Abstract
Water hampers the formation of strong and durable bonds between adhesive polymers and solid surfaces, in turn hindering the development of adhesives for biomedical and marine applications. Inspired by mussel adhesion, a mussel foot protein homologue (mfp3S-pep) is designed, whose primary sequence is designed to mimic the pI, polyampholyte, and hydrophobic characteristics of the native protein. Noticeably, native protein and synthetic peptide exhibit similar abilities to self-coacervate at given pH and ionic strength. 3,4-dihydroxy-l-phenylalanine (Dopa) proves necessary for irreversible peptide adsorption to both TiO2 (anatase) and hydroxyapatite (HAP) surfaces, as confirmed by quartz crystal microbalance measurements, with the coacervate showing superior adsorption. The adsorption of Dopa-containing peptides is investigated by attenuated total reflection infrared spectroscopy, revealing initially bidentate coordinative bonds on TiO2, followed by H-bonded and eventually long-ranged electrostatic and Van der Waals interactions. On HAP, mfp3s-pep-3Dopa adsorption occurs predominantly via H-bond and outer-sphere complexes of the catechol groups. Importantly, only the Dopa-bearing compounds are able to remove interfacial water from the target surfaces, with the coacervate achieving the highest water displacement arising from its superior wetting properties. These findings provide an impetus for developing coacervated Dopa-functionalized peptides/polymers adhesive formulations for a variety of applications on wet polar surfaces.
1. Introduction
The adhesion of polymers to wet surfaces is frustrated by a weak boundary layer of adsorbed water that obstructs molecular contact and hence the formation of strong and stable bonds between the polymer and the surface. Among sessile organisms, mussels routinely attach to diverse underwater surfaces by secreting a complex fluid that consists of adhesive proteins, which eventually harden into a solid byssus.[1] The catecholic amino acid known as Dopa (3,4-dihydroxyphenyl-l-alanine) is a key chemical signature of mussel adhesive proteins and implicated in promoting underwater interfacial adhesion, as well as bulk cohesion, via a variety of adaptive surface interactions and covalent crosslinking reactions, e.g., diDopa[2] and cysteinyl DOPA.[3] Catechol, a weak diprotic acid with pKa1 and pKa2 of ≈9.4 and ≈13.7, respectively,[4] exhibits high affinity for wet mineral and metal oxide surfaces.[5] It forms bidentate innersphere complexes on the {001} facets of TiO2 (anatase) between Ti(IV) atoms and its two o-hydroxyl groups,[6] whereas catechol interacts only through weaker hydrogen-bonds on mica.[7] As a result, the adhesion of the mussel foot protein (mfp-3) to mica is significantly weaker than to TiO2 (0.09 vs 0.51 µJ × m−2 molDopa−1), due to hydrogen bonds competing with water molecules for adsorption on the mica surface sites. Single-molecule atomic force microscopy measurements[7a] previously revealed a dissociation energy of 22 kcal mol−1 for Dopa in contact with wet TiO2, which constitutes one of the strongest reversible binding interactions known. Tyrosine (Tyr) binding to the same surface is much weaker, suggesting that the two o-hydroxyl groups in Dopa provide significantly stronger binding than a single OH group. Therefore, converting Tyr side chains to Dopa is a crucial step in the maturation of adhesive mfps and, more generally, for the formulation of mussel inspired synthetic adhesives.[8]
Dopa-containing mfp-3S[9] is the most abundant directly extractable protein from mussel adhesive plaques of the California mussel (Mytilus californianus). Mfp-3S is capable of promoting both adhesion and cohesion within the adhesive plaque and provides an efficient shielding environment against Dopa oxidation by dint of its abundant aromatic and hydrophobic residues (Tyr+Dopa+Trp ≈35%). Another intriguing aspect of mfp-3S is its ability to form single-component coacervates[10] – the first direct evidence of mussels using coacervation as a delivery strategy. To date, among all mfps tested, mfp-3S is the only protein that displays this ability. A biological model of coacervation as a delivery strategy for underwater adhesives has also been proposed for the sandcastle worm (Phragmatopoma californica) (complex coacervation between two oppositely charged proteins),[11] and may be more widespread among other fouling species than previously thought.[12] On the technological side, numerous reports have proposed to process and deliver synthetic adhesives/coatings as complex coacervates.[13]
Notwithstanding previous work, little effort has been dedicated to understanding and translating Dopa-containing coacervates. Important questions concerning self-coacervation still remain and cannot be addressed by investigating the native protein. These include (1) the contribution of Dopa to mfp-3S coacervation; (2) the influence of the primary amino acid sequence or composition on coacervation; and (3) the need for post-translational conversion of Tyr residues into Dopa for maximizing adsorption of the mfp-3S coacervate to wet substrates. To address these questions, as well as to provide a synthetic platform for potential applications, we designed a peptide analogue (mfp3S-pep) comprised of 25 amino acids to mimic the native protein. The peptide was then enzymatically modified to provide the desired level Tyr to Dopa conversion (mfp3S-pep-Dopa), including the ≈10 mol% Dopa content of the native protein.[3] The coacervation behavior of the modified and unmodified peptides was investigated over a range of pH and ionic strength to determine the optimal experimental conditions for peptide self-coacervation. Hydroxyapatite (HAP) and TiO2 (anatase) surfaces were used for adsorption studies owing to their obvious relevance to biomedical applications. Peptide and coacervate adsorption was investigated by quartz crystal microbalance with dissipation (QCM-D) and attenuated total reflection-infrared spectroscopy (ATR-IR). Dopamine was initially used in the ATR-IR experiments as a model catechol to aid in interpreting the IR spectra of the more complex Dopa-containing peptides and coacervates. mfp-3S-inspired self-coacervating peptides exhibited strong and irreversible binding to hydrated HAP and TiO2. Given the established biocompatibility[14] of mfps, our results suggest promising opportunities for the next generation adhesive technologies in tissue engineering, orthopedic, and dental applications.
2. Results and Discussion
2.1. Peptide Design
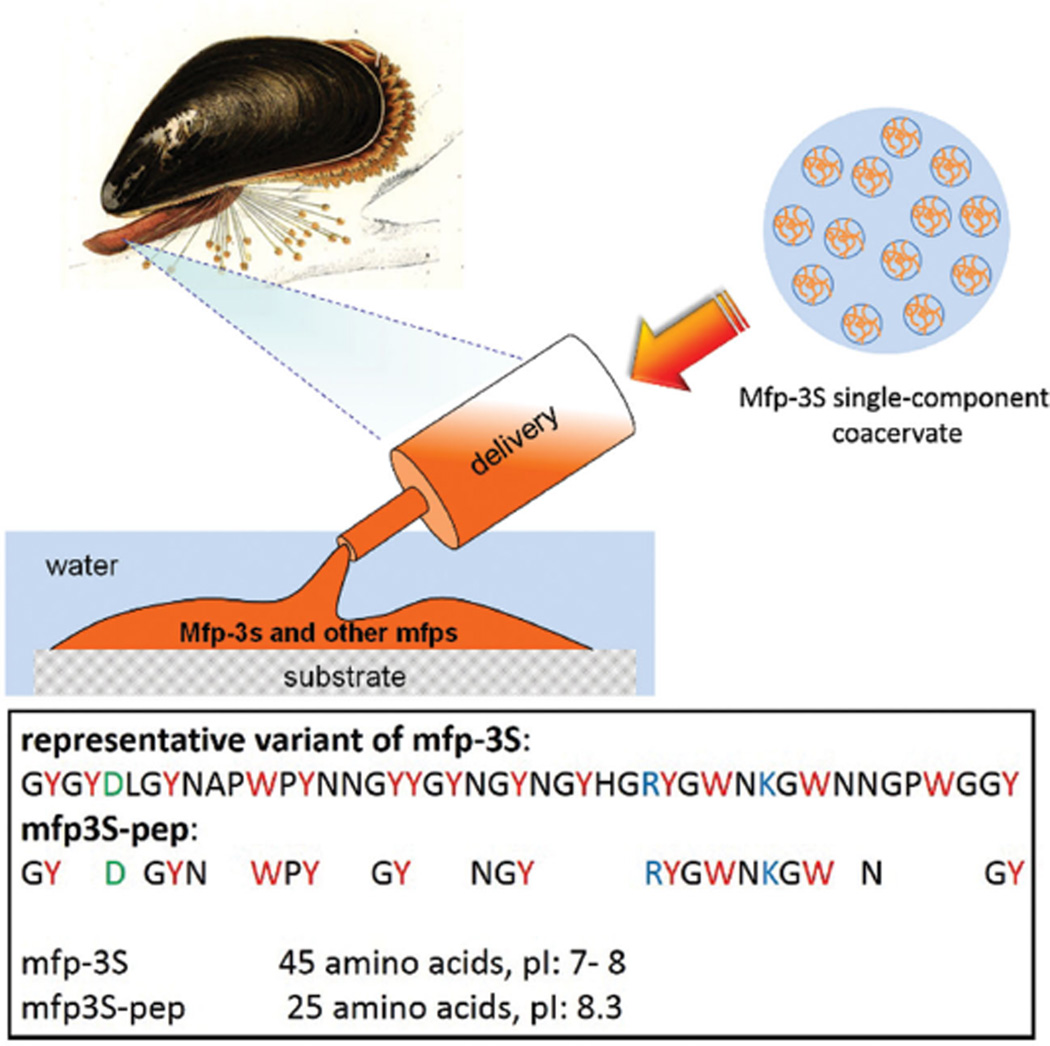
To address questions that would not be possible with a microgram supply of purified native mfps and to explore the potential of the unique mfp-3S system, we prepared a shorter peptide sequence (shown in Figure 1) modeled after the native protein by applying the following criteria: All of the charged amino acids as well as most aromatic residues including Tyr/Dopa in the native sequence were maintained. Furthermore, some flanking residues, such as Gly and Asn, were retained to maintain the peptide’s polarity and flexibility. For ease of synthesis and cost efficiency, the peptide length was limited to about half of the native protein mfp-3S. The above mentioned design criteria are based on our knowledge of the protein’s composition/sequence and property/function relationships. Notably, the amphoteric characteristic and the high content of hydrophobic/aromatic residues, including Dopa, are hallmarks for the adhesion, cohesion, and coacervation performance of mfp-3S.[10] The resulting peptide sequence (mfp3S-pep) exhibited a virtual isoelectric point (pI) of 8.3, which was slightly above that of the native mfp-3S (pI between 7 and 8).
Figure 1.
Mussel derived phase-separating adhesive proteins and their peptide mimics. Upper: schematic illustration of mfp-3S being delivered onto a wet substrate surface in the form of a single-component coacervate followed by its hardening into the mussel’s adhesive plaque together with other mfps. Lower: sequences of native mfp-3S and truncated peptide mfp3S-pep with their corresponding pI. Letters in red, blue, and green denote aromatic, basic, and acidic amino acids, respectively.
2.2. Incorporation of Dopa into Peptide Sequence
Mushroom tyrosinase is known to o-hydroxylate peptidyl Tyr to Dopa and was hence used to hydroxylate Tyr residues in mfp3s-pep.[15] This enzymatic reaction is typically conducted at pH 7 in a phosphate aqueous buffer. However, the peptide had very low solubility under these conditions, therefore, 10% acetonitrile was added to improve its solubility without sacrificing significant enzyme activity. Borate was used to reversibly capture Dopa immediately after it formed, hence minimizing the formation of undesirable side products such as 3,4,5-tri-hydroxyphenylalanine (Topa) and Dopaquinone. Reaction time and enzyme/substrate ratio were adjusted to achieve different Dopa contents. After stopping the reaction by addition of acetic acid to bring the pH down to 3, modified peptides were subjected to reverse phase HPLC, followed by size exclusion chromatography to purify mfp3S-pep with the desired Dopa content. Matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (MALDI-TOF-MS) was used to measure the peptide masses in the eluting fractions from HPLC and Shodex (Figure S1, Supporting Information). Those fractions with masses matching the calculated desired masses, namely 3119 Da and 3167 Da for peptide comprising 3 and 6 Dopa units, respectively, were pooled for amino acid analysis (Figure S2, Supporting Information). The peptide mass increases by 16 Da for each oxygen atom introduced as Tyr are converted into Dopa. Thus, the 48 Da mass differences in Figure S3 (Supporting Information) reflect a peptide with no Dopa (m/z 3071 Da), 3 Dopa residues (3119 Da), and 6 Dopa residues (3167 Da).
2.3. Coacervate Characterization by Turbidity Measurements and Imaging
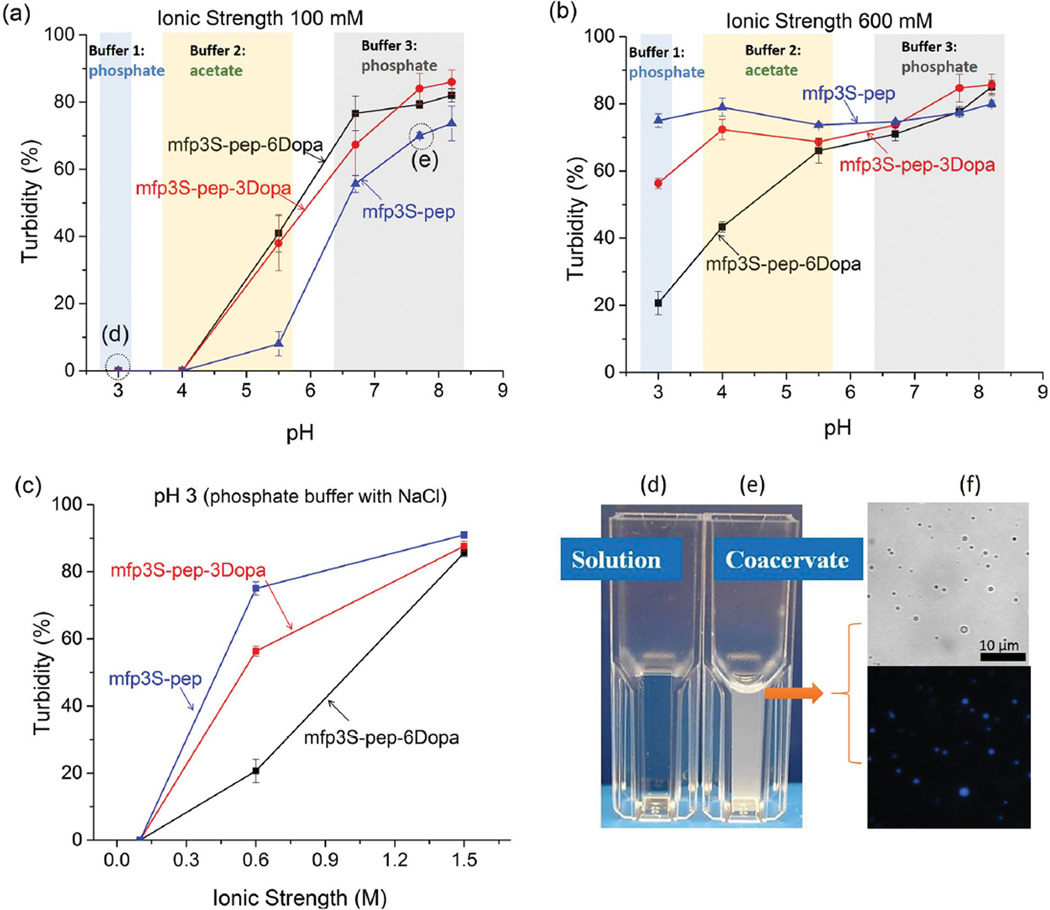
To confirm that turbidity (Figure 2) of mfp3S-pep, mfp3S-pep-3Dopa, and mfp3S-pep-6Dopa was due to coacervation and not precipitation, microscopic images of the peptides in different buffers were taken. Spherical droplets in the resulting mixtures indicated the existence of liquid microphases in the bulk solution, namely, coacervates. A representative image of mfp3S-pep coacervate in pH 6.5 buffer at 100 × 10−3 m ionic strength is shown in Figure 2f, along with a UV micrograph. The blue emission at 450 nm can be attributed to peptidyl-tryptophan residues and confirms that the droplets originated from mfp3S-pep coacervation (see Figure 1). All three peptides were able to form single-component coacervates by adjusting buffer pH and ionic strength. The general trend of coacervation for the three peptides was similar, with the peptide mixtures transitioning from solution to coacervate as pH and/or ionic strength increased. Whereas at 100 × 10−3 m ionic strength, the turbidity versus pH profiles of the three peptides were comparable (Figure 2a), a more distinct separation occurred in the low pH range (from pH 3 to 4) at 600 × 10−3 m ionic strength. We believe that the driving forces for the coacervation of these peptides are similar to those of the native protein mfp-3S,[10] and comprise hydrophobic, electrostatic as well as Dopa-mediated H-bonding interactions with other side chains, e.g., Asn and Tyr. At low pH, where the peptides are positively charged (pI = 8.3), long-range electrostatic repulsions keep peptide molecules apart. As the pH approaches the isoelectric point of the peptide and therefore net neutral charge, electrostatic repulsion decreases, while hydrophobic interaction and short-range electrostatic attraction increase, thereby leading to a net attractive interaction between the peptide chains and, ultimately, to coacervation.
Figure 2.
Turbidity of mfp3S-pep, mfp3S-pep-3Dopa, mfp3S-pep-6Dopa under different a+c) pH and c) ionic strength conditions. Comparison of mfp3S-pep (d) solution and e) coacervate, conditions of which are indicated in (a). f) Representative bright field (top) and UV microscope images (bottom) of mfp3S-pep coacervates.
Incorporation of Dopa into the peptide had a twofold effect on coacervation. First, it rendered the peptide more hydrophilic, thus decreasing inter/intra molecular hydrophobic interactions and helps explain why the peptide with the higher Dopa content exhibited lower turbidity at low pH (Figure 2b). Second, at short range, increased H-bonding between Dopa residues and peptide backbone/other side chains of the peptide (e.g., Asn and Tyr) resulted in increased inter/intramolecular interactions. Dopa involvement in H-bond formation came into play at pH near the peptide pI, where the peptide molecules are already driven in close proximity by hydrophobic interactions and short-range electrostatic attractions. At relatively high pH, H-bonding from Dopa counter-balanced the partial loss of hydrophobic interactions. Therefore, the turbidity values of three peptides merged at around pH 8.
2.4. Acidic Version of mfp3S-pep
Both mfp3S and the mfp3S-pep contain two negatively charged and 3 positively charged side chains (including the N- and C-termini), such that the sequences possess a +1 net positive charge. We tested the hypothesis that this +1 net charge helps drive coacervation by binding to aromatic residues via a π-cation interaction.[16] A new peptide was thus prepared by replacing Arg with Asp (pI = 4.2) (Figure S3a, Supporting Information), and this acidic version of mfp3s-pep was still able to self-coacervate. Not surprisingly, compared with mfp3S and mfp3S-pep, the optimum coacervation condition of the acidic peptide shifted to lower pH (Figure S3b, Supporting Information). This experiment proved that the positive charge of Arg in mfp3S-pep is not essential for the formation of a coacervate, and its replacement with an acidic residue, such as Asp, could trigger coacervation at acidic conditions. However, at pH 4 (close to their pI) and high ionic strength (600 × 10−3 m), the acidic peptides precipitated instead of coacervating, due to stronger intra/intermolecular attractive interactions, namely electrostatic, hydrophobic, as well as H-bonding interactions.
2.5. Mfp3S-pep with Randomized Sequences
In order to investigate the role of the primary amino acid sequence on the peptide coacervation behavior, the sequence of mfp3S-pep was randomized and three new peptides, termed mfp3S-pep-random #1, #2, and #3, were generated as shown in Figure S4 (Supporting Information). Mfp3S-pep-random #1 and #3’s solubilities in 10 × 10−3 m acetic acid were 0.2 mg mL−1 and ≈1 mg mL−1, respectively, which were both less soluble than mfp3S-pep. Under the experimental conditions used for mfp3S and mfp3S-pep, neither mfp3S-pep-random #1 nor #3 could form a coacervate. Only mfp3S-pep-random #2 showed similar coacervation behavior to mfp3S-pep under the tested conditions. In order to determine whether mfp3-pep and its randomized versions varied significantly in their secondary structures, we conducted circular dichroism (CD) measurements (Figure S5, Supporting Information). Mfp3s-pep-random #1 spectrum exhibited a significantly different spectral signature from the other peptides, with features characteristic of β-sheet. On the other hand, a random coil dominates the others, mfp3s-pep, random #2 and #3. The broad and rarely encountered positive peak centered at 230 nm in all 4 peptides’ spectra, can be attributed to the high content of Tyr.[17] These CD results are broadly consistent with the predicted conformation using the I-Tasser homology modeling package[18] (Figure S6, Supporting Information): Mfp3s-pep-random #1 shows the highest tendency of forming β-sheet in the region enriched with aromatic residues, whereas the other peptides were mainly predicted to adopt random coil conformation. The tightly packed region of β-sheet in mfp3s-pep-random #1 could be due to enhanced local π– π and quadrupole interactions,[19] which is also consistent with its lowest solubility in aqueous solution and its highest tendency to precipitate among all 4 peptides. A more comprehensive structural study of the peptides is in progress. The above results provide useful insights for designing self-coacervating peptides: the flanking distribution of small amino acids, such as glycine and asparagine, around tyrosine residues is essential to trigger the coacervate formation, since these side chains enhance the flexibility of the peptide backbone.[20]
2.6. Mfp3S-pep and mfp3S-pep-3Dopa Adsorption Behavior on HAP and TiO2 Surfaces by QCM-D
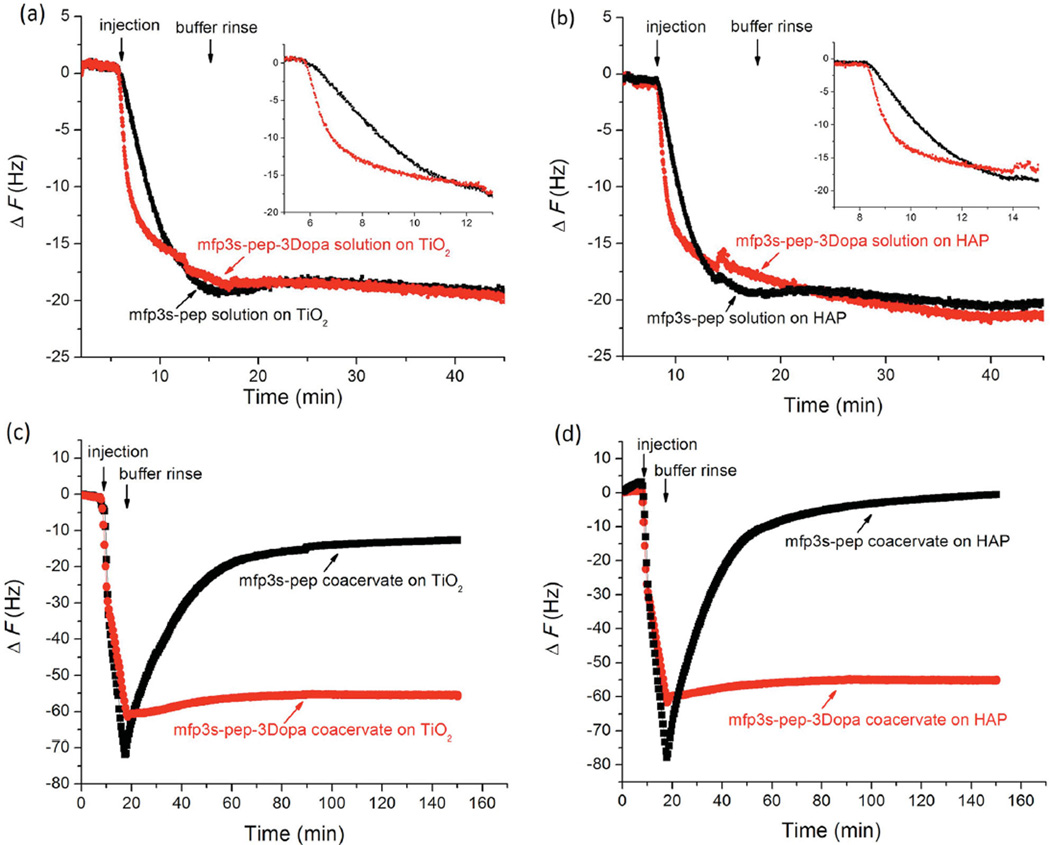
QCM-D was used to study the peptide adsorption behavior to HAP and TiO2 surfaces (Figure 3). We compared the adsorption behavior of mfp3S-pep and mfp3S-pep-3Dopa solutions at 0.1 mg mL−1 in 100 × 10−3 m acetic acid on TiO2 (Figure 3a) and HAP (Figure 3b) surfaces. Mfp3S-pep-3Dopa was chosen because its Dopa content is similar to that of the native protein (≈10 mol%). The ultimate adsorption in terms of frequency change of two peptides on each surface was similar (ΔF = −20 Hz), with negligible desorption when rinsing the surfaces with buffer. Interestingly, mfp3S-pep-3Dopa showed faster initial (≈1 min) adsorption (14.3 ΔF min−1 on TiO2; 16.7 ΔF min−1 on HAP) than mfp3S-pep (3.7 ΔF min−1 on TiO2; 5.3 ΔF min−1 on HAP) (see the insets in Figure 3a,b), indicating that Dopa accelerates the kinetics of adsorption on these two surfaces. We attribute this behavior to the longer binding lifetime of Dopa bidentate chelation and bidentate hydrogen-bonding on TiO2 and HAP, respectively, as compared with monodentate binding by Tyr.[21]
Figure 3.
Adsorption behavior of mfp3S-pep and mfp3S-pep-3Dopa on TiO2 and HAP surfaces by QCM. a) solution on TiO2 surface; b) solution on HAP surface; c) coacervate on TiO2 surface; d) coacervate on HAP surface. Inset figures in (a) and (b) show the initial adsorption. The concentration of peptides and coacervates was 0.1 mg mL−1, and buffers were 100 × 10−3 m acetic acid for peptides and PBS at pH 6.5 and 100 × 10−3 m ionic strength for coacervates.
Figure 3c,d shows the adsorption and desorption behavior of mfp3S-pep and mfp3S-pep-3Dopa coacervates formed at pH 6.5 buffer and 100 × 10−3 m ionic strength on TiO2s and HAP surfaces. Both coacervates attained similar maximum adsorption plateaus (ΔF between −70 and −80 Hz), showing a greater adsorptive ability (about fourfold increase in frequency change) than their corresponding peptide solutions. The other significant distinction between the two coacervates occurred upon rinsing with buffer. The desorbed mass, which is associated with an increase in frequency in QCM experiments, was negligible for the Dopa-containing coacervate, whereas mfp3-pep coacervate was substantially desorbed from both TiO2 and HAP. We attribute the different adsorption behavior to stronger interfacial bonding between the Dopa-containing coacervate and the substrates, as well as to enhanced Dopa-mediated intra/intermolecular H-bonding interactions within the coacervate adsorbed layer.
2.7. Adsorption Studies on TiO2 and HAP by ATR-IR Spectroscopy
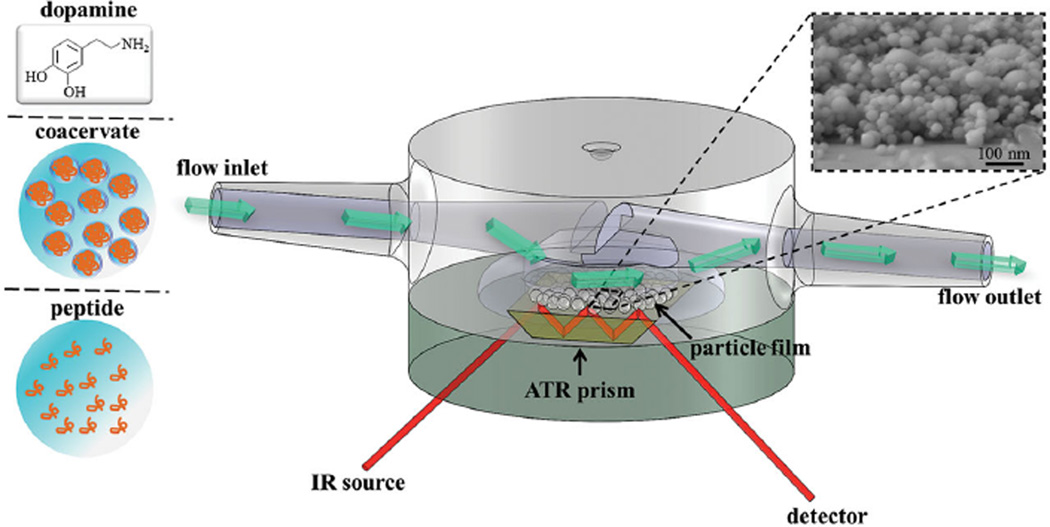
Figure 4 shows a schematic diagram of the flow cell setup used in ATR-IR adsorption experiments. A field emission scanning electron microscope (FESEM) image of the ≈100 nm thick HAP nanoparticle film used in this work is also presented along with the chemical structure of dopamine, the catechol model compound used to assign catechol vibrational modes in ATR-IR adsorption investigations of more complex peptide and coacervates. Micrographs of the ≈100 nm TiO2 (anatase) nanoparticle film have been reported previously.[22]
Figure 4.
Schematic diagram of the flow cell and the 3-internal reflection ZnSe prism adopted in this work for ATR-IR measurements of adsorbed molecules on a nanoparticle film-coated ZnSe prism. The inset shows an FESEM image of an ≈100 nm thick HAP nanoparticle film and the chemical structure of dopamine used as a model compound for adsorption studies of the catechol-containing peptide and its coacervate phase.
The ATR-IR spectrum of 100 mg mL−1 dopamine in 100 × 10−3 m acetic acid solution on the bare ZnSe prism was compared to those of dopamine adsorbed on nanoparticulate film of TiO2 and HAP from more dilute solutions (0.1, 0.5, 1, and 10 mg mL−1) (Figure S8, Supporting Information). The absorbance increased with dopamine concentration and was already detectable on TiO2 at the lowest concentration of 0.1 mg mL−1. Weaker, yet discernible absorbance was recorded on HAP at 0.5 mg mL−1 dopamine concentration. Of particular interest is the doublet observed on the TiO2 surface at 1492 and 1274 cm−1 arising from ν(CC) of the aromatic ring and ν(CO) modes, respectively, which is indicative of catechol-Ti(IV) bidentate binuclear coordinative bond.[23] Dopamine has been shown to complex metal cations and metal oxides via either a monodentate coordination (one catechol OH group binds to a surface metal ion, whereas the second OH group forms a hydrogen bond to the adjacent metal ion) or bidentate coordination known as chelation.[6c, 24] Such bands were not observed in the spectra of dopamine in solution and adsorbed on HAP. The ATR-IR spectra of dopamine adsorbed on HAP showed similarities in terms of bands number and position to the spectrum of catechol in solution. The major absorbance peaks at 1520, 1286, and ≈1250 cm−1 were thus assigned to the ν(CC)+δ(CH) and ν(CO) modes of catechol in solution. Although the HAP particle size was on average eightfold larger than that of TiO2 (<200 vs <25 nm), thus consequently exhibiting a smaller surface area, the catechol absorption on HAP was of considerable intensity, indicating its high affinity for HAP. The binding of catechol species to HAP has been previously ascribed to the formation of divalent hydrogen bonds.[25] However, water molecules also interact with HAP via hydrogen bonding, thus potentially competing with catechols via the same bond type. Substantial adsorption of catechol to HAP may therefore arise from a divalent hydrogen-bonding formation, which should have a longer binding lifetime than the corresponding monovalent bond of water molecules. Given the much greater number of water molecules hydrating the HAP surface, it can also be argued that other surface-bound structures are likely for catechol interacting with HAP. Strong noncovalent π-cation interactions between the aromatic ring of catechol and Ca+/2+ ions of HAP are possible yet rarely considered. Comprehensive quantum mechanic calculations have shown that Ca2+ ions likely do not bind directly to the aromatic ring because of the substantial energetic penalty of dehydrating a Ca2+ ion. However, the complex of a Ca2+ ion with its inner hydration shell, Ca2+(H2O)6, can interact electrostatically with the catechol aromatic ring.[26]
2.7.1. Adsorption on TiO2
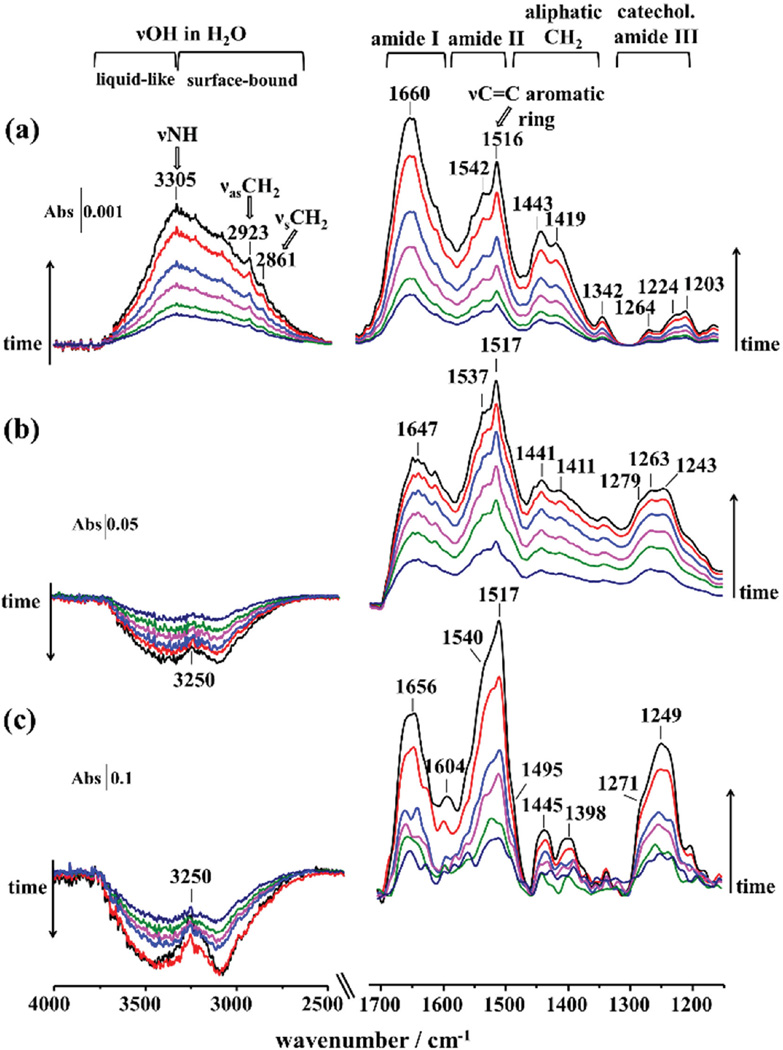
ATR-IR spectra recorded during the time-course of mfp3S-pep (all at 0.1 mg mL−1), mfp3S-pep-3Dopa solution, and coacervate adsorption to the TiO2-coated ZnSe prism are shown in Figure 5. The infrared spectra revealed peptide bonds, namely, amide I (≈1650 cm−1) and amide II (≈1540 cm−1), and a major absorbance peaking at 1517 cm−1 due to the ν(CC)+δ(CH) mode of aromatic rings. The amide I was the dominant absorption in the spectra of the adsorbed mfp3S-pep, whereas amide II region was the most intense for mfp3S-pep-3Dopa and its coacervate phase. This difference arises from the absorption associated with aromatic ring modes at 1517 cm−1, providing evidence for a greater involvement of Dopa/Tyr side chains in the adsorption process of the Dopa-bearing peptide and coacervate on the metal oxide surface, as compared to mfp3S-pep. Initially, the absorbance at 1517 cm−1 for the adsorbed coacervate (Figure 5c) was comparable with that of the amide II mode, but after ≈6 min the former increased with time to eventually become the dominant peak. Concomitantly, a peak due to aromatic residues at 1604 cm−1 became apparent after 10 min of adsorption.[27] Other vibrations in the ≈1450− 1400 cm−1 region are associated with CH2 aliphatic vibrations. In the catechol-related vibrational region (1300 to 1200 cm−1) a broad band with peaks at 1279, 1263, and 1243 cm−1 was recorded for mfp3S-pep-3Dopa (Figure 5b). The absorption peak at 1279 cm−1 is close to the peak associated with catechol-Ti(IV) coordination mode at 1274 cm−1 as observed for dopamine (see Figure S8b, Supporting Information). Both peaks originate from the in-plane δOH ring modes of the tyrosine present in mfp3S-pep-3Dopa.[28] The intensity of the catechol characteristic region in the adsorbed coacervate is markedly more intense than in the case of mfp3S-pep-3Dopa (see Figure 5), pointing to a greater interaction of the catechol residues in the coacervate phase to the substrate. Interestingly, the peak at 1249 cm−1 (δOH ring mode), which presents a distinct shoulder at 1279 cm−1, became dominant in this region over time, indicating that tyrosine and Dopa are preferentially located in the proximity of the TiO2 surface as compared to other side chains.
Figure 5.
ATR-IR spectra of adsorbed a) mfp3S-pep, b) mfp3S-pep-3Dopa, and c) mfp3S-pep-3Dopa coacervate on TiO2 from 0.1 mg mL−1 solutions recorded at ≈2 min intervals. Backgrounds were from (a) and (b) 100 × 10−3 m acetic acid and (c) buffer at pH 6.5 on TiO2-coated ZnSe prism.
The 4000−2500 cm−1 spectral region of the broad O—H stretching band (νOH) reveals insightful discrepancies in the adsorption mechanism of the mfp3S-pep versus the Dopa-containing peptide and its coacervate. In particular, the broad absorbance of νOH increased with time for mfp3S-pep (Figure 5a), with a peak at 3305 cm−1 arising from the N—H stretch (νNH) of peptide bonds. In addition, peaks from the characteristic aliphatic absorptions of CH2 asymmetric (νas CH2) and symmetric stretching (νs CH2) modes are visible at 2923 and 2861 cm−1, respectively. Adsorbed mfp3S-pep-3Dopa and its coacervate showed a broad negative band decreasing with time due to the absorbance reduction of the νOH mode associated with water molecules on the TiO2 nanoparticle surface.[29] A positive peak at 3250 cm−1 due to the νNH mode of the peptide bonds further confirms that the adsorption of peptide molecules (positive absorbance) causes the loss of interfacial water (negative absorbance). Surface-bound water molecules are also present as a hydration layer around the adsorbate and dislodged by direct adsorbate–surface interactions. Generally, the νOH envelope is located between ≈3650 and ≈3000 cm−1 and originates from water molecules embedded in distinct hydrogen-bonded environments, ranging from weakly (liquid-like) to strongly (surface-bound) H-bonded molecules at higher and lower wavenumbers, respectively. As H-bonds become stronger and more dominant, the νOH band center shifts toward lower wavenumbers.[30] The negative νOH band was previously observed for the adsorption of catechol[6a] and phosphate species onto TiO2 (anatase)[31] and was ascribed to the displacement of interfacial H2O molecules and to the concurrent loss of terminal hydroxyl groups from the anatase surface. In fact, it has been proposed that on TiO2 anatase, only the {001} facet is chemically active, since it contains both terminal and bridging OH groups. Connor et al.[31b] have assigned the absorption at ≈3400 cm≈1 to Ti—OH vibrational mode, with the Ti—OH2 vibration found at ≈3200 cm−1, and Ti—OH+—Ti and Ti—OH2+ vibrations at ≈3000 cm−. These surface hydroxyl groups are reactive toward catechol with a ligand-exchange model of specific adsorption to metal oxides and contribute to the observed loss of νOH band. Hence, mfp3S-pep-3Dopa is able to displace interfacial water molecules from the TiO2 substrate surface and additionally interacts with surface OH groups via a ligand exchange reaction. In contrast to other techniques characterizing surface hydration, e.g., overhauser dynamic nuclear polarization (ODNP),[32] which requires extensive surface preparation/modification, ATR-FTIR provides a noninvasive in situ real-time determination of both liquid-like and surface bond water.[33]
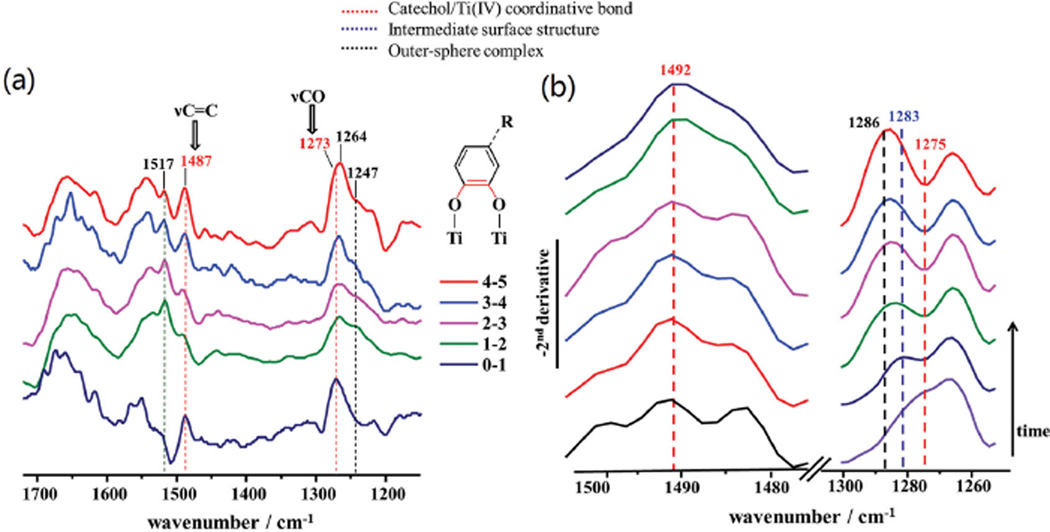
The characteristic doublet of catechol coordinating Ti(IV) atoms, as recorded in the adsorption experiments with dopamine (Figure S8a, Supporting Information), becomes evident when the ATR-IR spectra (Figure 5b) were normalized at the position of their most intense absorbance at 1517 cm−1 and subsequently subtracted between two consecutive time intervals (see Figure 6a). By doing so, the doublet peaking at 1487 and 1273 cm−1 is evident in all difference spectra, with the latter shifting to lower wavenumbers with time (1264 cm−1). Concurrently, the absorptions at 1247 and 1517 cm−1 due to aromatic ring modes increased, indicating that peptide species in solution become more abundant in the proximity of the nanoparticle film/solution interface, as they interact with the TiO2 surface via outersphere complexes.
Figure 6.
a) Difference spectra for mfps-pep-3Dopa adsorbed on TiO2 obtained by subtracting every two consecutive normalized ATR-IR spectra in Figure 5b, showing the emergence of specific absorptions over the course of the adsorption process, i.e., aromatic ring vibrations at 1517 and 1247 cm−1. The doublet due to catechol/Ti coordination bond is visible at 1487 and 1273 cm−1 (in red), and the specific bonds associated with these vibrations are presented in the schematic complex structure (bonds in red). b) Evolution of the negative second-derivative spectra of mfps-pep-3Dopa adsorbed on TiO2 from Figure 5c in the 1505−1475 and 1300−1255 cm−1 spectral regions. Peaks characteristic of the catechol-Ti coordinative bond (red), intermediate surface complex (blue), and outersphere complex (black) are indicated in the figure.
The overlap of catechol-characteristic vibrations in the original absorbance spectra in Figure 5 is significant especially in the 1300−1200 cm−1 region, and it is thus challenging to resolve and assign absorptions with confidence to specific surface structures. Additionally, the amide III mode lies in the same spectral region (1400−1200 cm−1), however, its intensity is particularly weak and generally negligible compared to the strong absorbance of the amide I and amide II modes,[34] as also observed in Figure 5a for the adsorbed mfp3S-pep. Besides the previously adopted method of obtaining difference spectra, the negative second-derivative of the ATR-IR spectra in Figure 6b revealed maxima at 1492 and 1275 cm−1 associated with catechol-Ti(IV) coordination, which is evident from the initial spectrum after ≈2 min. The latter peak shifted gradually to higher frequencies, indicating that coordination is, at first, the predominant binding mode, but later, the maximum at 1286 cm−1 due to catechol in solution and outersphere complexes became dominant. The peak at 1283 cm≈1 observed after ≈4 min appears to originate from a surface structure intermediate between catechol coordinated to Ti(IV) atoms and an outersphere complex, and can be assigned to an intermediate surface-bound structure, such as hydrogen-bonded catechol on the TiO2s surface. Hence, the evolution of the catechol binding modes during the adsorption of mfps-pep-3Dopa on TiO2s appears to follow a sequential pattern as the surface sites progressively become saturated: (1) displacement of interfacial surface-bound water molecules; (2) initial chemisorption via Ti(IV)-catechol covalent coordination; (3) intermediate innersphere complex formation, such as hydrogen-bond; and (4) outersphere complexes. By contrast, mfp3S-pep lacked this ability, hence confirming that the presence of Dopa is critical to dislodge interfacial water and to eventually trigger innersphere complexes to achieve irreversible adsorption.
The difference spectra between normalized ATR-IR spectra (Figure 5c) of mfp3S-pep-3Dopa coacervate at consecutive time intervals (Figure S9a, Supporting Information) did not reveal the sharp doublet associated with the catechol-Ti(IV) coordination bond, as previously observed for mfp3S-pep-3Dopa on the same surface. Weaker absorptions were observed at 1496 and 1273 cm−1 in the initial time intervals, but in the subsequent difference spectra the overlap between 1300 and 1200 cm−1 was considerable. To reveal the underlying overlapping contributions the second-derivative approach was employed and the results are displayed in Figure S9b (Supporting Information). The analysis indicates that upon displacement of interfacial water molecules, the initial interaction of catechol with the TiO2 substrate occurred via covalent coordination (doublet at 1492 and 1275 cm−1), with subsequent coacervates forming an intermediate innersphere surface complex, likely H-bonds (1280 cm−1), and eventually outersphere complexes (1285 cm−1) ensued (schematic Figure 7).
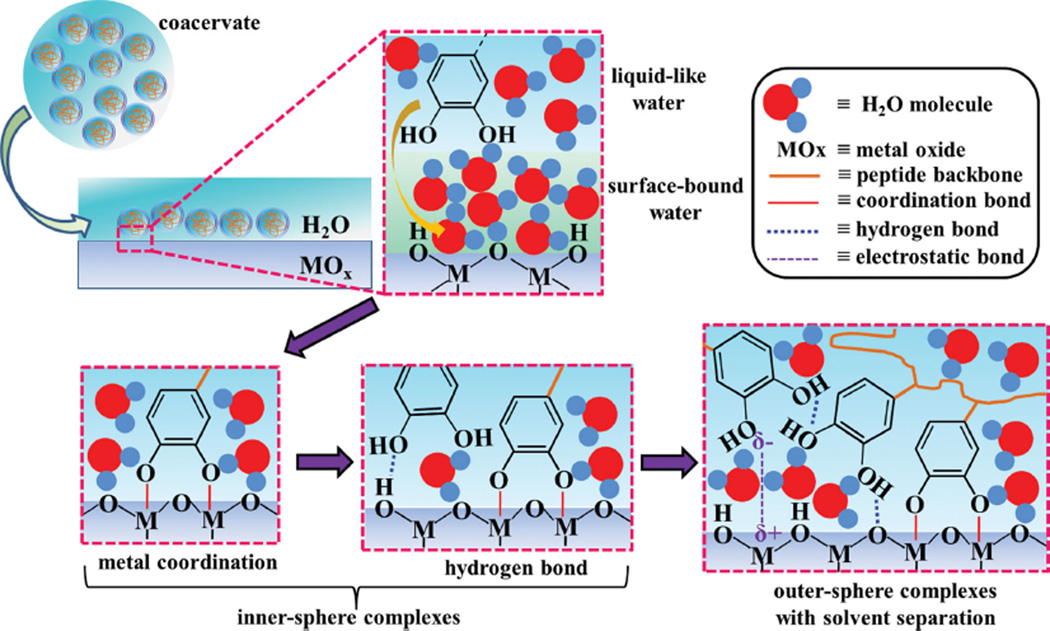
Figure 7.
Schematic of the adsorption mechanism of catechol groups in mfps-pep-3Dopa coacervate on a wet metal (hydroxide surface (MOx). The initial displacement of liquid-like and surface-bound water molecules is followed by catechol-metal coordination and hydrogen bond formation (innersphere complexes). Finally, as the surface sites become saturated, electrostatic interactions occur with solvent separation (outersphere complexes).
2.7.2. Adsorption on HAP
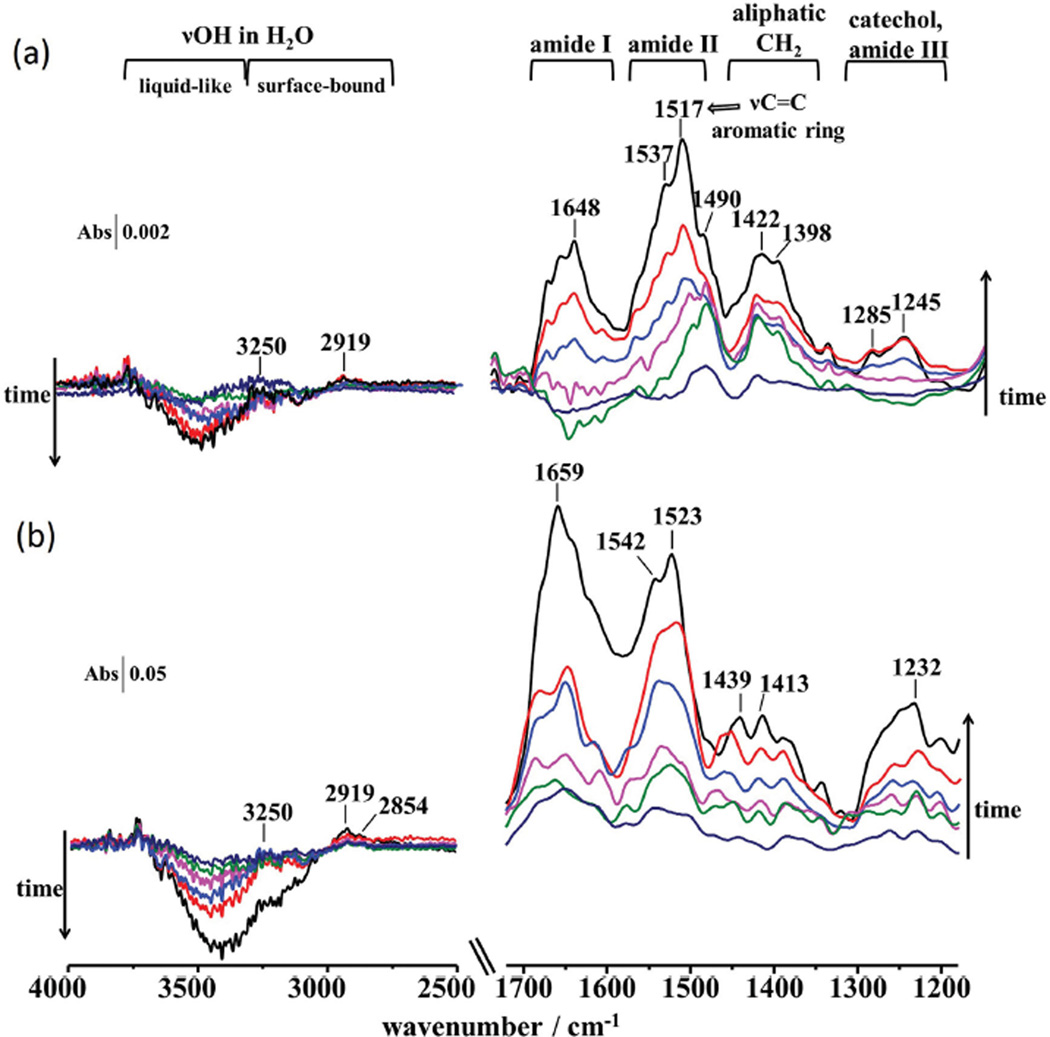
The adsorption of mfp3S-pep, mfp3S-pep-3Dopa, and mfp3S-pep-3Dopa coacervate was also investigated on HAP by ATR-IR (Figure 8). The absorption fingerprint (Figure 8a) for mfp3S-pep was comparable to that on TiO2, although the intensity was weaker. As on TiO2, the broad and positive intensity of the νOH mode indicates that mfp3S-pep was unable to remove interfacial water molecules from the HAP surfaces. Absorptions peaking at 1285 and 1245 cm−1 in the spectra of adsorbed mfp3S-pep-3Dopa are assigned to catecholaq and to the in-plane δOH mode, respectively. Absorptions in the 1300−1200 cm−1 catechol vibrational region were visible from the initial spectra of the adsorbed coacervate (Figure 8b) and appeared more enhanced than those in the corresponding ATR-IR spectra for the adsorbed mfp3-pep-3Dopa. This result suggests a greater involvement of the catechol groups in the surface interactions with HAP in its coacervate phase as compared to the solution peptide. Notably, the absorbance of the catechol characteristic region relative to that of the amide modes was weaker than for the adsorption on TiO2 (see Figure 5). This observation suggests a weaker involvement of catechol groups during the adsorption on HAP as compared to TiO2 in terms of direct adsorbate–adsorbent interactions. Furthermore, the amide I/amide II ratio of adsorbed mfp3S-pep-3Dopa is lower than 1, indicating that the interactions of aromatic functionalities on HAP arising from catechol and tyrosine residues are greater than in the case of the mfp3S-pep. Conversely, the adsorbed mfp3S-pep-3Dopa coacervate showed comparable intensities for amide I and II absorptions in the initial minutes of adsorption, with aromatic vibrations temporarily prevailing after 10 min, followed by the final predominant absorbance of the amide I band. The contribution of other interactions related to the peptide backbone appeared to be enhanced in the coacervate, likely due to their closer proximity (higher wettability) on the HAP surface as compared with the peptide solution.
Figure 8.
ATR-IR spectra of adsorbed a) mfp3S-pep-3Dopa and b) mfp3S-pep-3Dopa coacervate on HAP from 0.1 mg mL−1 solutions recorded at ≈2 min intervals. Backgrounds were from (a) 100 × 10−3 m acetic acid and (b) buffer at pH 6.5 and 100 × 10−3 m ionic strength on HAP-coated ZnSe prism.
The spectral region between 4000 and 2500 cm−1 showed a broad negative band associated with the νOH, which decreased with time for both mfp3S-pep-3Dopa and the coacervate. The νNH absorption at 3250 cm−1 from the peptide bond and a peak at 2919 cm−1 from the νas CH2 mode of the aliphatic moieties are visible. Additionally, the νs CH2 mode at 2854 cm−1 was observed in the spectra of the adsorbed coacervate. The signal loss between ≈3250 and 3000 cm−1 indicates that the coacervate was able to displace surface-bound interfacial water from HAP to a greater extent than mfp3S-pep-3Dopa, thus showing an overall enhanced absorbance.
The evolution of the spectral bands over the duration of the adsorption experiment on HAP was evaluated with the method of the negative second-derivative (see Figures S10 and S11, Supporting Information). Although a peak associated with the doublet of catechol coordinative bond at 1492 cm−1 was observed for both mfp3S-pep-Dopa3 and its coacervate, the corresponding peak at 1275 cm−1 was absent in all second-derivative spectra. As previously established for dopamine, both peaks are present if the innersphere coordination of catechol groups is present. Therefore, Dopa is unable to form coordinative bonds on the HAP surface as it does on TiO2.
The adsorbed mfp3S-pep-3Dopa showed a peak at 1280 cm−1 assigned to an intermediate innersphere surface structure, which was detected only in the first second-derivative spectrum, and subsequently vanished as peaks associated with outersphere catechol complexes emerged. The second-derivative analysis of adsorbed mfp3S-pep-3Dopa coacervate revealed an initial peak at 1289 cm−1 due to outersphere catechol complexes. After ≈8 min a component at 1283 cm−1 appeared, originating from an intermediate innersphere surface structure. It is likely that inner sphere complexes were not initially detected for the adsorbed coacervate because the adsorption kinetics were too fast to be detected with the time resolution of conventional ATR-IR spectroscopy. To summarize, the sequence of interactions of the mfp3S-pep-3Dopa and its coacervate on HAP proceeded as follows: (1) displacement of liquid-like interfacial water molecules from the HAP surface, with the coacervate displacing surface-bound water to a greater extent; (2) formation of innersphere surface complexes, such as H-bonding between catechols and the hydroxyl groups of HAP; (3) outer-sphere complex formation.
The isoelectric point (IEP) of TiO2 anatase is between pH 5 and 6,[6b] whereas HAP has an IEP between pH 7 and 8.[35] Therefore, when the peptide is adsorbed from 100 × 10−3 m acetic acid solution at pH ≈3, both surfaces are positively charged. The coacervate is adsorbed from a buffer solution at pH 6.5, hence slightly above the IEP of TiO2 and below that of HAP. It can be assumed that at pH 6.5 both surfaces approach neutrality, thus exhibiting comparable surface charges toward the adsorbing coacervate. Although adsorption rates are often pH-dependent, Gulley-Stahl et al.[23b] showed that catechol adsorption is relatively constant in the 3–7 pH interval and increases at pH above the catechol pKa1 of 9.4.[4] Therefore, both the enhanced spectral absorbance and the relative amount of displaced interfacial water on both surfaces are primarily attributed to the coacervate’s superior wetting properties compared to the solution peptide. Furthermore, the ability of catechol to coordinate Ti(IV) atoms on TiO2 provides an additional driving force to adsorption. On HAP, the greater adsorption of the Dopa-containing peptide and its coacervate appears to arise from divalent hydrogen-bonding, as previously suggested.[25] We also propose that the synergistic effect of multiple hydrogen bonds between the peptide and HAP can account for an enhanced adsorption capacity. As discussed for dopamine adsorbed on HAP, besides hydrogen-bonding other interactions are expected to account for the displacement of surface-bound water from the HAP surface. We suggest that another type of surface complex may arise from π-cation interactions between the catechol aromatic ring and Ca2+ ions of HAP.
3. Conclusion
Mfp-3S plays a central role in enabling mussel (M. californianus) adhesion, performing multiple functions including priming and dehydrating wet surfaces, interfacial adhesion, and connecting the adhesive layer with other proteins within the mussel plaque. In addition, mfp-3S forms a single-component coacervate, which provides a critical vehicle for the delivery of adhesives under wet conditions. To mimic mfp-3S’s property/function, a novel peptide (mfp3S-pep) corresponding to half the native protein’s length was designed by truncating mfp3S’s sequence based on known protein composition/property relationships. The shorter peptide length is desirable in applied research to improve cost-efficiency and increase production yields. Mfp3S-pep is demonstrated to form single-component coacervates over a defined range of pH and ionic strengths, and is driven by electro static and hydrophobic interactions, as well as H-bonding. By simply substituting one positively charged residue in mfp3s-pep sequence with a negative one, the coacervation conditions can be expanded. Dopa was introduced into the sequence by enzymatic conversion of tyrosine, with Dopa content tuned by adjusting reaction conditions. Thus, we were able to investigate Dopa’s effect on peptide coacervation and adsorption on biomedically-relevant surfaces, namely TiO2 (anatase) and HAP. Dopa influences peptide coacervates by increasing their hydrophilicity, with a parallel decrease of hydrophobic interactions by promoting molecular H-bonds.
Adsorption studies performed by QCM on HAP and TiO2 confirmed that Dopa promotes faster initial adsorption kinetics, with the coacervate dramatically enhancing the quantity of adsorbed peptide. ATR-IR spectroscopic analysis revealed catechol/Ti(IV) coordination during initial adsorption of Dopa-containing peptides on TiO2, followed by an intermediate surface complex and ultimately by outersphere catechol complexes. By contrast, no coordination was observed on the HAP surface. Only intermediate surface structures and outersphere complexes were detected and we postulate that multiple H-bonds synergistically displace surface-bound water molecules. Other surface interactions may also account for the significant adsorptive capacity of catechol-bearing compounds on hydroxyapatite, such as π-cation complexes between the aromatic ring of Dopa/Tyr and Ca2+ in HAP.
Compared to the widely studied polydopamine-based coatings that have found some success in wet adhesion, coacervates exhibit several specific advantages associated with their high concentration in the liquid phase. A thick layer of coacervate can be easily coated on target substrates, which is required for substrates of high surface roughness. Furthermore, because coacervates do not disperse in aqueous solutions their “on-site” delivery, for instance for soft tissues adhesion, is more convenient.[36] Additionally, coacervates exhibit an extremely low interfacial energy,[13a] which results in easy wetting and spreading onto wet hydrated surfaces. The simultaneous presence of Dopa and of a coacervate phase in our synthetic peptide significantly enhances adhesion under wet conditions by displacing surface-bound water molecules – the nemesis of wet adhesion – thereby allowing intimate interaction with the substrate surface. Peptide analogues of mussel adhesive proteins provide molecular level bio-inspiration for the development of synthetic adhesives in wet environments, such as in dental, orthopedic, and other biomedical applications.
4. Experimental Section
Incorporation of Dopa by Enzymatic Hydroxylation
Peptides were commercially synthesized by solid phase peptide synthesis method (GenScript Inc.). Tyr residues in the peptides were then hydroxylated in-house to Dopa by using commercially avail-able mushroom tyrosinase (Sigma-Aldrich), and the yield of Dopa conversion was controlled by tuning enzyme/substrate ratio and reaction time. Typical enzymatic hydroxylation and purification procedure proceeded as follows: (1) reaction buffer was made by dissolving 0.1 m Na2HPO4 and 50 × 10−3 m Na2B4O7 in water, with ascorbic acid powder subsequently added to adjust the pH to 7.0; (2) 4 mg of mfp3S-pep was first dissolved in 2 mL of 50:50 v/v acetonitrile/reaction buffer solution (acetonitrile helps to enhance solubility of mfp3-pep in the reaction buffer), following which 8 mL of reaction buffer was further added to obtain a final peptide solution (10% acetonitrile). The solution typically turns cloudy at this point; (3) 1 mg of mushroom tyrosinase was dissolved in 0.2 mL buffer and added to the peptide solution from step 2; (4) the obtained mixture from step 3 was constantly stirred for 2 h at room temperature, and in the end 0.5 mL glacial acetic acid was added to stop the reaction; (5) the resulting solution was freeze dried and re-dissolved in 2 mL of 5% acetic acid solution containing 8 m urea; (6) the suspension from step 5 was then subjected to C18 reverse phase High Performance Liquid Chromatography. The fractions with the desired degree of hydroxylation (as analyzed by mass spectroscopy) were collected and freeze dried; (7) the freeze dried samples were then dissolved in 5% acetic acid and subjected to Shodex size exclusion HPLC to remove residual tyrosinase-associated protein; (8) fractions containing the modified peptides were collected and freeze dried before re-dissolving in 10 × 10−3 m acetic acid for the other experiments. By following this procedure, mfp3S-pep-3Dopa and mfp3S-pep-6Dopa, which on average contain 3 and 6 Dopa units per peptide molecule, were synthesized and purified. Dopa content was confirmed by amino acid analysis and mass spectrometry.
Mass Spectroscopy
MALDI-TOF-MS (Bruker Microflex LRF) was used to characterize and assure purity of the peptides. 1–2 µL of sample peptide was spotted on the gold-plated sample plate and vacuum dried, on top of which 1 µL of matrix solution (α-cyano-3-hydroxycinnamic acid in aqueous 50% acetonitrile and 0.1% trifluoroacetic acid) was added. External calibrant was peptide calibration standard mixture from Bruker containing seven standard peptides with a mass range between ≈1000 and 3500 Da. The N2 laser (337 nm) was set up at 60 Hz.
Amino Acid Analysis
The amino acid composition of the hydrolyzed peptides was determined on a Hitachi L8900 amino acid analyzer system with ninhydrin-based chemistry. Polypeptides were hydrolyzed in 100 µL of 6 m HCl with 8% phenol, in vacuum at 158 °C for 40 min. After being washed with water and methanol, the hydrolyzed products were dissolved in 0.2 m HCl and injected into the amino acid analyzer.
Mfp3S-pep Self-Coacervation and Turbidity Measurements
Stock solutions of 1 mg mL−1 mfp3S-pep were prepared in 10 × 10−3 m acetic acid buffer. The final protein concentration was fixed at 0.1 mg mL−1 by adding stock solution to buffer at volume ratio of 1/9 (stock/buffer). Coacervation of mfp3S-pep at different buffer conditions was measured by turbidometry at 600 nm by UV–vis spectrophotometry. Mfp3S-pep absorbance was negligible at 600 nm. The relative turbidity is defined as 100 − 100 × 10−A (600).
Microscopy
The turbidity associated with coacervate droplet formation was visually inspected by inverted light microscopy. Protein distribution was also investigated with an Olympus DSU Fluorescent Microscope (model IX81 DSU, Olympus, Tokyo, Japan). Images were taken with an ImagEM camera (C9100-13, Hamamatsu, Shizuoka, Japan) under the control of MetaMorph software (Olympus). Desired excitation (350 nm) and emission wavelengths (450 nm) were obtained by mercury bulb combined with 89000 Sedat Quad Filter Set (Chroma Tech. Corp.)
Circular Dichroism
Peptides were dissolved in 10 × 10−3 m phosphoric acid at a concentration of 0.5 mg mL−1, and their secondary structure was determined by circular dichroism (CD) using a Chriascan spectropolarimeter (Model 420, AVIV Biomedical Inc.). Measurements were conducted in triplicate at wavelengths ranging from 190–260 nm, with 1 nm step size and 1 nm bandwidth. The spectra were smoothed by the Savitzky–Golay method with a polynomial order of 2.
QCM-D
HAP and TiO2 sensors were purchased from Biolin Scientific and cleaned according to the protocol suggested before use. QCM-D experiments were carried out in a Q-Sense E4 system using two flow modules in parallel. Samples were introduced into the modules at a 0.1 mL min−1 flow-rate using a 4-channel Ismatec IPC-N 4 peristaltic pump. In QCM-D, changes in resonance frequency (ΔF) and dissipation (ΔD) of a quartz crystal were recorded to measure the amount and viscoelastic property of the material deposited onto the sensor, respectively. The crystal was excited at its fundamental frequency, ≈5 MHz, and changes can be observed at the fundamental (n = 1) as well as overtone frequencies (n = 3, 5, 7, 9, 11). Readings taken at the fundamental frequency were typically ignored as they are prone to artifacts from the sensor clamp.
TiO2 and HAP Nanoparticle Films
Anatase TiO2 and HAP nanoparticles (both purchased from Sigma-Aldrich) measured <25 and <200 nm in diameter on average, respectively. TiO2 and HAP nanoparticle films were prepared by drying in air overnight 50 µL of 10−3 m sonicated aqueous suspensions deposited on the horizontal surface of the ZnSe ATR crystal (Pike Technologies) in the dark at room temperature. The thickness of the nanoparticle films was estimated by drying 50 µL of 10−3 m sonicated suspen-sions on glass, which were subsequently coated with 10 nm thick gold (JFC-16000 sputter coater, JEOL) and viewed in a FESEM (JSM-7600F, JEOL) at 5.0 kV accelerating voltage and at 13.6 mm working distance.
Attenuated Total Reflection-Infrared Spectroscopy
ATR-IR spectra were recorded with a MIRacleTM accessory (Pike Technologies) equipped with a 3-reflection ZnSe prism (6 mm in diameter sam-pling area) in an infrared spectrometer (Vertex 70, Bruker) equipped with a KBr beam splitter and a liquid nitrogen-cooled mercury cadmium telluride (MCT) detector. Nitrogen gas was used to continuously purge the optical bench. Opus software (version 6.5) was used to analyze the ATR-IR spectra recorded from 128 co-added scans at 4 cm−1 resolution. ATR-IR spectra were corrected for the frequency dependence of the penetration depth with the ATR correction program available in the Opus 6.5 software. The ZnSe ATR prism was coated with a thin film of metal oxide nanoparticles as previously described. A detailed description of the ATR-IR analysis of adsorption to metal oxide particle films can be found elsewhere.[37] The penetration depth (dp of the ATR-IR technique with a ZnSe prism (refractive index 2.4) and 45° angle of incidence in the mid-IR is about 1 µm.[38] The ≈100 nm thick TiO2 and HAP nanoparticle films were thus entirely probed by the evanescent wave exponentially decaying away from the ATR prism/nanoparticle film interface. The ATR-IR spectra were recorded from aqueous solutions of the investigated peptides on bare ZnSe prism, using the peptide-free aqueous solutions on ZnSe as background. Adsorption experiments on the nanoparticle film-coated ZnSe prism were conducted under flow from a peristaltic pump (Ismatec) at a flow rate of 50 µL min−1 through a 50 µL custom-made chamber sealed with an O-ring to the surface of the ZnSe prism (Figure 6). Prior to each experiment, the surface of the ZnSe prism was polished with an aqueous slurry of 0.05 µm Al2O3 powder (CH Instrument) and then rinsed with deionized water (MilliQ, Millipore). The mathematical method of the second-derivative analysis was applied in this work as an ancillary tool for the identification of overlapping catechol bands in the 1505−1475 cm−1 and 1300−1255 cm−1 regions. The second-derivative analysis of infrared spectra is routinely used to determine the number and the position of secondary structure components in peptides and proteins by deconvoluting the amide I band.[39] In brief, this analysis involves the principle of the line-narrowing of the infrared bands, thus allowing resolving overlapping bands, with negative peaks corresponding to inflections and shoulders of the original absorbance band, thus providing the position of the component bands.[40] Note that in this work the second-derivative spectra were multiplied by −1 and thus point upward for ease of illustration.
Supplementary Material
Acknowledgments
W.W. and L.P. contributed equally to this work. The authors thank Emeritus Prof. Jim McQuillan for the insightful discussions, Sharhouz Amini for preparing the ATR-IR setup figure, and Dr. Bram Cantaert for his help with FESEM. This research was supported by the Materials Research Science and Engineering Centers Program of the National Science Foundation under Award DMR 1121053 and the National Institutes of Health under Grant R01-DE018468. L.P. and A.M. were supported by the Singapore Maritime Institute (Grant No. SMI-2013-MA-03) and the Singapore National Research Foundation (NRF) through an NRF Fellowship awarded to A.M.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Wei Wei, Materials Research Lab, University of California, Santa Barbara, CA 93106, USA.
Luigi Petrone, School of Materials Science and Engineering, Nanyang Technological University, 639798, Singapore.
YerPeng Tan, Biomolecular Science and Engineering Program, University of California, Santa Barbara, CA 93106, USA.
Hao Cai, School of Materials Science and Engineering, Nanyang Technological University, 639798, Singapore.
Jacob N. Israelachvili, Materials Research Lab, University of California, Santa Barbara, CA 93106, USA Biomolecular Science and Engineering Program, University of California, Santa Barbara, CA 93106, USA.
Ali Miserez, School of Materials Science and Engineering, Nanyang Technological University, 639798, Singapore; School of Biological Sciences, Nanyang Technological University, 639798, Singapore.
J. Herbert Waite, Email: herbert.waite@lifesci.ucsb.edu, Materials Research Lab, University of California, Santa Barbara, CA 93106, USA; Biomolecular Science and Engineering Program, University of California, Santa Barbara, CA 93106, USA.
References
- 1.Lee BP, Messersmith PB, Israelachvili JN, Waite JH. Annu. Rev. Mater. Res. 2011;41:99. doi: 10.1146/annurev-matsci-062910-100429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu ME, Hwang JY, Deming TJ. J. Am. Chem. Soc. 1999;121:5825. [Google Scholar]
- 3.Zhao H, Waite JH. J. Biol. Chem. 2006;281:26150. doi: 10.1074/jbc.M604357200. [DOI] [PubMed] [Google Scholar]
- 4.He LH, Fullenkamp DE, Rivera JG, Messersmith PB. Chem. Commun. 2011;47:7497. doi: 10.1039/c1cc11928a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) McWhirter MJ, Bremer PJ, Lamont IL, McQuillan AJ. Langmuir. 2003;19:3575. [Google Scholar]; b) Mcbride MB, Wesselink LG. Environ. Sci. Technol. 1988;22:703. [Google Scholar]; c) Simpson SL, Powell KJ, Sjoberg S. J. Colloid Interface Sci. 2000;229:568. doi: 10.1006/jcis.2000.6999. [DOI] [PubMed] [Google Scholar]; d) Ramakrishna G, Ghosh HN. Langmuir. 2003;19:505. [Google Scholar]; e) Petrone L. Adv. Colloid Interface Sci. 2013;195:1. doi: 10.1016/j.cis.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 6.a) Connor PA, Dobson KD, Mcquillan AJ. Langmuir. 1995;11:4193. [Google Scholar]; b) Rodriguez R, Blesa MA, Regazzoni AE. J. Colloid Interface Sci. 1996;177:122. doi: 10.1006/jcis.1996.0012. [DOI] [PubMed] [Google Scholar]; c) Rodenstein M, Zurcher S, Tosatti SGP, Spencer ND. Langmuir. 2010;26:16211. doi: 10.1021/la100805z. [DOI] [PubMed] [Google Scholar]
- 7.a) Lee H, Scherer NF, Messersmith PB. Proc. Natl. Acad. Sci. USA. 2006;103:12999. doi: 10.1073/pnas.0605552103. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ye Q, Zhou F, Liu WM. Chem. Soc. Rev. 2011;40:4244. doi: 10.1039/c1cs15026j. [DOI] [PubMed] [Google Scholar]
- 8.Zhong C, Gurry T, Cheng AA, Downey J, Deng ZT, Stultz CM, Lu TK. Nat. Nanotechnol. 2014;9:858. doi: 10.1038/nnano.2014.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei W, Yu J, Broomell C, Israelachvili JN, Waite JH. J. Am. Chem. Soc. 2013;135:377. doi: 10.1021/ja309590f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei W, Tan YP, Rodriguez NRM, Yu J, Israelachvili JN, Waite JH. Acta Biomater. 2014;10:1663. doi: 10.1016/j.actbio.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart RJ, Weaver JC, Morse DE, Waite JH. J. Exp. Biol. 2004;207:4727. doi: 10.1242/jeb.01330. [DOI] [PubMed] [Google Scholar]
- 12.Yer Peng Tan SH, Guerette Paul A, Wei Wei, Ghadban Ali, Hao Cai, Miserez Ali, Waite J Herbert. Nat. Chem. Biol. 2015;11:488. doi: 10.1038/nchembio.1833. [DOI] [PubMed] [Google Scholar]
- 13.a) Hwang DS, Zeng HB, Srivastava A, Krogstad DV, Tirrell M, Israelachvili JN, Waite JH. Soft Matter. 2010;6:3232. doi: 10.1039/C002632H. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shao H, Stewart RJ. Adv. Mater. 2010;22:729. doi: 10.1002/adma.200902380. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Shao H, Bachus KN, Stewart RJ. Macromol. Biosci. 2009;9:464. doi: 10.1002/mabi.200800252. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kaur S, Weerasekare GM, Stewart RJ. ACS Appl. Mater. Interfaces. 2011;3:941. doi: 10.1021/am200082v. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Hunt JN, Feldman KE, Lynd NA, Deek J, Campos LM, Spruell JM, Hernandez BM, Kramer EJ, Hawker CJ. Adv. Mater. 2011;23:2327. doi: 10.1002/adma.201004230. [DOI] [PubMed] [Google Scholar]; f) Ortony JH, Choi SH, Spruell JM, Hunt JN, Lynd NA, Krogstad DV, Urban VS, Hawker CJ, Kramer EJ, Han S. Chem. Sci. 2014;5:58. [Google Scholar]
- 14.Hwang DSC, Choi YS, Cha HJ. In: Marine Biomaterials Characterization, Isolation and Applications. Kim SK, editor. Boca Raton: CRC Press; 2013. p. 289. [Google Scholar]
- 15.Taylor SW. Anal. Biochem. 2002;302:70. doi: 10.1006/abio.2001.5522. [DOI] [PubMed] [Google Scholar]
- 16.Mecozzi S, West AP, Dougherty DA. Proc. Natl. Acad. Sci. USA. 1996;93:10566. doi: 10.1073/pnas.93.20.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan MY, Villanueva G, Newman SA. J. Biol. Chem. 1989;264:2139. [PubMed] [Google Scholar]
- 18.Roy A, Kucukural A, Zhang Y. Nat. Protoc. 2010;5:725. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burley SK, Petsko GA. Science. 1985;229:23. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- 20.Creighton TE. Proteins Structures and Molecular Properties. New York: W. H. Freeman; 1994. p. 14. [Google Scholar]
- 21.Yu J, Wei W, Danner E, Ashley RK, Israelachvili JN, Waite JH. Nat. Chem. Biol. 2011;7:588. doi: 10.1038/nchembio.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.a) Petrone L, McQuillan AJ. Appl. Spectrosc. 2011;65:1162. doi: 10.1366/11-06236. [DOI] [PubMed] [Google Scholar]; b) Beaussart A, Petrone L, Mierczynska-Vasilev A, McQuillan AJ, Beattie DA. Langmuir. 2012;28:4233. doi: 10.1021/la204652f. [DOI] [PubMed] [Google Scholar]
- 23.a) Lana-Villarreal T, Rodes A, Perez JM, Gomez R. J. Am. Chem. Soc. 2005;127:12601. doi: 10.1021/ja052798y. [DOI] [PubMed] [Google Scholar]; b) Gulley-Stahl H, Hogan PA, Schmidt WL, Wall SJ, Buhrlage A, Bullen HA. Environ. Sci. Technol. 2010;44:4116. doi: 10.1021/es902040u. [DOI] [PubMed] [Google Scholar]
- 24.a) Li SC, Chu LN, Gong XQ, Diebold U. Science. 2010;328:882. doi: 10.1126/science.1188328. [DOI] [PubMed] [Google Scholar]; b) Li SC, Wang JG, Jacobson P, Gong XQ, Selloni A, Diebold U. J. Am. Chem. Soc. 2009;131:980. doi: 10.1021/ja803595u. [DOI] [PubMed] [Google Scholar]
- 25.Chirdon WM, O’Brien WJ, Robertson RE. J. Biomed. Mater. Res. B. 2003;66B:532. doi: 10.1002/jbm.b.10041. [DOI] [PubMed] [Google Scholar]
- 26.Santarelli VP, Eastwood AL, Dougherty DA, Ahern CA, Horn R. Biophys. J. 2007;93:2341. doi: 10.1529/biophysj.107.106856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poljansek I, Krajnc M. Acta Chim. Slov. 2005;52:238. [Google Scholar]
- 28.Grace LI, Cohen R, Dunn TM, Lubman DM, de Vries MS. J. Mol. Spectrosc. 2002;215:204. [Google Scholar]
- 29.a) Israelachvili J, Wennerstrom H. Nature. 1996;379:219. doi: 10.1038/379219a0. [DOI] [PubMed] [Google Scholar]; b) Mante PA, Chen CC, Wen YC, Chen HY, Yang SC, Huang YR, Chen IJ, Chen YW, Gusev V, Chen MJ, Kuo JL, Sheu JK, Sun CK. Sci. Rep. 2014;4:6249. doi: 10.1038/srep06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marechal Y. The Hydrogen Bond and the Water Molecule: the Physics and Chemistry of Water, Aqueous and Bio Media. Amsterdam: Elsevier; 2007. [Google Scholar]
- 31.a) Connor PA, McQuillan AJ. Langmuir. 1999;15:2916. [Google Scholar]; b) Connor PA, Dobson KD, McQuillan AJ. Langmuir. 1999;15:2402. [Google Scholar]
- 32.Akdogan Y, Wei W, Huang KY, Kageyama Y, Danner EW, Miller DR, Rodriguez NRM, Waite JH, Han S. Angew Chem. Int. Ed. 2014;53:11253. doi: 10.1002/anie.201406858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrone L, Kumar A, Sutanto CN, Patil NJ, Kannan S, Palaniappan A, Amini S, Zappone B, Verma C, Miserez A. Nat. Commun. 2015;6:1. doi: 10.1038/ncomms9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.a) Manning MC. Expert Rev. Proteomics. 2005;2:731. doi: 10.1586/14789450.2.5.731. [DOI] [PubMed] [Google Scholar]; b) Petrone L, Ragg NLC, McQuillan AJ. Biofouling. 2008;24:405. doi: 10.1080/08927010802339970. [DOI] [PubMed] [Google Scholar]
- 35.Kosmulski M. J. Colloid Interf. Sci. 2009;337:439. doi: 10.1016/j.jcis.2009.04.072. [DOI] [PubMed] [Google Scholar]
- 36.Kim HJ, Hwang BH, Lim S, Choi BH, Kang SH, Cha HJ. Biomaterials. 2015;72:104. doi: 10.1016/j.biomaterials.2015.08.055. [DOI] [PubMed] [Google Scholar]
- 37.a) Goormaghtigh E, Raussens V, Ruysschaert JM. Bba-Rev Biomembranes. 1999;1422:105. doi: 10.1016/s0304-4157(99)00004-0. [DOI] [PubMed] [Google Scholar]; b) McQuillan AJ. Adv. Mater. 2001;13:1034. [Google Scholar]
- 38.Vigano C, Manciu L, Ruysschaert JM. Acc. Chem. Res. 2005;38:117. doi: 10.1021/ar040021o. [DOI] [PubMed] [Google Scholar]
- 39.a) Bouhekka A, Burgi T. Appl. Surf. Sci. 2012;261:369. [Google Scholar]; b) Kong J, Yu S. Acta Bioch. Bioph. Sin. 2007;39:549. doi: 10.1111/j.1745-7270.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- 40.Barth A, Zscherp C. Q Rev. Biophys. 2002;35:369. doi: 10.1017/s0033583502003815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.