblueth and Wiener 1945), and so, just as we began by turning to the classical neurophysiological methods of Hodgkin and Huxley, now we turn to the classical philosophical method. Let us eavesdrop on a dialog between the great neurophilosopher Neurates and his aspiring student Modelophon. But first a note to the reader. The Socratic dialogue is a logical tool in which two persons (often a teacher and a student) disagree about a given subject. To drive the discussion properly, one of them must be wrong at least in part. Obviously, we decided that the student would be naive and express opinions we all too often hear from undergraduate and graduate students. Furthermore, in many Socratic dialogues, the teacher uses irony and sarcasm to drive the discussion. Thus, clearly, some of the statements are exaggerated. You may be annoyed by some of the statements made by Neurates or Modelphon. Pray forgive us in advance, the dialogue is not intended to upset you, it is only a logical algorithm attempting to stimulate discussion (Kreeft and Dougherty 2010).
Modelophon: We should create realistic compartmental models of neurons and use them to generate realistic neural networks.
Neurates: My dear Modelophon, what is a realistic model of a neuron?
Modelophon: It is a model of a neuron containing a realistic morphology, passive parameters, voltage-gated channels, and ligand-gated channels. The model behaves the same as the neuron does. Therefore, it is realistic and will perform the same calculation as the real neuron when inserted into the network.
Neurates: A useful definition! Now I know what it should be made of, and already, my clever student, you've surprised me. For wouldn't you say that the neuron and the channels, being realistic, must be modeled at the atomic level? Perhaps it must take into account also the quantum realm? Or should we know only the three-dimensional structure of all the molecules involved?
Modelophon: Not at all. We do not need so many details. The morphology of the neuron is reduced to a set of cylinders that act as capacitors and resistors, and the channels are modeled according to the time-honored formalism of Hodgkin and Huxley.
Neurates: But Modelophon, I do not understand. If the model is not made from atoms and bonds, how can it be realistic?
Modelophon: Well, when I say, it's realistic, what I mean is that the model simulates the behavior of the neuron accurately.
Neurates: Ah, thank you for making that clear. But do you think it's possible?
Modelophon: What do you mean?
Neurates: Take, for example, the kinetic formalism suggested by Hodgkin and Huxley which you wish to use; is it a realistic model for voltage-gated channels?
Modelophon: No, it is approximate, and studies have shown that Markov models better describe ion channel kinetics, but even Markov models are approximate.
Neurates: How approximate, I wonder. Did you record and analyze the ion channel kinetics yourself using the voltage-clamp technique?
Modelophon: No, I downloaded an existing kinetic script from the model database.
Neurates: Then how do you know if this computer representation of the channel is a good one? Did you verify that the database entry correctly describes the channel you need for your simulation?
Modelophon: We trust the scientist who contributed these kinetic models to the database.
Neurates: Do we?
Modelophon: If they are not good, then certainly they would have been routed out of the database.
Neurates: Unless everyone made that same reasonable assumption…. So you are not using a realistic model of the channels you introduce into the model, only one that you believe is accurate enough (let's call it a model of some biological detail), and you have not had personal encounter of the third kind with the ionic current generated by the channel, so you do not know how far the model is from reality [to the extent we can define reality in science (Rosenblueth and Wiener 1945)]. But still you want your model to be realistic. Even if your belief that the Hodgkin and Huxley kinetics are sufficiently accurate is true, it is unjustified. What test do you propose to justify the use of Hodgkin and Huxley's kinetics so that you may call your model realistic?
Modelophon: The test is whether the model reproduces the behavior of real neurons. I quantitatively compare the model with the behavior of the real neuron. If I see that they do not match, then I modify the model so that they do. Then I test the model against real neurons again, and so on, 'til after reiterating the procedure, I have a model that behaves as reality does.
Neurates: So you define a realistic model as one that faithfully mimics behavior. Do you compare the model to all the behaviors of the real neuron?
Modelophon: No, we compare the model to a few membrane potential traces recorded from the soma and in a few cases from the apical dendrite as well. But these are representative of the behaviors we care about for our model.
Neurates: By Zeus–you are like the god himself. You make things whatever you wish. You said that you would make a realistic model, and then you defined realistic as being behaviorally accurate. Now I have asked you about being accurate, and you have told me that it is only being accurate about what you care for. This is quite a trick, let's see if you can do it in reverse. When you are searching for the best model, do you compare the behaviors of the real neurons to all the possible models?
Modelophon: I cannot simulate and compare all possible permutations. But fortunately the task is no longer entrusted to humans. We trust in large high-performance parallel computer searches of the model's parameter space. That's how we find the model that behaves best.
Neurates: Wonderful. No doubt high-performance parallel computing will solve many of humanity's most important problems. But if you, my excellent student, cannot search all possible parameter combinations, how can a silicon machine which you program?
Modelophon: Well, there are many parameters of unknown value, and so there is a practically huge solution space. The computer applies smart search algorithms to sample this space wisely.
Neurates: A supercomputer wisely searching an infinite space–a smart computer indeed. But can these algorithms overcome the curse of dimensionality?
Modelophon: Oh, no. In general not even the computers can try all permutations.
Neurates: Pity. But perhaps you only need more data to more rigorously constrain your wandering among parameter combinations. You said that the target data set was recorded from the soma and occasionally also from the dendrite. Is this data sufficient for computer optimization of a full realistic model for the neuron?
Modelophon: Alas no. It has been shown by Keren et al. (2005) and by Huys et al. (2006) that many more recordings along the axonal-somato-dendritic axis should be performed simultaneously and included in the target data set to allow the optimization algorithm to find the optimal solution.
Neurates: I see. How unfortunate. But dauntless you wish to use imperfect kinetic models for ion channels, optimization algorithms that are susceptible to the curse of dimensionality, and incomplete data sets, and you believe you can still provide a realistic model for the neuron?
Modelophon: Yes, because, despite these shortcomings, my optimized model can faithfully reproduce the behavior of the neuron. Therefore, it is realistic and may be used as a building block in realistic neural networks.
Neurates: Are you not confusing two different matters–reproducing the behavior of the single neuron and using it as a building block in a network?
Modelophon: But they are one and the same. If the individual neurons are realistic, then they must be sufficient to build a network.
Neurates: Yet when Almog and Korngreen had three compartmental models compete for the laurel, none passed the third and final challenge. Thus, regardless if you are an acolyte of rate or temporal coding, they do not reproduce the computation performed by the neuron.
Modelophon: It's true, Neurates.
Neurates: Let us discuss the third challenge. Following a dendritic calcium spike, the apical dendrite and the soma hyperpolarizes (Fig. 7), and so there is a spike-induced refractory period (Larkum and Zhu 2002). Might this refractory period affect the firing rate and pattern of the neuron?
Modelophon: Yes, this refractory period is most likely to reduce action potential firing at the axon initial segment. In fact, Almog and Korngreen concur. They induced neurons to fire regularly, and found that, following the dendritic calcium spike, the axonal firing rate decreased (it is not shown in their figures).
Neurates: Might such a change in the firing rate be an important feature of the computational contribution made by layer 5 pyramidal neurons to their networks?
Modelophon: It is possible.
Neurates: Can you propose an experiment to determine which features in the cellular model are relevant for network computation and which are not? For without determining what matters, by what standard will we judge whether a model represents the behaviors essential for its being a network building block?
Modelophon: No, I do not know of such experiment.
Neurates: Can you then know from your model what is the input-output transformation of the neuron you are modeling?
Modelophon: Not really, I assume that by modeling the neuron with enough detail I ensure accurate synaptic integration. But, alas, I have no test to verify this assumption.
Neurates: Then however effective compartmental models may be, they cannot be justified as building blocks for network computation.
Modelophon: Master Neurates, I will not try to sway your opinion that compartmental models are limited as network building blocks, but why do you say that they are effective? Have you not dismissed them completely?
Neurates: Is it not the case, Modelophon, that each of the models mentioned in this dialog's prolog contributed valid information to the papers in which it appeared? The Schaefer model was instrumental in explaining the contribution of morphology to the observed variability in dendritic calcium spikes. The Hay model successfully used experimental variability to generate a library of possible neuronal models. The Almog model suggested an ionic mechanism for the generation of a dendritic spike. And all the models describing the generation of action potentials in the axon initial segment were instrumental in testing working hypotheses and suggesting biophysical mechanisms relevant to the study they were part of. So each model reviewed by Almog and Korngreen in the context of the study for which it was created was successful and effective.
Modelophon: Then wherein lies the problem? Why can we not use current compartmental models for network computation, nor call them realistic?
Neurates: The problem grows from an old and deep root. The brain–nature in general–has many layers of detail. There are many parameters we agree that are needed to define a model for describing the physiology of a single neuron, but we do not know, nor do we presently have the tools to discover, which of these manifold features is relevant to the function of the neuron in the network.
Modelophon: Then … nothing to be done?
Neurates: If you choose. But I am less pessimistic and defeatist. Perhaps Almog and Korngreen may offer a few suggestions.
Contra the rising tide aiming at massive use of compartmental models for network computation (Markram 2006, 2012; Markram et al. 2015), we here suggest that these proposals are mastodonic and cannot deliver the fantasies they promise; they cannot as long as our state-of-the-art remains ad hoc, unrealistic compartmental models which function poorly out of their contexts. We may take heart that the problem we face shares its basic nature with deep problems throughout systems biology and other fields: how to cross safely between levels of nature, preserving features of the “lower,” elemental level which are important to what emerges on the “higher,” whole system level while discarding second-order details? This is no trivial request, and we have yet to acquire our visas.
Let us consider a more specific and practical form of the question: how may we advance toward regularly generating reusable realistic models? Progress over the past decade in automatic optimization may provide a preliminary answer. As we showed above, models optimized using genetic algorithms responded better than models otherwise optimized. One might deduce from this that computer-optimized models should be used exclusively from now, and that hand-tuning is no good. We are not of this opinion. Hand-tuned models have been, and will continue to be, instrumental in a plethora of projects as long as their assumptions are not violated and their results are interpreted within the context for which they were created. It is important to remember that every parameter should be reviewed during hand-tuning and, even if it is set subjectively, should be able to be scientifically rationalized. Computer-optimized models represent only a fraction of the total number of compartmental models published to date. Furthermore, the handful of optimization methods used to date to optimize models is just a small fraction of the toolbox available for modeling in science. In addition, there are statistical methods including regression, machine-learning methods, some of which providing important estimates of variation. Furthermore, to probe model dynamics, systems identification methods exist. Thus computer-optimized models may in the future become building blocks for more complex simulations, but, before this becomes realistic, many steps are necessary. One such step should be, for example, the quantification of the impact made by the initial selection of parameters by the modeler prior to computer optimization, since even computer optimization is not free from informed human decisions.
One trivial but important conclusion to draw from all the work cited above is that there is an intimate link between the model being optimized, the target data set, and the cost function. The selection of these elements has a great impact on the outcome of the optimization process. (Fig. 8 presents a possible integrated view of the relationships between the differ parts of the automatic optimization process, but many of the possible interactions depicted in this figure have yet to be investigated.) It is should now be possible, with the increase in computing power, to quantitatively investigate the link between model, cost function, and target data set. We are still unable to answer what is the proper data set required to optimize a specific model. This is a cardinal problem since the optimization studies we cited in this review used data sets recorded from quiescent neurons in brain slices that were treated pharmacologically to block synaptic activity. Thus the data set may help optimize a neuron functioning outside of its realistic activity zone. Much more work using surrogate data may be required, at least phenomenologically, to address this issue. Surrogate data can be used to further investigate the impact of experimental and biological noise on the optimization process and to expand our arsenal of sophisticated optimization algorithms.
Fig. 8.

Interleaved interactions between components of automatic model optimization.
While in the immediate future individual laboratories can perform these simple, although time-consuming tasks, collaborative effort may greatly enhance our progress toward more detailed neuronal models. Currently, compartmental models and ion channel models are contributed to databases on a voluntary basis following the publication of a manuscript. The most extensive database is ModelDB (Crasto et al. 2007; Hines et al. 2004; McDougal et al. 2015; Migliore et al. 2003; Morse 2008; Peterson et al. 1996; Ranjan et al. 2011). It is common practice in the modeling community to scavenge within this and other public databases for code snippets and even full models. This practice is typical of the attitude displayed by programmers all over the world who scavenge the internet for available code. Personal experience has taught us that this worldwide trawling greatly reduces the time required for code development.
That said, when downloading a full or partial compartmental model, we must acknowledge that we are not downloading just a simple piece of computer code. Rather, to generate this model, the scientist made a series of assumptions that must be taken into account when developing our model. The downloaded code is a description of a physical system. Downloading and reusing a model from a public database launders the model clean of its physical structure, since the assumptions are not explicitly mentioned in the code or the database entry, and are often absent from the original paper. The laundered piece of code is then used within new models without regard to its original biological and physical limitations. This fault line between the original study and the database is severe. To compound this problem, it is very difficult, when using a downloaded model, to verify that the code is well written. There are practically no model validation techniques or tools available. This is especially problematic in case of large and complex models containing many lines of code. Consequentially, many assume the downloaded model to be correct, since the paper describing it was peer reviewed. However, current review process does not include scrutiny of the code. Papers are reviewed only based on their text and figures. This basic flaw in peer review of computational papers must be addressed. Finally, since relatively few models have been validated over the years, the same models are reused in a large number of studies without regard to context and documentation.
Basic structural changes in the model sharing system are needed. Database entries must better link to the studies for which the models were created, including explicitly listing the assumptions made during the modeling processes. Furthermore, as we advance toward wider use of computer optimization for neuronal models, the database should contain the data sets with which models were optimized. Without these target data sets, it is not possible to reproduce optimization results. And yet, if all this work is loaded upon scientists, we may receive considerably fewer database entries. In contrast with the fields of genomics and proteomics where contributing data sets to a central database is acceptable and common practice, electrophysiologists tend to keep their data and models inside their laboratories: there is simply nothing to gain by taking the pains to format the data, document it, and submit it to a database. One powerful incentive to comply with stricter database rules would be to have the data set and model contributions considered as scientific publications in and of themselves. These contributions would be peer-reviewed and, once online, citable, thus contributing to the publication list of the scientist. This can help in evaluating the quality of the data set by the number of times it will be used and cited by the community.
Publicly available databases for electrophysiological data can be used not only as hunting grounds for data-hungry theoreticians. Such data sets can be used as online seeds for model improvement. A model produced from a public data set may be reviewed as an addendum to that data set, uploaded to the same database, and linked to the original entry. For high-quality data sets, several models should be presented. Thus related models will be scored and provide a natural, community-based evolutionary mechanism for improving models. If, 60 years ago, Hodgkin and Huxley could have uploaded their data set, how many more models would have been linked to that database entry today? Imagine the fun we could have had. Take as a contemporary example the ongoing search for the mechanism of action potential generation in the axon initial segment. As we mentioned above, it seems that one crucial mechanism in action potential generation is the kinetic complexity of the two types of voltage-gated sodium channels in the axon initial segment, Nav1.2 and Nav1.6, both displaying fast and sustained kinetic components (Astman et al. 2006; Fleidervish et al. 1996; Fleidervish and Gutnick 1996; Hu et al. 2009; Rush et al. 2005). Constraining a detailed Markov model for these important conductances may be a vital step in understanding the biophysical mechanism of action potential initiation at the axon initial segment. If we were to use a high-grade data set as a seed for model development and link contending Markov models to the same database entry, we would have a tool for incrementally approaching more realistic descriptions of the axon initial segment.
Moreover, once contribution of data to the database becomes a standard procedure, the data sets will be used to greatly improve compartmental models. For example, it is possible to tentatively identify ill-constrained parameters by optimizing a model on several different data sets and then calculating the variance of the constrained parameters across data sets (Almog and Korngreen 2014; Gurkiewicz et al. 2011; Keren et al. 2005, 2009). It has even been shown that, if we could use a data set containing membrane potential traces from each compartment of the neuron to be modeled, we could use a simple gradient decent optimization algorithm instead of a stochastic one (Huys et al. 2006). Ten years ago that thought might have seemed far-fetched, but today, with the rapid advances in voltage-sensitive dye imaging (Hochbaum et al. 2014; Kralj et al. 2012), we may soon have much better data sets required for single-neuron optimization. In addition, using one or more additional data sets in which one (Nowotny et al. 2007) or more (Almog and Korngreen 2014; Keren et al. 2009) ion channels have been pharmacologically blocked greatly increases the ability of the optimization algorithm to fit the data to the model. Publicly available data sets will prove invaluable also for the ongoing process of developing optimization algorithms, testing score functions, and investigating the interplay between these two components and the data set (Fig. 8).
The third and likely the most important foundation stone of model optimization is the model itself, but it is also the weakest element of the process. Even with the best data set and excellent optimization algorithm, a model failing to describe an essential feature of the data will not be able to fit the data. If Modelophon is lucky, the optimization algorithm will not fit the model to the data and thus suggest a change in models, but, if Modelophon is unlucky, which is much more often the case, the model will fit the data nicely and produce a biophysically invalid parameter set. Such ill-fitted model is among the leading causes of computational neuroscientists waking up screaming in the middle of the night. It is hard to detect these fitting problems (where they be over- or underfitting of the model to the data). Global fitting of the model to a large enough data set, as we discussed above, may help. Fitting the model to several pharmacologically perturbed data sets, testing the optimized model using surrogate data, and challenging the model to reproduce a pharmacologically perturbed data set may also be the path to freedom from the trap and shackles of bad fitting and the curse of dimensionality.
Even with future advances in coding languages dedicated to compartmental modeling, a basic problem in generating a compartmental model remains: human bias. There are many ways to write a computer program, and there are many small decisions each one of us makes while developing a compartmental model. How to express channel kinetics? Should we use Hodgkin-Huxley formalism or Markov chains? How do we parameterize channel kinetics? What assumptions should we make regarding channel gradients and densities? Often we miss several parameters and adjust channel kinetics manually. Even the idiosyncratic way each one of us names variables and constants is a barrier for model sharing. It is no wonder that hand-tuned models fair so badly when challenged. Indeed, the mere partial success of the computer-optimized models may derive from the fact that the computers have optimized a model hand-crafted by a human programmer. It is fair to assume that every model contains human-induced errors, and so it is probable that automatic curve fitting will misadjust some of the model's parameters to fit the model to the data. Consequently, we arrive at nice fits with less-than-optimal models, as we demonstrated above by challenging these computer-optimized models. This over-fitting is almost impossible to detect. Since compartmental models are complex it is relatively easy to obtain a good fit between the data and the model. A partial solution to this problem may be to automatically modify also the structure of the model. This has been successfully implemented with Markov models of ion channels (Menon et al. 2009) but not yet with compartmental models of neurons.
So, can we realistically model single neurons? Sadly, we must answer that, at present, we cannot. However, the progress made over a single decade is promising. Peer-reviewed databases containing electrophysiological recordings alongside with standard computer languages describing compartmental models need to be updated (or reinvented). Development of code should be upgraded and standardized to include all the modern code development tools. Compartmental models should be developed in such a way that code will be modular, readable, and sharable. International collaborations, such and the Open Source Brain, the Human Brain Project, and the American BRAIN initiative, are striving to address these issues. Code review should be an integral part of the scientific review. Model optimization should be performed on data sets which are as large as possible, and several models ought to be tested on the same data set. Conversely, each model should be tested using several data sets and challenged to display behavior for which it was not optimized. Finally, we must transcend the boundaries between the electro-centric field of compartmental modeling and the chemo-centric field of system biology (De Schutter 2008) (Fig. 8).
Beyond all these important tasks awaits one challenge surpassing them all. We need to prove that compartmental models are valid within the context of network simulations. Without such proof, the field of compartmental models will be forever bound to single neurons. In constructing a compartmental model for a single neuron, all you need to do is to make assumptions about morphology, passive parameters, and ion channels. However, and this is a big however, you have not proven that your creation does what it is supposed to do at the network level, realistically transform synaptic input to spike trains. Scientists using compartmental model for network computation implicitly make this assumption. They claim that since they have with great detail recreated the cellular behavior of the neuron, then it is possible to use it as a building block in the network. This hypothesis remains to be proven. A starting point on the way to address this question may be to map the cellular parameters that affect spiking activity (Zerlaut et al. 2016) and those that do not.
Should We Realistically Model Single Neurons?
This, again, is a philosophical question. The answer may take several forms and depend on the point of view of the investigator. It is our opinion that not only should we attempt to generate realistic models for neurons, we must. Consider the multilevel structure of biological systems. We all agree that a neuron is a cell having many common features with other cells in our body. Like other cells, it contains a genetic code compacted in its nucleus. Expression of proteins from this genetic code happens all the time and is vital for the neuron. Is protein expression important for the computational properties of the neuron? Yes it is. Should we then add it explicitly to compartmental models? Probably not, it has no short-term impact on the electrical activity of the neuron. Should we ignore it completely? Not in the least. There is a vast literature clearly linking the genetic code to neuronal physiology. Modeling long-term processes such as learning and memory has to include a reference to genetic and proteomic processes.
One level above this we find the complex biochemical network controlling the response of the neuron to neuromodulators and second messengers. Should we then add these processes explicitly to compartmental models? The impact of the biochemical network on the computational properties of the neuron is substantial and in many cases shouldn't be ignored (Marder et al. 2014; Marder and Goaillard 2006; Marder and Prinz 2002). Moreover, it has been shown that neuromodulation of a single neuron may impact network activity (Caplan et al. 2014; Marder et al. 2014; O'Leary et al. 2013). Thus a bridge between the biochemical functionality of the neuron and its electrical activity is, in our eyes, a primary goal for compartmental modeling.
Looking sideways, we find the kinetic properties of voltage- and ligand-gated channels and their nonhomogenous distribution along the axonal-somatic-dendritic axis. It is now standard practice to extract channel kinetics and distributions using voltage-clamp recordings, usually with the patch-clamp technique (Hamill et al. 1981; Sakmann and Neher 1995). Given that we have a good approximation for channel kinetics, can we approximate the activity of the neuron to a point-computing device? Many experimental and theoretical studies over the past three decades have clearly demonstrated that a simple integrate-and-fire neuron cannot describe a substantial fraction of both vertebrate and invertebrate neurons properly. The neuron we focused on in this review, the layer 5 pyramidal neuron, is a case in point: we hope that the models we surveyed in this review adequately demonstrated that the computational properties of layer 5 pyramidal neurons cannot be modeled with a single compartment. And so defining the computational properties of single neurons engenders compartmental modeling. Thus experimental and theoretical studies show that realistic modeling of neurons is not optional: it is a must.
Conclusion
Despite great improvements in automated model optimization, compartmental models remain ad hoc unrealistic models. This is due to both intrinsic problems and the curse of dimensionality. That said, the models which we have examined here, as well as numerous others, have all contributed important information within the scope of the study for which they were created. They are not appropriate, however, for general reuse to address other scientific questions. Furthermore, due to their nature and their less-than-realistic virtual axons which they include, their validity within the context of the neuronal network remains to be determined. We share the belief of many scientists that realistic, or biologically detailed, modeling of single neurons is the gateway for realistic modeling of the nervous system, but we cannot ignore that the current state-of-the-art in this key field remains far from satisfactory. The great progress made over the past two decades and the immense expansion in computer power gives us hope that realistic neuronal models may become a reality in the future.
GRANTS
The research in our laboratory is generously supported by the Israel Science Foundation (no. 38/12), the German Israeli Foundation (no. 1091-27.1/2010), and the Bi-national Science Foundation/National Science Foundation Collaborative Research in Computational Neuroscience program (no. 2013905).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
M.A. and A.K. conception and design of research; M.A. and A.K. performed experiments; M.A. and A.K. analyzed data; M.A. and A.K. interpreted results of experiments; M.A. and A.K. prepared figures; M.A. and A.K. drafted manuscript; M.A. and A.K. edited and revised manuscript; M.A. and A.K. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank Dr. Izhar Bar-Gad for helpful comments and suggestions. The authors thank Ziv Almog for graphical design, and Meir-Simchah Panzer for editorial services.
Footnotes
Comprehensive lists of neural simulation tools can be found at http://home.earthlink.net/∼perlewitz/sftwr.html and https://grey.colorado.edu/emergent/index.php/Comparison_of_Neural_Network_Simulators.
REFERENCES
- Abeles M. Corticonics: Neural Circuits of the Cerebral Cortex. Cambridge, UK: Cambridge University Press, 1991, p. xiv, 280. [Google Scholar]
- Achard P, De Schutter E. Complex parameter landscape for a complex neuron model. PLoS Comput Biol 2: e94, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acker CD, Antic SD. Quantitative assessment of the distributions of membrane conductances involved in action potential backpropagation along basal dendrites. J Neurophysiol 101: 1524–1541, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almog M, Korngreen A. A quantitative description of dendritic conductances and its application to dendritic excitation in layer 5 pyramidal neurons. J Neurosci 34: 182–196, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitai Y, Friedman A, Connors BW, Gutnick MJ. Regenerative activity in apical dendrites of pyramidal cells in neocortex. Cereb Cortex 3: 26–38, 1993. [DOI] [PubMed] [Google Scholar]
- Ananthanarayanan R, Modha DS. Anatomy of a cortical simulator. In: Proceedings of the 2007 ACM/IEEE Conference on Supercomputing. New York: ACM, 2007, p. 411–422. [Google Scholar]
- Antic S, Major G, Zecevic D. Fast optical recordings of membrane potential changes from dendrites of pyramidal neurons. J Neurophysiol 82: 1615–1621, 1999. [DOI] [PubMed] [Google Scholar]
- Antic SD. Action potentials in basal and oblique dendrites of rat neocortical pyramidal neurons. J Physiol 550: 35–50, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T, Otani T. Response of single motoneurons to direct stimulation in toad's spinal cord. J Neurophysiol 18: 472–485, 1955. [DOI] [PubMed] [Google Scholar]
- Ariav G, Polsky A, Schiller J. Submillisecond precision of the input-output transformation function mediated by fast sodium dendritic spikes in basal dendrites of CA1 pyramidal neurons. J Neurosci 23: 7750–7758, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong CM, Bezanilla F. Inactivation of the sodium channel. II. Gating current experiments. J Gen Physiol 70: 567–590, 1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascoli GA, Donohue DE, Halavi NeuroMorpho M. Org: a central resource for neuronal morphologies. J Neurosci 27: 9247–9251, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astman N, Gutnick MJ, Fleidervish IA. Persistent sodium current in layer 5 neocortical neurons is primarily generated in the proximal axon. J Neurosci 26: 3465–3473, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer SM, Rinzel J. Propagation of dendritic spikes mediated by excitable spines: a continuum theory. J Neurophysiol 65: 874–890, 1991. [DOI] [PubMed] [Google Scholar]
- Bahl A, Stemmler MB, Herz AV, Roth A. Automated optimization of a reduced layer 5 pyramidal cell model based on experimental data. J Neurosci Methods 210: 22–34, 2012. [DOI] [PubMed] [Google Scholar]
- Baranauskas G, David Y, Fleidervish IA. Spatial mismatch between the Na+ flux and spike initiation in axon initial segment. Proc Natl Acad Sci U S A 110: 4051–4056, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battefeld A, Tran BT, Gavrilis J, Cooper EC, Kole MH. Heteromeric Kv7.2/7.3 channels differentially regulate action potential initiation and conduction in neocortical myelinated axons. J Neurosci 34: 3719–3732, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkers JM. Distribution and activation of voltage-gated potassium channels in cell-attached and outside-out patches from large layer 5 cortical pyramidal neurons of the rat. J Physiol 525: 611–620, 2000a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkers JM. Properties of voltage-gated potassium currents in nucleated patches from large layer 5 cortical pyramidal neurons of the rat. J Physiol 525: 593–609, 2000b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkers JM, Delaney AJ. Modulation of excitability by alpha-dendrotoxin-sensitive potassium channels in neocortical pyramidal neurons. J Neurosci 21: 6553–6560, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellman R. Adaptive Control Processes: A Guided Tour. Princeton, NJ: Princeton University Press, 1961, p.94. [Google Scholar]
- Bellman R. Dynamic Programming. Princeton, NJ: Princeton University Press, 1957, p. xxv, 342. [Google Scholar]
- Ben-Shalom R, Aviv A, Razon B, Korngreen A. Optimizing ion channel models using a parallel genetic algorithm on graphical processors. J Neurosci Methods 206: 183–194, 2012. [DOI] [PubMed] [Google Scholar]
- Ben-Shalom R, Liberman G, Korngreen A. Accelerating compartmental modeling on a graphical processing unit. Front Neuroinform 7: 4, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender KJ, Trussell LO. The physiology of the axon initial segment. Annu Rev Neurosci 35: 249–265, 2012. [DOI] [PubMed] [Google Scholar]
- Bezanilla F, Armstrong CM. Inactivation of the sodium channel. I. Sodium current experiments. J Gen Physiol 70: 549–566, 1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla US, Bower JM. Exploring parameter space in detailed single neuron models: simulations of the mitral and granule cells of the olfactory bulb. J Neurophysiol 69: 1948–1965, 1993. [DOI] [PubMed] [Google Scholar]
- Bischofberger J, Jonas P. Action potential propagation into the presynaptic dendrites of rat mitral cells. J Physiol 504: 359–365, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower JM, Beeman D. The Book of GENESIS: Exploring Realistic Neural Models with the GEneral NEural SImulation System. Santa Clara, CA: TELOS, 1995, p. xx, 409. [Google Scholar]
- Branco T, Clark BA, Hausser M. Dendritic discrimination of temporal input sequences in cortical neurons. Science 329: 1671–1675, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco T, Hausser M. The single dendritic branch as a fundamental functional unit in the nervous system. Curr Opin Neurobiol 20: 494–502, 2010. [DOI] [PubMed] [Google Scholar]
- Brette R. What is the most realistic single-compartment model of spike initiation? PLoS Comput Biol 11: e1004114, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody M, Korngreen A. Simulating the effects of short-term synaptic plasticity on postsynaptic dynamics in the globus pallidus. Front Syst Neurosci 7: 40, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumberg JC, Nowak LG, McCormick DA. Ionic mechanisms underlying repetitive high-frequency burst firing in supragranular cortical neurons. J Neurosci 20: 4829–4843, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunow B, Segev I, Fleshman JW. Modeling the electrical behavior of anatomically complex neurons using a network analysis program: excitable membrane. Biol Cybern 53: 41–56, 1985. [DOI] [PubMed] [Google Scholar]
- Burrone J, Murthy VN. Synaptic gain control and homeostasis. Curr Opin Neurobiol 13: 560–567, 2003. [DOI] [PubMed] [Google Scholar]
- Bush PC, Sejnowski TJ. Reduced compartmental models of neocortical pyramidal cells. J Neurosci Methods 46: 159–166, 1993. [DOI] [PubMed] [Google Scholar]
- Cannon RC, Gleeson P, Crook S, Ganapathy G, Marin B, Piasini E, Silver RA. LEMS: a language for expressing complex biological models in concise and hierarchical form and its use in underpinning NeuroML 2. Front Neuroinform 8: 79, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon RC, O'Donnell C, Nolan MF. Stochastic ion channel gating in dendritic neurons: morphology dependence and probabilistic synaptic activation of dendritic spikes. PLoS Comput Biol 6: e1000886, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan JS, Williams AH, Marder E. Many parameter sets in a multicompartment model oscillator are robust to temperature perturbations. J Neurosci 34: 4963–4975, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso MF, Salcedo RL, de Azevedo SF. The simplex-simulated annealing approach to continuous non-linear optimization. Comput Chem Eng 20: 1065–1080, 1996. [Google Scholar]
- Carnevale NT, Hines ML. The NEURON Book. Cambridge, UK: Cambridge University Press, 2006, p. xix, 457. [Google Scholar]
- Carnevale NT, Woolf TB, Shepherd GM. Neuron simulations with SABER. J Neurosci Methods 33: 135–148, 1990. [DOI] [PubMed] [Google Scholar]
- Casella G, George EI. Explaining the Gibbs sampler. Am Stat 46: 167–174, 1992. [Google Scholar]
- Cash S, Yuste R. Input summation by cultured pyramidal neurons is linear and position-independent. J Neurosci 18: 10–15, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash S, Yuste R. Linear summation of excitatory inputs by CA1 pyramidal neurons. Neuron 22: 383–394, 1999. [DOI] [PubMed] [Google Scholar]
- Chen S, Montgomery J, Bolufe-Rohler A. Measuring the curse of dimensionality and its effects on particle swarm optimization and differential evolution. Appl Intell 42: 514–526, 2015. [Google Scholar]
- Chen WR, Midtgaard J, Shepherd GM. Forward and backward propagation of dendritic impulses and their synaptic control in mitral cells. Science 278: 463–467, 1997. [DOI] [PubMed] [Google Scholar]
- Chen WR, Shen GY, Shepherd GM, Hines ML, Midtgaard J. Multiple modes of action potential initiation and propagation in mitral cell primary dendrite. J Neurophysiol 88: 2755–2764, 2002. [DOI] [PubMed] [Google Scholar]
- Clark BA, Monsivais P, Branco T, London M, Hausser M. The site of action potential initiation in cerebellar Purkinje neurons. Nat Neurosci 8: 137–139, 2005. [DOI] [PubMed] [Google Scholar]
- Clements JD, Redman SJ. Cable properties of cat spinal motoneurones measured by combining voltage clamp, current clamp and intracellular staining. J Physiol 409: 63–87, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewley R. Hybrid models and biological model reduction with PyDSTool. PLoS Comput Biol 8: e1002628, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert CM, Johnston D. Axonal action-potential initiation and Na+ channel densities in the soma and axon initial segment of subicular pyramidal neurons. J Neurosci 16: 6676–6686, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert CM, Pan E. Ion channel properties underlying axonal action potential initiation in pyramidal neurons. Nat Neurosci 5: 533–538, 2002. [DOI] [PubMed] [Google Scholar]
- Coombs JS, Curtis DR, Eccles JC. The generation of impulses in motoneurones. J Physiol 139: 232–249, 1957a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs JS, Curtis DR, Eccles JC. The interpretation of spike potentials of motoneurones. J Physiol 139: 198–231, 1957b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasto CJ, Marenco LN, Liu N, Morse TM, Cheung KH, Lai PC, Bahl G, Masiar P, Lam HY, Lim E, Chen H, Nadkarni P, Migliore M, Miller PL, Shepherd GM. SenseLab: new developments in disseminating neuroscience information. Brief Bioinform 8: 150–162, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crill WE. Persistent sodium current in mammalian central neurons. Annu Rev Physiol 58: 349–362, 1996. [DOI] [PubMed] [Google Scholar]
- Crook S, Gleeson P, Howell F, Svitak J, Silver RA. MorphML: level 1 of the NeuroML standards for neuronal morphology data and model specification. Neuroinformatics 5: 96–104, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuntz H, Borst A, Segev I. Optimization principles of dendritic structure. Theor Biol Med Model 4: 21, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuntz H, Forstner F, Borst A, Hausser M. One rule to grow them all: a general theory of neuronal branching and its practical application. PLoS Comput Biol 6: e1000877, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuntz H, Forstner F, Borst A, Hausser M. The TREES toolbox–probing the basis of axonal and dendritic branching. Neuroinformatics 9: 91–96, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter E. Computer software for development and simulation of compartmental models of neurons. Computers in biology and medicine 19: 71–81, 1989. [DOI] [PubMed] [Google Scholar]
- De Schutter E. Why are computational neuroscience and systems biology so separate? PLoS Comput Biol 4: e1000078, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb K, Pratap A, Agarwal S, Meyarivan T. A fast and elitist multiopjective genetic algorithm: NSGA-II. IEEE Trans Evol Comput 6: 182–197, 2002. [Google Scholar]
- Denk W, Delaney KR, Gelperin A, Kleinfeld D, Strowbridge BW, Tank DW, Yuste R. Anatomical and functional imaging of neurons using 2-photon laser scanning microscopy. J Neurosci Methods 54: 151–162, 1994. [DOI] [PubMed] [Google Scholar]
- Destexhe A. Simplified models of neocortical pyramidal cells preserving somatodendritic voltage attenuation. Neurocomputing 38–40: 167–173, 2001. [Google Scholar]
- Devor A, Yarom Y. Coherence of subthreshold activity in coupled inferior olivary neurons. Ann N Y Acad Sci 978: 508, 2002. [DOI] [PubMed] [Google Scholar]
- Dodge FA, Cooley JW. Action potential of the motorneuron. IBM J Res Dev 17: 219–229, 1973. [Google Scholar]
- Druckmann S, Banitt Y, Gidon A, Schurmann F, Markram H, Segev I. A novel multiple objective optimization framework for constraining conductance-based neuron models by experimental data. Front Neurosci 1: 7–18, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druckmann S, Berger TK, Hill S, Schurmann F, Markram H, Segev I. Evaluating automated parameter constraining procedures of neuron models by experimental and surrogate data. Biol Cybern 99: 371–379, 2008. [DOI] [PubMed] [Google Scholar]
- Druckmann S, Berger TK, Schurmann F, Hill S, Markram H, Segev I. Effective stimuli for constructing reliable neuron models. PLoS Comput Biol 7: e1002133, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druckmann S, Feng L, Lee B, Yook C, Zhao T, Magee JC, Kim J. Structured synaptic connectivity between hippocampal regions. Neuron 81: 629–640, 2014. [DOI] [PubMed] [Google Scholar]
- Eliasmith C, Stewart TC, Choo X, Bekolay T, DeWolf T, Tang C, Rasmussen D. A large-scale model of the functioning brain. Science 338: 1202–1205, 2012. [DOI] [PubMed] [Google Scholar]
- Eliasmith C, Trujillo O. The use and abuse of large-scale brain models. Curr Opin Neurobiol 25: 1–6, 2014. [DOI] [PubMed] [Google Scholar]
- Engert F, Bonhoeffer T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature 399: 66–70, 1999. [DOI] [PubMed] [Google Scholar]
- Feldman ML. Morphology of the neocortical pyramidal neuron. In: Cerebral Cortex, edited by Peters A, Jones EG. New York: Plenum, 1984, p. 123–200. [Google Scholar]
- Fleidervish IA, Friedman A, Gutnick MJ. Slow inactivation of Na+ current and slow cumulative spike adaptation in mouse and guinea-pig neocortical neurones in slices. J Physiol 493: 83–97, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleidervish IA, Gutnick MJ. Kinetics of slow inactivation of persistent sodium current in layer V neurons of mouse neocortical slices. J Neurophysiol 76: 2125–2130, 1996. [DOI] [PubMed] [Google Scholar]
- Fleidervish IA, Lasser-Ross N, Gutnick MJ, Ross WN. Na+ imaging reveals little difference in action potential-evoked Na+ influx between axon and soma. Nat Neurosci 13: 852–860, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust A, Popovic M, Zecevic D, McCormick DA. Action potentials initiate in the axon initial segment and propagate through axon collaterals reliably in cerebellar Purkinje neurons. J Neurosci 30: 6891–6902, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuortes MG, Frank K, Becker MC. Steps in the production of motoneuron spikes. J Gen Physiol 40: 735–752, 1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini S, Magee JC. State-dependent dendritic computation in hippocampal CA1 pyramidal neurons. J Neurosci 26: 2088–2100, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini S, Migliore M, Magee JC. On the initiation and propagation of dendritic spikes in CA1 pyramidal neurons. J Neurosci 24: 11046–11056, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken W, Purvis L, Butera R. Genetic algorithm for optimization and specification of a neuron model. In: Proceedings of the 27th Annual International Conference of the Engineering in Medicine and Biology Society. New York: IEEE-EMBS, 2005, p. 4321–4323. [DOI] [PubMed] [Google Scholar]
- Gerstein GL, Mandelbrot B. Random walk models for the spike activity of a single neuron. Biophys J 4: 41–68, 1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewaltig MO, Cannon R. Current practice in software development for computational neuroscience and how to improve it. PLoS Comput Biol 10: e1003376, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson P, Crook S, Cannon RC, Hines ML, Billings GO, Farinella M, Morse TM, Davison AP, Ray S, Bhalla US, Barnes SR, Dimitrova YD, Silver RA. NeuroML: a language for describing data driven models of neurons and networks with a high degree of biological detail. PLoS Comput Biol 6: e1000815, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson P, Steuber V, Silver RA. neuroConstruct: a tool for modeling networks of neurons in 3D space. Neuron 54: 219–235, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding NL, Spruston N. Dendritic sodium spikes are variable triggers of axonal action potentials in hippocampal CA1 pyramidal neurons. Neuron 21: 1189–1200, 1998. [DOI] [PubMed] [Google Scholar]
- Golding NL, Staff NP, Spruston N. Dendritic spikes as a mechanism for cooperative long-term potentiation. Nature 418: 326–331, 2002. [DOI] [PubMed] [Google Scholar]
- Goldman MS, Golowasch J, Marder E, Abbott LF. Global structure, robustness, and modulation of neuronal models. J Neurosci 21: 5229–5238, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golowasch J, Goldman MS, Abbott LF, Marder E. Failure of averaging in the construction of a conductance-based neuron model. J Neurophysiol 87: 1129–1131, 2002. [DOI] [PubMed] [Google Scholar]
- Gould E, Woolley CS, Frankfurt M, McEwen BS. Gonadal steroids regulate dendritic spine density in hippocampal pyramidal cells in adulthood. J Neurosci 10: 1286–1291, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe BF, Bonnan A, Frick A. Back-propagation of physiological action potential output in dendrites of slender-tufted L5A pyramidal neurons. Front Cell Neurosci 4: 13, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienberger C, Chen X, Konnerth A. Dendritic function in vivo. Trends Neurosci 38: 45–54, 2015. [DOI] [PubMed] [Google Scholar]
- Grienberger C, Chen X, Konnerth A. NMDA receptor-dependent multidendrite Ca(2+) spikes required for hippocampal burst firing in vivo. Neuron 81: 1274–1281, 2014. [DOI] [PubMed] [Google Scholar]
- Grubb MS, Burrone J. Activity-dependent relocation of the axon initial segment fine-tunes neuronal excitability. Nature 465: 1070–1074, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulledge AT, Kampa BM, Stuart GJ. Synaptic integration in dendritic trees. J Neurobiol 64: 75–90, 2005. [DOI] [PubMed] [Google Scholar]
- Gurkiewicz M, Korngreen A. A numerical approach to ion channel modelling using whole-cell voltage-clamp recordings and a genetic algorithm. PLoS Comput Biol 3: e169, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurkiewicz M, Korngreen A. Recording, analysis, and function of dendritic voltage-gated channels. Pflügers Arch 453: 283–292, 2006. [DOI] [PubMed] [Google Scholar]
- Gurkiewicz M, Korngreen A, Waxman SG, Lampert A. Kinetic modeling of Nav1.7 provides insight into erythromelalgia-associated F1449V mutation. J Neurophysiol 105: 1546–1557, 2011. [DOI] [PubMed] [Google Scholar]
- Halavi M, Polavaram S, Donohue DE, Hamilton G, Hoyt J, Smith KP, Ascoli GA. NeuroMorpho.Org implementation of digital neuroscience: dense coverage and integration with the NIF. Neuroinformatics 6: 241–252, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflügers Arch 391: 85–100, 1981. [DOI] [PubMed] [Google Scholar]
- Harris KM, Jensen FE, Tsao B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation. J Neurosci 12: 2685–2705, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausser M, Mel B. Dendrites: bug or feature? Curr Opin Neurobiol 13: 372–383, 2003. [DOI] [PubMed] [Google Scholar]
- Hausser M, Spruston N, Stuart GJ. Diversity and dynamics of dendritic signaling. Science 290: 739–744, 2000. [DOI] [PubMed] [Google Scholar]
- Hausser M, Stuart G, Racca C, Sakmann B. Axonal initiation and active dendritic propagation of action potentials in substantia nigra neurons. Neuron 15: 637–647, 1995. [DOI] [PubMed] [Google Scholar]
- Hay E, Hill S, Schurmann F, Markram H, Segev I. Models of neocortical layer 5b pyramidal cells capturing a wide range of dendritic and perisomatic active properties. PLoS Comput Biol 7: e1002107, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay E, Schurmann F, Markram H, Segev I. Preserving axosomatic spiking features despite diverse dendritic morphology. J Neurophysiol 109: 2972–2981, 2013. [DOI] [PubMed] [Google Scholar]
- Helmchen F, Svoboda K, Denk W, Tank DW. In vivo dendritic calcium dynamics in deep-layer cortical pyramidal neurons. Nat Neurosci 2: 989–996, 1999. [DOI] [PubMed] [Google Scholar]
- Herde MK, Iremonger KJ, Constantin S, Herbison AE. GnRH neurons elaborate a long-range projection with shared axonal and dendritic functions. J Neurosci 33: 12689–12697, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz AV, Gollisch T, Machens CK, Jaeger D. Modeling single-neuron dynamics and computations: a balance of detail and abstraction. Science 314: 80–85, 2006. [DOI] [PubMed] [Google Scholar]
- Hines M. A program for simulation of nerve equations with branching geometries. Int J Biomed Comput 24: 55–68, 1989. [DOI] [PubMed] [Google Scholar]
- Hines ML, Carnevale NT. Expandin g NEURON's repertoire of mechanisms with NMODL. Neural Comput 12: 995–1007, 2000. [DOI] [PubMed] [Google Scholar]
- Hines ML, Carnevale NT. The NEURON simulation environment. Neural Comput 9: 1179–1209, 1997. [DOI] [PubMed] [Google Scholar]
- Hines ML, Carnevale NT. NEURON: a tool for neuroscientists. Neuroscientist 7: 123–135, 2001. [DOI] [PubMed] [Google Scholar]
- Hines ML, Davison AP, Muller E. NEURON and Python. Front Neuroinform 3: 1, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines ML, Morse T, Migliore M, Carnevale NT, Shepherd GM. ModelDB: a database to support computational neuroscience. J Comput Neurosci 17: 7–11, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochbaum DR, Zhao Y, Farhi SL, Klapoetke N, Werley CA, Kapoor V, Zou P, Kralj JM, Maclaurin D, Smedemark-Margulies N, Saulnier JL, Boulting GL, Straub C, Cho YK, Melkonian M, Wong GK, Harrison DJ, Murthy VN, Sabatini BL, Boyden ES, Campbell RE, Cohen AE. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods 11: 825–833, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin AL. Chance and design in electrophysiology: an informal account of certain experiments on nerve carried out between 1934 and 1952. J Physiol 263: 1–21, 1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF. The components of membrane conductance in the giant axon of Loligo. J Physiol 116: 473–496, 1952a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF. Currents carried by sodium and potassium ions through the membrane of the giant axon of Loligo. J Physiol 116: 449–472, 1952b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF. The dual effect of membrane potential on sodium conductance in the giant axon of Loligo. J Physiol 116: 497–506, 1952c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol 117: 500–544, 1952d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF, Katz B. Measurement of current-voltage relations in the membrane of the giant axon of Loligo. J Physiol 116: 424–448, 1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DA, Magee JC, Colbert CM, Johnston D. K+ channel regulation of signal propagation in dendrites of hippocampal pyramidal neurons. Nature 387: 869–875, 1997. [DOI] [PubMed] [Google Scholar]
- Hoffman F, Hargrove W. Parallel computing with Linux. Crossroads (De Kalb) 6: 23–27, 1999. [Google Scholar]
- Horn R, Lange K. Estimating kinetic constants from single channel data. Biophys J 43: 207–223, 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Tian C, Li T, Yang M, Hou H, Shu Y. Distinct contributions of Na(v)1.6 and Na(v)1.2 in action potential initiation and backpropagation. Nat Neurosci 12: 996–1002, 2009. [DOI] [PubMed] [Google Scholar]
- Huys QJ, Ahrens MB, Paninski L. Efficient estimation of detailed single-neuron models. J Neurophysiol 96: 872–890, 2006. [DOI] [PubMed] [Google Scholar]
- Huys QJ, Paninski L. Smoothing of, and parameter estimation from, noisy biophysical recordings. PLoS Comput Biol 5: e1000379, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhikevich EM. Dynamical Systems in Neuroscience: the Geometry of Excitability and Bursting. Cambridge, MA: MIT Press, 2007, p. xvi, 441. [Google Scholar]
- Izhikevich EM, Edelman GM. Large-scale model of mammalian thalamocortical systems. Proc Natl Acad Sci U S A 105: 3593–3598, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson ME, Cauller LJ. Evaluation of simplified compartmental models of reconstructed neocortical neurons for use in large-scale simulations of biological neural networks. Brain Res Bull 44: 7–17, 1997. [DOI] [PubMed] [Google Scholar]
- Jacobson GA, Diba K, Yaron-Jakoubovitch A, Oz Y, Koch C, Segev I, Yarom Y. Subthreshold voltage noise of rat neocortical pyramidal neurones. J Physiol 564: 145–160, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaslove SW. The integrative properties of spiny distal dendrites. Neuroscience 47: 495–519, 1992. [DOI] [PubMed] [Google Scholar]
- John ES, Jan S, David JH, Kirby LV, Wen-mei WH, Klaus S. High performance computation and interactive display of molecular orbitals on GPUs and multi-core CPUs. In: Proceedings of 2nd Workshop on General Purpose Processing on Graphics Processing Units. Washington, DC: ACM, 2009. [Google Scholar]
- Johnston D. Functional properties of hippocampal dendrites. Dev Neuropsychol 16: 309–310, 1999. [Google Scholar]
- Johnston D, Christie BR, Frick A, Gray R, Hoffman DA, Schexnayder LK, Watanabe S, Yuan LL. Active dendrites, potassium channels and synaptic plasticity. Philos Trans R Soc Lond B Biol Sci 358: 667–674, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston D, Magee JC, Colbert CM, Cristie BR. Active properties of neuronal dendrites. Annu Rev Neurosci 19: 165–186, 1996. [DOI] [PubMed] [Google Scholar]
- Kamondi A, Acsady L, Buzsaki G. Dendritic spikes are enhanced by cooperative network activity in the intact hippocampus. J Neurosci 18: 3919–3928, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastellakis G, Cai DJ, Mednick SC, Silva AJ, Poirazi P. Synaptic clustering within dendrites: an emerging theory of memory formation. Prog Neurobiol 126: 19–35, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy JF, Eberhart RC, Shi Y. Swarm intelligence. San Francisco, CA: Morgan Kaufmann, 2001, p. xxvii, 512. [Google Scholar]
- Keren N, Bar-Yehuda D, Korngreen A. Experimentally guided modelling of dendritic excitability in rat neocortical pyramidal neurones. J Physiol 587: 1413–1437, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren N, Peled N, Korngreen A. Constraining compartmental models using multiple voltage recordings and genetic algorithms. J Neurophysiol 94: 3730–3742, 2005. [DOI] [PubMed] [Google Scholar]
- Khaliq KM, Misra SN, George AL Jr. Impaired inactivation gate stabilization predicts increased persistent current for an epilepsy-associated SCN1A mutation. J Neurosci 26: 10958–10966, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaliq ZM, Raman IM. Relative contributions of axonal and somatic Na+ channels to action potential initiation in cerebellar Purkinje neurons. J Neurosci 26: 1935–1944, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HG, Connors BW. Apical dendrites of the neocortex: correlation between sodium- and calcium-dependent spiking and pyramidal cell morphology. J Neurosci 13: 5301–5311, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Guzman SJ, Hu H, Jonas P. Active dendrites support efficient initiation of dendritic spikes in hippocampal CA3 pyramidal neurons. Nat Neurosci 15: 600–606, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick S, Gelatt CD, Vecchi MP. Optimization by simulated annealing. Science 220: 671–680, 1983. [DOI] [PubMed] [Google Scholar]
- Knight BW. Dynamics of encoding in a population of neurons. J Gen Physiol 59: 734–766, 1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C. Biophysics of Computation. Oxford, UK: Oxford University Press, 1999. [Google Scholar]
- Koch C. Computation and the single neuron. Nature 385: 207–210, 1997. [DOI] [PubMed] [Google Scholar]
- Koch C, Segev I. The role of single neurons in information processing. Nat Neurosci 3, Suppl: 1171–1177, 2000. [DOI] [PubMed] [Google Scholar]
- Kole MH. First node of Ranvier facilitates high-frequency burst encoding. Neuron 71: 671–682, 2011. [DOI] [PubMed] [Google Scholar]
- Kole MH, Ilschner SU, Kampa BM, Williams SR, Ruben PC, Stuart GJ. Action potential generation requires a high sodium channel density in the axon initial segment. Nat Neurosci 11: 178–186, 2008. [DOI] [PubMed] [Google Scholar]
- Kole MH, Stuart GJ. Is action potential threshold lowest in the axon? Nat Neurosci 11: 1253–1255, 2008. [DOI] [PubMed] [Google Scholar]
- Kole MH, Stuart GJ. Signal processing in the axon initial segment. Neuron 73: 235–247, 2012. [DOI] [PubMed] [Google Scholar]
- Kording KP, Konig P. Learning with two sites of synaptic integration. Network 11: 25–39, 2000. [PubMed] [Google Scholar]
- Kording KP, Konig P. Neurons with two sites of synaptic integration learn invariant representations. Neural Comput 13: 2823–2849, 2001a. [DOI] [PubMed] [Google Scholar]
- Kording KP, Konig P. Supervised and unsupervised learning with two sites of synaptic integration. J Comput Neurosci 11: 207–215, 2001b. [DOI] [PubMed] [Google Scholar]
- Korngreen A, Sakmann B. Voltage-gated K+ channels in layer 5 neocortical pyramidal neurones from young rats: subtypes and gradients. J Physiol 523: 621–639, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korogod SM, Kopysova IL, Bras H, Gogan P, Tyc-Dumont S. Differential back-invasion of a single complex dendrite of an abducens motoneuron by N-methyl-d-aspartate-induced oscillations: a simulation study. Neuroscience 75: 1153–1163, 1996. [DOI] [PubMed] [Google Scholar]
- Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat Methods 9: 90–95, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreeft P, Dougherty T. Socratic Logic: A Logic Text using Socratic Method, Platonic Questions & Aristotelian Principles. South Bend, IN: St. Augustine's, 2010, p. x, 399. [Google Scholar]
- Lampert A, Korngreen A. Markov modeling of ion channels: implications for understanding disease. Prog Mol Biol Transl Sci 123: 1–21, 2014. [DOI] [PubMed] [Google Scholar]
- Larkum ME, Kaiser KM, Sakmann B. Calcium electrogenesis in distal apical dendrites of layer 5 pyramidal cells at a critical frequency of back-propagating action potentials. Proc Natl Acad Sci U S A 96: 14600–14604, 1999a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkum ME, Nevian T, Sandler M, Polsky A, Schiller J. Synaptic integration in tuft dendrites of layer 5 pyramidal neurons: a new unifying principle. Science 325: 756–760, 2009. [DOI] [PubMed] [Google Scholar]
- Larkum ME, Zhu JJ. Signaling of layer 1 and whisker-evoked Ca2+ and Na+ action potentials in distal and terminal dendrites of rat neocortical pyramidal neurons in vitro and in vivo. J Neurosci 22: 6991–7005, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkum ME, Zhu JJ, Sakmann B. Dendritic mechanisms underlying the coupling of the dendritic with the axonal action potential initiation zone of adult rat layer 5 pyramidal neurons. J Physiol 533: 447–466, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkum ME, Zhu JJ, Sakmann B. A new cellular mechanism for coupling inputs arriving at different cortical layers. Nature 398: 338–341, 1999b. [DOI] [PubMed] [Google Scholar]
- Lasser-Ross N, Miyakawa H, Lev-Ram V, Young SR, Ross WN. High time resolution fluorescence imaging with a CCD camera. J Neurosci Methods 36: 253–261, 1991. [DOI] [PubMed] [Google Scholar]
- Lavian H, Korngreen A. Inhibitory short-term plasticity modulates neuronal activity in the rat entopeduncular nucleus in vitro. Eur J Neurosci 43: 870–884, 2016. [DOI] [PubMed] [Google Scholar]
- Lavzin M, Rapoport S, Polsky A, Garion L, Schiller J. Nonlinear dendritic processing determines angular tuning of barrel cortex neurons in vivo. Nature 490: 397–401, 2012. [DOI] [PubMed] [Google Scholar]
- LeMasson G, Maex R. Introduction ot equation solving and parameter fitting. In: Computational Neuroscience: Realistic Modelling for Experimentalists, edited by De Schutter E. London: CRC, 2001, p. 1–25. [Google Scholar]
- Lewis AH, Raman IM. Resurgent current of voltage-gated Na+ channels. J Physiol 592: 4825–4838, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Tian C, Scalmani P, Frassoni C, Mantegazza M, Wang Y, Yang M, Wu S, Shu Y. Action potential initiation in neocortical inhibitory interneurons. PLoS Biol 12: e1001944, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Maskell DL, Schmidt B. CUDASW++: optimizing Smith-Waterman sequence database searches for CUDA-enabled graphics processing units. BMC Res Notes 2: 73, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llinas R, Sugimori M. Electrophysiological properties of in vitro Purkinje cell dendrites in mammalian cerebellar slices. J Physiol 305: 197–213, 1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London M, Hausser M. Dendritic computation. Annu Rev Neurosci 28: 503–532, 2005. [DOI] [PubMed] [Google Scholar]
- Lorincz A, Nusser Z. Cell-type-dependent molecular composition of the axon initial segment. J Neurosci 28: 14329–14340, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz A, Nusser Z. Molecular identity of dendritic voltage-gated sodium channels. Science 328: 906–909, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losonczy A, Magee JC. Integrative properties of radial oblique dendrites in hippocampal CA1 pyramidal neurons. Neuron 50: 291–307, 2006. [DOI] [PubMed] [Google Scholar]
- Losonczy A, Makara JK, Magee JC. Compartmentalized dendritic plasticity and input feature storage in neurons. Nature 452: 436–441, 2008. [DOI] [PubMed] [Google Scholar]
- Magee J, Hoffman D, Colbert C, Johnston D. Electrical and calcium signaling in dendrites of hippocampal pyramidal neurons. Annu Rev Physiol 60: 327–346, 1998. [DOI] [PubMed] [Google Scholar]
- Magee JC. Dendritic integration of excitatory synaptic input. Nat Rev Neurosci 1: 181–190, 2000. [DOI] [PubMed] [Google Scholar]
- Magee JC. Dendritic lh normalizes temporal summation in hippocampal CA1 neurons. Nat Neurosci 2: 508–514, 1999. [DOI] [PubMed] [Google Scholar]
- Magee JC, Johnston D. Characterization of single voltage-gated Na+ and Ca2+ channels in apical dendrites of rat CA1 pyramidal neurons. J Physiol 487: 67–90, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee JC, Johnston D. Plasticity of dendritic function. Curr Opin Neurobiol 15: 334–342, 2005. [DOI] [PubMed] [Google Scholar]
- Magee JC, Johnston D. A synaptically controlled, associative signal for Hebbian plasticity in hippocampal neurons. Science 275: 209–213, 1997. [DOI] [PubMed] [Google Scholar]
- Magleby KL. Ion channels. Preventing artifacts and reducing errors in single-channel analysis. Methods Enzymol 207: 763–791, 1992. [DOI] [PubMed] [Google Scholar]
- Mainen ZF, Sejnowski TJ. Influence of dendritic structure on firing pattern in model neocortical neurons. Nature 382: 363–366, 1996. [DOI] [PubMed] [Google Scholar]
- Major G, Larkman AU, Jonas P, Sakmann B, Jack JJ. Detailed passive cable models of whole-cell recorded CA3 pyramidal neurons in rat hippocampal slices. J Neurosci 14: 4613–4638, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major G, Larkum ME, Schiller J. Active properties of neocortical pyramidal neuron dendrites. Annu Rev Neurosci 36: 1–24, 2013. [DOI] [PubMed] [Google Scholar]
- Makara JK, Magee JC. Variable dendritic integration in hippocampal CA3 pyramidal neurons. Neuron 80: 1438–1450, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino H, Malinow R. Compartmentalized versus global synaptic plasticity on dendrites controlled by experience. Neuron 72: 1001–1011, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maletic-Savatic M, Malinow R, Svoboda K. Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science 283: 1923–1927, 1999. [DOI] [PubMed] [Google Scholar]
- Manavski SA, Valle G. CUDA compatible GPU cards as efficient hardware accelerators for Smith-Waterman sequence alignment. BMC Bioinformatics 9, Suppl 2: S10, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor Y, Gonczarowski J, Segev I. Propagation of action potentials along complex axonal trees. Model and implementation. Biophys J 60: 1411–1423, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E, Goaillard JM. Variability, compensation and homeostasis in neuron and network function. Nat Rev Neurosci 7: 563–574, 2006. [DOI] [PubMed] [Google Scholar]
- Marder E, O'Leary T, Shruti S. Neuromodulation of circuits with variable parameters: single neurons and small circuits reveal principles of state-dependent and robust neuromodulation. Annu Rev Neurosci 37: 329–346, 2014. [DOI] [PubMed] [Google Scholar]
- Marder E, Prinz AA. Modeling stability in neuron and network function: the role of activity in homeostasis. Bioessays 24: 1145–1154, 2002. [DOI] [PubMed] [Google Scholar]
- Marder E, Taylor AL. Multiple models to capture the variability in biological neurons and networks. Nat Neurosci 14: 133–138, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H. The blue brain project. Nat Rev Neurosci 7: 153–160, 2006. [DOI] [PubMed] [Google Scholar]
- Markram H. The human brain project. Sci Am 306: 50–55, 2012. [DOI] [PubMed] [Google Scholar]
- Markram H, Lubke J, Frotscher M, Sakmann B. Regulation of synaptic efficacy by coincidence of postsynaptic APs and EPSPs. Science 275: 213–215, 1997. [DOI] [PubMed] [Google Scholar]
- Markram H, Muller E, Ramaswamy S, Reimann MW, Abdellah M, Sanchez CA, Ailamaki A, Alonso-Nanclares L, Antille N, Arsever S, Kahou GA, Berger TK, Bilgili A, Buncic N, Chalimourda A, Chindemi G, Courcol JD, Delalondre F, Delattre V, Druckmann S, Dumusc R, Dynes J, Eilemann S, Gal E, Gevaert ME, Ghobril JP, Gidon A, Graham JW, Gupta A, Haenel V, Hay E, Heinis T, Hernando JB, Hines M, Kanari L, Keller D, Kenyon J, Khazen G, Kim Y, King JG, Kisvarday Z, Kumbhar P, Lasserre S, Le Be JV, Magalhaes BR, Merchan-Perez A, Meystre J, Morrice BR, Muller J, Munoz-Cespedes A, Muralidhar S, Muthurasa K, Nachbaur D, Newton TH, Nolte M, Ovcharenko A, Palacios J, Pastor L, Perin R, Ranjan R, Riachi I, Rodriguez JR, Riquelme JL, Rossert C, Sfyrakis K, Shi Y, Shillcock JC, Silberberg G, Silva R, Tauheed F, Telefont M, Toledo-Rodriguez M, Trankler T, Van Geit W, Diaz JV, Walker R, Wang Y, Zaninetta SM, DeFelipe J, Hill SL, Segev I, Schurmann F. Reconstruction and simulation of neocortical microcircuitry. Cell 163: 456–492, 2015. [DOI] [PubMed] [Google Scholar]
- Martina M, Vida I, Jonas P. Distal initiation and active propagation of action potentials in interneuron dendrites. Science 287: 295–300, 2000. [DOI] [PubMed] [Google Scholar]
- McBride TJ, Rodriguez-Contreras A, Trinh A, Bailey R, Debello WM. Learning drives differential clustering of axodendritic contacts in the barn owl auditory system. J Neurosci 28: 6960–6973, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougal RA, Morse TM, Hines ML, Shepherd GM. ModelView for ModelDB: online presentation of model structure. Neuroinformatics 13: 459–470, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus OB, Magleby KL. Kinetic time constants independent of previous single-channel activity suggest Markov gating for a large conductance Ca-activated K channel. J Gen Physiol 94: 1037–1070, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus OB, Spivak CE, Blatz AL, Weiss DS, Magleby KL. Fractal models, Markov models, and channel kinetics. Biophys J 55: 383–385, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks JP, Mennerick S. Action potential initiation and propagation in CA3 pyramidal axons. J Neurophysiol 97: 3460–3472, 2007. [DOI] [PubMed] [Google Scholar]
- Mel BW. Synaptic integration in an excitable dendritic tree. J Neurophysiol 70: 1086–1101, 1993. [DOI] [PubMed] [Google Scholar]
- Menon V, Spruston N, Kath WL. A state-mutating genetic algorithm to design ion-channel models. Proc Natl Acad Sci U S A 106: 16829–16834, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliore M, Morse TM, Davison AP, Marenco L, Shepherd GM, Hines ML. ModelDB: making models publicly accessible to support computational neuroscience. Neuroinformatics 1: 135–139, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliore M, Shepherd GM. Emerging rules for the distributions of active dendritic conductances. Nat Rev Neurosci 3: 362–370, 2002. [DOI] [PubMed] [Google Scholar]
- Milescu LS, Akk G, Sachs F. Maximum likelihood estimation of ion channel kinetics from macroscopic currents. Biophys J 88: 2494–2515, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M. An Introduction to Genetic Algorithms. Cambridge, MA: MIT Press, 1996. [Google Scholar]
- Moore JW, Stockbridge N, Westerfield M. On the site of impulse initiation in a neurone. J Physiol 336: 301–311, 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse TM. ModelDB in computational neuroscience education–a research tool as interactive educational media. Brains Minds Media 3: bmm1409, 2008. [PMC free article] [PubMed] [Google Scholar]
- Motulsky H, Christopoulos A. Fitting Models to Biological Data Using Linear and Nonlinear Regression: A Practical Guide to Curve Fitting. Oxford, UK: Oxford University Press, 2004, p. 351. [Google Scholar]
- Nageswaran JM, Dutt N, Krichmar JL, Nicolau A, Veidenbaum AV. A configurable simulation environment for the efficient simulation of large-scale spiking neural networks on graphics processors. Neural Netw 22: 791–800, 2009. [DOI] [PubMed] [Google Scholar]
- Nettleton JS, Spain WJ. Linear to supralinear summation of AMPA-mediated EPSPs in neocortical pyramidal neurons. J Neurophysiol 83: 3310–3322, 2000. [DOI] [PubMed] [Google Scholar]
- Nevian T, Larkum ME, Polsky A, Schiller J. Properties of basal dendrites of layer 5 pyramidal neurons: a direct patch-clamp recording study. Nat Neurosci 10: 206–214, 2007. [DOI] [PubMed] [Google Scholar]
- Nowotny T, Szucs A, Levi R, Selverston AI. Models wagging the dog: are circuits constructed with disparate parameters? Neural Comput 19: 1985–2003, 2007. [DOI] [PubMed] [Google Scholar]
- O'Leary T, Williams AH, Caplan JS, Marder E. Correlations in ion channel expression emerge from homeostatic tuning rules. Proc Natl Acad Sci U S A 110: E2645–E2654, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oviedo HV, Reyes AD. Integration of subthreshold and suprathreshold excitatory barrages along the somatodendritic axis of pyramidal neurons. PLoS One 7: e33831, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer LM, Clark BA, Grundemann J, Roth A, Stuart GJ, Hausser M. Initiation of simple and complex spikes in cerebellar Purkinje cells. J Physiol 588: 1709–1717, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer LM, Shai AS, Reeve JE, Anderson HL, Paulsen O, Larkum ME. NMDA spikes enhance action potential generation during sensory input. Nat Neurosci 17: 383–390, 2014. [DOI] [PubMed] [Google Scholar]
- Palmer LM, Stuart GJ. Site of action potential initiation in layer 5 pyramidal neurons. J Neurosci 26: 1854–1863, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashut T, Magidov D, Ben-Porat H, Wolfus S, Friedman A, Perel E, Lavidor M, Bar-Gad I, Yeshurun Y, Korngreen A. Patch-clamp recordings of rat neurons from acute brain slices of the somatosensory cortex during magnetic stimulation. Front Cell Neurosci 8: 145, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashut T, Wolfus S, Friedman A, Lavidor M, Bar-Gad I, Yeshurun Y, Korngreen A. Mechanisms of magnetic stimulation of central nervous system neurons. PLoS Comput Biol 7: e1002022, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Garci E, Larkum ME, Nevian T. Inhibition of dendritic Ca2+ spikes by GABAB receptors in cortical pyramidal neurons is mediated by a direct Gi/o-beta-subunit interaction with Cav1 channels. J Physiol 591: 1599–1612, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Proskauer CC, Kaiserman-Abramof IR. The small pyramidal neuron of the rat cerebral cortex. The axon hillock and initial segment. J Cell Biol 39: 604–619, 1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BE, Healy MD, Nadkarni PM, Miller PL, Shepherd GM. ModelDB: an environment for running and storing computational models and their results applied to neuroscience. J Am Med Inform Assoc 3: 389–398, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto N, Doukhan D, DiCarlo JJ, Cox DD. A high-throughput screening approach to discovering good forms of biologically inspired visual representation. PLoS Comput Biol 5: e1000579, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirazi P, Brannon T, Mel BW. Arithmetic of subthreshold synaptic summation in a model CA1 pyramidal cell. Neuron 37: 977–987, 2003a. [DOI] [PubMed] [Google Scholar]
- Poirazi P, Brannon T, Mel BW. Pyramidal neuron as two-layer neural network. Neuron 37: 989–999, 2003b. [DOI] [PubMed] [Google Scholar]
- Poirazi P, Mel BW. Impact of active dendrites and structural plasticity on the memory capacity of neural tissue. Neuron 29: 779–796, 2001. [DOI] [PubMed] [Google Scholar]
- Polsky A, Mel BW, Schiller J. Computational subunits in thin dendrites of pyramidal cells. Nat Neurosci 7: 621–627, 2004. [DOI] [PubMed] [Google Scholar]
- Popovic MA, Foust AJ, McCormick DA, Zecevic D. The spatio-temporal characteristics of action potential initiation in layer 5 pyramidal neurons: a voltage imaging study. J Physiol 589: 4167–4187, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouille F, Cavelier P, Desplantez T, Beekenkamp H, Craig PJ, Beattie RE, Volsen SG, Bossu JL. Dendro-somatic distribution of calcium-mediated electrogenesis in purkinje cells from rat cerebellar slice cultures. J Physiol 527: 265–282, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press WH. Numerical Recipes in C: The Art of Scientific Computing. Cambridge, UK: Cambridge University Press, 1992, p. xxvi, 994. [Google Scholar]
- Prinz AA, Billimoria CP, Marder E. Alternative to hand-tuning conductance-based models: construction and analysis of databases of model neurons. J Neurophysiol 90: 3998–4015, 2003. [DOI] [PubMed] [Google Scholar]
- Prinz AA, Bucher D, Marder E. Similar network activity from disparate circuit parameters. Nat Neurosci 7: 1345–1352, 2004. [DOI] [PubMed] [Google Scholar]
- Qin F, Auerbach A, Sachs F. A direct optimization approach to hidden Markov modeling for single channel kinetics. Biophys J 79: 1915–1927, 2000a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Auerbach A, Sachs F. Hidden Markov modeling for single channel kinetics with filtering and correlated noise. Biophys J 79: 1928–1944, 2000b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rall W. Branching dendritic trees and motorneuron membrane resistivity. Exp Neurobiol 1: 491–527, 1959. [DOI] [PubMed] [Google Scholar]
- Rall W. Membrane potential transients and membrane time constant of motoneurons. Exp Neurol 2: 503–532, 1960. [DOI] [PubMed] [Google Scholar]
- Rall W. Membrane time constant of motoneurons. Science 126: 454, 1957. [DOI] [PubMed] [Google Scholar]
- Rall W. Theoretical significance of dendritic trees for neuronal input-output relations. In: Neural Theory and Modeling, edited by Reiss RF. Palo Alto, CA: Stanford University Press, 1964, p. 73–97. [Google Scholar]
- Rall W, Segev I, Rinzel J, Shepherd GM. The Theoretical Foundation of Dendritic Function: Selected Papers of Wilfrid Rall with Commentaries. Cambridge, MA: MIT Press, 1995, p. vii, 456. [Google Scholar]
- Raman IM, Bean BP. Inactivation and recovery of sodium currents in cerebellar Purkinje neurons: evidence for two mechanisms. Biophys J 80: 729–737, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman IM, Bean BP. Resurgent sodium current and action potential formation in dissociated cerebellar Purkinje neurons. J Neurosci 17: 4517–4526, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman IM, Sprunger LK, Meisler MH, Bean BP. Altered subthreshold sodium currents and disrupted firing patterns in Purkinje neurons of Scn8a mutant mice. Neuron 19: 881–891, 1997. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Markram H. Anatomy and physiology of the thick-tufted layer 5 pyramidal neuron. Front Cell Neurosci 9: 233, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan R, Khazen G, Gambazzi L, Ramaswamy S, Hill SL, Schurmann F, Markram H. Channelpedia: an integrative and interactive database for ion channels. Front Neuroinform 5: 36, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp M, Yarom Y, Segev I. Modeling back propagating action potential in weakly excitable dendrites of neocortical pyramidal cells. Proc Natl Acad Sci U S A 93: 11985–11990, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Bhalla US. PyMOOSE: interoperable scripting in Python for MOOSE. Front Neuroinform 2: 6, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy S, Csicsvari J, Beck H. Activity-dependent control of neuronal output by local and global dendritic spike attenuation. Neuron 61: 906–916, 2009. [DOI] [PubMed] [Google Scholar]
- Rettig J, Heinemann SH, Wunder F, Lorra C, Parcej DN, Dolly JO, Pongs O. Inactivation properties of voltage-gated K+ channels altered by presence of beta-subunit. Nature 369: 289–294, 1994. [DOI] [PubMed] [Google Scholar]
- Rhodes KJ, Keilbaugh SA, Barrezueta NX, Lopez KL, Trimmer JS. Association and colocalization of K+ channel alpha- and beta-subunit polypeptides in rat brain. J Neurosci 15: 5360–5371, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes KJ, Monaghan MM, Barrezueta NX, Nawoschik S, Bekele-Arcuri Z, Matos MF, Nakahira K, Schechter LE, Trimmer JS. Voltage-gated K+ channel beta subunits: expression and distribution of Kvβ1 and Kvβ2 in adult rat brain. J Neurosci 16: 4846–4860, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CB, O'Boyle MP, Suter KJ. Dendrites determine the contribution of after depolarization potentials (ADPs) to generation of repetitive action potentials in hypothalamic gonadotropin releasing-hormone (GnRH) neurons. J Comput Neurosci 26: 39–53, 2009. [DOI] [PubMed] [Google Scholar]
- Rosenblueth A, Wiener N. The role of models in science. Philos Sci 14: 316–321, 1945. [Google Scholar]
- Rossant C, Goodman DFM, Platkiewicz J, Brette R. Automatic fitting of spiking neuron models to electrophysiological recordings. Front Neuroinform 4: 2, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Bahl A. Divide et impera: optimizing compartmental models of neurons step by step. J Physiol 587: 1369–1370, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Häusser M. Compartmental models of rat cerebellar Purkinje cells based on simultaneous somatic and dendritic patch-clamp recordings. J Physiol 535: 445–472, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royeck M, Horstmann MT, Remy S, Reitze M, Yaari Y, Beck H. Role of axonal NaV1.6 sodium channels in action potential initiation of CA1 pyramidal neurons. J Neurophysiol 100: 2361–2380, 2008. [DOI] [PubMed] [Google Scholar]
- Ruppersberg JP, Schroter KH, Sakmann B, Stocker M, Sewing S, Pongs O. Heteromultimeric channels formed by rat brain potassium-channel proteins. Nature 345: 535–537, 1990. [DOI] [PubMed] [Google Scholar]
- Rush AM, Dib-Hajj SD, Waxman SG. Electrophysiological properties of two axonal sodium channels, Nav1.2 and Nav1.6, expressed in mouse spinal sensory neurons. J Physiol 564: 803–815, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakmann B, Neher E. (Editors). Single Channel Recording. New York: Plenum, 1995. [Google Scholar]
- Schaefer AT, Helmstaedter M, Sakmann B, Korngreen A. Correction of conductance measurements in non-space-clamped structures. 1. Voltage-gated K+ channels. Biophys J 84: 3508–3528, 2003a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AT, Helmstaedter M, Schmitt AC, Bar-Yehuda D, Almog M, Ben-Porat H, Sakmann B, Korngreen A. Dendritic voltage-gated K+ conductance gradient in pyramidal neurones of neocortical layer 5B from rats. J Physiol 579: 737–752, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AT, Larkum ME, Sakmann B, Roth A. Coincidence detection in pyramidal neurons is tuned by their dendritic branching pattern. J Neurophysiol 89: 3143–3154, 2003b. [DOI] [PubMed] [Google Scholar]
- Schiller J, Major G, Koester HJ, Schiller Y. NMDA spikes in basal dendrites of cortical pyramidal neurons. Nature 404: 285–289, 2000. [DOI] [PubMed] [Google Scholar]
- Schiller J, Schiller Y, Stuart G, Sakmann B. Calcium action potentials restricted to distal apical dendrites of rat neocortical pyramidal neurons. J Physiol 505: 605–616, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Hieber C, Bischofberger J. Fast sodium channel gating supports localized and efficient axonal action potential initiation. J Neurosci 30: 10233–10242, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwindt PC, Crill WE. Properties of a persistent inward current in normal and TEA-injected motoneurons. J Neurophysiol 43: 1700–1724, 1980. [DOI] [PubMed] [Google Scholar]
- Schwindt PC, Crill WE. Synaptically evoked dendritic action potentials in rat neocortical pyramidal neurons. J Neurophysiol 79: 2432–2446, 1998. [DOI] [PubMed] [Google Scholar]
- Segev I, Fleshman JW, Miller JP, Bunow B. Modeling the electrical behavior of anatomically complex neurons using a network analysis program: passive membrane. Biol Cybern 53: 27–40, 1985. [DOI] [PubMed] [Google Scholar]
- Sejnowski TJ, Koch C, Churchland PS. Computational neuroscience. Science 241: 1299–1306, 1988. [DOI] [PubMed] [Google Scholar]
- Serodio P, Rudy B. Differential expression of Kv4 K+ channel subunits mediating subthreshold transient K+ (A-type) currents in rat brain. J Neurophysiol 79: 1081–1091, 1998. [DOI] [PubMed] [Google Scholar]
- Shakespeare W. Julius Caesar. 1599. [Google Scholar]
- Sheng M, Tsaur ML, Jan YN, Jan LY. Contrasting subcellular localization of the Kv1.2 K+ channel subunit in different neurons of rat brain. J Neurosci 14: 2408–2417, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver RA. Neuronal arithmetic. Nat Rev Neurosci 11: 474–489, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloper JJ, Powell TP. A study of the axon initial segment and proximal axon of neurons in the primate motor and somatic sensory cortices. Philos Trans R Soc Lond B Biol Sci 285: 173–197, 1979. [DOI] [PubMed] [Google Scholar]
- Smith SL, Smith IT, Branco T, Hausser M. Dendritic spikes enhance stimulus selectivity in cortical neurons in vivo. Nature 503: 115–120, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruston N. Pyramidal neurons: dendritic structure and synaptic integration. Nat Rev Neurosci 9: 206–221, 2008. [DOI] [PubMed] [Google Scholar]
- Spruston N, Schiller Y, Stuart G, Sakmann B. Activity-dependent action potential invasion and calcium influx into hippocampal CA1 dendrites. Science 268: 297–300, 1995. [DOI] [PubMed] [Google Scholar]
- Stafstrom CE, Schwindt PC, Crill WE. Negative slope conductance due to a persistent subthreshold sodium current in cat neocortical neurons in vitro. Brain Res 236: 221–226, 1982. [DOI] [PubMed] [Google Scholar]
- Stafstrom CE, Schwindt PC, Crill WE. Repetitive firing in layer V neurons from cat neocortex in vitro. J Neurophysiol 52: 264–277, 1984a. [DOI] [PubMed] [Google Scholar]
- Stafstrom CE, Schwindt PC, Flatman JA, Crill WE. Properties of subthreshold response and action potential recorded in layer V neurons from cat sensorimotor cortex in vitro. J Neurophysiol 52: 244–263, 1984b. [DOI] [PubMed] [Google Scholar]
- Stone JE, Phillips JC, Freddolino PL, Hardy DJ, Trabuco LG, Schulten K. Accelerating molecular modeling applications with graphics processors. J Comput Chem 28: 2618–2640, 2007. [DOI] [PubMed] [Google Scholar]
- Stuart G, Hausser M. Initiation and spread of sodium action potentials in cerebellar Purkinje cells. Neuron 13: 703–712, 1994. [DOI] [PubMed] [Google Scholar]
- Stuart G, Sakmann B. Amplification of EPSPs by axosomatic sodium channels in neocortical pyramidal neurons. Neuron 15: 1065–1076, 1995. [DOI] [PubMed] [Google Scholar]
- Stuart G, Schiller J, Sakmann B. Action potential initiation and propagation in rat neocortical pyramidal neurons. J Physiol 505: 617–632, 1997a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart G, Spruston N. Determinants of voltage attenuation in neocortical pyramidal neuron dendrites. J Neurosci 18: 3501–3510, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart G, Spruston N, Hausser M. Dendrites. Oxford, UK: Oxford University Press, 1999. [Google Scholar]
- Stuart G, Spruston N, Sakmann B, Hausser M. Action potential initiation and backpropagation in neurons of the mammalian CNS. Trends Neurosci 20: 125–131, 1997b. [DOI] [PubMed] [Google Scholar]
- Stuart GJ, Dodt HU, Sakmann B. Patch-clamp recordings from the soma and dendrites of neurons in brain slices using infrared video microscopy. Pflügers Arch 423: 511–518, 1993. [DOI] [PubMed] [Google Scholar]
- Stuart GJ, Palmer LM. Imaging membrane potential in dendrites and axons of single neurons. Pflügers Arch 453: 403–410, 2006. [DOI] [PubMed] [Google Scholar]
- Stuart GJ, Sakmann B. Active propagation of somatic action potentials into neocortical pyramidal cell dendrites. Nature 367: 69–72, 1994. [DOI] [PubMed] [Google Scholar]
- Stuhmer W, Ruppersberg JP, Schroter KH, Sakmann B, Stocker M, Giese KP, Perschke A, Baumann A, Pongs O. Molecular basis of functional diversity of voltage-gated potassium channels in mammalian brain. EMBO J 8: 3235–3244, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szigeti B, Gleeson P, Vella M, Khayrulin S, Palyanov A, Hokanson J, Currie M, Cantarelli M, Idili G, Larson S. OpenWorm: an open-science approach to modeling Caenorhabditis elegans. Front Comput Neurosci 8: 137, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak J, Moore LE. Simulation and parameter estimation study of a simple neuronal model of rhythm generation: role of NMDA and non-NMDA receptors. J Comput Neurosci 5: 209–235, 1998. [DOI] [PubMed] [Google Scholar]
- Tabak J, Murphey CR, Moore LE. Parameter estimation methods for single neuron models. J Comput Neurosci 9: 215–236, 2000. [DOI] [PubMed] [Google Scholar]
- Talley EM, Cribbs LL, Lee JH, Daud A, Perez-Reyes E, Bayliss DA. Differential distribution of three members of a gene family encoding low voltage-activated (T-type) calcium channels. J Neurosci 19: 1895–1911, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AL, Goaillard JM, Marder E. How multiple conductances determine electrophysiological properties in a multicompartment model. J Neurosci 29: 5573–5586, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AL, Hickey TJ, Prinz AA, Marder E. Structure and visualization of high-dimensional conductance spaces. J Neurophysiol 96: 891–905, 2006. [DOI] [PubMed] [Google Scholar]
- Thome C, Kelly T, Yanez A, Schultz C, Engelhardt M, Cambridge SB, Both M, Draguhn A, Beck H, Egorov AV. Axon-carrying dendrites convey privileged synaptic input in hippocampal neurons. Neuron 83: 1418–1430, 2014. [DOI] [PubMed] [Google Scholar]
- Torben-Nielsen B, De Schutter E. Context-aware modeling of neuronal morphologies. Front Neuroanat 8: 92, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimmer JS, Rhodes KJ. Localization of voltage-gated ion channels in mammalian brain. Annu Rev Physiol 66: 477–519, 2004. [DOI] [PubMed] [Google Scholar]
- Tripathy SJ, Burton SD, Geramita M, Gerkin RC, Urban NN. Brain-wide analysis of electrophysiological diversity yields novel categorization of mammalian neuron types. J Neurophysiol 113: 3474–3489, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaur ML, Chou CC, Shih YH, Wang HL. Cloning, expression and CNS distribution of Kv4.3, an A-type K+ channel alpha subunit. FEBS Lett 400: 215–220, 1997. [DOI] [PubMed] [Google Scholar]
- Tsien RY. Fluorescent probes of cell signaling. Annu Rev Neurosci 12: 227–253, 1989. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG, Nelson SB. Hebb and homeostasis in neuronal plasticity. Curr Opin Neurobiol 10: 358–364, 2000. [DOI] [PubMed] [Google Scholar]
- van Elburg RA, van Ooyen A. Impact of dendritic size and dendritic topology on burst firing in pyramidal cells. PLoS Comput Biol 6: e1000781, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanier MC, Bower JM. A comparative survey of automated parameter-search methods for compartmental neural models. J Comput Neurosci 7: 149–171, 1999. [DOI] [PubMed] [Google Scholar]
- Vella M, Cannon RC, Crook S, Davison AP, Ganapathy G, Robinson HP, Silver RA, Gleeson P. libNeuroML and PyLEMS: using Python to combine procedural and declarative modeling approaches in computational neuroscience. Front Neuroinform 8: 38, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataramanan L, Sigworth FJ. Applying hidden Markov models to the analysis of single ion channel activity. Biophys J 82: 1930–1942, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter P, Roth A, Häusser M. Propagation of action potentials in dendrites depends on dendritic morphology. J Neurophysiol 85: 926–937, 2001. [DOI] [PubMed] [Google Scholar]
- Weaver CM, Wearne SL. Neuronal firing sensitivity to morphologic and active membrane parameters. PLoS Comput Biol 4: e11, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CM, Wearne SL. The role of action potential shape and parameter constraints in optimization of compartment models. Neurocomputing 69: 1053–1057, 2006. [Google Scholar]
- Williams SR. Encoding and decoding of dendritic excitation during active states in pyramidal neurons. J Neurosci 25: 5894–5902, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SR. Spatial compartmentalization and functional impact of conductance in pyramidal neurons. Nat Neurosci 7: 961–967, 2004. [DOI] [PubMed] [Google Scholar]
- Williams SR, Stuart GJ. Mechanisms and consequences of action potential burst firing in rat neocortical pyramidal neurons. J Physiol 521: 467–482, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SR, Stuart GJ. Voltage- and site-dependent control of the somatic impact of dendritic IPSPs. J Neurosci 23: 7358–7367, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won-Ki J, Beyer J, Hadwiger M, Blue R, Law C, Vazquez-Reina A, Reid RC, Lichtman J, Pfister H. Ssecrett and NeuroTrace: interactive visualization and analysis tools for large-scale neuroscience data sets. IEEE Comput Graph Appl 30: 58–70, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wybo WA, Boccalini D, Torben-Nielsen B, Gewaltig MO. A sparse reformulation of the Green's function formalism allows efficient simulations of morphological neuron models. Neural Comput 27: 2587–2622, 2015. [DOI] [PubMed] [Google Scholar]
- Yadav A, Gao YZ, Rodriguez A, Dickstein DL, Wearne SL, Luebke JI, Hof PR, Weaver CM. Morphologic evidence for spatially clustered spines in apical dendrites of monkey neocortical pyramidal cells. J Comp Neurol 520: 2888–2902, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R. Dendritic spines and distributed circuits. Neuron 71: 772–781, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerlaut Y, Telenczuk B, Deleuze C, Bal T, Ouanounou G, Destexhe A. Heterogeneous firing rate response of mice layer V pyramidal neurons in the fluctuation-driven regime. J Physiol 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JJ. Maturation of layer 5 neocortical pyramidal neurons: amplifying salient layer 1 and layer 4 inputs by Ca2+ action potentials in adult rat tuft dendrites. J Physiol 526: 571–587, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]