Abstract
The effects of changing solution conditions on the strength of triple helical complexes formed between pyrimidine-rich circular DNA oligonucleotides and their homopurine complements are examined. The complexes display properties different from those seen for other types of DNA triplexes.
Perhaps the majority of DNA in nature exists in circular form, and it is clear that the circular topology leads to properties different from those of linear DNA.1 Despite these facts, synthetic circular oligonucleotides have received only limited attention to date. A few studies have been carried out on circles which are internally base-paired, giving dumbbell-shaped structures.2–7 Synthetic studies have also been reported addressing the problem of construction of very small circles up to ~14 nucleotides in size.8–10 Studies of larger synthetic circular DNAs may have been hindered in part by lack of a practical method for their construction.
We recently reported the synthesis of a new class of oligonucleotide ligand for single-stranded polynucleotides.11–16 Certain pyrimidine-rich circular oligonucleotides were shown to bind strongly and sequence-selectively to single-stranded DNA by forming a pyrimidine•purine•pyrimidine triple helical complex. In this complex, two pyrimidine binding domains on opposite sides of a circle form a sandwich-type complex17–19 with the central polynucleotide substrate, with the binding domains being connected by two bridging loops. Four or five hydrogen bonds are formed with each base in the target strand, as compared with two or three for Watson-Crick complexation. This approach allows formation of complexes with very high affinity12,13 and sequence selectivity.11 Such circular oligomers have the ability to accelerate kinetically the unwinding of a target strand in duplex DNA,14 and in addition, they are highly resistant against degradation in human Plasma.15
In this report are described studies carried out to explore further the properties of two such circle•single strand complexes: one containing T-A-T base triads alone, and a second with both T-A-T and C-G-C base triads. Thermal denaturation experiments for the two complexes under varied conditions of pH, ethanol content, and sodium chloride and polycation concentrations show that they display properties consistent with their triple helical structure. Because these complexes are bimolecular, however, they share some properties more common to duplex DNA, including melting with a single transition and destabilization by ethanol.
Experimental Procedures
Oligonucleotide synthesis
DNA oligomers were synthesized on a Pharmacia LKB automated synthesizer using the standard phosphoramidite method.20 The linear precursors of the circles were also 5'-phosphorylated on the synthesizer using a commercially available reagent purchased from Cruachem. Oligomers were purified by preparative 20% denaturing polyacrylamide gel electrophoresis, isolated as the sodium salt, and quantitated by absorbance at 260 nm. Extinction coefficients for the oligomers were calculated by the nearest neighbor method.21 The construction and characterization of the cyclic oligonucleotides in this study were carried out as previously described.11,13
Melting studies
Solutions for the thermal denaturation studies contained a 1:1 ratio of circle and complementary oligomer (3 µM each). Solutions were buffered with 10 mM Tris•HCI, Na•MES (2-N-morpholinoethanesulfonate), Na•BES (N,N-bis(2-hydroxyethyl)2-aminoethanesulfonate), Na•CHES (2-(N-cyclohexylamino)ethanesulfonate) or sodium phosphate, as specified. Buffers were purchased from Sigma. The buffer pH is that of a 500 mM stock solution at 25°C; after dilution the final solution pH was shown to be within 0.1 unit of the buffer stock. In the ethanol addition experiments the listed pH is that of the aqueous stock solution. Ethanol, NaCl, MgCl2, or spermine hydrochloride (Sigma) were also added in various experiments. After the solutions were prepared they were heated to 90°C and allowed to cool slowly to room temperature prior to melting. The melting experiments were carried out in sealed 0.1 cm pathlength quartz microcells under nitrogen atmosphere on a Gilford 250 UV-vis spectrophotometer equipped with a Gilford 2527 thermoprogrammer. Absorbance was monitored while temperature was raised from 0.2° to 90°C at a rate of 1°/min. In all cases the circle complexes displayed sharp transitions with apparently two-state melting from bound complex to free oligomers. Melting temperatures (Tm) were taken as the inflection point of transition, as determined by computer fit from plots of absorbance vs. temperature.22 Uncertainty in Tm is estimated to be ± 0.5°C, and in ΔH°, ΔS° and ΔG°, ±10%, based on variance from repeated experiments.
Results and Discussion
Thermodynamics of circle complexation
To obtain thermodynamic parameters, melting curves for DNA complexes, measured in the presence of 100 mM Na+ and 10 mM Mg2+, were fit to theoretical curves using a two-state model.22 Four different oligomers, two linear and two circular (forming complexes [1] and [2]; sequences given in Table I), were complexed to two separate purine sequences, dA12 and dAAGAAAAGAAAG. Linear and circular oligomers were hybridized to the same purine oligomer for comparison of binding affinities at pH 7. This was done for two separate sequences, to check how differences between the circular and linear oligomers might depend on base composition. Since the second circular oligomer forms a triple helical complex containing C-G-C triads, it was also examined at pH 5.8, where it is fully protonated (vide infra).
Table I.
Thermodynamic Parameters, Association Constants, and Melting Temperatures for Complexation of 12-base Complementary DNA Strands by Circular and Linear DNA Oligomers (pH = 7.0 unless otherwise noted).
| complex | −ΔH° (kcal/mol) | −TΔS° (kcal/mol) | −ΔG°37 (kcal/mol) | Kassoc (M−1) | Tm (°c) |
|---|---|---|---|---|---|
| 78 | 70 | 8.1 | 5.1 × 105 | 36.9 | |
 |
140 | 124 | 16.1 | 5.9 × 1011 | 57.4 |
| 105 | 95 | 10.3 | 1.8 × 107 | 43.8 | |
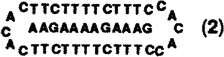 |
111 | 94 | 16.4 | 3.6 × 1011 | 62.3 |
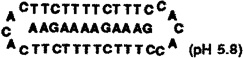 |
129 | 108 | 20.6 | 3.3 × 1014 | 70.3 |
Results show that the circles bind single strands with much higher affinity than linear Watson-Crick oligomers do, and that the effect is both enthalpic and entropic in origin. For the first target sequence, dA12, the enthalpy of complexation by the circle is −140 kcal/mol, nearly twice that of the linear dT12, which is consistent with the greater number of H-bonds and stacked bases in the circle complex. The loss of entropy is also greater for the circle, although it is not twice as high: TΔScomplex is −124 kcal/mol for the circle and −70 kcal/mol for the linear strand at 37°C. This result is consistent with the expected entropic benefit of circularity, since the circle has more than twice the number of nucleotides undergoing restriction of conformational freedom. For the mixed target sequence the effect is similar: Comparing the mixed-sequence duplex and circle complex [2] at pH 7, we find that the added affinity of the circle complex arises largely from differences in the enthalpic term (~6 kcal), while the entropy loss is nearly the same in both complexes. This may again reflect the entropic advantage of circularity, since a 34 nucleotide oligomer would ordinarily be expected to have a much greater entropy loss in helix formation than the 12 nucleotides of the linear strand.
When both Hoogsteen H-bonds are formed (pH 7 for [1] and pH 5.8 for [2]), the free energies measured for complexation by the two circles are very nearly twice those for the Watson-Crick strands. The corresponding association constants at 37°C for the circles are 1011 to 1014 M−1, while the duplexes have association constants of 105 to 107 M−1. The circle which binds to the mixed sequence has an affinity which is pH-dependent: at pH 7 it binds with a free energy (37°C) of −16.4 kcal/mol, somewhat less strongly than at pH 5.8, where it has a free energy of −20.6 kcal/mol. It is clear that both circle complexes are stronger than the simple addition of Watson-Crick and Hoogsteen strand affinities. The free energy of the third (Hoogsteen) strand binding in a T-A-T termolecular triplex has been measured at ~0.5 times that of the underlying duplex base pairs.23,24 Thus, the addition of the Watson-Crick and Hoogsteen bonds should result in free energies ~1.5 times that of the duplex alone. By contrast, both circle complexes studied here are ~2.0 times stronger in free energy than the corresponding Watson-Crick duplexes (Table I). This effect is likely due in part to the entropic benefit of combining both domains in one closed molecule. We have shown that "nicked" circles, which are linear and still can form triplexes with a single strand, do not bind as strongly as their covalently closed counterparts,13,17 a finding consistent with this idea. It is also likely that some enthalpic stabilization for these complexes is gained from base stacking of the loop bases with the binding domains. The overall data show that the free energies are derived from large counterbalancing enthalpy and entropy terms,25 and it is undoubtedly true that the parameters depend both on the DNA itself (and the sequences involved) as well as the solvating water molecules and cations around the complexes.6
Effects of ethanol
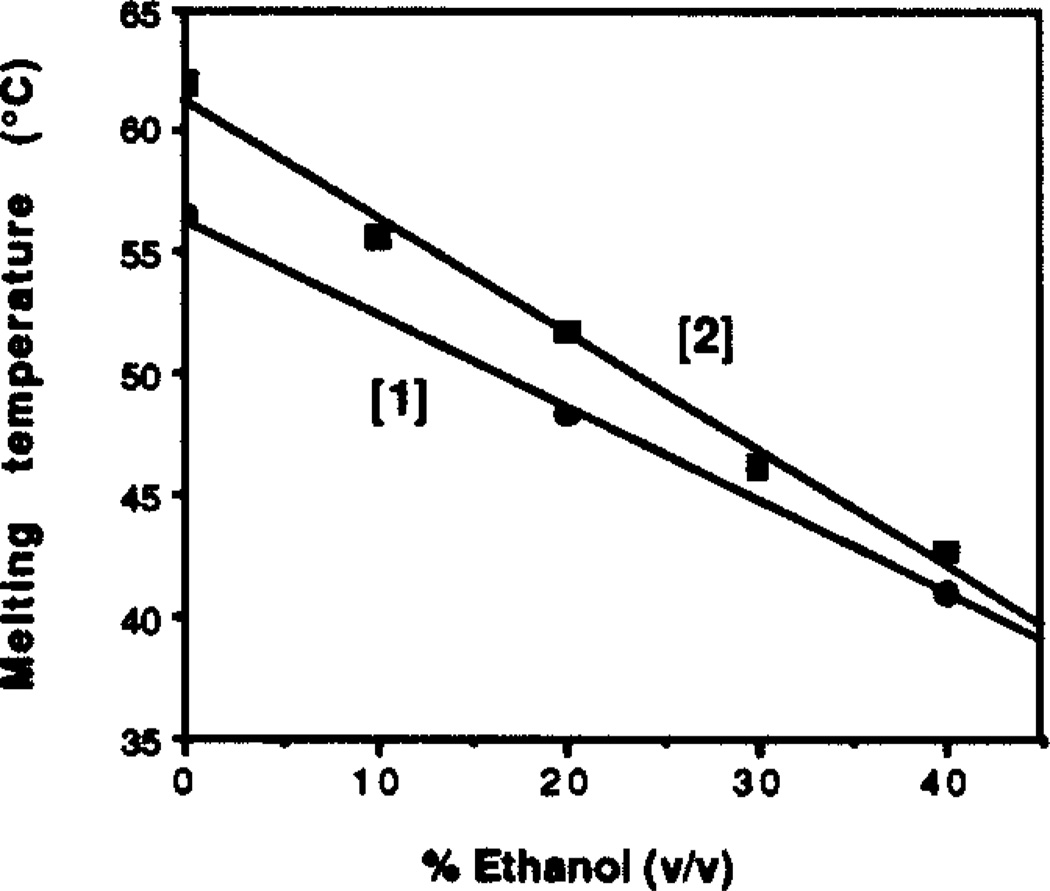
The effect of added ethanol on the melting transition was measured for 0–40% (v/v) ethanol, using both complexes [1] and [2] at pH 7.0 (Tris•HCl, 10 mM) with 100 mM NaCl and 10 mM MgCl2. Experiments with higher ethanol contents showed signs of precipitation. Results show (Fig. 1) that for both complexes there is a strong downward trend in thermal stability with increasing ethanol, with the Tm dropping 15–19°C with an increase in ethanol content from 0–40%. The plots appear linear over the range studied. Plots of free energy vs. percent ethanol show a similar trend (data not shown).
Figure 1.
The destabilization of the complexes by ethanol is unusual for a triple helical structure. Ethanol is known to increase the stability of the complexation of a T15 third strand with duplex DNA in termolecular triple helices, and to have little effect on binding of a mixed T,C strand.26 The stabilization has been attributed to the fact that B-form duplex DNA changes conformation to the A form at high ethanol concentrations;27 since DNA triple helices may exist in a conformation with A-form charactetistics,28,29 it is possible that added ethanol favors the triplex by stabilizing this conformation. Watson-Crick duplexes, on the other hand, are destabilized by ethanol at these concentrations,30 probably due in part to increased interstrand charge repulsion. For the circle complex it appears that the observed effect is similar to the destabilization effect which occurs with Watson-Crick duplexes. Prior to binding, the uncomplexed circle and substrate in the present case are both single stranded, and so the binding event would not benefit from an ethanol-induced conformational change.
Effects of changing PH
The effects of changing pH were measured for both the complexes [1] and [2] in the presence of 10 mM MgCl2 and 100 mM NaCl with 10 mM buffer. Four buffer systems, Tris•HCl, MES•Na, BES•Na and sodium phosphate were used to cover the pH range; overlapping data from different buffers showed identical trends. It was noted that some buffers stabilized the complexes better than others; for example, data with BES buffer showed the same pH-independent trend for complex [1], but the Tm's were about 5°C lower than for data with phosphate or Tris buffer.
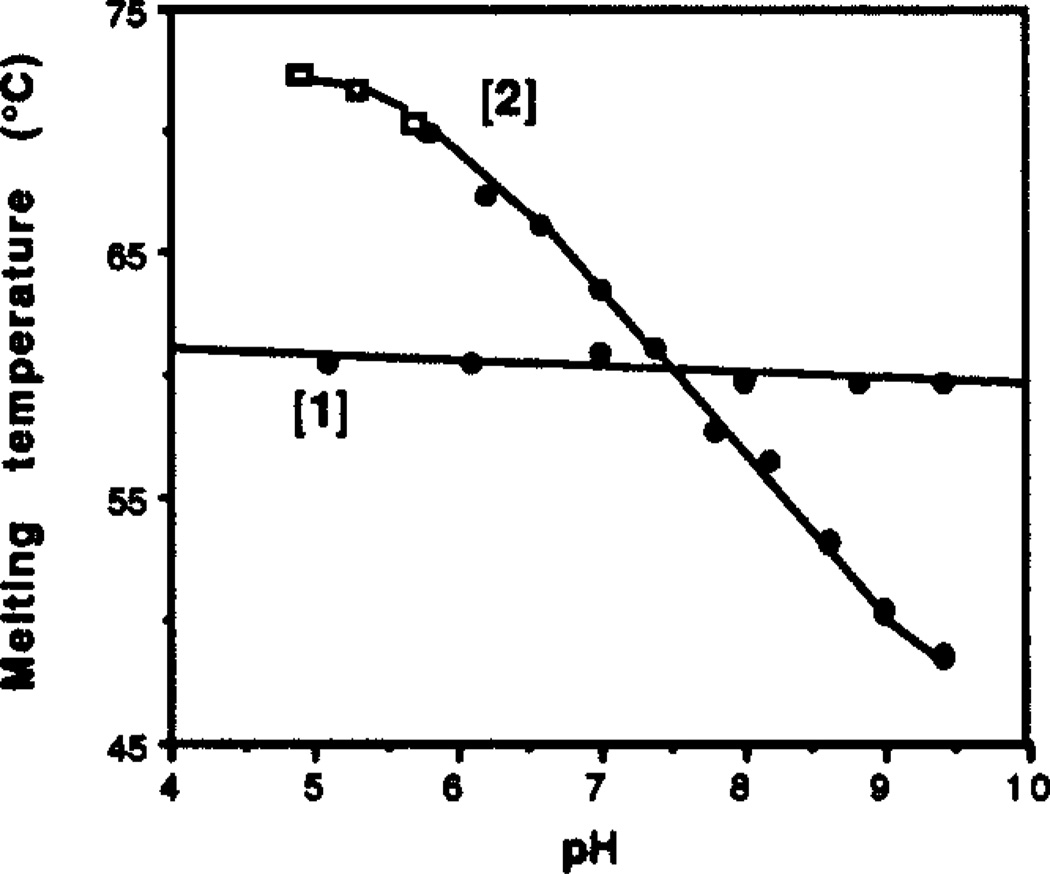
For clarity, Figure 2 gives data obtained with MES (squares) and Tris (circles) as buffers. Results show the contrasting behavior of the two complexes with changing PH. Complex [1] is insensitive to varied pH over the range 5.2 to 9.4, with little or no change in Tm (<2°C). Complex [2] has a relatively high Tm of 72°C at pH 5.2, but the Tm drops with increasing pH. At pH 9.4 the Tm is 49°C. At the lowest pH values the slope of the plot flattens, suggesting nearly complete protonation of basic groups in the complex.
Figure 2.
The effects of changing pH on the two complexes are consistent with their triple helical structure. The insensitivity of complex [1] over the four pH unit range is expected for a triplex composed of T-A-T triads. Complex [2], however, is quite sensitive to pH, at pH 5.2 it has a Tm 11°C higher than [1], and at pH 9.4 its Tm is 11°C lower than [1]. This finding is consistent with the triple helical structure of [2] involving protonation of C-G-C base triads.31 The crossing point for the two complexes, where the thermal stabilities are approximately equal, appears at pH 7.5. By contrast, the corresponding Watson-Crick duplex (d(AAGAAAAGAAAG)•d(CTTTCTTTTCTT)) is not affected significantly by changing pH; for example, at pH 5.8 its Tm is 42°C, about the same as at pH 7.0.12
The data in Fig. 2 represent a partial titration curve for complex [2]. Based on the leveling of Tm for the complex at low pH, we can place a lower limit of 7.0 for the pKa of an average cytosine in the helix, which is in reasonable agreement with values reported in other studies.32 The pKa of cytosine in a given complex undoubtedly depends on the number of, and distance between, C+G-C triads in a triplex. Interestingly, complex [1] contains six cytosines in the loop regions (as does [2]), but these unpaired cytosines apparently have a considerably lower pKa than those in the helical domain of [2] and thus do not affect stability over this range. A pKa value of 4.3 is reported for deoxycytidine 5'-monophosphate;33 this value may be a reasonable estimate for the cytosines in the loop portions of [1] and [2]. Thus, the ~2–3 pKa unit difference between the loop and helix cytosines in these complexes presents a striking demonstration of the effect of local environment on their basicity.
Effects of ionic strength
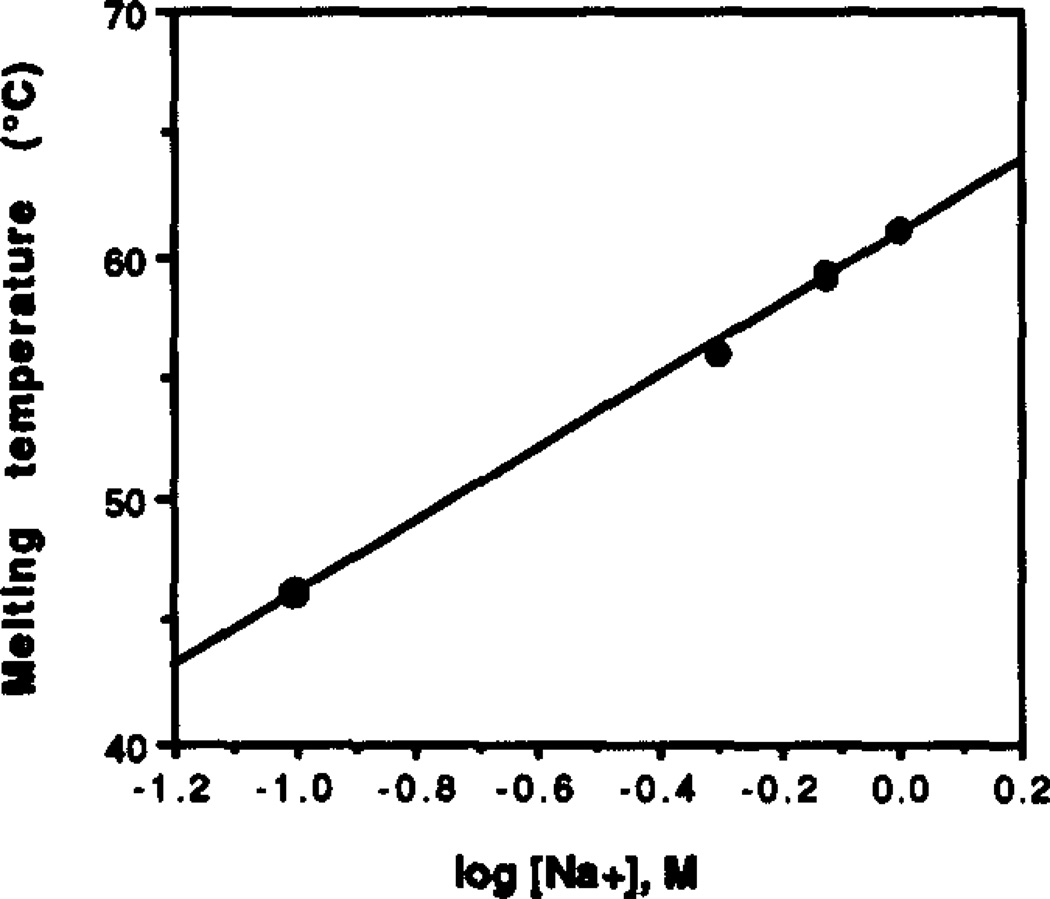
The effects of increasing NaCl concentration on the melting transition were studied with complex [2] (Figure 3). Experiments were performed at pH 7.0 with 10 mM Tris•HCl and in the absence of other added metal cations. Results show an increase in Tm as [NaCl] is raised from 0.0 to 1.0 M, giving a Tm of 43°C with no added salt and 61°C at 1.0 M concentration. A plot of log[NaCl] vs. Tm (Fig. 3) is linear with a slope indicating a 15°C increase in Tm per decade of rise in salt concentration.
Figure 3.
The stability of the circle complexes is raised by increasing ionic strength and by added cations, consistent with the increase in negative charge density on complexation for nucleic acids.34 Quite unusual, however, is the fact that these complexes can form in the absence of any added cations, except for the 10 mM Tris buffer. With only the buffer present,35 we measured a Tm of 43 °C for complex [2] at pH 7. Triple helices require significant added salt or polycations for stability;34,36 the close association of three intertwined polyanion strands necessitates the presence of considerable positive charge in the vicinity of the helix. It has been shown that between 100 mM and 2 M NaCl is required for the formation of termolecular triple helices.23,24 This difference is apparently due to the high stability of the circle complexes.
Effects of polycations
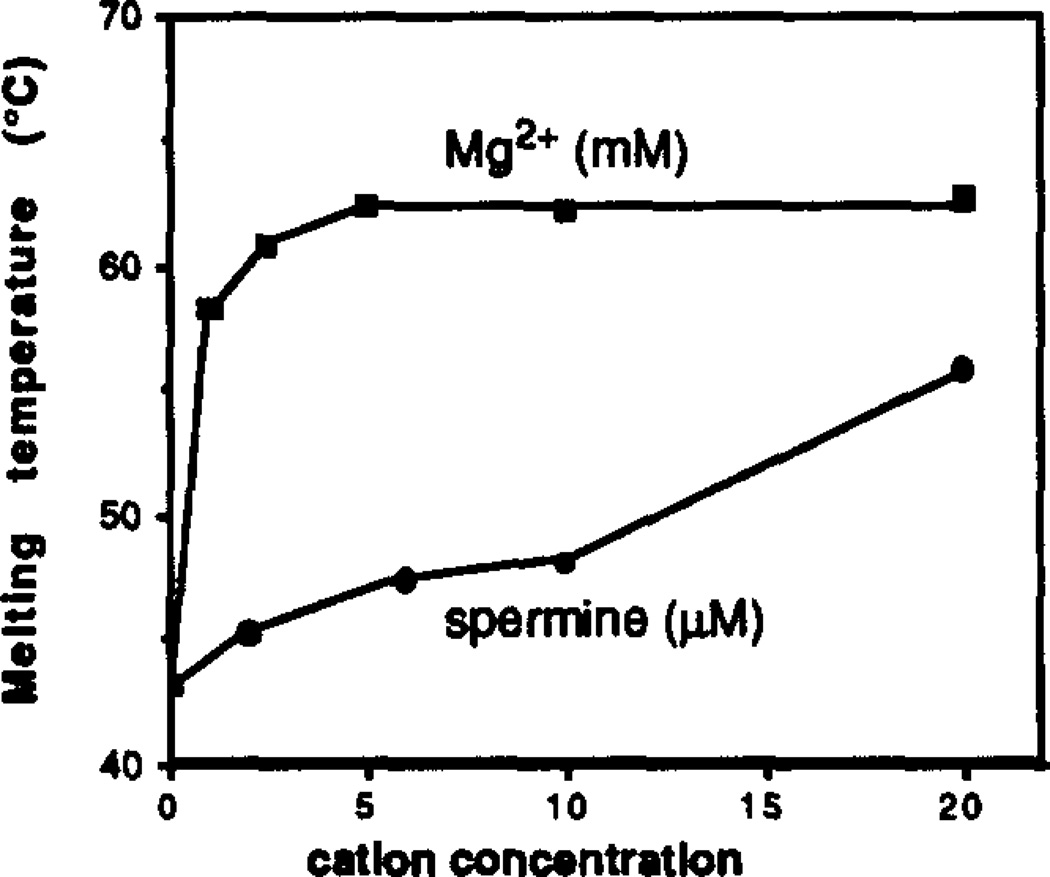
Spermine and Mg2+ were also examined separately for their ability to stabilize complex [2] at pH 7.0 in the absence of added NaCl. The results are displayed in Figure 4. Magnesium was studied over the range 0 – 20 mM; the plot shows a very sharp initial rise in Tm at the lowest concentrations, and almost no rise above 5 mM. The majority of the stabilization effect is seen by 1 mM Mg2+, with relatively little increase thereafter. Spermine was studied at the lower concentrations of 0 to 20 µM. An upward trend is again seen. with Tm rising from 44°C to 58°C over this range.
Figure 4.
Based on these results, the polycations Mg2+ and spermine appear to be more effective at ameliorating interstrand anion-anion repulsion, and lower concentrations are required to achieve the same effect as Na+. In the present study, spermine is ~100 times more efficient at supporting the complex than Mg2+ on a molar basis, and Mg2+ is ~300 times more efficient than Na+ (compare Figs. 3 and 4). Intracellular magnesium levels in humans are 5–30 mM,38 and spermine has been found in mammalian tissue at a level of 38 mg/kg,39 so complexes such as [1] and [2] are likely to be quite stable under physiological ionic conditions.
In summary, observation of the stabilities of circle complexes under varying conditions reveals that while they are triple helical, they have properties which differ from those of more conventional DNA duplexes or triplexes, including stronger binding, higher sequence specificity, and resistance to degradation. This type of complex thus represents a potentially useful motif in the recognition of single-stranded polynucleotides.
Acknowledgments
We thank the National Institutes of Health (RO1-GM46625) for support, Prof. D. Turner for use of the spectrophotometer, and S. Wang for providing additional data. E.T.K acknowledges a Camille and Henry Dreyfus Teacher-Scholar Award, and awards from the Young Investigator Programs of the Arnold and Mabel Beckman Foundation, the Office of Naval Research, and the Army Research office.
Reference
- 1.Watson JD, Hopkins NH, Roberts JW, Steitz JA, Weiner AM. Molecular Biology of the Gene. 4th. Menlo Park: Benjamin/Cummings; 1987. pp. 193–195.pp. 257 [Google Scholar]
- 2.Olivera BM, Scheffler IE, Lehman IR. J. Mol. Biol. 1968;36:275. doi: 10.1016/0022-2836(68)90381-1. [DOI] [PubMed] [Google Scholar]
- 3.Wemmer DE, Benight AS. Nucleic Acids. Res. 1985;13:8611. doi: 10.1093/nar/13.23.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalisch BW, Krawetz SA, Schoenwaelder K-H, van de Sande JH. Gene. 1986;44:263. doi: 10.1016/0378-1119(86)90190-3. [DOI] [PubMed] [Google Scholar]
- 5.Erie DA, Jones RA, Olson WK, Sinha NK, BresIauer KJ. Biochemistry. 1989;28:268. doi: 10.1021/bi00427a037. [DOI] [PubMed] [Google Scholar]
- 6.Ashley GW, Kushlan DM. Biochemistry. 1991;30:2927. doi: 10.1021/bi00225a028. [DOI] [PubMed] [Google Scholar]
- 7.Chu BCF, Orgel LE. Nucleic Acids Res. 1992;20:2497. doi: 10.1093/nar/20.10.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbato S, DeNapoli L, Picciali G, Santacroce C. Tetrahedron Lett. 1987;28:5727. [Google Scholar]
- 9.de Vroom E, Broxterman HJG, Sliedregt LAJM, van der Marel GA, van Boom JH. Nucleic Acids Res. 1988;16:4607. doi: 10.1093/nar/16.10.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeNapoli L, Messere A, Montesarchio D, Piccialli G, Santacroce C. Nucleosides & Nucleotides. 1993;12:21. [Google Scholar]
- 11.Kool ET. J. Am. Chem. Soc. 1991;113:6265. [Google Scholar]
- 12.Prakash G, Kool ET. J. Chem. Soc., Chem. Commun. 1991:1161. doi: 10.1039/C39910001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prakash G, Kool ET. J. Am. Chem. Soc. 1992;114:3523. doi: 10.1021/ja00035a056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perkins TA, Goodman JL, Kool ET. J. Chem. Soc., Chem. Commun. 1993:215. [Google Scholar]
- 15.Rumney S, Kool ET. Angew. Chem. Int. Ed. Engl. 1992;31:1617. doi: 10.1002/anie.199216171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubin E, McKee TL, Kool ET. J. Am. Chem. Soc. 1993;115:360. doi: 10.1021/ja00054a060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xodo LE, Manzini G, Quadrifoglio F. Nucleic Acids Res. 1990;18:3557. doi: 10.1093/nar/18.12.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Souza DJ, Kool ET. J. Biomol. Struct. Dyn. 1992;10:141. doi: 10.1080/07391102.1992.10508634. [DOI] [PubMed] [Google Scholar]
- 19.GiovannangeIi C, Montenay-Garestier T, Rougée M, Chassignol M, Thuong NT, Héléne C. J. Am. Chem. Soc. 1991;113:7775. [Google Scholar]
- 20.Beaucage SL, Caruthers MH. Tetrahedron Lett. 1981;22:1859. [Google Scholar]
- 21.Borer PN. In: Handbook of Biochemistry and molecule Biology. 3rd. Fasman GD, editor. I. Cleveland: CRC Press; p. 589. [Google Scholar]
- 22.Petersheim M, Turner DH. Biochemistry. 1983;22:256. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- 23.Pilch DS, Levenson C, Brousseau R, Shafer RH. Proc. Natl. Acad. Sci. (USA) 1990;87:1942. doi: 10.1073/pnas.87.5.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilch DS, Brousseau R, Shafer RH. Nucleic Acids Res. 1990;18:5743. doi: 10.1093/nar/18.19.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Searle MS, Williams DH. Nucleic Acids Res. 1993;21:2051. doi: 10.1093/nar/21.9.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moser HE, Dervan PB. Science. 1987;238:645. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 27.Zimmerman SB, Pheiffer BH. J. Mol. Biol. 1979;135:1023. doi: 10.1016/0022-2836(79)90526-6. [DOI] [PubMed] [Google Scholar]
- 28.Arnott S, Selsing E. J. Mol. Biol. 1974;88:509. doi: 10.1016/0022-2836(74)90498-7. [DOI] [PubMed] [Google Scholar]
- 29.RajagopaI P, Feigon J. Nature. 1989;339:637. doi: 10.1038/339637a0. [DOI] [PubMed] [Google Scholar]
- 30.Hickey DR, Turner DH. Biochemistry. 1985;24:2086. doi: 10.1021/bi00329a042. [DOI] [PubMed] [Google Scholar]
- 31.Lipsett MN. J. Biol. Chem. 1964;239:1256. [PubMed] [Google Scholar]
- 32.Callahan DE, Trapane TL, Miller PS, Ts'o POP, Kan L-S. Biochemistry. 1991;30:1650. doi: 10.1021/bi00220a030. [DOI] [PubMed] [Google Scholar]
- 33.Gray DM, Ratliff RL, Antao VP, Gray CW. In: Structure and Expression: DNA and its Drug Complexes. Sarma RH, Sarma MH, editors. II. New York: Adenine Press; 1987. pp. 147–166. [Google Scholar]
- 34.Manning GS. Quart. Rev. Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 35.Also present are the sodium counterions of the phosphates on the DNA from gel purification.
- 36.Felsenfeld G, Miles HT. Annu. Rev. Biochem. 1967;36:407–448. doi: 10.1146/annurev.bi.36.070167.002203. [DOI] [PubMed] [Google Scholar]
- 37.Glaser R, Gabbay EJ. Biopolymers. 1968;6:243–254. doi: 10.1002/bip.1968.360060207. [DOI] [PubMed] [Google Scholar]
- 38.Mudge GH. In: The Pharmacological Basis of Therapeutics. Goodman AG, Goodman LS, Rall TW, Murad F, editors. New York: MacMillan; 1985. p. 874. [Google Scholar]
- 39.Rosenthal SM, Tabor CW. J. Pharmacol. Exper. Therap. 1956;116:131. [PubMed] [Google Scholar]