Abstract
Late embryogenesis abundant (LEA) proteins have been identified in a wide range of organisms and are believed to play a role in the adaptation of plants to stress conditions. In this study, we performed genome-wide identification of LEA proteins and their coding genes in Moso bamboo (Phyllostachys edulis) of Poaceae. A total of 23 genes encoding LEA proteins (PeLEAs) were found in P. edulis that could be classified to six groups based on Pfam protein family and homologous analysis. Further in silico analyses of the structures, gene amount, and biochemical characteristics were conducted and compared with those of O. sativa (OsLEAs), B. distachyon (BdLEAs), Z. mays (ZmLEAs), S. bicolor (SbLEAs), Arabidopsis, and Populus trichocarpa. The less number of PeLEAs was found. Evolutionary analysis revealed orthologous relationship and colinearity between P. edulis, O. sativa, B. distachyon, Z. mays, and S. bicolor. Analyses of the non-synonymous (Ka) and synonymous (Ks)substitution rates and their ratios indicated that the duplication of PeLEAs may have occurred around 18.8 million years ago (MYA), and divergence time of LEA family among the P. edulis-O. sativa and P. edulis–B. distachyon, P. edulis-S. bicolor, and P. edulis-Z. mays was approximately 30 MYA, 36 MYA, 48 MYA, and 53 MYA, respectively. Almost all PeLEAs contain ABA- and (or) stress-responsive regulatory elements. Further RNA-seq analysis revealed approximately 78% of PeLEAs could be up-regulated by dehydration and cold stresses. The present study makes insights into the LEA family in P. edulis and provides inventory of stress-responsive genes for further functional validation and transgenic research aiming to plant genetic improvement of abiotic stress tolerance.
Introduction
As sessile organisms, plants have evolved a wide spectrum of adaptations to cope with the inevitable challenges of environmental stress, such as drought, high salinity, and cold, etc. Many aspects of these adaptation processes, including developmental, physiological and biochemical changes, are regulated or achieved by stress-responsive gene expression. The late embryogenesis abundant (LEA) proteins constitute of a family of hydrophilic proteins that are presumed to play a protective role during exposure to different abiotic stresses. They were first described to highly accumulate during the late stages of cotton seed development, when the embryo becomes desiccation tolerant [1]. They were not only found in the seeds of many other plants, but also detected in vegetative organs. More importantly, they are usually induced under stress conditions such as cold, drought, or high salinity [2, 3].
LEA proteins were initially classified to six subgroups on the basis of specific domains [4]. With increasing information on family members, expression profile differences, derived organisms and also the development of bioinformatic tools, the classification has been subjected to different rearrangements [5–10]. Many studies have been performed to characterize their functions, especially the roles in stress responses. LEA25, a group 4 LEA protein from tomato (Solanum lycopersicum), can improve tolerance against high salinity and freezing when expressed in Saccharomyces cerevisiae [11]; Overexpression of barley (Hordeum vulgare) HVA1 in wheat (Triticum aestivum) and rice (Oryza sativa) confers enhanced drought tolerance [12, 13]. Virus-induced silence of HVA1 and DHN6 resulted in significant decrease in drought tolerance [14]. JcLEA, a abscisic acid (ABA) and stress-induced group 5 LEA protein of Jatropha curcas, could enhance tolerance to drought and salt stress in Arabidopsis [15]. Overexpression of a maize group 3 LEA gene, ZmLEA3, in tobacco and yeast conferred tolerance to osmotic and oxidative stresses [16]. Results from these studies suggest that LEA family could be considered as a reservoir for stress-responsive genes, which have great potential in genetic improvement of stress tolerance in plants.
The rapid generation of plant whole genome sequences provides opportunities to genome-widely identify and classify the genes encoding LEA proteins in plant, which will not only provide insights into evolution of LEA family, but also provide basis for further systematically expression profiling, and in-depth biochemical, functional and physiological studies. To date, the genome-wide characterization of LEA family has been performed in several genome-sequenced plant species, such as Arabidopsis [6, 8], Populus trichocarpa [17], legumes [10], O. sativa [18], and Brachypodium distachyon [19].
Bambusoideae, generally called bamboo, belongs to grass family (Poaceae) and is comprised of more than 1,400 species. Unlike other herbaceous species of Poaceae, the major components of Bambusoideae are arborescent and perennial woody species, which live exclusively in forests and grow large woody culms up to 30 cm in diameter and 12 m in height [20]. Fast growing, high productivity, strong regeneration capability make it one of the most important non-timber forest resources in the world. According to the statistics, about 2.5 billion people depend economically on bamboo, and the annual international trade in bamboo amounts to over 2.5 billion US dollars [21]. Despite of its economic importance, little is known about its responses to abiotic stress and underlying mechanism at molecular level. This might be partly due to the lack of genomic resources. Recently, the genome of Moso bamboo (Phyllostachys edulis), a large woody bamboo with high ecological and economic values, were decoded [22]. In this study, we searched the P. edulis genome to identify the genes encoding LEA proteins (PeLEAs). In silico analyses of promoter elements, biochemical properties, and evolutionary features were performed. Further RNA-seq based expression profiling was also conducted to investigated their responses to dehydration and cold.
Materials and Methods
Data resources
The whole genome dataset, full length cDNA, and EST of P. edulis are downloaded from the Bamboo Genome Database (www.bamboogdb.org) and National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/). They consist of 31,987 protein-coding genes predicted from whole genome sequences, and 10,608 low redundant full-length cDNA (FLcDNA) sequences and 38,000 ESTs from leaf, shoot, and seedling libraries. The LEA genes in A. thaliana genome were according to Hundertmark and Hincha [8], and their sequences were obtained from The Arabidopsis Information Resource (www.arabidopsis.org). The O. sativa and B. distachyon genome data were also obtained from the website of Rice Genome Annotation Project (O. sativa.plantbiology.msu.edu), and the genome data of Zea mays and Sorghum bicolor was obtained from EnsemblPlants (plants.ensembl.org/)
Identification of genes encoding LEA protein from P. edulis genome
All LEA protein sequences of A. thaliana [8] were used as queries to Blast search against whole genome dataset of P. edulis with expectation value of 0.01. The resulted sequences were analyzed by Pfam database [23] to characterize obtained sequences into Pfam family. The full length cDNA and EST datasets were queried for further evidences for the obtained genes.
In silico analyses of of LEAs
The grand average of hydropathicity index (GRAVY), theoretical isoelectric point (pI) and molecular weight were analyzed by using the ProtParam Tool (web.expasy.org/protparam/).
The coordinates of exon and intron of LEA genes of P. edulis were extracted from their corresponding scaffolds and exon-intron structures were illustrated using Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/) [24].
Sequences of 1500 nt upstream of the coding sequences were retrieved from bamboo genome database (http://www.bamboogdb.org/). The putative cis-acting elements related to abiotic stress response were analyzed by querying the PLACE database ((http://www.dna.affrc.go.jp/PLACE/) [25].
Evolutionary analyses of the paralogues and orthologues in four grass species
Reciprocal BLASTP was carried out to establish orthologous relationship among P. edulis, O. sativa and B. distachyon. The hits threshold values were set as E-value <1e-10, score >200, and positive >70%. The paralogous relationship within P. edulis was also analyzed with more stringent parameters of E-value <1e-50, score >200, and positive >80%. The synonymous (Ks) and non-synonymous (Ka) substitution rates of paralogues and orthologues were analyzed by Ka_Ks calculator 2.0 [26]. Time (million years ago, MYA) of duplication and divergence was calculated using a synonymous mutation rate of one substitutions per synonymous site per year as T = Ks/2λ (λ = 6.5×10−9) [27, 28].
Stress treatment and expression profiling
To evaluate expression patterns of PeLEA under abiotic stress, dehydration and cold treatments were conducted. The full young unexpanded leaves were detached from different P. edulis plants with similar growth status. For dehydration treatment, the whole leaves were placed on the dry filter paper and treated under room temperature (20°C and 50% humidity). For cold treatment, the leaves were put into a chamber set to 0°C without light. At 2h and 8h after each treatment, ten individual leaves were immediately frozen in liquid nitrogen and the total RNA of was extracted according to the manual of the TRIZOL RNA Kit (TIANGEN, Beijing, China). The same amount of untreated leaves were also sampled and used as control. The qualities and quantities of extracted nucleotide were measured by NanoDrop 2000 Spectrophotometer (Thermo Fisher, USA) and Agilent 2100 RNA 6000 Nano kit. The threshold of the quality of extracted RNA was RIN ≥ 7 with concentration ≥ 150 ng/ul and amount ≥ 5 ug.
The cDNA library construction and sequencings on Illumina HiSeq™ 4000 platform were performed by Onmath Co.(Chengdu, China), following the manufacturer’s standard protocol. The 150 bp sequences by pair-end sequencing were generated as raw data. The filtered clean reads were mapped to all obtained PeLEA by using TopHat v2.0.9. HTSeq v0.6.1 was used to count the reads numbers mapped to each gene. And then RPKM of each gene was calculated based on the length of the gene and reads count mapped to this gene. RPKM, Reads Per Kilobase of exon model per Million mapped reads, considers the effect of sequencing depth and gene length for the reads count at the same time, and is currently the most commonly used method for estimating gene expression levels [29].
Results and Discussions
Identification and classification of LEA genes in P. edulis
By blast query of gene model of P. edulis genome, 23 putative LEA proteins were found. By Pfam family domain analysis, 21 candidates could be assigned to LEA family, in which 6 were supported by known FLcDNA or EST (S1 Table). Six groups of LEA protein were identified by homology to known LEA proteins. We here classified them based on the Pfam nomenclature, as it is specifically related to conserved domains [8] and integrate sequences from diverse species. The correspondence of different nomenclatures or classifications of LEA family was present in S2 Table. Only one gene was found in groups LEA_1 and LEA_6 (also known as PvLEA18); Four LEA2 proteins were detected; Both group LEA_3 and group LEA_4 were comprised of five proteins, and six proteins are assigned as dehydrins. However, no gene was identified as groups LEA_5, SMP, or AtM (S1 Table) in P. edulis.
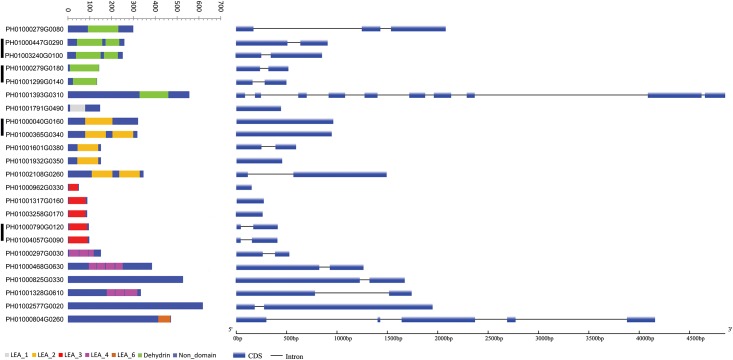
Members of groups LEA_1, LEA_3, and LEA_6, and four members of dehydrin, and three LEA_2 members have a single LEA domain; two members of dehydrin and LEA_2 contain two repetitive domains, and three members of LEA_4 contain three or four domains (Fig 1a). No conserved domains were found in two candidates PH01000825G0330 and PH01002577G0020. However, they showed high degree of homology to LEA_4 proteins from Arabidopsis, AT1G72100.1 (E-value = 6e-062) and AT2G42560.1 (E-value = 1e-025), respectively. Therefore, we also assigned the two PeLEA proteins into group LEA_4. Seven LEAs of P. edulis are comprised of a single uninterrupted coding region, whereas 16 members are composed of two to ten exons and one to nine introns (Fig 1b).
Fig 1. Protein domain organization (left column) and exon-intron structure (right column) of PeLEAs.
The present domains were identified by Pfam database. Different color boxes indicate the conserved domain or non-domain region on LEA proteins in of LEA groups. Black bars indicate paralogous gene pairs.
By BLAST search, we found five PeLEAs are homologous to at least 13 functionally known LEA genes associated with abiotic stress tolerance (S3 Table), indicating that they might be considered as candidate genes for drought and cold tolerance.
Comparison of gene amount and protein characteristics
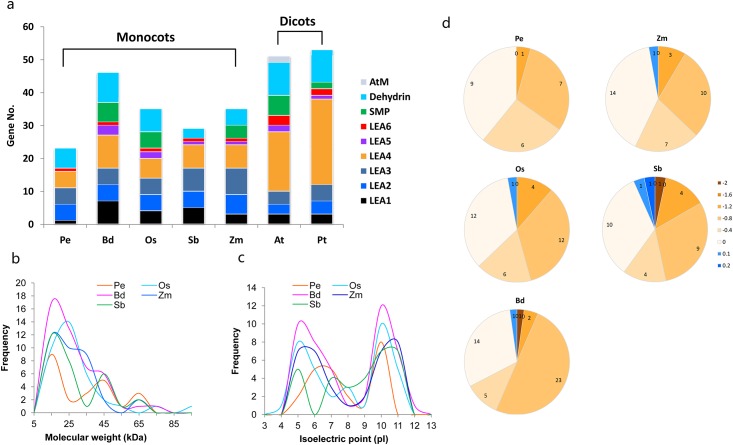
P. edulis belongs to grass family (Poaceae). It is interesting to compare the LEA families among the sequenced species of Poaceae. Filiz et al. identified 36 LEA genes in B. distachyon [19]. However, these genes were obtained by BLAST search using only a representative of LEA2-LEA6 of Arabidopsis as queries. This might lose sight on some LEAs, such as seed mature protein (SMP). Additionally, the LEA gene number of O. sativa varies in different studies [17, 18]. Therefore, we re-searched LEAs in the two species by the same strategy used in this study. As a result, 46 LEA genes were identified in B. distachyon (S1 Table), and 35 LEA genes were found in O. sativa (S1 Table), respectively. Additionally, we also search the high quality genome data of Z. mays and S. bicolor, by which 35 and 30 ZmLEAs and SbLEAs were found. They were also classified by Pfam and homology analyses according to Pfam family nomenclature (S1 Table).
We then compared the gene amount of different LEA groups between P. edulis, and four Poaceae species, as well as two well studied dicot representatives A. thaliana [6, 8] and P. trichocarpa [17]. Comparing to five monocot species, two dicots have more LEA proteins in their genome. P. edulis contains the least number of LEAs among the seven species; O. sativa and Z. mays share similar LEAs number, and B. distachyon contains the most abundant LEA proteins in five monocots analyzed (Fig 2a). Dehydrin, LEA_1, LEA_2, LEA_3, LEA_4, and LEA_6 are common groups in all species. Dicots rich in LEA_4, accounting for more than 35% and 49% of LEA family members in A. thaliana and P. trichocarpa, respectively. As mentioned above, no LEA_5, SMP, or AtM were found in P. edulis. The AtM are only found in A. thaliana.
Fig 2. Comparisons of gene amount (a), molecular weight (b), isoelectric point (c), and grand average of hydropathicity index (GRAVY, d).
Most of the PeLEA genes encode rather small proteins, in which the deduced molecular weights (MW) of ~61% members are less than 35 kDa. Similarly, ~70%- ~88% of LEAs in other four monocots are smaller than 35 kDa (Fig 2b). The theoretic pI values of PeLEAs range from 4.81 to 9.86, and the other four monocots show similar pI ranges from ~4.0 to 11. Approximately ~50% of the LEA proteins of P. edulis, O. sativa, B. distachyon, and Z. mays are more than 7.0. Whereas 70% of SbLEAs have pI more than 7.0, which is significantly higher than those in other four monocot species (Fig 2c).
We also calculated the grand average hydropathicity (GRAVY) index of LEAs. All PeLEAs show negative values, indicating that PeLEAs are all hydrophobic. Among the OsLEAs, BdLEAs, ZmLEAs, and SbLEAs, only one or two LEA proteins show GRAVY values larger than 0 (Fig 2d, S1 Table). This is similar as those reported in dicots previously [8,17], suggesting the apparently hydrophobic characteristic of LEA proteins in plants.
Orthologous and paralogous relationship among Poaceae
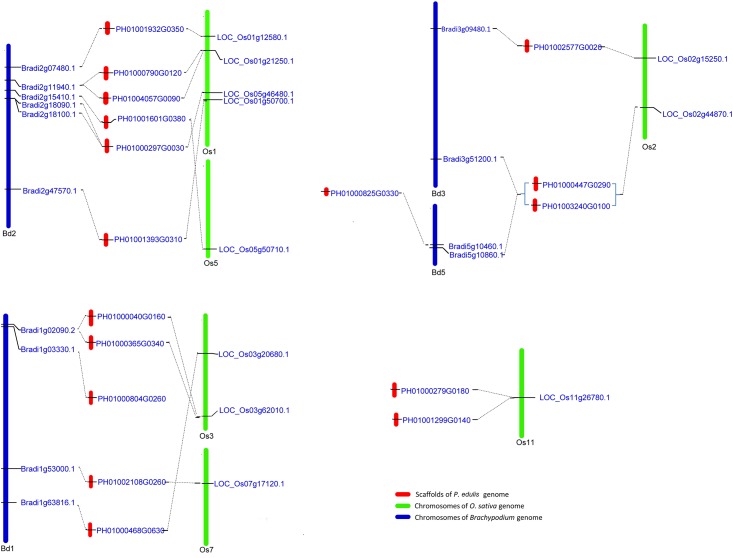
We conducted reciprocal BLASTP analysis of the orthologous relationship between five monocot species. A total of 11 OsLEAs (~31.4%), 14 BdLEAs (~30.4%), 14 ZmLEAs (~40%), and 11 SbLEAs (~36.7%) were found to be orthologous to 18 PeLEAs (~78.3%) (S4 Table), respectively. Most of the orthologues of PeLEAs and OsLEAs were located in the P. edulis-O. sativa colinearity regions [22]. The orthologous OsLEAs and BdLEAs were mainly distributed on six and four chromosomes, respectively, and also shared same syntenic patterns as revealed previously [30] (Fig 3).
Fig 3. Genome organization, orthologous relationship and colinearity of LEAs among P. edulis (Pe), O. sativa (Os), and B. distachyon (Bd).
Only the orthologues are present. The blue and green bars indicate chromosomes of B. distachyon and O. sativa, respectively. Their corresponding chromosomes numbers were showed at the bottom of chromosomes. The scaffolds on which PeLEAs are located are represented by red bars.
Genome duplications, such as tandem and segmental duplications usually give rise to gene copy numbers. The reciprocal BLASTP analysis identified four paralogous pairs of PeLEAs (S4 Table). Previous study proposed that the monocot chromosomes, such as O. sativa (12 pairs of chromosomes) and B. distachyon (five pairs chromosomes), were derived from an intermediate with 12 pairs of chromosomes [30]. P. edulis has 24 pairs of chromosomes and sequencing of its genome revealed that it carries as two duplicates as that of rice gene model sets [22]. However, only four duplicated PeLEAs to orthologous OsLEAs were detected. Additionally, P. edulis contains the least PeLEAs gene number among the species analyzed. These results suggested that LEA family in P. edulis may have undergone significantly gene loss during evolution.
Divergence rates and selection
In order to evaluate the timing of intragenomic gene duplication events, as well as divergence of orthologues, the synonymous substitution rate (Ks) was calculate. The paralogous gene pairs exhibited mean Ks of 0.24. Then estimated by universal substitution rate of 6.5 × 10−9 mutations per site per year, the duplications of PeLEAs may occur around 18.8 million years ago (MYA) (Fig 4a and 4b, S5 Table). This is different from the estimated timing of whole genome duplication at 7-12MYA [22], as well as divergence time of AP2/ERF transcription factors superfamily of around 15 MYA [31].
Fig 4. Ks (a), divergence time (a) and Ka/Ks (b) distributions of paralogous of P. edulis, and between P. edulis-O. sativa (Os), P. edulis-B. distachyon, P. edulis-Z. mays, and P. edulis-S. bicolor orthologue pairs.
Among the P. edulis-O. sativa, P. edulis–B. distachyon, P. edulis-S. bicolor and P. edulis-Z. mays orthologous gene pairs, the mean Ks of are ~0.40, ~0.47, ~0.62, and ~0.69, indicating that the divergent time of LEAs families among these species was approximately 30 MYA, 36 MYA, 48 MYA, and 53MYA, respectively (S5 Table, Fig 4a). This result is also different from previous estimation of divergence time within Poaceae. By using whole-genome sequences of chloroplasts, Wu and Ge estimated the divergence time of three subfamilies, Bambusoideae (three bamboo species), Pooideae (nine species including O. sativa, O. nivara, etc.) and Ehrhartoideae (five species including Triticum aestivum, H. vulgare, B. distachyon, etc.) of Poaceae [32]. They showed that Ehrhartoideae diverged from the clade of Bambusoideae and Pooideae at approximately 46.98 (40.80–51.60) MYA, whereas the Bambusoideae and Pooideae clades split at ~42.80 (36.61–48.80) MYA [32]. Similar results obtained by calculating the Ks of 968 single-copy gene clusters, indicating that the mean Ks for B. distachyon–P. edulis, O. sativa–P. edulis, Sorghum–P. edulis and Z. mays–P. edulis are 0.61, 0.63, 0.76, and 0.84, and their divergence time is around 46.9MYA, 48.6MYA, 58.8MYA, and 64.6 MYA, respectively [22]. These differences may be due to that the LEA proteins are not highly conserved and might not be essential for surviving, and therefore they may have different substitution rates to the universal rate.
We also calculated the ratios of non-synonymous (Ka) versus synonymous (Ks) substitution rate (Ka/Ks) for duplicated gene-pairs as well as the orthologues of O. sativa, B. distachyon, Z. mays, and S. bicolor (Fig 5c). The Ka/Ks ratio is a measure of the selection pressure to which a gene pair is subjected. Ka/Ks < 1 means purifying or negative selection, Ka/Ks = 1 stands for neutral selection, and Ka/Ks > 1 indicates positive selection [27]. The Ka/Ks for paralogous gene pair of PeLEAs is 0.16 to 0.30 with mean of ~0.24. Those for orthologous gene pairs of PeLEAs-OsLEAs, PeLEAs-BdLEAs, PeLEAs-SbLEAs, and PeLEAs-ZmLEAs are 0.13 to 0.67 with mean of ~0.34, 0.15 to 0.58 with mean of ~0.34, 0.12 to 0.57 with mean of 0.28, and 0.14 to 0.57 with mean of 0.32, respectively (Fig 4b). These results indicated that they appear to have undergone extensive purifying selection during evolution.
Fig 5. Expression profiling of PeLEA family under dehydration and cold stresses.
a, Heatmap of expression values (showed by log2 RPKM values) in control, and in treated samples 2h and 8h after treatment; b, Percentage of genes in different expression levels under normal conditions; c, Percentage of genes exhibiting different responses to dehydration and cold; d, Comparison of fold changes of co-upregulated genes under dehydration and cold stresses. The expression data is obtained through two biological replicates. The relatedness of two replicates was present in S1 Fig. The black bars on the left indicate paralogous genes.
The cis-acting regulatory elements and stress-induced expression
The cis-acting elements in the promoter region are short motifs on which the transcription factors could bind on to regulate their expressions. The ABRE (ABA responsive element) plays a key role in ABA signaling during seed development and under abiotic stresses, while the DRE/CRT/LTRE (drought responsive/C-repeat/low temperature response) is well known to be involved in drought-, cold- and high-salt-responsive gene expression regulated by CBF/DREB1 transcription factors [33, 34]. The two motifs are predominantly present in LEA genes [18, 8]. To identify putative stress-responsive LEAs in P. edulis, we queried both motifs in the -1500 nt promoter region of 23 PeLEAs by PLACE database. Almost all the PeLEAs contain both of motifs in their promoters.
We further performed RNA-seq to evaluate dynamic expression levels of PeLEA genes under dehydration and cold stresses (Fig 5a). Approximately 4 giga bases high quality data (Q30>92%) for each sample were generated and used to calculate RPKM values of PeLEAs. It indicated that 26% of PeLEAs highly expressed (log2 RPKM value >4) in leaf under normal conditions, 22% showed moderate expression (log2 RPKM value between 1 to 4), and the remaining 52% are in very low expression level or not expressed (Fig 5a and 5b). In Arabidopsis, 22 LEAs showed highly expression levels in non-seed tissue under non-stressful condition, in which 10 (19.6%) highly expressed in leaf [8]. Three out of six highly-expressed PeLEAs belong to LEA_2 group, and one of dehydrin, LEA_3 and LEA_4 exhibited high expression levels. This results suggest that members of group LEA_2 in P. edulis might play important roles in development.
A total of 17 PeLEAs were upregulated (RPKM fold change>2) by dehydration, and none was downregulated. Under cold treatment, 11 genes were upregulated and only no gene was downregulated. Ten genes cold be upregulated by both dehydration and cold stresses. Only five genes kept in constant expression level or didn’t expressed (Fig 5c) under the two treatments. Interestingly, most of the co-upregulated genes are much more sensitive to the dehydration than to cold (Fig 5d), but opposite condition appears in Arabidopsis. These results suggest that the stress-induced responses of LEA family may be divergent between P. edulis and Arabidopsis.
We analyzed expression patterns of paralogous PeLEAs. Gene duplication is one of the resources of pseudogenes, as an intact functional copy still exists and then the function loss of a duplicated gene only has little effect on an organism's fitness. According to the transcriptome data, all four paralogous gene pairs showed detectable expression levels, indicating that they might not be pseudogenes Three out of the four paralogous gene pairs share highly conserved expression patterns during dehydration and cold stresses (Fig 5a). We also noticed that these paralogues also contain conserved intron/exon structures (Fig 1). These results indicated the functional and structural conservation during evolution.
Dehydrin is the most characterized group of LEA proteins and accumulate during seed desiccation and in response to water deficit induced by drought, low temperature or salinity [35–37]. All six dehydrins of P. edulis were induced by dehydration, and four of them were also induced by cold ((Fig 5a). This is quite different from those in Arabidopsis, that five of 10 dehydrins are responsive to cold and only two are induced by drought [8].
Both P. edulis and Arabidopsis (At5g06760, previously known as LEA4-5) has one LEA of LEA_1 group which is more sensitive to dehydration or dehydration. This gene has been reported to be responsive to water deficit and its overexpression leads to tolerance to severe drought in Arabidopsis. Four of five genes in LEA_3 group are positively responsive to dehydration, and 3 of them are also responsive to cold. Arabidopsis has three stress-induced LEA_3: At1g02820 and At4g15910 (Drought-induced 21, DI21) are responsive to both of drought and cold, At4g02380 (Senescence-associated gene 21, SAG21) is regulated by cold, respectively. Two genes of LEA_4 (PH01000297G0030 and PH01001328G0610) are significantly upregulated by dehydration, in which later is more sensitive and is also induced by cold. In Arabidopsis, LEA_4 is in predominant number among the AtLEA family (18 of 51), but only two of them, At2g42530 (Cold-regulated 15B, COR15B) and At2g42540 (COR15B) are upregulated by cold [8]. P. edulis has only one LEA_6 (also known as PvLEA18) and its expression is not detectable in leaf both in normal and stressful conditions.
Three of five LEA_2 are upregulated by dehydration and two of them are also responsive to cold. Previous studies classified this group as 'atypical' LEA proteins because of their more hydrophobic character [38, 39]. Although little is known of their function, some reports indicated that they will accumulate in response to diverse stresses in plants, such as cotton (LEA14-A) [38], Craterostigma plantagineum (PcC27-45) [40], soybean (D95-4) [41], tomato (ER5) [42], and Arabidopsis (LEA14) [43], etc. Overexpression of CaLEA6 in tobacco improves tolerance to dehydration and NaCl [44]; Transgenic sweetpotato non-embryogenic calli that overexpressed IbLEA14 showed increased tolerance to drought and salt stress by enhancing lignification [45]; Overexpression of SiLEA14 foxtail millet improved tolerance to salt and drought [46]. All these results suggest that LEAs of group LEA_2 proteins are also closely associated to the resistance to multiple abiotic stresses.
Conclusion
In this study, we identified 23 LEA proteins and their coding genes from Moso bamboo genome and classified them to six groups. We performed comparative analyses of structures, gene amount, biochemical characteristics, and evolutionary features of PeLEAs with those of O. sativa (OsLEAs), B. distachyon (BdLEAs), Z. mays (ZmLEAs), S. bicolor (SbLEAs), Arabidopsis (AtLEAs), and P. trichocarpa (PtLEAs). RNA-seq based expression profile revealed genes involved in responses to dehydration and cold stresses. The results present here provide comprehensive insights into the LEA family in P. edulis and the expression altas under dehydration and cold stresses, which will help to cope with the increasing environmental challenges in the future.
Supporting Information
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We wish to thank the anonymous reviewers for helpful comments and constructive suggestions that improved the manuscript.
Data Availability
The raw data is deposited into The Sequence Read Archive (SRA) of National Center of Biotechnology Information (NCBI) with accession No. of SRR4450542 - SRR4450551.
Funding Statement
This work was Financially supported by Department of Science and Technology of Sichuan Province, China (2015JY0085) and Science and Technology Support Project of Sichuan Province, China (16ZC2871).
References
- 1.Dure L, Greenway SC, and Galau GA (1981) Developmental biochemistry of cotton seed embryogenesis and germination: changing messenger ribonucleic acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry. 20, 4162–4168. [DOI] [PubMed] [Google Scholar]
- 2.Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 47, 377–403. 10.1146/annurev.arplant.47.1.377 [DOI] [PubMed] [Google Scholar]
- 3.Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 50, 571–599. 10.1146/annurev.arplant.50.1.571 [DOI] [PubMed] [Google Scholar]
- 4.Dure L, Crouch M, Harada J, Ho TH, Mundy J, Quatrano R, et al. (1989) Common Amino Acid Sequence Domains among the LEA Proteins of Higher Plants. Plant Molecular Biology. 12, 475–486. 10.1007/BF00036962 [DOI] [PubMed] [Google Scholar]
- 5.Tunnacliffe A, and Wise M (2007) The Continuing Conundrum of the LEA Proteins. Naturwissenschaften. 94, 791–812. 10.1007/s00114-007-0254-y [DOI] [PubMed] [Google Scholar]
- 6.Bies-Ethève N, Gaubier-Comella P, Debures A, Lasserre E, Jobet E, Raynal M, et al. (2008) Inventory, Evolution and Expression Profiling Diversity of the LEA (Late Embryogenesis Abundant) Protein Gene Family in Arabidopsis thaliana. Plant Molecular Biology. 67, 107–124. 10.1007/s11103-008-9304-x [DOI] [PubMed] [Google Scholar]
- 7.Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, and Covarrubias AA (2008) The Enigmatic LEA Proteins and Other Hydrophilins. Plant Physiology.148, 6–24. 10.1104/pp.108.120725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hundertmark M, and Hincha DK (2008) LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 9, 118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shih M-D, Hoekstra F A, and Hsing Y-I C (2008) Late Embryogenesis Abundant Proteins. Advances in Botanical Research. 48, 211–255. [Google Scholar]
- 10.Battaglia M, and Covarrubias AA (2013) Late Embryogenesis Abundant (LEA) Proteins in Legumes. Front Plant Science.; 25, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imai R, Chang L, Ohta A, Bray EA, and Takagi M (1996) A lea-class gene of tomato confers salt and freezing tolerance when expressed in Saccharomyces cerevisiae. Gene. 170, 243–248. [DOI] [PubMed] [Google Scholar]
- 12.Xu D, Duan X, Wang B, Hong B, Ho T and Wu R (1996) Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 110, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sivamani E, Bahieldin A, Wraith J.M, Al-Niemi T, Dyer WE, Ho TD. et al. (2000) Improved biomass productivity and water use efficiency under water deficit conditions in transgenic wheat constitutively expressing the barley HVA1 gene. Plant Sci. 155, 1–9. [DOI] [PubMed] [Google Scholar]
- 14.Liang J, Deng G, Long H, Pan Z, Wang C, Cai P, et al. (2012) Virus-induced silencing of genes encoding LEA protein in Tibetan hulless barley (Hordeum vulgare ssp. vulgare) and their relationship to drought tolerance. Mol Breeding. 30, 441–451. [Google Scholar]
- 15.Liang J, Zhou M, Zhou X, Jin Y, Xu M, Lin J (2013) JcLEA, a Novel LEA-Like Protein from Jatropha curcas, Confers a High Level of Tolerance to Dehydration and Salinity in Arabidopsis thaliana. PLoS ONE. 8(12), e83056 10.1371/journal.pone.0083056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Wang L, Xing X, Sun L, Pan J, Kong X, et al. (2013) ZmLEA3, a Multifunctional Group 3 LEA Protein from Maize (Zea mays L.), is Involved in Biotic and Abiotic Stresses. Plant Cell Physiol.54(6), 944–959. 10.1093/pcp/pct047 [DOI] [PubMed] [Google Scholar]
- 17.Lan T, Gao J, Zeng QY (2013) Genome-wide analysis of the LEA (late embryogenesis abundant) protein gene family in Populus trichocarpa. Tree Genetics & Genomes. 9, 253–264. [Google Scholar]
- 18.Wang XS, Zhu HB, Jin GL, Liu HL, Wu WR, Zhu J (2007) Genome-scale identification and analysis of LEA genes in rice (Oryza sativa L.) Plant Science.172(2), 414–420 [Google Scholar]
- 19.Filiz E, Ozyigit II, Tombuloglu H, Koc I (2013) In silico comparative analysis of LEA (Late Embryogenesis Abundant) proteins in Brachypodium distachyon L. Plant Omics J. 6(6), 433–440. [Google Scholar]
- 20.Barker NP, Clark LG, Davis JI, Duvall M.R, Guala GF, Hsiao C, et al. (2001) Phylogeny and subfamilial classification of the grasses (Poaceae) Ann Missouri Bot Garden. 88, 373–457. [Google Scholar]
- 21.Lobovikov M, Paudel S, Piazza M, Ren H, and Wu J (2007) World Bamboo Resources: A Thematic Study Prepared in the Framework of the Global Forest Resources Assessment 2005, Food and Agriculture Organization of the United Nations, Rome [Google Scholar]
- 22.Peng ZH, Lu Y, Li LB, Zhao Q, Feng Q, Gao Z, et al. (2013) The draft genome of the fast-growing non-timber forest speciesmoso bamboo (Phyllostachys heterocycla) Nat Genet. 45, 456–461. 10.1038/ng.2569 [DOI] [PubMed] [Google Scholar]
- 23.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. (2014) The Pfam protein families database. Nucleic Acids Research. Database Issue 42, 222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu B, Jin J, Guo A.Y, Zhang H, Luo J, and Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 31(8), 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, Li J, Zhao XQ, Wang J, Wong K.S, Yu J (2006) KaKs_Calculator: Calculating Ka and Ks Through Model Selection and Model Averaging. Genomics, Proteomics & Bioinformatics. 4(4), 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science. 290, 1151–1155. [DOI] [PubMed] [Google Scholar]
- 28.Yang Z, Gu S, Wang X, Li W, Tang Z, Xu C (2008) Molecular evolution of the cpp-like gene family in plants: insights from comparative genomics of Arabidopsis and rice. J Mol Evol. 67, 266–277. 10.1007/s00239-008-9143-z [DOI] [PubMed] [Google Scholar]
- 29.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Meth. 5, 621–628. [DOI] [PubMed] [Google Scholar]
- 30.The International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 463, 763–768. 10.1038/nature08747 [DOI] [PubMed] [Google Scholar]
- 31.Wu H, Lv H, Li L, Liu J, Mu S, Li X, et al. (2015) Genome-Wide Analysis of the AP2/ERF Transcription Factors Family and the Expression Patterns of DREB Genes in Moso Bamboo (Phyllostachys edulis) PLoS ONE. 10(5): e0126657 10.1371/journal.pone.0126657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu ZQ and Ge S (2012) The phylogeny of the BEP clade in grasses revisited: Evidence from the whole-genome sequences of chloroplasts. Mol Phylogen Evo. 62: 573–578. [DOI] [PubMed] [Google Scholar]
- 33.Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci. 24; 23–58. [Google Scholar]
- 34.Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 10, 88–94. 10.1016/j.tplants.2004.12.012 [DOI] [PubMed] [Google Scholar]
- 35.Ismail A, Hall A, and Close T (1999) Purification and Partial Characterization of a Dehydrin Involved in Chilling Tolerance during Seedling Emergence of Cowpea. Plant Physiology. 120, 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nylander M, Svensson J, Palva ET, and Welin BV (2001) Stress-Induced Accumulation and Tissue-Specific Localization of Dehydrins in Arabidopsis thaliana. Plant Molecular Biology.45, 263–279. [DOI] [PubMed] [Google Scholar]
- 37.Brini F, Hanin M, Lumbreras V, Amara I, Khoudi H, Hassairi A. et al. (2007) Overexpression of Wheat Dehydrin DHN-5 Enhances Tolerance to Salt and Osmotic Stress in Arabidopsis thaliana. Plant Cell Reports. 11, 2017–2026. [DOI] [PubMed] [Google Scholar]
- 38.Galau GA, Wang HY-C, Hughes DW (1993) Cotton Lea5 and Lea14 encode atypical late embryogenesis-abundant proteins. Plant Physiol. 101, 695–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker J, Steele C, Dure L (1988) Sequence and characterization of 6 LEA proteins and their genes from cotton. Plant Mol. Bio. 11, 277–291. [DOI] [PubMed] [Google Scholar]
- 40.Piatkowski D, Schneider K, Salamini F, Bartels D (1990) Characterization of five abscisic acid-responsive cDNA clones isolated from the desiccation-tolerant plant Craterostigma plantagineum and their relationship to other water-stress genes. Plant Physiol. 94, 1682–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maitra N, and Cushman J (1994) Isolation and Characterization of a Drought-Induced Soybean cDNA Encoding a D95 Family Late-Embryogenesis-Abundant Protein. Plant Physiology. 106, 805–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zegzouti H, Jones B, Marty C, Lelievre J.M, Latche A, Pech JC, et al. (1997) ER5, a tomato cDNA encoding an ethylene-responsive LEA-like protein: characterization and expression in response to drought, ABA and wounding. Plant Mol Biol. 35, 847–854. [DOI] [PubMed] [Google Scholar]
- 43.Kimura M, Yamamoto YY, Seki M, Sakurai T, Abe T, Yoshida S, et al. (2003) Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochem Photobiol. 77, 226–233. [DOI] [PubMed] [Google Scholar]
- 44.Kim HS, Lee JH, Kim JJ, Kim CH, Jun SS, Hong YN (2005) Molecular and functional characterization of CaLEA6, the gene for a hydrophobic LEA protein from Capsicum annuum. Gene. 344, 115–123. 10.1016/j.gene.2004.09.012 [DOI] [PubMed] [Google Scholar]
- 45.Park SC, Kim YH, Jeong JC, Kim CY, Lee HS, Bang JW, et al. (2011) Sweetpotato late embryogenesis abundant 14 (IbLEA14) gene influences lignifications and increases osmotic- and salt stress-tolerance of transgenic calli. Planta. 233, 621–634. 10.1007/s00425-010-1326-3 [DOI] [PubMed] [Google Scholar]
- 46.Wang M, Li P, Li C, Pan Y, Jiang X, Zhu D, et al. (2014) SiLEA14, a novel atypical LEA protein, confers abiotic stress resistance in foxtail millet. BMC Plant Biology. 14, 290 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The raw data is deposited into The Sequence Read Archive (SRA) of National Center of Biotechnology Information (NCBI) with accession No. of SRR4450542 - SRR4450551.