Abstract
The gut microbiota plays a crucial role in the maturation of the intestinal mucosal immune system of its host1,2. Within the thousand bacterial species present in the intestine, the symbiont Segmented Filamentous Bacterium (SFB) is unique in its ability to potently stimulate the post-natal maturation of the B and T cell compartments and induce a striking increase in the small intestinal Th17 responses3–5. Unlike other commensals, SFB intimately attaches to absorptive epithelial cells in the ileum and cells overlying Peyer's patches6,7. This colonization does not result in pathology but rather protects the host from pathogens4. Yet, little is known about the host-SFB interaction that underlies the important immunostimulatory properties of SFB as SFB has resisted in vitro culturing for over 50 years. Here we succeeded in growing mouse SFB outside of its host in an SFB-host cell co-culturing system. Single-celled SFB isolated from monocolonized mice undergo filamentation, segmentation, and differentiation to release viable infectious particles, the intracellular offsprings or IOs, that can colonize mice to induce signature immune responses. In vitro, IOs can attach to mouse and human host cells and recruit actin. In addition, SFB can potently stimulate the up-regulation of host innate defence genes, inflammatory cytokines, and chemokines. In vitro culturing thereby mimics the in vivo niche, provides novel insights into SFB growth requirements and its immunostimulatory potential, and opens the door for the investigation of the complex developmental stages of SFB and the detailed dissection of the unique SFB-host interaction on a cellular and molecular level.
SFB or “Candidatus Arthromitus” are anaerobic, Clostridia-related, spore-forming commensals found in the gut of many vertebrate species, including mice and likely humans8,9. SFB has garnered much interest due to its unique ability to educate the gut immune system and to induce a healthy state of physiological inflammation3,4. SFB colonization leads to a maturation of the gut mucosal lymphoid tissue, induces a strong and broad IgA response, stimulates the T cell compartment, and upregulates intestinal innate defence mediators4,5,10. In addition, SFB colonization exerts an adjuvant effect on systemic responses and can thus exacerbates pathologies in mouse models of encephalitis and arthritis, while protecting genetically predisposed mice against the development of type I diabetes11–15. Recent sequencing of the rat and mouse SFB genomes revealed the highly auxotrophic needs of SFB and placed SFB between obligate and facultative symbionts. These findings suggest that SFB obtains at least some of its nutritional requirements from its interaction with the host 16–19.
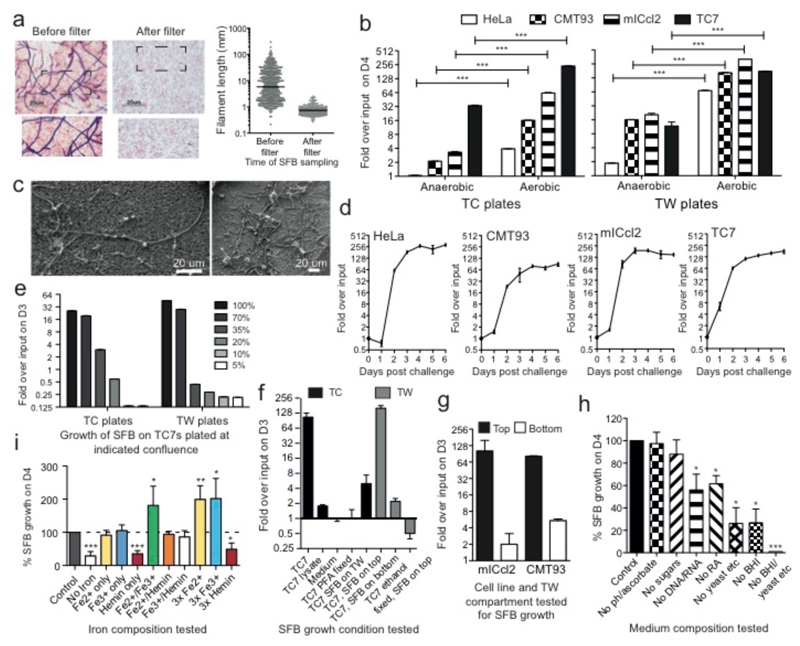
To culture SFB, we aimed to mimic its replicative niche. We therefore designed an SFB-host cell co-culturing system whereby SFB20 isolated from monoassociated mice are cultured with eukaryotic cells grown at low but physiological oxygen conditions21 in a rich tissue culture medium containing bacterial medium components and additional supplements. SFB from monoassociated mice were collected, filtered through a 100-μm mesh, separated from most other fecal matter using a Nycodenz column and passed through a 5-μm filter to obtain a pure culture of unicellular IOs (average 0.7 μm) (Fig. 1a). Eukaryotic cells grown on tissue culture (TC) wells or transwells (TWs) were placed in a humidified anaerobic cabinet, challenged with IOs, and kept at either strict anaerobic conditions within a sealed box or left in the anaerobic cabinet where the oxygen concentration was maintained at low levels (0.5-1.4% O2). After 4 days, bacterial growth in the culture supernatant was quantitated by qPCR using SFB-specific 16S rDNA primers (Fig. 1b) and confirmed using scanning electron microscopy (SEM) (Fig. 1c). SFB growth was observed in all conditions for most cell lines assayed but growth was usually enhanced on TWs compared to TC wells and growth was significantly better in the presence of oxygen, revealing SFB to be a relatively aerotolerant anaerobe. Both human (TC7/ HeLa) and mouse (mICcl2/CMT93) cell lines supported growth but TC7s was the most resilient cell line generally supporting SFB growth most robustly. Based on temporal analysis, the highest exponential growth phase occurred between days 1 and 3 with an average maximum doubling time of 5.0 hours (Fig. 1d).
Figure 1. Growth and growth requirements of SFB in vitro.
a, Gram stain and length of SFB before and after 5-μm filtration; b, QPCR quantitation of SFB growth on host cells at the indicated conditions; c, SEM images of 4-day old SFB filaments grown anaerobically on TC7s on TWs; d-i, SFB growth at low oxygen concentrations d, on the indicated cell line seeded on TWs; e, on TC7s at indicated cell confluence; f/g, under various conditions and with different cell lines; h/i, on TC7s on TWs with indicated medium composition. f-i: values are Means +/- SEM from three experiments.
Next we dissected the growth requirements for SFB. SFB growth had a striking dependence on host cell number, decreasing in number with decreasing cell density (Fig. 1e). In addition, negligible growth occurred in medium alone, medium supplemented with cell lysate, or when cells were fixed prior to SFB challenge (Fig. 1f), indicating that live host cells are required for SFB proliferation. SFB also required close contact for efficient growth as only little growth (0-6%) occurred when IOs were added to the bottom chamber of TWs or when bacteria were placed in transwells above cells in TC wells (Fig. 1f/g). Yet, as host cell contact was not an absolute requirement, it suggests that host cells may release a soluble factor that promotes SFB growth. To address the requirement for medium supplementation, SFB was grown on TC7s in complete medium or medium missing individual additives. Brain heart infusion, a yeast/peptone/casein amino acid mixture and particularly iron supplementation were critical for SFB growth (Fig. 1h/i).
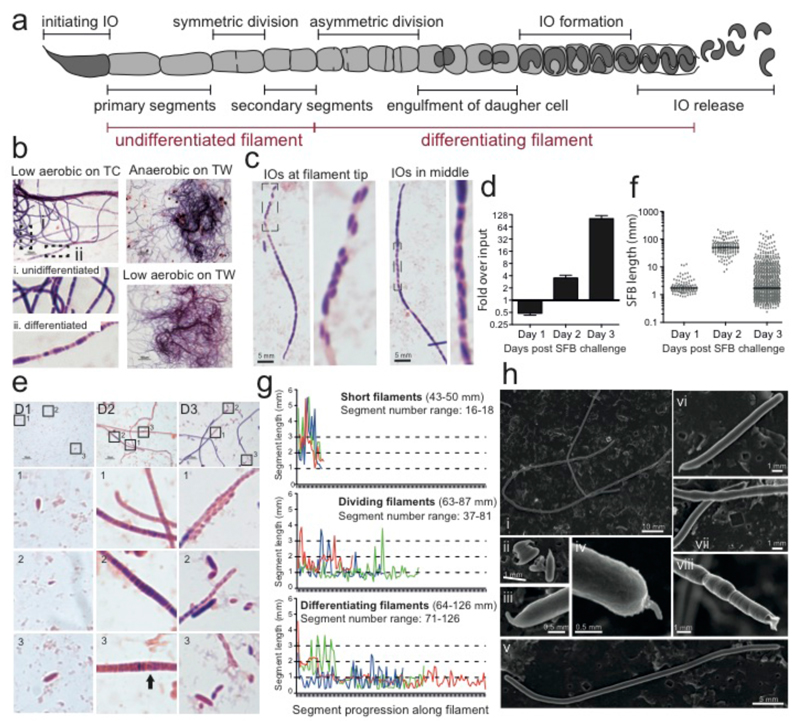
In the 1970s, transmission electron microscopy studies of SFB present in the murine gut lead to a proposed life-cycle whereby attachment via the holdfast at the IO tip is followed by filamentation and then a complex developmental progression that commences at the distal tip and ultimately leads to IO formation and release (Fig. 2a)7,22. According to this model, when filaments grow beyond 50 μm in length, the large primary filament segments start to undergo a symmetric division to form smaller secondary segments. These differentiate by dividing asymmetrically to form a mother/daughter cell. The daughter cell becomes engulfed and subsequently divides to form two IOs within the surrounding mother cell segment. IOs are then released from the filament by breakdown of the filament septa and cell wall and reattach to the host.
Figure 2. Differentiation of SFB from filaments to IOs during in vitro growth.
a, Schematic of an SFB filament highlighting stages of its growth and differentiation. Gram stain of SFB after 4 days of growth on b, TC7s incubated at the indicated condition, and c, TC7s grown on TWs in low oxygen. Analysis of SFB grown on mICcl2 cells (d-g) and TC7s (h) on TWs at 1-2.5% O2: d, qPCR quantitation; e, length of individual SFBs; f, Gram stain; and g, Segment length analysis of representative 2-day old SFB filaments. h, SEM of SFB after 4 days of growth.
In vitro, growth of SFB on TC7s cells often yielded considerable quantities of long filaments that could be clumped together in a hairball-like phenotype easily seen with the naked eye (Fig. 2b). The majority of these filaments were undifferentiated after four days with only some filaments showing a characteristic heterogeneous staining of differentiating filaments (Fig. 2b i/ii). IOs could be seen located at the filament tip or occasionally in the central part of a filament (Fig. 2c), similar to filaments recovered from SFB-monoassociated mice (data not shown). Differentiation of SFB occurred on all four cell lines tested but we noticed that differentiation was more pronounced when higher oxygen concentrations (1-2.5%) were used. In time course analysis at this higher oxygen concentration (Fig. 2d), only short bacteria were detected one day after challenge (Fig. 2e/f). After two days, only long filaments were present (Fig. 2e) and the bacterial septa could be identified quite clearly using the Gram stain (Fig. 2f). Three types of filaments were found (Fig 2f/g); short filaments with long intracellular segments of ~2.6 μm, medium sized filaments with smaller intracellular segments of ~1.2 μm, and medium to long filaments that had a more heterogeneous distribution of segment lengths including very small segments (Fig. 2f/g) and rare segments with half-circular structures (Fig. 2fD2-3, arrow) that resembled the engulfment of a daughter cell by a mother cell. After three days of growth, most filaments had differentiated, at least in part, to the final IO stage and IOs could be seen in a characteristic doublet orientation at the filament tip where the filament cell wall appeared to have lost its structure (Fig. 2e/f). Many IOs of varying lengths were also no longer associated with filaments. SEM confirmed the presence of the needle-like holdfast structure on IOs and at the tip of filaments (Fig. 2h (ii-vii)) and could clearly distinguish between undifferentiated thin and smooth filaments (Fig. 2h (i, vi-vii)) and those that are broader with a heterogeneous and bulbous morphology that correspond to differentiating filaments (Fig. 2h (i, vii-viii)7,23. In addition, cell wall remnants could be detected at the distal tip of differentiating filaments where IOs had been released (Fig. 2h viii, ED Fig. 1b)7. Together, these data demonstrate that in vitro culturing of SFB supports the full differentiation of in vitro-grown SFB filaments to the IO stage and confirms the SFB life-cycle inferred from in vivo observations.
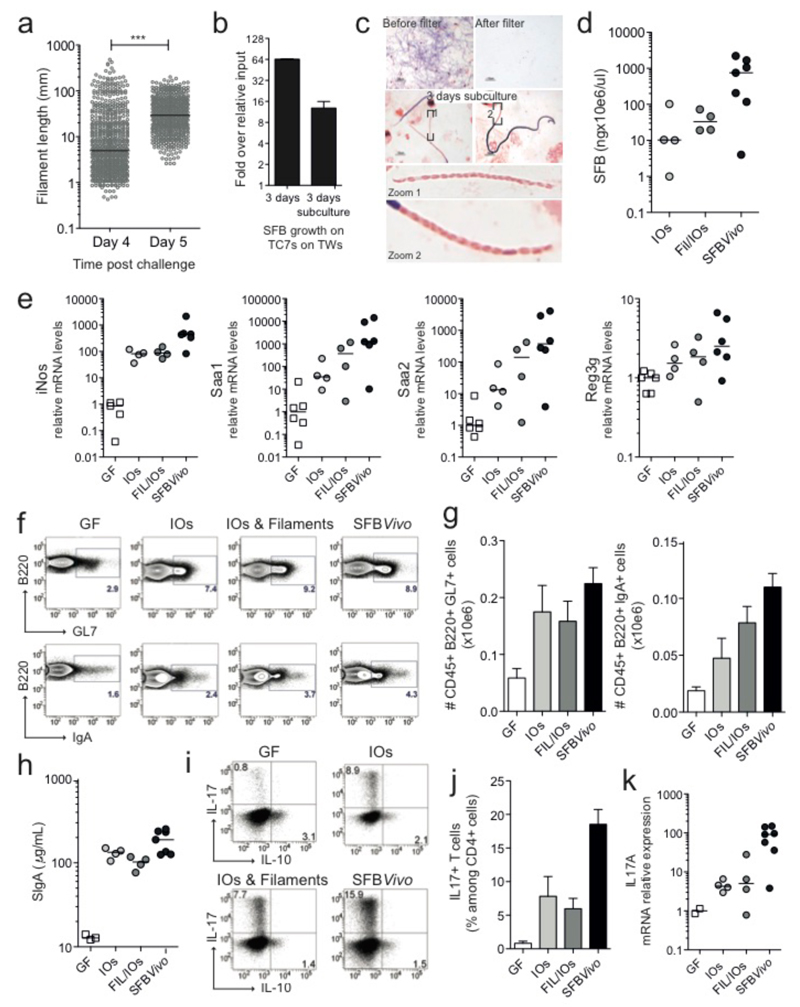
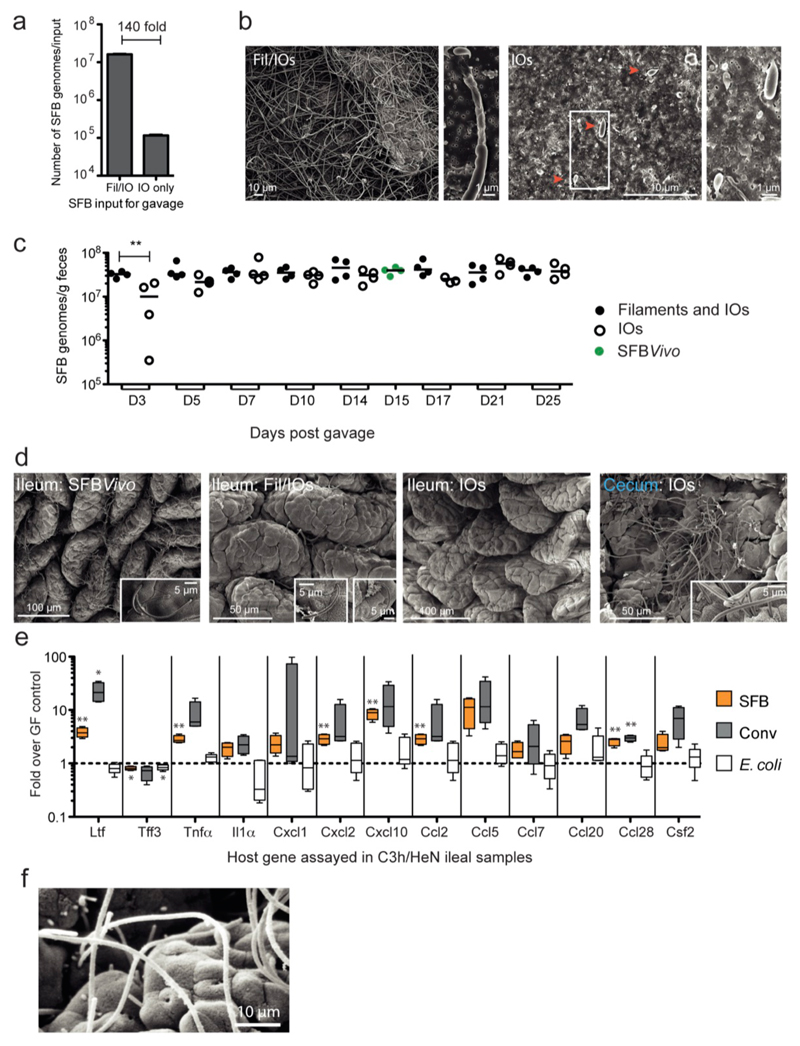
The viability and infectivity of in vitro-formed IOs was then tested. SFB were first grown on TC7s under aerobic conditions (Fig. 3a) until many IOs appeared. After one additional day, IOs were no longer present and the average bacterial length was significantly longer, indicating IO outgrowth into filaments. Similarly, when IOs grown in vitro were separated from filaments by filtration through a 5-μm filter and added to newly plated cells, SFB numbers increased and the newly formed filaments differentiated into IOs at the filament tip (Fig. 3b/c), demonstrating filamentation and differentiation of in vitro-formed IOs. To assess whether SFB grown in vitro retained their ability to colonize mice and stimulate the characteristic innate and acquired immune responses, SFB grown in vitro for three days were divided into a Filament/IOs fraction and a pure IOs fraction (ED Fig. 1a/b) and gavaged into germfree mice. Colonization was firmly established by both inputs at 5 days post gavage (ED Fig. 1c) despite a 140-fold input difference for Fil/IOs and IOs. However, unlike mice gavaged with SFB derived from fecal samples (SFBVivo), which generally showed good colonization of the ileum, mice gavaged with in vitro-grown SFB (SFBVitro) had much lower numbers of SFB colonizing the ileum (Fig. 3d, ED Fig.1d) but instead showed heavy colonization of the cecum (ED Fig. 1d). Thus, while SFBVitro clearly can attach to the ileum, these results suggest that ileal colonization may be more efficient by IOs released from spores found in the fecal input, possibly due to the expression of flagella at this particular stage of the life-cycle19. Notably, the magnitude of the innate host response was proportional to the colonization level of the ileum (Fig. 3e) and not the overall SFB fecal load (ED Fig. 1c), revealing the requirement for ileal attachment of SFB to induce the innate host response. Similar to SFBVivo, albeit with less potency, SFBVitro were also able to stimulate the B cell compartment in Peyer's patches (Fig. 3f/g), enhance IgA secretion in the feces (Fig. 3h) and increase the number of Th17 cells and the level of IL-17A mRNA in the small intestine lamina propria (Fig. 3i-k).
Figure 3. Viability, colonization and immunostimulatory potential of in vitro-grown SFB.
a, SFB length after growth on TC7s on TWs; b, Quantitation and c, Gram stain of SFB growth on TC7s on TWs before and after a 3-day sub-culturing of the 5-μm filtrate; d-k, Analysis of germfree C57BL/6 mice gavaged with either Fil/IOs mix, IOs, or feces of SFB-monoassociated mice (SFBVivo); d, Quantification of ileum-associated SFB; e/k, Host gene expression in the ileal lamina propria; Representative flow cytometry plots and quantitation of B220+B-cell (f/g) and CD45+CD3+CD4+T-cell (i/j) frequencies of the indicated markers; h, fecal secretory IgA quantitation by ELISA.
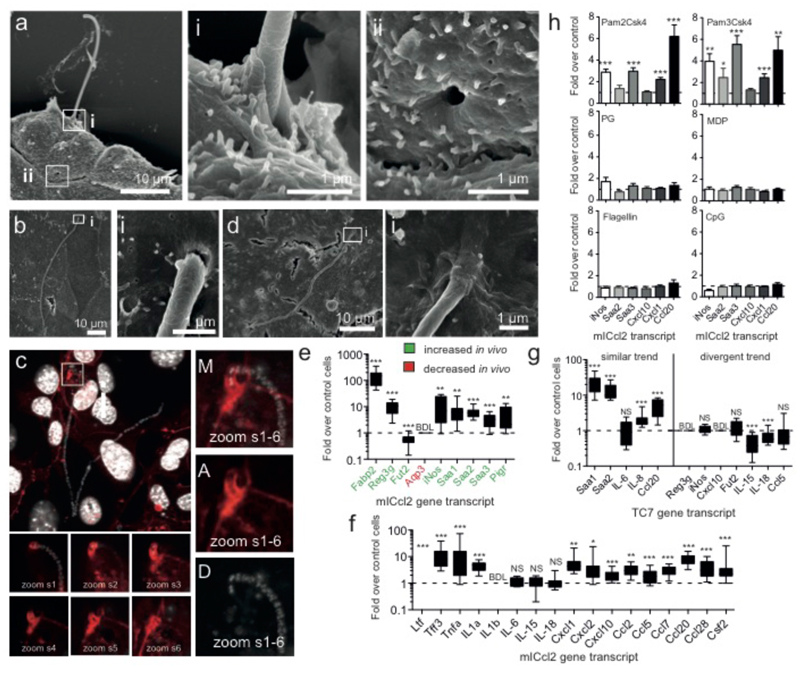
Lastly, we investigated the SFB-host interaction and the host response to SFB growth. Despite the apparent requirement for close contact between host cells and SFB for efficient growth, a tight interaction was not readily observed. However, when the interaction was promoted by gently spinning IOs onto cells, SFB filaments were found attached to mICcl2 cells (Fig. 4a-c). This stable interaction was accompanied by actin accumulation surrounding the filament tip (Fig. 4c) and could leave structurally intact vacant attachment sites (Fig. 4a ii) similar to those observed in the ileum of mice (ED Fig. 1f). Attached filaments included both undifferentiated ones and ones that had reached the final IO stage. Contrary to in vivo results24, attachment in vitro was not species-specific (Fig. 4d). To assess the similarity in the host response to SFB in vitro and in vivo, we analysed the gene expression profile of epithelial-derived host factors known to be regulated by SFB colonization4,5,25–27. We found that the gene regulation in vitro closely recapitulates gene regulation in vivo (Fig. 4e), thereby further supporting our in vitro model system. Going beyond previously implicated epithelial factors, we tested a number of additional cytokines, chemokines and host defence genes (Fig. 4f). Our data show that SFB growth leads to a strong inflammatory host response with the induction of pleiotropic inflammatory mediators such as TNFα, IL1α, and Saa1-3, induction of a number of innate host defence mechanisms (Reg3γ, iNos, and lactoferrin) and an immunological environment that is conducive for the recruitment of B cells, the transmigration of IgA, recruitment and activation of T cells, and recruitment of neutrophils, dendritic cells, as well as monocytes. In agreement, we observed that the transcript level of most immune genes that are up-regulated during co-culture with mICcl2 cells in vitro was also increased by SFB, and not by E. coli, during colonization experiments in vivo (ED Fig. 1e). Conversely, the transcriptional response in the human TC7 cell line was divergent from that observed in mICcl2 cells and less consistent with in vivo results (Fig. 4g). Lastly, by using an array of microbe-associated molecular patterns (MAMPs), we demonstrate that the inflammatory response to SFB in vitro is likely shaped by the activation of TLR2 (Fig. 4h).
Figure 4. SFB-host cell interaction and host response.
SEM of SFB attached to subconfluent (a) and confluent (b) mICcl2 cells after 2 days of IO challenge. c, Merge (M) of Dapi (D) and Actin (A) stain of mICcl2 cells challenged with IOs for 2 days showing confocal slices (s) and Z-projections. d, SEM of 3-day old SFB filament attached to TC7s. Host response using qPCR analysis of mICcl2 cells (e/f/h) and TC7 cells (g) after 3 days post challenge with SFB (e-g) or various MAMPs (h). NS: not significant; BDL: below detection limit.
We thereby demonstrate the successful culturing of SFB in vitro and provide novel insights into SFB growth requirements and the host response to SFB challenge. Our data suggests that in vivo, attachment of SFB to the ileal surface is an important feature to elicit epithelial cell responses while in vitro, where attachment remained infrequent, the close proximity of SFB and cells appears to largely bypass the need for attachment to deliver the stimulating signal(s). These findings highlight the importance of the privileged location of the replicative niche of SFB at the ileal epithelial surface in mediating the stimulatory potential of SFB. Future investigations using the in vitro culturing system will be needed to dissect the contribution of bacterial factors, including MAMPs, and attachment per se to the unique immunostimulatory properties of this unusual and still enigmatic commensal.
Methods
Cell culture and SFB-specific culture medium
TC7, CMT93 and HeLa cells were cultured in DMEM (Gibco 31885) with 10% inactivated fetal calf serum (FCS; AbCys CVFSVF00-0U) and non-essential amino acids (Invitrogen 11140-035) while mICcl2 were maintained in DMEM/F12 advanced medium (Gibco 12634) with 2% inactivated FCS and Glutamax (Gibco 35050) and supplemented with 10 nM hEGF (Sigma E9644), 50 nM Dexamethasone (Sigma D4902) and 1 nM triiodothyronine (Sigma T5516). HeLa cells were obtained from ATCC; CMT93 cells were provided by Hervé Blottière, INRA, UMR 1319 Micalis, France; TC7s from Alain Servin, Faculte de Pharmacie de Chatenay Malabry and mICcl2 cells from the laboratory of Alain Vandewalle, Faculté de Médecine Xavier Bichát, Paris, France. All cell lines were tested for mycoplasma every 2 weeks and always found to be negative. Cells were plated form 1 to 3 days before the experiment such that a monolayer was present at the start of the experiment. SFB-medium was made up of: DMEM/F12 advanced medium with 2% FCS, Glutamax and 12.5 mM HEPES (Sigma H0887) with the following supplementation: 1 in 100 dilution of: 1. Brain heart infusion (BD Difco 237500) at 5x concentrated; 2. Peptone/yeast (BBL Biosafe 211862) at 10% and Casein amino acids at 5% (DIFCO 0320-01-1); 3. Ribose/Cellobiose/Mannose (Sigma: R9629, C7252, M6020) at 200 mM. 1 in 1,000 dilution of: 1. Ferrous sulphate (Merck 3965) at 10 mM; 2. Ferric ammonium citrate (LabGuard 0658) at 12.5 mM; 3. Hemin (Sigma 51280) at 1.5 mM in 50% ethanol with 1.4 N NH4OH; 4. Sodium ascorbate at 10 mg/ml with 1-phosphoascorbate at 500 mM (Sigma: A4034, 49752). 1 in 10,000 dilution of: Retinoic acid (Sigma R2625) at 30 mg/ml in DMSO. 1 in 500 dilution of: 1. Sperm DNA (Life technologies 15632-011) at 10 mg/ml digested for 1 hour with 10 ul DNaseI (Roche 04 716 728 001) at 37°C and heat inactivated at 75°C for 30 min; 2. RNA at 10 mg/ml (Sigma R6750) undigested. SFB-medium specific medium supplements, except for the nucleotides and hemin were prepared fresh every two weeks and otherwise stored at 4 deg C or -20 deg C for retinoic acid, and nucleotides. When mICcl2 cells were used, SFB-medium was further supplemented with hEGF, Dexamethasone and triiodothyronine. Notably, additional supplementation with 0.2% yeast extract (BD 212750; 1/100 from 20%) has since further improved SFB growth, particularly for mICcl2 cells. Cells were plated either on regular 12-well tissue culture plates or on Costar transwell plates with a 0.4 μm filter (Sigma CLS3460 and Fisher Scientific W2127P).
Purification of IOs from SFB monoassociated mice and infection protocol
All liquids used for the isolation of IOs were pre-equilibrated overnight in an anaerobic chamber set to 0% oxygen. SFB monoassociated JH mice were sacrificed aseptically in a tissue culture hood and then placed in an anaerobic cabinet for dissection. The ileal, ceacal and colonic contents were resuspended in 50 ml PBS and homogenized by vortexing. Homogenates were filtered through a 100-μm mesh to remove large fecal debris. The filtrate was spun at 8,000xg for 5 min to pellet bacteria and insoluble material and the pellet was resuspended in 3 ml PBS per mouse sacrificed, layered onto 3 ml 50% and 2 ml 30% Nycodenz (AbCYS 1002424) solution made with PBS in 15 ml Falcon tubes and spun for 10 minutes at 4,000xg. SFB within the 30% fraction were collected, diluted in PBS and bacteria were pelleted for 10 minutes at 8,500xg. Pellets were resuspended in 15 ml pre-equilibrated PBS by pipetting/vortexing and filtered through a 5 μm filter (Sigma Z612502). The filtrate was again centrifuged for 5 min at 8,000xg and the pellet was resuspended in an appropriate amount of pre-equilibrated culture medium. Usually one mouse was sacrificed for every four 12-well plates used and 50 μl of bacterial suspension was added to each well. To facilitate adhesion, cells challenged with SFB were sealed in ziplock bags within the cabinets, removed from the cabinet and spun for 10 min at 300xg.
SFB recovery and quantification and analysis of SFB growth
To recover SFB, the culture supernatant was collected and centrifuged for 4 minutes at 8,000xg and the pellet was resuspended in 100 μl PBS of which 20 μl was spotted on glass slides for the Gram stain, 30 μl was mixed with an equal volume of 50% glycerol and frozen at -80°C, and the remaining 50 μl was used for DNA extraction. The DNA was isolated with the Qiagen stool kit (51504; without the use of the inhibitor tablet) and diluted 1 in 20. SFB growth was enumerated by qPCR analysis of the 16S rRNA genes using the following primer pairs: the SFB specific F: 5’-AGGAGGAGTCTGCGGCACATTAGC-3’; and the universal R: 5’-TCCCCACTGCTGCCTCCCGTAG-3’. For quantitative PCR, 6 ul of diluted SFB DNA was mixed with 1.5 μl of a 4 mM primer mix and 7.5 μl of Power SybrGreen Master mix (Applied 4368708) and run on an ABI 7900HT machine in a 384 well plate. All experiments were performed in a minimum of three independent experiments and representative experiments with the Mean +/- SD are shown unless otherwise indicated. Statistical analysis was performed by the two-tailed Student T-test (* p=< 0.05, ** p=< 0.01, *** p=< 0.001). SFB segment length analysis was performed using ImageJ. For SEM analysis, SFB-containing supernatants were washed with PBS and suctioned onto 0.1 μm filters (Watman 110405) and fixed in 0.1M Cacodylate buffer containing 2.5% glutaraldehyde before being processed. Cells for SEM were fixed before in PHEM buffer (18.14g PIPES, 6.5g HEPES, 3.8g EGTA, 0.99g MgSO4 per liter with 10M KOH to pH 7.0) containing 4% sucrose and 2.5% glutaraldehyde and processed for SEM. For fluorescence, cells were fixed in PBS/3.7% PFA, permeabilized with PBS/0.1% Tx100, stained with DAPI and A568-phalloidin, and stacks were taken on a Leica Sp5 confocal microscope.
Colonization of GF mice with in vitro-grown SFB
This experiment was performed two independent times with similar results; one experiment is shown. SFB grown in vitro for 3 days on TC7 and mICcl2 cells on TWs were divided in equal parts and one half was filtered through a 5-μm filter to obtain an IO only fraction. Bacteria were concentrated by centrifugation to obtain 0.25 ml of bacteria in PBS per mouse. The number of animals used followed availability of animals, isolators and input quantities. Randomization or blinding was otherwise not performed. Two groups of 4 11-week old C57BL/6 male and female mice maintained at the germfree facility at Institut Pasteur were starved for one night, gavaged with 0.25 ml 400 mM Sodium Bicarbonate, followed by 0.25 ml of in vitro-grown SFB. Age-matched control mice were colonized with in vivo-derived SFB as described below. Fecal samples were collected for each mouse at various times during a 3 week period, SFB DNA was extracted using the Qiagen stool kit and quantitated based on qPCR analysis of 16S rDNA and comparison to an SFB DNA sample of known SFB genome concentration as determined using Illumina sequencing. To monitor SFB associated with the ileum, DNA was extracted from frozen ileal biopsies using the method by Godon et al. (1997)28.
Isolation and staining of lamina propria lymphocytes from C57BL/6 mice
Age matched GF B6 mice and mice colonized with either in vitro or in vivo-grown SFB for 3 weeks were analysed for their innate and adaptive immune response as described in Gaboriau-Routhiau et al. 20095. Briefly, after excision of PP, the mouse small intestine was washed in PBS, ileal samples were placed in RNAlater for RNA extraction, cDNA synthesis and qPCR analysis using SYBR or Taqman technologies (Applied Biosystems) using a QuantStudio7 qPCR machine. Values were normalized to TfrC.
Lamina propria lymphocytes (LPL) were prepared as previously described5. The remaining small intestine was incubated 4-times in 60 mL of PBS-3 mM EDTA (Sigma) for 10 min at 37°C, and digested in 60 mL of RPMI 1640 added with 20% FCS (Gibco), 100 U/mL of collagenase (Sigma) and 175 U/mL of DNase I (Sigma) for 40 min at 37°C. LPL were then purified on a 40-80% Percoll gradient run for 15 min at 2,000 g and resuspended in DMEM-Glutamax added with 8% FCS, 1mM HEPES, 0.02 mM folic acid, 0.67 mM L-Arginine and 0.27 mM L-Asparagine (all from Sigma).
Analysis of LPL for surface antigens and intracellular expression of IL-17 and IL-10 was performed by flow cytometry as described5. Briefly, LPL were stimulated for 4 hours with 100 ng/mL phorbol 12-myristate 13-acetate and 1μg/mL ionomycin, in the presence of Brefeldin A (10μg/mL) (all from Sigma). Cells used for surface analysis were left unstimulated. For surface staining, LPL were labeled for 20 min at 4°C with a cocktail of the following antibodies: FITC-anti-GL7 (clone GL7), PerCP-anti-CD8a (clone 53-6.7), APC-H7-anti CD4 (clone GK1.5), AF647-anti-B220 (clone RA3-6B/2) (all from BD Pharmingen), PE-anti-IgA (Southern Biotech), eFluor450-anti-CD45 (clone 30-F11) and PECy7-anti-CD3 (clone 145-2C11) (both from eBioscience).
For intracellular cytokine staining, cells were further fixed in 2% PFA for 20 min at room temperature, washed and stained overnight at 4°C with PE-anti-IL-17 (clone TC11-18H10) and APC-anti-IL-10 (clone JES5-16E3) (BD Pharmingen) diluted in PBS-1% FCS-0.5% saponin (Sigma). Labeled cells were analyzed with a FACS CANTO II and Diva software (BD Biosciences). Gates were set on living cells after Aqua live/dead dye exclusion (Invitrogen).
For qPCR analysis, the median value of GF mice was calculated and used as the reference value of 1 for comparison of the median value of the test samples.
Colonization of GF mice with SFB from feces and E. coli
Germfree male and female C3H/HeN mice were obtained from INRA (Anaxem plateform, Jouy-en-Josas) germfree facilities. 8-9 week-old germfree mice were gavaged with 0.5 mL of either fresh anaerobic cultures of E. coli MG1655 or fecal homogenate from SFB-monoassociated mice5. The data set includes 7 GF mice, 4 SFB-colonized mice, 5 E. coli-colonized mice and 4 conventional mice obtained from 2 independent experiments. Colonization by SFB was monitored in feces through bacterial DNA extraction and 16S rDNA amplification by qPCR using specific primer pairs for SFB. Values were normalized to Ccl25, a constitutively expressed epithelial cell marker, and compared to GF control mice Data are presented as box plots of 25-75% percentiles with median and min/max whiskers. Statistical significance was calculated using the two-tailed Student t-test (* p=< 0.05, ** p=< 0.01, *** p=< 0.001). Germfree and gnotobiotic mice were maintained in plastic isolators and fed ad libitum on a commercial diet sterilized by γ-irradiation (40 kGy). The number of animals used followed availability of animals, isolators and were performed at minimum in duplicate. Randomization or blinding was otherwise not performed. Gnotobiotic mice were sacrificed on day 21 post colonization in parallel with age-matched germfree controls. All procedures were carried out in accordance with European guidelines for the care and use of laboratory animals and with permission of the French veterinary services.
Host response in vitro to SFB growth and MAMPs stimulation
Host response to SFB and MAMPs includes pooled results from 5 and 3 independent experiments, respectively, of 2 to 3 technical samples for a total of 16 and 8 samples, respectively. Generally a minimum of triplicate biological replicates was used and increased if trends were clear but significance not. After 3 days of in vitro growth of SFB on either mICcl2 or TC7s on TWs in SFB medium lacking hemin and sodium ascorbate, at 1-2.5% oxygen, cells were lysed and RNA was extracted using the Nucleospin RNA kit (Macherey-Nagel). cDNA was synthesized using RNA superscript II, oligo dT, RNAseout and dNTPs (Invitrogen) and pPCR was performed on ABI 7900HT and QuantStudio7 (Life Technologies) qPCR machine using the protocol described in Schnupf et al.29. TaqMan assays were performed as suggested by the supplier. Values were normalized to B2M and Ct values for Reg3γ, Tnfα and Fabp2 were set to 41 in control cells due to the lack of transcript detection. MAMPs stimulation was performed with the following agonists at the highest concentrations recommended by the supplier (Invivogen): Pam2CSK4 (tlrl-pm2s-1) at 100 ng/ml, Pam3CSK4 (tlrl-pms) at 300 ng/ml, peptidoglycan of E. coli K12 (Tlr-ksspgn) at 10 μg/ml, MDP (tlrl-mdp) at 10 μg/ml, CpG (tlr-1584) at 3 μg/ml, flagellin (tlrl-pstfla-5) at 100 ng/ml. 10x excess for flagellin was also tested and found to be similar. Statistical analysis was performed using the two-tailed Student t-test (* p=< 0.05, ** p=< 0.01, *** p=< 0.001) and center values are Mean values ± s.e.m.
Mouse and human qPCR primers used
| Mouse Taqman® assays | Mouse Sybr® primers | ||
|---|---|---|---|
| B2M | Mm00437762_m1 | B2M | F:tcagtcgtcagcatggctcgc;R:tccggtgggtggcgtgagtatac |
| TfrC | Mm00441941_m1 | iNos | F:cagctgggctgtacaaacctt;R:cattggaagtgaagcgtttcg |
| Ccl25 | Mm00436443_m1 | Saa1 | F:catttgttcacgaggctttcc;R:gtttttccagttagcttccttcatgt |
| Fabp2 | Mm00433188_m1 | Saa2 | F:tgtgtatcccacaaggtttcaga;R:ttattaccctctcctcctcaagca |
| Reg3γ | Mm01181783_g1 | Saa3 | F:cgcagcacgagcaggat;R:ccaggatcaagatgcaaagaatg |
| Fut2 | Mm00490152_S1 | Cxcl1 | F:tggctgggattcacctcaag;R:caagcctcgcgaccattct |
| Aqp3 | Mm01208559_m1 | Cxcl10 | F:gccgtcattttctgcctcat;R:gcttccctatgcccctcatt |
| Pigr | Mm00465049_m1 | Human Taqman® assays | |
| Ltf | Mm00434787_m1 | B2M | Hs00984230_m1 |
| Tff3 | Mm00495590_m1 | Saa1 | Hs00761940_s1 |
| Tnfα | Mm00443258_m1 | Saa2 | Hs01667582_m1 |
| IL1α | Mm00439620_m1 | IL6 | Hs00985639_m1 |
| IL1β | Mm00434228_m1 | IL8 | Hs99999034_m1 |
| IL6 | Mm00446190_m1 | Ccl20 | Hs01011368_m1 |
| IL15 | Mm00434210_m1 | Reg3γ | Hs00417999_m1 |
| IL18 | Mm00434225_m1 | iNos | Hs01075529_m1 |
| Cxcl2 | Mm00436450_m1 | Cxcl10 | Hs00171042_m1 |
| Ccl2 | Mm00436450_m1 | Fut2 | Hs00382834_m1 |
| Ccl5 | Mm01302428_m1 | IL15 | Hs01003716_m1 |
| Ccl7 | Mm01308393_g1 | IL18 | Hs01038788_m1 |
| Ccl20 | Mm01268754_m1 | Ccl5 | Hs00982282_m1 |
| Ccl28 | Mm00445039_m1 | ||
| Csf2 | Mm01290062_m1 | ||
Extended Data
Extended Data Figure 1. Intestinal colonization of in vitro-grown SFB and host response.
QPCR quantitation (a) and SEM (b) of in vitro-grown SFB used for gavage; c, QPCR quantitation of SFB in fecal samples; d, SEM of SFB attachment in vivo at 25 days post gavage of C57BL/6 mice; e, Host gene expression in the ileal lamina propria in conventional or germfree mice colonized with either SFB or E. coli for 21 days; f, SEM of 21-day SFB-colonized germfree C3H/HeN mouse ileum showing vacant attachment sites.
Acknowledgments
We are grateful to Benoît Marteyn, F-X Campbell-Valois and Claude Parsot for helpful discussions and Marion Picard and Sabine Rakotobe for help with in vivo experiments. This work was supported by INSERM, Institut Pasteur, College de France and INRA, and grants TORNADO-FP7-KBBE-2007-2A-222720, ANR-2010-BLAN1317, ERC-2009-AG-232798-HOMEOPITH, ERC-2013-AdG-339579-DECRYPT and ERC-2013-AdG-339407-IMMUNOBIOTA and the Investissement d’Avenir ANR-10-IAHU-01 and LabEX IBEID. PSa is a HHMI Foreign Scholar.
Footnotes
The authors declare no conflict of interest.
Author Contributions:
NCB, VGR, PSa. and PSc conceived the project and discussed experiments. PSc. designed and performed all in vitro experiments. VGR and MG performed the in vivo challenge experiments, VGR maintained SFB mice and MG analysed the TC7 and RF the mICl2 host response. MMN processed SEM samples and took images with PS. GN assisted in vitro experiments. PSc wrote the paper and PSa, NCB and VGR edited the manuscript.
References
- 1.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol. 2010;10:735–744. doi: 10.1038/nri2850. [DOI] [PubMed] [Google Scholar]
- 3.Schnupf P, Gaboriau-Routhiau V, Cerf-Bensussan N. Host interactions with Segmented Filamentous Bacteria: An unusual trade-off that drives the post-natal maturation of the gut immune system. Seminars in Immunology. 2013;25:342–351. doi: 10.1016/j.smim.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 6.Jepson MA, Clark MA, Simmons NL, Hirst BH. Actin accumulation at sites of attachment of indigenous apathogenic segmented filamentous bacteria to mouse ileal epithelial cells. Infection and Immunity. 1993;61:4001–4004. doi: 10.1128/iai.61.9.4001-4004.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chase DG, Erlandsen SL. Evidence for a complex life cycle and endospore formation in the attached, filamentous, segmented bacterium from murine ileum. J Bacteriol. 1976;127:572–583. doi: 10.1128/jb.127.1.572-583.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klaasen H, Koopman JP, Poelma F. Intestinal, segmented, filamentous bacteria. FEMS Microbiology. 1992;88:165–180. doi: 10.1111/j.1574-6968.1992.tb04986.x. [DOI] [PubMed] [Google Scholar]
- 9.Yin Y, et al. Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J. 2012;7:615–621. doi: 10.1038/ismej.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schnupf P, Gaboriau-Routhiau V, Cerf-Bensussan N. Host interaction with Segmented filamentous Bacteria: An unusual trade-off that drives the post-natal maturation of the gut immune system. Seminars in Immunology. 2013;25:342–351. doi: 10.1016/j.smim.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010 doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chappert P, Bouladoux N, Naik S, Schwartz RH. Specific gut commensal flora locally alters T cell tuning to endogenous ligands. Immunity. 2013;38:1198–1210. doi: 10.1016/j.immuni.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kriegel MA, et al. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci USA. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yurkovetskiy L, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prakash T, Oshima K, Morita H, Fukuda S, Imaoka A. Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of th17 cell differentiation. Cell host & microbe. 2011;10:273–284. doi: 10.1016/j.chom.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Sczesnak A, et al. The Genome of Th17 Cell-Inducing Segmented Filamentous Bacteria Reveals Extensive Auxotrophy and Adaptations to the Intestinal Environment. Cell Host & Microbe. 2011;10:260–272. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuwahara T, et al. The Lifestyle of the Segmented Filamentous Bacterium: A Non-Culturable Gut-Associated Immunostimulating Microbe Inferred by Whole-Genome Sequencing. DNA Research. 2011;18:291–303. doi: 10.1093/dnares/dsr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pamp SJ, Harrington ED, Quake SR, Relman DA, Blainey PC. Single-cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB) Genome Res. 2012;22:1107–1119. doi: 10.1101/gr.131482.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolotin A, et al. Genome Sequence of ‘Candidatus Arthromitus’ sp. Strain SFB-Mouse-NL, a Commensal Bacterium with a Key Role in Postnatal Maturation of Gut Immune Functions. Genome Announc. 2014;2:1–2. doi: 10.1128/genomeA.00705-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He G, et al. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc Natl Acad Sci USA. 1999;96:4586–4591. doi: 10.1073/pnas.96.8.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferguson DJ, Birch-Andersen A. Electron microscopy of a filamentous, segmented bacterium attached to the small intestine of mice from a laboratory animal colony in Denmark. Acta Pathol Microbiol Scand B. 1979;87:247–252. doi: 10.1111/j.1699-0463.1979.tb02434.x. [DOI] [PubMed] [Google Scholar]
- 23.Ferguson DJP, Birch-Andersen A. Electron Microscopy of a Filamentous, Segmented Bacterium Attached to the Small Intestine of Mice from a Laboratory Animal Colony in Denmark. Acta Pathologica Microbiologica Scandinavica Section B Microbiology. 2009;87B:247–252. doi: 10.1111/j.1699-0463.1979.tb02434.x. [DOI] [PubMed] [Google Scholar]
- 24.Tannock GW, Miller JR, Savage DC. Host specificity of filamentous, segmented microorganisms adherent to the small bowel epithelium in mice and rats. Appl Environ Microbiol. 1984;47:441–442. doi: 10.1128/aem.47.2.441-442.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goto Y, et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 2014;345:1254009. doi: 10.1126/science.1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shima T, et al. Differential effects of two probiotic strains with different bacteriological properties on intestinal gene expression, with special reference to indigenous bacteria. FEMS Immunology & Medical Microbiology. 2008;52:69–77. doi: 10.1111/j.1574-695X.2007.00344.x. [DOI] [PubMed] [Google Scholar]
- 27.LEcuyer E, et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Godon JJ, Zumstein E, Dabert P, Habouzit F, Moletta R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol. 1997;63:2802–2813. doi: 10.1128/aem.63.7.2802-2813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnupf P, Sansonetti PJ. Quantitative RT-PCR profiling of the rabbit immune response: assessment of acute Shigella flexneri infection. PLoS ONE. 2012;7:e36446. doi: 10.1371/journal.pone.0036446. [DOI] [PMC free article] [PubMed] [Google Scholar]