Abstract
Intravascular optical coherence tomography (IV-OCT) allows evaluation of atherosclerotic plaques; however, plaque characterization is performed by visual assessment and requires a trained expert for interpretation of the large data sets. Here, we present a novel computational method for automated IV-OCT plaque characterization. This method is based on the modeling of each A-line of an IV-OCT data set as a linear combination of a number of depth profiles. After estimating these depth profiles by means of an alternating least square optimization strategy, they are automatically classified to predefined tissue types based on their morphological characteristics. The performance of our proposed method was evaluated with IV-OCT scans of cadaveric human coronary arteries and corresponding tissue histopathology. Our results suggest that this methodology allows automated identification of fibrotic and lipid-containing plaques. Moreover, this novel computational method has the potential to enable high throughput atherosclerotic plaque characterization.
OCIS codes: (170.6935) Tissue characterization, (170.4500) Optical coherence tomography, (170.3880) Medical and biological imaging, (100.2960) Image analysis, (100.5010) Pattern recognition
1. Introduction
Globally, atherosclerosis is the leading cause of death and morbidity. The different stages involved in atherosclerotic plaque formation must be studied and characterized in order to develop new therapies for atherosclerosis. In a stable plaque, the accumulation of collagen forming a fibrous plaque can lead to progressive stenosis, while in a vulnerable plaque, the build-up of lipids and necrotic core can lead to sudden thrombosis and heart attack [1]. Most vulnerable plaques are characterized by having a large necrotic core covered by a thin inflamed fibrous cap, the hallmarks of the thin cap fibroatheroma. To improve our understanding of the mechanisms that lead to a thrombotic event, it is relevant to characterize both the morphology and the composition of the plaque with the potential to identify the more deadly vulnerable plaques.
Since its development in 1991 and its implementation for vascular imaging in 1996 [2], OCT has shown high potential for imaging of atherosclerotic plaques. OCT has rapidly evolved in the past decade as a clinically useful diagnostic modality and has become very useful for intravascular (IV) imaging due its high spatial resolution and sensitivity [3–6]. A standard nomenclature has been generated to provide a consensus in the usage guidelines, measurement methodology, and visual image interpretation criteria for IV-OCT image characterization [6]. Under this consensus, the normal vessel wall or intimal thickening is characterized by a highly backscattering or signal-rich intima layer, a low backscattering and signal-poor media layer, and a heterogeneous and highly backscattering adventitia layer. High backscattering and homogeneous IV-OCT signal characterize a fibrous plaque. A calcific plaque is usually surrounded by fibrous tissue, and present a signal-poor region with sharply delineated borders. Superficial-Lipid plaques appear as a signal-poor region with poorly delineated borders and fast IV-OCT signal decay. An OCT thin-capped fibroatheroma presents a delineated necrotic core overlaid by a fibrous cap with a thickness less than a predetermined threshold. Macrophage accumulations are characterized as signal-rich regions with high standard deviation, distinct, and/or confluent punctate spots that surpass the intensity of background speckle noise.
Fibrous cap thickness is an important parameter of plaque vulnerability, which IV-OCT is able to accurately assess, owing to its high axial resolution [7]. However, visual assessment [8–10] of OCT cross-sectional images (B-scans) is cumbersome, extremely time consuming, and requires trained experts to classify diverse features present in each stage of the disease development. Furthermore, the inherent effect of speckle and the scattering attenuation of the optical scheme limit the direct interpretation of OCT images.
Although OCT is becoming very popular for intravascular imaging of atherosclerotic plaques, a single pullback generates hundreds of B-scans; interpreting this large amount of data makes the characterization process very tedious. To overcome these difficulties, different automatic characterization methods have been developed based on either texture segmentation or attenuation analysis. Random Forests classification using intensity and texture-based features extracted from regions in the B-scan corresponding to the atherosclerotic plaque area has been used to classify four types of plaque: calcium, lipid pool, fibrous tissue and mixed plaque [11]. The main limitation of this method is the identification of lipid pool and mixed plaque since the classification accuracy reported was lower than 80%. A texture based segmentation method uses Spatial Gray Level Dependence Matrices Method, second order statistical texture features extraction, to automatically detect plaque and non plaque regions from OCT images [12]. The drawback of this method is the clusters, distance metric, initialization of cluster centroids and convergence criterion parameters that need to be set correctly to ensure that the algorithm converges and produces satisfying results. Even so, it may not reliably detect all the different plaque types. Windows of variable length can be used to estimate the backscatter and the extinction coefficients from transverse sections resulting in a quantitative estimation of these coefficients for every A-line [13]. This approach was intended to identify five different tissue types: fibrous and smooth muscle cells, early necrotic core, advanced necrotic core, calcification, and hemorrhage. Yet, it is prone to erroneously compute the backscatter coefficient due tissue heterogeneity and the lack of an intensity calibration. Similarly, estimation of the optical attenuation coefficient combined with a supervised classification of image pixels according to textural features was proposed to improve the automated and systematic characterization with IV-OCT of the following atherosclerotic plaque types: fibrotic, calcified, and lipid-rich; yet, this procedure requires user input for the selection of regions of interest (ROIs) for plaque assessment [14]. Recently, a method was proposed to estimate the attenuation coefficient as a function of depth for each A-line in order to to identify calcifications, necrotic core, and mixed plaque [15]. The estimation of optical parameters from volumes of interest has also been attempted in order to classify calcified, fibrotic and lipid plaques; however, this volumetric analysis can be computationally expensive [16]. A combined depth resolved spectral and attenuation model showed the potential of predicting lipid content in OCT images of atherosclerotic plaques [17]. This classification method is based on a quadratic discriminant analysis model utilized on phantoms of known chemical mixtures, such as collagen, calcium and cholesterol, as well as simulated tissue of a lipid-rich plaque. However, the inherent heterogeneity of the fibroatheromas might affect the performance of the proposed approach. A novel method intended for lipid-rich plaque characterization in IV-OCT images used the spectroscopic absorption properties of lipids to evaluate lipid phantom models and in-vivo atherosclerotic rabbit models [18]. In this study the Gaussian center of mass was used as a metric to visualize the property of the spectrum with a high sensitivity, specificity, and correlation of 94.3%, 76.7%, and R=0.8572, respectively. However, this method focused only in the composition of lipids, yet the heterogeneous plaque composition must be evaluated for further validation. A recent study have showed the potential birefringence images of polarization-sensitive optical frequency domain imaging (PS-OFDI) as a source of contrast to evaluate tissue composition [19]. Birefringence is elevated in areas with thick collagen fibers or smooth muscle cells, while in the fibrous cap region both at the rupture site and proximally present low birefringence. This optical tissue property can be used as a feature for an automated computational characterization method.
In this paper, we present a novel computational method for plaque tissue characterization that does not require estimating optical properties. Instead, this method allows for tissue characterization based on the intrinsic morphological characteristics of the A-lines in IV-OCT scans. Since this method is fully automated, it has the potential for high throughput atherosclerotic plaque assessment.
2. Methods
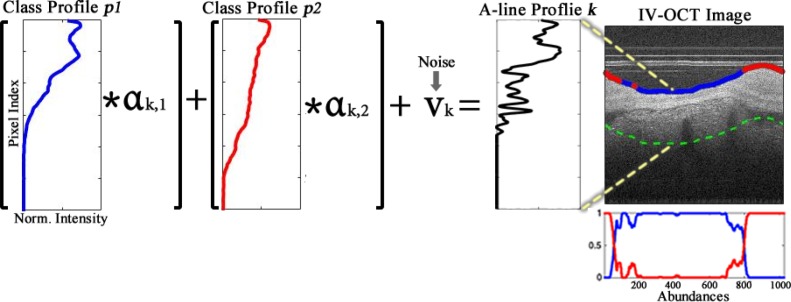
The main idea behind our method for automated plaque characterization is modeling each A-line of the IV-OCT B-scan as a linear combination of N depth profiles (p1, . . . , pN). These profiles are estimated anew from each B-scan or a set of consecutive B-scans, and are then further classified to a specific tissue type, based on morphological features. This is illustrated in Fig. 1, in which two profiles (p1, p2) estimated from the sample B-scan are modeling any given A-line of the B-scan. In this particular example, the blue profile resembles an Intimal-Thickening profile having two prominent peaks and one valley, which represent the intima, media, and adventitia layers; those peaks are faded in the red profile due to thickening caused by fibrous tissue build up, while the decay of the signal is slower due to the characteristic high backscattering of fibrotic plaque. Once the profiles are estimated for a given B-scan, they are automatically classified into one of several potential tissue classes based on a set of morphological features. Since the k-th A-line in the B-scan has a characteristic weight or abundance αk,n for each profile pn, their magnitudes can be used to assign each A-line to a specific tissue type. This is illustrated in Fig. 1, in which a sample A-line at the position k = 400 is composed by the Blue-class profile times the weight or abundance αk,1, plus the Red-class profile times the abundance αk,2, with the addition of noise vk. Since the Blue-class profile has the most dominant abundance, the resulting A-line profile must be more akin to this class. Thus, the lumen is color-coded as blue indicating Intimal-Thickening in that particular A-line position. A detailed description of our proposed methodology, as summarized in Fig. 2, is presented below.
Fig. 1.
Illustration of the k-th A-line profile (black) being modeled as the linear combination of two main profiles (p1 in blue and p2 in red) each with corresponding abundances (αk,1, αk,2) for that particular A-line, plus a noise component vk. The color on the lumen of the OCT image indicate the most dominant class at that specific A-line. The dotted green line marks the maximum depth considered for the profile in the region of interest. The right-bottom plot depicts the abundances of both profiles at each A-line.
Fig. 2.
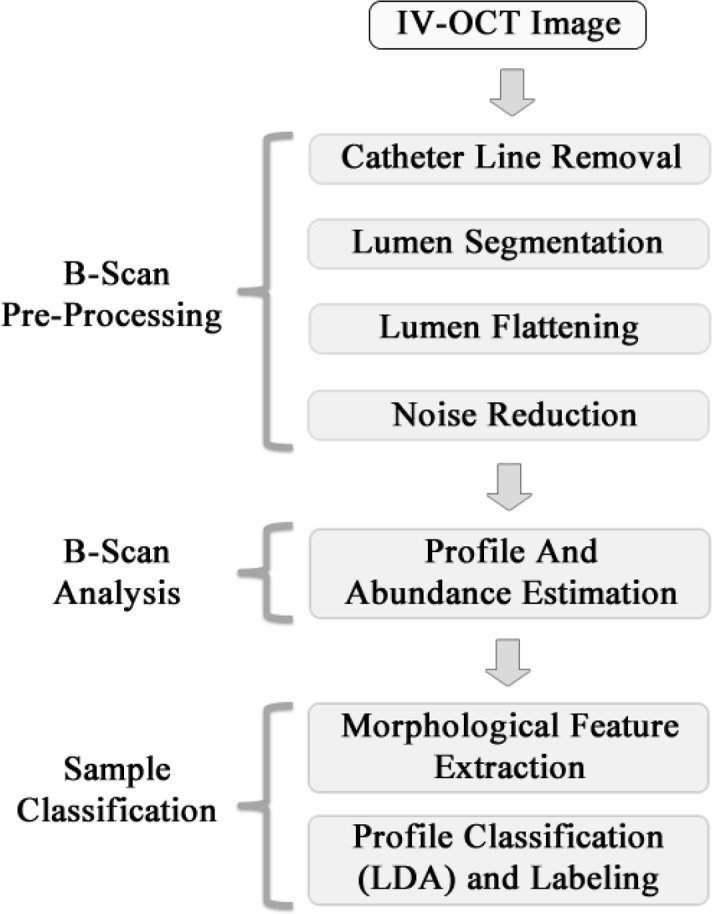
Block diagram of the automatic classification process.
2.1. B-scan pre-processing
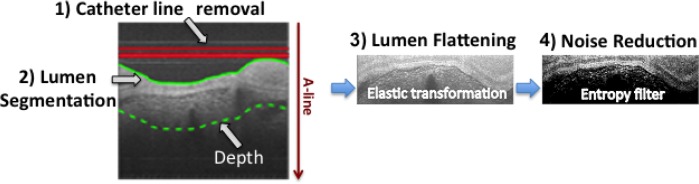
Each IV-OCT B-scan was first pre-processed as follows. As shown in Fig. 3, the lines produced by the catheter ball lens and sheath signal are removed, and the lumen surface is segmented. Next, the lumen surface is flattened using an elastic transformation method [20] and a fix amount of pixels are selected from the lumen surface to certain depth where the signal has decayed to the background (dotted line in Fig. 3); a constant depth of 300 pixels (300 × 4.8 μm = 1, 440 μm, see section 2.5 for system resolution details) was used throughout this study. An Entropy filter with a window size of 5 × 5 pixels is applied to the segmented image to reduce the speckle effect; although other spatial filters such as Median, Wiener, Lee, and Frost with different window sizes can be also used [21, 22]. A logarithmic transformation looks to enhance the small details in the signals, which is especially important for visualization purposes. However, since we are applying a linear combination model of the profiles by A-line, the processing of the B-scans in logarithmic scale would induce a multiplicative effect in the linear scale, which is not suitable for the posterior interpretation and classification stage. As a result, the B-scan pre-processing is performed on the normalized linear signal (0–1) and the Log-compressed image is used only for visualization purposes.
Fig. 3.
B-scan pre-processing steps.
2.2. B-scan analysis
Our proposed B-scan analysis is based on the following considerations:
A set of OCT A-lines can be represented as a non-negative matrix of L pixels by K A-lines (In this study the set of A-lines was taken from 9 consecutive B-scans).
Each measured A-line is composed by a linear mixture of N profiles (p1, . . . , pN), and these profiles are invariant over the whole set of A-lines.
Therefore, considering a linear mixture of profiles at the k-th measured A-line yk with random noise vk, the following observation model is assumed:
| (1) |
where the coefficients αk,n > 0 are interpreted as the abundance or weight parameters of the n-th profile at the k-th A-line. By considering a matrix notation, the observation model can be written as
| (2) |
where Y = [y1 . . . yK] is a matrix of size L × K, representing the set of measured A-lines, P = [p1 . . . pN] is a matrix of size L × N, representing the unknown profiles, A = [α1 . . . αK] is a matrix of size N × K containing the unknown abundances, and V = [v1 . . . vK] is the matrix gathering noise components. In this representation, the vector of abundances at the k − th A-line is denoted as αk = [αk,1 . . . αk,N]⊤. In this way, the simultaneous estimation of the unknown sets of profiles P and abundances A is formulated as a nonlinear quadratic optimization process: [23–25]
| (3) |
with the following restrictions
| (4) |
i.e. A⊤1 = 1 is equivalent to normalization condition of each column of matrix A as required by (1). This optimization problem is nonlinear in the product of the matrices P and A, so an alternating least squares strategy is pursued to find the optimal solutions [26–28].
The first term in (3), is related to the approximation error by the observation model in (2); the second term is a regularization component in the cost function linked to the estimated profiles that penalizes the distance among them by using the weight ρ > 0; finally the last term is another regularization component now focused on the abundances that maximizes its values by A-line and this condition is controlled by the weight μ > 0. Since the abundances are always positive and normalize to one per A-line, the last term in (3), could produce a low entropy condition in the estimated abundances. In this way, the tradeoff between reducing the approximation error, controlling the distance among the estimated profiles or achieving a low entropy condition is regulated by the parameters ρ and μ.
This alternating least squares strategy begins with a matrix P0 that is initially defined for the iterative scheme. For this initialization process, the approach in [26] is followed, where the A-line with the largest energy in the B-scan is selected, as well as the A-line with the largest norm error with respect to the first one. Then, the system is solved for A. Once A is found, the system is solved again for P, and so on. Fixing either P or A in (3), the optimization problem becomes quadratic and it is solved over a convex domain. Nevertheless, the alternating least squares iteration can only assure convergence to a local minimum.
2.3. Automated profile classification
Once the profiles (p1, . . . , pN) and their abundances (α1, . . . , αK) are estimated, the resulting profiles are automatically classified and labeled to identify four different tissue types: Intimal-Thickening (IT), Fibrotic, Superficial-Lipid, and Fibrotic-Lipid. To achieve this, each profile pn is normalized to its maximum intensity value and a set of morphological features are estimated from it, as summarized in Table 1. The Intimal-Thickening profiles are identified based on the first four features in Table 1, and are characterized as having two prominent peaks and one valley with restricted widths and within the first part of the profile (blue region in Fig. 4, before the signal decay). If a profile is not identified as Intimal-Thickening based on the previous criteria, it is further classified into one of the other tissue types based on a Linear Discriminant Analysis (LDA) algorithm that uses the remaining morphological features defined in Table 1.
Table 1.
Profile morphological features
| Feature | Description |
|---|---|
| η | Number of prominent peaks |
| ω(Peakn) | Width of the n-th peak |
| ψ(Peakn) | Location of the n-th peak |
| ϕ(Peakn) | Prominence of the n-th peak |
| ; | GOF of Gaussian fit of the signal decay * |
| Ia | Index of the beginning of the signal decay |
| Ib | Index of the end of the signal decay * |
| d = |Ia − Ib| | Distance from Ia to Ib |
| Area under the signal signal decay * | |
| Area under the total profile signal * | |
| C(x) = c1x2 + c2x + c3 | Quadratic polynomial fitting * |
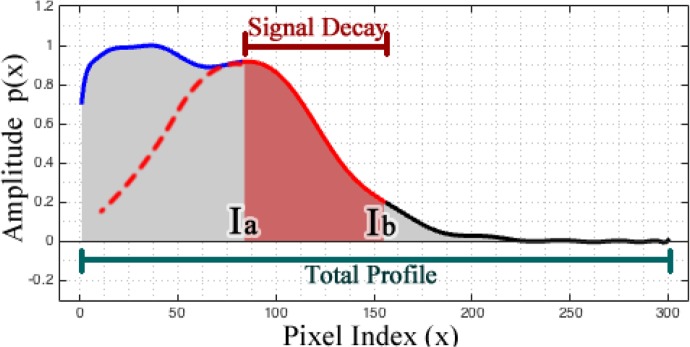
Fig. 4.
Profiles partition for feature extraction. The profile is divided into two sections: first for the region of the profile that is not highly attenuated (blue line) and second for the part where the most significant signal decay occurs (red solid line).
From all the morphological features explored and summarized in Table 1, only the features marked with an (*) were used in the LDA classifier. These features were selected based on the exhaustive search strategy described in section 3.3. The first feature is the Goodness-of-fit (GOF) of a single term Gaussian model applied only to the signal decay section. The assumption is that the attenuation of lipids resembles a Gaussian model (red line in Fig. 4); thus, the GOF should be better (lower values) for the Superficial-Lipid and Fibrotic-Lipid profiles than for the Fibrotic profiles. The second feature is the index of the end of the signal decay, point Ib in the second section, indicating the signal has decreased to a value close to the background. The third feature is the area under the signal decay (AR). The fourth feature is the area under the total profile signal (AT). The last features are taken from the coefficients of a second polynomial fitting to the signal decay section, which assess the nonlinearity of the signal decay. Finally, once the profiles are classified and labeled, each A-line is linked to a specific label that was assigned to the profile with the highest abundance at that specific A-line.
2.4. Automated sample classification
In the context of sample level classification, the method evaluates whether or not a sample has accumulation of lipids. Thus, samples containing any profile classified as Superficial-Lipid or Fibrotic-Lipid are catalogued as Lipid-containing plaques; likewise, sample containing only profiles classified as Intimal-Thickening or Fibrotic are attributed a No-Lipid classification. The algorithm highlights only the segments of the lumen where lipids are detected.
2.5. Database
Cadaveric human coronary arteries were imaged with a fiber-based intravascular Optical Frequency Domain Imaging (OFDI) system [3, 29], having a swept range of 115 nm, centered at 1320 nm and a repetition rate of 54 kHz; resulting in axial and lateral resolutions of 9.4 μm and 30 μm, respectively. It is important to state that this system takes polarization sensitive measurements, detecting two orthogonal polarization channels in parallel, and alternating the polarization state of the light incident on the sample in between A-lines. The sum of the squared norm of the complex-valued tomogram of each channel was computed, ignoring the modulation of the polarization states in between A-lines. The database consisted of a total of 57 datasets of IV-OCT images, each consisting of 9 consecutive frames spaced by 100 μm in the longitudinal dimension. Each frame consists of 1024 axial depth-points and 1024 A-lines, corresponding to an angular sampling step of 0.35°. The axial resolution was 4.8 μm, assuming a refractive index of 1.34 of the tissue, for a total depth of 1024 × 4.8 μm = 4915.2 μm. The fifth of the 9 frames (central frame) was matched with one histological section, stained with Masson’s Trichrome.
2.6. Validation
Each histological section was visually assessed, blinded to the IV-OCT images, and the lumen was partitioned into segments, labeled as one of the following tissue types: Intimal-Thickening, Fibrous, Fibro-lipid, or Superficial-Lipid. The two, N=2, most abundant tissue types found in histopathology served as the ground-truth for training of the LDA classifier, to be predicted by the morphological features of the estimated profiles found in the corresponding IV-OCT data set. For sample-level classification, sections that contained any amount of lipid (at least 25% of the tissue lumen) were classified as Lipid-containing. Only sections without any Fibro-Lipid or Superficial-Lipid segments were classified as No-lipid. Analogously, a trained IV-OCT observer, blinded to the histopathology, classified all the IV-OCT cross-sections, by segmenting them into the same tissue categories, and for the sample-level classification, any lipid segment resulted in a plaque to be attributed to the Lipid-containing category. The automatic sample-level classification was compared to both the visual IV-OCT and histopathological diagnosis. The performance of the classification method was quantified with a leave-one-out-validation strategy on the entire histopathology data-base. For each iteration of the leave-one-out validation, one dataset was assigned for testing, and the other 56 data sets for training.
3. Results
In this section, we present the results of applying the proposed method to the aforementioned database assuming the presence of two most significant profiles (N=2) per B-scan or volume. In fact, N can take values > 2, however, if the number of profiles is too large, the decomposition algorithm could estimate profiles that are either not linearly independent or with insignificant abundances throughout the B-scan. The results presented here were obtained using the 9 consecutive B-scans for each histopathology section as input. The A-lines of the 9 B-scans, 1024 A-lines each, were appended together to form one single set of 9,216 A-lines.
3.1. B-scan pre-processing results
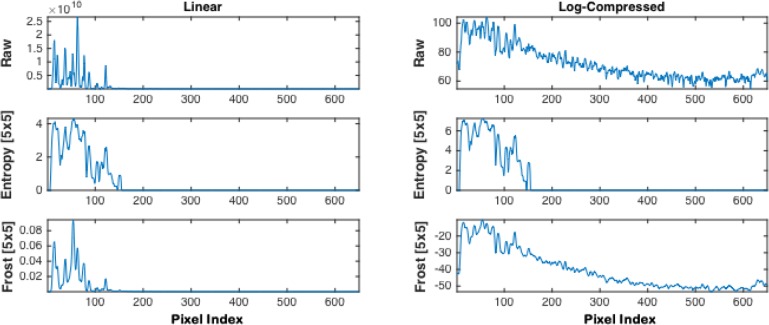
In the preprocessing stage, the following common speckle noise reduction filters were assessed with the IV-OCT images: Entropy, Median, Wiener, Lee, and Frost. Figure 5 shows a comparison of an A-line signal after applying the Entropy and Frost speckle reduction filters to an IV-OCT image. The first and second columns display plots of the linear and Log-compressed signal, respectively, for the raw and filtered signals. The first row shows the raw signal, the second row shows the result of applying the Entropy filter using a window of 5 × 5, and the last row shows the analogous version filtered with Frost. The Median, Wiener, and Lee filters are not shown in the figure since they produce similar results to the Frost filter. It is noticeable that raw Log-compressed image amplifies the signal from the sample but also from the background, while the Entropy minimizes the speckle and amplifies the signal from the sample, while suppressing the background signal. The Frost filter reduces the speckle noise, however, the background signal is not reduced significantly. Hence, for simplicity, speed, and efficiency, the Entropy filter with a size of 5 × 5 was adopted for this study.
Fig. 5.
Comparison of an A-line signal after the Entropy and Frost filters were applied using a window size of 5 × 5. The first column shows the linear signal, while the second column shows the log-compressed version. The raw Linear and Log-compressed signals are plotted as reference in the first row, the second row shows the Entropy filtered A-line, and the third row depicts the Frost filtered version.
3.2. B-scan analysis results
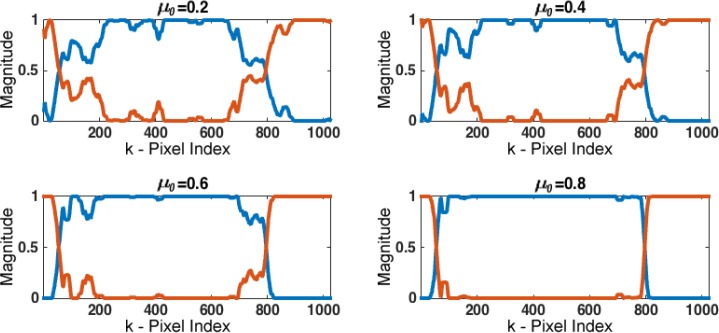
In the B-scan analysis step, the parameters ρ and μ were tuned to determine their optimal values. As stated in section 2.2, changes in ρ values directly affect the difference between the profiles, hence keeping this parameter to lower values leads to more distinct profiles. Similarly, μ controls the amplitude relation among the abundances. However, selecting the right scaling depends on the problem at hand, by the formulation of the quadratic optimization in (3), and this parameter is selected as μ = μ0 λmin(P⊤P), where λmin(·) denotes the minimum eigenvalue of the matrix. As a consequence, μ0 ∈ (0, 1) to avoid a singular problem during the computation of the abundances. Then, as shown in Fig. 6, the magnitudes of the abundances αk,n separate with larger values of μ0. For the analysis presented here, ρ was set to 0.25 to coerce the resultant profiles to be different to each other, while μ0 was set to an intermediate value of 0.6 to maintain a balanced separation between abundances.
Fig. 6.
Abundances estimated with different μ0 values. By increasing the values of μ0, the magnitudes of the abundances αk,n separate; e.g. at the A-line k = 200 the higher abundance (blue) increases while the lower abundance (orange) decreases separating them from each other.
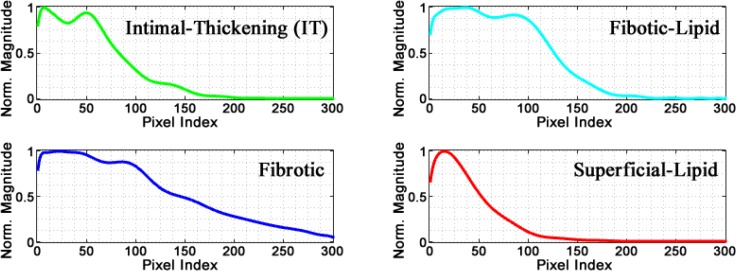
Figure 7 shows an example of the typical profile morphology for each of the four main classes; a color code is used to differentiate each class. It was found that for Intimal-Thickening profiles, green plot in Fig. 7, the main characteristics are the two prominent peaks and one valley within the first 90 pixels. For the Fibrotic profiles (blue), the signal decays slower due to high scattering, while in Superficial-Lipid profiles (red) the decay is faster due high attenuation. For the Fibrotic-Lipid profiles (cyan), the first segment of the profile resembles the first part of a Fibrotic profile and the second part is very similar to the decay of the Superficial-Lipid class.
Fig. 7.
Typical morphology of different class profiles.
3.3. Morphological features selection results
The feature selection was based on an exhaustive search strategy, which selects the combination of features, described in Table 1, that leads to the higher classification accuracy estimated following a leave-one-out validation on the entire database. Thus, other features that were considered, such as index of the beginning of the signal decay (Ia), and the distance (d) from Ia to Ib, were discarded using this strategy.
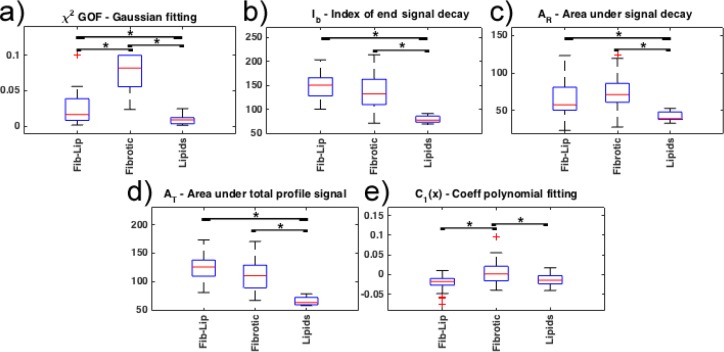
Figure 8 shows a comparison of the distribution between the features described in section 2.3. The GOF of Gaussian fit (χ2) values for Fibrotic profile are significantly higher (worse Gaussian fitting) than for Superficial-Lipid and Fibrotic-Lipid profiles. This is expected, since a better Gaussian fitting of the signal decay region was observed for the Superficial-Lipid and Fibrotic-Lipid profiles (Fig. 8(a)). The index of the end of the signal decay (Ib) shows lower values for Superficial-Lipid profiles than both Fibrotic-Lipid and Fibrotic profiles due to higher attenuation of Superficial-Lipid profiles (Fig. 8(b)). For the signal decay area (AR), Fibrotic and Fibrotic-Lipid profiles exhibit larger areas as the result of a slower decay. On the contrary, Superficial-Lipid profiles present significantly lower signal decay area due to high signal attenuation (Fig. 8(c)). Similarly, the total area AT is higher for Fibrotic-Lipid and Fibrotic than Superficial-Lipid profiles (Fig. 8(d)). The values for the quadratic coefficient of the polynomial fitting (C1(x)) are close to zero for Fibrotic and different from zero for Superficial-Lipid and Fibrotic-Lipid profiles. Thus, Fibrotic profiles can be closely fitted to a linear function, while Superficial-Lipid and Fibrotic-Lipid profiles can be fitted to a quadratic function (Fig. 8(e)).
Fig. 8.
Comparison of features considered for the LDA classification. Multiple comparison with Bonferroni correction (p < 0.05).
3.4. Profile classification results
For the profile classification stage, the morphological features were analyzed to select those that will be used in the LDA classifier. The LDA classifier was tested using leave-one-out validation for the following types of discriminant function: Linear, Diagonal-linear, Quadratic, Diagonal-quadratic, and Mahalanobis. According to our testing, the Linear, Diagonal-linear, Quadratic, Diagonal-quadratic, and Mahalanobis led to an accuracy of 83.6%, 82.7%, 85.4%, 88.2%, and 85.4%, respectively, from which the Diagonal-quadratic gave the highest accuracy. Finally, each A-line is color-labeled in accordance with the classified profile with highest abundance.
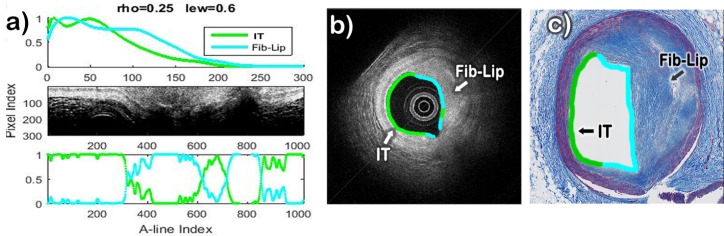
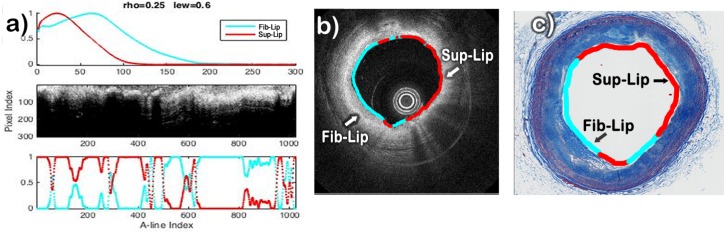
The results of the classification method applied to three datasets characterized by histopathology as having regions corresponding to Intimal-Thickening, Fibrotic, Fibrotic-Lipid, and/or Superficial-Lipid plaques are shown in Figures 9–11. In each of these figures, the estimated profiles (pn) and abundances (αk,n) are shown on the top-left and bottom-left panels. The two profiles are color-coded based on the result of the automated classification as either Intimal-Thickening (green), Fibrotic (blue), Fibrotic-Lipid (cyan), or Superficial-Lipid (red). The middle-left panel shows the corresponding B-scan in Cartesian representation, in which the lumen is also color-coded based on the most dominant abundance for each A-line. The right panel shows the corresponding histopathology section, in which the lumen is also color-coded based on the histopathological evaluation.
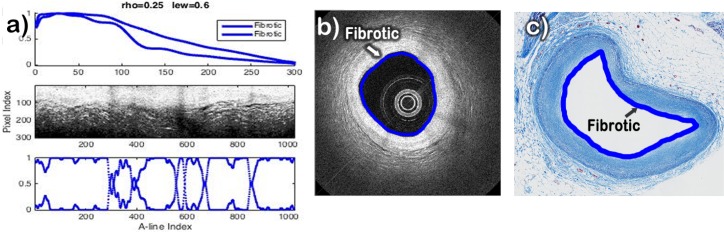
Fig. 9.
Automatic classification of a sample characterized by histopathology as being a Fibrotic plaque. a) Profiles plot (top), IV-OCT image with lumen flattened (center), and Abundances plot (Bottom); b) IV-OCT image with the lumen automatically colored according to the automatic classification; c) histological with the lumen colored manually according to the histological classification.
Fig. 11.
Automatic classification of a sample having regions corresponding to Intimal-Thickening (IT) and Fibrotic-Lipid. a) Profiles plot (top), IV-OCT image with lumen flattened (center), and Abundances plot (Bottom); b) IV-OCT image with the lumen automatically colored according to the automatic classification; c) histological with the lumen colored manually according to the histological classification.
The results of the B-scan analysis for the case of a Fibrotic plaque are shown in Fig. 9(a). The B-scan image (Fig. 9(b)) exhibits high intensity and low attenuation for all the artery tissue, which indicates that only Fibrotic plaque is present. Since both profiles were classified as Fibrotic, the lumen was color coded blue all around in the B-scan, as shown in Fig. 9(b). The corresponding histology section (Fig. 9(c)) clearly indicates a homogeneous fibrotic plaque with no apparent lipid accumulation.
The results of the B-scan analysis for a particular sample presenting both Superficial-Lipid and Fibrotic-Lipid regions are shown in Fig. 10. The two profiles estimated from this B-scan are shown in Fig. 10(a). The red profile showed an immediate fast decay and was correctly classified as a Superficial-Lipid profile. The cyan profile showed an initial low attenuation region followed by a fast decay and was correctly classified as a Fibrotic-Lipid profile. The B-scan (Fig. 10(b)) illustrates areas with high attenuation at deeper depths and higher attenuation close to the surface suggesting the presence of a mix of Fibrotic-Lipid and Superficial-Lipid areas, respectively. The lumen was color-coded based on the values of the estimated abundances as either Fibrotic-Lipid (cyan) or Superficial-Lipid profiles (red), as shown in the B-scan (Fig. 10(b)). The corresponding histology section (Fig. 10(c)) clearly indicates various regions with superficial and deep accumulation of lipids.
Fig. 10.
Automatic classification of a sample having regions of Superficial-Lipid and Fibrotic-Lipid plaques. a) Profiles plot (top), IV-OCT image with lumen flattened (center), and Abundances plot (Bottom); b) IV-OCT image with the lumen automatically colored according to the automatic classification; c) histological with the lumen colored manually according to the histological classification.
The results of the B-scan analysis for a plaque having both Intimal-Thickening and Fibrotic-Lipid regions are shown in Fig. 11. The two profiles estimated from this B-scan are shown in Fig. 11(a). The green profile showed two prominent peaks and one valley and was correctly classified as a Intimal-Thickening profile. The cyan profile showed an initial low attenuation region followed by a fast decay and was correctly classified as a Fibrotic-Lipid profile. In fact, the two profiles in Fig. 10(a) and the second profile (cyan) in Fig. 11(a) have very similar signal decay. The B-scan shows two distinct areas, in which the three artery layers are noticeable in one side of the artery and the other side has attenuation at certain depth where fibrotic tissue and lipids are mixed. The lumen was color-coded based on the values of the estimated abundances as either Intimal-Thickening (green) or Fibrotic-Lipid profiles (cyan), as shown in the B-scan (Fig. 11(b)). The corresponding histology section (Fig. 11(c)) clearly indicates one Intimal-Thickening area and a fibrotic plaque with deep lipid accumulations.
The performance of the automated profile classification method assessed following a leave-one-out validation approach is summarized by the confusion matrix in Table 2. In our evaluation, the overall accuracy for this classification tests was 88.2%. The sensitivity and specificity for both the Fibrotic and Fibrotic-Lipid classes were approximately 84% and 90%, respectively. The sensitivity and specificity for both the Intimal-Thickening and Superficial-Lipid classes were 100%.
Table 2.
Profile level confusion matrix.
| Leave-One-Out Classification | |||||||
|---|---|---|---|---|---|---|---|
| IT | Fib-Lip | Fibrotic | Sup-Lipid | Sensitivity % | Specificity % | ||
| Histopathology | IT | 13 | 0 | 0 | 0 | 100 | 100 |
| Fib-Lip | 0 | 39 | 7 | 0 | 84.8 | 90.6 | |
| Fibrotic | 0 | 6 | 30 | 0 | 83.3 | 90.5 | |
| Sup-Lipid | 0 | 0 | 0 | 15 | 100 | 100 | |
Notice that in Table 2 there are a total of 110 profiles and 114 where expected from 57 samples. This method tries to find two profiles, N=2, from each sample, yet the estimated profiles are compared at each iteration of the alternating least squares to decide if they are alike so they can be considered as only one class. Thus, if a sample has only one class and the two estimated profiles are similar to certain extent, the result would be a single profile for that dataset (in Fig. 9, the two profiles did not match this criterion and, therefore, the method gave two different fibrotic profiles as result). In this database, there were 53 samples with two profiles (106 profiles) and 4 samples with only one profile; most of these cases came from datasets having only Intimal-Thickening or Fibrous tissue having similar thickness through all the artery section.
3.5. Sample classification results
In the context of sample level classification, the method evaluated whether or not a sample had accumulation of lipids. Thus, either a Superficial-Lipid profile or a Fibrotic-Lipid profile counts as Lipid-containing classification.
The visual classification provided by the IV-OCT observer was compared to the histopathological gold standard classification leading to the confusion matrix in Table 3 and exhibiting and accuracy of 80.7%. This classification showed sensitivity of 77.3% and specificity of 92.3% for detecting lipid plaques.
Table 3.
Visual sample level classification confusion matrix
| Visual Classification | |||||
|---|---|---|---|---|---|
| Lipids | No Lipids | Sensitivity % | Specificity % | ||
| Histopathology | Lipids | 34 | 10 | 77.3 | 92.3 |
| No Lipids | 1 | 12 | |||
Table 4 shows the confusion matrix for the automatic sample level classification with an accuracy of 85.6%. The sensitivity and specificity for detecting plaque with accumulation of lipids were 86.4% and 84.6%, respectively.
Table 4.
Automatic sample level classification confusion matrix
| Leave-One-Out classification | |||||
|---|---|---|---|---|---|
| Lipids | No Lipids | Sensitivity % | Specificity % | ||
| Histopathology | Lipids | 38 | 6 | 86.4 | 84.6 |
| No Lipids | 2 | 11 | |||
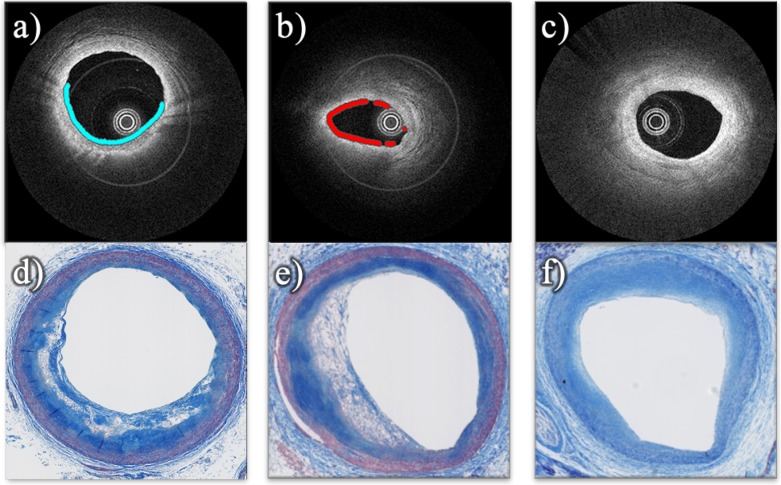
For this case, the proposed classification algorithm automatically plots a line only on a specific section of the lumen where lipids where detected, as shown in Figure 12. In these examples, the proposed algorithm draws a cyan and red line on the lumen of the arteries in the two first IV-OCT images (a and b) having Fibrotic-Lipid and Superficial-Lipid plaques build up, respectively. For these cases, it is noticeable that the histological images show white areas revealing he presence of extracellular lipids and/or foam cells. The last IV-OCT image (c) corresponding to an artery with Fibrous plaque and intimal thickening does not have any line on the lumen. Thus, no accumulation of lipids was detected. The histological image (f) illustrates a homogeneous Fibrotic plaque.
Fig. 12.
Automatic sample level classification. IV-OCT images (first row) with a plot on the section of lumen and the corresponding histological images (second row); where (a) and (b) presenting Lipid plaque, and (c) without lipids.
4. Discussion
Visual plaque characterization requires trained experts, and is complicated by variation in the tissue attenuation and speckle effects. Moreover, large data sets, consisting of hundreds of B-scans, make visual plaque classification impractical, especially in a clinical setting. Therefore, the proposed automatic algorithm could offer an improved plaque classification by analyzing entire pullbacks and assist in clinical diagnosis and therapy guidance.
Methods for texture-based plaque characterization automatically classify each pixel with intensity and texture features, which tend to mislead the correct identification of some pixels where the speckle and attenuation effect is high. On the other hand, methods correlating the backscattering and attenuation coefficients tend to misclassify mixed fibrolipid plaques. Also, the classification using these coefficients can be misinterpreted due to a lack of tissue heterogeneity and intensity calibration.
This work shows that our automated classification method can effectively identify and label A-line profiles based on morphological features. Using the polar IV-OCT images, each A-line is classified to one of the main types of tissue present in atherosclerotic tissue namely: Intimal-Thickening, Fibrotic, Superficial-Lipid, and Fibrotic-Lipid plaque.
The histological evaluation of many of the plaques of our database showed mixed pathological categories. We focused on the two most prominent histopathological categories per plaque. In all plaques included, these two most prominent histopathological categories were always included among the four categories defined for the automated classification algorithm. However, we had to exclude a few plaques in which the histology was not clear for identifying the tissue types. In addition, two samples with artifacts produced by blood in the lumen were also excluded, since the resulting blood absorption would produce atypical profiles. In the future, we could try to characterize profiles that would allow identifying the presence of blood clots.
Also, macrophages/foam-cells were co-localized with lipids accumulation, so there were present in our Lipids and Fibro-lipids plaques. Calcification and cholesterol crystals were present in a few samples included in our database, but they were not the predominant histological category. Therefore, the presence of macrophages/foam-cells, calcification and cholesterol crystals did not affect significantly the performance of our classification algorithm, in spite of the fact that we did not consider a specific category for these histopathological features. In the future, we could try to characterize profiles that would allow identifying these other histopathological features.
The detection of superficial lipid, and especially the presence of necrotic cores would be clinically relevant. However, to date, OCT has not been shown to be able to differentiate between these two categories. Although the presented algorithm may be able to be trained to identify these two types, there are not enough examples of these two categories in our data set; a larger and more complete data set would be needed to investigate this aspect.
The algorithm was programmed in MATLAB R2016a and tested in a computer with the following characteristics: Windows 7 Enterprise 64-bit, CPU Intel i7 with 6 cores at 3.2GHz, and 48GB RAM. The average processing time for a dataset with 9 B-scans is 9.5s, from which the optimization process to calculate the profile and abundances described in section 2.2 takes 9s. The average time of processing one single B-scan is 1.5s, from which the optimization process takes 1s. Thus, the convergence time of the optimization process is the bottleneck of the algorithm. It is worth mentioning that the code has not been optimized and a parallelized algorithm could allow high throughput atherosclerotic plaque characterization.
The high classification accuracy of 88% demonstrates the reliability of this method. Additionally, the high sensitivity and specificity for the detection of Fibrotic, Fibrotic-Lipid, and Superficial-Lipid plaques confirm the ability to detect lipid-containing plaques with high accuracy, and quantify this along the pullback segment. Moreover, the estimation of the abundances may give a quantitative indicator of the contribution of a profile class at each A-line. However, there is still a need for the automatic estimation of the optimal value of the regularization parameters (μ and ρ), which in this case were fixed and manually defined by the user for all datasets. Furthermore, the identification of only two class profiles when more than two classes are present in the sample, causes estimation of a combined profile that contains morphological information of mixed classes, which mislead the classification. Further improvements will focus on automatically identifying more than two class profiles and combine those that belong to the same class. Also, employment of additional morphological features combined with texture features might enhance the accuracy of the proposed method.
In a clinical setting, this algorithm will have to deal with the guide wire signal and its shadow, which was not considered in this study. This issue could be resolved by adding a third profile, N=3, to identify the guide wire features reliably. Also, other image processing approaches could be used to identify the guide wire, such as standard deviation filtering or image convolution. Moreover, since the lumen surface is flattened by the elastic transformation method, the distance from the ball-lens to the lumen surface varies between A-lines. This effect might induce signal fluctuations in the A-lines intensities. However, the attenuation by the light propagation from the ball-lens to the lumen surface will be minimal with respect to the tissue effect. Furthermore, these signal fluctuations have minor effect since the profiles are normalized during the synthesis scheme, and the classification depends on the morphology neglecting the amplitude of the profiles.
5. Conclusion
In summary, we have presented a novel computational method for automated tissue characterization of atherosclerotic plaques imaged with IV-OCT. This method is based on the modeling of each A-line of an IV-OCT data set as a linear combination of N depth profiles. The profiles are estimated by an alternating least square optimization strategy for an entire pullback segment, and are further automatically classified to a predefined tissue type. In this study, our proposed method was applied to classify each A-line on an IV-OCT B-scan as Intimal-Thickening, Fibrotic, Fibro-Lipid, or Superficial-Lipid. Such profile classification also allowed the automated classification of IV-OCT B-scans as either Lipid-containing or No-Lipid plaques (overall accuracy of 85%, compared to 80% based on visual interpretation by a trained IV-OCT reader). Our encouraging results validated on ex-vivo IV-OCT suggest that this methodology has the potential to allow high throughput atherosclerotic plaque assessment through automated tissue characterization.
Funding
National Heart, Lung, and Blood Institute (NHLBI) (1R01HL111361); National Institute of Biomedical Imaging and Bioengineering (NIBIB)(Center for Biomedical OCT Research and Translation, P41EB015903).
References and links
- 1.Virmani R., Burke A. P., Farb A., Kolodgie F. D., “Pathology of the vulnerable plaque,” J. Am. Coll. Cardiol. 47(8), C13–C18 (2006). 10.1016/j.jacc.2005.10.065 [DOI] [PubMed] [Google Scholar]
- 2.Brezinski M.E., Tearney G. J., Bouma B. E., Boppart S. A., Hee M. R., Swanson E. A., Southern J. F., Fujimoto J. G., “Imaging of coronary artery microstructure (in vitro) with optical coherence tomography,” Am. J. Cardiol. 77(1), 92–93 (1996). 10.1016/S0002-9149(97)89143-6 [DOI] [PubMed] [Google Scholar]
- 3.Yun S. H., Tearney G. J., Vakoc B. J., Shishkov M., Oh W. Y., Desjardins A. E., Suter M. J., Chan R. C., Evans J. A., Jang I. K., Nishioka N. S., de Boer J. F., Bouma B. E., “Comprehensive volumetric optical microscopy in vivo,” Nat. Med. 12, 1429–1433 (2006). 10.1038/nm1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drexler W., Liu M., Kumar A., Kamali T., Unterhuber A., Leitgeb R. A., “Optical coherence tomography today: speed, contrast, and multimodality,” J. Biomed. Opt. 19(7), 071412 (2014). 10.1117/1.JBO.19.7.071412 [DOI] [PubMed] [Google Scholar]
- 5.Sun Z., “Atherosclerosis and atheroma plaque rupture: Imaging modalities in the visualization of vasa vasorum and atherosclerotic plaques,” Sci. World J. 2014, 312764 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tearney G. J., Regar E., Akasaka T., Adriaenssens T., Barlis P., Bezerra H. G., Bouma B., Bruining N., Cho J., Chowdhary S., Costa M. A., de Silva R., Dijkstra J., Di Mario C., Dudeck D., Falk E., Feldman M. D., Fitzgerald P., Garcia H., Gonzalo N., Granada J. F., Guagliumi G., Holm N. R., Honda Y., Ikeno F., Kawasaki M., Kochman J., Koltowski L., Kubo T., Kume T., Kyono H., Lam C. C. S., Lamouche G., Lee D. P., Leon M. B., Maehara A., Manfrini O., Mintz G. S., Mizuno K., Morel M., Nadkarni S., Okura H., Otake H., Pietrasik A., Prati F., Räber L., Radu M. D., Rieber J., Riga M., Rollins A., Rosenberg M., Sirbu V., Serruys P. W. J. C., Shimada K., Shinke T., Shite J., Siegel E., Sonada S., Suter M., Takarada S., Tanaka A., Terashima M., Troels T., Uemura S., Ughi G. J., van Beusekom H. M. M., van der Steen A. F. W., van Es G., van Soest G., Virmani R., Waxman S., Weissman N. J., Weisz G., “Consensus standards for acquisition, measurement, and reporting of intravascular optical coherence tomography studies: a report from the International Working Group for Intravascular Optical Coherence Tomography Standardization and Validation,” J. Am. Coll. Cardiol. 59(12), 1058–1072 (2012). 10.1016/j.jacc.2011.09.079 [DOI] [PubMed] [Google Scholar]
- 7.Kume T., Akasaka T., Kawamoto T., Okura H., Watanabe N., Toyota E., Neishi Y., Sukmawan R., Sadahira Y., Yoshida K., ”Measurement of the thickness of the fibrous cap by optical coherence tomography,” Am. Heart. J. 152(4), 755 (2006). 10.1016/j.ahj.2006.06.030 [DOI] [PubMed] [Google Scholar]
- 8.Yabushita H., Bouma B. E., Houser S. L., Aretz H. T., Jang I., Schlendorf K. H., Kauffman C. R., Shishkov M., Kang D., Halpern E. F., Tearney G. J., “Characterization of Human Atherosclerosis by Optical Coherence Tomography,” Circulation 106(13), 1640–1645 (2002). 10.1161/01.CIR.0000029927.92825.F6 [DOI] [PubMed] [Google Scholar]
- 9.Herrero-Garibi J., Cruz-González I., Parejo-Díaz P., Jang I., “Optical Coherence Tomography: Its Value in Intravascular Diagnosis Today,” Rev. Esp. Cardiol. 63(8), 951–962 (2010). 10.1016/S0300-8932(10)70207-4 [DOI] [PubMed] [Google Scholar]
- 10.Soeda T., Uemura S., Saito Y., Mizuno K., Jang I., “Optical Coherence Tomography and Coronary Plaque Characterization,” J. Jpn. Coron. Assoc. 19(4), 307–314 (2013). 10.7793/jcoron.19.033 [DOI] [Google Scholar]
- 11.Athanasiou L.S., Exarchos T.P., Naka K.K., Michalis L.K., Prati F., Fotiadis D.I., “Atherosclerotic plaque characterization in optical coherence tomography images,” Conf. Proc. IEEE Eng. Med. Biol. Soc. 32, 4485–4488 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Prakash A., Hewko M., Sowa M., Sherif S., “Texture based segmentation method to detect atherosclerotic plaque from optical tomography images,” Proc. SPIE 8802, 88020S (2013). 10.1117/12.2032197 [DOI] [Google Scholar]
- 13.van Soest G., Goderie T., Regar E., Koljenović S., van Leenders G. L., Gonzalo N., van Noorden S., Okamura T., Bouma B. E., Tearney G. J., Oosterhuis J. W., Serruys P. W., van der Steen A. F., “Atherosclerotic tissue characterization in vivo by optical coherence tomography attenuation imaging,” J. Biomed. Opt. 15(1), 011105 (2010). 10.1117/1.3280271 [DOI] [PubMed] [Google Scholar]
- 14.Ughi G. J., Adriaenssens T., Sinnaeve P., Desmet W., D’hooge J., “Automated tissue characterization of in vivo atherosclerotic plaques by intravascular optical coherence tomography images,” Biomed. Opt. Express 4(7), 1014–1030 (2013). 10.1364/BOE.4.001014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karanasos A, Simsek C, Gnanadesigan M, van Ditzhuijzen NS, Freire R, Dijkstra J, Tu S, Van Mieghem N, van Soest G, de Jaegere P, Serruys PW, Zijlstra F, van Geuns RJ, Regar E, “OCT assessment of the long-term vascular healing response 5 years after everolimus-eluting bioresorbable vascular scaffold,” J. Am. Coll. Cardiol. 64(22), 2343–2356 (2014). 10.1016/j.jacc.2014.09.029 [DOI] [PubMed] [Google Scholar]
- 16.Gargesha M., Shalev R., Prabhu D., Tanaka K., Rollins A. M., Costa M., Bezerra H. G., Wilson D. L., “Parameter estimation of atherosclerotic tissue optical properties from three-dimensional intravascular optical coherence tomography,” J. Med. Imaging 2(1), 016001 (2015). 10.1117/1.JMI.2.1.016001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleming C. P., Eckert J., Halpern E. F., Gardecki J. A., Tearney G. J., “Depth resolved detection of lipid using spectroscopic optical coherence tomography,” Biomed. Opt. Express 4, 1269–1284 (2013). 10.1364/BOE.4.001269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nam H. S., Song J. W., Jang S.-J., Lee J. J., Oh W.-Y., Kim J. W., Yoo H., “Characterization of lipid-rich plaques using spectroscopic optical coherence tomography,” J. Biomed. Opt. 21(7), 75004 (2016). 10.1117/1.JBO.21.7.075004 [DOI] [PubMed] [Google Scholar]
- 19.van der Sijde J. N., Karanasos A., Villiger M., Bouma B. E., Regar E., “First-in-man assessment of plaque rupture by polarization-sensitive optical frequency domain imaging in vivo,” Eur. Heart. J. May 37(24), 1932 (2016). 10.1093/eurheartj/ehw179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marques O., Practical Image and Video Processing Using MATLAB (John Wiley & Sons, 2011). 10.1002/9781118093467 [DOI] [Google Scholar]
- 21.Lopes A., Touzi R., Nezry E., “Adaptive speckle filters and scene heterogeneity,” IEEE Geosci. Remote. Sens. Mag. 28(6), 992–1000 (2002). 10.1109/36.62623 [DOI] [Google Scholar]
- 22.Maity A., Pattanaik A., Sagnika S., Pani S., “A Comparative Study on Approaches to Speckle Noise Reduction in Images,” Comput. Intell. and Netw. (CINE) 5, 148–155 (2015). 10.1109/CINE.2015.36 [DOI] [Google Scholar]
- 23.Rivera M., Ocegueda O., Marroquin J. L., “Entropy-controlled quadratic Markov measure field models for efficient image segmentation,” IEEE Trans. Image Process. 16(12), 3047–3057 (2007). 10.1109/TIP.2007.909384 [DOI] [PubMed] [Google Scholar]
- 24.Rivera M., Dalmau O., “Variational Viewpoint of the Quadratic Markov Measure Field Models: Theory and Algorithms,” IEEE Trans. Image Process. 21(3), 1246–1257 (2012). 10.1109/TIP.2011.2168409 [DOI] [PubMed] [Google Scholar]
- 25.Gutierrez-Navarro O., Campos-Delgado D., Arce-Santana E., Mendez M., Jo J., “Blind end-member and abundance extraction for multi-spectral fluorescence lifetime imaging microscopy data,” IEEE J. Biomed. Health Inform. 18(2), 606–617 (2014). 10.1109/JBHI.2013.2279335 [DOI] [PubMed] [Google Scholar]
- 26.Bioucas-Dias J., Condessa F., Kovacevic J., “Alternating direction optimization for image segmentation using hidden Markov measure field models,” Proc. of SPIE-IS&T Electronic Imaging 9019, 90190P (2014). [Google Scholar]
- 27.Jaumot J., Gargallo R., de Juan A., Tauler R., “A graphical user-friendly interface for MCR-ALS: a new tool for multivariate curve resolution in matlab,” Chemometr. Intell. Lab 76(1), 101–110 (2005). 10.1016/j.chemolab.2004.12.007 [DOI] [Google Scholar]
- 28.Young F. W., de Leeuw J., Takane Y., “Regression with qualitative and quantitative variables: An alternating least squares method with optimal features,” Psychometrika 41(4), 505–529 (1976). 10.1007/BF02296972 [DOI] [Google Scholar]
- 29.Villiger M., Zhang E. Z., Nadkarni S. k., Oh W., Vakoc B. J., Bouma B. E., “Spectral binning for mitigation of polarization mode dispersion artifacts in catheter-based optical frequency domain imaging,” Opt. Express 21(14), 16353–16369 (2013). 10.1364/OE.21.016353 [DOI] [PMC free article] [PubMed] [Google Scholar]