Abstract
Thymic epithelial cells (TECs) provide essential signals for αβT‐cell development, and medullary TECs (mTECs) control T‐cell tolerance through both negative selection and Foxp3+ regulatory T (Treg) cell development. Although heterogeneity within the mTEC compartment is well studied, the molecular regulators of specific stages of mTEC development are still poorly understood. Given the importance of the RANK‐RANKL axis in thymus medulla formation, we have used RANK Venus reporter mice to analyze the ontogeny of RANK+ TECs during development and correlated RANK expression with mTEC stem cells defined by SSEA‐1. In addition, we have investigated how requirements for the key regulators Foxn1 and Relb map to specific stages of mTEC development. Here, we show SSEA‐1+ mTEC stem cells emerge prior to RANK expression and are present in both nude and Relb −/− mice, providing direct evidence that mTEC lineage specification occurs independently of Foxn1 and Relb. In contrast, we show that Relb is necessary for the effective production of downstream RANK+ mTEC progenitors. Collectively, our work defines stage‐specific requirements for critical TEC regulators during medulla development, including the timing of Relb dependency, and provides new information on mechanisms controlling mTEC specification.
Keywords: Foxn1, Stem cell, Thymic epithelium, Thymic microenvironments, Thymus, TNF receptor superfamily
Introduction
The formation of phenotypically and functionally distinct cortical and medullary thymic epithelial cells (cTECs and mTECs) is initiated during embryonic development, and provides essential support for T‐cell precursors. While cTECs regulate early phases of thymocyte development including positive selection 1, mTECs mediate both negative selection and Foxp3+ regulatory T (Treg) cell development, with several studies highlighting the importance of mTECs in tolerance induction 2. For example, lack of mTEC development in Relb −/− mice 3, 4 and compromised tissue‐restricted antigen (TRA) expression in Aire −/− mice 5, 6 both result in T‐cell‐mediated autoimmunity. Thus, development of mTECs represents a key stage in the generation of self‐tolerant αβT‐cells.
We, and others, have shown that both cTECs and mTECs derive from a common bipotent cell 7, 8, 9, 10. Further studies demonstrated that cells with multiple cTEC traits including β5t and CD205 give rise to both cTECs and mTECs in the embryo, providing a phenotypic profile of bipotent TECs 11, 12, 13, 14. Recently, Sekai et al. 15 showed that Cld3,4HighSSEA‐1+ TECs (where Cld is claudin) possess self‐renewal properties and are capable of the specific and long‐term generation of mTECs. Based on these findings such cells were termed mTEC stem cells suggesting that the thymus, as is the case with other epithelial tissues such as mammary gland 16, contains unipotent stem cells. Importantly, given their ability to generate mTECs but not cTECs, such cells are likely downstream of bipotent TECs and so provide a clear starting point for analysis of mTEC lineage. Consistent with this, mTECs in the postnatal thymus arise from a pool of mTEC‐committed progenitors 17, which have been reported in other studies to be located at the corticomedullary junction 18. Collectively, these findings provide a framework for stages in mTEC development. However, little is known about how these stages are controlled at the molecular level 19. For example, the temporal requirement for the key TEC transcription factors Foxn1 and Relb in the mTEC pathway described above is unknown. Moreover, while the TNFRSF member RANK is a known regulator of mTEC progenitors 20, the relationship between RANK expression and mTEC lineage development has not been addressed.
Here, we have examined how and when a series of key TEC regulators influence mTEC lineage development in the embryonic thymus, and used RANK reporter mice to provide the first analysis of RANK expression during mTEC development. We show that SSEA‐1+ mTEC stem cells lack RANK expression and can be generated in the absence of Foxn1, demonstrating that mTEC specification of bipotent TECs can occur independently of this key transcription factor. In addition, we correlate expression of RANK with the requirement for Relb during mTEC development, and show that Relb operates downstream of mTEC stem cells and is required for the effective generation of RANK+ mTEC progenitors. Collectively, our findings identify the timing of RANK expression during mTEC lineage emergence, and reveal a stage‐specific requirement for Relb in the molecular regulation of thymus medulla formation.
Results and discussion
RANK expression in embryonic thymus development defines a TEC subset distinct from mTEC stem cells
Thymus medulla development is initiated during fetal life, taking place following the generation of Cld3,4HighSSEA‐1+ mTEC stem cells 15. Importantly, while RANKL+ hemopoietic cells drive the RANK‐mediated development of mTEC progenitors 2, the timing of RANK expression in relation to mTEC stem cell availability during thymus development is unknown. To address this issue, we used RANK reporter mice 21 to analyze RANK expression during mTEC development.
Initially, embryonic CD45−EpCAM‐1+ TECs were analyzed for expression of Cld3,4, SSEA‐1 and RANK Venus. Cld3,4HighSSEA‐1+ mTEC stem cells were readily detectable at embryonic day of gestation (E) 13 to E15 (Fig. 1A), and their frequency decreased with increasing age (Fig. 1B). Analysis of RANK Venus expression showed that while RANK+ cells were virtually absent at E13, they were detectable by E14, and in contrast to mTEC stem cells, their frequency increased with age (Fig. 1A and B). Importantly, within Cld3,4High TEC, simultaneous analysis of SSEA‐1 and Rank Venus expression identified nonoverlapping SSEA‐1+RANK Venus− and SSEA‐1−RANK Venus+ subsets that were most notable at E15. Interestingly, at this stage of gestation, a low level of RANK Venus expression was observed in a small proportion of Cld3,4− TECs (Fig. 1A). Whether these cells represent the first mature mTEC to arise in ontogeny is not clear. Whatever the case, our findings demonstrate for the first time that SSEA‐1+ mTEC stem cells are RANK−, and suggest that RANK expression identifies a subset of Cld3,4High mTEC progenitors that first emerge around E13 of gestation.
Figure 1.
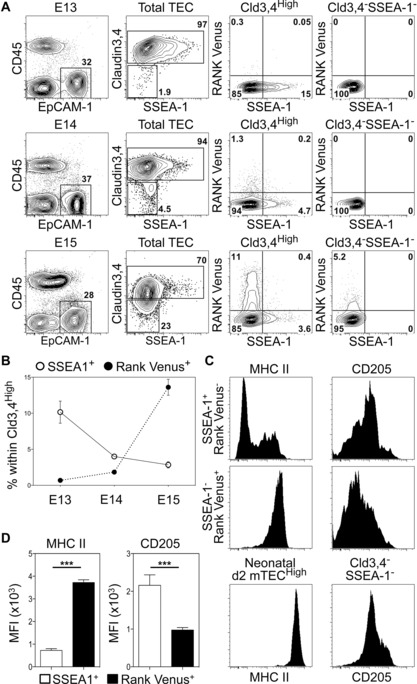
RANK+ TEC progenitors are distinct from, and arise after, mTEC stem cells. (A) Analysis of SSEA‐1+ mTEC stem cells and RANK Venus expression at E13 (upper panels), E14 (middle panels) and E15 (lower panels). Right‐hand panels are gated on either total Cld3,4High TEC or Cld3,4−SSEA‐1− TEC as indicated. (B) Proportions of SSEA‐1+ mTEC stem cells and Cld3,4HighSSEA‐1−RANK Venus+ TEC during ontogeny. (C) Expression of MHCII and CD205 in SSEA‐1+ mTEC stem cells and Cld3,4HighSSEA‐1−RANK Venus+ TEC at E15. For comparison, MHCII and CD205 levels are shown on postnatal day 2 mTECHigh and E15 EpCAM1+Cld3,4−SSEA‐1−, respectively. (D) Graphs show MFI of MHCII and CD205 in E15 SSEA‐1+ mTEC stem cells and Cld3,4HighSSEA‐1−RANK Venus+ TEC. Data shown are representative of a minimum of two separate experiments, where n = 11 for E13, n = 9 for E14, n = 16 for E15, and n = 11 for postnatal day 2. Error bars represent the SEM using an unpaired Student two‐tailed t‐test; ***p < 0.001.
To further define RANK+ TEC in relation to mTEC stem cells, we determined the levels of MHCII that increase with TEC maturation. We also analyzed levels of the cTEC marker CD205, which defines embryonic TECs able to give rise to both cTECs and mTECs 22. While MHCII levels on E15 RANK+ TECs were higher than those seen on mTEC stem cells, they were lower than that of the neonatal mTECHigh subset (Fig. 1C and D). Moreover, CD205 levels on RANK+ TECs were lower than that of mTEC stem cells (Fig. 1C and D). Collectively, these observations suggest that during thymus medulla development, the generation of mTEC stem cells is followed by the induction of RANK expression in a distinct subset of mTEC progenitors, and that this coincides with MHCII upregulation and CD205 downregulation.
RANK Venus+ TECs preferentially give rise to mTECs
To investigate the developmental potential of embryonic Cld3,4HighRank Venus+ TECs, we purified these cells from E15 thymuses (Fig. 2A). Cells were sorted on the basis of lack of CD80 expression, to formally exclude the few mature CD80+ mTEC present at this stage (Fig. 2A). As with previous analysis of mTEC progenitors, purified “spike” cells were mixed with total digested E15 WT “filler” thymus cells to make reaggregate thymus organ cultures (RTOCs) 15, 22. After 4 days, cultures were digested and the spiked progeny of RANK Venus+ TECs were identified by IAq expression, while IAb identified WT filler TECs. A readily detectable population of IAq+ TECs was evident in harvested RTOCs that was distinct from IAb+ filler cells (Fig. 2B). As expected, IAb+ filler TECs consisted of both Ly51+ cTECs and Ly51− mTECs (Fig. 2B and D), at an mTEC:cTEC ratio of approximately 5:1 (Fig 2C). In contrast, RANK Venus+ spiked cells generated few if any Ly51+ cTECs, but generated both Ly51−MHCIILowCD80Low and Ly51−MHCIIHighCD80High mTEC subsets, with mTECs/cTECs detectable at a ratio of over 40:1 (Fig. 2B–D). Thus, embryonic RANK Venus+ TECs showed a strong bias toward the generation of mTECs. Given that SSEA‐1+ mTEC stem cells are RANK−, this finding suggests that within the Cld3,4High population, RANK expression identifies mTEC‐committed progenitors that are downstream of SSEA‐1+ mTEC stem cells. Interestingly, we found that the frequency of IAq+ spike cells recovered from RTOCs was small. Whether this is due to technical issues such as post‐sort cell viability and/or successful incorporation into RTOCs is not clear. Perhaps a more interesting possibility is that mTEC progenitors within the filler TEC preparations compete with the introduced Rank Venus+ cells for limited niches that are required to support their survival and further maturation. Further experiments are required to test this possibility.
Figure 2.
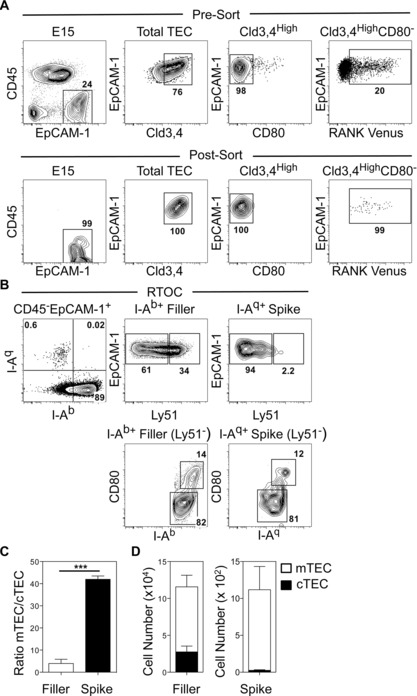
Embryonic Cld3,4hiRANK+ progenitors preferentially give rise to mTECs. (A) E15 RANK Venus thymuses before (upper panels) and after (lower panels) sorting of CD45−EpCAM1+Cld3,4High CD80−RANK Venus+ TEC. (B) Analysis of 4‐day RTOCs formed from E15 IAq+ RANK Venus+ TEC (spike) and E15 IAb+ total WT thymus (filler). Ratios (C) and numbers (D) of mTECs/cTECs generated from E15 IAq+ RANK Venus+ TEC spike and E15 IAb+ total WT filler populations. Data are typical of three separate experiments. In the experiment shown 6000 RANK Venus+ TECs were mixed with 5 × 105 WT thymus cells. Error bars represent SEM using an unpaired Student two‐tailed t‐test; ***p < 0.001.
Relb controls the generation of RANK+ mTEC progenitors but not mTEC lineage specification
The molecular regulators controlling cTEC/mTEC development from progenitor/stem cells are largely unknown. While both Foxn1 and Relb represent key transcription factors for thymus development, the timing of their requirement in TEC lineage specification is poorly understood. For example, although previous studies indicate some aspects of mTEC development can occur in nude mice 9, 23, they did not directly assess the requirement for Foxn1 in relation to recently described mTEC stem cells. Similarly, although mature mTECs are absent in Relb−/− mice 3, 4, the timing of requirement for Relb during mTEC stem/progenitor cell development is unknown.
To address these issues, we compared mTEC stem cell availability in WT and nude embryonic thymuses. While Cld3,4HighSSEA‐1+ mTEC stem cells were detectable in nude thymus rudiments, their numbers were significantly reduced and they expressed lower levels of MHCII compared to WT (Fig. 3A). In contrast, SSEA‐1+ mTEC stem cells were present in normal numbers in Relb−/− embryonic thymuses Moreover, no differences were noted in the MHCII levels on mTEC stem cells in WT and Relb−/− mice (Fig. 3B). Collectively, such findings indicate that while Foxn1 is not required for emergence of the mTEC lineage, its absence appears to impact the size of the mTEC stem cell compartment, perhaps consistent with Foxn1 mRNA expression in mTEC stem cell colonies 15. In contrast, both initial mTEC lineage development and mTEC stem cell pool size appear to operate independently of Relb.
Figure 3.
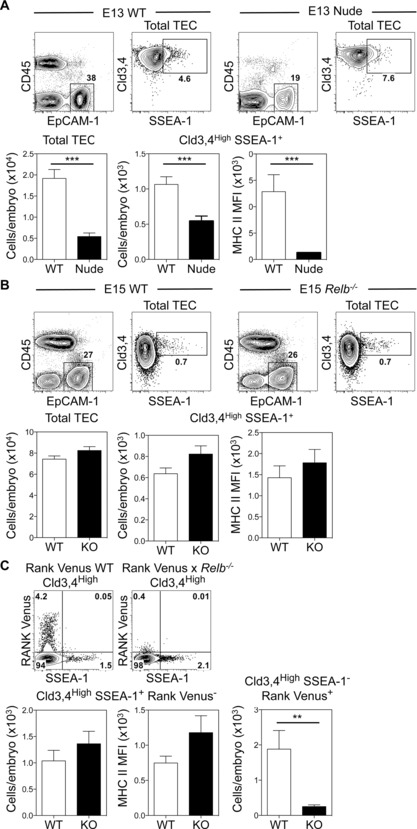
Relb controls the generation of RANK+ mTEC progenitors but is dispensable for mTEC lineage emergence. (A) Proportions, numbers and MHCII expression of Cld3,4HighSSEA‐1+ mTEC stem cells in E13 WT and nude thymuses. WT n = 9, nude n = 11. (B) E15 WT and Relb−/− thymuses were analyzed as in (A), WT n = 9, Relb−/− n = 11. (C) Analysis of SSEA‐1 and Venus expression in Cld3,4High TEC from E15 RANK Venus and RANK Venus x Relb−/− thymuses. Lower graphs show numbers and MHCII MFI of EpCAM1+Cld3,4HighSSEA‐1+RANK Venus− cells, and numbers of EpCAM1+Cld3,4HighSSEA‐1−RANK Venus+ cells in RANK Venus (white bars) and RANK Venus × Relb−/− (black bars) thymuses. RANK Venus n = 4, RANK Venus × Relb−/− n = 7. Error bars represent SEM using an unpaired Student two‐tailed t‐test; ***p < 0.001 and **p < 0.01.
Finally, given the presence of mTEC stem cells in Relb−/− mice, we examined at which downstream stages in the mTEC pathway Relb is required. Specifically, we investigated the importance of Relb during the emergence of RANK+ mTEC progenitors by crossing RANK Venus mice with Relb−/− mice. Consistent with our findings in Relb−/− mice (Fig. 3B), SSEA‐1+ mTEC stem cells were present at a normal frequency in E15 RANK Venus × Relb−/− mice, and expressed MHCII at levels comparable to WT (Fig. 3C). Strikingly however, Cld3,4HighRANK Venus+ TECs were virtually undetectable in the absence of Relb, both in terms of proportion and absolute number (Fig. 3C). Collectively, these findings identify Relb as an important regulator for the generation of RANK+ TEC progenitors, and so pinpoint the timing of its requirement in the mTEC lineage.
Concluding remarks
Generation of the mTEC lineage is an important step in the ability of the thymus to impose tolerance. However, an inability to directly visualize RANK‐expressing TECs has prevented analysis of how key regulators such as Relb might correlate with developmental stages that include mTEC stem cells. The timing of RANK expression reported here extends previous ontogenetic analyses of thymus medulla formation. For example, that mTEC stem cells lack RANK provides a temporal context for its requirement at later stages of mTEC development. Indeed, our detection of RANK+ mTEC progenitors at E14 and E15 fits well with the presence of close associations between RANKL+ cells and developing mTECs at these developmental stages 24, which are required to trigger RANK‐mediated mTEC development. Moreover, while our observations in nude mice support the idea that Foxn1 is not essential for mTEC lineage emergence, they also suggest that Foxn1 plays an important role in controlling the size of the mTEC stem cell compartment. In contrast, we found no evidence for a role for Relb in mTEC lineage specification. Instead, our data suggest that Relb operates downstream of mTEC stem cells by regulating the generation of RANK+ mTEC progenitors. Interestingly, a recent study shows that in both the steady state and following injury, thymus medulla development occurs via a lineage‐restricted progenitor 17. Whether this involves the RANK+ progenitors described here is not clear. Equally, given that mTEC‐committed progenitors are thought to reside at the corticomedullary junction 18, 25, 26 it will be interesting to define the positioning of RANK+ mTEC progenitors in the fetal and adult thymus. In conclusion, our work provides new insight into the molecular regulation of mTEC development, and further work using RANK Venus mice will enable the characterization and isolation of RANK+ mTEC progenitors for a better understanding of the mechanisms controlling thymic tolerance.
Materials and methods
Mice
RANK Venus BAC transgenic mice (CD45.2+IAq+) expressing the fluorescent protein Venus under the control elements of the murine Tnfrsf11a gene have been described 21. RANK Venus mice, Relb −/− 3, CD45.2+IAb+ C57BL/6 WT and nude mice were all bred and maintained at the Biomedical Services Unit at the University of Birmingham under local and national Home Office regulations. In timed matings, vaginal plug detection was designated day 0.
Antibodies and flow cytometry
Digested embryonic thymuses 27 were stained with the following reagents (eBioscience unless otherwise stated): anti‐CD45 APC eFlour780 (30‐F11), anti‐CD80 APC (16‐10A1), anti‐CD80 Brilliant Violet 605 (16‐10A1, Biolegend), anti‐EpCAM‐1 PerCP‐Cy5.5 (G8.8, Biolegend), anti‐EpCAM‐1 PE‐Cy7 (G8.8), anti‐IA/IE Pacific Blue (M5/114.15.2, Biolegend), anti‐IAb Pacific Blue (AF6‐120.1), anti‐IAq APC (KH116, Biolegend), anti‐CD205 PE‐Cy7 (205yekta), anti‐Ly51 APC (6C3, Biolegend), anti‐SSEA‐1 PE (MC‐480), and biotinylated C‐CPE. Streptavidin PE‐Cy7/PE was used to reveal biotinylated antibodies. Flow cytometry was performed using a Becton Dickinson Fortessa (BD Biosciences), and data were analyzed using FloJo software (v. 8.7, Tree Star). RANK Venus+ TEC were sorted from freshly digested E15 thymus lobes using a Beckman Coulter MoFlo Astrios EQ cell sorter.
Reaggregate thymus organ cultures
RTOCs 27 were formed by mixing purified E15 CD45−EpCAM1+Cld3,4HighRANK Venus+CD80− TECs with total suspensions of E15 WT thymuses. Typically, 6 × 103 RANK Venus+ TECs were mixed with 5 × 105 WT E15 thymus cells. Cell mixtures were centrifuged, and the pellet was deposited on the surface of a 0.8 μm Nucleopore filter in organ culture 27. After 4 days, cultures were digested with 0.25% trypsin and analyzed by flow cytometry.
Statistical analysis
In all experiments, Prism software (GraphPad) was used for statistical analysis, which was performed using an unpaired Student two‐tailed t‐test where ***p < 0.001; **p < 0.01. Non‐significant difference is not specified. In all figures, bar charts and error bars represent the mean and SEM, respectively.
Conflict of interest
The authors declare no commercial or financial conflict of interest.
Abbreviations
- Cld
claudin
- cTEC
cortical thymic epithelial cell
- E
embryonic day of gestation
- mTEC
medullary thymic epithelial cell
- RTOCs
reaggregate thymus organ cultures
- TEC
thymic epithelial cell
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting information
Acknowledgments
This work was supported by an MRC Programme Grant to G.A. and a BBSRC Project Grant to W.E.J. We thank Jorge Caamano for providing Relb−/− mice originally generated at Bristol‐Myers Squibb, and Dr. Matthew MacKenzie for expert FACS sorting.
References
- 1. Takada, K. and Takahama, Y. , Positive‐selection‐inducing self‐peptides displayed by cortical thymic epithelial cells. Adv. Immunol. 2015. 125: 87–110. [DOI] [PubMed] [Google Scholar]
- 2. Cowan, J. E. , Jenkinson, W. E. and Anderson, G. , Thymus medulla fosters generation of natural Treg cells, invariant gammadelta T cells, and invariant NKT cells: what we learn from intrathymic migration. Eur. J. Immunol. 2015. 45: 652–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weih, F. , Carrasco, D. , Durham, S. K. , Barton, D. S. , Rizzo, C. A. , Ryseck, R. P. , Lira, S. A. et al, Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF‐kappa B/Rel family. Cell 1995. 80: 331–340. [DOI] [PubMed] [Google Scholar]
- 4. Burkly, L. , Hession, C. , Ogata, L. , Reilly, C. , Marconi, L. A. , Olson, D. , Tizard, R. et al, Expression of relB is required for the development of thymic medulla and dendritic cells. Nature 1995. 373: 531–536. [DOI] [PubMed] [Google Scholar]
- 5. Abramson, J. and Goldfarb, Y. , AIRE: from promiscuous molecular partnerships to promiscuous gene expression. Eur. J. Immunol. 2015. 46: 22–33. [DOI] [PubMed] [Google Scholar]
- 6. Sansom, S. N. , Shikama‐Dorn, N. , Zhanybekova, S. , Nusspaumer, G. , Macaulay, I. C. , Deadman, M. E. , Heger, A. et al, Population and single‐cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self‐antigen expression in thymic epithelia. Genome Res. 2014. 24: 1918–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rossi, S. W. , Jenkinson, W. E. , Anderson, G. and Jenkinson, E. J. , Clonal analysis reveals a common progenitor for thymic cortical and medullary epithelium. Nature 2006. 441: 988–991. [DOI] [PubMed] [Google Scholar]
- 8. Bleul, C. C. , Corbeaux, T. , Reuter, A. , Fisch, P. , Monting, J. S. and Boehm, T. , Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature 2006. 441: 992–996. [DOI] [PubMed] [Google Scholar]
- 9. Ucar, A. , Ucar, O. , Klug, P. , Matt, S. , Brunk, F. , Hofmann, T. G. and Kyewski, B. , Adult thymus contains FoxN1(‐) epithelial stem cells that are bipotent for medullary and cortical thymic epithelial lineages. Immunity 2014. 41: 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wong, K. , Lister, N. L. , Barsanti, M. , Lim, J. M. , Hammett, M. V. , Khong, D. M. , Siatskas, C. et al, Multilineage potential and self‐renewal define an epithelial progenitor cell population in the adult thymus. Cell Rep. 2014. 8: 1198–1209. [DOI] [PubMed] [Google Scholar]
- 11. Shakib, S. , Desanti, G. E. , Jenkinson, W. E. , Parnell, S. M. , Jenkinson, E. J. and Anderson, G. , Checkpoints in the development of thymic cortical epithelial cells. J. Immunol. 2009. 182: 130–137. [DOI] [PubMed] [Google Scholar]
- 12. Ohigashi, I. , Zuklys, S. , Sakata, M. , Mayer, C. E. , Zhanybekova, S. , Murata, S. , Tanaka, K. et al, Aire‐expressing thymic medullary epithelial cells originate from beta5t‐expressing progenitor cells. Proc. Natl. Acad. Sci. USA 2013. 110: 9885–9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alves, N. L. , Takahama, Y. , Ohigashi, I. , Ribeiro, A. R. , Baik, S. , Anderson, G. and Jenkinson, W. E. , Serial progression of cortical and medullary thymic epithelial microenvironments. Eur. J. Immunol. 2014. 44: 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ribeiro, A. R. , Rodrigues, P. M. , Meireles, C. , Di Santo, J. P. and Alves, N. L. , Thymocyte selection regulates the homeostasis of IL‐7‐expressing thymic cortical epithelial cells in vivo. J. Immunol. 2013. 191: 1200–1209. [DOI] [PubMed] [Google Scholar]
- 15. Sekai, M. , Hamazaki, Y. and Minato, N. , Medullary thymic epithelial stem cells maintain a functional thymus to ensure lifelong central T cell tolerance. Immunity 2014. 41: 753–761. [DOI] [PubMed] [Google Scholar]
- 16. van Keymeulen, A. , Rocha, A. S. , Ousset, M. , Beck, B. , Bouvencourt, G. , Rock, J. , Sharma, N. et al, Distinct stem cells contribute to mammary gland development and maintenance. Nature 2011. 479: 189–193. [DOI] [PubMed] [Google Scholar]
- 17. Ohigashi, I. , Zuklys, S. , Sakata, M. , Mayer, C. E. , Hamazaki, Y. , Minato, N. , Hollander, G. A. et al, Adult thymic medullary epithelium is maintained and regenerated by lineage‐restricted cells rather than bipotent progenitors. Cell Rep. 2015. 13: 1432–1443. [DOI] [PubMed] [Google Scholar]
- 18. Onder, L. , Nindl, V. , Scandella, E. , Chai, Q. , Cheng, H. W. , Caviezel‐Firner, S. , Novkovic, M. et al, Alternative NF‐kappaB signaling regulates mTEC differentiation from podoplanin‐expressing presursors in the cortico‐medullary junction. Eur. J. Immunol. 2015. 45: 2218–2231. [DOI] [PubMed] [Google Scholar]
- 19. Hamazaki, Y. , Adult thymic epithelial cell (TEC) progenitors and TEC stem cells: Models and mechanisms for TEC development and maintenance. Eur. J. Immunol. 2015. 45: 2985–2993. [DOI] [PubMed] [Google Scholar]
- 20. Anderson, G. and Takahama, Y. , Thymic epithelial cells: working class heroes for T cell development and repertoire selection. Trends Immunol. 2012. 33: 256–263. [DOI] [PubMed] [Google Scholar]
- 21. McCarthy, N. I. , Cowan, J. E. , Nakamura, K. , Bacon, A. , Baik, S. , White, A. J. , Parnell, S. M. et al, Osteoprotegerin‐mediated homeostasis of Rank+ thymic epithelial cells does not limit Foxp3+ regulatory T cell development. J. Immunol. 2015. 195: 2675–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baik, S. , Jenkinson, E. J. , Lane, P. J. , Anderson, G. and Jenkinson, W. E. , Generation of both cortical and Aire(+) medullary thymic epithelial compartments from CD205(+) progenitors. Eur. J. Immunol. 2013. 43: 589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nowell, C. S. , Bredenkamp, N. , Tetelin, S. , Jin, X. , Tischner, C. , Vaidya, H. , Sheridan, J. M. et al, Foxn1 regulates lineage progression in cortical and medullary thymic epithelial cells but is dispensable for medullary sublineage divergence. PLoS Genet. 2011. 7: e1002348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roberts, N. A. , White, A. J. , Jenkinson, W. E. , Turchinovich, G. , Nakamura, K. , Withers, D. R. , McConnell, F. M. et al, Rank signaling links the development of invariant gammadelta T cell progenitors and Aire(+) medullary epithelium. Immunity 2012. 36: 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anderson, G. and Jenkinson, W. E. , Border control: Anatomical origins of the thymus medulla. Eur. J. Immunol. 2015. 45: 2203–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mayer, C. E. , Žuklys, S. , Zhanybekova, S. , Ohigashi, I. , Teh, H. Y. , Sansom, S. N. , Shikama‐Dorn, N. et al, Dynamic spatio‐temporal contribution of single β5t+ cortical epithelial precursors to the thymus medulla. Eur. J. Immunol. 2015. doi: 10.1002/eji.201545995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rossi, S. W. , Kim, M. Y. , Leibbrandt, A. , Parnell, S. M. , Jenkinson, W. E. , Glanville, S. H. , McConnell, F. M. et al, RANK signals from CD4(+)3(‐) inducer cells regulate development of Aire‐expressing epithelial cells in the thymic medulla. J. Exp. Med. 2007. 204: 1267–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting information


