Summary
Many eukaryotic green algae possess biophysical carbon‐concentrating mechanisms (CCMs) that enhance photosynthetic efficiency and thus permit high growth rates at low CO 2 concentrations. They are thus an attractive option for improving productivity in higher plants. In this study, the intracellular locations of ten CCM components in the unicellular green alga Chlamydomonas reinhardtii were confirmed. When expressed in tobacco, all of these components except chloroplastic carbonic anhydrases CAH3 and CAH6 had the same intracellular locations as in Chlamydomonas. CAH6 could be directed to the chloroplast by fusion to an Arabidopsis chloroplast transit peptide. Similarly, the putative inorganic carbon (Ci) transporter LCI1 was directed to the chloroplast from its native location on the plasma membrane. CCP1 and CCP2 proteins, putative Ci transporters previously reported to be in the chloroplast envelope, localized to mitochondria in both Chlamydomonas and tobacco, suggesting that the algal CCM model requires expansion to include a role for mitochondria. For the Ci transporters LCIA and HLA3, membrane location and Ci transport capacity were confirmed by heterologous expression and H14 CO 3 ‐ uptake assays in Xenopus oocytes. Both were expressed in Arabidopsis resulting in growth comparable with that of wild‐type plants. We conclude that CCM components from Chlamydomonas can be expressed both transiently (in tobacco) and stably (in Arabidopsis) and retargeted to appropriate locations in higher plant cells. As expression of individual Ci transporters did not enhance Arabidopsis growth, stacking of further CCM components will probably be required to achieve a significant increase in photosynthetic efficiency in this species.
Keywords: photosynthesis improvement, carbon‐concentrating mechanism, bicarbonate transporter, Arabidopsis thaliana, tobacco, Chlamydomonas reinhardtii
Introduction
Most plants, including the major grain crops rice and wheat, assimilate carbon using the C3 photosynthetic pathway. C3 plants rely on passive diffusion to deliver carbon dioxide (CO2) from the atmosphere (ca. 400 ppm) to the chloroplasts inside leaf mesophyll cells, wherein CO2 photoassimilation proceeds via the primary carboxylase enzyme, ribulose‐1,5‐bisphosphate carboxylase/oxygenase (RuBisCO, EC 4.1.1.39). Diffusive resistances result in a gradient (ca. 40% under high irradiance) between CO2 levels in the substomatal cavity of the leaf and the steady‐state level of dissolved CO2 in chloroplasts (10–20 μm at 25 °C) (Price et al., 2011). Here, CO2 is not saturating for RuBisCO and oxygen (O2; ca. 250 μm at 25 °C) competes at the RuBisCO active sites, resulting in both loss of assimilated carbon and nitrogen and energy consumption in the photorespiratory pathway that recycles the product of RuBP oxygenation (Sharkey, 1988). The productivity of C3 crops is thus limited by the efficiency of CO2 photoassimilation, even when grown under elevated CO2 levels (up to 650 ppm) (Long et al., 2006). Generating C3 crop plants with increased photosynthetic efficiencies is a major target for improving yields and safeguarding future food security. Strategies under consideration and development include modifying canopies to increase light interception, enhancing repair mechanisms to overcome lags associated with photoprotection, increasing the efficiency of RuBisCO and eliminating photorespiration by introducing molecular components of microbial carbon‐concentrating mechanisms (CCM) (Lin et al., 2014a,b; Long et al., 2015; Parry et al., 2013; Whitney et al., 2011; Zhu et al., 2010).
Many photosynthetic organisms including cyanobacteria, most green algae and a single group of land plants, the hornworts, have evolved biophysical CCMs that actively increase the CO2 concentration around RuBisCO, thus suppressing RuBisCO oxygenase activity and associated photorespiration. In eukaryotic algae, CCMs involve inorganic carbon (Ci) transporters at the plasma membrane and chloroplast envelope and carbonic anhydrases, which act in concert to deliver above ambient concentrations of CO2 to RuBisCO, usually within a chloroplast microcompartment called the pyrenoid. The pyrenoid is mainly composed of densely packaged RuBisCO (Engel et al., 2015). Whilst not all algae with a CCM have a pyrenoid, the microcompartment enhances the efficiency of CO2 assimilation (Morita et al., 1998). Theoretical modelling approaches have demonstrated the requirement for a pyrenoid in algal systems (Badger et al., 1998) and shown that some form of microcompartment containing RuBisCO would also be needed for a successful CCM in higher plant systems (Price et al., 2013).
The best‐characterized algal CCM is that of the model green alga Chlamydomonas reinhardtii (Chlamydomonas throughout). To date, a large number of molecular components have been implicated in the Chlamydomonas CCM through mutant screens, transcriptomic studies and functional homology with components in other photosynthetic organisms (Brueggeman et al., 2012; Fang et al., 2012; Meyer and Griffiths, 2013). At least 14 genes are thought to be important in maintaining a fully functional CCM under ambient or below ambient CO2 concentrations (see Wang et al. (2015) for review). Briefly, these include five Ci transporters, four CAs, two pyrenoid peripheral proteins, a putative methyl transferase and two nuclear transcription regulators. The precise function and importance of these components remains only partly understood. In addition, the Chlamydomonas CCM requires a RuBisCO that can be targeted to the pyrenoid, a property known to be dependent on sequence elements of the RuBisCO small subunit (Genkov et al., 2010; Meyer et al., 2012).
To examine the feasibility of enhancing photosynthetic efficiency in C3 plants by introducing CCM components from Chlamydomonas, we chose ten components that are considered essential or potentially important for CCM functionality (Table 1). These included intracellular components that mediate the transport and conversion of Ci from the external environment to the active sites of RuBisCO. LCI1 and HLA3 are putative Ci transporters reportedly in the plasma membrane; LCIA, CCP1 and CCP2 are putative chloroplast envelope Ci transporters, although only the former has been localized in vivo. CAH1, CAH3 and CAH6 are carbonic anhydrases, thought to be located in the periplasmic space (CAH1), the thylakoid lumen (CAH3) and the chloroplast stroma (CAH6). LCIB and LCIC are proteins of unclear function that are reported to surround the pyrenoid when CO2 becomes limiting.
Table 1.
Chlamydomonas CCM genes used in this study. Locus name refers to the gene ID as supplied by Phytozome v5.5 (http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Creinhardtii)
| Gene | Locus name | Protein length, size of precursor | Putative function in native algal CCM | Examples of experimental evidence for function | Mutant phenotype |
|---|---|---|---|---|---|
| HLA3 | Cre02.g097800 | 1325 aa, 147 kDa | Ci uptake into cytosol | RNAi lines grown under alkaline conditions (main Ci species is ) have a HCR (1); synergistic effect with LCIA (1;2); 14Ci uptake assay (2;3); affinity for Ci when controlling gene expression with dTALE (3); + work presented here | Reduction in Ci accumulation when CO2 <0.02%; HCR |
| LCI1 | Cre03.g162800 | 192 aa, 21 kDa | Ci uptake into cytosol | 14Ci uptake assay (4) | No published mutant; overexpression in CCM regulatory mutant promotes uptake |
| LCIA | Cre06.g309000 | 336 aa, 35 kDa | Ci transport from cytosol to stroma | Synergistic effect with HLA3 (2;3); electrophysiology in Xenopus oocytes (5); reduced affinity for Ci at alkaline pH (6); + work presented here | Reduction in Ci accumulation when CO2 <0.02%; HCR |
| CCP1 | Cre04.g223300 | 358 aa, 38 kDa | Ci transport from cytosol to stroma | RNAi lines support role in Ci transport (7); putative localization inferred bioinformatically | No published mutant |
| CCP2 | Cre04.g222750 | 355 aa, 38 kDa | Ci transport from cytosol to stroma | RNAi lines support role in Ci transport (7); putative localization inferred bioinformatically | No published mutant |
| CAH1 | Cre04.g223100 | 377 aa, 42 kDa | CO2 and at cell surface | Absence of growth effect in the presence of membrane impermeable CA inhibitors (9) | No apparent deleterious effect on growth |
| CAH6 | Cre12.g485050 | 264 aa, 28 kDa | Recapture of CO2 leaking from the pyrenoid | No experimental evidence of function; putative role in CCM inferred from putative localization | No published mutant |
| CAH3 | Cre09.g415700 | 310 aa, 33 kDa | Terminal dehydration of to CO2, to saturate RuBisCO in the pyrenoid | Low CO2‐induced phosphorylation relocalizes CAH3 preferentially to pyrenoid tubules (13); alternative CCM‐unrelated function (regulation of water oxydation at PSII) has been proposed (14) | Mutation produces overaccumulation of Ci |
| LCIB | Cre10.g452800 | 448 aa, 48 kDa | CO2 uptake or trapping of stromal CO2, pyrenoid localization | Synergetic role with Ci pumps (lcia/lciB double mutant lethal when CO2 <0.02%) (1); MS‐identification of LCIB‐FLAG pull‐down and gel filtration showed that LCIB‐C form a 360 kDa hetero‐hexamer, which localizes around the pyrenoid when CO2 <0.02% (16) | Lethal under air‐level CO2 but rescued when CO2 <0.02% |
| LCIC | Cre06.g307500 | 443 aa, 48 kDa | CO2 uptake or trapping of stromal CO2, pyrenoid localization | MS‐identification of LCIB‐FLAG pull‐down and gel filtration showed that LCIB‐C form a 360 kDa hetero‐hexamer, which localizes around the pyrenoid when CO2 <0.02% (16) | No published mutant |
| Gene | Locus name | Functional annotation | Location as per latest CCM model (Wang et al., 2015) | Examples of experimental evidence for location; work presented here for all ten genes | References |
|---|---|---|---|---|---|
| HLA3 | Cre02.g097800 | ABC transporter superfamily | Plasmamembrane | Immunofluorescence (2); immunoblot on membrane fractions (2;3) | (1) Duanmu et al. (2009a); (2) Yamano et al. (2015); (3) Gao et al. (2015) |
| LCI1 | Cre03.g162800 | Unknown | Plasmamembrane | Immunofluorescence + GFP fusion + immunoblot on membrane fractions (4) | (4) Ohnishi et al. (2010 |
| LCIA | Cre06.g309000 | Formate/nitrite transporter | Chloroplast membrane | Immunofluorescence (2;6) | (5) (Mariscal et al. (2006); (6) Wang and Spalding, (2014a) |
| CCP1 | Cre04.g223300 | Mitochondrial carrier protein | Chloroplast membrane | Immunoblot on membrane fractions (8)); + work presented here | (7) Pollock et al. (2004); (8) Ramazanov et al. (1993) |
| CCP2 | Cre04.g222750 | Mitochondrial carrier protein | Chloroplast membrane | Immunoblot on membrane fractions (8); + work presented here | (7) Pollock et al. (2004); (8) Ramazanov et al. (1993) |
| CAH1 | Cre04.g223100 | α carbonic anhydrase | Periplasmic space | Cell wall lysis + CA assay on supernatant (10); immunogold (11) | (9) Moroney and Tolbert, (1985); (10) Kimpel et al. (1983); (11) Ynalvez et al. (2008) |
| CAH6 | Cre12.g485050 | β carbonic anhydrase | Stroma | Immunogold (12) | (12) Mitra et al. (2004) |
| CAH3 | Cre09.g415700 | α carbonic anhydrase | Thylakoid lumen | Immunogold (12;15) | (13) Blanco‐Rivero et al. (2012); (14) Villarejo et al. (2002); (15) Markelova et al. (2009) |
| LCIB | Cre10.g452800 | Unknown | Stroma | Immunogold (16); GFP fusion (16,17); immunofluorescence (6;18) | (16) Yamano et al. (2010); (17) Yamano et al. (2014); (18) Wang and Spalding (2014b) |
| LCIC | Cre06.g307500 | Unknown | Stroma | Immunogold + immunofluorescence (16) |
CA, carbonic anhydrase; Ci, inorganic carbon; HCR: high CO2‐requiring phenotype, a nonlethal mutation rescued by growth under elevated CO2 (2–5% [v/v]).
We first expressed fluorescently tagged versions of all these proteins in Chlamydomonas cells to obtain definitive information about their locations and then expressed them transiently in tobacco (Nicotiana benthamiana L.) leaves. With one exception, proteins localized to identical compartments in the two organisms. Subsequent analyses focussed on the putative Ci transporters LCIA and HLA3, which have been shown to cooperatively drive bicarbonate uptake from the extracellular environment to the chloroplast stroma (Yamano et al., 2015). We showed that both function as Ci transporters when expressed in the outer membrane of Xenopus oocytes. We also expressed these proteins stably in transgenic Arabidopsis plants, which grew as well as wild‐type plants. Our results show that CCM components from Chlamydomonas can be expressed in appropriate locations in higher plant cells without compromising growth, although – consistent with modelling predictions – additional elements of the algal CCM will need to be co‐expressed to achieve enhanced productivity.
Results
Subcellular localization of native CCM components in Chlamydomonas
The locations of CCM components in Chlamydomonas were investigated by transforming Chlamydomonas cells with constructs encoding these proteins fused to a fluorescent tag (Venus) at the C‐terminus (Figures 1a and S1a). Full‐length open reading frames were cloned from genomic DNA and constitutively expressed from the PsaD promoter. Most of the proteins had the subcellular locations expected from previous studies (Table 1). LCIA: Venus was confined to the chloroplast envelope; CAH3: Venus, LCIB: Venus and LCIC: Venus were in the chloroplast, with LCIC: Venus and LCIB: Venus producing the distinctive circular pattern around the pyrenoid in CO2‐starved cells. LCI1: Venus and HLA3: Venus were in the plasma membrane. In the absence of an available Chlamydomonas strain expressing tagged CAH1, we examined the location of the structurally related isozyme CAH2 (contiguous to CAH1 on chromosome 4, probably resulted from a gene duplication event, 91.8% identical amino acid sequences) (Fujiwara et al., 1990). CAH2: Venus was at the cell periphery, consistent with the expected periplasmic locations of CAH1 and CAH2 (Ynalvez et al., 2008). Unexpectedly, signals for CCP1: Venus and CCP2: Venus showed punctate subcellular localization, consistent with location in mitochondria. Furthermore, the location of fluorescence for CCP1: Venus and CCP2: Venus overlapped with that of a mitochondrial marker dye (Mitotracker Red CMXRos), indicating that both of these putative Ci transporters were located in mitochondria (Figure S2). There were no obvious signals for CAH6: Venus inside the Chlamydomonas cell (not shown).
Figure 1.
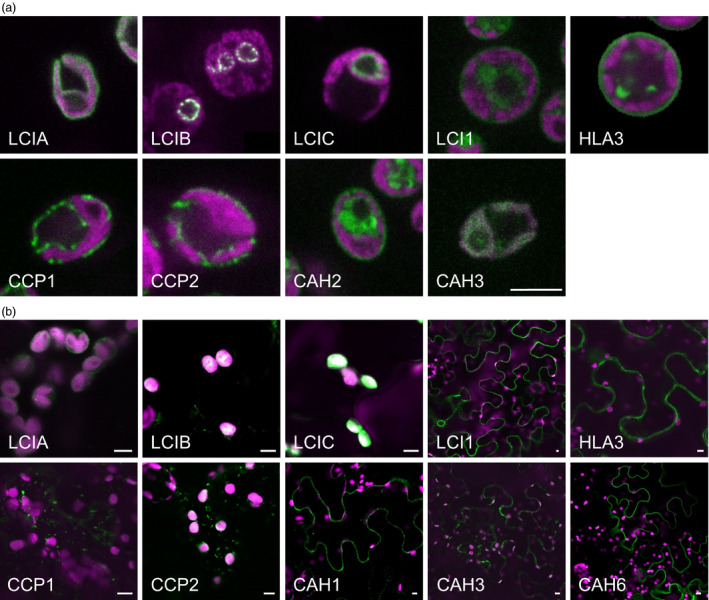
Expression of fluorescent‐tagged CCM components in Chlamydomonas and tobacco. Expression of Venus‐fused CCM components in Chlamydomonas reinhardtii (a). Expression in tobacco of GFP‐fused CCM components from Chlamydomonas (b). Green and purple signals are Venus or GFP fluorescence and chlorophyll autofluorescence, respectively. Overlaid images of these signals are shown: overlaps are white. Scale bar = 5 μm (all 5 μm for Chlamydomonas images). For images of separate signals see Figure S1.
Chlamydomonas CCM proteins can be expressed in tobacco leaves
The CCM components localized in Chlamydomonas cells were selected for expression in tobacco leaves. Binary expression vectors carrying each CCM gene individually were generated by PCR amplification of cDNA and subsequent Gateway cloning (Karimi et al., 2002). Gene expression in tobacco was under the control of the constitutive 35S promoter and nopaline synthase (nos) terminator. Stop codons were removed to allow in‐frame C‐terminal fusion to a sequence encoding GFP. For CAH1, the N‐terminal sequence (17 aa) was replaced with a leader sequence (22 aa) from tobacco, as described by Roberts and Spalding (1995), to facilitate processing and secretion to the apoplast.
The GFP‐fused CCM components were expressed transiently in tobacco leaves by agro‐infiltration. The locations of eight of the components were consistent with the demonstrated location in Chlamydomonas (Figures 1b and S1b). Fluorescent signals for LCIA: GFP were in the chloroplast envelope, and LCIB: GFP and LCIC: GFP signals were stromal. LCI1: GFP, HLA3: GFP and CAH1: GFP were at the cell periphery. The location of fluorescence for LCI1: GFP and HLA3: GFP overlapped with that of an integral plasma membrane transporter protein (NPSN12, AT1G48240) fused to mCherry (Geldner et al., 2009), indicating that both of these putative Ci transporters were associated with the plasma membrane (Figure 2). Although co‐expression of CAH1: GFP with NPSN12: mCherry indicated that CAH1 was located at the cell periphery, we could not resolve whether the enzyme, which is periplasmic in Chlamydomonas, was expressed discretely in the intercellular space. The fluorescence signals for CCP1: GFP and CCP2: GFP appeared predominantly in numerous discrete structures much smaller than chloroplasts. This distribution is consistent with the locations of these proteins in mitochondria, as was the case in Chlamydomonas (Figure 1a). Furthermore, fluorescence signals for CCP1: GFP and CCP2: GFP overlapped with that of a mitochondrial marker (the targeting sequence of yeast cytochrome oxidase IV [COX4] fused to mCherry [Nelson et al., 2007]), indicating that both of these putative Ci transporters were associated with the mitochondria (Figure 2).
Figure 2.
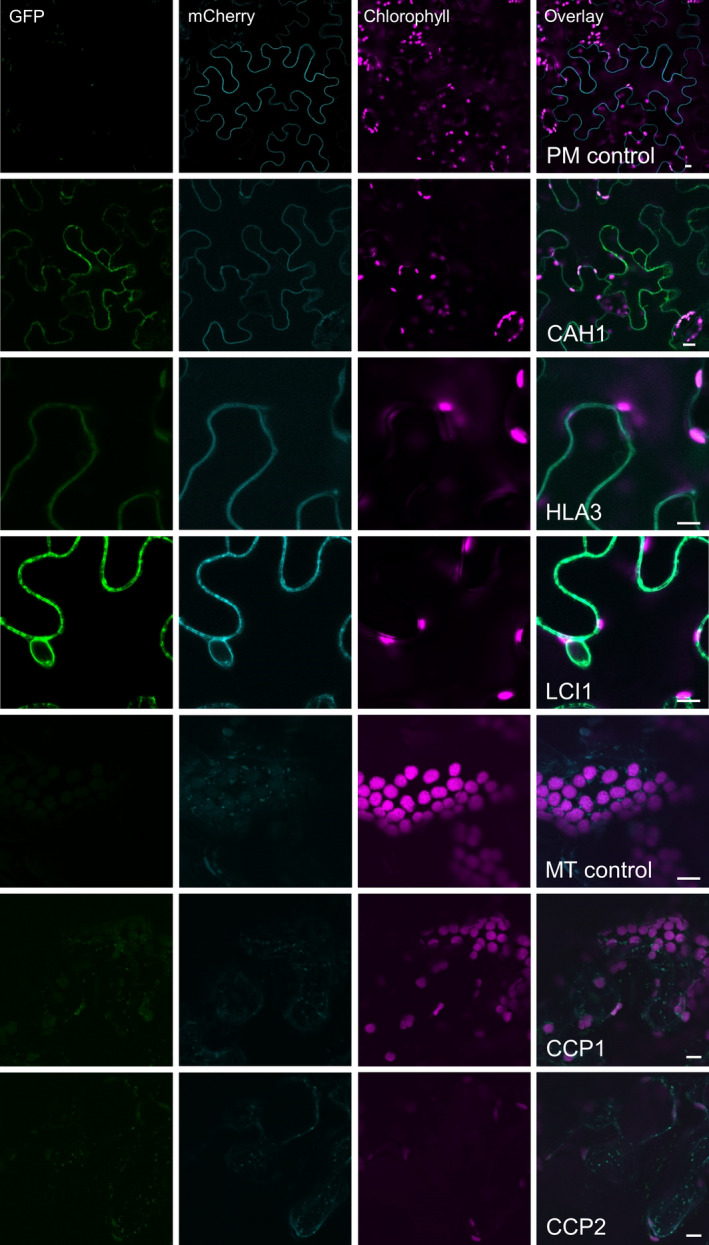
Co‐expression of GFP‐fused CCM components with a mCherry‐fused plasma membrane transporter NPSN12 or a known mitochondrial marker (the targeting sequence of yeast cytochrome oxidase IV [COX4] fused to mCherry) in tobacco. Purple, green and cyan signals are chlorophyll autofluorescence, GFP and mCherry fluorescence, respectively. Overlaid images of these signals are shown: overlaps of GFP and mCherry are pale green. PM, plasma membrane; MT, mitochondria. Scale bar = 10 μm.
The fluorescent signal for CAH3: GFP was present in both cytosol and chloroplasts when this fusion protein was expressed in tobacco. In contrast, CAH6: GFP appeared to be located exclusively in the cytosol (Figure 1b). Our own and previous data suggest that CAH3 is chloroplastic in Chlamydomonas and probably contained within the thylakoid lumen where it is suggested to play a pivotal role in the supply of CO2 to RuBisCO (Figure 1) (Blanco‐Rivero et al., 2012; Duanmu et al., 2009b; Karlsson et al., 1998; Sinetova et al., 2012). CAH6 has a predicted chloroplast transit peptide (TP), and there is some evidence for a chloroplast location in Chlamydomonas (Mitra et al., 2004). However, our CAH6: Venus protein did not have a clear intracellular location. Thus, CAH3 appears to be mistargeted when expressed in tobacco, and there is insufficient information on CAH6 to determine whether this protein is correctly localized in tobacco.
Modification of target peptides for direction of CCM components to tobacco chloroplasts
We tested whether plasma membrane localized Ci transporters from Chlamydomonas could be retargeted to the chloroplast envelope in tobacco leaves. Screening of the plant membrane protein database (Aramemnon, http://aramemnon.botanik.uni-koeln.de/) identified a chloroplast envelope transporter belonging to the ATP‐binding cassette (ABC) superfamily: ABCI13 of Arabidopsis (AT1G65410). The predicted N‐terminal TP of ABCI13 (ABC‐TP, 60 aa) was attached to the N‐termini of HLA3 and LCI1. As controls, we used constructs encoding a Chlamydomonas protein: GFP fusion already shown to localize to the tobacco chloroplast envelope (LCIA, see above) that were modified either to remove the TP or to replace it with the ABC‐TP.
When expressed transiently in tobacco, LCIA: GFP lacking the predicted native LCIA‐TP (73 aa) (Miura et al., 2004) localized to the cytosol instead of the chloroplast (Figures 3 and S3). Chloroplast envelope targeting was recovered with a N‐terminal ABC‐TP, similarly to full‐length LCIA: GFP. This result showed that the ABC‐TP could target some transporter proteins to the chloroplast envelope. However, the addition of the ABC‐TP to LCI1: GFP or to HLA3: GFP was not sufficient to retarget these two proteins to the chloroplast envelope. The fluorescence signal for ABC‐TP: LCI1: GFP indicated that the protein was in the chloroplast stroma, whereas the signal for ABC‐TP: HLA3: GFP was in the cytosol.
Figure 3.
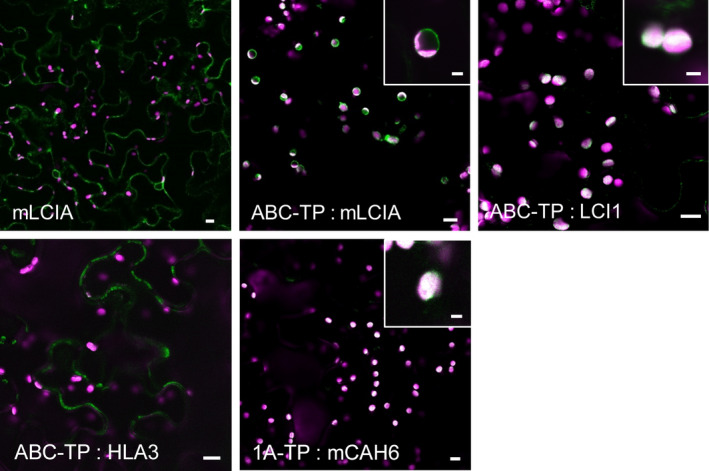
Expression of GFP‐fused CCM components carrying native Arabidopsis chloroplast transit peptides in tobacco. Green and purple signals are GFP fluorescence and chlorophyll autofluorescence, respectively. Overlaid images of these signals are shown: overlaps are white. 1A‐TP, RuBisCO small subunit RBCS1A (AT1G67090) transit peptide; ABC‐TP, ABC transporter ABCI13 (AT1G65410) transit peptide; mCAH6, mature CAH6; mLCIA, mature LCIA. Main image scale bar = 10 μm, inset image scale bar = 3 μm. For images of separate signals see Figure S3.
Having found that CAH6: GFP did not localize to the chloroplast stroma, we replaced the native TP with the TP of the Arabidopsis RuBisCO small subunit, RBCS1A (AT1G67090). By placing the first 80 aa of RBCS1A (1A‐TP) upstream of the mature CAH6 (mCAH6) to generate 1A‐TP: mCAH6: GFP, the CAH6: GFP protein was retargeted to the chloroplast stroma (Figure 3). Thus, 1A‐TP is a suitable sequence to direct the localization of soluble Chlamydomonas proteins to the stroma.
LCIA and HLA3 are located in the plasma membrane in Xenopus oocytes and increase Ci uptake rates
To investigate the putative function of LCIA and HLA3 as transmembrane Ci transporters, we expressed these proteins in Xenopus oocytes, with or without N‐terminal GFP fusions. Oocytes injected with mRNA of either mature LCIA: GFP (mLCIA; lacking the N‐terminal TP) or HLA3: GFP displayed a fluorescent signal on the cell surface after 3 d, indicating protein expression and incorporation into the plasma membrane (Figure 4a). mRNA‐injected cells were assayed for Ci uptake using H14CO3 ‐ as a tracer. Oocytes transformed with mLCIA: GFP or HLA3: GFP accumulated 2.0‐ and 2.7‐fold more 14C than water‐injected controls, respectively (Figure 4b). The presence of a GFP‐tag had no adverse effect on H14CO3 ‐ uptake.
Figure 4.
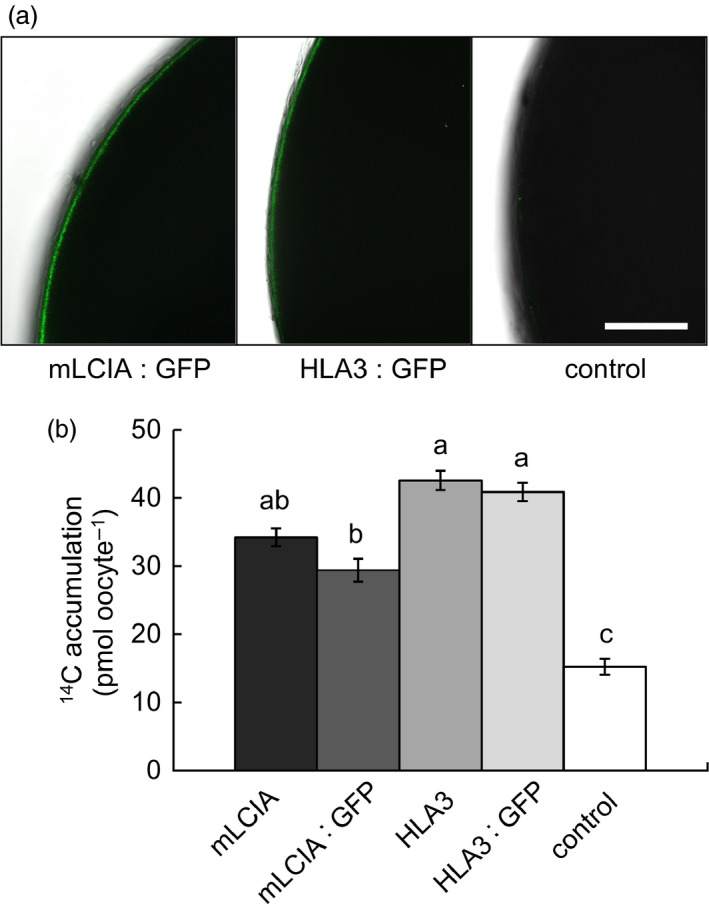
Chlamydomonas CCM components LCIA and HLA3 facilitate increased accumulation of inorganic carbon in Xenopus oocytes. Confocal images of oocytes expressing GFP fused to mature LCIA (LCIA lacking a chloroplast transit peptide, mLCIA) or HLA3 3 d after injection (a). 14C accumulation in oocytes expressing mLCIA or HLA3 either untagged or fused to GFP following 10‐min incubation in MBS containing 0.12 mM NaH14 CO 3 (b). Values are means of measurements on 20 oocytes; bars are means ± standard error (SE). Letters above the bars indicate a difference or between values; where a, b and c indicate significant difference (P < 0.05) as determined by analysis of variance (ANOVA) followed by Tukey's honestly significant difference (HSD) tests.
LCIA and HLA3 express in appropriate locations in Arabidopsis leaf cells following stable transformation
Following transformation by floral dip, three Arabidopsis thaliana (ecotype Columbia; Col‐0) homozygous T3 lines with stable expression of either LCIA: GFP or HLA3: GFP were selected for further study. Both fusion proteins resulted in fluorescent signals in the same subcellular locations as in tobacco leaves (Figures 5a and S4). Leaf proteins were separated on SDS‐PAGE and probed after blotting with a commercial antibody raised against GFP. Polypeptides corresponding to 54 kDa for LCIA: GFP and 170 kDa for HLA3: GFP were resolved (Figure 5b). These masses are consistent with those expected for GFP (27 kDa) fusions of LCIA after TP cleavage (27.5 kDa) and HLA3 (147 kDa) (Yamano et al., 2015).
Figure 5.
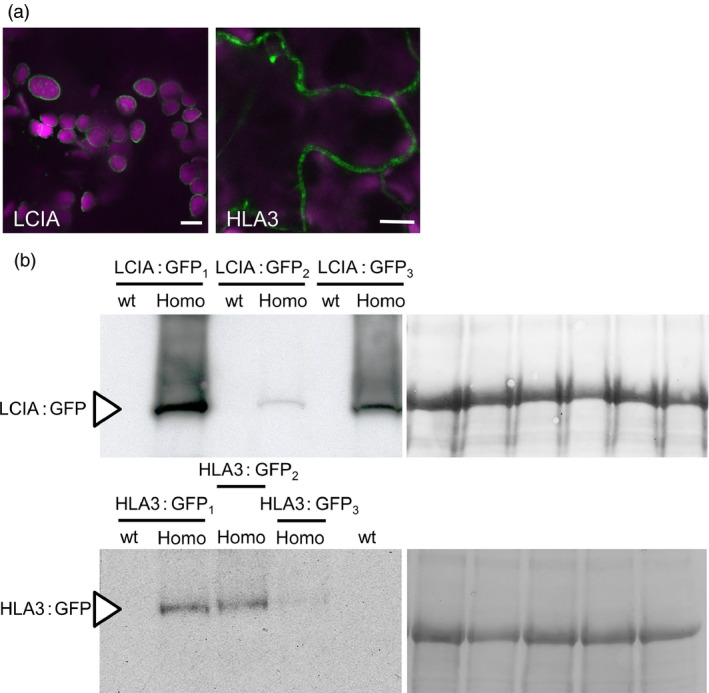
Stable expression of LCIA: GFP and HLA3: GFP in Arabidopsis. Representative confocal images of LCIA and HLA3 fused to GFP (a). Green and purple signals are GFP fluorescence and chlorophyll autofluorescence, respectively. Overlaid images of these signals are shown: overlaps are white. Scale bar = 10 μm. For images of separate signals see Figure S4. Immunoblots of rosette extracts (10 μg protein) from LCIA: GFP‐ and HLA3: GFP‐expressing lines probed with an antibody against GFP (b). LCIA: GFP is present in three separate homozygous T3 insertion lines (LCIA: GFP 1‐3), but not in segregating wild‐type lines. HLA3: GFP is visible in HLA3: GFP 1‐3 but not in the segregating wild‐type for HLA3: GFP 1 or a wild‐type equivalent for HLA3: GFP 2 and HLA3: GFP23. LCIA: GFP and HLA3: GFP have approximate masses of 54 and 170 kDa, respectively (arrow). Ponceau stains of each blot (right) show the band attributable to the RuBisCO large subunit (RbcL, 55 kD) as a loading control.
Transgenic Arabidopsis have normal growth and photosynthetic characteristics
Wild‐type plants and LCIA: GFP‐ or HLA3: GFP‐expressing lines were grown together under ambient CO2 (ca. 400 μmol/mol) and a light intensity of 100 μmol photons/m2/s (Figure 6a). Growth rate was compared by measuring rosette expansion, fresh weight and dry weight. No differences were observed between wild‐type and transgenic lines. Furthermore, there was no difference in leaf chlorophyll content (Figure S5). To investigate whether the presence of algal Ci transporters affected growth rate under conditions in which chloroplastic CO2 concentration is expected to be a major limitation on photosynthesis, plants were grown under low CO2 (250 μmol/mol) and high light (350 μmol photons/m2/s) (Figure 6b). All genotypes had higher growth rates and lower specific leaf areas under these conditions (Figure S6), but again, there were no significant differences in growth between wild‐type and transgenic lines.
Figure 6.
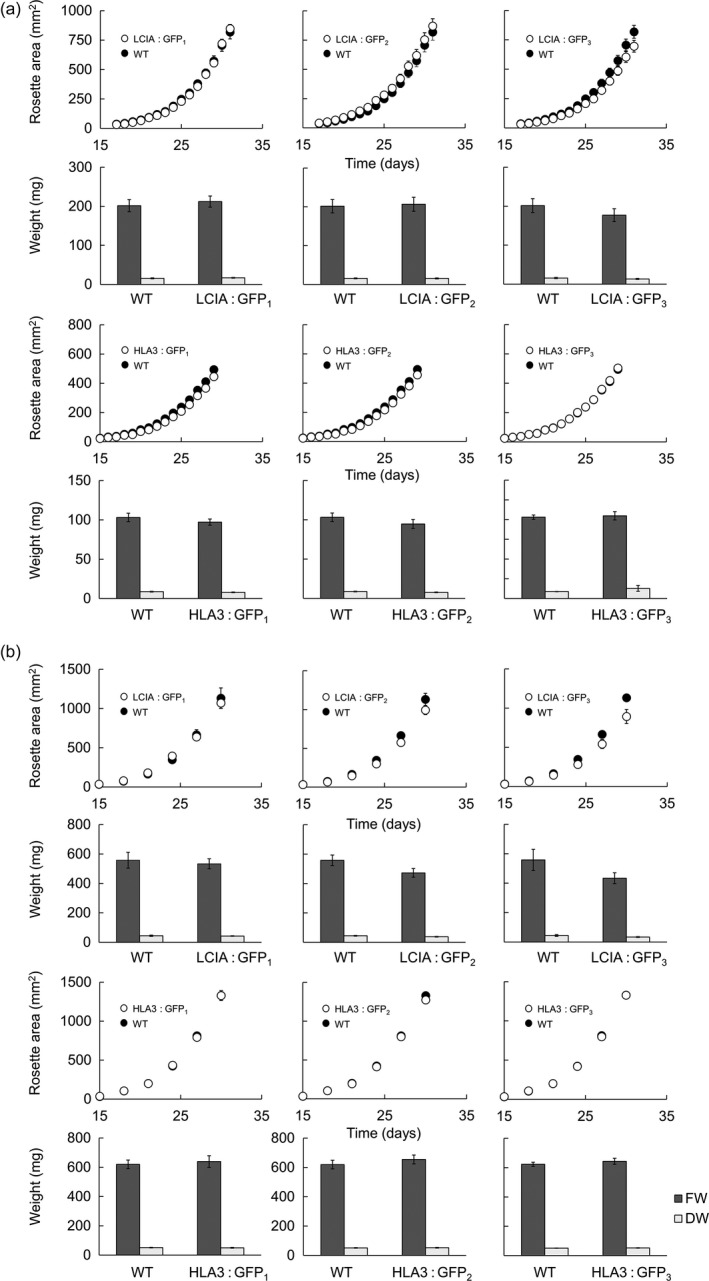
Growth of phenotypes in different environmental conditions of transgenic Arabidopsis plants expressing LCIA or HLA3. Plants were grown under ambient CO 2 (ca. 400 μmol/mol) and 100 μmol photons/m2/s (a) or low CO 2 (250 μmol/mol) and 350 μmol photons/m2/s (b). Growth rates (1st and 3rd row) and fresh weight (FW) and dry weight (DW) (2nd and 4th row) are shown for LCIA and HLA3, respectively. HLA3 transgenic lines had a lower FW and DW compared to LCIA when grown under ambient CO 2, as plants were harvested slightly earlier (at 29 days vs 31 days). All plants grown under low CO 2 were harvested at 30 days. Values are the means ± SE of measurements made on 24 rosettes.
We checked whether expression of LCIA: GFP or HLA3: GFP affected the steady‐state rate of photosynthetic CO2 assimilation at ambient CO2 (A n) (Table 2). We also measured the relationship between the rate of photosynthesis and the CO2 concentration in the leaf substomatal cavity (A/C i, Figure 7) and used this to infer other photosynthetic parameters including the diffusion of CO2 from substomatal cavity to chloroplast (mesophyll conductance, g m) (Griffiths and Helliker, 2013) and the maximum RuBisCO carboxylation rate (V c,max). There were no significant differences in any of the parameters between transgenic lines expressing LCIA: GFP or HLA3: GFP and wild‐type plants.
Table 2.
Photosynthetic parameters determined from gas exchange analysis of LCIA or HLA3 transgenic plants. Values are the mean ± SE of measurements made on four leaves, each from a different plant (as shown in Figure 7)
| Wild type | HLA3: GFP1 | HLA3: GFP2 | HLA3: GFP3 | wild type | LCIA: GFP1 | LCIA: GFP2 | LCIA: GFP3 | |
|---|---|---|---|---|---|---|---|---|
| A n (μmol CO2/m2/s) | 9.4 ± 0.6 | 11 ± 0.9 | 10 ± 0.8 | 9.9 ± 0.8 | 9.7 ± 0.2 | 9.4 ± 0.5 | 9.7 ± 1.2 | 8.8 ± 0.2 |
| g s (mol CO2/m2/s) | 0.21 ± 0.04 | 0.29 ± 0.03 | 0.21 ± 0.03 | 0.25 ± 0.04 | 0.26 ± 0.03 | 0.24 ± 0.04 | 0.25 ± 0.05 | 0.24 ± 0.01 |
| g m (mol CO2/m2/s) | 0.047 ± 0.003 | 0.048 ± 0.005 | 0.047 ± 0.003 | 0.044 ± 0.005 | 0.04 ± 0.001 | 0.038 ± 0.002 | 0.04 ± 0.004 | 0.034 ± 0.001 |
| V c,max (μmol CO2/m2/s) | 30 ± 3.2 | 30.4 ± 2.4 | 29.3 ± 1.8 | 27.3 ± 2.2 | 28.7 ± 1.1 | 29.2 ± 2.8 | 31.4 ± 2.3 | 27 ± 0.7 |
| J max (μmol e‐/m2/s) | 64.5 ± 4.8 | 71.6 ± 5.6 | 67 ± 4.7 | 64.3 ± 5.2 | 64.5 ± 1.9 | 65.1 ± 4.6 | 69.5 ± 6.6 | 61.5 ± 1.1 |
| Γ (μmol CO2/mol) | 39.5 ± 2.5 | 44 ± 2.6 | 37.9 ± 1.4 | 36.1 ± 2.6 | 33.2 ± 0.6 | 31.5 ± 2.4 | 32.9 ± 0.9 | 29.1 ± 1 |
| Initial slope (A n/C i) | 0.044 ± 0.002 | 0.046 ± 0.004 | 0.044 ± 0.003 | 0.043 ± 0.004 | 0.038 ± 0.001 | 0.035 ± 0.002 | 0.038 ± 0.005 | 0.036 ± 0.001 |
A n, net photosynthesis at ambient CO2; g s, stomatal conductance to CO2; g m, mesophyll conductance to CO2; V c,max, maximum velocity of RuBisCO carboxylation; J max, maximum capacity of electron transport; Γ, CO2 compensation point.
ANOVA revealed that there were no statistically significant differences between samples (P < 0.05).
Figure 7.
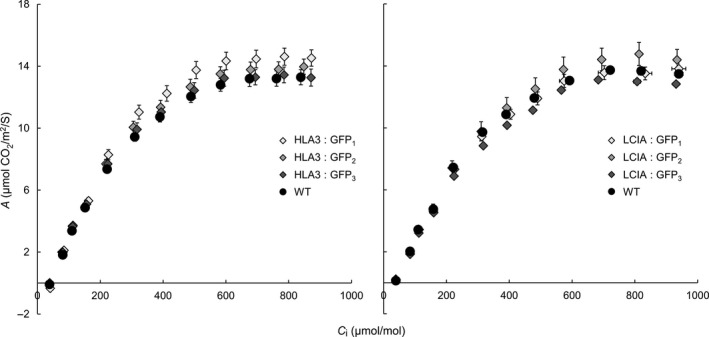
Photosynthetic responses of transgenic plants. Photosynthetic rates were determined as a function of increasing substomatal CO 2 concentrations (A/C i) at saturating light levels (1500 μmol photons/m2/s). Each curve represents the means ± SE of values from four leaves, each on a different plant.
Discussion
The introduction of a microbial eukaryotic CCM into crop plants will require robust methods to ensure that the proteins of interest perform their intended function in the foreign host system. To that effect, we developed a pipeline to (i) determine the native subcellular location in Chlamydomonas, (ii) test the targeting efficiency of constructs by transient expression in tobacco and (iii) provide a platform for physiological characterization of transgenic lines by stably transforming Arabidopsis. Through this approach, we showed that eight CCM components from Chlamydomonas can be successfully transferred into appropriate locations in leaves of higher plants. Confirmation of subcellular localization of CCM components is an important step in systematic introduction of a fully operational CCM into higher plants (Pengelly et al., 2014). For example, successful expression and assembly of components of the cyanobacterial carboxysome microcompartment in tobacco chloroplasts will help to guide future attempts to assemble the cyanobacterial CCM into higher plants (Lin et al., 2014a,b). In contrast, transfer of ictB, a putative cyanobacterial CCM component, into Arabidopsis and tobacco plants has resulted in enhanced biomass accumulation (Lieman‐Hurwitz et al., 2003; Simkin et al., 2015), but lack of information on ictB protein structure, function and cellular localization in transgenic plants limits our understanding of this growth response (Simkin et al., 2015).
The need to establish robustly the native location of CCM components, prior to incorporation into higher plants, is further supported by our work on two putative chloroplast membrane transporters. The suggestion that CCP1 and CCP2 are in the chloroplast envelope was based on their enrichment in chloroplast preparations and in preparations of envelope membranes (Mason et al., 1990; Ramazanov et al., 1993). The possibility of a mitochondrial location was not explicitly examined in these studies. Here, our robust co‐expression approach revealed that tagged CCP1/2 proteins were associated with mitochondria rather than the chloroplast envelope in both Chlamydomonas and tobacco (Figures 1 and 2; Figures S1 and S2). This location is also consistent with the high degree of similarity between CCP1/2 and the mitochondrial carrier protein superfamily (Pollock et al., 2004; Pfam 00153; KOG0758). The expression of CCP1 and CCP2 is strongly induced by low CO2, both genes are under the control of the CCM ‘master switch’ Cia5/Ccm1, and reducing CCP1/2 expression (through an RNAi approach) resulted in slower growth rates at low CO2. However, cells with reduced CCP1/2 proteins did not lose CCM or photosynthetic capacity at low CO2 (Pollock et al., 2004). Taken together, these results suggest that transport across mitochondrial membranes plays an important role in coordinating the CCM with growth‐related metabolism. In Chlamydomonas, mitochondria are known to relocate from a central position within the cup of the chloroplast to a peripheral position close to the plasma membrane when cells grown in high CO2 are exposed to limiting CO2 (Geraghty and Spalding, 1996). It has been suggested that mitochondrial relocation may be important either for energization of plasma membrane Ci transporters (to date, plasma membrane HLA3 is the only ATP‐dependent Ci transporter candidate) or for scavenging glycolate produced in photorespiration during acclimation to limiting CO2 (Spalding, 2009). A better understanding of the role of CCP1/2 in Chlamydomonas will inform future engineering strategies for higher plants.
The location of the carbonic anhydrases CAH3 and CAH6 in tobacco also differed from predicted results (Figure 2). The partial cytosolic localization of CAH3 is likely an effect of the failure of the higher plant import machinery to recognize the dual signalling peptide of this protein. In Chlamydomonas (Figure 1), results were consistent with location of the protein in the thylakoid lumen (Duanmu et al., 2009b; Sinetova et al., 2012). In particular, the presence of fluorescence in the centre of the pyrenoid is consistent with the suggestion that CAH3 is concentrated in the transpyrenoidal thylakoid tubules following low CO2‐induced phosphorylation (Figure 1) (Blanco‐Rivero et al., 2012). In contrast, there is no clear evidence of a role for CAH6 in the Chlamydomonas CCM, and very little is known about its function. CAH6 represents the only apparent stromal carbonic anhydrase activity and has been adopted into the current CCM model as a speculative mechanism for Ci uptake and/or recovery of CO2 escaping from the pyrenoid (Wang et al., 2015; Yamano et al., 2010). Evidence for chloroplast location in Chlamydomonas is based on a predicted stroma‐signalling peptide and on immunogold labelling (Mitra et al., 2004), but we did not observe an intracellular location for a CAH6: Venus protein.
Our attempts to relocate CCM components in tobacco cells by modifying targeting sequences were successful for some components. CAH6 could be redirected from the cytosol to the chloroplast stroma in tobacco by removal of the predicted TP and fusion to the TP of the Arabidopsis RuBisCO small subunit 1A, 1A‐TP (Figure 3). The plasma membrane protein LCI1 could be relocated to the chloroplast by fusion with the TP of an Arabidopsis chloroplast envelope transporter, and the same TP could substitute for the native TP of the Chlamydomonas chloroplast envelope protein LCIA. However, this TP did not redirect the plasma membrane protein HLA3 to the chloroplast envelope. Moving larger transmembrane proteins like HLA3 to the chloroplast may require further modifications to remove potential competing signal motifs and to ensure correct orientation in the case of directional channels.
The putative Ci transporters HLA3 and LCIA localized to the plasma membrane and chloroplast envelope, respectively, in Chlamydomonas, tobacco and Arabidopsis (Figures 1 and 5). Recent work has confirmed these locations in Chlamydomonas (Yamano et al., 2015). We showed that both proteins can facilitate uptake of Ci into Xenopus oocytes, consistent with previous demonstrations that LCIA can facilitate Ci movement across membranes (Mariscal et al., 2006) and that HLA3 overexpression contributes to increased Ci uptake in Chlamydomonas (Gao et al., 2015; Yamano et al., 2015). Further kinetic analyses will be required to determine the mode of action of these proteins (i.e. active vs. passive transport). This information will be crucial for modelling approaches and rational engineering strategies. For example, rational approaches for engineering the cyanobacterial CCM into higher plants are based on a good understanding of the catalytic characteristics of cyanobacterial Ci transporters BicA and SbtA (Du et al., 2014; McGrath and Long, 2014; Price et al., 2011, 2013).
The addition of LCIA or HLA3 did not confer a growth advantage to Arabidopsis plants (Figures 6 and 7, Table 2). This is not entirely unexpected, as biophysical CCMs, in both algae and cyanobacteria, require additional features to function, including a microcompartment containing RuBisCO and additional components to reduce Ci leakage (Badger et al., 1998; Price et al., 2013; Yamano et al., 2010). Furthermore, it is likely that native carbonic anhydrase activity, particularly in the stroma, would hinder Ci accumulation. Biophysical CCMs rely on the capacity to accumulate bicarbonate; thus, specific localization and control of carbonic anhydrase activity appear to play an important role in functionality. For example, ectopic expression of carbonic anhydrase within the cytoplasm of cyanobacterial cells leads to a debilitating leakage of Ci due to rapid equilibration between bicarbonate and CO2 in the cytosol (Price and Badger, 1989). Predictive models indicate that the removal of native stromal carbonic anhydrases is a key target for introducing biophysical CCMs into higher plants (McGrath and Long, 2014; Price et al., 2013). Our results do demonstrate that higher plant cells can express Ci transporters from Chlamydomonas in appropriate locations, without deleterious effect. Both LCIA and HLA3 showed appropriate transmembrane integration and increased Ci uptake when expressed in Xenopus oocytes (Figures 1, 4 and 5). Furthermore, we were able to detect both proteins in transgenic Arabidopsis leaves (Figure 5). Together, these data suggest that LCIA and HLA3 were active, but that activity was not sufficient to drive up CO2 levels around RuBisCO and hence improve the rate of CO2 assimilation.
Recent work has shown that expression and function of HLA3 and LCIA may be closely coordinated in Chlamydomonas (Yamano et al., 2015). Enhanced Ci uptake was observed when both proteins were overexpressed in wild‐type Chlamydomonas at high CO2. However, negligible changes in photosynthesis and Ci uptake were observed when either HLA3 or LCIA was overexpressed, indicating that, in the absence of other CCM components or a microcompartment such as a pyrenoid, these proteins are not able to enhance chloroplastic CO2 concentrations (Gao et al., 2015; Yamano et al., 2015). These data suggest that subsequent engineering strategies in higher plants should focus on co‐expression of HLA3 and LCIA. Additional modifications will probably be required to establish a functional biophysical CCM in higher plants, including the removal of native carbonic anhydrases, which would otherwise short circuit active uptake mechanisms and eliminate any stromal pool of bicarbonate (Price et al., 2013). The next challenge will be to stack key CCM components, and develop a strategy to retarget stromal RuBisCO to a chloroplast microcompartment, a predicted requirement for an algal‐type CCM (Badger et al., 1998).
Experimental procedures
Plant material and growth conditions
Mutant and transgenic plants of Arabidopsis (Arabidopsis thaliana) were in the Col‐0 wild‐type background. Arabidopsis seeds were sown on compost, stratified for 3 days at 4 °C and grown at 20 °C, under ambient CO2 (ca. 400 μmol/mol), at 70% relative humidity and under 100 μmol photons/m2/s in 12‐h light/12‐h dark cycles, unless otherwise stated.
For analyses of transgenic lines, homozygous insertion or wild‐type out‐segregant lines (T3) were compared. Where wild‐type out‐segregants were not available, homozygous insertion lines were compared with Col‐0 plants from seed stocks of the same age generated under similar conditions. Tobacco plants (Nicotiana benthamiana L.) were cultivated under glass house conditions (minimum 20 °C, natural light supplemented to give at least 12‐h light). Venus‐tagged proteins were expressed in wild‐type Chlamydomonas strain cMJ030 (CC‐4533)(Zhang et al., 2014). Cells were maintained in constant low light (~10 μmol photons/m2/s) at RT on 1.5% (w/v) agar plates containing Tris–acetate–phosphate (TAP) (Kropat et al., 2011). For imaging, cells were grown in liquid TAP media to a concentration of 106 cells/mL, pelleted by centrifugation (1000 g, 4 min), resuspended in Tris–phosphate (T‐P) minimal media (Kropat et al., 2011) and grown for 24 h in ambient CO2 before imaging.
Cloning and expression of CCM components in Chlamydomonas
The open reading frames (ORFs) of Chlamydomonas genes were expressed in frame with Venus from the PsaD promoter using the pLM005 vector. ORFs were amplified from genomic DNA using Phusion Hotstart II polymerase (Thermo Fisher Scientific, www.thermofisher.com) with the respective oligos in Table S1. HpaI‐cut pLM005 vector and PCR products were gel purified and assembled by Gibson assembly (Gibson et al., 2009). Due to the large gene length of HLA3, it was cloned in two fragments then assembled in the pLM005 vector by Gibson assembly. The pLM005 vector contains the AphVIII gene for paromomycin resistance in Chlamydomonas and ampicillin resistance for bacterial selection. All construct junctions were verified by Sanger sequencing. Constructs were transformed into Chlamydomonas by electroporation as in Zhang et al. (2014). Briefly, 250 μL of 2 × 108 cells/mL was transformed with 14.5 ng/kbp of EcoRV‐cut plasmid at 16 °C. Cells were spread on 86 mL TAP agar plates containing paromomycin (20 μg/mL) and kept in low light (~10 μmol photons/m2/s) until colonies were ~2–3 mm in diameter. Plates were screened for fluorescent colonies using a Typhoon TRIO fluorescence scanner (GE Healthcare, www.gelifesciences.com) with excitation/emission wavelengths 532 nm/520–555 nm for Venus and 633 nm/630–670 nm for chlorophyll autofluorescence.
Cloning and expression of CCM components in tobacco and Arabidopsis
Genes were cloned from cDNA derived from Chlamydomonas (strain CC‐4886, Chlamydomonas Resource Center). Primers were designed from sequences available on Phytozome v10.2 (Chlamydomonas reinhardtii v5.5 [Augustus u11.6], http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Creinhardtii) (see Table S1 for oligo details). Gene sequences for LCIA and HLA3 were codon‐optimized for expression in higher plants and synthesized de novo (DNA2.0, CA, USA) (Figure S7), then cloned into Gateway entry vectors (pCR®8/GW/TOPO®TA Cloning® Kit) using Platinum® Taq DNA Polymerase High Fidelity according to the manufacturer's instructions (Invitrogen™ Life Technologies, www.lifetechnologies.com) and subsequently cloned into the destination binary vectors pK7FWG2,0 (Karimi et al., 2002) or pGWB5 (Nakagawa et al., 2009). Gibson assembly was used to generate transit peptide gene fusions. Binary vectors were transformed into Agrobacterium tumefaciens (AGL1) for transient gene expression in tobacco leaves (Schöb et al., 1997) or stable insertion in Arabidopsis plants by floral dipping (Clough and Bent, 1998). Co‐expression studies were performed with the WAVE131 vector of the ‘wave’ marker set (Geldner et al., 2009) and the mt‐rb vector (Nelson et al., 2007) for plasma membrane and mitochondria localization, respectively.
DNA extraction, PCR and protein analysis
For screening transgenic Arabidopsis lines, genomic DNA was extracted from mature, nonflowering rosettes of T1 plants as described in Li and Chory (Li and Chory, 1998). PCRs were performed as in McCormick and Kruger (2015). Where possible, the location of gene inserts was confirmed by TAIL PCR as described by Liu et al. (1995). Homozygous insertion lines were identified in the T2 generation either by PCR or by seedling segregation ratios on kanamycin‐containing Murashige and Skoog (MS) medium (0.5x) plates.
Relative levels of LCIA: GFP and HLA3: GFP proteins in leaves were confirmed by immunoblot. Approximately 10 μg protein from whole 28‐d‐old rosettes (100 mg fresh weight) was fractionated by SDS‐PAGE on a 10% (w/v) acrylamide: bisacrylamide (40:1) gel transferred to PVDF membrane, probed with mouse anti‐GFP IgG2a at 1:1,000 dilution (Santa Cruz, http://www.scbt.com/) and visualized using an HRP‐conjugated goat anti‐mouse IgG2a at 1:50 000 dilution. HRP activity was detected using Supersignal Ultra (Pierce, www.piercenet.com) according to the manufacturer's instructions.
Oocyte expression and bicarbonate uptake assays
Synthesized gene sequences for mature LCIA (mLCIA) or HLA3 lacking a stop codon were cloned into the expression vector Vivid Colors™ pcDNA™ 6.2/EmGFP (Invitrogen™ Life Technologies) to generate mLCIA‐ or HLA3‐GFP fusions. For LCIA, the N‐terminal transit sequence peptide (73 aa) was removed and replaced with the sequence ‘GACATG’ to add a Kozak sequence and new start codon. For HLA3, ‘GAC’ was added immediately upstream of the start codon. To generate equivalent vectors lacking a fluorescent tag, Gibson assembly was used to add a stop codon to mLCIA or HLA3 and remove the GFP sequence (720 bp). Plasmids were linearized by AvrII or StuI (Roche, www.roche.co.uk) and capped mRNA was synthesized using the mMESSAGE mMACHINE® T7 Transcription Kit (Ambion®; Life Technologies) according to the manufacturer's instructions. Mature LCIA or HLA3 mRNA was expressed in oocytes as described by Feng et al. (2013). Xenopus oocytes were injected with 50 nL of mRNA (1 μg/μL) or diethyl pyrocarbonate (DEPC)‐treated water as a control. Bicarbonate uptake assays and confocal imaging were performed 3 d after injection in a protocol adapted from Mariscal et al. (2006). Oocytes were incubated in fresh MBS media (88 mm NaCl, 1 mm KCl, 2.4 mm NaHCO3, 0.71 mm CaCl2, 0.82 mm MgSO4 and 15 mm HEPES, pH 7.4), containing 0.12 mm NaHCO3 (1.85 GBq/mol NaH14CO3). After 10 min, the oocytes were washed three times with ice‐cold MBS and lysed in 200 μL SDS (10% [w/v]), and the radioactivity retained in individual oocytes was measured.
Chlorophyll quantification
Chlorophyll was extracted from powdered leaf discs in ice‐cold 80% (v/v) acetone and 10 mm Tris–HCl, and concentration was measured according to Porra et al. (1989).
Measurement of photosynthetic parameters
Gas exchange rates were determined using a LI‐6400 portable infrared gas analyser (LI‐COR Biosciences, http://www.licor.com/) on either the sixth or seventh leaf of 35‐ to 45–d‐old mature, nonflowering rosettes grown in large pots to generate leaf area sufficient for gas exchange measurements (Flexas et al., 2007). The response of net photosynthetic CO2 assimilation (A) to substomatal CO2 concentration (C i) was measured by varying the external CO2 concentration from 0 to 1000 μmol/mol under a constant photosynthetic active radiation of 1500 μmol photons/m2/s (provided by a red–blue light source attached to the leaf chamber). Gas exchange data were corrected for CO2 diffusion as in Bellasio et al. (2015). Leaf temperature and chamber relative humidity were maintained at 21 °C and 70%, respectively. To calculate maximum carboxylation rate (V c,max), maximum electron transport flow (J max) and mesophyll conductance (g m), the A/Ci data were fitted to the C3 photosynthesis model (Farquhar et al., 1980) with modifications to include estimations for g m as described by Ethier and Livingston (2004).
Confocal laser scanning microscopy
A Leica TCS SP2 laser scanning confocal microscope (Leica Microsystems) with a water immersion objective lens (HCX IRAPO 25.0x0.95) was used for imaging leaves and oocytes. Excitation/emission wavelengths were 488 nm/500–530 nm for GFP, 543 nm/590–620 nm for mCherry and 488 nm/680–750 nm for chlorophyll autofluorescence. Images were acquired using Leica LAS AF software (http://www.leica-microsystems.com/). Prior to imaging Venus‐tagged proteins in Chlamydomonas, 15 μL of cells were added to a well of a 96‐well optical plate (Brooks Life Science Systems, http://www.brooks.com) and covered with 150 μL of 1.5% low melting point agarose containing T‐P (~35 °C). For mitochondria staining, CCP1: Venus‐ and CCP2: Venus‐expressing lines were grown in liquid T‐P media with paromomycin (2 μg/mL) to a concentration of 2–4 × 106 cells/mL. Cultures were incubated with MitoTracker Red CMXRos (Thermo Fisher Scientific) to a final concentration of 1 μm for 10 min, then spotted on a polylysine‐coated slide for imaging. Cells were imaged using a custom adapted confocal microscope (Leica DMI6000) with settings at 514 nm/532–555 nm for Venus, 561 nm/573–637 nm for staining by Mitotracker Red CMXRos and 561 nm/665–705 nm for chlorophyll autofluorescence. Images were analysed using Fiji software (http://fiji.sc/Fiji).
Statistical analysis
Variations in response between genotypes were assessed by either analysis of variance (ANOVA) or Student's t‐tests followed by Tukey's honest significant difference (HSD) post hoc test (SPSS Statistics 18, http://www.ibm.com/). Differences for which P < 0.05 are considered significant.
Supporting information
Figure S1 Expression of fluorescent‐tagged CCM components in Chlamydomonas and tobacco (from Figure 1).
Figure S2 Co‐expression of Venus‐fused CCM components with Mitotracker Red CMXRos in Chlamydomonas.
Figure S3 Expression of GFP‐fused CCM components carrying native Arabidopsis chloroplast transit peptides in tobacco (from Figure 3).
Figure S4 Stable expression of LCIA: GFP and HLA3: GFP in Arabidopsis (from Figure 5a).
Figure S5 Chlorophyll content of transgenic Arabidopsis plants expressing LCIA or HLA3.
Figure S6 Specific leaf area (area/DW) of transgenic LCIA or HLA3 Arabidopsis plants.
Figure S7 DNA sequences of codon‐optimized LCIA and HLA3.
Table S1 Sequences of synthetic oligonucleotides used in this study.
Acknowledgements
We thank Livia Scheunemann, Xiaorong Fan, Paloma Menguer and Anthony Miller (John Innes Centre) for advice and help with oocyte experiments, Mark Fricker (University of Oxford) for use of LeafLab software and Fani Ntana (University of Edinburgh) for technical contributions. AJM, DF and AMS were funded by grants from the Biotechnology and Biological Sciences Research Council (BBSRC), UK: BB/I024453/1 and Institute Strategic Programme Grant BB/J004561/1 to the John Innes Centre. NA was funded by BBSRC: BB/M006468/1, HG and MM were funded by BBSRC: BB/M007693/1. LM and MJ were funded by US National Science Foundation (NSF): 1359682.
Conflict of interest
The authors declare that they have no competing interests.
References
- Badger, M.R. , Andrews, T.J. , Whitney, S.M. , Ludwig, M. , Yellowlees, D.C. , Leggat, W. and Price, G.D. (1998) The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast‐based CO2‐concentrating mechanisms in algae. Can. J. Bot. 76, 1052–1071. [Google Scholar]
- Bellasio, C. , Beerling, D.J. and Griffiths, H. (2015) An Excel tool for deriving key photosynthetic parameters from combined gas exchange and chlorophyll fluorescence: theory and practice. Plant, Cell Environ. doi: 10.1111/pce.12560. [DOI] [PubMed] [Google Scholar]
- Blanco‐Rivero, A. , Shutova, T. , Roman, M.J. , Villarejo, A. and Martinez, F. (2012) Phosphorylation controls the localization and activation of the lumenal carbonic anhydrase in Chlamydomonas reinhardtii . PLoS ONE, 7, e49063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueggeman, A.J. , Gangadharaiah, D.S. , Cserhati, M.F. , Casero, D. , Weeks, D.P. and Ladunga, I. (2012) Activation of the carbon concentrating mechanism by CO2 deprivation coincides with massive transcriptional restructuring in Chlamydomonas reinhardtii . Plant Cell, 24, 1860–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Du, J. , Forster, B. , Rourke, L. , Howitt, S.M. and Price, G.D. (2014) Characterisation of cyanobacterial bicarbonate transporters in E. coli shows that SbtA homologs are functional in this heterologous expression system. PLoS ONE, 9, e115905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu, D. , Miller, A.R. , Horken, K.M. , Weeks, D.P. and Spalding, M.H. (2009a) Knockdown of limiting‐CO2‐induced gene HLA3 decreases HCO3 ‐ transport and photosynthetic Ci affinity in Chlamydomonas reinhardtii . Proc. Natl Acad. Sci. USA, 106, 5990–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu, D. , Wang, Y. and Spalding, M.H. (2009b) Thylakoid lumen carbonic anhydrase (CAH3) mutation suppresses air‐Dier phenotype of LCIB mutant in Chlamydomonas reinhardtii . Plant Physiol. 149, 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel, B.D. , Schaffer, M. , Kuhn Cuellar, L. , Villa, E. , Plitzko, J.M. and Baumeister, W. (2015) Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo‐electron tomography. eLife, 4, e04889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethier, G.J. and Livingston, N.J. (2004) On the need to incorporate sensitivity to CO2 transfer conductance into the Farquhar‐von Caemmerer‐Berry leaf photosynthesis model. Plant, Cell Environ. 27, 137–153. [Google Scholar]
- Fang, W. , Si, Y. , Douglass, S. , Casero, D. , Merchant, S.S. , Pellegrini, M. , Ladunga, I. et al. (2012) Transcriptome‐wide changes in Chlamydomonas reinhardtii gene expression regulated by carbon dioxide and the CO2‐concentrating mechanism regulator CIA5/CCM1. Plant Cell, 24, 1876–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar, G.D. , von Caemmerer, S. and Berry, J.A. (1980) A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta, 149, 78–90. [DOI] [PubMed] [Google Scholar]
- Feng, H. , Xia, X. , Fan, X. , Xu, G. and Miller, A.J. (2013) Optimizing plant transporter expression in Xenopus oocytes. Plant Methods, 9, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flexas, J. , Ortuno, M.F. , Ribas‐Carbo, M. , Diaz‐Espejo, A. , Florez‐Sarasa, I.D. and Medrano, H. (2007) Mesophyll conductance to CO2 in Arabidopsis thaliana . New Phytol. 175, 501–511. [DOI] [PubMed] [Google Scholar]
- Fujiwara, S. , Fukuzawa, H. , Tachiki, A. and Miyachi, S. (1990) Structure and differential expression of two genes encoding carbonic anhydrase in Chlamydomonas reinhardtii . Proc. Natl Acad. Sci. USA, 87, 9779–9783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, H. , Wang, Y. , Fei, X. , Wright, D.A. and Spalding, M.H. (2015) Expression activation and functional analysis of HLA3, a putative inorganic carbon transporter in Chlamydomonas reinhardtii . Plant J. 82, 1–11. [DOI] [PubMed] [Google Scholar]
- Geldner, N. , Dénervaud‐Tendon, V. , Hyman, D.L. , Mayer, U. , Stierhof, Y.‐D. and Chory, J. (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59, 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genkov, T. , Meyer, M. , Griffiths, H. and Spreitzer, R.J. (2010) Functional hybrid Rubisco enzymes with plant small subunits and algal large subunits: engineered rbcS cDNA for expression in Chlamydomonas . J. Biol. Chem. 285, 19833–19841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraghty, A.M. and Spalding, M.H. (1996) Molecular and structural changes in Chlamydomonas under limiting CO2 (a possible mitochondrial role in adaptation). Plant Physiol. 111, 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, D.G. , Young, L. , Chuang, R.Y. , Venter, J.C. , Hutchison, C.A. 3rd and Smith, H.O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods, 6, 343–345. [DOI] [PubMed] [Google Scholar]
- Griffiths, H. and Helliker, B.R. (2013) Mesophyll conductance: internal insights of leaf carbon exchange. Plant, Cell Environ. 36, 733–735. [DOI] [PubMed] [Google Scholar]
- Karimi, M. , Inze, D. and Depicker, A. (2002) Gateway vectors for Agrobacterium‐mediated plant transformation. Trends Plant Sci. 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Karlsson, J. , Clarke, A.K. , Chen, Z.Y. , Hugghins, S.Y. , Park, Y.I. , Husic, H.D. , Moroney, J.V. et al. (1998) A novel alpha‐type carbonic anhydrase associated with the thylakoid membrane in Chlamydomonas reinhardtii is required for growth at ambient CO2 . EMBO J. 17, 1208–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimpel, D.L. , Togasaki, R.K. and Miyachi, S. (1983) Carbonic anhydrase in Chlamydomonas reinhardtii I. Localization. Plant Cell Physiol. 24, 255–259. [Google Scholar]
- Kropat, J. , Hong‐Hermesdorf, A. , Casero, D. , Ent, P. , Castruita, M. , Pellegrini, M. , Merchant, S.S. et al. (2011) A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii . Plant J. 66, 770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. and Chory, J. (1998) Preparation of DNA from Arabidopsis. Methods Mol. Biol. 82, 55–60. [DOI] [PubMed] [Google Scholar]
- Lieman‐Hurwitz, J. , Rachmilevitch, S. , Mittler, R. , Marcus, Y. and Kaplan, A. (2003) Enhanced photosynthesis and growth of transgenic plants that express ictB, a gene involved in HCO3 ‐ accumulation in cyanobacteria. Plant Biotechnol. J. 1, 43–50. [DOI] [PubMed] [Google Scholar]
- Lin, M.T. , Occhialini, A. , Andralojc, P.J. , Devonshire, J. , Hines, K.M. , Parry, M.A.J. and Hanson, M.R. (2014a) β‐Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J. 79, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, M.T. , Occhialini, A. , Andralojc, P.J. , Parry, M.A.J. and Hanson, M.R. (2014b) A faster Rubisco with potential to increase photosynthesis in crops. Nature, 513, 547–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.‐G. , Mitsukawa, N. , Oosumi, T. and Whittier, R.F. (1995) Efficient isolation and mapping of Arabidopsis thaliana T‐DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Long, S.P. , Zhu, X.G. , Naidu, S.L. and Ort, D.R. (2006) Can improvement in photosynthesis increase crop yields? Plant, Cell Environ. 29, 315–330. [DOI] [PubMed] [Google Scholar]
- Long, S.P. , Marshall‐Colon, A. and Zhu, X.G. (2015) Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell, 161, 56–66. [DOI] [PubMed] [Google Scholar]
- Mariscal, V. , Moulin, P. , Orsel, M. , Miller, A.J. , Fernandez, E. and Galvan, A. (2006) Differential regulation of the Chlamydomonas Nar1 gene family by carbon and nitrogen. Protist, 157, 421–433. [DOI] [PubMed] [Google Scholar]
- Markelova, A.G. , Sinetova, M.P. , Kupriyanova, E.V. and Pronina, N.A. (2009) Distribution and functional role of carbonic anhydrase Cah3 associated with thylakoid membranes in the chloroplast and pyrenoid of Chlamydomonas reinhardtii . Russ. J. Plant Physiol. 56, 761–768. [Google Scholar]
- Mason, C.B. , Manuel, L.J. and Moroney, J.V. (1990) A new chloroplast protein is induced by growth on low CO2 in Chlamydomonas reinhardtii . Plant Physiol. 93, 833–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, A.J. and Kruger, N.J. (2015) Lack of fructose 2,6‐bisphosphate compromises photosynthesis and growth in Arabidopsis in fluctuating environments. Plant J. 81, 670–683. [DOI] [PubMed] [Google Scholar]
- McGrath, J.M. and Long, S.P. (2014) Can the cyanobacterial carbon‐concentrating mechanism increase photosynthesis in crop species? A theoretical analysis. Plant Physiol. 164, 2247–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, M. and Griffiths, H. (2013) Origins and diversity of eukaryotic CO2‐concentrating mechanisms: lessons for the future. J. Exp. Bot. 64, 769–786. [DOI] [PubMed] [Google Scholar]
- Meyer, M.T. , Genkov, T. , Skepper, J.N. , Jouhet, J. , Mitchell, M.C. , Spreitzer, R.J. and Griffiths, H. (2012) Rubisco small‐subunit alpha‐helices control pyrenoid formation in Chlamydomonas . Proc. Natl Acad. Sci. USA, 109, 19474–19479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, M. , Lato, S.M. , Ynalvez, R.A. , Xiao, Y. and Moroney, J.V. (2004) Identification of a new chloroplast carbonic anhydrase in Chlamydomonas reinhardtii . Plant Physiol. 135, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K. , Yamano, T. , Yoshioka, S. , Kohinata, T. , Inoue, Y. , Taniguchi, F. , Asamizu, E. et al. (2004) Expression profiling‐based identification of CO2‐responsive genes regulated by CCM1 controlling a carbon‐concentrating mechanism in Chlamydomonas reinhardtii . Plant Physiol. 135, 1595–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, E. , Abe, T. , Tsuzuki, M. , Fujiwara, S. , Sato, N. , Hirata, A. , Sonoike, K. et al. (1998) Presence of the CO2‐concentrating mechanism in some species of the pyrenoid‐less free‐living algal genus Chloromonas (Volvocales, Chlorophyta). Planta, 204, 269–276. [DOI] [PubMed] [Google Scholar]
- Moroney, J.V. and Tolbert, N.E. (1985) Inorganic carbon uptake by Chlamydomonas reinhardtii . Plant Physiol. 77, 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T. , Ishiguro, S. and Kimura, T. (2009) Gateway vectors for plant transformation. Plant Biotechnol. 26, 275–284. [Google Scholar]
- Nelson, B.K. , Cai, X. and Nebenführ, A. (2007) A multicolored set of in vivo organelle markers for co‐localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Ohnishi, N. , Mukherjee, B. , Tsujikawa, T. , Yanase, M. , Nakano, H. , Moroney, J.V. and Fukuzawa, H. (2010) Expression of a low CO2‐inducible protein, LCI1, increases inorganic carbon uptake in the green alga Chlamydomonas reinhardtii . Plant Cell, 22, 3105–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry, M.A. , Andralojc, P.J. , Scales, J.C. , Salvucci, M.E. , Carmo‐Silva, A.E. , Alonso, H. and Whitney, S.M. (2013) Rubisco activity and regulation as targets for crop improvement. J. Exp. Bot. 64, 717–730. [DOI] [PubMed] [Google Scholar]
- Pengelly, J.J. , Forster, B. , von Caemmerer, S. , Badger, M.R. , Price, G.D. and Whitney, S.M. (2014) Transplastomic integration of a cyanobacterial bicarbonate transporter into tobacco chloroplasts. J. Exp. Bot. 65, 3071–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock, S.V. , Prout, D.L. , Godfrey, A.C. , Lemaire, S.D. and Moroney, J.V. (2004) The Chlamydomonas reinhardtii proteins Ccp1 and Ccp2 are required for long‐term growth, but are not necessary for efficient photosynthesis, in a low‐CO2 environment. Plant Mol. Biol. 56, 125–132. [DOI] [PubMed] [Google Scholar]
- Porra, R.J. , Thompson, W.A. and Kriedemann, P.E. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta, 975, 384–394. [Google Scholar]
- Price, G.D. and Badger, M.R. (1989) Expression of human carbonic anhydrase in the cyanobacterium Synechococcus PCC7942 creates a high CO2‐requiring phenotype ‐ evidence for a central role for carboxysomes in the CO2 concentrating mechanism. Plant Physiol. 91, 505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, G.D. , Badger, M.R. and von Caemmerer, S. (2011) The prospect of using cyanobacterial bicarbonate transporters to improve leaf photosynthesis in C3 crop plants. Plant Physiol. 155, 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, G.D. , Pengelly, J.J. , Forster, B. , Du, J. , Whitney, S.M. , von Caemmerer, S. , Badger, M.R. et al. (2013) The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species. J. Exp. Bot. 64, 753–768. [DOI] [PubMed] [Google Scholar]
- Ramazanov, Z. , Mason, C.B. , Geraghty, A.M. , Spalding, M.H. and Moroney, J.V. (1993) The Low CO2‐Inducible 36‐kilodalton protein is localized to the chloroplast envelope of Chlamydomonas reinhardtii . Plant Physiol. 101, 1195–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, C.S. and Spalding, M.H. (1995) Post‐translational processing of the highly processed, secreted periplasmic carbonic anhydrase of Chlamydomonas is largely conserved in transgenic tobacco. Plant Mol. Biol. 29, 303–315. [DOI] [PubMed] [Google Scholar]
- Schöb, H. , Kunz, C. and Meins, F. Jr . (1997) Silencing of transgenes introduced into leaves by agroinfiltration: a simple, rapid method for investigating sequence requirements for gene silencing. Mol. Gen. Genet. 256, 581–585. [DOI] [PubMed] [Google Scholar]
- Sharkey, T.D. (1988) Estimating the rate of photorespiration in leaves. Physiol. Plant. 73, 147–152. [Google Scholar]
- Simkin, A.J. , McAusland, L. , Headland, L.R. , Lawson, T. and Raines, C.A. (2015) Multigene manipulation of photosynthetic carbon assimilation increases CO2 fixation and biomass yield in tobacco. J. Exp. Bot. 66, 4075–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinetova, M.A. , Kupriyanova, E.V. , Markelova, A.G. , Allakhverdiev, S.I. and Pronina, N.A. (2012) Identification and functional role of the carbonic anhydrase Cah3 in thylakoid membranes of pyrenoid of Chlamydomonas reinhardtii . Biochim. Biophys. Acta, 1817, 1248–1255. [DOI] [PubMed] [Google Scholar]
- Spalding, M.H. (2009) The CO2‐concentrating mechanism and carbon assimilation. In The Chlamydomonas Sourcebook: Organellar and Metabolic Processes ( Harris, E.H. and Stern, D.B. , eds), pp. 257–301. Oxford: Academic. [Google Scholar]
- Villarejo, A. , Shutova, T. , Moskvin, O. , Forssen, M. , Klimov, V.V. and Samuelsson, G. (2002) A photosystem II‐associated carbonic anhydrase regulates the efficiency of photosynthetic oxygen evolution. EMBO J. 21, 1930–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. and Spalding, M.H. (2014a) Acclimation to very low CO2: contribution of limiting CO2 inducible proteins, LCIB and LCIA, to inorganic carbon uptake in Chlamydomonas reinhardtii . Plant Physiol. 166, 2040–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. and Spalding, M.H. (2014b) LCIB in the Chlamydomonas CO2‐concentrating mechanism. Photosynth. Res. 121, 185–192. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Stessman, D.J. and Spalding, M.H. (2015) The CO2 concentrating mechanism and photosynthetic carbon assimilation in limiting CO2: how Chlamydomonas works against the gradient. Plant J. 82, 429–448. [DOI] [PubMed] [Google Scholar]
- Whitney, S.M. , Houtz, R.L. and Alonso, H. (2011) Advancing our understanding and capacity to engineer nature's CO2‐sequestering enzyme, Rubisco. Plant Physiol. 155, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano, T. , Tsujikawa, T. , Hatano, K. , Ozawa, S. , Takahashi, Y. and Fukuzawa, H. (2010) Light and low‐CO2‐dependent LCIB‐LCIC complex localization in the chloroplast supports the carbon‐concentrating mechanism in Chlamydomonas reinhardtii . Plant Cell Physiol. 51, 1453–1468. [DOI] [PubMed] [Google Scholar]
- Yamano, T. , Asada, A. , Sato, E. and Fukuzawa, H. (2014) Isolation and characterization of mutants defective in the localization of LCIB, an essential factor for the carbon‐concentrating mechanism in Chlamydomonas reinhardtii . Photosynth. Res. 121, 193–200. [DOI] [PubMed] [Google Scholar]
- Yamano, T. , Sato, E. , Iguchi, H. , Fukuda, Y. and Fukuzawa, H. (2015) Characterization of cooperative bicarbonate uptake into chloroplast stroma in the green alga Chlamydomonas reinhardtii . Proc. Natl Acad. Sci. USA, 112, 7315–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ynalvez, R.A. , Xiao, Y. , Ward, A.S. , Cunnusamy, K. and Moroney, J.V. (2008) Identification and characterization of two closely related beta‐carbonic anhydrases from Chlamydomonas reinhardtii . Physiol. Plant. 133, 15–26. [DOI] [PubMed] [Google Scholar]
- Zhang, R. , Patena, W. , Armbruster, U. , Gang, S.S. , Blum, S.R. and Jonikas, M.C. (2014) High‐throughput genotyping of green algal mutants reveals random distribution of mutagenic insertion sites and endonucleolytic cleavage of transforming DNA. Plant Cell, 26, 1398–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X.G. , Long, S.P. and Ort, D.R. (2010) Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 61, 235–261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Expression of fluorescent‐tagged CCM components in Chlamydomonas and tobacco (from Figure 1).
Figure S2 Co‐expression of Venus‐fused CCM components with Mitotracker Red CMXRos in Chlamydomonas.
Figure S3 Expression of GFP‐fused CCM components carrying native Arabidopsis chloroplast transit peptides in tobacco (from Figure 3).
Figure S4 Stable expression of LCIA: GFP and HLA3: GFP in Arabidopsis (from Figure 5a).
Figure S5 Chlorophyll content of transgenic Arabidopsis plants expressing LCIA or HLA3.
Figure S6 Specific leaf area (area/DW) of transgenic LCIA or HLA3 Arabidopsis plants.
Figure S7 DNA sequences of codon‐optimized LCIA and HLA3.
Table S1 Sequences of synthetic oligonucleotides used in this study.


