Abstract
Purpose
A growing body of evidence suggests that the vasoactive peptides endothelins (ETs) and their receptors (primarily the ETB receptor) are contributors to neurodegeneration in glaucoma. However, actions of ETs in retinal ganglion cells (RGCs) are not fully understood. The purpose of this study was to determine the effects of ETs on gene expression in primary RGCs.
Methods
Primary RGCs isolated from rat pups were treated with 100 nM of ET-1, ET-2, or ET-3 for 24 hours. Total RNA was extracted followed by cDNA synthesis. Changes in gene expression in RGCs were detected using Affymetrix Rat Genome 230 2.0 microarray and categorized by DAVID analysis. Real-time PCR was used to validate gene expression, and immunocytochemistry and immunoblotting to confirm the protein expression of regulated genes.
Results
There was more than 2-fold upregulation of 328, 378, or 372 genes, and downregulation of 48, 33, or 28 genes with ET-1, ET-2, or ET-3 treatment, respectively, compared to untreated controls. The Bcl-2 family, S100 family, matrix metalloproteinases, c-Jun, and ET receptors were the major genes or proteins that were regulated by endothelin treatment. Immunocytochemical staining revealed a significant increase in ETA receptor, ETB receptor, growth associated protein 43 (GAP-43), phosphorylated c-Jun, c-Jun, and Bax with ET-1 treatment. Protein levels of GAP-43 and c-Jun were confirmed by immunoblotting.
Conclusions
Expression of key proteins having regulatory roles in apoptosis, calcium homeostasis, cell signaling, and matrix remodeling were altered by treatment with endothelins. The elucidation of molecular mechanisms underlying endothelins' actions in RGCs will help understand endothelin-mediated neurodegenerative changes during ocular hypertension.
Keywords: endothelins, glaucoma, gene expression, retinal ganglion cells
Endothelin peptides are involved in glaucomatous neurodegeneration. In the current study, endothelin-mediated changes in gene expression in rat retinal ganglion cells were investigated by gene microarray and validated by real-time PCR, immunocytochemistry, and immunoblot analysis.
Glaucoma, the second leading cause of blindness, is an optic neuropathy characterized by degeneration of the optic nerve, cupping of the optic disc, and apoptosis of retinal ganglion cells (RGCs). The detailed mechanisms underlying the pathogenesis of glaucoma still are not clear. Among several risk factors associated with glaucoma, such as age, race, sex, and systemic hypertension, increased IOP is the most significantly correlated with glaucoma, especially primary open angle glaucoma. Endothelin-1 (ET-1) is a 21-amino acid peptide belonging to the endothelin family that includes the vasoactive peptides, ET-1, ET-2, and ET-3. Endothelin-1 concentration at basal levels is approximately 1.5 to 2 pg/mL in normal human plasma and increases several fold in many pathological conditions,1 including myocardial infarction, angina pectoris, acute ischemic cerebral stroke, and systemic sclerosis. Studies in the past two decades have shown that ET-1 levels also are elevated in the aqueous humor and circulation of glaucoma patients as well as in animal models of glaucoma.2–6 There is increasing evidence to show the involvement of ET-1 in several cellular processes, including development, cell differentiation, cell proliferation, tumor metastasis, angiogenesis, electrolyte balance, extracellular matrix formation, mitogenesis, acute or neuropathic pain, apoptosis, and cell survival.7–20 The diverse actions of ET-1 also are illustrated by its expression in various cell types within tissues, and activation of a variety of signaling pathways upon binding to ET receptors. The main source of ET-1 in circulation is the endothelial cells; however, it also is produced by many other types of cells, including epithelial cells, macrophages, fibroblasts, cardiomyocytes, brain neurons, and ocular cells, including retinal pigment epithelial cells, ciliary epithelial cells, trabecular meshwork, and optic nerve head astrocytes.14,16,17,21
Our laboratory has shown that ET-1 levels are elevated in aqueous humor in the Morrison's rat model of elevated IOP, and associated with increased expression of glial fibrillary acidic protein (GFAP) in astrocytes and ETB receptor immunoreactivity in optic nerve.5 Using RT-PCR, Ahmed et al.22 demonstrated a significant increase in mRNA levels of ET receptors, metallopeptidase inhibitor–1 (timp1), matrix metalloproteinase–3 (mmp3), ET-2, and fibronectin following IOP elevation in Brown Norway rats. Furthermore, intravitreal injection or perfusion of ET-1 into eyes in many animal models, such as primates, rabbits, and rats, caused optic nerve head damage similar to that seen in glaucoma, including increased disk cupping, axon loss, astrogliosis,23–26 as well as disruption of axonal transport through the optic nerve axons.27,28 Taken together, these data suggest that ET-1 is an important factor that contributes to the pathogenesis of glaucoma and progression of glaucomatous damage.16,17 Endothelin-1 acts through two main classes of receptors namely, endothelin A (ETA) and endothelin B (ETB) receptors. Expression of the ETB receptor is increased in retinas of eyes in a rat model with elevated IOP.29 Moreover, ET-1 treatment has been shown to produce apoptotic changes in primary RGCs in culture.29 However, the mechanisms underlying these effects are not completely understood. The aim of this study was to investigate the effects of ETs on gene expression in isolated, purified rat RGCs.
Methods
Primary RGC Isolation
Female timed-pregnant Sprague-Dawley rats were purchased from Charles River Laboratories (Wilmington, MA, USA). Primary rat RGCs were isolated from postnatal days 4 to 6 rat pups using a Thy-1.1 antibody-panning method described by Barres et al.30 All procedures were done in accordance with the ARVO Statement on the Use of Animals in Ophthalmic and Vision Research under the guidelines of the Institutional Animal Care and Use Committee (IACUC) at University of North Texas Health Science Center at Fort Worth, TX, USA. Retinal ganglion cell cultures of purity greater than 95% were routinely obtained by using this method.30,31 In brief, rat pups (postnatal days 4–6) obtained from the pregnant rats (Charles River Laboratories) were killed, and the isolated retinas were treated with 4.5 units/mL of papain solution (Worthington, Lakewood, NJ, USA) to dissociate the cells. Cell suspensions were incubated for 10 minutes with a rabbit anti-macrophage antibody (Cedarlane, Burlington, Ontario, Canada) and transferred to a 150-mm petri dish coated with a goat anti-rabbit IgG (H+L chain) antibody (Jackson ImmunoResearch, West Grove, PA, USA) for 30 minutes. Cells that did not get attached to the coated anti-rabbit IgG were transferred to a 100-mm dish coated with anti-Thy1.1 antibody (generated from hybridoma T11D7; American Type Culture Collection, Rockville, MD, USA) for 60 minutes with intermittent shaking. Cells then were dissociated by trypsin treatment (1250 units/mL; Sigma-Aldrich Corp., St. Louis, MO, USA) from the Thy-1.1 antibody-coated dish and seeded on glass coverslips coated with mouse-laminin (Trevigen, Inc., Gaithersburg, MD, USA) in serum-free Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Grand Island, NY, USA) containing brain-derived neurotrophic factor (50 ng/mL; Peprotech, Rocky Hill, NJ, USA), ciliary neurotrophic factor (10 ng/mL; Peprotech), and forskolin (5 ng/mL; Sigma-Aldrich Corp.). Cells were incubated at 37°C in a humidified atmosphere of 10% CO2 and 90% air. One-half volume of the culture medium was changed every two days.
Affymetrix Rat Genome 230 2.0 Microarray
Primary RGCs were isolated from postnatal days 4 to 6 rats by panning with Thy-1.1 antibody as described above (section on primary RGC isolation) and approximately 300,000 RGCs were seeded in a 60-mm dish. After 7 days in culture, isolated RGCs were treated with 100 nM ET-1, ET-2, or ET-3 for 24 hours, while untreated RGCs served as the control group. Total RNA was extracted using Qiagen RNeasy Mini kit (Catalog #74104; Qiagen, Valencia, CA, USA). The quality and quantity of total RNA was determined by a bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). The extracted total RNA was used to synthesize biotinylated cDNA and the cDNA product was tested for its integrity, quantity, and quality using a bioanalyzer. Gene microarray analyses were done at UT Southwestern Medical Center Microarray Core Facility. Affymetrix Rat Genome 230 2.0 Microarray (Affymetrix, Santa Clara, CA, USA) was used to analyze the changes in gene expression in RGCs in response to 100 nM ET-1, ET-2, and ET-3 treatments. A single chip contained more than 31,000 probe sets, which can analyze over 30,000 transcripts and variants from over 28,000 well-substantiated rat genes. There were 11 pairs of probes to detect each gene sequence. Each pair of probe set contained a perfect-matched probe and a mismatched probe. Based on the hybridization signal from each probes, detection P value were calculated by a 1-sided Wilcoxon's signed rank test to reflect the statistical significance of the differences between perfect-matched and mismatched using Affymetrix Gene Chip Operating Software (GCOS). If the detection P value is less than 0.05, the detected gene is considered as Present (“P”); the detection P value is close to 0.05, the detected gene is considered as Marginally Present (“M”); and the detection P value is greater than 0.05, the detected gene is considered as Absent (“A”). Only genes labeled with P and M were selected for analysis. Three sets of experiments (biological replicates) were performed, including RNA extractions following endothelin treatments and microarray analysis, and each microarray contained two separate arrays (technical replicates). The relative fold change was calculated and normalized by setting the untreated (control) value as 1. The genes flagged as “P” and “M” with at least 2-fold change compared to control were selected for further DAVID analysis (available in the public domain at http://david.abcc.ncifcrf.gov/, an online bioinformatic tool to systematically and integratively analyze the large gene lists), which was used to categorize the regulated genes into different functional clusters.32,33 The threshold of the Expression Analysis Systematic Explorer (EASE) scores, which represent the enrichment of a group of genes, was set at 1.3. The microarray analysis was used as a screening method and further confirmation of changes in gene expression of candidate genes was done using quantitative PCR and immunocytochemical analysis.
Real-Time PCR
Total RNA was extracted from RGCs using Qiagen RNeasy Mini kit (Qiagen). The quality and yield of the RNA were monitored using a bioanalyzer as described above. Total cDNA was prepared from equal amount of total RNA from different samples using iScript Reverse Transcription Kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA). To get a quantitative estimate of changes in gene expression, real-time PCR (SsoAdvanced SYBR Green Supermix) was performed using cDNA as template to detect gene expression (primers are listed in Supplementary Table S1). Real-time PCR running conditions were as follows: 95°C 30 seconds followed 45 two-step cycles of 95°C 10 seconds and 58°C 20 seconds. PCR primers were validated by using cDNA produced from the whole retina and detection of melting temperature. Three individual sets of samples were used in real-time PCR, the assay was repeated 2 to 4 times for some genes. Cyclophilin A served as an internal control to normalize for equal loading of cDNA template. The results were presented as relative fold change compared to untreated control. The statistical significance was calculated by Student's t-test, and P values <0.05 were considered significant.
Immunofluorescent Staining
RGCs were cultured for 7 days following isolation from rat retinas, to allow them to attach and extend neurites. RGCs were then treated with 100 nM ET-1 or ET-3 for 24 hours and subsequently fixed with 4% paraformaldehyde in PBS for 10 minutes. Permeabilization was performed using 0.1% Triton X-100, the coverslips were washed with PBS and nonspecific binding was blocked by incubation with 5% bovine serum albumin and 5% normal donkey serum in PBS for 1 hour. The coverslips were incubated overnight at 4°C with combinations of primary antibodies against specific proteins (Bax, #sc-526, Santa Cruz Biotechnology, Inc., Dallas, TX, USA; Endothelin B receptor [ETBR], #Ab117529, Abcam, Cambridge, United Kingdom; Endothelin A receptor [ETAR], #E3651, Sigma-Aldrich Corp.; c-Jun, #sc-1694, Santa Cruz Biotechnology, Inc.; p-c-Jun, #sc-822, Santa Cruz Biotechnology, Inc.; GAP-43, #G9264, Sigma-Aldrich; RNA-Binding Protein With Multiple Splicing [RBPMS], #GTX118619, GeneTex, Inc., Irvine, CA, USA; β-tubulin III, #T8578, Sigma-Aldrich Corp.; Neurofilament-L, #2837s, Cell Signaling, Danvers, MA, USA; Brn-3b, custom polyclonal antibody from rabbit, Antibody Research Corporation, St. Charles, MO, USA). Primary antibody incubations were followed by PBS washes and the corresponding secondary antibody (donkey anti-mouse Alexa 546/488 or donkey anti-rabbit 647-conjugate) incubations. Control sections incubated with only secondary antibodies served as background control. The coverslips were mounted on slides with antifade medium (FluorSave; Calbiochem, La Jolla, CA, USA). Serial images captured using Zeiss 510meta confocal microscope with Z-scan were stacked except the images for analysis of protein co-localization.
Immunoblotting
Approximately, 300,000 RGCs were seeded in each well of 6-well plates and cultured for 7 days. The cultured RGCs were treated with 100 nM ET-1 for 24 hours, and untreated RGCs served as control. After washing twice with PBS, RGCs were lysed using 100 μL of RIPA buffer (50 mM Tris-HCl, pH 8.0, with 150 mM sodium chloride, 1.0% Igepal CA-630 (equivalent to NP-40), 0.5% sodium deoxycholate, and 0.1% SDS). Equal amount of cell lysates were subjected to 4% to 15% SDS-PAGE gels (Bio-Rad Laboratories, Inc.). Antibodies c-Jun, p-c-Jun, and GAP-43 (listed above) were used to probe the specific proteins and calnexin (Cat #: ADI-SPA-860, Enzo Life Sciences, Inc., Farmingdale, NY, USA) served as loading control. The densitometry analyses of immunoblotting signals were performed using Bio-Rad Image Lab software, and the results from 3 to 5 individual experiments were presented as the bar graph after normalization with calnexin.
Results
Upregulated and Downregulated Genes in Response to Treatments of RGCs With ET-1, ET-2, and ET-3
Purified RGCs (cultured for 7 days following isolation) were either untreated (control) or treated with 100 nM ET-1, ET-2, or ET-3 for 24 hours. Total RNA was extracted from the cells and cDNA was synthesized using equal amount of total RNA (quantitated using a bioanalyzer). The yield and quality of total RNA was summarized in Supplementary Table S2.
Affymetrix Rat Genome 230 2.0 Microarray was performed, and the microarray data were validated by the obtained flag value using Affymetrix GCOS. The results obtained from three biological replicates and two technical replicates from microarray assay were averaged and resultant mean was used to evaluate the gene responses. The raw data of microarrays have been submitted to GEO database of the National Center for Biotechnology Information (NCBI) and the access number is GSE71021. Genes with a Detection P value ≤0.05 by a 1-sided Wilcoxon's signed rank test and with at least 2-fold increase or decrease compared to control were selected for DAVID analysis. There was more than 2-fold upregulation of 328, 378, or 372 genes and downregulation of 48, 33, or 28 genes with ET-1, ET-2, or ET-3 treatment, respectively, compared to untreated controls. In the microarray analysis, mRNA levels of several house-keeping genes, including GAPDH, cyclophilin A, and β-actin, displayed very stable levels and ET treatment did not alter the expression of these genes, which provided important normalization standards for validating the results (data not shown).
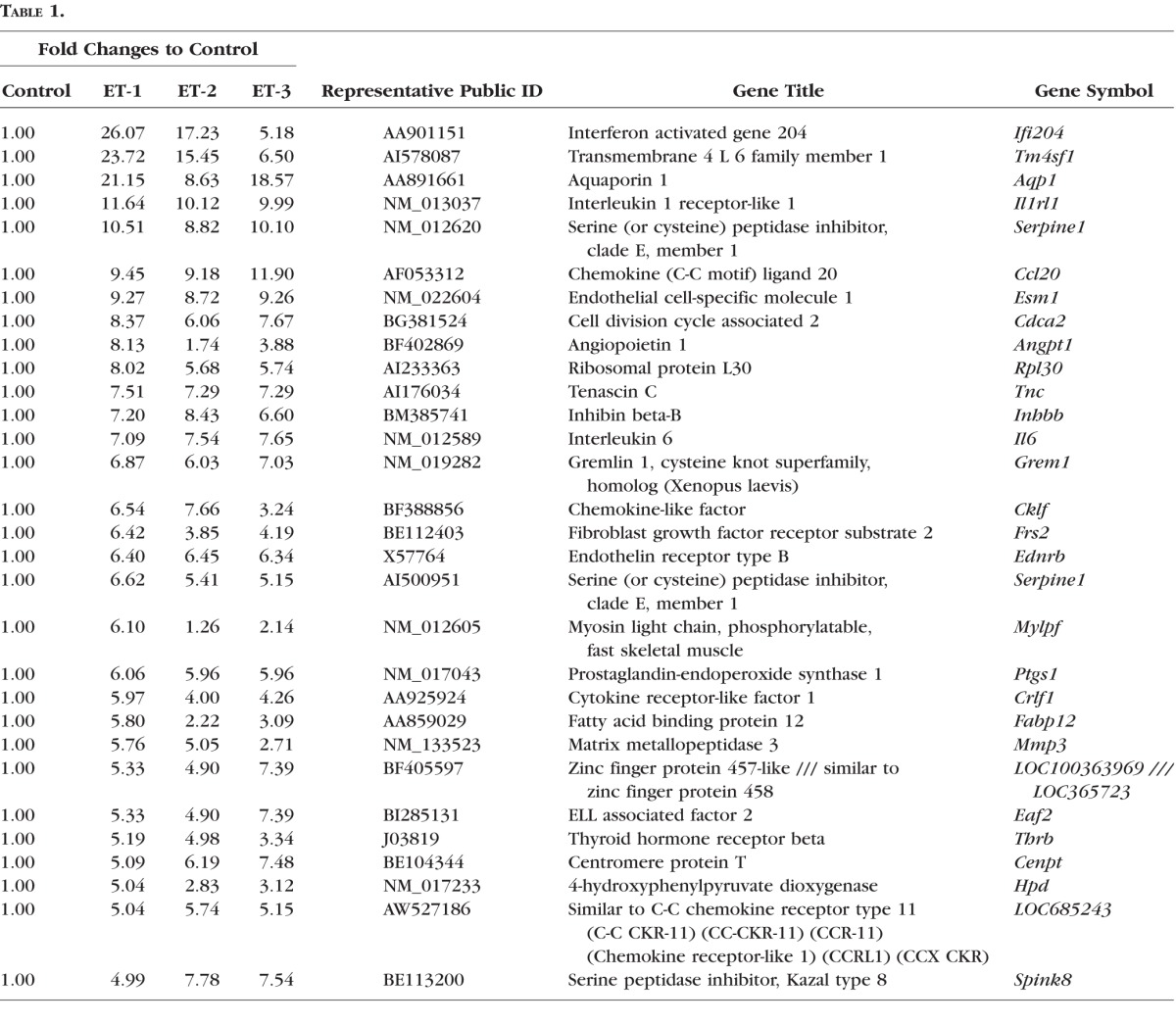
There were 30 genes that were highly upregulated in RGCs in response to ET-1 treatment for 24 hours, which are shown in Table 1. Among these genes, mRNA levels of tenascin C (tnc), IL-6, ETBR, and mmp3 were selected and confirmed using real-time PCR. A similar fold increase in gene expression was observed in these 30 genes in response to treatments of ET-1, ET-2, or ET-3.
Table 1.
A total of 30 Genes With Most Upregulation by ET-1 in Gene Microarray in RGCs
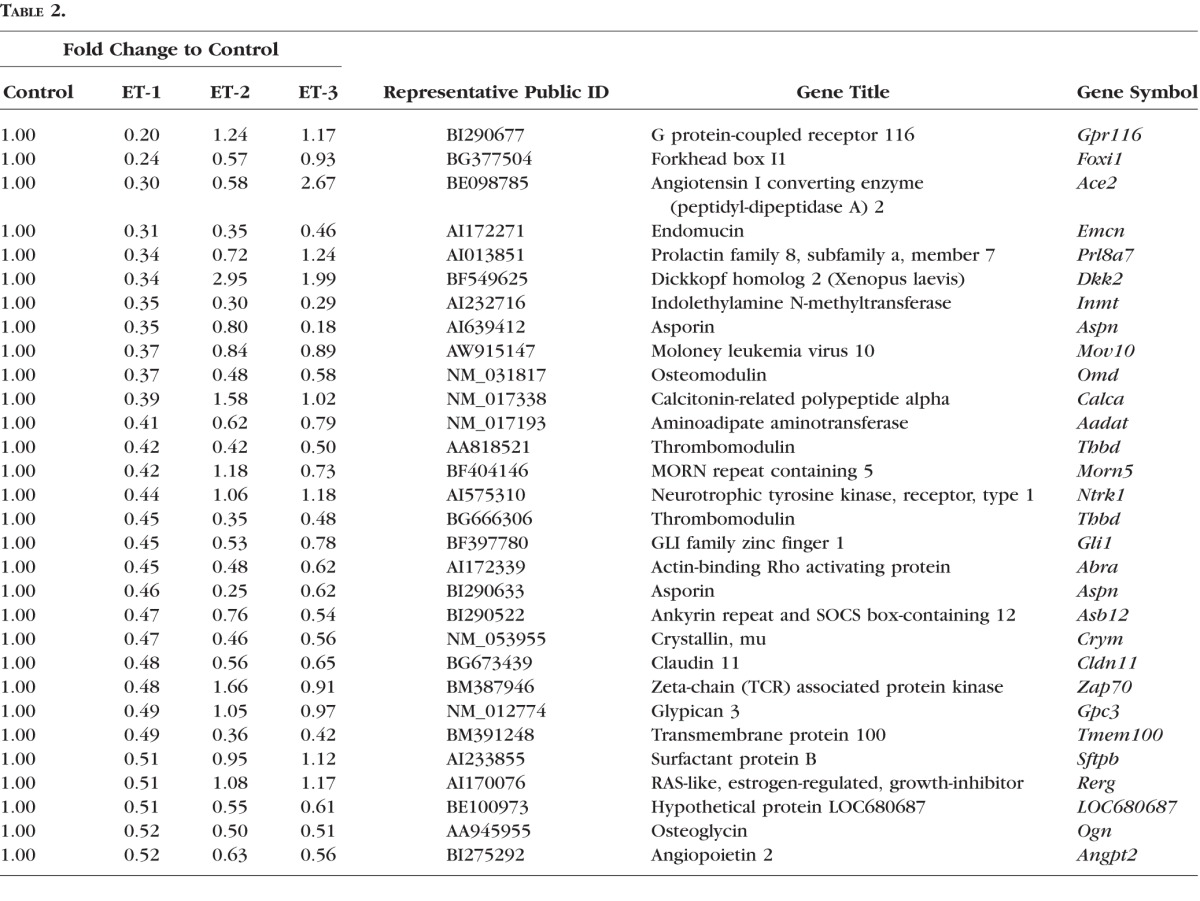
Moreover, 30 genes most downregulated in RGCs in response of ET-1 treatment for 24 hours are shown in Table 2. Asporin (aspn) was selected for further analysis and its expression was confirmed by real-time PCR. Among these 30 downregulated genes, 10 genes showed different response to ET-2 and/or ET-3 treatment compared to ET-1, others displayed the similar trend (Table 2).
Table 2.
A Total of 30 Genes With Most Downregulation by ET-1 in Gene Microarray in RGCs
Gene Clusters Identified by DAVID Analysis
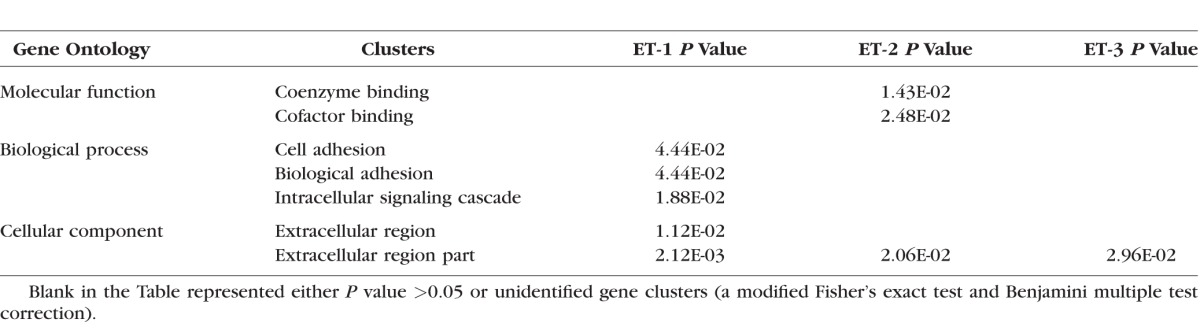
To summarize the biological information from the microarray data, the upregulated and downregulated genes with a detection P value ≤0.05 and a relative fold ≥2 were screened by instruction of DAVID analysis. The three gene lists were extracted based on the ranking of fold change in response to ET-1, ET-2, and ET-3 treatments, respectively, from the upregulated, screened genes. Similarly, three gene lists were generated from the genes downregulated by treatment with the ET-1, ET-2, and ET-3. All six gene lists were analyzed using DAVID individually, and gene clusters were selected by EASE scores ≥1.3 and P ≤ 0.05 (a modified Fisher's exact test and Benjamini multiple test correction). Gene clusters were categorized into three gene ontologies (GO) including molecular function, biological process, and cellular component; in addition, KEGG pathways also were determined. The results are summarized in Tables 3 and 4. From identification of gene clusters in the biological process, the most significantly regulated gene classes were those of cell cycle, cell growth, cell migration, and responses to stimuli, and in accordance with this, synthesis of microtubule, cytoskeleton parts, and extracellular matrix was most affected by ET treatment (Tables 3, 4).
Table 3.
Gene Clusters of Upregulated Genes in Response to ETs Identified by DAVID Analysis
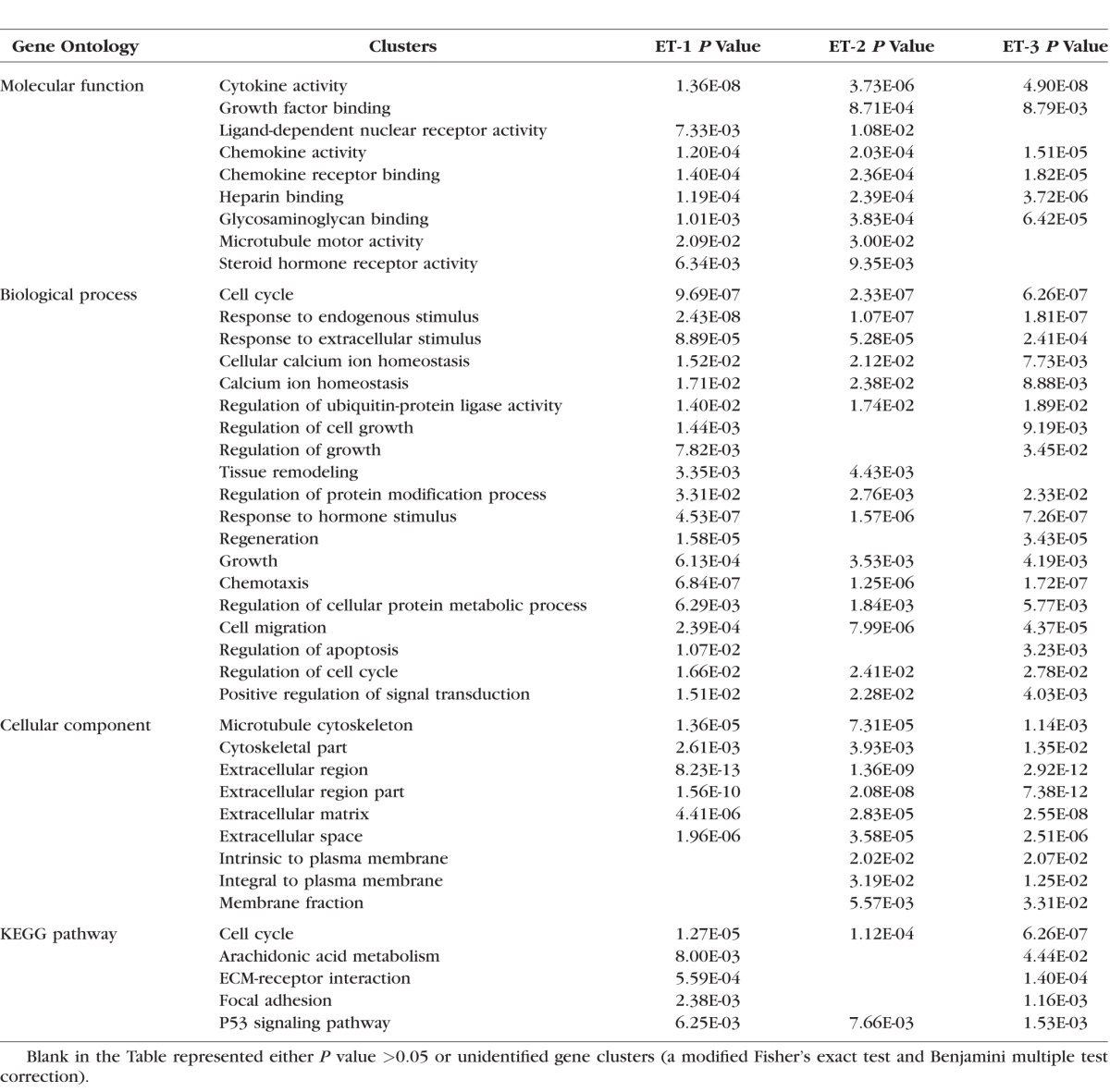
Table 4.
Gene Clusters of Downregulated Genes in Response to ETs Identified by DAVID Analysis
Calcium Pathway-Associated Genes, Cyclin-Dependent Kinase 1, and Interleukins
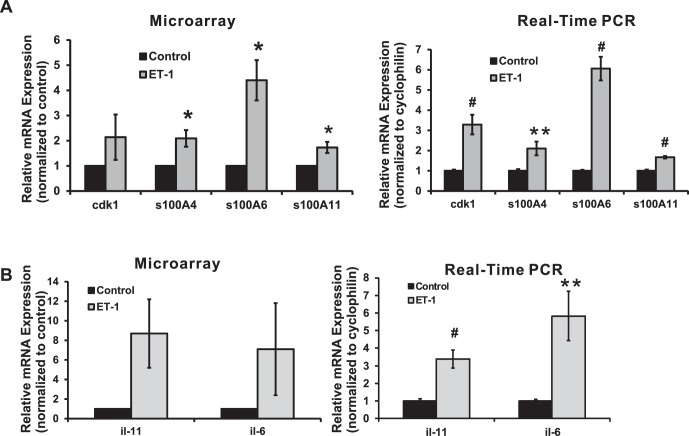
Cyclin-dependent kinase 1 (cdk1) has an important role in controlling cell cycles by regulation of G1/S and G2/M phase transitions in eukaryotic cells.34 Microarray analysis revealed that there was a 2.1-fold increase in expression of cdk1 detected in ET-1–treated RGCs, and several gene clusters associated with cell cycle in biological process also were screened by DAVID criteria (shown in Table 3) from gene ontology analysis of microarray. As a further confirmation, real-time PCR data showed a 3.3-fold increase, which was statistically significant (P < 0.001, t-test, n = 9; Fig. 1A).
Figure 1.
Calcium-associated genes, cdk1, and interleukins in response to ET-1 treatment in RGCs. Expression levels of genes were detected by microarray and real-time PCR using cDNA template synthesized from total RNA, which was extracted from RGCs following ET-1 treatment. (A) Expression of cdk1 and S100 family genes was detected by microarray and real-time PCR. An increased expression of 2.1-, 4.4-, and 1.7-fold in ET-1 treatment was detected in microarray for s100A4, s100A6, and s100A11, the same trend also was observed as an increase of 2.1-fold (n = 6, t-test, P = 0.01), 6.1-fold (n = 12, t-test, P ≤ 0.001), and 1.7-fold (n = 6, t-test, P ≤ 0.001) from real-time PCR, respectively (A). Statistical analysis was performed using t-test. (*P < 0.05, **P ≤ 0.01, #P ≤ 0.001). (B) Interleukin-6 and il-11 were detected by microarray and real-time PCR. Statistical analysis was performed using t-test. (*P < 0.05, **P ≤ 0.01, #P ≤ 0.001). Endothelin-1 treatment of RGC induced a 7.1- and 8.7-fold increase of il-6 and il-11 in microarray, although there was a large variation detected from three sets of data (B); real-time PCR confirmed this significant upregulation of mRNA level for il-6 and il-11 at 5.8- and 3.4-fold with t-test P < 0.01 (both with n = 12; relative mRNA levels (mean ± SEM) are represented in bar graph; in microarray, the vehicle control served as control; in real-time PCR, results were normalized to the expression of cyclophilin A, and values were compared to the vehicle-treated control cells).
The S100 proteins belonging to the EF-hand family with 2 binding sites for calcium are involved in a variety of cell processes, including cell proliferation, apoptosis, and cell metabolism.35,36 Several members of this family were shown to be upregulated in RGCs with ET-1 treatment. There was a 2.1-, 4.4-, and 1.7-fold increase detected in microarray for the expression of s100A4, s100A6, and s100A11. A similar trend of 2.1-, 6.1-, and 1.7-fold upregulation of these genes, respectively, also was found by real-time PCR analysis (Fig. 1A). The upregulation of all three genes in both assays were statistically significant. In ontology analysis, there were two gene clusters, including calcium ion homeostasis and cellular calcium ion homeostasis, that were upregulated and enriched by ET treatment, identified by DAVID (Table 3).
Interleukins
Treatment of RGCs with ET-1 induced a 7.1- and 8.7-fold increase in expression of interleukin-6 (il-6) and il-11 determined by microarray analysis; although a large variation was detected from three sets of data (Fig. 1B). The results from real-time PCR confirmed this significant upregulation of mRNA levels for il-6 and il-11 (5.8- and 3.4-fold respectively) with statistical significance (P < 0.01, n = 12) using the t-test.
Extracellular Matrix Proteins
No change was detected in the mRNA level of mmp2 in RGCs with ET-1 treatment from microarray and real-time PCR data; however, a significant upregulation of mmp3 expression was detected in both assays (8.2- and 5.4-fold, respectively) with ET-1 treatment (Fig. 2). An appreciable 2.2-fold increase (P = 0.065, t-test, n = 3) in timp1, an endogenous inhibitor of MMPs, was found in microarray and a 2.4-fold increase (P < 0.001, t-test, n = 12) also was found by real-time PCR in RGCs with ET-1 treatment. Tenascin C (tnc), the extracellular matrix protein regulated by a variety of factors, including TGFβ and TNFα, has been shown to have an important role in neuronal development. In the current study, mRNA level of tnc in RGCs treated with ET-1 was significantly elevated by 7.5- and 17.0-fold in microarray and real-time PCR, respectively. Apart from the MMP family, mRNA levels of other ECM molecules also were altered in response to ET treatment. Asporin (aspn), an extracellular protein belonging to the members of the small leucine-rich proteoglycan, displays a regulatory role by inhibiting TGFβ1-induced signaling.37 Our results showed a 2.4- and 5.3-fold decrease of the mRNA level for this gene detected from microarray and real-time PCR following a 24-hour treatment of RGCs with ET-1 (Fig. 2).
Figure 2.
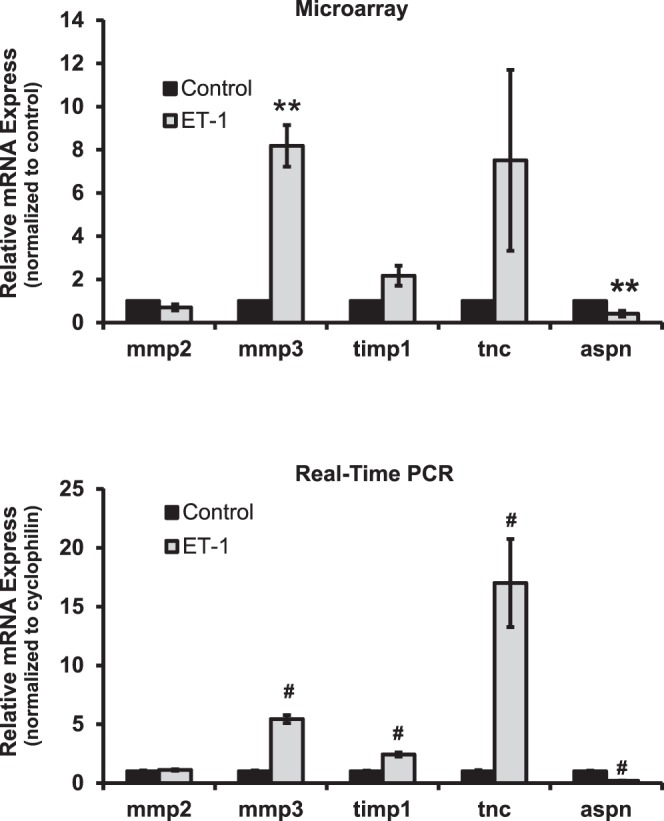
Changes in expression of genes encoding extracellular matrix proteins in response to ET-1 treatment in RGCs. Expression of mmp2, mmp3, timp1, tnc, and aspn was detected by microarray and real-time PCR. There was no change detected in mRNA level of mmp2 in RGCs with ET-1 treatment from microarray and real-time PCR; however, a significant upregulation of mmp3 expression was detected in both assays (8.2-fold, n = 3 and 5.4-fold, n = 12, respectively) with ET-1 treatment. An appreciable 2.2-fold increase (P = 0.065, t-test, n = 3) of timp1 (an endogenous inhibitor of MMPs) was found by microarray, and a 2.4-fold increase (P < 0.001, t-test, n = 12) of timp1 was found in real-time PCR in RGCs treated with ET-1. The mRNA level of tnc in RGCs in response to ET-1 treatment was significantly upregulated to 7.5- and 17.0-fold in microarray and real-time PCR, respectively. There was a 2.4- and 5.3-fold decrease of mRNA of asporin (aspn) detected from microarray and real-time PCR after 24-hour treatment of ET-1 in RGCs. (mRNA levels [mean ± SEM] are represented in bar graph; in microarray, the vehicle-treated cells served as control; in real-time PCR, results were normalized to the expression of cyclophilin A, and values were compared to vehicle-treated control cells). Statistical analysis was performed using t-test (*P < 0.05, **P ≤ 0.01, #P ≤ 0.001).
ER-Stress Associated Genes
To investigate if ET-induced apoptosis acts through the ER stress pathways, the mRNA expression of several key elements involved in ER stress was determined by microarray and real-time PCR analysis (Fig. 3). No appreciable changes were detected in the mRNA level of DNA-damage-inducible transcript 3 (ddit3), eukaryotic translation initiation factor 2-α kinase 3 (eif2ak3 or perk), endoplasmic reticulum to nucleus signaling 1 (ern1), and X-Box binding protein 1 (xbp1) in ET-1–treated groups from microarray compared to control (no data for ern1 were obtained from microarray). Also, mRNA of ern1 and xbp1 did not change in real-time PCR. However, ddit3 and eif2ak3 expression was decreased to 79% and 70% of control in real-time PCR. The only statistically significant decrease in mRNA expression were detected for eif2ak3 in real-time PCR assay (P < 0.005, n = 6, t-test).
Figure 3.
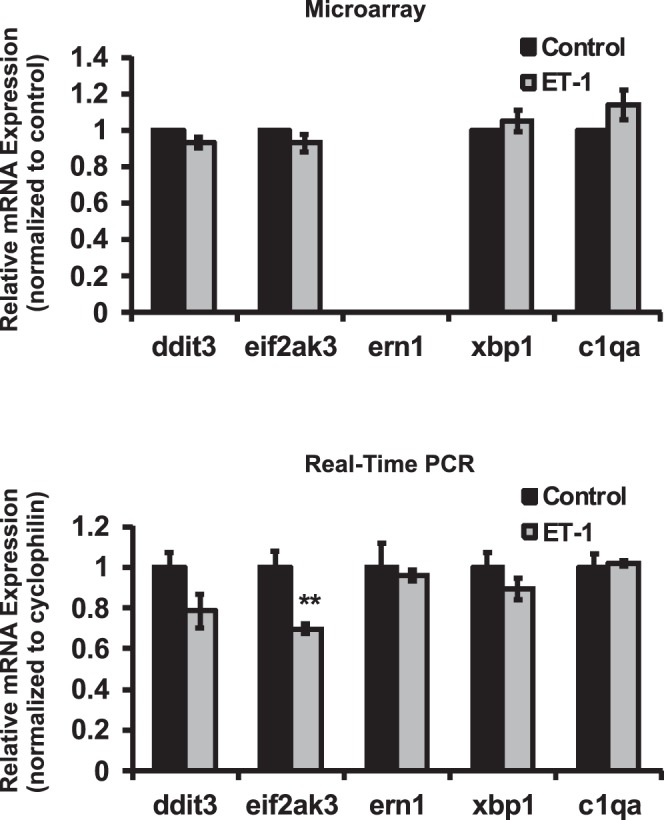
Alterations in expression of ER-stress associated genes in RGCs treated with ET-1. Genes involved in ER stress pathways were determined by microarray and real-time PCR analysis. No appreciable changes of ddit3, eif2ak3, ern1, and xbp1 were detected at the mRNA level in ET-1–treated groups from microarray (no data of ern1 obtained from microarray). mRNA of ern1 and xbp1 also did not change in real-time PCR. However, ddit3 and eif2ak3 expression was decreased to 79% and 70% of control in real-time PCR. Only change of eif2ak3 in real-time PCR was statistically significant (0.005, n = 6, t-test). *P < 0.05, **P ≤ 0.01, #P ≤ 0.001; mRNA levels (mean ± SEM) are represented in bar graph; in microarray, the vehicle control served as control; in real-time PCR, results were normalized to the expression of cyclophilin A, and compared to vehicle-treated control cells.
Endothelin Receptors
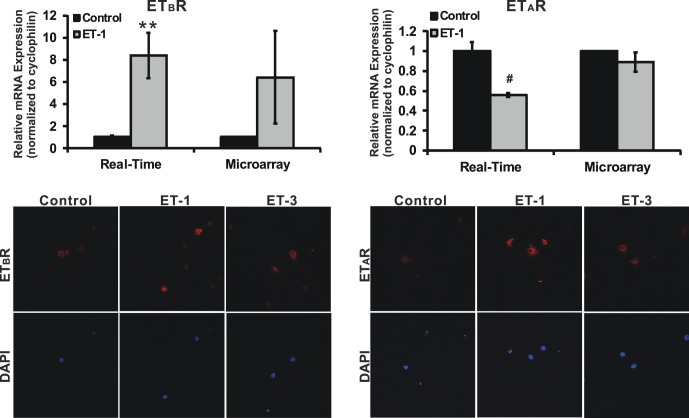
Previously, our laboratory reported an upregulation of ETB receptor mRNA and protein levels in retinas of rats with elevated IOP. Furthermore, RGC loss and axon degeneration were attenuated in ETB-deficient rats.29,38 Taken together, the data suggested that ETB receptor is involved in neurodegeneration in glaucoma. Therefore, mRNA levels of ETB receptor were investigated by microarray and real-time PCR (Fig. 4). There was a 6.4- and 8.4-fold increase in the mRNA levels of ETB receptor in RGCs treated with ET-1 detected by microarray and real-time PCR, respectively. In addition, an enhanced immunostaining of ETB receptor also was found in RGCs treated with ET-1 or ET-3 (ET-1 binds to ETA and ETB receptor with the same affinity, whereas ET-3 has a higher affinity of binding to the ETB receptor). On the other hand, there was no change or a decrease to 56% of control in the mRNA levels of ETA receptor in ET-1–treated RGCs determined by microarray or real-time PCR, respectively; however, protein levels of ETA receptor determined by immunostaining were increased in RGCs with ET-1 as well as with ET-3 treatment, compared to untreated controls. The staining of ETA and ETB receptors was detected mainly in soma of RGC cells. ETA receptor also was colocalized with β-tubulin III (a neuronal marker) in RGCs (data shown in Supplementary Fig. S1).
Figure 4.
Endothelin receptor expression in RGCs treated with ETs. mRNA and protein levels of ETB receptor and ETA receptor were investigated by microarray, real-time PCR, and immunocytochemistry. There was a 6.4-fold (n = 3) and 8.4-fold (n = 12) increase in mRNA of ETB receptor detected by microarray and real-time PCR respectively; however, there was either no change or a decrease to 56% of control (n = 9) in mRNA levels of ETA receptor in both assays. On the other hand, an enhanced staining of ETB receptor and ETA receptor was detected in RGCs treated with ET-1 or ET-3 compared to untreated controls. The staining of ETA and ETB receptors was detected mainly in soma of RGC cells. t-test, *P < 0.05, **P ≤ 0.01, #P ≤ 0.001; mRNA levels (mean ± SEM) are represented in bar graph; in microarray, the vehicle control served as normalization control; in real-time PCR, results were normalized to the expression of cyclophilin A, and values were compared to vehicle-treated control cells.
Transcription Factors
Brn-3b (POU4F2), which belongs to POU domain transcription factor family (that includes Brn-3a, Brn-3b, and Brn-3c), has a regulatory role in the development of RGCs in the retina.39,40 To test the effects of ETs on expression, the mRNA expression of brn-3b was tested by real-time PCR, since no hybridization oligos were present in the microarray. ET-1 treatment in RGCs resulted in significant downregulation of the mRNA of brn-3b to 70% of control (P < 0.001, n = 6, t-test, Fig. 5). Activating transcription factor 3 (atf3), a member of ATF/cAMP responsive element-binding protein family, is an immediate early response gene that is induced by a varieties of stimuli and stress.41 It also is related to components of the AP-1 transcription factor that affects ETB receptor expression. In a rat model of glaucoma, atf3 expression was boosted 15-fold in RGCs in response to elevated IOP.42 However, no appreciable change was detected in ET-1–treated RGCs using microarray and real-time PCR. Signal transducer and activator of transcription 3 (stat3) is a transcription factor that acts downstream of Janus Kinase (Jak) to mediate the cellular reaction in response to interleukins and many growth factors, such as IL-6, EGF, and BMP2.43–45 Since a significant upregulation of il-6 mRNA was detected in ET-1–treated RGCs (Fig. 1B), stat3 was a potential target gene to be studied; however, there was no appreciable change of stat3 mRNA detected in microarray, and a slight decrease was found in real-time PCR with no statistical difference (P = 0.197, n = 3, t-test, Fig. 5).
Figure 5.
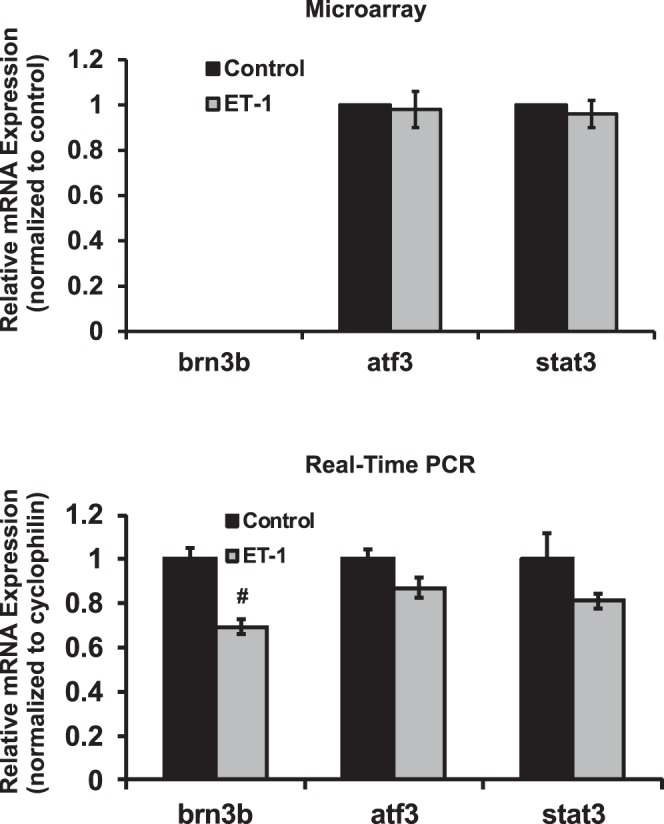
Expression of transcription factors brn3b, atf3 and stat3 in RGCs treated with ET-1. ET-1 treatment in RGCs decreased the expression of brn-3b to 70% of control in real-time PCR (P < 0.001, n = 6, t-test) and no data were obtained for brn-3b because of no hybridization oligos in microarray chip. There were no significant changes of atf3 and stat3 detected in ET-1–treated RGCs using microarray. *P < 0.05, **P ≤ 0.01, #P ≤ 0.001; mRNA levels (mean ± SEM) are represented in bar graph; in microarray, the vehicle control served as normalization control; in real-time PCR, results were normalized to the expression of cyclophilin A, and then compared to vehicle-treated control cells.
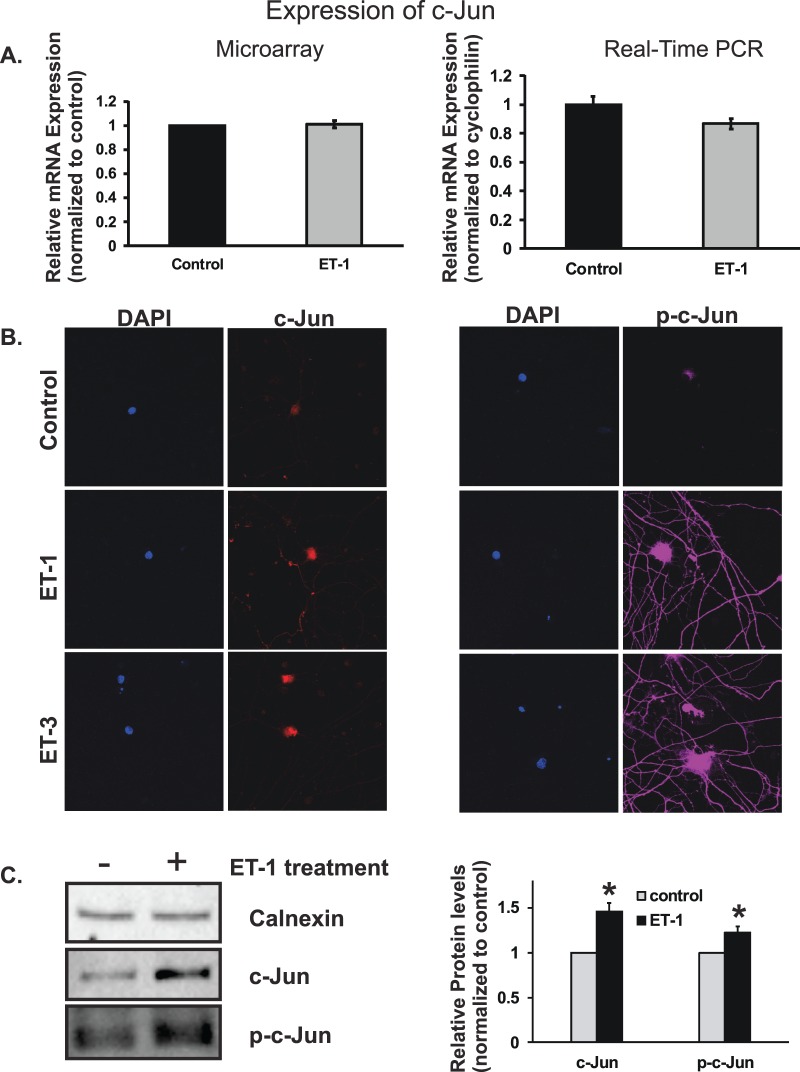
mRNA and Protein Levels of c-Jun
c-Jun has been shown to be upregulated at the mRNA and protein levels in retinas of rats with elevated IOP,38,46 and phosphorylated c-Jun also was detected as early as 1 week after IOP elevation.46 Surprisingly, c-Jun mRNA was not changed in RGCs treated with ET-1 in microarray assay, and even slightly decreased in real-time PCR (Fig. 6A); however, the results of c-Jun protein levels were different. Immunostaining was used to determine the protein level of c-Jun and its phosphorylated form. Staining of c-Jun was appreciably increased in RGCs treated with 100 nM ET-1 or ET-3 for 24 hours (Fig. 6B), the staining was detected mainly in soma of RGCs. There was no discernible difference in the intensity of c-Jun immunostaining between ET-1 and ET-3 treatment. A visibly enhanced staining of phosphorylated c-Jun (p-c-Jun) also was detected in RGCs treated ET-1 or ET-3, the staining was observed not only in the soma of RGCs, but also in the neurites. There was no appreciable difference in the staining intensity for p-c-Jun between ET-1 and ET-3 treatments. ET-1–mediated upregulation of c-Jun and p-c-Jun in protein levels also was confirmed by immunoblotting (Fig. 6C).
Figure 6.
Messenger RNA and protein levels of c-Jun in RGCs treated with ET-1. Microarray, real-time PCR, and immunocytochemistry were used to determine mRNA and protein levels of c-Jun. c-Jun mRNA was not changed in RGC cells treated with ET-1 in microarray assay, even decreased in real-time PCR without statistical significance (A). Staining of c-Jun was increased in RGC cells treated with 100 nM ET-1 or ET-3 for 24 hours, and the staining was detected mainly in soma of RGCs (B). An enhanced staining of phosphorylated c-Jun (p-c-Jun) also was detected in RGCs treated ET-1 or ET-3, the staining was observed not only in the soma of RGC, but also in the neurites. There was no appreciable difference in the staining intensity for c-Jun and p-c-Jun between ET-1 and ET-3 treatments (*P < 0.05, **P < 0.01, #P < 0.001). The increased protein levels of c-Jun and p-c-Jun in response to ET-1 treatment in RGCs also was confirmed using immunoblotting and bar graph represents the densitometric analysis of the specific bands of c-Jun (n = 5, P < 0.05, t-test) and p-c-Jun (n = 3, P < 0.05, t-test; C). mRNA levels (mean ± SEM) are represented in bar graph; in microarray, the vehicle control served as normalization control; in real-time PCR, the data were normalized to the expression of cyclophilin A, and then the vehicle-treated cells served as control.
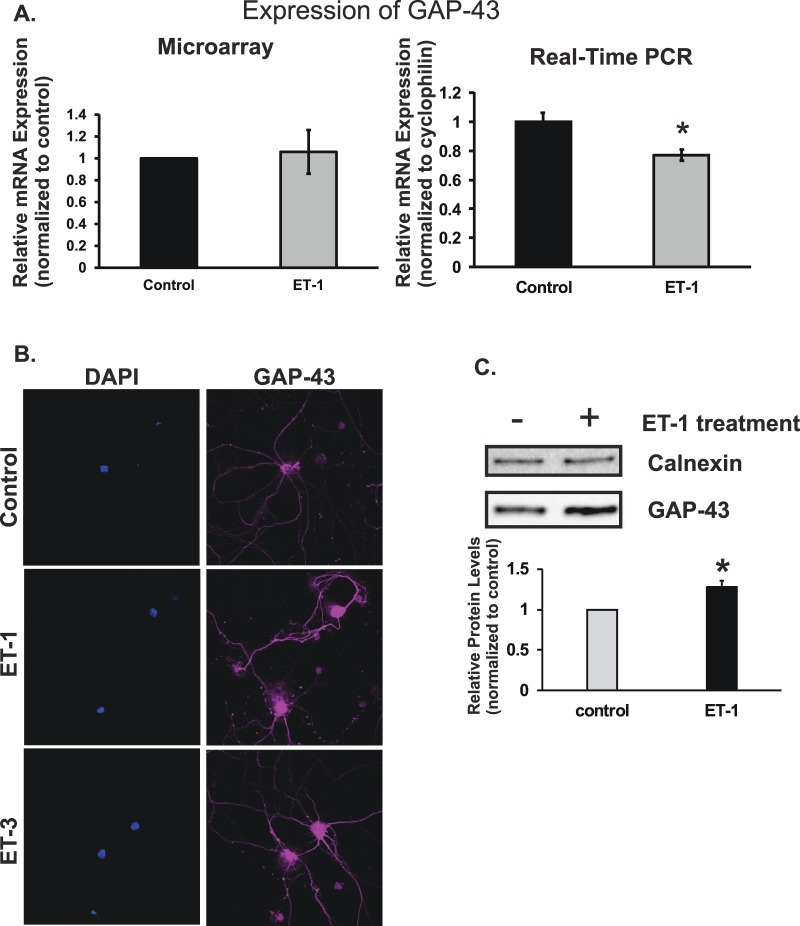
Growth Associated Protein 43 Response to ET-1 Treatment
Growth Associated Protein 43 (GAP-43) is a protein that is highly expressed in axonal growth cone during development and axon neuroregeneration. Levels of GAP-43 mRNA in RGCs treated with ET-1 were unchanged when determined by microarray and decreased to 77% of control when analyzed by real-time PCR, respectively (Fig. 7A). The decrease of mRNA expression in ET-1 treatment detected by real-time PCR was statistically different (P < 0.05, n = 6, t-test). Levels of GAP-43 protein also were tested by immunocytochemistry. The staining intensity was greatly enhanced in RGCs treated with ET-1 and ET-3, and increased staining was observed in somas and neurites (Fig. 7B). Furthermore, immunoblotting of GAP-43 in RGCs showed that ET-1 treatment promoted increased protein expression of GAP-43 as seen in the immunostaining (Fig. 7C).
Figure 7.
Levels of GAP-43 in response to ET-1 treatment; GAP-43 mRNA levels in RGCs treated with ET-1 were unchanged in microarray analysis and decreased to 77% of control by real-time PCR, respectively (A). The decrease of mRNA expression in ET-1 treatment detected by real-time PCR was statistically significant (P < 0.05, n = 6, t-test). Levels of GAP-43 protein tested by immunohistochemistry showed the higher intensity of staining in RGCs treated with ET-1 and ET-3 compared to vehicle control, and the enhanced staining was observed in somas and neurites (*P < 0.05, **P < 0.01, #P < 0.001, t-test; B). Endothelin-1–mediated upregulation of GAP-43 protein in RGCs was confirmed using immunoblotting and the densitometry analysis of the specific bands of GAP-43 was represented as bar graph (n = 4, P < 0.01, t-test, C). Messenger RNA levels (mean ± SEM) are represented in bar graph; in microarray, the vehicle control served as control; in real-time PCR, results were normalized to the expression of cyclophilin A, and compared to vehicle-treated control cells.
Bcl-2 Gene Family
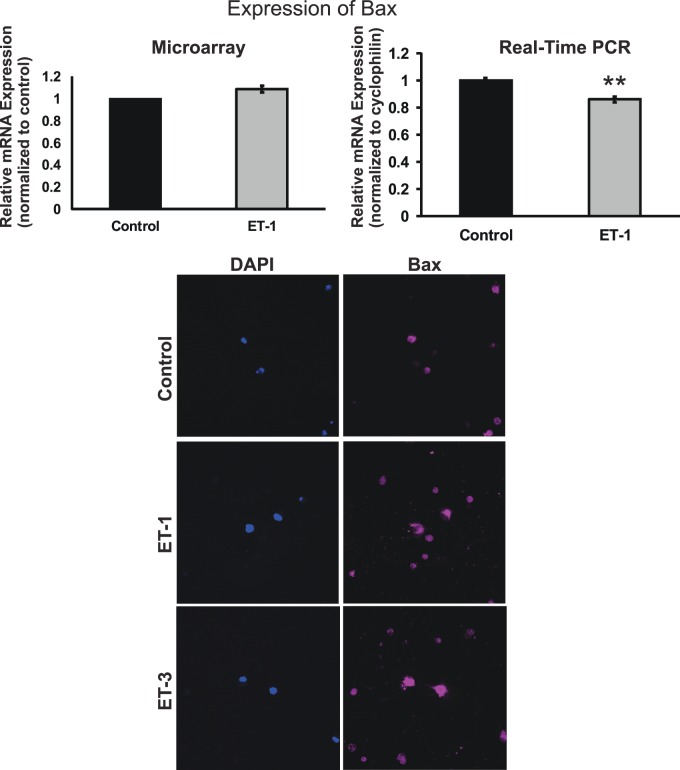
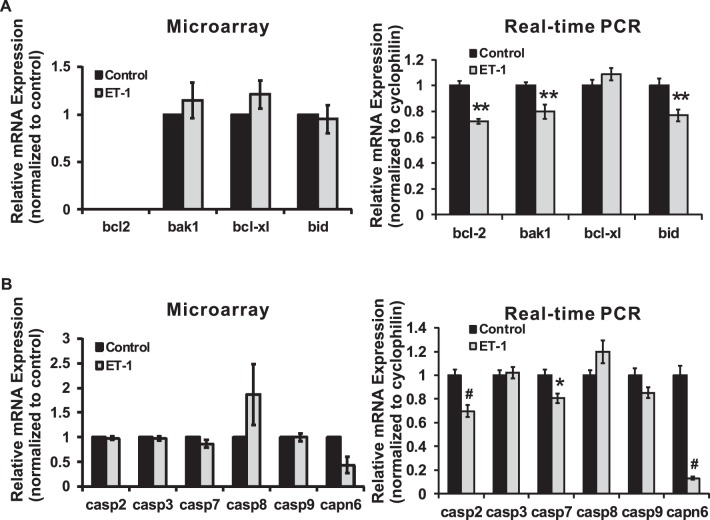
Bcl-2 gene family consists of proapoptotic genes (bax, bad, bak, bid, and so forth) and antiapoptotic genes (bcl-2, bcl-XL, and so forth). These genes have the important roles in controlling the fate of survival of cells. Among the proapoptotic genes, analysis of bax gene expression by microarray and real-time PCR yielded different results. No change of bax mRNA expression was detected in ET-1–treated RGCs from microarray; however, a significant decrease (P = 0.001, t-test, n = 6) to 86% of control in ET-1–treated RGCs was found by real-time PCR (Fig. 8). On the other hand, ET-1 or ET-3 treatment of RGCs increased the protein level of Bax detected by immunocytochemistry, and the staining intensity of Bax was higher in the endothelin treated group compared to control. In addition, Bax staining was detected mainly in somas of RGCs. mRNA levels of bcl-2, bak1, bcl-xl, and bid were investigated using both arrays (Fig. 9). Among these genes, ET-1 treatment of RGCs induced a significant decrease in bcl-2, bak1, and bid (P < 0.01, t-test, n = 3–6) shown in real-time PCR. The Bcl-2 gene family comprises of upstream regulatory factors (survival and cell death promoting) controlling the apoptotic pathway. The caspase family, consisting of a group of cysteine-dependent aspartate-directed proteases, functions as initiators and effectors controlling apoptosis, inflammation, and cell cycle. mRNA levels of caspases also were determined by microarray and real-time PCR. The results from both assays were consistent. A statistically significant downregulation of mRNA levels of casp2 and casp7 in RGCs treated with ET-1 was detected by real-time PCR. Calpain is another family of calcium-dependent cysteine proteases having diverse roles in cell cycle and cell death. Within this family, capn6 was downregulated in ET-1–treated RGCs in microarray, and the same trend also was detected by real-time PCR. In the latter experiment, the mRNA level of capn6 was decreased to 12.9% of control following ET-1 treatment in RGCs (P < 0.001, t-test, n = 9).
Figure 8.
Messenger RNA and protein levels of Bax in RGCs. There was no change in the mRNA levels of bax detected in ET-1–treated RGCs from microarray, but a decrease to 86% of control was observed in ET-1–treated RGCs by real-time PCR, which was statistically significant (P = 0.001, t-test, n = 6). Endothelin-1 or ET-3 treatment in RGC boosted the protein level of Bax, the staining intensity of Bax was higher in ET-treated group compared to control (*P < 0.05, **P < 0.01, #P < 0.001); mRNA levels (mean ± SEM) are represented in bar graph; in microarray, the vehicle control served as control; in real-time PCR, results were normalized to the expression of cyclophilin A, and values were compared to vehicle-treated control cells.
Figure 9.
Endothelin-1 mediated changes in expression of members of the Bcl-2 and caspase gene family. Endothelin-1 treatment of RGCs induced a significant decrease in the mRNA levels of bcl-2, bak1, and bid (P < 0.01, t-test, n = 3–6) determined by real-time PCR (A). A downregulation of mRNA expression of casp2 and casp7 was found by real-time PCR in RGCs treated with ET-1, which was statistically significant (B). Expression of capn6 was downregulated in ET-1–treated RGCs determined by microarray analysis, and the same trend also was detected in real-time PCR. Messenger RNA level of capn6 in ET-1 treatment in real-time was 12.9% of control (P < 0.001, t-test, n = 9 [B]). *P < 0.05, **P < 0.01, #P < 0.001; mRNA levels (mean ± SEM) are represented in bar graph; in microarray, the vehicle control served as control; in real-time PCR, results were normalized to the expression of cyclophilin A, and compared to vehicle-treated control cells.
Discussion
Previous studies have shown that ETs induced apoptosis of RGCs that were demonstrated in various experimental paradigms, including ET treatment of cultured RGC cells,29 intravitreal injection in rats,47 and continuous administration using implanted osmotic mini-pumps in rats.25 However, the mechanisms by which ETs induce apoptosis of RGCs still are not clear. In the current study, ET-induced changes in gene expression and protein levels were investigated using a genome-wide screening by microarray, and validated by real-time PCR, immunocytochemistry, and immunoblotting. A diverse set of gene responses were found in RGCs treated with endothelins, including calcium homeostasis, cell cycle, cell growth, and neurite outgrowth and projection. Since ET treatment of primary RGCs results in apoptotic changes,29 the study provides an important insight into changes in gene expression that contribute to cell death following treatment with various endothelins.
ET-1 Actions in Apoptosis
Receptors ETA and ETB are expressed in many types of cells in the central nervous system (CNS) with ETB being predominant in the CNS,48 including neurons and astrocytes.49,50 Endothelin B (ETB) receptors are upregulated at the mRNA and protein levels in many ocular tissues, including optic nerve head, nerve fiber layer, RGCs, and inner plexiform layer of the retina in animal models of glaucoma.29,38,51,52 Our immunocytochemistry data showed that ETB and ETA receptors were upregulated at the protein level in RGCs treated with 100 nM ET-1, compared to those of untreated control cells (Fig. 4). However, only the ETB receptor was increased at the mRNA level. The changes in ET-1 expression and/or regulation of ET receptors may have an important role in neuronal death in RGCs. Minton et al. 29 demonstrated that 100 nM ET-1 and ET-3 treatment triggers cell death of cultured RGCs via apoptosis. A significant loss of RGCs was found in association with increased immunostaining of the ETB receptor in wild-type rats with IOP elevation for 4 weeks. Moreover, RGC loss was attenuated and a significant protection of RGCs from cell death was found in the ETB receptor-deficient rats. This suggests that the ETB receptor has a causative role in RGC loss during ocular hypertension. Furthermore, intravitreal administration of ET-1 induced apoptosis in wild-type rats, which was attenuated in ETB receptor-deficient rats.47 In addition, administration of an ETA/ETB dual antagonist, bosentan, diminished neurodegeneration in the congenital DBA/2J mice model of glaucoma that has a spontaneous elevation of IOP.53 Taken together, the data suggest that activation of ETB receptors by ET-1 induces neurodegenerative effects in ET-1–treated RGCs in culture as well as in rat models with ocular hypertension. The precise role of ETA receptor in glaucomatous neurodegeneration remains to be understood.
c-Jun and GAP-43 Upregulation in RGCs Following ET-1 Treatment
c-Jun is an immediate early gene whose expression is elevated in the early stage of neuronal damage and degeneration,54,55 and also associated with axonal regeneration of neurons and neuronal survival.54,56 Our laboratory previously reported that ET-1 and ET-3 produced apoptotic changes in isolated RGCs.29 In the current study, protein levels of c-Jun and phosphorylated c-Jun were greatly increased in ET-1 and ET-3–treated RGCs, although there was no apparent change in mRNA level of c-Jun (Fig. 6). c-Jun N-terminal kinase (JNK) is a stress induced MAPK kinase that is involved in several pathological pathways in neurodegenerative diseases.57 c-Jun is a direct target of phosphorylation by JNK and is a component of the AP-1 transcription factor. The neuroprotective effect of attenuation of phosphorylation of c-Jun (by administration of JNK inhibitor, SP600125, in mice) and in JNK knock-out mice further proves the degenerative role of c-Jun in neuronal damage under glaucomatous conditions. A significant protection of RGCs from cell death was found in JNK2/3 deficient mice following optic nerve crush.58 Furthermore, administration of JNK inhibitor SP600125 significantly protected RGCs from cell death and reduced axon loss in mice with elevated IOP.59 These observations suggest that c-Jun and phosphorylation of c-Jun by upstream JNK have a critical role in RGC death. It is possible that one mechanism by which JNK/c-Jun induces cell death of RGCs is through upregulation of ETB receptors. Our previous study showed that overexpression of c-Jun produced increased expression of ETB receptors in human nonpigmented ciliary epithelial cells (HNPE).38 Increased c-Jun and AP-1 immunostaining also was observed in conjunction with elevated ETB receptors in the RGC layer of retina with elevated IOP.38 The present results show that ET-1 treatment in RGCs promoted not only increased expression of the ETB receptor at the mRNA and protein levels (Fig. 4), but also increased the expression and phosphorylation of c-Jun. A key question to address is whether c-Jun is an upstream regulatory transcription factor of the ETB receptor gene. Previous data indicated that there were six AP-1 and 40 C/EBPβ binding sites in the upstream promoter region of the human ETB receptor gene.38 Mutations or deletions at the AP-1 binding sites significantly attenuated gene transcription of the ETB receptor. Since ET levels are elevated in glaucomatous conditions in human patients as well as in animal models, it is plausible that ET's actions could contribute to c-Jun activation and elevated mRNA expression of ETA receptor, ETB receptor, in RGCs of rats with IOP elevation (McGrady N, et al. IOVS 2014;55:ARVO E-Abstract 1917 and Refs. 29, 38).
GAP-43 is highly upregulated in developmental stages of neurons, and its expression also is increased in regenerating axons of RGCs.60 GAP-43 is identified in the mammalian brain,60–62 RGCs and optic nerves of toads and goldfish.63,64 Interestingly, GAP-43 expression is not limited to neuronal cells, but also found in astrocytes and microglia.65,66 GAP-43 has an important role in reconstruction of synapses in axons or neurites, and its phosphorylated form is present in the presynaptic membranes.67 In the current study (Fig. 7), GAP-43 protein was detectable in somas and neurites of RGCs by immunocytochemistry, and ET-1 or ET-3 treatment for 24 hours increased the protein levels of GAP-43 in somas and neurites, as shown by immunocytochemistry and immunoblotting. Based on our previous report29 that ET-treatment induced RGC apoptosis, it appears that upregulation of GAP-43 could be a protective response of neurons to the external stimuli (albeit not sufficient to block apoptotic changes). The changes in GAP-43 expression show tremendous variation depending upon the tissues and the nature of the stimulus. Ischemia generated by cerebral artery occlusion induced the downregulation of GAP-43 in most damaged neurons and upregulation in some intact neurons depending upon the stage of ischemia or its duration.68 Increased GAP-43 protein was detected in spinal cord, brainstem, and optic nerve with neuronal trauma, brain damage, or axon injuries.69–72 These observations suggested that elevated GAP-43 protein is a protective response to compensate and restore structural integrity and improve neuronal functions. Interestingly, several AP-1 transcription factor binding sites were identified at the promoter region of GAP-43,73 and GAP-43 and c-Jun were expressed in RGCs in rats subjected to optic nerve transection.74 In the current study, protein levels of c-Jun and its phosphorylated isoform were shown to be significantly upregulated in RGCs treated with ET-1 or ET-3, suggesting the involvement of c-Jun in the increased expression of GAP-43 in RGCs. The possibility of c-Jun–mediated upregulation of GAP43 expression must be further investigated.
ET-Induced Calcium Signaling
ET-1–mediated signaling through G-protein coupled receptor (GPCR), acting primary through Gq coupling, activate phospholipase C (PLCβ), generating inositol 1,4,5-trisphosphate (IP3), and diacylglycerol (DAG) by hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2). Inositol 1,4,5-trisphosphate acts on IP3 receptors in calcium stores, including the endoplasmic reticulum releasing Ca2+ and activates protein kinase C (PKC) through the involvement of DAG. Protein kinase C could act upstream of the MAPK pathway via activation of Ras and Raf, to influence cell proliferation through a classical Ras-Raf-MEK-ERK pathway by the activation of growth factor receptors.75–78 The increased intracellular Ca2+ also could directly trigger the activation of Ca2+-dependent kinases (CaMK, and so forth), as well as phospholipase A2, and promote the release of arachidonic acid.1
The S100 protein family has an important role in controlling cell proliferation, apoptosis, calcium concentration, through diverse mechanisms. The proteins of this family contain a EF-hand helix-loop-helix domain, which is crucial for calcium binding.35,36 Several members of the S100 family gene were found to be significantly upregulated at the mRNA level following ET treatment (Fig. 1). In real-time PCR, mRNA expression of s100A4, s100A6, and s100A11 was elevated 2.1-, 6.0-, and 1.7-fold with ET-1 treatment, respectively. The results from microarray were consistent with real-time PCR. Leclerec et al.79 have shown that s100A6 in micromolar concentrations has the ability to induce apoptosis of cultured human SH-SY5Y neuroblastoma cells. On similar grounds, upregulation of s100A6 in Hep3B cells increased caspase 3 expression at the transcriptional level and enhanced cell death.80 Thus, it is possible that s100A6 also may be a contributor to RGC cell death, but the detailed mechanisms must be addressed. Endothelins could affect calcium homeostasis through S100 family, possibly through Gq protein coupling of the ETA receptor, IP3 production, leading to elevation of intracellular calcium.
Comparison of Gene Expression at the mRNA and Protein Level
In the current study, real-time PCR was used to confirm the mRNA expression of selected genes. In addition, immunocytochemistry also was used to determine the corresponding protein levels of these genes. Herein, the regulation of mRNA expression of some genes and their corresponding proteins were not synchronized in response to ET-1 treatment. Although no appreciable changes were detected for mRNA of c-jun and even a decreased in expression was found for gap-43, ETAR, and bax. However, at the protein level significantly higher staining intensity was found by immunocytochemistry for their corresponding proteins. This suggests that there are distinct mechanisms controlling protein expression and posttranslational modifications, which operate independent of the regulation of gene expression at the level of mRNA. Several steps in protein translation could be responsible for the discrepancy between mRNA and protein levels of gene expression: (1) translation of protein from mRNA itself could be controlled by more complicated mechanisms which are not fully understood; (2) some proteins undergo posttranslational modifications to exert their functions, such as phosphorylation, glycosylation, and so forth; (3) some proteins undergo cleavage to smaller forms to be activated. For instance, members of the caspase family and MMP family are initially translated as proproteins, cleavage of the precursor proteins produces the mature forms of these proteins; (4) the half-life of some proteins also is under the precise control of synthesis and degradation mechanisms. Treatment with ET-1 produced an increase in c-Jun protein levels in RGCs, although no appreciable changes were found at the mRNA level. Moreover, a robust increase of phosphorylated c-Jun was detected in somas and neurites of RGCs (Fig. 6), suggesting that the c-Jun protein and its phosphorylated form are critical protein targets of endothelin actions. In a study conducted in vasopressin-treated kidney cells, there were 188 proteins with a significant change in abundance, which was evident from a large-scale quantitative protein mass spectrometry, whereas more than 1/3 of these proteins did not show the corresponding changes in mRNA levels.81 The correlation of mRNA and protein levels appears to be poor to moderate under some circumstances.
The Extracellular Matrix (ECM)
Extensive remodeling of ECM occurs in ocular tissues both in glaucoma patients and in animal models of the disease.82–86 Matrix metalloproteinases (MMPs) and their endogenous tissue inhibitors of TIMPs have important roles in the regulation of ECM turnover and maintenance of the cell matrix. Primarily, MMP gene expression is regulated by growth factors, cytokines and other factors, like stress, and neurotrauma.87,88 Endothelin-1 not only is known as a potent vasoactive peptide, but also is involved in ECM remodeling by shifting the balance between MMPs and TIMPs. In addition, a number of studies showed that ET-1 also induced ECM protein expression, including collagens, laminin, and fibronectin in cell culture and in vivo models.89–93 We reported earlier that ET-1 regulated ECM remodeling in primary human optic nerve head astrocytes and lamina cribrosa cells.94,95 These observations suggest that ET-1 could be involved in ECM remodeling of the optic nerve head in glaucoma through astrogliosis and reactivation of astrocytes. However, a detailed assessment of the ET-1 mediated changes in activity of MMPs and TIMPs and ECM remodeling in rat models of glaucoma has not been done to the best of our knowledge. Guo et al.96 reported that increased MMP-9 immunostaining was found in the RGC layer following 3 months of elevated IOP. An increase in mRNA levels of ET receptors, timp1, mmp3, ET-2, fibronectin, and GFAP, also was detected by gene microarray analysis and RT-PCR in Brown Norway rats with elevated IOP.22 The results of the current study showed that ET treatment induced the regulation of transcription of a variety of ECM genes (Fig. 2), including mmp3, timp1, tnc, and aspn. The categorized results of gene clusters using DAVID analysis also showed that tissue and ECM remodeling, synthesis of ECM components, and ECM-receptor interaction were most regulated in RGCs in response to ETs (Tables 3, 4). Overall, the data suggest that endothelins exert their regulatory roles in ECM remodeling not only through astrocytes and other cell types, but also directly through RGCs. Extensive ECM remodeling is correlated with RGC apoptosis and axon loss, possibly through increased MMP activity, which could enhance ECM degradation, including collagen degradation, as a mechanism to facilitate the migration of astrocytes into optic nerve bundles.
ET-1, ET-2, and ET-3 Displayed Similar Actions
All the three endothelins (ET-1, ET-2, and ET-3) induced profound alterations in expression of a variety of genes, including cytokines, structural proteins, signaling pathways, transcription factors, and matrix molecules in RGCs, and also triggered significant changes in neuronal gene expression.
Endothelin-1 and ET-2 bind to the ETA receptor with high affinities, whereas ET-3 binds with 70 to 100 times lower affinity. Interestingly, all three ET peptides bind to the ETB receptor with similar high affinities.14 Protein kinase C, MAPK, and PI3K-Akt are involved in signaling induced by ET-1 through its receptors ETAR and/or ETBR. Activation of these pathways is a key step in triggering downstream signaling and potential activation of transcriptional factors, such as c-Myc, Elk-1, c-Fos.97–101 It has been suggested that some transcription factors, such Elk-1, c-Fos, AP-1, are controlled simultaneously by MAPK-ERK and PKC,13,97,102 whereas p38 and JNK share different transcription factors. In this study, ET-1, ET-2, and ET-3 induced a similar pattern of gene expression for most of the genes, but there still were some exceptions. For instance, there was no change in the mRNA level of neurotrophic tyrosine kinase receptor type 1 (Ntrk1) in ET-2- and ET-3-treated RGCs, whereas there was a 2.3-fold downregulation of this gene in ET-1–treated RGCs (Table 2). Endothelin-2 and ET-3 treatment induce a slight increase of the expression of Mylpf gene in RGCs; however, ET-1 treatment produced a 6-fold elevation in the mRNA level of this gene compared to control (Table 1). A detailed investigation of these differential responses to ETs will reveal the mechanisms and signaling pathways by which the activation of different ET receptors is triggered.
Taken together, several studies have shown that endothelin peptides ET-1/ET-2/ET-3 are involved in the pathogenesis of glaucoma. Endothelins mediate profound cellular responses at the mRNA and protein level through activation of ETAR and/or ETBR in RGCs. In the current study, based on the data obtained from microarray, some classes of genes were selected and investigated by real-time PCR and immunocytochemistry. The Bcl-2 family, S100 family, MMPs, c-Jun and ET receptors are some of the major genes or proteins that are regulated by ET-mediated signaling. These proteins have important roles in apoptosis, calcium homeostasis, cell signaling, and matrix remodeling. The exploration of the roles of endothelins in glaucoma will help understand some key molecular mechanisms underlying neurodegenerative changes during ocular hypertension.
Supplementary Material
Acknowledgments
The authors thank Alena Z. Minton (University of North Texas Health Science Center) for reviewing the manuscript and helpful discussions.
Supported by National Institutes of Health (Bethesda, MD, USA) Grant RO1 EY019952 (RRK).
Disclosure: S. He, None; Y.H. Park, None; T. Yorio, None; R.R. Krishnamoorthy, None
References
- 1. Sokolovsky M. Endothelin receptor subtypes and their role in transmembrane signaling mechanisms. Pharmacol Ther. 1995; 68: 435–471. [DOI] [PubMed] [Google Scholar]
- 2. Noske Whensen J Wiederholt M. Endothelin-like immunoreactivity in aqueous humor of patients with primary open-angle glaucoma and cataract. Graefes Arch Clin Exp Ophthalmol. 1997; 235: 551–552. [DOI] [PubMed] [Google Scholar]
- 3. Tezel G,, Kass M,, Kolker A,, Becker B,, Wax M. Plasma and aqueous humor endothelin levels in primary open-angle glaucoma. J Glaucoma. 1997. ; 6: 83–89. [PubMed] [Google Scholar]
- 4. Kallberg ME,, Brooks DE,, Garcia-Sanchez GA,, Komaromy AM,, Szabo NJ,, Tian L. Endothelin 1 Levels in the aqueous humor of dogs with glaucoma. J Glaucoma. 2002; 11: 105–109. [DOI] [PubMed] [Google Scholar]
- 5. Prasanna G,, Hulet C,, Desai D,, et al. Effect of elevated intraocular pressure on endothelin-1 in a rat model of glaucoma. Pharmacol Res. 2005. ; 51: 41–50. [DOI] [PubMed] [Google Scholar]
- 6. Oku H,, Sugiyama T,, Kojima S,, Watanabe T,, Azuma I. Experimental optic cup enlargement caused by endothelin-1-induced chronic optic nerve head ischemia. Surv Ophthalmol. 1999; 44: S74–S84. [DOI] [PubMed] [Google Scholar]
- 7. Felx M,, Guyot M-C,, Isler M,, et al. Endothelin-1 (ET-1) promotes MMP-2 and MMP-9 induction involving the transcription factor NF-kB in human osteosarcoma. Clinic Sci. 2006. ; 110: 645–654. [DOI] [PubMed] [Google Scholar]
- 8. Liu S,, Premont RT,, Kontos CD,, Huang J,, Rockey DC. Endothelin-1 activates endothelial cell nitric-oxide synthase via heterotrimeric G-protein βγ subunit signaling to protein kinase B/Akt. J Biol Chem. 2003; 278: 49929–49935. [DOI] [PubMed] [Google Scholar]
- 9. Grant K,, Loizidou M,, Taylor I. Endothelin-1: a multifunctional molecule in cancer. Br J Cancer. 2003. ; 88: 163–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nambi P,, Clozel M,, Feuerstein G. Endothelin and heart failure. Heart Fail Rev. 2001. ; 6: 335–340. [DOI] [PubMed] [Google Scholar]
- 11. Nelson J,, Bagnato A,, Battistini B, Nisen P. The endothelin axis: emerging role in cancer. Nature Rev Cancer. 2003; 3: 110–116. [DOI] [PubMed] [Google Scholar]
- 12. Sokolovsky M. Endothelin receptor heterogeneity, G-proteins, and signaling via cAMP and cGMP cascades. Cell Mol Neurobiol. 1995. ; 15: 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sugden PH,, Clerk A. Endothelin Signalling in the Cardiac Myocyte And Its Pathophysiological Relevance. Curr Vascu Pharmacol. 2005; 3: 343–351. [DOI] [PubMed] [Google Scholar]
- 14. Kedzierski RM,, Yanagisawa M. Endothelin system: the double-edged sword in health and disease. Ann Rev Pharmacol Toxicol. 2001; 41: 851–876. [DOI] [PubMed] [Google Scholar]
- 15. Rosendorff C. Endothelin vascular hypertrophy, and hypertension. Cardiovascu Drugs Ther. 1997. ; 10: 795–802. [DOI] [PubMed] [Google Scholar]
- 16. Yorio T,, Krishnamoorthy R,, Prasanna G. Endothelin: is it a contributor to glaucoma pathophysiology? J Glaucoma. 2002. ; 11: 259–270. [DOI] [PubMed] [Google Scholar]
- 17. Prasanna G,, Narayan S,, Krishnamoorthy RR,, Yorio T. Eyeing endothelins: a cellular perspective. Mol Cell Biochem. 2003; 253: 71–88. [DOI] [PubMed] [Google Scholar]
- 18. Goraca A. New views on the role of endothelin. Endocri Regul. 2002; 36: 161–167. [PubMed] [Google Scholar]
- 19. Sugden P. An overview of endothelin signaling in the cardiac myocyte. J Mol Cell Cardiol. 2003. ; 35: 871–886. [DOI] [PubMed] [Google Scholar]
- 20. Bagnato A,, Natali PG. Targeting endothelin axis in cancer. Cancer Treat Res. 2004. ; 119: 293–314. [DOI] [PubMed] [Google Scholar]
- 21. Narayan S,, Prasanna G,, Krishnamoorthy RR,, Zhang X,, Yorio T. Endothelin-1 Synthesis and Secretion in Human Retinal Pigment Epithelial Cells (ARPE-19): differential regulation by cholinergics and TNF-{alpha}. Invest Ophthalmol Vis Sci. 2003; 44: 4885–4894. [DOI] [PubMed] [Google Scholar]
- 22. Ahmed F,, Brown KM,, Stephan DA,, Morrison JC,, Johnson EC,, Tomarev SI. Microarray analysis of changes in mRNA levels in the rat retina after experimental elevation of intraocular pressure. Invest Ophthalmol Vis Sci. 2004; 45: 1247–1258. [DOI] [PubMed] [Google Scholar]
- 23. Orgul S,, Cioffi G,, Bacon D,, Van Buskirk E. An endothelin-1-induced model of chronic optic nerve ischemia in rhesus monkeys. J Glaucoma. 1996. ; 5: 135–138. [PubMed] [Google Scholar]
- 24. Orgul S,, Cioffi G,, Wilson D,, Bacon D,, Van Buskirk E. An endothelin-1 induced model of optic nerve ischemia in the rabbit. Invest Ophthalmol Vis Sci. 1996. ; 37: 1860–1869. [PubMed] [Google Scholar]
- 25. Chauhan BC,, LeVatte TL,, Jollimore CA,, et al. Model of endothelin-1-induced chronic optic neuropathy in rat. Invest Ophthalmol Vis Sci. 2004. ; 45: 144–152. [DOI] [PubMed] [Google Scholar]
- 26. Cioffi G,, Sullivan P. The effect of chronic ischemia on the primate optic nerve. Eur J Ophthalmol. 1999; (suppl 1): S34–S36. [DOI] [PubMed]
- 27. Cioffi G,, Orgul S,, Onda E,, Bacon D,, Van Buskirk E. An in vivo model of chronic optic nerve ischemia: the dose-dependent effects of endothelin-1 on the optic nerve microvasculature. Curr Eye Res. 1995. ; 14: 1147–1153. [DOI] [PubMed] [Google Scholar]
- 28. Stokely ME,, Brady ST,, Yorio T. Effects of endothelin-1 on components of anterograde axonal transport in optic nerve. Invest Ophthalmol Vis Sci. 2002; 43: 3223–3230. [PubMed] [Google Scholar]
- 29. Minton AZ,, Phatak NR,, Stankowska DL,, et al. Endothelin B receptors contribute to retinal ganglion cell loss in a rat model of glaucoma. PLoS One. 2012; 7: e43199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barres BA,, Silverstein BE,, Corey DP,, Chun LL. Immunological morphological, and electrophysiological variation among retinal ganglion cells purified by panning. Neuron. 1988. ; 1: 791–803. [DOI] [PubMed] [Google Scholar]
- 31. Mueller BH,, II,, Park Y,, Daudt DR,, III,, et al. Sigma-1 receptor stimulation attenuates calcium influx through activated L-type voltage gated calcium channels in purified retinal ganglion cells. Exp Eye Res. 2013; 107: 21–31. [DOI] [PubMed] [Google Scholar]
- 32. Huang da W Sherman BT Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 33. Huang da W Sherman BT Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009; 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pagano M,, Theodoras AM,, Tam SW,, Draetta GF. Cyclin D1-mediated inhibition of repair and replicative DNA synthesis in human fibroblasts. Genes Dev. 1994. ; 8: 1627–1639. [DOI] [PubMed] [Google Scholar]
- 35. Donato R. Functional roles of S100 proteins calcium-binding proteins of the EF-hand type. Biochim Biophys Acta. 1999. ; 1450: 191–231. [DOI] [PubMed] [Google Scholar]
- 36. Donato R,, Cannon BR,, Sorci G,, et al. Functions of S100 proteins. Curr Mol Med. 2013. ; 13: 24–57. [PMC free article] [PubMed] [Google Scholar]
- 37. Kou I,, Nakajima M,, Ikegawa S. Expression and regulation of the osteoarthritis-associated protein asporin. J Biol Chem. 2007. ; 282: 32193–32199. [DOI] [PubMed] [Google Scholar]
- 38. He S,, Minton AZ,, Ma H-Y,, Stankowska DL,, Sun X,, Krishnamoorthy RR. Involvement of AP-1 and C/EBPβ in upregulation of endothelin B (ETB) receptor expression in a rodent model of glaucoma. PLoS One. 2013; 8: e79183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mu X,, Beremand PD,, Zhao S,, et al. Discrete gene sets depend on POU domain transcription factor Brn3b/Brn-3.2/POU4f2 for their expression in the mouse embryonic retina. Development. 2004. ; 131: 1197–1210. [DOI] [PubMed] [Google Scholar]
- 40. Wang SW,, Mu X,, Bowers WJ,, et al. Brn3b/Brn3c double knockout mice reveal an unsuspected role for Brn3c in retinal ganglion cell axon outgrowth. Development. 2002. ; 129: 467–477. [DOI] [PubMed] [Google Scholar]
- 41. Nilsson M,, Ford J,, Bohm S,, Toftgard R. Characterization of a nuclear factor that binds juxtaposed with ATF3/Jun on a composite response element specifically mediating induced transcription in response to an epidermal growth factor/Ras/Raf signaling pathway. Cell Growth Differ. 1997. ; 8: 913–920. [PubMed] [Google Scholar]
- 42. Guo Y,, Johnson EC,, Cepurna WO,, Dyck JA,, Doser T,, Morrison JC. Early gene expression changes in the retinal ganglion cell layer of a rat glaucoma model. Invest Ophthalmol Vis Sci. 2011; 52: 1460–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Atsumi T,, Singh R,, Sabharwal L,, et al. Inflammation amplifier, a new paradigm in cancer biology. Cancer Res. 2014. ; 74: 8–14. [DOI] [PubMed] [Google Scholar]
- 44. Kumar J,, Ward AC. Role of the interleukin 6 receptor family in epithelial ovarian cancer and its clinical implications. Biochim Biophys Acta. 2014. ; 1845: 117–125. [DOI] [PubMed] [Google Scholar]
- 45. Siveen KS,, Sikka S,, Surana R,, et al. Targeting the STAT3 signaling pathway in cancer: role of synthetic and natural inhibitors. Biochim Biophys Acta. 2014. ; 1845: 136–154. [DOI] [PubMed] [Google Scholar]
- 46. Levkovitch-Verbin H,, Quigley HA,, Martin KRG,, et al. The transcription factor c-jun is activated in retinal ganglion cells in experimental rat glaucoma. Exp Eye Res. 2005. ; 80: 663–670. [DOI] [PubMed] [Google Scholar]
- 47. Krishnamoorthy RR,, Rao VR,, Dauphin R,, Prasanna G,, Johnson C,, Yorio T. Role of the ETB receptor in retinal ganglion cell death in glaucoma. Can J Physiol Pharmacol. 2008. ; 86: 380–393. [DOI] [PubMed] [Google Scholar]
- 48. Morga E,, Faber C,, Heuschling P. Stimulation of Endothelin B receptor modulates the inflammatory activation of rat astrocytes. J Neurochem. 2000; 74: 603–612. [DOI] [PubMed] [Google Scholar]
- 49. Rogers S,, Peters C,, Pomonis J,, Hagiwara H,, Ghilardi J,, Mantyh P. Endothelin B receptors are expressed by astrocytes and regulate astrocyte hypertrophy in the normal and injured CNS. Glia. 2003. ; 41: 180–190. [DOI] [PubMed] [Google Scholar]
- 50. Peters CM,, Rogers SD,, Pomonis JD,, et al. Endothelin receptor expression in the normal and injured spinal cord: potential involvement in injury-induced ischemia and gliosis. Exp Neurol. 2003. ; 180: 1–13. [DOI] [PubMed] [Google Scholar]
- 51. Rogers SD,, Demaster E,, Catton M,, et al. Expression of Endothelin-B receptors by gliain vivois increased after CNS injury in rats, rabbits, and humans. Exp Neurol. 1997; 145: 180–195. [DOI] [PubMed] [Google Scholar]
- 52. Wang X,, LeVatte TL,, Archibald ML,, Chauhan BC. Increase in endothelin B receptor expression in optic nerve astrocytes in endothelin-1 induced chronic experimental optic neuropathy. Exp Eye Res. 2009. ; 88: 378–385. [DOI] [PubMed] [Google Scholar]
- 53. Howell GR,, Macalinao DG,, Sousa GL,, et al. Molecular clustering identifies complement and endothelin induction as early events in a mouse model of glaucoma. J Clin Invest. 2011. ; 121: 1429–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Herdegen T,, Skene P,, Bähr M. The c-Jun transcription factor – bipotential mediator of neuronal death, survival and regeneration. Trends Neurosci. 1997. ; 20: 227–231. [DOI] [PubMed] [Google Scholar]
- 55. Raivich G,, Behrens A. Role of the AP-1 transcription factor c-Jun in developing, adult and injured brain. Prog Neurobiol. 2006. ; 78: 347–363. [DOI] [PubMed] [Google Scholar]
- 56. Raivich G,, Bohatschek M,, Da Costa C,, et al. The AP-1 Transcription Factor c-Jun is required for efficient axonal regeneration. Neuron. 2004; 43: 57–67. [DOI] [PubMed] [Google Scholar]
- 57. Borsello T,, Forloni G. JNK signalling: a possible target to prevent neurodegeneration. Curr Pharm Des. 2007. ; 13: 1875–1886. [DOI] [PubMed] [Google Scholar]
- 58. Fernandes KA,, Harder JM,, Fornarola LB,, et al. JNK2 and JNK3 are major regulators of axonal injury-induced retinal ganglion cell death. Neurobiol Dis. 2012. ; 46: 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sun H,, Wang Y,, Pang I-H,, et al. Protective effect of a JNK inhibitor against retinal ganglion cell loss induced by acute moderate ocular hypertension. Mol Vis. 2011. ; 17: 864–875. [PMC free article] [PubMed] [Google Scholar]
- 60. Skene JH,, Willard M. Changes in axonally transported proteins during axon regeneration in toad retinal ganglion cells. J Cell Biol. 1981. ; 89: 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Woolf CJ,, Reynolds ML,, Molander C,, O'Brien C,, Lindsay RM,, Benowitz LI. The growth-associated protein gap-43 appears in dorsal root ganglion cells and in the dorsal horn of the rat spinal cord following peripheral nerve injury. Neuroscience. 1990; 34: 465–478. [DOI] [PubMed] [Google Scholar]
- 62. Neve RL,, Finch EA,, Bird ED,, Benowitz LI. Growth-associated protein GAP-43 is expressed selectively in associative regions of the adult human brain. Proc Natl Acad Sci U S A. 1988. ; 85: 3638–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Benowitz LI,, Routtenberg A. A membrane phosphoprotein associated with neural development, axonal regeneration, phospholipid metabolism, and synaptic plasticity. Trends Neurosci. 1987. ; 10: 527–532. [Google Scholar]
- 64. Benowitz LI,, Perrone-Bizzozero NI,, Finklestein SP. Molecular properties of the growth-associated protein GAP-43 (B-50). J Neurochem. 1987. ; 48: 1640–1647. [DOI] [PubMed] [Google Scholar]
- 65. da Cunha A,, Vitkovic L. Regulation of immunoreactive GAP-43 expression in rat cortical macroglia is cell type specific. J Cell Biol. 1990. ; 111: 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vitković L,, Steisslinger HW,, Aloyo VJ,, Mersel M. The 43-kDa neuronal growth-associated protein (GAP-43) is present in plasma membranes of rat astrocytes. Proc Natl Acad Sci U S A. 1988; 85: 8296–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Neve RL,, Coopersmith R,, McPhie DL,, et al. The neuronal growth-associated protein GAP-43 interacts with rabaptin-5 and participates in endocytosis. J Neurosci. 1998. ; 18: 7757–7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li Y,, Jiang N,, Powers C,, Chopp M. Neuronal damage and plasticity identified by microtubule-associated protein 2 growth-associated protein 43, and cyclin D1 immunoreactivity after focal cerebral ischemia in rats. Stroke. 1998; 29: 1972–1981. [DOI] [PubMed] [Google Scholar]
- 69. Doster SK,, Lozano AM,, Aguayo AJ,, Willard MB. Expression of the growth-associated protein GAP-43 in adult rat retinal ganglion cells following axon injury. Neuron. 1991. ; 6: 635–647. [DOI] [PubMed] [Google Scholar]
- 70. Li GL,, Farooque M,, Holtz A,, Olsson Y. Increased expression of growth-associated protein 43 immunoreactivity in axons following compression trauma to rat spinal cord. Acta Neuropathol. 1996. ; 92: 19–26. [DOI] [PubMed] [Google Scholar]
- 71. Michler SA,, Illing RB. Acoustic trauma induces reemergence of the growth- and plasticity-associated protein GAP-43 in the rat auditory brainstem. J Comp Neurol. 2002. ; 451: 250–266. [DOI] [PubMed] [Google Scholar]
- 72. Hulsebosch CE,, DeWitt DS,, Jenkins LW,, Prough DS. Traumatic brain injury in rats results in increased expression of Gap-43 that correlates with behavioral recovery. Neurosci Lett. 1998. ; 255: 83–86. [DOI] [PubMed] [Google Scholar]
- 73. Eggen BJL,, Nielander HB,, Rensen-de Leeuw MGA,, Schotman P,, Gispen WH,, Schrama LH. Identification of two promoter regions in the rat B-50/GAP-43 gene. Mol Brain Res. 1994. ; 23: 221–234. [DOI] [PubMed] [Google Scholar]
- 74. Schaden H,, Stuermer CA,, Bahr M. GAP-43 immunoreactivity and axon regeneration in retinal ganglion cells of the rat. J Neurobiol. 1994. ; 25: 1570–1578. [DOI] [PubMed] [Google Scholar]
- 75. Clerk A,, Bogoyevitch M,, Anderson M,, Sugden P. Differential activation of protein kinase C isoforms by endothelin-1 and phenylephrine and subsequent stimulation of p42 and p44 mitogen- activated protein kinases in ventricular myocytes cultured from neonatal rat hearts. J Biol Chem. 1994. ; 269: 32848–32857. [PubMed] [Google Scholar]
- 76. Robin P,, Boulven I,, Desmyter C,, Harbon S,, Leiber D. ET-1 stimulates ERK signaling pathway through sequential activation of PKC and Src in rat myometrial cells. Am J Physiol Cell Physiol. 2002; 283: C251–C260. [DOI] [PubMed] [Google Scholar]
- 77. Robin P,, Boulven I,, Bole-Feysot C,, Tanfin Z,, Leiber D. Contribution of PKC-dependent and -independent processes in temporal ERK regulation by ET-1 PDGF, and EGF in rat myometrial cells. Am J Physiol Cell Physiol. 2004; 286: C798–C806. [DOI] [PubMed] [Google Scholar]
- 78. He S,, Dibas A,, Yorio T,, Prasanna G. Parallel signaling pathways in endothelin-1–induced proliferation of U373MG astrocytoma cells. Exp Biol Med. 2007; 232: 370–384. [PubMed] [Google Scholar]
- 79. Leclerc E,, Fritz G,, Weibel M,, Heizmann CW,, Galichet A. S100B and S100A6 differentially modulate cell survival by interacting with distinct RAGE (receptor for advanced glycation end products) immunoglobulin domains. J Biol Chem. 2007; 282: 31317–31331. [DOI] [PubMed] [Google Scholar]
- 80. Joo JH,, Yoon SY,, Kim JH,, et al. S100A6 (calcyclin) enhances the sensitivity to apoptosis via the upregulation of caspase-3 activity in Hep3B cells. J Cell Biochem. 2008. ; 103: 1183–1197. [DOI] [PubMed] [Google Scholar]
- 81. Khositseth S,, Pisitkun T,, Slentz DH,, et al. Quantitative protein and mRNA profiling shows selective post-transcriptional control of protein expression by vasopressin in kidney cells. Mol Cell Proteomics. 2011. ; 10:M110 004036. [DOI] [PMC free article] [PubMed]
- 82. Hernandez M,, Pena J. The optic nerve head in glaucomatous optic neuropathy. Arch Ophthalmol. 1997. ; 115: 389–395. [DOI] [PubMed] [Google Scholar]
- 83. Hernandez MR. The optic nerve head in glaucoma: role of astrocytes in tissue remodeling. Prog Retinal Eye Res. 2000. ; 19: 297–321. [DOI] [PubMed] [Google Scholar]
- 84. Hernandez M,, Andrzejewska W,, Neufeld A. Changes in the extracellular matrix of the human optic nerve head in primary open-angle glaucoma. Am J Ophthalmol. 1990. ; 109: 180–188. [DOI] [PubMed] [Google Scholar]
- 85. Johnson EC,, Morrison JC,, Farrell S,, Deppmeier L,, Moore CG,, McGinty MR. The effect of chronically elevated intraocular pressure on the rat optic nerve head extracellular Matrix. Exp Eye Res. 1996; 62: 663–674. [DOI] [PubMed] [Google Scholar]
- 86. Morrison JC,, Johnson EC,, Cepurna W,, Jia L. Understanding mechanisms of pressure-induced optic nerve damage. Prog Retinal Eye Res. 2005. ; 24: 217–240. [DOI] [PubMed] [Google Scholar]
- 87. Westermarck J,, Kahari V-M, Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J. 1999. ; 13: 781–792. [PubMed] [Google Scholar]
- 88. Gardner J,, Ghorpade A. Tissue inhibitor of metalloproteinase (TIMP)-1: the TIMPed balance of matrix metalloproteinases in the central nervous system. J Neurosci Res. 2003; 74: 801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tian X,, Tang G,, Chen Y. The effects of endothelin-1 and selective endothelin receptor-type A antagonist on human renal interstitial fibroblasts in vitro. Zhonghua Yi Xue Za Zhi. 2002. ; 82: 5–9. [PubMed] [Google Scholar]
- 90. Fraccarollo D,, Bauersachs J,, Kellner M,, Galuppo P,, Ertl G. Cardioprotection by long-term ETA receptor blockade and ACE inhibition in rats with congestive heart failure: mono- versus combination therapy. Cardiovascul Res. 2002. ; 54: 85–94. [DOI] [PubMed] [Google Scholar]
- 91. Fraccarollo D,, Galuppo P,, Bauersachs J,, Ertl G. Collagen accumulation after myocardial infarction: effects of ETA receptor blockade and implications for early remodeling. Cardiovascu Res. 2002. ; 54: 559–567. [DOI] [PubMed] [Google Scholar]
- 92. Gomez-Garre D,, Ruiz-Ortega M,, Ortego M,, et al. Effects and interactions of endothelin-1 and angiotensin II on matrix protein expression and synthesis and mesangial cell growth. Hypertension. 1996. ; 27: 885–892. [DOI] [PubMed] [Google Scholar]
- 93. Nakamura T,, Ebihara I,, Tomino Y,, Koide H. Effect of a specific endothelin A receptor antagonist on murine lupus nephritis. Kidney Int. 1995. ; 47: 481–489. [DOI] [PubMed] [Google Scholar]
- 94. He S,, Prasanna G,, Yorio T. Endothelin-1-mediated signaling in the expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases in astrocytes. Invest Ophthalmol Vis Sci. 2007; 48: 3737–3745. [DOI] [PubMed] [Google Scholar]
- 95. Rao VR,, Krishnamoorthy RR,, Yorio T. Endothelin-1 mediated regulation of extracellular matrix collagens in cells of human lamina cribrosa. Exp Eye Res. 2008. ; 86: 886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Guo L,, Moss SE,, Alexander RA,, Ali RR,, Fitzke FW,, Cordeiro MF. Retinal ganglion cell apoptosis in glaucoma is related to intraocular pressure and IOP-induced effects on extracellular matrix. Invest Ophthalmol Vis Sci. 2005; 46: 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Schinelli S,, Zanassi P,, Paolillo M,, Wang H,, Feliciello A,, Gallo V. Stimulation of endothelin B receptors in astrocytes induces cAMP response element-binding protein phosphorylation and c-fos expression via multiple mitogen-activated protein kinase signaling pathways. J Neurosci. 2001; 21: 8842–8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Shichiri M,, Sedivy JM,, Marumo F,, Hirata Y. Endothelin-1 is a potent survival factor for c-Myc-dependent apoptosis. Mol Endocrinol. 1998; 12: 172–180. [DOI] [PubMed] [Google Scholar]
- 99. Chen S,, Mukherjee S,, Chakraborty C,, Chakrabarti S. High glucose-induced, endothelin-dependent fibronectin synthesis is mediated via NF-kappa B and AP-1. Am J Physiol Cell Physiol. 2003; 284: C263–C272. [DOI] [PubMed] [Google Scholar]
- 100. Iwai-Kanai E,, Hasegawa K. Intracellular signaling pathways for norepinephrine- and endothelin-1-mediated regulation of myocardial cell apoptosis. Mol Cell Biochem. 2004. ; 259: 163–168. [DOI] [PubMed] [Google Scholar]
- 101. Chintalgattu V,, Katwa LC. Role of protein kinase Cδ in endothelin-induced type I collagen expression in cardiac myofibroblasts isolated from the site of myocardial infarction. J Pharmacol Exp Ther. 2004; 311: 691–699. [DOI] [PubMed] [Google Scholar]
- 102. Hua H,, Goldberg HJ,, Fantus IG,, Whiteside CI. High glucose-enhanced mesangial cell extracellular signal-regulated protein kinase activation and α1(IV) collagen expression in response to endothelin-1: role of specific protein kinase C isozymes. Diabetes. 2001. ; 50: 2376–2383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.