Abstract
Background
Even though virtually all Ewing sarcoma patients achieve a radiographic complete response, up to 30% of patients who present with localized disease and up to 90% of those who present with metastases suffer a metastatic relapse, highlighting our inability to identify patients with residual disease at the end of therapy. Up to 95% of Ewing sarcomas carry a driving EWS-ETS translocation which has an intronic breakpoint that is specific to each tumor, and we developed a system to quantitatively detect the specific breakpoint DNA fragment in patient plasma.
Methods
We used a long range multiplex PCR technique to identify tumor-specific EWS-ETS breakpoints in Ewing sarcoma cell lines, patient derived xenografts, and patient tumors, and this sequence was used to design tumor specific primer sets to detect plasma tumor DNA (ptDNA) by droplet digital PCR (ddPCR) in xenograft bearing mice and patients.
Results
Tumor specific breakpoint DNA fragments were detected in the plasma of xenograft bearing mice, and signal correlated with tumor burden during primary tumor growth, following surgical resection, and upon metastatic relapse. Furthermore, we were able to detect the specific breakpoint in plasma DNA obtained from 3 patients with Ewing sarcoma, and in 2 patients, we were able to detect ptDNA when there was radiographically undetectable disease present.
Conclusions
Use of ddPCR to detect tumor specific EWS-ETS fusion gene breakpoint ptDNA fragments can be developed into a highly personalized biomarker of relapse that can be optimized in animal studies for ultimate use in patients.
Keywords: cell free DNA, plasma tumor DNA, circulating tumor DNA, metastasis, EWS-FLI1
Introduction
With the introduction of conventional intensified chemotherapy, long term survival for patients with localized Ewing sarcoma has dramatically improved, from the 8–20% seen in the pre-chemotherapy era to an event free survival of 73% in the most recently published COG study AEWS00311–3. Although this is one of the exemplary success stories of intensive chemotherapy in solid tumors, these numbers indicate that up to 20% of patients would not have required systemic therapy, and as many as 30% will not be cured despite multimodal treatment. Similarly, although the majority of Ewing sarcoma patients presenting with metastatic disease achieve a complete remission, cure rates hover at about 20%. Our inability to distinguish patients who are cured at the end of therapy from those who are destined to relapse highlights the limitations of current radiographic techniques to assess disease status, and led us to develop a personalized and sensitive molecular marker of disease burden.
Cell free circulating tumor DNA (ctDNA) has been explored as a biomarker in multiple cancer types in both plasma and serum, and most applications of this technology have focused on developing a “liquid biopsy”, using ctDNA to detect de novo mutations and clonal evolution in heterogeneous tumors in order to direct therapy4. While this is an attractive application for carcinomas with high mutational loads, especially to detect actionable rare mutations within a tumor, pediatric sarcomas have “quiet” genomes, and are driven by a relatively small number of mutations, making “liquid biopsy” less attractive for clinical use5, 6. In patients with carcinomas, this approach has been used to detect the development of secondary mutations before clinical progression, as well as to monitor metastatic disease burden, and while the results have been promising, these efforts have been focused on patients with radiographically detectable disease4, 7–11. With the introduction of droplet digital PCR (ddPCR), the sensitivity of plasma tumor DNA (ptDNA) analysis has dramatically improved12, 13, making detection of very low disease burden a possibility. In order to identify a tumor-specific genetic event to monitor, current methods use next generation sequencing (NGS) followed by detection of a tumor specific genetic signature in the ptDNA to detect occult metastatic disease14. NGS has become more readily available in the clinical setting, but remains costly and time consuming, and for monitoring disease burden in a tumor driven by a translocation oncogene (as opposed to identifying clonal evolution or targetable mutations), NGS might not be necessary, and a more rapid, economical, and sensitive platform would be of value.
Ewing sarcoma is driven by a reciprocal translocation between the EWS gene and an ETS family member (FLI1 in 85% of tumors and ERG in another 10%)15, 16. The translocation oncogene is essential for the initiation and maintenance of the transformed phenotype, making this an ideal molecular marker, since it will not be down-regulated or lost during tumor progression, in addition to being present in every single tumor cell as the essential driver17, 18. While there are a number of variations of the resulting fusion mRNA transcript, the translocation breakpoints are intronic, with no evolutionary advantage for a particular breakpoint, and thus are unique in each patient. We have taken advantage of the high specificity of the breakpoint and here describe the development of a novel, highly personalized biomarker of disease burden based on detection of ptDNA containing the patient specific translocation breakpoint. The method has been successfully used in a preclinical mouse model, demonstrating a correlation between ptDNA and both disease burden and metastatic relapse. Furthermore, we piloted the use of our method in 3 patients in whom the breakpoint specific ptDNA was successfully and quantitatively detected, supporting the potential for developing quantitative assessment of ptDNA as a biomarker of disease burden and risk of metastatic relapse in patients with Ewing sarcoma.
Results
Rapid identification of the individual breakpoint
Utilizing a multiplex PCR method (previously described by Berger et al19 and illustrated in Figure 1), individual breakpoints were first verified using cell lines with known breakpoint sequences, TC71, TC32, and SK-ES-1 using the primers EWS12-22 and FLI12-22. Additionally, we developed an ERG primer set to expand this methodology to EWS-ERG translocation tumors (Supplemental Table 1). In two steps, the most 3’ EWS forward primer in combination with the most 5’ FLI1 or ERG reverse primer is identified. The resultant PCR product is Sanger sequenced, revealing the individual breakpoint (Table 1). Using the previously described EWS-FLI1 primer set, as well as our novel ERG primer set, breakpoints were identified for additional Ewing sarcoma cell lines, including TTC-5838, A4573, MHH-ES-1, CHLA9, CHLA10, CHLA25, COG-E-352, for our patient derived xenografts (PDXs) designated EWS1 and EWS4, and for 3 patients with either EWS-FLI1 or EWS-ERG translocations. This process was simple, rapid (from DNA isolation to breakpoint identification takes only 48 hours), and affordable, significant advantages over other methods such as whole genome sequencing19.
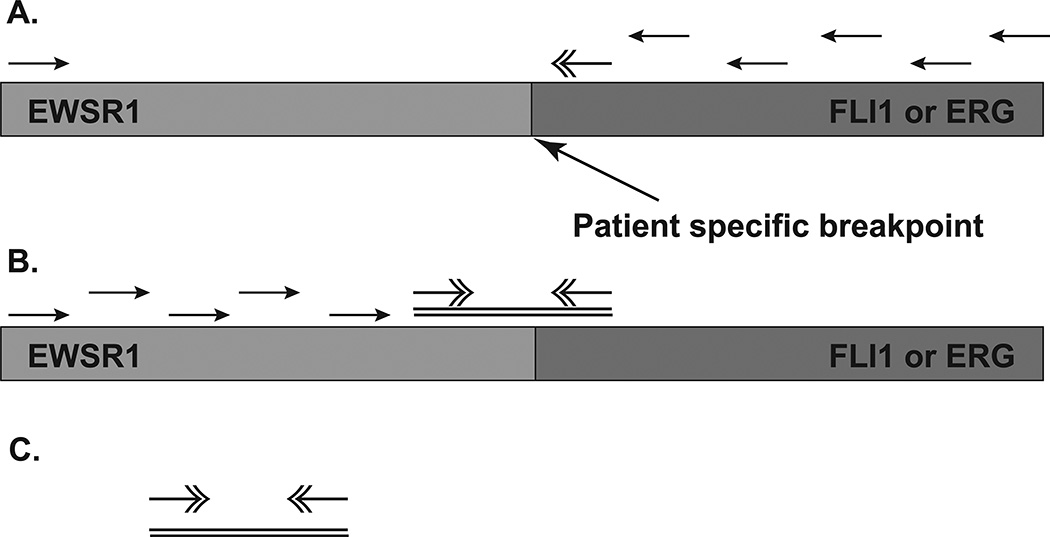
Fig. 1. Schematic of the MLR PCR Procedure.
(A) Genomic DNA isolated from the tumor is amplified first with a single primer at the 5’ end of the EWSR1 gene and a series of primers dispersed along the FLI1 gene. (B) Next, genomic DNA is amplified using the 5’ most successful FLI1 primer and a series of primers dispersed along the EWSR1 gene. (C) The 3’- most EWSR1 primer and the 5’-most FLI1 primer are then used to amplify the intronic junction of the fusion gene, and Sanger sequencing is used to pinpoint the break points.
Table 1.
Location of specific breakpoints of EWS-ETS translocation. 10 cell lines (TC71, TC32, SK-ES-1, A4573, MHH-ES-1, CHLA-9, CHLA-10, CHLA-24, COG-E-352, TTC5838), 2 PDX models (EWS1, EWS4), and 3 patient tumors had the individual translocation breakpoint identified using our long range PCR assay.
| Name | Sequence 5’-3’ | Breakpoint location | |
|---|---|---|---|
| TC71 | EWS | aaagagcctacctatt | 20448–20449 |
| aaagagcccactgcct | |||
| FLI1 | gcgatatgcactgcct | 103088–103089 | |
| TC32 | EWS | gctcacttcctactgg | 21373–21374 |
| gctcacttagttgaaa | |||
| FLI1 | actttcaaagttgaaa | 95776–95777 | |
| SK-ES-1 | EWS | agaaagtctttagtga | 21823–21824 |
| agaaagtcaataaaaa | |||
| FLI1 | tgtttcaaaataaaaa | 108543–108544 | |
| A4573 | EWS | agagcctacctattaa | 20448–20449 |
| agagcccactgcctta | |||
| FLI1 | tcagtccactgcctta | 104174–104175 | |
| MHH-ES-1 | EWS | ccagtgtgatattctt | 24261–24262 |
| ccagtgtggagagaaa | |||
| FLI1 | acaaactagagagaaa | 99670–99671 | |
| CHLA-9 | EWS | catatattaagtggca | 20948–20949 |
| catatattttccatca | |||
| FLI1 | acagggaattccatca | 116309–116310 | |
| CHLA | EWS | catatattaagtggca | 20948–20949 |
| (same patient as CHLA | catatattttccatca | ||
| FLI1 | acagggaattccatca | 116309–116310 | |
| CHLA-25 | EWS | agagcctacctattaa | 20450–20451 |
| agagcctaaggcacct | |||
| ERG | atggtggcaggcacct | 267281–267282 | |
| COG-E-352 | EWS | cttggttagtgccttg | 20499–20500 |
| cttggttataaaggaa | |||
| ERG | tttcatcagaaaggaa | 269916–269917 | |
| TTC-5838 | EWS | gcatgggtgtttatgg | 20632–20633 |
| gcatgggttatagctt | |||
| ERG | cagaagggtatagctt | 262167–262168 | |
| EWS1 (PDX) | EWS | tagatccagatgaaga | 24996–24497 |
| tagatccaaaaataat | |||
| FLI1 | ctgtctgcaaaataat | 109796–109797 | |
| EWS4 (PDX) | EWS | aattaatttctgcatt | 20871–20872 |
| aattaattaatcagcc | |||
| FLI1 | cctgtagccatcagcc | 106383–106384 | |
| Patient 1 | EWS | ggaggacgcggtggaa | 20767–20768 |
| ggaggacgcaagtgtc | |||
| ERG | ggaatacacaagtctc | 262345–232346 | |
| Patient 2 | EWS | gggcatggcattccag | 19579–19580 |
| gggcatggagttcctt | |||
| FLI1 | ggatattgagttcctt | 100313–100314 | |
| Patient 3 | EWS | tgatattcttgctgtc | 24267–24268 |
| tgatattccagttcct | |||
| FLI1 | atattatccagttcct | 122313–122314 |
Detected ptDNA events correlate with disease burden and predict relapse in a preclinical model
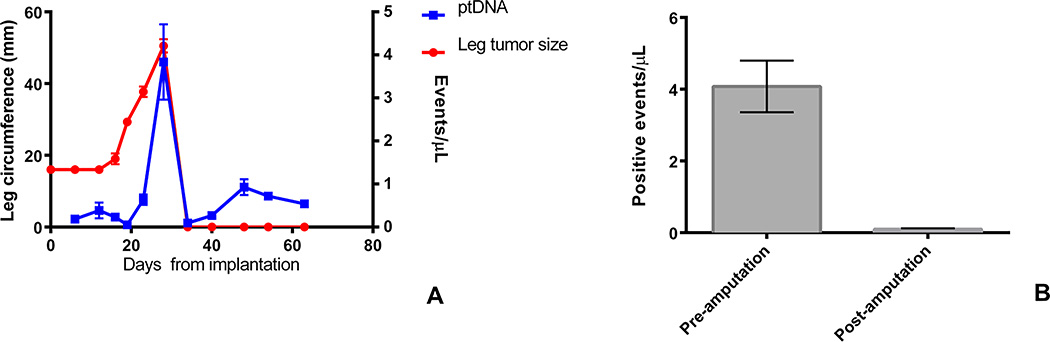
Utilizing our previously described orthotopic implantation/amputation (OIA) model20, 4 mice were implanted with a TC71 xenograft in the pretibial space. The mice had 50 µL of blood drawn every 7 days until the affected leg reached a diameter of 15 mm, at which point the leg was amputated. Blood continued to be collected every 7 days after amputation until mice were sacrificed 35 days following amputation. Cell free ptDNA was isolated from the collected blood and analyzed for the presence of the tumor-specific EWS-FLI1 breakpoint. The quantity of breakpoint specific ptDNA fragments increased with tumor growth, peaking at the time of amputation (Figure 2A). The number of detected events dropped sharply after amputation to levels equivalent to background signal (Figure 2B). Breakpoint-specific ptDNA fragments became detectable again by day 45, and remained positive for all mice until day 65, when the experiment was concluded (Figure 2A). At day 65, all mice were euthanized and were found to have lung metastases by necropsy.
Fig. 2. ptDNA Parallels Disease Burden in a TC71 Xenograft Model.
(A) 3 mm fragments of TC71 tumor were implanted in the pretibial space of NSG mice. Leg circumference (in red) and ptDNA events (in blue) were assessed weekly. (B) ptDNA events in the serum of mice pre- and post-amputation were quantified by ddPCR. The difference is statistically significant (p<0.0001 by Student’s t test).
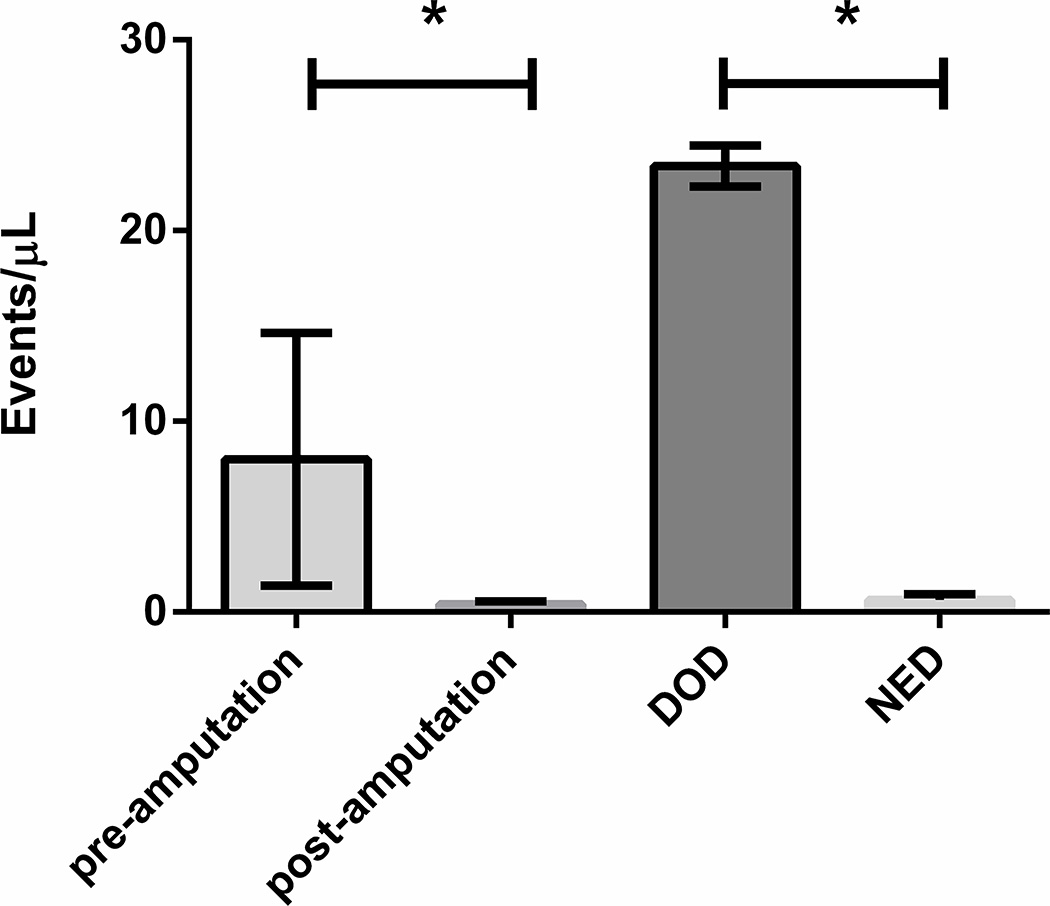
To validate these findings in a second animal model, we performed a similar experiment using a PDX designated EWS1. EWS1 tumors have lower metastatic potential than other cell line xenografts and PDXs and was selected as an ideal model to compare ptDNA levels in mice that develop metastatic disease after amputation with those that do not20. EWS1 tumor fragments were implanted in the pretibial space of 5 NSG mice, and 50 µL of blood was collected every 14–21 days, just prior to amputation, the week following amputation, and when the mice had a clinical indication for euthanasia. Cell free ptDNA was isolated and the breakpoint specific DNA fragments were detected and quantified using ddPCR with a primer set specific for the EWS1 xenograft EWS-FLI1 breakpoint. In this experiment, we were able to detect a significant breakpoint specific ptDNA burden pre-amputation, which rapidly declined following amputation (Figure 3). Following the amputation, 2 mice developed metastatic disease and had to be euthanized, while 3 mice died with no evidence of disease (NED). In this model, the breakpoint specific ptDNA burden was significantly higher in the mice who died with metastases than in the mice who died with NED. (Table 2).
Fig. 3. ptDNA Kinetics in the EWS1 PDX Model.
3 mm fragments of the EWS1 PDX were implanted in the pretibial space of 5 NSG mice. ptDNA was quantified before and after amputation when the affected leg circumference reached 15mm, and when the mice developed extensive metastasis or had to be euthanized. The ptDNA burden showed significant decline after amputation, which increased significantly for those mice which died of relapsed disease, but remained low for those mice who did not have relapsed disease. (*p<0.05 by Student’s t test)
Table 2. ptDNA Events/µL in the EWS1 PDX Model.
Mice with large primary tumors had a significant decrease of ptDNA events/µL following amputation. At terminal collection, mice 1 and 2, which had extensive metastasis, had a large ptDNA burden detected, while mice 3–5, which had no evidence of disease, continued to have a low ptDNA burden.
| Pre-amputation | Post-amputation | Terminal collection |
Terminal condition | |
|---|---|---|---|---|
| Mouse 1 | 19.4 | 0.56 | 22.62 | Extensive metastasis |
| Mouse 2 | 3.52 | 0.48 | 24.14 | Extensive metastasis |
| Mouse 3 | 5.62 | 0.4 | 0.5 | NED |
| Mouse 4 | 3.52 | 0.48 | 0.42 | NED |
| Mouse 5 | 3.48 | 0.16 | 0.98 | NED |
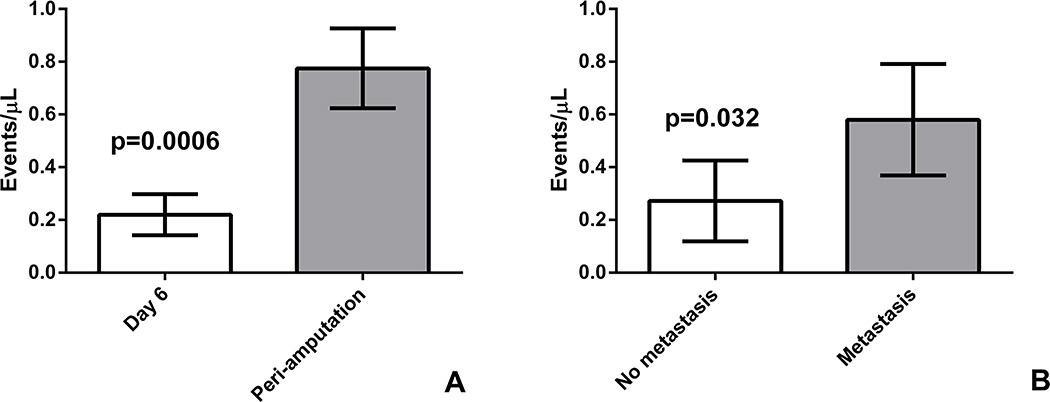
We used a third model, the EWS4 PDX, which has an intermediate metastatic potential, to confirm our findings that a) ptDNA burden increases as the primary tumor grows and b) that mice who die with metastatic disease have a higher ptDNA level that those that die without metastases. In one cohort of mice, fragments of the EWS4 PDX were implanted in the pretibial space, and 4 mice had 50 µL of blood collected on Day 6 post-implantation and again in the peri-amputation period. In the second cohort, 8 mice had fragments of the EWS4 PDX similarly implanted in the pretibial space, but had only one blood collection, which was done terminally at 45 days following amputation. This design was selected to let a larger cohort of mice survive long enough to be able to test whether ptDNA can distinguish between mice with metastatic relapse and those without. Cell free ptDNA was isolated and the breakpoint specific DNA fragments were detected and quantified using ddPCR with a primer set specific for the EWS4 xenograft EWS-FLI1 breakpoint. In the first cohort, the detected breakpoint specific ptDNA burden was significantly increased when the primary tumor had reached amputation criteria, compared to day 6 when the tumor was not palpable yet (Figure 4A). In the second cohort, mice with lung metastasis had significantly more specific breakpoint fragments detected by ddPCR than mice with no evidence of metastasis (Figure 4B).
Fig. 4. ptDNA Kinetics in the EWS4 PDX Model.
(A) 3 mm fragments of the EWS4 PDX were implanted in the pretibial space of 4 NSG mice. ptDNA was quantified 6 days after implantation, when the tumor was not palpable, and when the tumor was large enough for amputation, which showed significant increase in ptDNA burden. (p=0.0006 by Student’s t test) (B) 3 mm fragments of the EWS4 PDX were implanted in the pretibial space of 8 NSG mice. Tumors were amputated when leg circumference reached 15 mm and mice were euthanized on post-amputation day 45. ptDNA was quantified by ddPCR from serum collected during euthanasia. Mice found to have metastatic disease on necropsy had statistically significantly more ptDNA than mice without evidence of disease (p=0.03 by Student’s t test).
Breakpoint specific ptDNA fragments can be detected in patients with localized and metastatic Ewing sarcoma
Next, we proceeded to test the feasibility of this method in patients with Ewing sarcoma. Quantitative PCR based methods are known to suffer from background positivity, so prior to utilizing this method in patient samples, we performed a preliminary experiment to determine background signal when no DNA is present in the reaction. Twenty-two separate water samples (no input DNA) were analyzed on the ddPCR platform, and showed a mean of 3.591 events/reaction (95% confidence interval 2.6452 to 4.5368). For patient blood samples, the ptDNA was extracted from 1 mL of blood, divided into 2 aliquots which were run as duplicate ddPCR reactions, and detected events were summed and reported as events/mL. Therefore the background event threshold was determined to be a mean of 7.182 events/mL (95% confidence interval 5.294 to 9.0736; double the number of events/reaction).
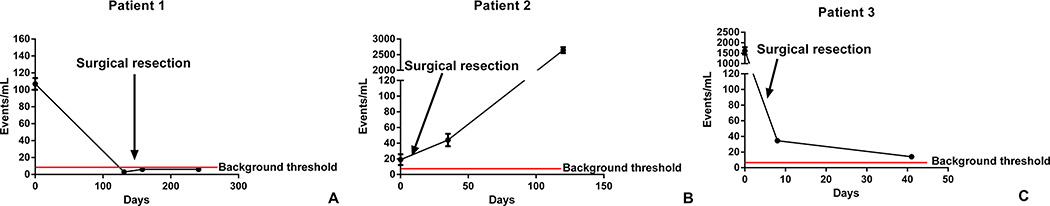
We collected fresh tumor from 3 patients either during initial diagnostic biopsy or at the time of local tumor resection. The EWS-FLI breakpoint (patients 2 and 3) or the EWS-ERG breakpoint (patient 1) were identified (Table 1). Patient 1 presented with a localized chest wall Ewing sarcoma. Blood was collected prior to any therapy, after neoadjuvant chemotherapy, after tumor resection, and at the conclusion of adjuvant chemotherapy. Tumor was collected at the time of resection, and the EWS-ERG breakpoint was identified (Table 1). This patient had a significant EWS-ERG breakpoint specific ptDNA burden detected prior to therapy which was no longer discernable from background after neoadjuvant therapy, after tumor resection, or at the conclusion of adjuvant therapy (Figure 5A). This patient currently remains with no evidence of disease radiographically after completing standard therapy. Resected tumor demonstrated over 99% necrosis with negative margins. Patient 2 had a treatment refractory, localized para-spinal Ewing sarcoma. Blood was drawn prior to complete resection of the tumor, 1 month and 3 months after resection. Tumor was collected at the time of resection, and the EWS-FLI1 breakpoint was identified (Table 1). The specific breakpoint ptDNA fragment was detectable prior to surgery, and remained detectable 1 month after resection, even though the patient had no evidence of metastatic disease on a CT scan performed immediately post-operatively and had no clinical signs or symptoms suggestive of metastatic disease. Three months following resection, the patient was found to have bilateral lung metastasis, and was also found to have significant specific breakpoint ptDNA fragment burden (Figure 5B). Patient 3 had a localized right femoral Ewing sarcoma, and was referred to our center during neoadjuvant chemotherapy with a concern for primary refractory disease. Blood was collected at the conclusion of neoadjuvant chemotherapy, after tumor resection which showed 40% necrosis with negative margins, and after 1 cycle of adjuvant chemotherapy. Tumor was collected during resection, and the EWS-FLI1 breakpoint was identified (Table 1). The patient’s EWS-FLI1 breakpoint specific ptDNA fragment was detected at the conclusion of neoadjuvant chemotherapy, and although the ptDNA burden was lower post-operatively (consistent with resection of a large tumor), and seemed to be decreasing over time, detectable ptDNA persisted even after tumor resection and after his first cycle of post-operative chemotherapy (in contrast to patient 1; Figure 5C). Although the patient had no evidence of metastatic disease by PET scan immediately prior to surgery, the patient developed increasing bone pain after 1 cycle of adjuvant chemotherapy, and was found to have a previously undetected contralateral femoral bone metastasis. Patients 2 and 3 thus demonstrate the ability to detect ptDNA when there is clinically and radiographically undetectable disease.
Fig. 5. ptDNA Measurement in Patients.
Graphs show ptDNA events, measured by ddPCR at the indicated time points. The arrow indicates the timing of surgery. The horizontal bar indicates the identified background signal threshold. Patient 1 presented with a large chest wall Ewing sarcoma which was resected after neoadjuvant chemotherapy and found to be 99% necrotic. Patient 2 had refractory, but localized, paraspinal Ewing sarcoma, was rendered NED by surgery, but was found to have pulmonary metastases 4 months later. Patient 3 presented with a refractory right femoral localized Ewing sarcoma, was rendered NED by surgery, but after a single cycle of post-operative chemotherapy was found to have a new metastatic focus in the contralateral femur.
Discussion
In this study, we describe a novel method of detection and quantification of the EWS-ETS fusion gene ptDNA fragment in Ewing sarcoma patients using ddPCR. The biology of Ewing sarcoma presents a unique opportunity in that, although the tumors have been found to have “quiet” genomes with few mutations, the EWS-ETS fusion gene is the driver of oncogenesis, is present in every tumor cell, and the intronic breakpoint is highly patient specific5, 19. This allows us to explore ptDNA detection not for the increasingly common use as a “liquid biopsy”, but as a potential molecular biomarker of radiographically undetectable disease and of metastatic risk Exploiting a previously reported multiplex long range PCR method19, we were able to identify the specific EWS-FLI1 and EWS-ERG breakpoints in multiple tumors with relative ease and rapid turnaround. Using this information, we were able to sensitively detect tumor specific translocation breakpoint ptDNA fragments from patients and in a murine preclinical model. In the animal model, the specific breakpoint ptDNA fragment burden parallels disease burden and distinguishes between mice which develop metastatic disease and those which do not. In two patients, we were able to detect radiographically undetectable disease months before it was clinically or radiographically evident. The successful translation to Ewing sarcoma patients undergoing routine care demonstrates the feasibility of advancing this method in a larger multi-center study.
There are many methods to detect the patient specific EWS-ETS breakpoint, and each has advantages and disadvantages compared to the others. The rapid turnaround and relative ease of the simple long range PCR method presents a significant advantage in cost and time. While NGS can provide additional genetic information about the tumor, this still requires specialized equipment and bioinformatics expertise, and may be difficult to perform in all patients in a large multi-center study. Moreover, since mutations detected by NGS are heterogeneous within tumors and can be affected by clonal evolution, there is advantage to focusing on the EWS-ETS breakpoint if the goal is to develop a biomarker of disease burden and to identify patients with radiographically undetectable disease. One weakness of the PCR method is the necessity for fresh tumor samples to detect the breakpoint, because DNA fragmentation during fixation in formalin makes the long range PCR assay difficult. Although initial, diagnostic biopsy specimens are often small and immediately fixed in formalin for histologic evaluation, we have been able to recover sufficient DNA even from tumors with good histologic response (Patient 1) resected after neoadjuvant chemotherapy to allow breakpoint identification and analysis of frozen plasma obtained at diagnosis. In situations where only formalin fixed tissue is available, capture sequencing or NGS would be viable alternatives.
Although the use of ptDNA as a “liquid biopsy” has been studied by multiple groups in the past, the kinetics and quantification of translocation breakpoint specific ptDNA fragments is largely unexplored. Our preclinical data using genetically distinct xenografts suggests that there may be tumor specific differences in the release of ptDNA. The ability to measure ptDNA in a mouse model of Ewing sarcoma is another strength of our approach. Given the rarity of this disease (approximately 300 cases per year in the US21), many aspects of the kinetics of ptDNA and the relationship between ptDNA and response to therapy will be challenging to evaluate in patients. These issues can be addressed in future preclinical work, generating hypotheses that can be tested in clinical samples as more are collected and analyzed.
Our pilot data supports the hypothesis that the presence or absence of breakpoint specific ptDNA can distinguish between patients with radiographically undetectable disease and those with no disease. Our data represent a pilot study with only 3 patients, and there are many aspects of the clinical utility of this technique that need to be addressed in larger studies. Do patients who convert from positive to negative ptDNA have less risk of relapse? Identification of patients with no risk of post-surgical relapse could spare those who are already cured from the toxicity associated with adjuvant chemotherapy. For example, although patient 1 had no detectable ptDNA after neoadjuvant chemotherapy, and histologic evaluation of the resected tumor showed >99% necrosis and wide negative margins, he still received intensive adjuvant chemotherapy per standard of care. Perhaps if this test were sufficiently sensitive, patients like him could be observed and monitored after resection, with additional chemotherapy reserved for those patients with detectable ptDNA. Does positive ptDNA at the end of therapy correlate with impending disease relapse, and will this early detection allow us to alter the outcome with earlier intervention? And finally, how well does a decrease in ptDNA correlate with histologic response to therapy? These questions can be addressed in animal models, but also need to be explored in larger prospective clinical studies. More accurate identification of patients with residual disease despite achieving a radiographic complete remission, as well as a more rapid blood test assessment of response to therapy, will have profound effects on the way patients with Ewing sarcoma are managed in the future.
Methods
Cell lines and Xenografts
ESFT cell lines TC71, TC32, MHH-ES-1, A4573, SK-ES-1 were kind gifts from Dr. Jeffery Toretsky (Georgetown University, Washington, DC), cell line TTC-5838 was obtained from Dr. Timothy Triche (University of Southern California, Los Angeles, CA), and cell lines CHLA9, CHLA10, CHLA25, COG-E-352 were obtained from the Children’s Oncology Group Cell Culture and Xenograft Repository (Lubbock, TX). Cells were cultured at 50–70% confluence in RPMI-1640 medium supplemented with 10% fetal bovine serum (Invitrogen, Grand Island, NY). All cell lines were verified by STR profiling at the Genetic Resources Core Facility, Johns Hopkins Institute of Genetic Medicine (Baltimore, MD). Transplantable xenografts of primary human Ewing sarcoma were a kind gift from Dr. Chand Khanna (National Institutes of Health, Bethesda, MD). The creation of these xenografts was approved by the Institutional Review Board of the National Institutes of Health, and patients gave informed consent for their creation.
Animal experiments
NOD/SCID/IL-2Rγ-null (NSG) mice were implanted with 3 mm fragments of either a TC71 xenograft or patient derived xenograft (PDX) EWS 1 or EWS4 in the pre-tibial space as previously described20. The circumference of the affected limb was measured every 3–4 days, and 50 µL of blood was collected by tail prick in 100 µL of PBS with 1 mM EDTA. This blood sample was processed within 1 hour with double centrifugation, first at 4,000 rpm for 10 min at 4°C, then at 13,000 rpm for 10 min at 4°C, and DNA was extracted from the resultant plasma using the DNA micro kit (Qiagen) as previously described22. Plasma was stored at −80°C until further use. When the leg circumference reached 15 mm, mice had their affected limbs amputated as previously described20.
Patients and sample collection
Blood specimens and fresh tumor samples were collected under a protocol approved by the Johns Hopkins University Institutional Review Board, after obtaining informed consent. A total of 10 samples were collected from 3 patients who presented to Johns Hopkins University Sidney Kimmel Comprehensive Cancer Center with a diagnosis of Ewing sarcoma.
Identification of the translocation breakpoint
Multiplex long range PCR was performed on genomic DNA isolated from freshly frozen tumor samples using the AccuPrime Taq Polymerase System (Invitrogen) and a EWS-FLI1 primer set as previously described by Berger et al19, or with an EWS-ERG specific primer set (Supplemental Table 1). PCR products were sequenced by the 3730xl DNA analyzer (Applied Biosystems) at the Genetic Resources Core Facility, Johns Hopkins Institute of Genetic Medicine (Baltimore, MD), and the sequence was aligned to the NCBI reference sequences for EWS, FLI1, and ERG, revealing the specific translocation breakpoint for each tumor. Using this specific translocation breakpoint sequence, a primer set was designed containing the exact breakpoint for use in ddPCR.
Isolation and quantification of ptDNA
Blood samples were obtained from patients in an EDTA blood collection tube and were processed within 1 hour of collection. Blood was centrifuged twice, first 4,000 rpm for 10 min at 4°C, then the collected plasma was centrifuged at 13,000 rpm for 10 min at 4°C. The resultant cell free plasma was stored in 1 ml aliquots and frozen at -80°C until use. DNA was extracted from 1 ml of plasma using the Qiagen Circulating Nucleic Acid kit (Qiagen) per the manufacturer’s protocol.
Droplet digital PCR detection
Extracted plasma cell free DNA was subjected to ddPCR amplification using the primers listed in Supplementary Table 1. ddPCR was performed using the QX200 Evagreen Supermix (Bio-Rad Technologies) per the manufacturer’s protocol using the QX200 Droplet Digital PCR System (Bio-Rad Technologies). Positive droplets were analyzed using the QuantaSoft program (Bio-Rad Technologies) to quantify detected breakpoint specific ptDNA, and were reported as positive events/mL for patients, and positive events/µL for mice considering body size and blood volume differences.
Statistical analysis
The statistical difference in ptDNA burden before and after amputation was tested using a two-sided Student’s t test. All statistical analyses were performed using Prism 6 software (GraphPad Software, Inc., La Jolla, CA).
Approval of human and animal studies
Blood samples were collected form patients at the Johns Hopkins University Sidney Kimmel Comprehensive Cancer Center who provided written, informed consent, on a protocol approved by the Institutional Review Board of Johns Hopkins University. All experiments with mice were carried out according to a study protocol that was approved by the Johns Hopkins University Institutional Animal Care and Use Committee.
Supplementary Material
Statement of significance.
By identifying the specific EWS-ETS breakpoint in patients with Ewing sarcoma, we were able to design a personalized plasma tumor DNA detection system using droplet digital PCR. The number of detected breakpoint fragments correlates with tumor burden and metastatic relapse in an animal model, and was piloted in 3 patients, showing strong translational potential.
Acknowledgments
Funding: This work was supported by grants from National Institutes of Health (1R01CA138212-01), The Rally Foundation for Childhood Cancer Research, The Pediatric Cancer Foundation, and the Liddy Shriver Sarcoma Initiative (all to D. M. L.) as well as The Giant Food Children’s Cancer Research Fund and the Core Grant supporting the Sidney Kimmel Comprehensive Cancer Center (2P30 CA006973). M. H. was supported by a grant from the Pablove Foundation and by the SARC Sarcoma SPORE Career Development Award (5U54CA168512-02).
Footnotes
Conflict of Interest: The authors have declared that no conflict of interest exists.
Author contributions: M.H., B.H.P., D.M.L. developed concepts, designed the experiments and wrote the manuscript. M.H., D.C. performed the experiments and analyzed the data. C.F.M., N.J.L., G.M., C.D.M., A.L., J-P.W., C.M.A., D.S., gave technical support and conceptual advice.
References
- 1.Falk S, Alpert M. Five-year survival of patients with Ewing's sarcoma. Surg Gynecol Obstet. 1967;124:319–324. [PubMed] [Google Scholar]
- 2.Rosen G, Wollner N, Tan C, et al. Proceedings: Disease-free survival in children with Ewing's sarcoma treated with radiation therapy and adjuvant four-drug sequential chemotherapy. Cancer. 1974;33:384–393. doi: 10.1002/1097-0142(197402)33:2<384::aid-cncr2820330213>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 3.Womer RB, West DC, Krailo MD, et al. Randomized controlled trial of interval-compressed chemotherapy for the treatment of localized Ewing sarcoma: a report from the Children's Oncology Group. J Clin Oncol. 2012;30:4148–4154. doi: 10.1200/JCO.2011.41.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tirode F, Surdez D, Ma X, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014;4:1342–1353. doi: 10.1158/2159-8290.CD-14-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shern JF, Chen L, Chmielecki J, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014;4:216–231. doi: 10.1158/2159-8290.CD-13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beaver JA, Jelovac D, Balukrishna S, et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res. 2014;20:2643–2650. doi: 10.1158/1078-0432.CCR-13-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 9.Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leary RJ, Sausen M, Kinde I, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4:162ra154. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7:302ra133. doi: 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 13.Chu D, Paoletti C, Gersch C, et al. ESR1 Mutations in Circulating Plasma Tumor DNA from Metastatic Breast Cancer Patients. Clin Cancer Res. 2015 doi: 10.1158/1078-0432.CCR-15-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsson E, Winter C, George A, et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol Med. 2015;7:1034–1047. doi: 10.15252/emmm.201404913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delattre O, Zucman J, Melot T, et al. The Ewing family of tumors--a subgroup of small-round-cell tumors defined by specific chimeric transcripts. N Engl J Med. 1994;331:294–299. doi: 10.1056/NEJM199408043310503. [DOI] [PubMed] [Google Scholar]
- 16.Sorensen PH, Lessnick SL, Lopez-Terrada D, Liu XF, Triche TJ, Denny CT. A second Ewing's sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nat Genet. 1994;6:146–151. doi: 10.1038/ng0294-146. [DOI] [PubMed] [Google Scholar]
- 17.Kovar H, Aryee DN, Jug G, et al. EWS/FLI-1 antagonists induce growth inhibition of Ewing tumor cells in vitro. Cell Growth Differ. 1996;7:429–437. [PubMed] [Google Scholar]
- 18.Ouchida M, Ohno T, Fujimura Y, Rao VN, Reddy ES. Loss of tumorigenicity of Ewing's sarcoma cells expressing antisense RNA to EWS-fusion transcripts. Oncogene. 1995;11:1049–1054. [PubMed] [Google Scholar]
- 19.Berger M, Dirksen U, Braeuninger A, et al. Genomic EWS-FLI1 fusion sequences in Ewing sarcoma resemble breakpoint characteristics of immature lymphoid malignancies. PLoS One. 2013;8:e56408. doi: 10.1371/journal.pone.0056408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldstein SD, Hayashi M, Albert CM, Jackson KW, Loeb DM. An orthotopic xenograft model with survival hindlimb amputation allows investigation of the effect of tumor microenvironment on sarcoma metastasis. Clin Exp Metastasis. 2015;32:703–715. doi: 10.1007/s10585-015-9738-x. [DOI] [PubMed] [Google Scholar]
- 21.Esiashvili N, Goodman M, Marcus RB., Jr Changes in incidence and survival of Ewing sarcoma patients over the past 3 decades: Surveillance Epidemiology and End Results data. J Pediatr Hematol Oncol. 2008;30:425–430. doi: 10.1097/MPH.0b013e31816e22f3. [DOI] [PubMed] [Google Scholar]
- 22.Rago C, Huso DL, Diehl F, et al. Serial assessment of human tumor burdens in mice by the analysis of circulating DNA. Cancer Res. 2007;67:9364–9370. doi: 10.1158/0008-5472.CAN-07-0605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.