Summary
In angiosperms, double fertilization of the egg and central cell of the megagametophyte leads to the development of the embryo and endosperm, respectively. Control of cell cycle progression in the megagametophyte is essential for successful fertilization and development. Central cell‐targeted expression of the D‐type cyclin CYCD7;1 (endCYCD7;1) using the imprinted FWA promoter overcomes cycle arrest of the central cell in the Arabidopsis female gametophyte in the unfertilized ovule, leading to multinucleate central cells at high frequency. Unlike FERTILIZATION‐INDEPENDENT SEED (fis) mutants, but similar to lethal RETINOBLASTOMA‐RELATED (rbr) mutants, no seed coat development is triggered. Unlike the case with loss of rbr, post‐fertilization endCYCD7;1 in the endosperm enhances the number of nuclei during syncytial endosperm development and induces the partial abortion of developing seeds, associated with the enhanced size of the surviving seeds. The frequency of lethality was less than the frequency of multinucleate central cells, indicating that these aspects are not causally linked. These larger seeds contain larger embryos composed of more cells of wild‐type size, surrounded by a seed coat composed of more cells. Seedlings arising from these larger seeds displayed faster seedling establishment and early growth. Similarly, two different embryo‐lethal mutants also conferred enlarged seed size in surviving siblings, consistent with seed size increase being a general response to sibling lethality, although the cellular mechanisms were found to be distinct. Our data suggest that tight control of CYCD activity in the central cell and in the developing endosperm is required for optimal seed formation.
Keywords: Arabidopsis, D‐type cyclins, seed size, cell cycle, endosperm, central cell
Significance Statement
Controlling cell cycle progression in the female gametophyte is essential for fertilization and development. Here we use targeted activation of a cyclin in the central cell to show that division of the central cell nucleus can be uncoupled from egg cell proliferation. Further phenotypes of these lines show that accurate control of CYCD activity is needed for seed development.
Introduction
Seeds are essential for the dispersal and survival of higher plants, and are central to human nutrition. Seeds consist of three compartments – the mature embryo, the endosperm and the encapsulating seed coat – carrying different balances of maternal and paternal genome (Goldberg et al., 1994). They arise from a double fertilization of the megagametophyte (ovule), in which one sperm cell fuses with the egg cell, forming a diploid embryo with equal contributions from each parental genome, whereas the other sperm cell fuses with the diploid central cell to form the triploid endosperm carrying a 2:1 ratio of maternal:paternal genome (Faure et al., 2002; Berger, 2008). The seed coat protecting the two zygotic tissues derives from the diploid maternal integument.
The success of seed production depends on the proper formation of the female or megagametophyte prior to fertilization (Drews and Koltunow, 2011) and the coordination of growth of the three seed compartments post‐fertilization (Garcia et al., 2005). The megagametophyte, embedded in the maternal integuments, arises from syncytial divisions of the haploid nucleus of the megaspore that produce eight nuclei and gives rise to seven cells, which include the egg cell and the single central cell containing two identical haploid nuclei. In the mature female gametophyte, these two haploid central cell nuclei fuse to form a polar diploid nucleus located towards the micropyle, closest to the egg cell (Drews and Koltunow, 2011). Upon delivery of two sperm cells by the pollen tube, fertilization results from the fusion of the two sperm cell nuclei with the nuclei of the egg cell and central cell of the gametophyte. This triggers cell division of the fertilized egg cell to generate the embryo, whereas mitosis of the central cell produces the initially multinucleate syncitial endosperm. Endosperm development, normally in response to fertilization, triggers ovule integuments to differentiate into the seed coat (Garcia et al., 2005), although parthenogenetic seed development in the fertilization‐independent seed (fis) mutants also triggers seed coat differentiation (Chaudhury et al., 1998; Ingouff et al., 2006). The development of both the megagametophyte and of the seed is therefore characterized by high mitotic activity (Garcia et al., 2005; Ingouff et al., 2006; Sabelli and Larkins, 2009).
Cell cycle progression in eukaryotes is controlled by the activity of cyclin‐dependent kinases (CDKs). In complex with their cyclin (CYC) regulatory subunit (Kono et al., 2007), CDKs form serine‐threonine protein kinases, the activity of which is further controlled by post‐translational modifications and regulatory interactions. In plants, key controllers of CDK/CYC activity are the ICK/KRP and SIM‐related inhibitors (De Veylder et al., 2007). The D‐type cyclins (CYCDs) are represented by 10 genes in seven groups in Arabidopsis, and these, associated with the A‐type CDK (CDKA), are responsible for the transition of cells from G1 into S phase of the mitotic cell cycle through the canonical RETINOBLASTOMA‐RELATED (RBR)‐E2F pathway (Boniotti and Gutierrez, 2001; Gutierrez et al., 2002). CDKA/CYCD kinases phosphorylate RBR modulating the RBR interaction with transcription factors, including those involved in cell cycle progression, such as E2Fs (Magyar et al., 2012), which promote the transcription of genes required for the S phase (Kuwabara and Gruissem, 2014).
Regulation of the cell cycle is required for the correct development of the megagametophyte and the subsequent seed development of Arabidopsis (Johnston et al., 2008, 2010; Collins et al., 2012). In Arabidopsis rbr loss‐of‐function mutants, the central cell of the mature female gametophyte has supernumerary nuclei, suggesting a failure of cell‐cycle arrest (Ebel et al., 2004; Johnston et al., 2008, 2010). Furthermore, RBR is required for the correct expression of imprinted genes, the transcriptional activity of which is dependent on the parental origin. During female gametophyte development, MET1, encoding a mediator of DNA methylation, is transcriptionally repressed by a complex of RBR and the Arabidopsis homologue of yeast MULTICOPY SUPPRESSOR OF IRA1 (MSI1), allowing the activation of imprinted genes in the maternal germ line (Johnston et al., 2008; Jullien et al., 2008).
The CYCD3 subfamily of CYCDs has three genes in Arabidopsis, and forms the CDKA‐CYCD3 kinase that phosphorylates RBR (Boniotti and Gutierrez, 2001). The cycd3‐1;2;3 triple mutant shows reduced cell proliferation in the shoot (Dewitte et al., 2007), and embryo development is delayed and the seed abortion is increased (Collins et al., 2012). The same authors showed that transactivation of either CYCD3;1 or CYCD7;1 in both embryo and endosperm triggered cell division and conferred often lethal defects on the embryo, especially in the case of CYCD3;1 overexpression; however, a role for CYCD activity in the megagametophyte has not been shown.
Here we report on the effects of targeted upregulation of the core cell cycle component CYCD7;1 in non‐embryonic tissues, namely the central cell and endosperm. We show that control of CYCD activity is required for cell‐cycle arrest in the central cell, and for the proper formation of the female gametophyte and subsequent seed development. Lethality in a subset of developing seeds leads to increased seed size, which appears to be a general phenomenon because of increased cell proliferation in the embryo. The arising larger seeds show enhanced growth, suggesting that partial seed abortion could be used as a tool for increasing seed size when faster seedling development is required.
Results
Central cell‐targeted CYCD7;1 overcomes cell‐cycle arrest in the central cell of female gametophyte
Ectopic expression of CYCD3;1 and CYCD7;1 under the control of the non‐specific RPS5A promoter in the proliferating tissues conferred developmental defects associated with over‐proliferation (Collins et al., 2012). As ectopic expression of CYCD3;1 led to severe phenotypes by preventing cell division arrest in the suspensor and embryos, we explored the effects of localized CYCD7;1 activation by using the FWA promoter (Kinoshita et al., 2004) to target CYCD7;1 expression specifically to the central cell and endosperm (end CYCD7;1). Previous work showed that a protein fusion of GFP to the FWA promoter fragment and the N‐terminal homeodomain and nuclear localization signal of the FWA protein (dFWA) is sufficient to target the GFP to the nuclei of the central cell in the female gametophyte, prior to fertilization, and the nuclei of developing endosperm, until cellularization (Kinoshita et al., 2004; Figure 1). We therefore tested whether this FWA promoter fragment alone is sufficient to target expression to the mature central cell and the developing endosperm. A reporter construct containing a 3.2‐kb fragment of FWA promoter driving an NLS‐3XVENUS fusion was examined in Arabidopsis ovules and seeds (Figure 1). Activity from both the dFWA‐GFP protein fusion and the FWA promoter reporter was restricted to the mature central cell and developing endosperm. In both transgenic lines, pFWA:NLS‐3xVENUS and pFWA:dFWA‐GFP, the signal decreased in the early cellularized endosperm, disappeared completely in the cellular endosperm surrounding a heart‐stage embryo (Figure 1) and was absent in a mature male gametophyte. These results corroborate the expression pattern reported by Kinoshita et al. (2004), and demonstrate that the promoter fragment alone is sufficient to confer specific expression.
Figure 1.
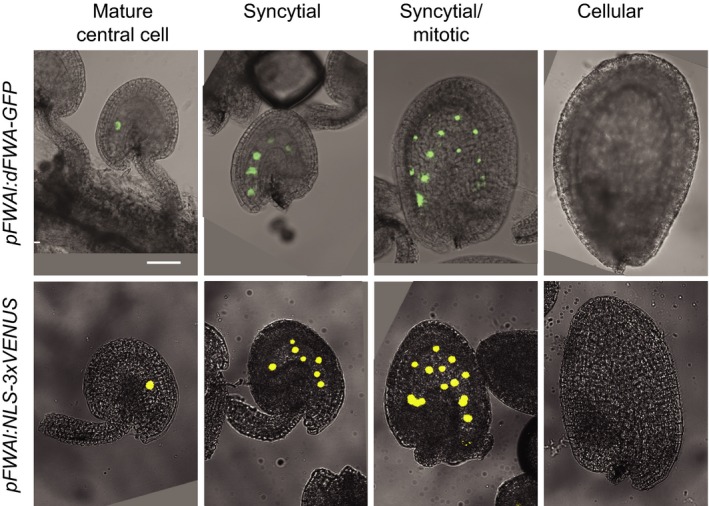
The FWA cis‐regulatory sequence drives expression in the central cell and syncytial endosperm. The FWA promoter fragment is sufficient to restrict dFWA‐GFP and 3XVENUS expression to the central cell in the mature female gametophyte, and to the syncytial endosperm after fertilization. GFP and 3XVENUS are targeted to the nucleus with the N‐terminal homeodomain fragment of the FWA protein that contains a nuclear‐targeting sequence and an NLS, respectively. Scale bar: 50 μm (shown only in the upper left‐hand panel).
In unfertilized wild‐type (WT) ovules, the gametophyte central cell contains a single nucleus, located towards the egg apparatus on the micropyle pole of the ovule (Figure 2a). In end CYCD7;1 lines, between 63 and 80% of ovules displayed supernumerary nuclei in the central cell of the unfertilized mature ovule, indicating a failure of cell‐cycle arrest (Figure 2b,c). In lines with higher levels of end CYCD7;1 transactivation (Figure 2e), the number of supernumerary nuclei was found to be higher compared with lines of lower CYCD7;1 expression (Figure 2d), displaying up to around 20 nuclei. In end CYCD7;1 ovules, the supernumerary nuclei are distributed not only towards the micropylar pole but throughout the gametophyte.
Figure 2.
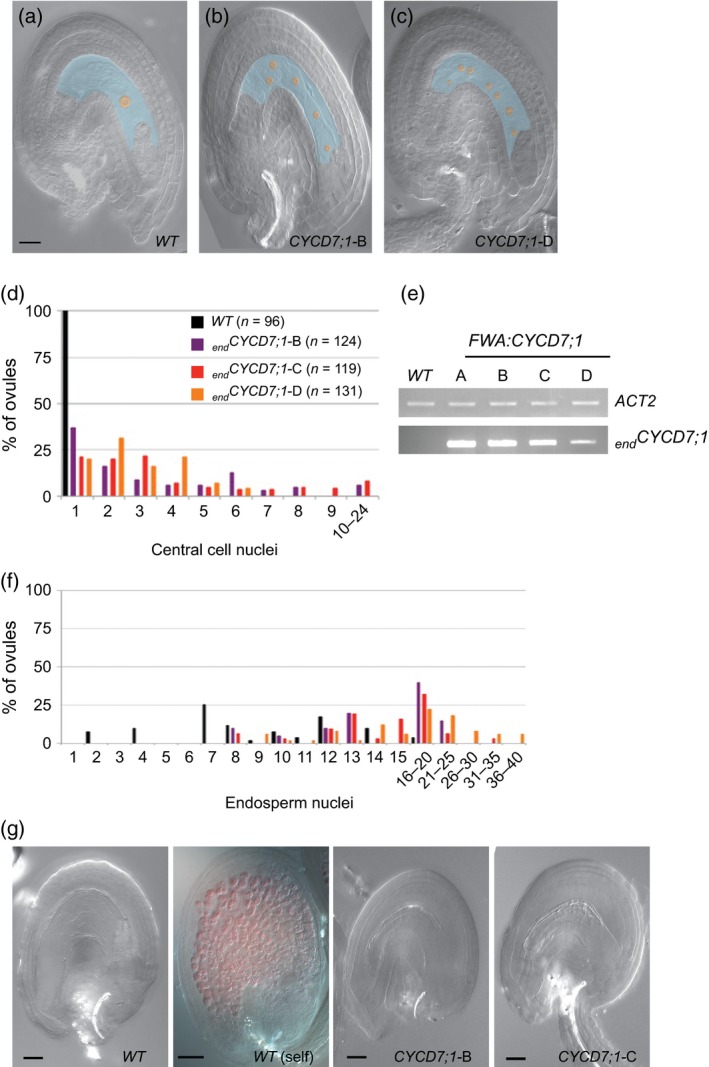
CYCD7;1 expression in the central cell of the female gametophyte overcomes cell‐cycle arrest.
(a–c) Digital interference contrast (DIC) micrographs of unfertilized ovules. Wild‐type (WT) ovules show a single nucleus (orange) in central cell (blue) (a), whereas several nuclei are visible in independent lines of endosperm‐targeted CYCD7;1 expression (end CYCD7;1; two representative examples are shown in b and c). Scale bar: 20 μm.
(d) Bar chart showing the number of nuclei in the central cell of ovules sampled from WT (top panel) and end CYCD7;1 (bottom panel) emasculated pistils.
(e) Estimation of expression level of end CYCD7;1 transcripts in four independent transgenic lines (end CYCD7;1 A–D) by semi‐quantitative RT‐PCR.
(f) Number of nuclei in the seed endosperm 24 h after pollination (n = 51).
(g) Fertilization event triggers the synthesis of proanthocyanidins (PAs) in the maternal integuments marking the acquisition of seed coat fate. PA‐accumulating cells are decorated red after vanillin staining. No PA was detected by this method in end CYCD7;1 ovules. Scale bars: 40 μm.
The multicellular endosperm in unfertilized ovules of fie mutants is a consequence of the fertilization‐independent proliferation of the central cell that occurs together with the acquisition of endosperm fate and the fertilization‐independent differentiation of the seed coat. We therefore examined ovules for evidence of premature seed coat differentiation using vanillin staining (Ingouff et al., 2006; Figure 2g), but saw no evidence of this. Hence, although end CYCD7;1 induces central cell nuclear proliferation, the absence of seed coat differentiation suggests that end CYCD7;1 does not trigger autonomous seed development as seen in fie. Mutants in the rbr gene also show spontaneous nuclear proliferation in female gametophytes in the absence of fertilization, producing up to 20 nuclei (Ingouff et al., 2006), very similar to CYCD7;1 ectopic expression. This confirms that nuclear proliferation does not itself trigger the seed coat differentiation seen in fie mutants, and further suggests that CYCD7;1 may be operating by inactivating RBR.
CYCD7;1 expression in the endosperm leads to accelerated seed development during the syncytial phase
After fertilization, the triploid endosperm undergoes several rounds of syncytial mitosis (Boisnard‐Lorig et al., 2001). Cellularization of the endosperm starts at the micropylar end when the embryo has reached the late globular stage. In end CYCD7;1 lines, the endosperm was observed to develop faster than the WT counterpart. By 24 h after pollination (24 HAP), a high proportion of end CYCD7;1 ovules contain 16–20 nuclei in the endosperm, compared with between seven and nine in the WT ovules (Figure 2f).
The rate of embryo development progression was also influenced by end CYCD7;1 (Figure 3). At 3 days after pollination (3 DAP), in WT plants, 20% of embryos reached the 16‐cell stage, 41% reached the 8‐cell stage, 24% reached the 2/4‐cell stage and 15% were in the embryo‐proper stage (immediately after the first division of the zygote). At this point, up to 60% of end CYCD7;1 embryos were already at the globular stage (line C), only 10% were at the 2/4‐cell stage and no embryos were still at the embryo‐proper stage. At 4 DAP, the majority of WT embryos (54%) were in the globular stage, only 27% had progressed beyond this point and 19% were still in the 16‐cell stage. end CYCD7;1 embryos were mainly at the triangular and globular stages. At 5 DAP, line A had 72% of embryos at the heart stage and 17% of embryos at the triangular stage. Line D had 28% at the heart stage and 41% at the triangular stage. A high proportion of end CYCD7;1 embryos reached the heart phase faster than in the WT, but the cellular patterning of the heart stage embryo was unaffected (Figure S1).
Figure 3.
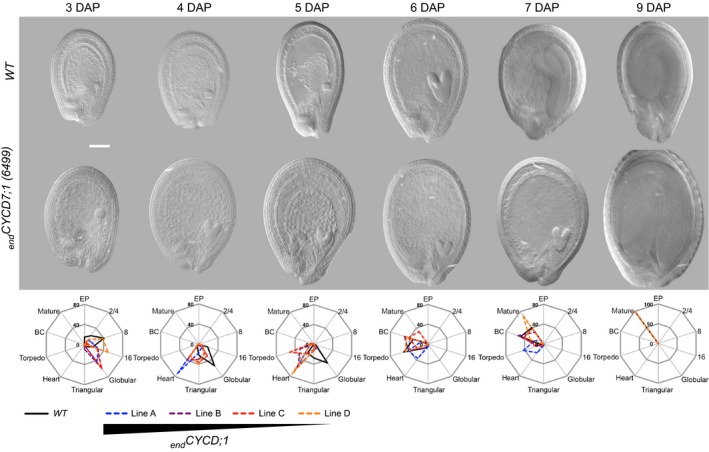
Seed development in wild type (WT) and endosperm‐targeted CYCD7;1 lines. Upper panel: representative examples of WT and end CYCD7;1‐A seeds sampled at different time points after pollination (3, 4, 5, 6, 7 and 9 days after pollination, DAP). Scale bar: 50 μm. Middle panel: radar plots depicting the proportion of seed with embryos at different stages at 3, 4, 5, 6, 7 and 9 DAP of WT and end CYCD7;1 lines. For each time point and genotype, the number of seeds scored ranged from 158 to 237. Lower panel: the relative expression of end CYCD7;1 in the different lines is indicated by the triangle.
Finally, from 6 DAP onwards, end CYCD7;1 seed development slowed, with a marked effect on the transition from the heart phase to the torpedo phase. This could be consistent with a reduction of CYCD7;1 expression in the endosperm at this stage; however, detailed analysis of the different lines showed that from 7 DAP onwards, the line with the strongest level of expression (line A) tends to develop more slowly than the WT, whereas seeds from the line with the lowest expression level are still somewhat further ahead in seed development than WT seeds (Figure 3).
In conclusion, although reaching heart phase faster, end CYCD7;1 embryos stay longer in this phase, with presumably reduced embryo growth at this stage, so that they reach the mature phase at a similar time to the WT. Taking into account the effect of end CYCD7;1 on the population of nuclei during the syncytial phase and pFWA‐driven expression in the central cell and syncytial endosperm (see above), it therefore appears that the accelerated endosperm development in end CYCD7;1 lines stimulates the early phases of embryo development, but may subsequently lead to a slower progression to later stages beyond the heart phase.
Endosperm‐targeted CYCD7;1 produces larger seeds accompanied by lethality
Several studies have shown that when genes expressed in the different tissues of the ovules and/or developing seeds are misregulated, an effect on the final seed size can be observed (Jofuku et al., 1994; Werner et al., 2003; Garcia et al., 2005; Luo et al., 2005; Riefler et al., 2006; Schruff et al., 2006; Adamski et al., 2009). Therefore, we investigated the effect of endosperm‐targeted CYCD7;1 on the size of mature seeds. The size of homozygous end CYCD7;1 seeds was compared with seeds from untransformed Col‐0 WT plants and from WT segregants derived from the primary transformants, all obtained from plants grown alongside each other under the same conditions (Figure 4a,b). Seed projected area was measured using a custom imagej plug‐in (see Experimental Procedures). A Col‐0 WT seed had an area of 118 (±18) × 103 μm2. WT segregant lines produce seeds with areas ranging from 110 × 103 to 128 × 103 μm2, and were no significantly different from the untransformed WT (anova, P = 0.08). In contrast, homozygous end CYCD7;1 plants produced significantly larger seeds (55 and 13% larger in lines A and D, respectively; anova, P = 7.17 × 10−19; Figure 4a). Although untransformed WT and WT segregants display a degree of variation, endCYCD7;1 OE seeds were consistently larger than their segregating WT counterparts.
Figure 4.
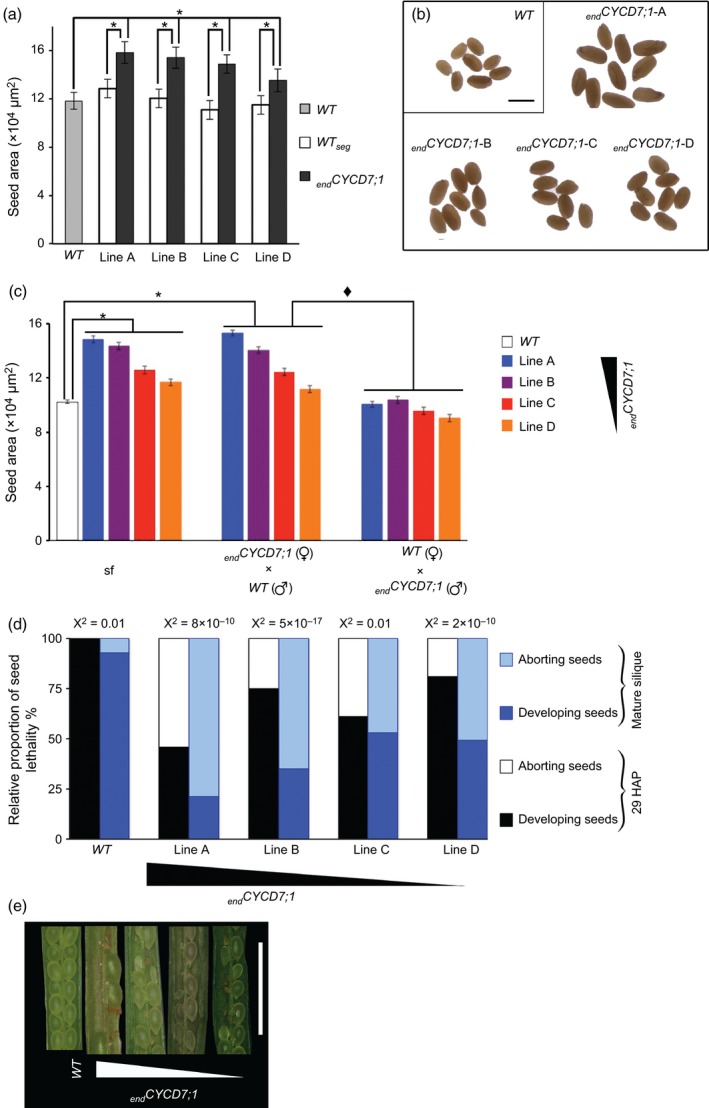
Endosperm‐targeted CYCD7;1 expression confers a larger seed phenotype that is maternally transmitted.
(a) Bar chart presenting average projected areas of mature dry seeds of wild type (WT) and end CYCD7;1 lines.
(b) Mature dried seeds from WT Col‐0 and end CYCD7;1 lines. Scale bar: 500 μm.
(c) Comparison of projected area of F1 seed generated by crossing WT and end CYCD7;1. end CYCD7;1 seeds derived by self‐pollination (sf) as well as F1 seeds from end CYCD7;1 (female) × Col‐0 (male) are larger then seeds from Col‐0 (female) × end CYCD7;1 (male). Diamonds indicate that seed size between the reciprocal crosses was significant, and the asterisk indicates a statistical difference when compared with the WT. The relative expression of end CYCD7;1 in lines A–D is indicated by the triangle.
(d) Relative proportion of developing/aborted seeds in siliques from WT and end CYCD7;1 plants (lines A–D).
(e) Aborted and developing seeds in WT (first on the top) and end CYCD7;1 siliques (from the second, top to bottom). Scale bar: 2 mm. Error bars represent standard errors on bar charts (n = 75).
The end CYCD7;1 transgenic lines in which CYCD7;1 expression was under the control of the FWA promoter thus produced seeds with an enlarged overall size. The imprinting of the FWA promoter region confers female gametophyte‐specific expression because of a lack of methylation of the maternal genome (Kinoshita et al., 2004). If the enlarged seed phenotype is caused by end CYCD7;1 expression, we would predict this will only occur from a maternally inherited gene copy. Reciprocal crosses between Col‐0 WT and end CYCD7;1 lines were performed, as provided that the imprinting is not disrupted, end CYCD7;1 should be expressed only in the female gametophyte. Hence a single maternal copy of end CYCD7;1 should be sufficient to confer the phenotype. WT self‐pollinated negative control seeds had a mean area of 102 × 103 μm2 (Figure 4c; Table S1). In the positive controls, where seeds were produced from manual self‐crossing of end CYCD7;1 plants, the seed areas were significantly larger, with average areas of 148 × 103 μm2 (line A, 45% increase), 143 × 103 μm2 (line B, 40% increase), 125 × 103 μm2 (line C, 23% increase), 116 × 103 μm2 (line D, 14% increase; anova, P = 3.4 × 10−47). The seed size increases were similar between manual and natural occurring end CYCD7;1 self‐crosses (Table S1). When end CYCD7;1 pistils were pollinated with WT pollen, seed sizes were similar to the seeds produced by end CYCD7;1 self‐fertilization (anova, P = 0.32), and for the four end CYCD7;1 lines, seeds were larger than WT × WT crosses (anova, P = 1.44 × 10−20). In contrast, when WT pistils were pollinated with end CYCD7;1 pollen, the average seed area did not significantly differ from the average for seeds from WT × WT (P = 0.35), and was significantly smaller than the for seeds from end CYCD7;1 self‐fertilized plants (P = 3.9 × 10−4).
Taken together, these results show that the production of enlarged seed is conferred by the end CYCD7;1 genotype of the female gametophyte.
A negative effect on silique length was observed in the end CYCD7;1 lines, however (Figure S2): WT mature siliques measured on average 1.61 ± 0.17 cm in length, whereas end CYCD7;1 siliques were significantly shorter (line A, 0.86 ± 0.17 cm; line B, 1.20 ± 0.22 cm; line C, 1.36 ± 0.17 cm; line D, 1.28 ± 0.2 cm; anova, n = 75, P = 3 × 10−6). As the siliques were shorter, the total number of healthy and aborted seed in the pods was recorded (Figure S2). In mature WT siliques, 5% of the total number of seeds produced were aborted, whereas in end CYCD7;1 siliques, 80% of seeds were aborted in line A, 65% in line B, and 50% in lines C and D (Figure 4d,e). Hence, in end CYCD7;1 plants, the relative proportion of seed lethality is substantially greater than in the WT (anova, n = 75, P = 1.33 × 10−13).
To understand better the stage at which this lethality arises, we examined siliques 29 h after pollination and found that a significant proportion of developing end CYCD7;1 embryos had aborted (Figure 4d), although this was a lower frequency of abortion than is observed in mature siliques. These siliques showed no aborted embryos (n = 128) that had progressed beyond globular phase, suggesting that the early stages of embryo development are particularly susceptible to the effects of end CYCD7;1.
Given that enlarged seed size was shown to be linked to the maternal genotype, we investigated the parental origin of seed lethality. Reciprocal crosses were performed between WT and end CYCD7;1 plants, and the proportion aborted was recorded in both mature siliques and during early stages (Table 1). A self‐fertilized cross by manual pollination between WT plants produced 49 ± 8 seeds per silique with 7 ± 8 seeds aborting, corresponding to 14% abortion. Manually selfed end CYCD7;1 plants produced 38 seeds per silique for line A, 43 seeds per silique for line B, 48 seeds per silique for line C, and 47 seeds per silique for line D. When end CYCD7;1 pistils were manually pollinated with WT pollen, the total number of seeds produced per silique and the proportion of lethality was comparable with end CYCD7;1 pistils pollinated with end CYCD7;1 pollen for the four end CYCD7;1 lines studied (Table 1). On the other hand, a WT pistil pollinated with end CYCD7;1 pollen produced 50 developing seeds with three aborting (for line A), similar to the number of seeds developing and the proportion aborting in the WT pistil pollinated by WT pollen (chi‐squared test, P = 0.54). Similar results were found for the three other end CYCD7;1 lines (lines B, C and D; Table 1). From these observations, we conclude that the maternal end CYCD7;1 genotype confers both enlarged seed size and elevated seed lethality.
Table 1.
Reciprocal crosses between Col‐0 wild type and end CYCD7;1 lines reveal a maternal origin of seed lethality
| ♀ × ♂ | n | Developing seeds | Aborted seeds | Total number of seed | Chi‐squared test Expected values are from the cross with | |
|---|---|---|---|---|---|---|
| end CYCD7;1 pistil | Col‐0 pistil | |||||
| Col × Col | 54 | 42 ± 13 | 7 ± 8 | 49 ± 8 | ||
| Col × A | 30 | 50 ± 5 | 3 ± 3 | 53 ± 4 | 0.544826851 | |
| Col × B | 27 | 44 ± 11 | 5 ± 8 | 50 ± 6 | 0.345231072 | |
| Col × C | 33 | 35 ± 10 | 7 ± 7 | 43 ± 5 | 0.236723571 | |
| Col × D | 30 | 40 ± 6 | 3 ± 3 | 43 ± 4 | 0.473200418 | |
| A × A | 32 | 11 ± 11 | 26 ± 11 | 38 ± 9 | ||
| A × Col | 36 | 15 ± 2 | 21 ± 5 | 36 ± 4 | 0.12 | |
| B × B | 52 | 8 ± 5 | 35 ± 8 | 43 ± 7 | ||
| B × Col | 31 | 8 ± 10 | 34 ± 7 | 42 ± 6 | 0.865772375 | |
| C × C | 52 | 27 ± 11 | 21 ± 9 | 48 ± 7 | ||
| C × Col | 30 | 21 ± 7 | 20 ± 8 | 41 ± 8 | 0.239938989 | |
| D × D | 56 | 22 ± 9 | 25 ± 9 | 47 ± 8 | ||
| D × Col | 30 | 24 ± 8 | 22 ± 9 | 47 ± 4 | 0.461680188 | |
Partial abortion conferred by embryo lethal mutant alleles increased seed size
Whether there is a general relationship between seed size and sibling lethality is unclear. In order to establish whether partial lethality reducing the number of seeds per silique leads to the enlargement of the surviving sibling seeds in other mutant backgrounds, we examined ovule and embryo development and measured seed size in surviving healthy progeny of heterozygous cdka1;1 and nclpp2 mutants (Figure S3). CDKA1;1 encodes the catalytic partner of CYCD and is required for embryo development (Nowack et al., 2006). nclpp2 is a mutant in ATP‐dependent Clp protease proteolytic subunit‐related protein 2, a chloroplastic protein unrelated to cell division control (Kim et al., 2009). These mutants were chosen for having a link to cell cycle control in the case of cdka1;1–1, and no link in the case of nclpp2.
In the unfertilized ovules of the cdka1;1–1 +/− mutant, no ovules were observed with more than one central cell nucleus in the gametophyte (Figure S3a–c). In the nclpp2 +/− mutant, 20% of ovules showed an abnormal ovule development. Of the properly developed ovules in nclpp2 +/−, 96% of ovules showed a single central cell nucleus and 4% displayed two or three nuclei in the central cell of the gametophyte. The presence of extra nuclei in the central cell of unfertilized ovules suggests a lack of fusion, uncoordinated cellularization or division in the formation of the female gametophyte and ovules in the nclpp2 background. These results were in contrast to the end CYCD7;1 ovules with the penetrant supernumerary central cell nucleus phenotype (Figure 2d).
The proliferation of the endosperm post‐fertilization was, on average, delayed in cdka1;1–1 +/− and nclpp2 +/− progeny when compared with WT and end CYCD7;1 developing seeds (Figure S3d). Although each of these mutant alleles conferred a significant lethality in the mature silique of 45 and 75%, respectively, the timing of the abortion appeared to differ. At 48 h after pollination, 75% of ovules in the nclpp2 +/− progeny were already degenerating and not progressing in embryogenesis, whereas 25% progressed, albeit with a delay in embryogenesis. In contrast, in the cdka1;1–1 +/− pistils no ovule degeneration was observed at this point in time, although embryogenesis was delayed (Figure S4b). Nevertheless, despite these substantial differences in the phenotypes leading to lethality, in both cases a substantial increase in average seed size was observed compared with either the progeny of a Col‐0 WT or the progeny of a WT sibling of the heterozygote mutant (Figure S4a), similar to end CYCD7;1 lines. These observations suggest that, at least under growth‐room conditions, sibling lethality has a strong effect on the size of the remaining seeds, consistent with either the removal of silique space constraints or a strong sink effect of the ovules normally limiting the growth of Arabidopsis Col‐0 seeds in the silique.
Larger end CYCD7;1 seeds have more cells in both embryo and seed coat, and display faster seedling establishment
In order to understand the larger seed phenotype and the effect of end CYCD7;1 expression, we examined the different seed compartments. A WT embryo had a volume of 25 (±4.7) × 106 μm3, whereas end CYCD7;1 embryos had volumes ranging from 61 (±10.1) × 106 μm3 (line A) to 38 (±11.3) × 106 μm3 (line D), correlated with levels of CYCD7;1 activation (Figure 5a,b). These results show a significant increase in volume, ranging from 50 to 139% (anova, 36 > n > 30, P = 2.8 × 10−17), indicating that the enlarged overall seed size is associated with larger embryos.
Figure 5.
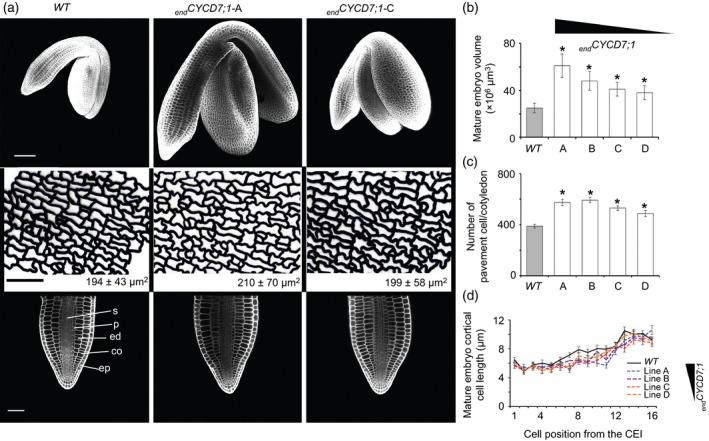
Endosperm‐targeted CYCD7;1 seeds contain elevated numbers of cells in the embryo and seed coat.
(a) Phenotype of embryos in mature seeds. Upper row: 3D reconstruction based on confocal Z‐stacks of mature embryos. end CYCD7;1 embryos are larger than the WT. Scale bar: 100 μm. Middle row: traces of cell outlines for the abaxial epidermis of the cotyledon. The average pavement cell area is shown in the insert (±SDs). Scale bar: 50 μm. Lower row: longitudinal confocal section of the radicle of mature embryo. Scale bar: 100 μm. Abbreviations: co, cortex; ed, endodermis; ep, epidermis; p, pericycle; s, stele.
(b) Quantification of mature embryo volume.
(c) Calculated numbers of pavement cells of cotyledons (ratio cotyledon area/epidermal cell area).
(d) Quantification of cortical cell length. Error bars show ±SEs. Asterisks indicate a statistical difference in variation of seed size parameters compared with the WT. The relative expression of end CYCD7;1 in the different lines is indicated by the triangle.
No defects in embryonic anatomy were observed (Figure 5a). Confocal examination of cleared mature embryos showed that the end CYCD7;1 radicles have no extra layers, and have a radial pattern similar to that of WT. The length of the first 16 cortical cells, upwards from the cortical–endodermis initial (CEI), was measured. The average length of first cortical cell in a WT root was 6.5 μm. In end CYCD7;1, cortical cell lengths ranged from 5.5 to 6 μm depending on the line observed (Figure 5d). The sixteenth cortical cell length measured was 9.2 μm in WT, and ranged between 8.8 and 10.6 μm for end CYCD7;1. In general, end CYCD7;1 cortical cells were slightly smaller than WT equivalents, but a two‐way anova test showed that cortical cell length did not significantly differ between WT and end CYCD7;1 (n > 74 cortical files, P = 0.23). Overall, the cell length increased up the root, and for both genotypes the cortical cell length is statistically different, depending on the position with respect to the CEI (n > 74 cortical files, P = 4.6 × 10−6).
The cotyledon areas and the average pavement cell areas were also measured in the mature embryo (Figure 5a), making it possible to estimate the number of pavement cells on the surface of the cotyledon. The WT cotyledon surface measured, on average, 75 × 103 μm2 (Figures 5a and S5a). end CYCD7;1 measured 120 × 103 μm2 (line A), 116 × 103 μm2 (line B), 106 × 103 μm2 (line C), and 97 × 103 μm2 (line D). As the embryo volume increased, the cotyledon area in end CYCD7;1 was significantly enlarged, with a 29–60% increase (anova, n > 74, P = 1.29 × 10−5). The average surface areas of epidermal pavement cells were 194 μm2 for the WT and 210, 196, 200 and 199 μm2 for end CYCD7;1 lines A, B, C and D, respectively (Figure S5b). A Bonferroni multi‐comparison test revealed that only from end CYCD7;1 line A were the pavement cells significantly larger, by 8% (n > 555, P = 0.3 × 10−4, other comparison P > 0.7). The ratio of cotyledon surface:epidermal pavement cell surface showed that the WT area consisted of 388 cells, and that the end CYCD7;1 area consisted area of 572, 589, 529 and 448 cells for lines A, B, C and D, respectively (Figure 5c). WT cotyledons therefore had fewer pavement cells than the end CYCD7;1 lines (anova, n > 74, P = 7.5 × 10−20). Given the small effect on cell size, but the large increase in cell number, we conclude that in end CYCD7;1 lines the major driver towards additional embryo growth is an increase in cell number in the mature embryo and hence cell production during embryo growth.
A similar effect was seen in the end CYCD7;1 seed coat. The size of cells in the outer layer of the outer integument was measured on mature dry seeds. Cells of the outer integuments had on average an area of 778 ± 220 μm2 for the WT (Figure S5e). The average cell areas for the end CYCD7;1 lines were 854 ± 256 μm2 (line A), 857 ± 261 μm2 (line B), 821 ± 285 μm2 (line C) and 807 ± 237 μm2 (line D), which do not significantly differ from the WT cell area (n = 450 for each genotype, anova, P = 0.18). The WT seed area was 12.3 × 104 μm2, whereas end CYCD7;1 seed areas were significantly larger, with averages of 17.4 × 104 μm2 (line A), 17.3 × 104 μm2 (line B), 16.2 × 104 μm2 (line C) and 14.3 × 104 μm2 (line D) (anova, n = 250, P = 5.9 × 10−4). Therefore, the estimated number of cells in the outer integument inferred by the ratio of seed and cell area was 159 for WT. The end CYCD7;1 lines contained significantly more cells, with averages of 204, 203, 197 and 181 for lines A, B, C and D, respectively (anova, n = 450, P = 6 × 10−4). Therefore, the larger end CYCD7;1 seed is linked to more cell proliferation in the various seed compartments rather than increased cell expansion.
Our analysis of surviving siblings in the other embryo‐lethal mutants examined, however, showed that an increase in cell number is not a prerequisite for larger seeds. The surviving seeds derived from heterozygote cdka1;1–1 +/− and nclpp2 +/− plants are larger, but on average contain fewer cells in the outer integuments, suggesting that cell expansion is underpinning the increase of seed size in these mutants (Figure S5).
In order to test whether or not the larger seed sizes of end CYCD7;1 plants would influence the early development of the seedlings, we compared germination rate and seedling development up to 7 days in WT and end CYCD7;1 lines (Figure 6). No major effect on germination rate was recorded, 50% of seeds in all lines germinated between 23 and 26 h, and after 30 h all seeds had germinated (Figure S6). Two days after stratification, primary root length did not differ between WT and end CYCD7;1 lines, but from 3 days after stratification the rate of primary root growth of end CYCD7;1 lines was faster (Figure 6C). end CYCD7;1 lines displayed larger cotelydons when measured 7 days after stratification (Figure 6a,b). It thus seems that the larger seeds and embryo size provide an advantage for the early establishment of the seedlings, potentially through the larger embryo and cotyledon size.
Figure 6.
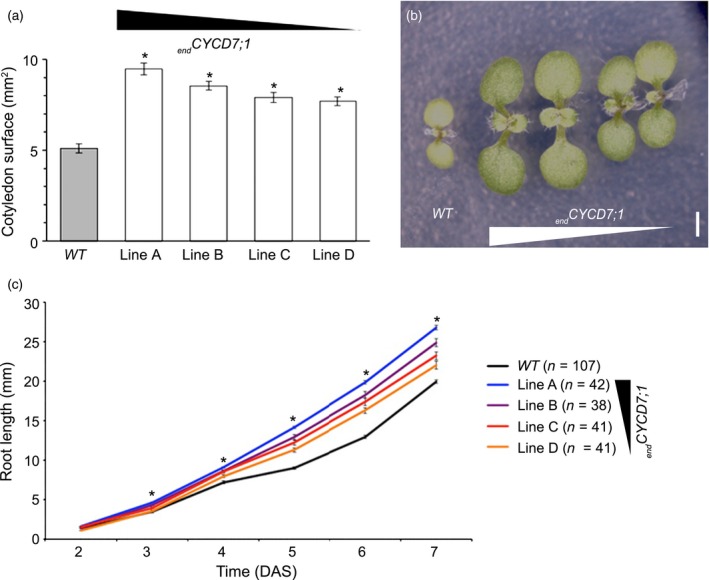
Endosperm‐targeted CYCD7;1 stimulates seedling establishment.
(a) Cotelydon surface areas from 7‐day‐old wild‐type (WT) and end CYCD7;1 seedlings (60 < n < 90).
(b) Seven‐day old seedlings: from left to right, WT and end CYCD7;1 lines A, B, C and D. Scale bar: 1 mm.
(c) Average primary root length in WT and end CYCD7;1 (lines A–D) seedlings 3–7 days after stratification (30 < n < 45). The relative expression of end CYCD7;1 in the different lines is indicated by the triangle. Asterisks show statistical difference in root length at each time point compared with the WT.
Discussion
The RBR pathway is a key controller of endosperm development in both Arabidopsis (Ebel et al., 2004) and Zea mays (maize) (Sieberer et al., 2009). Ovules carrying the rbr1 loss‐of‐function mutation in the RBR gene megagametophytes are aborted (Ebel et al., 2004), and in the absence of fertilization, spontaneous excessive proliferation is observed at the micropylar end that normally develops the egg apparatus and the central cell, producing up to 20 nuclei (Ingouff et al., 2006).
Mutants of the fis class can initiate parthenogenetic seed development in the absence of fertilization. This is characterized by an autonomous endosperm developing from the central cell, coupled with growth and differentiation of the seed coat (Debeaujon et al., 2003; Ingouff et al., 2006). This mutant class includes medea, fis2, fertilization independent endosperm (fie) and msi1 (Ohad et al., 1996; Chaudhury et al., 1998; Chaudhury and Berger, 2001; Kohler et al., 2003; Guitton et al., 2004); however, unlike the fis mutants, growth and differentiation of the seed coat does not occur in rbr1 (Ingouff et al., 2006).
D‐type cyclins can inactivate RBR by hyperphosphorylation (Boniotti and Gutierrez, 2001), and here we show that targeting CYCD7;1 to the central cell phenocopies aspects of the rbr mutant phenotype. FWA‐directed expression of CYCD7;1 expression in the central cell after the last mitosis overcomes cell‐cycle arrest and induces nuclear proliferation in the majority of unfertilized ovules to produce multinucleate ovules at high frequency, but does not affect the formation of the egg apparatus. Although RBR expression is detectable in mature ovules only in the central cell, it is expressed throughout megagametophyte development (Ingouff et al., 2006). Targeted inactivation of RBR in the central cell therefore allows the effect of overcoming cell‐cycle arrest to be separated from possible effects of RBR on megagametophyte development itself (Ingouff et al., 2006). As we only observe a specific effect of CYCD7;1 expression upon the induction of central cell divisions without ovule lethality to the same extent as observed with the loss of function of rbr1, we suggest that the lethality in rbr1 may be related to earlier functions of RBR in megagametophyte development. Although we have no direct evidence that CYCD7;1 expression specifically directs the phosphorylation of RBR in the central cell, the similarity of this aspect of the phenotype is consistent with this conclusion. Alternatively, CYCD7;1 might indirectly upregulate CDK activity, for example by sequestering and thereby inactivating the CDK kinase inhibitors of the SIM or ICK/KRP family. In any case, our data indicate that proliferation of the central cell nucleus can be uncoupled from the proliferation of the egg cell to some extent without impinging on the viability of the ovule. It is also important to note that the frequency of multinucleate central cell nuclei is much higher (Figure 2d) than the subsequent embryo lethality that we observe (discussed further below), and so the presence of multiple nuclei in the central cell is not in itself sufficient to cause later lethality.
Normally after fertilization the endosperm rapidly triggers sustained cell division and differentiation in the seed coat (Ingouff et al., 2006), and this same link between endosperm and seed coat development is seen in fis mutants, even in the absence of any parthenogenetic embryo, as in fis2; however, the excessive nuclear proliferation in the rbr1 female gametophyte does not trigger the growth and differentiation of a seed coat, and although the proliferative aspect of the rbr1 phenotype resulting in supernumerary nuclei is superficially similar to fis mutants, they appear to derive from continued proliferation of the central cell lineage, and not from an autonomous seed developmental programme (Ingouff et al., 2006). The absence of seed coat development before fertilization in end CYCD7;1 lines, based on the absence of vanillin staining, indicates that the initiation of the proliferation of central cell nuclei by CYCD7;1 is not sufficient to trigger other aspects of seed development, such as seed coat differentiation, as is also observed in rbr1 mutants.
The use of ectopic end CYCD7;1 expression allows us to examine their effect post‐fertilization, which is not possible in rbr1 mutants. An increase in nuclear proliferation in the syncytial phase was observed, in line with the canonical role of D‐type cyclins in the promotion of the mitotic cell cycle, presumably as a consequence of continued RBR inactivation. Only progression through the early stages of embryo development was stimulated by end CYCD7;1 in the endosperm, and after reaching heart phase, the development of end CYCD7;1 embryos slowed, so that WT and overexpressor reached maturity around the same time. These results show that increased proliferation within the endosperm strongly promotes early embryo growth. This is consistent with the conclusion that seed size is controlled by the endosperm (Garcia et al., 2003, 2005); however, previous analysis has suggested that integument cell proliferation and endosperm growth are largely independent, and cell elongation in the seed coat driven by growth in the endosperm appears to play the major role in the control of seed size (Garcia et al., 2005). This led to the suggestion that the maternal‐derived diploid seed coat regulates endosperm growth, which in turn affects the final seed size; however, the targeted expression of CYCD7;1 only to the endosperm led to increased endosperm nuclear proliferation, associated in the resulting larger seed with an increased number of integument cells. This is consistent with a more dynamic interplay of signals between endosperm and integument than previously suggested, with increased endosperm division triggering increased integument division.
Seed size has been reported to be affected by a range of genetic, epigenetic and environmental factors, particularly internal developmental signals originating from both maternal sporophytic and from post‐fertilization tissues of the endosperm and embryo (Li and Li, 2015). The maternal integuments around the ovule go on to form the seed coat post‐fertilization, and have been proposed to set an upper limit to final seed size acting through the ubiquitin receptor DAR1 (Jofuku et al., 1994; Adamski et al., 2009; Fang et al., 2012; Xia et al., 2013). Further maternally acting factors include the transcription factor APETALA 2, and other components of the ubiquitin pathway (Jofuku et al., 2005; Li et al., 2008; Adamski et al., 2009; Ohto et al., 2009; Xia et al., 2013). After fertilization, several factors have been identified that lead to small seeds through an effect on the regulation of endosperm growth in Arabidopsis (Garcia et al., 2003; Kesavan et al., 2013; Luo et al., 2005; Zhou et al., 2009; reviewed Sundaresan, 2005). These include HAIKU 1 (IKU1), IKU2, MINISEED 3 (MINI3) and SHORT HYPOCOTYL UNDER BLUE 1 (SHB1), which function in the same genetic pathway to promote endosperm growth (Garcia et al., 2003; Luo et al., 2005; Zhou et al., 2009). The IKU pathway regulates cytokinin levels in the endosperm through the cytokinin‐degrading enzyme CYTOKININ OXIDASE 2 (CKX2), and this is proposed to integrate genetic and epigenetic regulation of endosperm growth (Tanurdzic et al., 2008).
Our results showing that lethality in sibling embryos can, by itself, lead to the increased size of surviving seed may require some of these analyses to be reinterpreted. We found that surviving seeds in end CYCD7;1 lines produce larger seeds, as do two other heterozygote mutants that segregate lethal homozygous embryos. The timing of abortion does not seem to be critical to this effect, as end CYCD7;1 and nclpp2 abort before the globular stage, either without fertilization or in the pre‐globular phases of embryogenesis, whereas the cdka1;1 mutants abort later. Hence a mutant associated with reduced seed production may lead to the production of larger seeds through a general response of the plant, rather than a specific effect on seed development. Given the fewer but larger cells in the integuments in surviving siblings in lines with nclpp2 and cdka1;1 mutations, larger seeds are not necessarily composed of tissues with more cells, but can also arise from promoting cell expansion.
Besides the effects in the developing seed, there thus appears to be a reciprocal relationship between ovule numbers and seed size, suggesting a potential nutrient sink effect on seed size. Indeed, a similar effect was also found in other mutants with a reduction of seed numbers, either by abortion or reduced initiation. For example, an increase in seed size was found in other mutants with a reduced number of seeds in eod3‐1D and ap2 (Jofuku et al., 2005; Riefler et al., 2006; Ohto et al., 2009; Fang et al., 2012). Reduced CK signalling resulted in fewer yet larger seeds in the ahk2‐3‐4 mutants in multiple cytokinin receptors (Riefler et al., 2006), and mutants in the cytokinin transcription factors arr1‐10‐12 also have shorter siliques containing larger seeds (Lee et al., 2010). This could well reflect specific cytokinin effects, but may also be the result of altered numbers. Hence, we suggest that although an observed effect on seed size certainly does not exclude specific action of regulatory pathways during seed formation, the effect of sink size as a result of ovule number and/or lethality may provide an interfering influence that must be considered. Our analysis thus suggests that careful analysis is required before concluding that specific mutations lead to seed size increase if there is associated embryo lethality.
We expressed end CYCD7;1 under the FWA promoter, which is normally imprinted and only maternally expressed. Consistent with this, we saw lethality and the larger seed phenotype only when end CYCD7;1 was of maternal origin, indicating that the transgene is correctly controlled by imprinting and that the maternal allele is expressed in the developing seed. In this case, the increase in seed size was associated with increased nuclei in the endosperm, but also with elevated cell numbers in both the seed coat and mature embryo. In these seed compartments, which were not exposed to ectopic CYCD7;1 expression, the relationship between cell growth and cell division was unaltered, resulting in larger seeds containing larger embryos composed of more cells, without an effect on cell size. The reduction of the level of ICK/KRP CDK inhibitors in higher‐order krp mutants (Callard et al., 1996) also conferred larger seeds with larger embryos; however, in this case the relationship between cell growth and cell size was found to be altered, and embryos were composed of smaller cells, indicating that KRPs mediate cell size within the embryo itself. A similar increase in seed size was observed upon stimulating the CLE8‐WOX8 pathway, not cell‐autonomously involved in coordinating cell proliferation in the embryo and endosperm (Blom et al., 1992), and therefore an attractive hypothesis is that enhanced cell proliferation in the endosperm also stimulates cell proliferation in the embryo.
As the expression of end CYCD7;1 induced partial seed abortion, and partial ovule abortion in embryo‐lethal mutants also increased the size of surviving seeds in our studies, the sink effect potentially provides a confounding influence with respect to overall seed size. We cannot therefore distinguish whether the enlarged seed phenotype is the result of stimulation of endosperm cell division, triggered by ectopic CYCD7;1, or the result of additional resources being available to each developing seed as a consequence of sibling mortality.
In conclusion, ectopic expression of the D‐type cyclin CYCD7;1 in the central cells overcomes cell‐cycle arrest in the female gametophyte and stimulates the syncytial divisions in the endosperm upon fertilization. This negatively affects seed development because a subset of developing seeds aborts, indicating that tight regulation of cell division in the female gametophyte and developing endosperm is required. Nevertheless, the surviving seeds are larger and the resulting seedlings become established more quickly, possibly through the relocation of maternal resources.
Experimental Procedures
Plant material and growth conditions
Seeds were surface sterilized with 5 g L−1 Chlorifix (sodium dichloroisocyanurate; Bayrol) in 70% ethanol, washed three times with 70% ethanol. Sterilized seeds were dispersed on GM medium [4.4 g L−1 MS medium (Sigma‐Aldrich, http://www.sigmaaldrich.com), 1.5% sucrose and 1% agar]. The seeds were stratified (4°C for 48 h) and germinated on GM, then transferred onto soil. Plants were grown at 21°C under a 16‐h light/8‐h dark photoperiod.
Arabidopsis thaliana ecotype Columbia (Col‐0) was used. pFWA:NLS‐3xVENUS and pFWA:CYCD7;1 constructs were introduced using floral‐dipping methods (Clough and Bent, 1998). Primary transformants were selected on GM medium containing kanamycin (50 μg ml−1) or d,l‐phosphinothricin (15 μg ml−1). The transgenic line carrying pBCH2‐pFWA:dFWA‐GFP was used (Kinoshita et al., 2004). The embryo‐lethal mutants used were were cdka1;1+/− (SALK_106809; Nowack et al., 2006) and nclpp2 (SALK_016774; Rudella et al., 2006) in the Col‐0 A. thaliana background.
Plasmids and construct
A 3.2‐kb region upstream of the FWA (At4g25530) translational start was amplified by PCR from genomic Arabidopsis Col‐0 DNA and subcloned HindIII‐BamHI in the pBluescript SK‐II (pBSC_pFWA) (Addgene, http://www.addgene.org). The CYCD7;1 coding sequence was amplified by PCR from cDNA derived from Col‐0 seedlings. The CYCD7;1 coding sequence was then inserted in BamHI‐SacI in the pBSC_pFWA. The pFWA:CYCD7;1 cassette was excised from HindIII‐SacI and inserted in the plant binary vector pGPTV‐bar (Becker et al., 1992).
To construct the transcriptional reporter pFWA:3xVENUS, the FWA promoter fragment and 3xVENUS fragment were excised from pBSC_FWA (HindIII‐BamHI) and pCUC2:3xVENUS (BamHIII‐NotI), respectively (Heisler et al., 2005). The two fragments were inserted in the plant binary vector pGreenI‐0029 between HindIII and NotI sites (Rudella et al., 2006).
RT‐PCR
Total RNA was extracted from siliques emerging from flowers using the RNeasy Plant mini Kit (Qiagen, http://www.qiagen.com). Total RNA (0.5 μg) was used to perform a OneStep RT‐PCR (Qiagen). ACTIN2 transcripts were used as a reference. Primer sequences are available on request, and the intensity of the amplicons was compared on agarose gels.
Whole‐mount staining for the detection of seed coat development
Pistils were emasculated and left for 3 days before sampling. Emasculated pistils (1.5 days old) from the WT were pollinated and left for 1.5 days before sampling.
The ovules or seeds from WT and end CYCD7;1 were dissected from pistils, and siliques were incubated in an acidic solution (6 N HCl) of 1% (w/v) vanillin (Sigma‐Aldrich) at room temperature (20°C) for 20 min and rinsed in H2O prior to differential interference contrast (DIC) microscopy. Vanillin reacts with the proanthocyanidins synthesized by the seed coat of the fertilized ovules, producing a red reaction product (Debeaujon et al., 2003).
Microscopy of ovule and seeds
For examination with DIC optics, seeds and ovules were cleared and mounted in chloral hydrate/30% glycerol (1:1; 30 ml H2O, 80 g chloral hydrate; C8383, Sigma‐Aldrich). Cleared samples were observed using a Zeiss Axio Imager M1 microscope equipped with DIC optics (http://www.zeiss.com).
Seed developmental progression was recorded after hand pollination of emasculated flowers. For each time point (3, 4, 5, 6, 7 and 9 days after pollination, DAP), approximately 230 seeds were scored for each line. The percentage of seeds at each embryo stage was recorded (West and Harada, 1993).
Seed abortion frequency was also scored in hand‐pollinated siliques.
Mature dry seeds were photographed under a Leica microscope (MZ16F) using a LeicaFire Cam digital camera (http://www.leica.com).
For the detection of fluorescence by confocal microscopy (Zeiss LSM‐710), the following light‐path settings were used: YFP‐VENUS was excited with 514 nm and emission was recorded by a 519–621 nm bandpass filter; and for propidium iodide, excitation was set at 543 nm, and emission recorded through a 493–572 nm bandpass filter).
Cell wall staining of mature embryos and seeds
Dry seeds were imbibed for 2 days in the dark at 4°C. Mature embryos were extracted from the seed coat and fixed with 50% ethanol/10% acetic acid for 24 h, whereas dried mature seeds were fixed directly with 50% ethanol/10% acetic acid for 24 h (Forzani et al., 2014). Embryos or seeds were rinsed with water and incubated in 1% periodic acid for 40 min at room temperature. Embryos and seeds were rinsed and stained with Schiff reagent (100 mm sodium metabisulphite, 0.15 N HCl, propidium iodide 100 μg ml−1) for 1 h at room temperature. Embryos and seeds were rinsed with water and cleared with a chloral hydrate solution. Embryos and seeds were examined with a confocal laser microscope (Zeiss LSM 710).
Seed and embryo size measurements
Seed and embryo Z‐stack images were processed using ImageJ (Schneider et al., 2012). Projective areas of mature dry seeds were generated using Seed Measurer, a purpose‐written plug‐in. Embryonic volume was determined with an imagej plug‐in, embryo 3d, that reconstitutes the 3D structure of the embryo from confocal Z‐stacks and calculates the volume.
Statistical analysis using one‐ and two‐way anova and chi‐squared test were performed with the statistics package pasw 16.0 (SPSS Inc., now IBM, http://www.ibm.com). Differences between means for different populations were assumed to be significant when P > 0.05.
Supporting information
Figure S1. Heart‐stage embryos in WT and end CYCD7;1 lines.
Figure S2. CYCD7;1 expression under the activity of FWA induces an increase of seed abortion.
Figure S3. Developmental characterization of cdka1;1–1 +/− and nclpp2 +/− mutant‐derived ovules and seeds.
Figure S4. Characteristics of mutant and end CYCD7;1 seeds.
Figure S5. Features of enlarged end CYCD7;1 mature seeds.
Figure S6. Germination of seeds derived from end CYCD7;1, WT and nclpp2 +/−.
Table S1. Reciprocal crosses between Col‐0 WT and end CYCD7;1 lines reveal a maternal origin of seed size increase (m, manual; sf, self‐pollinated).
Acknowledgements
This work was supported by Bayer CropScience NV and BBSRC (BB/I004661/1, BB/E0223831 and BB/J009199/1) funding. The authors wish to thank Barend de Graaf and Marc Bots (Bayer CropScience NV) for advice, and Angela Marchbank and Joanne Kilby for expert technical support. The authors declare no conflicts of interest.
Accession numbers: Arabidopsis thaliana CYCD7;1 (AT5G02110); Arabidopsis thaliana FWA (AT4G25530).
References
- Adamski, N.M. , Anastasiou, E. , Eriksson, S. , O'Neill, C.M. and Lenhard, M. (2009) Local maternal control of seed size by KLUH/CYP78A5‐dependent growth signaling. Proc. Natl Acad. Sci. 106, 20115–20120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, D. , Kemper, E. , Schell, J. and Masterson, R. (1992) New plant binary vectors with selectable markers located proximal to the left T‐DNA border. Plant Mol. Biol. 20, 1195–1197. [DOI] [PubMed] [Google Scholar]
- Berger, F. (2008) Double‐fertilization, from myths to reality. Sex. Plant Reprod. 21, 3–5. [Google Scholar]
- Blom, T.J. , Kreis, W. , van Iren, F. and Libbenga, K.R. (1992) A non‐invasive method for the routine‐estimation of fresh weight of cells grown in batch suspension cultures. Plant Cell Rep., 11, 146–149. [DOI] [PubMed] [Google Scholar]
- Boisnard‐Lorig, C. , Colon‐Carmona, A. , Bauch, M. , Hodge, S. , Doerner, P. , Bancharel, E. , Dumas, C. , Haseloff, J. and Berger, F. (2001) Dynamic analyses of the expression of the HISTONE:YFP fusion protein in arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell, 13, 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniotti, M.B. and Gutierrez, C. (2001) A cell‐cycle‐regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J. 28, 341–350. [DOI] [PubMed] [Google Scholar]
- Callard, D. , Axelos, M. and Mazzolini, L. (1996) Novel molecular markers for late phases of the growth cycle of Arabidopsis thaliana cell‐suspension cultures are expressed during organ senescence. Plant Physiol. 112, 705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury, A.M. and Berger, F. (2001) Maternal control of seed development. Semin. Cell Dev. Biol. 12, 381–386. [DOI] [PubMed] [Google Scholar]
- Chaudhury, A.M. , Craig, S. , Dennis, E. and Peacock, W. (1998) Ovule and embryo development, apomixis and fertilization. Curr. Opin. Plant Biol. 1, 26–31. [DOI] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Collins, C. , Dewitte, W. and Murray, J.A. (2012) D‐type cyclins control cell division and developmental rate during Arabidopsis seed development. J. Exp. Bot. 63, 3571–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L. , Beeckman, T. and Inze, D. (2007) The ins and outs of the plant cell cycle. Nat. Rev. 8, 655–665. [DOI] [PubMed] [Google Scholar]
- Debeaujon, I. , Nesi, N. , Perez, P. , Devic, M. , Grandjean, O. , Caboche, M. and Lepiniec, L. (2003) Proanthocyanidin‐accumulating cells in arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell, 15, 2514–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte, W. , Scofield, S. , Alcasabas, A.A. et al. (2007) Arabidopsis CYCD3 D‐type cyclins link cell proliferation and endocycles and are rate‐limiting for cytokinin responses. Proc. Natl Acad. Sci. 104, 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews, G.N. and Koltunow, A.M. (2011) The female gametophyte. Arabidopsis Book, 9, e0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel, C. , Mariconti, L. and Gruissem, W. (2004) Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature, 429, 776–780. [DOI] [PubMed] [Google Scholar]
- Fang, W. , Wang, Z. , Cui, R. , Li, J. and Li, Y. (2012) Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana . Plant J. 70, 929–939. [DOI] [PubMed] [Google Scholar]
- Faure, J.E. , Rotman, N. , Fortune, P. and Dumas, C. (2002) Fertilization in Arabidopsis thaliana wild type: developmental stages and time course. Plant J. 30, 481–488. [DOI] [PubMed] [Google Scholar]
- Forzani, C. , Aichinger, E. , Sornay, E. , Willemsen, V. , Laux, T. , Dewitte, W. and Murray, J.A. (2014) WOX5 suppresses CYCLIN D activity to establish quiescence at the center of the root stem cell niche. Curr. Biol. 24, 1939–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, D. , Saingery, V. , Chambrier, P. , Mayer, U. , Jurgens, G. and Berger, F. (2003) Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 131, 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, D. , Fitz Gerald, J.N. and Berger, F. (2005) Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell, 17, 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, R.B. , de Paiva, G. and Yadegari, R. (1994) Plant embryogenesis: zygote to seed. Science, 266, 605–614. [DOI] [PubMed] [Google Scholar]
- Guitton, A.E. , Page, D.R. , Chambrier, P. , Lionnet, C. , Faure, J.E. , Grossniklaus, U. and Berger, F. (2004) Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana . Development, 131, 2971–2981. [DOI] [PubMed] [Google Scholar]
- Gutierrez, C. , Ramirez‐Parra, E. , Castellano, M.M. and del Pozo, J.C. (2002) G(1) to S transition: more than a cell cycle engine switch. Curr. Opin. Plant Biol. 5, 480–486. [DOI] [PubMed] [Google Scholar]
- Heisler, M.G. , Ohno, C. , Das, P. , Sieber, P. , Reddy, G.V. , Long, J.A. and Meyerowitz, E.M. (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15, 1899–1911. [DOI] [PubMed] [Google Scholar]
- Ingouff, M. , Jullien, P.E. and Berger, F. (2006) The female gametophyte and the endosperm control cell proliferation and differentiation of the seed coat in Arabidopsis. Plant Cell, 18, 3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku, K.D. , den Boer, B.G. , Van Montagu, M. and Okamuro, J.K. (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2 . Plant Cell, 6, 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku, K.D. , Omidyar, P.K. , Gee, Z. and Okamuro, J.K. (2005) Control of seed mass and seed yield by the floral homeotic gene APETALA2 . Proc. Natl Acad. Sci. 102, 3117–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, A.J. , Matveeva, E. , Kirioukhova, O. , Grossniklaus, U. and Gruissem, W. (2008) A dynamic reciprocal RBR‐PRC2 regulatory circuit controls arabidopsis gametophyte development. Curr. Biol. 18, 1680–1686. [DOI] [PubMed] [Google Scholar]
- Johnston, A.J. , Kirioukhova, O. , Barrell, P.J. , Rutten, T. , Moore, J.M. , Baskar, R. , Grossniklaus, U. and Gruissem, W. (2010) Dosage‐sensitive function of RETINOBLASTOMA RELATED and convergent epigenetic control are required during the arabidopsis life cycle. PLoS Genet. 6, e1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien, P.E. , Mosquna, A. , Ingouff, M. , Sakata, T. , Ohad, N. and Berger, F. (2008) Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol. 6, e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesavan, M. , Song, J.T. and Seo, H.S. (2013) Seed size: a priority trait in cereal crops. Physiol. Plant. 147, 113–120. [DOI] [PubMed] [Google Scholar]
- Kim, J. , Rudella, A. , Ramirez Rodriguez, V. , Zybailov, B. , Olinares, P.D. and van Wijk, K.J. (2009) Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell, 21, 1669–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, T. , Miura, A. , Choi, Y. , Kinoshita, Y. , Cao, X. , Jacobsen, S.E. , Fischer, R.L. and Kakutani, T. (2004) One‐way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science, 303, 521–523. [DOI] [PubMed] [Google Scholar]
- Kohler, C. , Hennig, L. , Bouveret, R. , Gheyselinck, J. , Grossniklaus, U. and Gruissem, W. (2003) Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22, 4804–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono, A. , Umeda‐Hara, C. , Adachi, S. , Nagata, N. , Konomi, M. , Nakagawa, T. , Uchimiya, H. and Umeda, M. (2007) The Arabidopsis D‐type cyclin CYCD4 controls cell division in the stomatal lineage of the hypocotyl epidermis. Plant Cell, 19, 1265–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara, A. and Gruissem, W. (2014) Arabidopsis Retinoblastoma‐related and Polycomb group proteins: cooperation during plant cell differentiation and development. J. Exp. Bot. 65, 2667–2676. [DOI] [PubMed] [Google Scholar]
- Lee, T.J. , Pascuzzi, P.E. , Settlage, S.B. et al. (2010) Arabidopsis thaliana chromosome 4 replicates in two phases that correlate with chromatin state. PLoS Genet. 6, e1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N. and Li, Y. (2015) Maternal control of seed size in plants. J. Exp. Bot. 66, 1087–1097. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Zheng, L. , Corke, F. , Smith, C. and Bevan, M.W. (2008) Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana . Genes Dev. 22, 1331–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M. , Dennis, E.S. , Berger, F. , Peacock, W.J. and Chaudhury, A. (2005) MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine‐rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl Acad. Sci. 102, 17531–17536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar, Z. , Horvath, B. , Khan, S. , Mohammed, B. , Henriques, R. , De Veylder, L. , Bako, L. , Scheres, B. and Bogre, L. (2012) Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR‐bound and RBR‐free complexes. EMBO J. 31, 1480–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack, M.K. , Grini, P.E. , Jakoby, M.J. , Lafos, M. , Koncz, C. and Schnittger, A. (2006) A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38, 63–67. [DOI] [PubMed] [Google Scholar]
- Ohad, N. , Margossian, L. , Hsu, Y.C. , Williams, C. , Repetti, P. and Fischer, R.L. (1996) A mutation that allows endosperm development without fertilization. Proc. Natl Acad. Sci. 93, 5319–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto, M.A. , Floyd, S.K. , Fischer, R.L. , Goldberg, R.B. and Harada, J.J. (2009) Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex. Plant Reprod. 22, 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler, M. , Novak, O. , Strnad, M. and Schmulling, T. (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell, 18, 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudella, A. , Friso, G. , Alonso, J.M. , Ecker, J.R. and van Wijk, K.J. (2006) Downregulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. Plant Cell, 18, 1704–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli, P.A. and Larkins, B.A. (2009) The contribution of cell cycle regulation to endosperm development. Sex. Plant Reprod. 22, 207–219. [DOI] [PubMed] [Google Scholar]
- Schneider, C.A. , Rasband, W.S. and Eliceiri, K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schruff, M.C. , Spielman, M. , Tiwari, S. , Adams, S. , Fenby, N. and Scott, R.J. (2006) The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development, 133, 251–261. [DOI] [PubMed] [Google Scholar]
- Sieberer, B.J. , Kieft, H. , Franssen‐Verheijen, T. , Emons, A.M. and Vos, J.W. (2009) Cell proliferation, cell shape, and microtubule and cellulose microfibril organization of tobacco BY‐2 cells are not altered by exposure to near weightlessness in space. Planta, 230, 1129–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan, V. (2005) Control of seed size in plants. Proc. Natl Acad. Sci. 102, 17887–17888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanurdzic, M. , Vaughn, M.W. , Jiang, H. , Lee, T.J. , Slotkin, R.K. , Sosinski, B. , Thompson, W.F. , Doerge, R.W. and Martienssen, R.A. (2008) Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol. 6, 2880–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, T. , Motyka, V. , Laucou, V. , Smets, R. , Van Onckelen, H. and Schmulling, T. (2003) Cytokinin‐deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell, 15, 2532–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, M. and Harada, J.J. (1993) Embryogenesis in higher plants: an overview. Plant Cell, 5, 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, T. , Li, N. , Dumenil, J. , Li, J. , Kamenski, A. , Bevan, M.W. , Gao, F. and Li, Y. (2013) The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell, 25, 3347–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Zhang, X. , Kang, X. , Zhao, X. , Zhang, X. and Ni, M. (2009) SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell, 21, 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Heart‐stage embryos in WT and end CYCD7;1 lines.
Figure S2. CYCD7;1 expression under the activity of FWA induces an increase of seed abortion.
Figure S3. Developmental characterization of cdka1;1–1 +/− and nclpp2 +/− mutant‐derived ovules and seeds.
Figure S4. Characteristics of mutant and end CYCD7;1 seeds.
Figure S5. Features of enlarged end CYCD7;1 mature seeds.
Figure S6. Germination of seeds derived from end CYCD7;1, WT and nclpp2 +/−.
Table S1. Reciprocal crosses between Col‐0 WT and end CYCD7;1 lines reveal a maternal origin of seed size increase (m, manual; sf, self‐pollinated).


